Abstract
The significant increase in foodborne outbreaks caused by contaminated fresh produce, such as alfalfa sprouts, lettuce, melons, tomatoes and spinach, during the last 30 years stimulated investigation of the mechanisms of persistence of human pathogens on plants. Emerging evidence suggests that Salmonella enterica and Escherichia coli, which cause the vast majority of fresh produce outbreaks, are able to adhere to and to form biofilms on plants leading to persistence and resistance to disinfection treatments, which subsequently can cause human infections and major outbreaks. In this review, we present the current knowledge about host, bacterial and environmental factors that affect the attachment to plant tissue and the process of biofilm formation by S. enterica and E. coli, and discuss how biofilm formation assists in persistence of pathogens on the plants. Mechanisms used by S. enterica and E. coli to adhere and persist on abiotic surfaces and mammalian cells are partially similar and also used by plant pathogens and symbionts. For example, amyloid curli fimbriae, part of the extracellular matrix of biofilms, frequently contribute to adherence and are upregulated upon adherence and colonization of plant material. Also the major exopolysaccharide of the biofilm matrix, cellulose, is an adherence factor not only of S. enterica and E. coli, but also of plant symbionts and pathogens. Plants, on the other hand, respond to colonization by enteric pathogens with a variety of defence mechanisms, some of which can effectively inhibit biofilm formation. Consequently, plant compounds might be investigated for promising novel antibiofilm strategies.
Introduction
The number of outbreaks of foodborne illness arising from the consumption of fresh and fresh-cut produce increased dramatically two decades ago and has, since then, continued to be high in both, absolute numbers of outbreaks and relative numbers compared with other foodborne outbreaks with an identified source (Anonymous, 2008; Olaimat and Holley, 2012; CDC, 2014). Microorganisms that have been frequently associated with illness related to consumption of fresh produce include bacteria as diverse as Salmonella enterica serovars, Escherichia coli pathovars, Listeria monocytogenes, Bacillus cereus, Vibrio cholerae, Shigella spp., Campylobacter spp., Yersinia enterocolitica, Aeromonas hydrophila and Clostridium spp.; viruses such as norovirus and hepatitis A; and protozoa such as Cyclospora cayetanensis and Cryptosporidium parvum. Specific types of fresh foods that have been identified as common sources in produce-associated outbreaks include sprouts, green leaves like lettuce and spinach, and fruits and vegetables like melons and tomatoes (Doyle and Erickson, 2008; Yaron, 2014). Salmonella enterica and E. coli are the two major species that cause large outbreaks of foodborne illness associated with fresh produce. Salmonella enterica is more frequent in outbreaks caused by fruits, seeds and sprouts, while E. coli O157:H7 is more frequent in leafy greens (Brandl, 2006).
Since fruits, vegetables and leafy greens are typically consumed without thermal treatment, outbreaks originating from such food sources usually affect a large number of individuals. An example is the recent E. coli O104:H4 outbreak in North Germany in 2011. A newly emerged E. coli O104:H4 strain caused the highest frequency of haemolytic uremic syndrome and death ever recorded in a single E. coli outbreak. Seeds of fenugreek imported from Egypt were likely the source of the outbreak (Mariani-Kurkdjian and Bingen, 2012).
Another problem associated with enteric pathogens linked to fresh produce is relating to the fact that washing of produce with chlorine or other antimicrobial solutions fails to significantly reduce the attached pathogens (Beuchat, 1997; Gandhi et al., 2001; Kondo et al., 2006). Most of the available literature regarding the use of chemicals for washing has concluded that each treatment reduces the pathogens associated with the produce by no more than 3 logs, and usually less than 1 log (Beuchat et al., 2004; Gonzalez et al., 2004; Allende et al., 2007; Shirron et al., 2009). Moreover, recent evidence has shown that enteric pathogens are less susceptible to common sanitizing agents like chlorine than the indigenous microorganisms, suggesting that after sanitizing, remaining pathogens can survive and regrow on the wet products with less competition (Shirron et al., 2009).
