Abstract
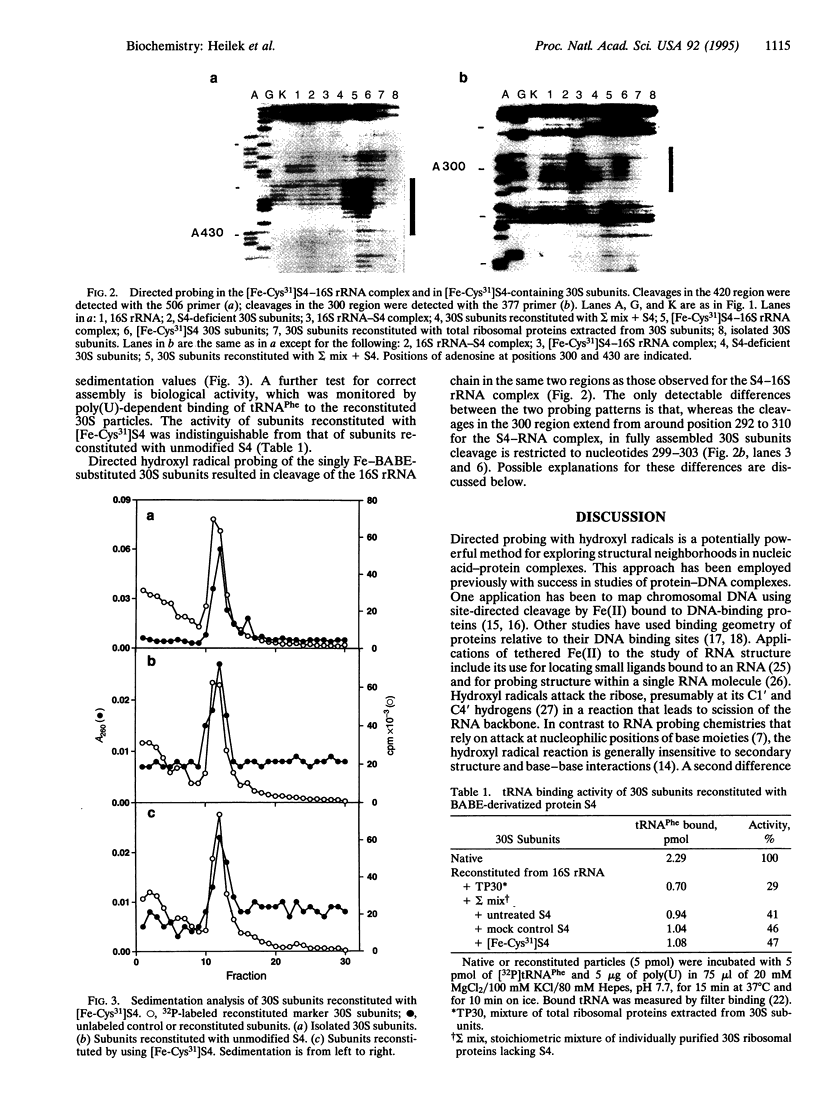
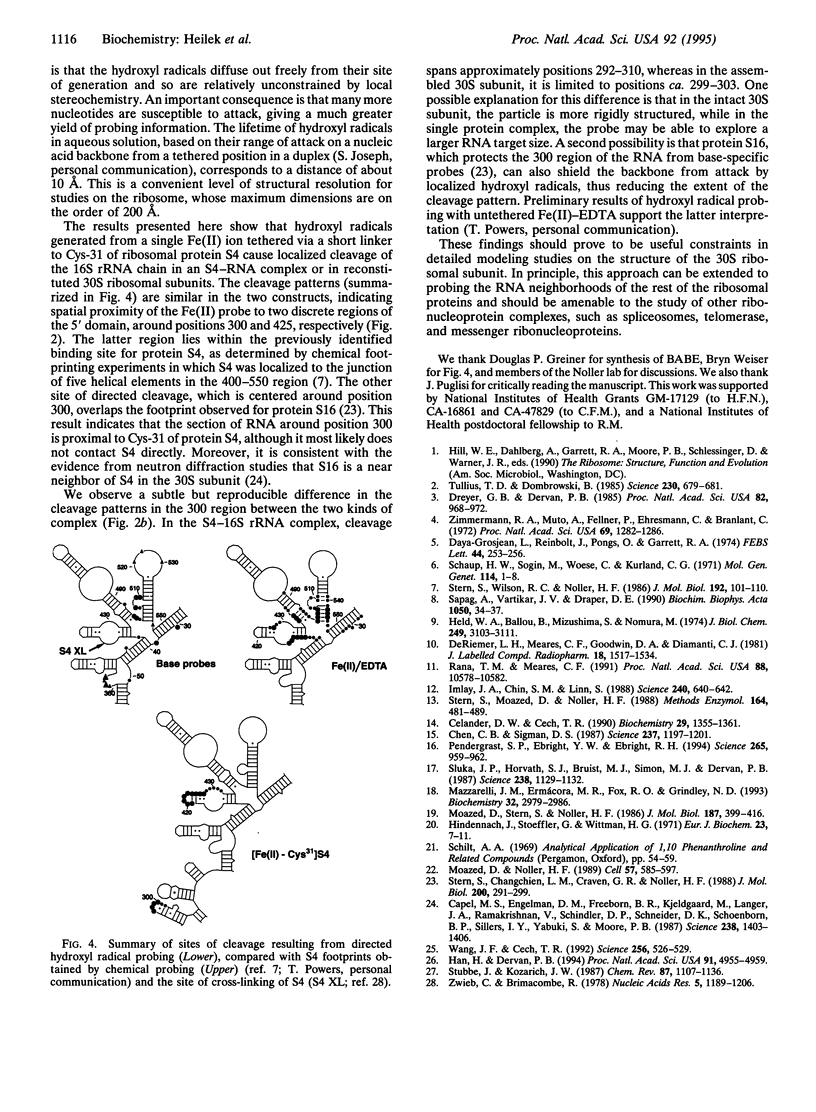
Localized hydroxyl radical probing has been used to explore the rRNA neighborhood around a unique position in the structure of the Escherichia coli 30S ribosomal subunit. Fe(II) was attached to ribosomal protein S4 at Cys-31 via the reagent 1-(p-bromoacetamidobenzyl)-EDTA. [Fe-Cys31]S4 was then complexed with 16S rRNA or incorporated into active 30S ribosomal subunits by in vitro reconstitution with 16S rRNA and a mixture of the remaining 30S subunit proteins. Hydroxyl radicals generated from the tethered Fe resulted in cleavage of the 16S rRNA chain in two localized regions of its 5' domain. One region spans positions 419-432 and is close to the multihelix junction previously placed at the RNA binding site of S4 by chemical and enzymatic protection (footprinting) and crosslinking studies. A second site of directed cleavage includes nucleotides 297-303, which overlap a site that is protected from chemical modification by protein S16, a near neighbor of S4 in the ribosome. These results provide useful information about the three-dimensional organization of 16S rRNA and indicate that these two regions of its 5' domain are in close spatial proximity to Cys-31 of protein S4.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Capel M. S., Engelman D. M., Freeborn B. R., Kjeldgaard M., Langer J. A., Ramakrishnan V., Schindler D. G., Schneider D. K., Schoenborn B. P., Sillers I. Y. A complete mapping of the proteins in the small ribosomal subunit of Escherichia coli. Science. 1987 Dec 4;238(4832):1403–1406. doi: 10.1126/science.3317832. [DOI] [PubMed] [Google Scholar]
- Celander D. W., Cech T. R. Iron(II)-ethylenediaminetetraacetic acid catalyzed cleavage of RNA and DNA oligonucleotides: similar reactivity toward single- and double-stranded forms. Biochemistry. 1990 Feb 13;29(6):1355–1361. doi: 10.1021/bi00458a001. [DOI] [PubMed] [Google Scholar]
- Chen C. H., Sigman D. S. Chemical conversion of a DNA-binding protein into a site-specific nuclease. Science. 1987 Sep 4;237(4819):1197–1201. doi: 10.1126/science.2820056. [DOI] [PubMed] [Google Scholar]
- Daya-Grosjean L., Reinbolt J., Pongs O., Garrett R. A. A study of the regions of ribosomal proteins S4, S8, S15 and S20 that interact with 16 S RNA of Escherichia coli. FEBS Lett. 1974 Aug 30;44(3):253–256. doi: 10.1016/0014-5793(74)81151-8. [DOI] [PubMed] [Google Scholar]
- Dreyer G. B., Dervan P. B. Sequence-specific cleavage of single-stranded DNA: oligodeoxynucleotide-EDTA X Fe(II). Proc Natl Acad Sci U S A. 1985 Feb;82(4):968–972. doi: 10.1073/pnas.82.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Dervan P. B. Visualization of RNA tertiary structure by RNA-EDTA.Fe(II) autocleavage: analysis of tRNA(Phe) with uridine-EDTA.Fe(II) at position 47. Proc Natl Acad Sci U S A. 1994 May 24;91(11):4955–4959. doi: 10.1073/pnas.91.11.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held W. A., Ballou B., Mizushima S., Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem. 1974 May 25;249(10):3103–3111. [PubMed] [Google Scholar]
- Hindennach I., Stöffler G., Wittmann H. G. Ribosomal proteins. Isolation of the proteins from 30S ribosomal subunits of Escherichia coli. Eur J Biochem. 1971 Nov 11;23(1):7–11. doi: 10.1111/j.1432-1033.1971.tb01584.x. [DOI] [PubMed] [Google Scholar]
- Imlay J. A., Chin S. M., Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988 Apr 29;240(4852):640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- Mazzarelli J. M., Ermácora M. R., Fox R. O., Grindley N. D. Mapping interactions between the catalytic domain of resolvase and its DNA substrate using cysteine-coupled EDTA-iron. Biochemistry. 1993 Mar 30;32(12):2979–2986. doi: 10.1021/bi00063a008. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989 May 19;57(4):585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Pendergrast P. S., Ebright Y. W., Ebright R. H. High-specificity DNA cleavage agent: design and application to kilobase and megabase DNA substrates. Science. 1994 Aug 12;265(5174):959–962. doi: 10.1126/science.8052855. [DOI] [PubMed] [Google Scholar]