Plants were commonly considered not to support the persistence and colonization of enteric pathogens. Until recently, the conventional view was that bacterial enteric pathogens such as E. coli O157:H7 and S. enterica survive poorly in the harsh environment encountered on plant surfaces. The raise in produce-borne outbreaks during the last decades has evoked intensive surveys of fresh produce products. These studies indicate that contamination of fresh produce with foodborne pathogens might occur more frequently than previously thought. For example, surveillance studies to determine the incidence of S. enterica serovars on farm and retail products have shown that the prevalence of S. enterica ranges from 0% to as high as 35.7% of the sampled foods (Doyle and Erickson, 2008). However, it seems that routine testing of fresh produce using standard recovery methods may fail in recognition of contaminations, because in cases of low abundance of the pathogens, such methods may not be sensitive enough to detect the presence of the pathogens, resulting in underestimation of the contamination frequency (Kisluk et al., 2012). Furthermore, it was reported that pathogens form aggregates or biofilms (Brandl and Mandrell, 2002), or alternatively can evolve into a viable but non-cultivable (VBNC) state on plants (Dinu and Bach, 2011). The limited ability to enumerate aggregated bacteria or to detect low levels of the pathogens, and the possibility of induction of VBNC cells in plants are a source of concern, since the infective dose in several large outbreaks was considered to be as low as a few cells (Lehmacher et al., 1995; Collignon and Korsten, 2010; Kisluk et al., 2012).
Recent analyses of outbreaks associated with identified contaminated sources showed that contamination of at least 20% of the products occurred on the farm, while the rest of the outbreaks was associated with improper handling of produce after leaving the farm (Yaron, 2014). Contamination of fresh produce is aided as enteric pathogens are able to survive on the produce in the field or post-harvest for long periods of time although their overall populations most often decline after inoculation (Brandl and Mandrell, 2002; Brandl, 2006; Kisluk and Yaron, 2012). For example, S. Typhimurium inoculated on parsley or basil survived for at least 100 days on the leaves (Kisluk and Yaron, 2012; Kisluk et al., 2013), E. coli O157:H7 survived on parsley 177 days (Islam et al., 2004) and E. coli O104:H4 survived even better than E. coli O157:H7 on spinach, basil and lettuce (Markland et al., 2012). In all of these examples, the bacteria survived without causing disease symptoms in planta. Although these microorganisms are considered to be adapted to colonize warm- and cold-blooded animals, enteric bacteria are usually exposed to a new host via contaminated foods or water, and excreted back to the environment through the animal feces. As these pathogens persist for a certain time in the environment, plants may serve as potential vehicles for their transfer from the environment to a new host (Ochman and Groisman, 1995). Consequently, enteric bacteria not only survive, but also replicate on the plants until the plant is consumed by a new potential host. Thus, it is reasonable to propose intimate interactions between the bacteria and the plant (Shirron and Yaron, 2011), interactions that recently have begun to be scrutinized (Hernandez-Reyes and Schikora, 2013).
One of the most fascinating strategies to gain fitness against the challenging conditions on or in the plant is the formation of biofilms. Microbial biofilms can be formed on leaves, on root surfaces and also within intercellular spaces of plant tissues. As a benefit, biofilm formation protects attached bacteria from desiccation, UV radiation and other environmental stresses, as well as from the plant immune response and from antimicrobial compounds produced by the plant or by indigenous microorganisms. The ability to form biofilms also provides enhanced protection against chemicals used for disinfection during processing of the food (Scher et al., 2005; Lapidot et al., 2006). This review will present the factors affecting the attachment to and the process of biofilm formation on plant tissue by foodborne pathogens, and will discuss the topic of how biofilm formation assists in persistence of pathogens on the plants. Although a variety of pathogens have been implicated in outbreaks arising from produce, this review will focus primarily on S. enterica and E. coli because of the high frequency of outbreaks associated with these pathogens and the relative depth to which these foodborne pathogens have been studied in relation to biofilm formation on plants.
The plant environment and bacterial survival strategies
In order to understand the fate of enteric pathogens on plants, it is important to be familiar with the conditions the bacteria face in the plant environment. Depending on the route of transmission (water, manure, improper handling and other measures), bacteria may be located in the rhizosphere or the phyllosphere. The root zone in the soil is relatively rich in nutrients, thus supporting the persistence of 106 to 109 bacteria per gram of roots (Hallmann et al., 2001). The rhizosphere contains root exudates including compounds released as a consequence of root cell metabolism or after lysis of plant cells. A major compound of root secretions is mucilage composed of hydrated polysaccharides, organic acids, vitamins and amino acids which are excellent substrates for microbial growth. Mucilage binds water and thus helps to form a well-hydrated environment for the roots and rhizosphere microorganisms. Some bacteria that colonize the root surface are able to infect the vascular parenchyma followed by invasion into the xylem vessels and transfer to the upper parts of the plants (Kutter et al., 2006; Klerks et al., 2007).
Unlike the rhizosphere, nutrients are scarce on the foliage surface. The few plant-derived nutrients on leaves probably originate from mesophyll and epidermal cell exudates leaking onto the surface as well as from wounds and broken trichomes. The distribution of these nutrients is highly heterogeneous. Moreover, the phyllosphere is subjected to large and rapid fluctuations in temperature, solar radiation and water availability, and therefore typically supports fewer than 103 to 107 bacteria per gram leaf (Hallmann et al., 2001). These environmental conditions differ significantly from the comparatively weak and buffered fluctuations of abiotic conditions prevailing in the rhizosphere or the rich and relatively stable environment in the intestine of animals. Foliar bacteria may follow two major strategies for their growth and survival on the plant surface: A tolerance strategy that requires the ability to resist exposure to environmental stresses on leaf surfaces or an avoidance strategy by which the bacteria seek sites that are protected from those stresses (Beattie and Lindow, 1999). Based on these strategies, a general model of leaf colonization was developed. According to this model, the bacteria that arrive on the leaf surface are randomly distributed. Some bacteria enter into the leaf via openings such as stomata, and those that stay on the surface modify their local environment. The bacteria adhere to the surface, start to multiple and form aggregates or microcolonies, which may be further developed into biofilms. Some bacteria continue to invade into internal spaces, in which they modify the habitat.
Knowledge about the behaviour of human enteric pathogens on plants has just begun to accumulate. It is however emerging that those ‘non-professional’ plant-interacting organisms use similar mechanisms with plants as described above (Brandl, 2006). Using a similar strategy for survival, the main difference between plant and human enteric pathogens is that no significant multiplication on leaves surfaces of mature plants is observed for enteric pathogens, though growth was observed under specific conditions such as on cut products (Pan and Schaffner, 2010) or during germination of sprouts (Gandhi et al., 2001). In addition, in most cases, enteric pathogens survive on or in the leaf without significant changes of the habitat, and thus, without visible symptoms. These bacteria rarely modify the plant structure, but tend to aggregate or to form biofilms as will be discussed in next sections.
Bacterial biofilms
Biofilms are complex communities of microorganisms in which cells are attached to a surface and to each other, and are embedded in a self-produced matrix of extracellular polymeric substances (EPS) (Costerton et al., 1999). The major component of biofilms is actually water (up to 97%) and bacterial cells make up to 35% of the dry weight. Apart of live and dead bacteria, a variety of secreted compounds such as polysaccharides, proteins, lipopolysaccharides (LPS), DNA and lipids contribute to the dry weight of the biofilm in addition to minerals and other components from lysed or dead cells or from the environment (like host components) that jointly form the biofilm matrix (Costerton et al., 1999).
Development of bacterial biofilms on surfaces typically involves several stages, which are likely to occur also on the surface of plants. The initial stages of biofilm formation depend on bacterial motility which enables the free-swimming bacteria to reach a suitable surface (Blair et al., 2008). Consequently, the flagella act as motility organelles that assist in arrival to favourable habitats and can be adhesion factors that promote attachment to the surface. Stringent regulation of flagella rotation and functionality is subsequently required for optimal biofilm formation. For example, in Bacillus subtilis, disengaging the flagellum from the rotor facilitated the transition to the biofilm state (Blair et al., 2008). Next, the bacteria adhere to the surface, irreversibly attach to it, form microcolonies and secrete EPS that are required for the interactions of the cells with the surface, with other cells and with alternative matrix components to develop the complex architecture of the biofilm. Proteins in the biofilm matrix carry out primarily both structural and physiological functions. Exopolysaccharides confer mechanical stability and have a role in water retention and nutrient availability. In late stages of biofilm development, the microcolonies develop into mature biofilms with complex three-dimensional structures. Bacteria may actively or passively detach from the biofilm, and dispersed individual cells or clumps may spread into a new environment. Environmental signals, quorum sensing and cyclic dimeric guanosine monophosphate (di-GMP) secondary messenger signalling are major components to regulate the different stages of the biofilm developmental process (Blair et al., 2008; Ahmad et al., 2013). Consequently, mature biofilms are dynamic heterogenic environments.