- Rana T. M., Meares C. F. Transfer of oxygen from an artificial protease to peptide carbon during proteolysis. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10578–10582. doi: 10.1073/pnas.88.23.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapag A., Vartikar J. V., Draper D. E. Dissection of the 16S rRNA binding site for ribosomal protein S4. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):34–37. doi: 10.1016/0167-4781(90)90137-q. [DOI] [PubMed] [Google Scholar]
- Schaup H. W., Sogin M., Woese C. Characterization of an RNA "binding site" for a specific ribosomal protein of Escherichia coli. Mol Gen Genet. 1972;114(1):1–8. doi: 10.1007/BF00268740. [DOI] [PubMed] [Google Scholar]
- Sluka J. P., Horvath S. J., Bruist M. F., Simon M. I., Dervan P. B. Synthesis of a sequence-specific DNA-cleaving peptide. Science. 1987 Nov 20;238(4830):1129–1132. doi: 10.1126/science.3120311. [DOI] [PubMed] [Google Scholar]
- Stern S., Changchien L. M., Craven G. R., Noller H. F. Interaction of proteins S16, S17 and S20 with 16 S ribosomal RNA. J Mol Biol. 1988 Mar 20;200(2):291–299. doi: 10.1016/0022-2836(88)90241-0. [DOI] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Stern S., Wilson R. C., Noller H. F. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J Mol Biol. 1986 Nov 5;192(1):101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- Wang J. F., Cech T. R. Tertiary structure around the guanosine-binding site of the Tetrahymena ribozyme. Science. 1992 Apr 24;256(5056):526–529. doi: 10.1126/science.1315076. [DOI] [PubMed] [Google Scholar]
- Zimmermann R. A., Muto A., Fellner P., Ehresmann C., Branlant C. Location of ribosomal protein binding sites on 16S ribosomal RNA. Proc Natl Acad Sci U S A. 1972 May;69(5):1282–1286. doi: 10.1073/pnas.69.5.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C., Brimacombe R. RNA-protein cross-linking in Eschericia coli 30S ribosomal subunits: a method for the direct analysis of the RNA regions involved in the cross-links. Nucleic Acids Res. 1978 Apr;5(4):1189–1206. doi: 10.1093/nar/5.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]