Cells in the biofilm are more resistant to chemicals, stress conditions and components of the host immune system (Costerton et al., 1999), and thus it was suggested that the formation of biofilms by bacterial cells on plant surfaces is a survival strategy to withstand the harsh conditions in this environment. Several mechanisms contribute to the enhanced resistance of biofilm-associated cells, which also depend on the property of the antimicrobial compound and the genetic potential of the bacterial strain (reviewed in del Pozo and Patel, 2007). For example, EPS can provide a physical barrier against the diffusion of antimicrobial agents and compounds of the defence response and offers protection against environmental stress factors such as UV radiation, osmotic stresses and desiccation.
Like other species, the ecological success of enteric pathogens such as S. enterica and E. coli in a variety of hosts, including plants, and in different niches in the environment is in part due to their ability to grow in biofilm (Costerton et al., 1995; Davey and O'Toole, 2000). These species form biofilms on abiotic surfaces such as stainless steel and glass (Joseph et al., 2001; Zogaj et al., 2001; Kim and Wei, 2007; Schlisselberg and Yaron, 2013), on surfaces in the host such as the epithelial cell layer and gallstones (Prouty and Gunn, 2003; Esteves et al., 2005), and on plant surfaces (Mahon et al., 1997; Campbell et al., 2001; Franz et al., 2007). Additional biofilms are pellicles at the air–liquid interface (Anriany et al., 2001; Scher et al., 2005; 2007), biofilms colonizing cancer tissue, food stuff, equipment in the food industry and biofilms occurring under many more circumstances (Thomas and McMeekin, 1981; Craven and Williams, 1998; Prouty et al., 2002; Winfield and Groisman, 2003; Chia et al., 2009; Vestby et al., 2009; Crull et al., 2011).
Reversible and irreversible attachment of native bacteria and enteric pathogens to plant tissue
As mentioned above, attachment is an initial step crucial for biofilm formation on the plant surface. Analysis of attachment of plant pathogens and symbionts such as Rhizobium and Agrobacterium to the root or leaf surfaces showed a biphasic process that occurs after bacterial contact with plant surfaces. In the first few seconds, the initial adhesion is characterized by a weak, reversible and unspecific binding that usually depends on hydrophobic and electrostatic interactions. In the second phase of binding, a strong irreversible attachment might occur (Dunne, 2002). This form of attachment has also been called ‘firm’ attachment, since removal of the attached bacteria cannot be readily achieved. In many symbionts, the second attachment step involves bacterial cellulose fibres (Laus et al., 2005).
Studies of the attachment of human enteric pathogens indicate that they can rapidly adhere to a variety of plant tissues (leaves, fruits, roots) of growing or harvested plants using a similar scheme of attachment. Attachment is irreversible, since bacteria are not removed by washing. Table 1 lists studies on the attachment of E. coli and S. enterica serovars to different plants types and plant tissues. Adhesion studies conducted for more than 4 h were excluded, because after a long time, attached bacteria may die, or, alternatively, particularly in sprouts or cut plant tissues, can grow, so it is impossible to discriminate attachment, from other processes such as survival and growth. Exemplified in Table 1, ubiquitously a firm attachment was obtained within few seconds to less than few hours as depending on the detection time. More than qualitative comparisons are however not applicable due to major differences in the experimental set-up, including preparation of the inoculum, concentration of the bacteria in the binding assay, type of liquid (water, saline, buffer, etc.), temperature of the assay, methods used to recover the attached bacteria and different reports of result output parameters.
Table 1.
Adhesion and attachment of Salmonella and E. coli to plants tissues
| Pathogen | Plant/part | Time | No. attached | Comments | Reference |
|---|---|---|---|---|---|
| E. coli O157:H7 | Cut green pepper | 2 h | 6.6–7.3 log cfu g−1 | Injured fruits also investigated | (Han et al., 2000) |
| E. coli O157:H7 Salmonella serovars | Alfalfa sprouts | 4 h | 2.8–3 log cfu per sprout 3.0–3.3 log cfu per sprout | (Barak et al., 2002) | |
| E. coli O157:H7 | Arugula leaves | 1 h | 2 log cfu cm−2 | Mutants investigated | (Shaw et al., 2008) |
| E. coli O157:H7 Salmonella serovars | Peach fruits | 60 s | 1.6 log cfu cm−2 4.1 log cfu cm−2 | (Collignon and Korsten, 2010) | |
| E. coli O157:H7 Salmonella serovars | Plum fruits | 30 s | 0.6 log cfu cm−2 3.2 log cfu cm−2 | (Collignon and Korsten, 2010) | |
| E. coli O157:H7 | Spinach leaves | < 1 h | 2.7–2.9 log cfu per leaf | Mutants investigated | (Macarisin et al., 2012) |
| E. coli O157:H7 E. coli K-12 | Lettuce leaves | 0.25–2 h | 1–2.5 log cfu cm−2 | Mutants investigated | (Fink et al., 2012) |
| S. Stanley | Cantaloupes | 10 min | 3.8 log cfu cm−2 | (Ukuku and Sapers, 2001) | |
| S. Chester | Cut green pepper | 30 s | 5.85 log cfu per disc (56 mm2) | (Liao and Cooke, 2001) | |
| S. Montevideo | Tomatoes | 1.5 h | 4–5.4 log cfu per fruit | After few sec the attachment is fourfold lower. Type of product, temperature and relative humidity affect attachment | (Iturriaga et al., 2003) |
| S. Newport | Alfalfa sprouts | 1 h | 700–800 cfu per sprout | Mutants investigated | (Barak et al., 2005) |
| S. Typhimurium | Parsley leaves | 1 h | 7.4 log cfu g−1 | Mutants investigated | (Lapidot et al., 2006) |
| S. Enteritidis | Alfalfa sprouts | 1 h | 2.3 log cfu per sprout | Mutants investigated | (Barak et al., 2007) |
| S. Typhimurium and Senftenberg | Basil, lettuce, rocket and spinach leaves | 1 h | 200–250 cfu mm−2 | Mutants investigated. Attachment is affected by temperature in Senftenberg. | (Berger et al., 2009) |
| S. Typhimurium | Parsley leaves Cucumber fruits | 30 min | 6.1 log cfu g−1 5.1 log cfu g−1 | (Shirron et al., 2009) | |
| S. serovars Negev, Newport, Tennessee, Thompson, Braenderup | Intact and cut lettuce (Romaine, Iceberg) and cabbage | 1–4 h | 2–4 log cfu cm−2 | Attached to Romaine lettuce at higher numbers than those attached to Iceberg lettuce or cabbage. Differences between serovars. Attached preferentially to cut surface of all produce. Attachment increased with time. | (Patel and Sharma, 2010) |
| S. Typhimurium | Cut Romaine lettuce leaves | 2 h | 6.4–7.7 log cfu g−1 | Levels depend on the leaf region and age. Maximal attachment is near the petiole or on older leaves. | (Kroupitski et al., 2011) |
| S. Typhimurium, Senftenberg and Thompson | Tomato fruits | 1 h | 4.9–5.1 log cfu mm−2 | Mutants investigated | (Shaw et al., 2011) |
| S. Typhimurium and Saintpaul | Intact spinach leaves Grape tomatoes | 10 min | 6–7 log cfu per 3 leaves 5–6 log cfu per 3 fruits | Mutants investigated | (Salazar et al., 2013) |
Besides the bacterial inoculum and exposure time, both host plants and bacterial properties influence the efficacy by which the enteric pathogens attach to plants. Attachment to basil, lettuce or spinach leaves differed among S. enterica serovars, as S. Senftenberg and S. Typhimurium showed higher attachment compared with S. Agona or S. Arizonae. Interestingly, the S. Senftenberg strain with highest adhesion capability to basil was a clinical isolate from a basil-derived outbreak in the UK in 2007 (Berger et al., 2009). Microscopic observations of three Salmonella serovars attached to tomato fruits show that although all investigated serovars were attached to tomatoes with similar efficiencies, serovars Senftenberg and Typhimurium adhered to the fruits in an aggregative pattern, while serovars Thompson adhered in a diffuse pattern (Shaw et al., 2011). Enteric pathogens such as E. coli, Salmonella and Listeria adhered more effectively to the peach fruit than plum surfaces attributed to the increased surface area of the peach fruits due to the presence of trichomes (Collignon and Korsten, 2010). Also, in line with epidemiological data, the affinity of Salmonella serovars to lettuce was significantly twofold to threefold higher than to cabbage (Patel and Sharma, 2010). Lettuce is very often associated with foodborne outbreaks, whereas outbreaks associated with cabbage are rare.
Environmental factors affect the attachment as well. The adhesion of pathogens in wash water to fresh cucumber surfaces depends on temperature, and is less extensive at lower temperatures. The effect of dewaxing of fruits on adhesion depends on the bacteria. While adhesion of Listeria to dewaxed fruits was higher than to waxed fruits, the opposite was reported for S. Typhimurium and Staphylococcus aureus (Reina et al., 2002).
Factors that play a role in attachment of S. enterica and E. coli to plants
Properties of plant surfaces
Most aerial plant surfaces are covered in cuticle, a hydrophobic material composed primarily of fatty acids, waxes and polysaccharides. The cuticle favours attachment of hydrophobic molecules. However, breaks in the cuticle may expose hydrophilic structures (Patel and Sharma, 2010). In this case and on the root surface, the bacteria are exposed to the plant cells, which are generally covered with glycoproteins and polysaccharides such as cellulose and pectins. Many of these molecules are hydrophilic and in some cases have negative charge (Torres et al., 2005). Plant surface charge correlates with the strength of attachment (Ukuku and Fett, 2002; 2006), but the exact receptors or binding sites, if existent, have not been identified. Investigation of attachment of S. Typhimurium to sliced potatoes indicated that the bacteria attach to cell wall junctions. In particular, the bacteria appeared to attach to the pectin layer at the junctions, indicating that pectin may be the bacterial attachment site (Saggers et al., 2008). In contrary, Tan and colleagues have shown that Salmonella attached in lower numbers to plant cell wall components when pectin was part of the composite, supporting that pectin is unfavourable for the bacterial attachment compared with cellulose (Neff et al., 1987; Tan et al., 2013).
Topography and architecture of the surface of the plant are also important factors in microbial adhesion. Roughness is important not only for adherence but also for survival on the plant tissue, as demonstrated for E. coli O157:H7 adhesion on leaves of different spinach cultivars (Macarisin et al., 2013). The surface roughness of the plant organs such as leaves depends on the nature of the plant and on the age of the leaves. Indeed, the affinity of Salmonella to artificially contaminated old lettuce leaves was higher compared with young leaves. Moreover, higher numbers of S. Typhimurium were localized close to the petiole, and the bacteria displayed higher affinity towards the abaxial side compared with the adaxial side of the leaves (Kroupitski et al., 2011). Fissures in the cantaloupe netting provided attachment sites for cells of Salmonella which aid bacterial survival when in contact with aqueous sanitizers (Annous et al., 2004; 2005).
The plant microflora is not homogenously distributed on the leaf surface, rather bacterial cells have been shown to attach and colonize at specific sites in and on leaf surfaces, including the base of trichomes, at stomata, epidermal cell wall junctions, as well as in grooves along veins and depressions or beneath in the cuticle (Beattie and Lindow, 1999). These habitats apparently constitute stress-protected, rich-in-water and rich-in-nutrients sites. Plant appendages such as secretory cavities or ducts may release plant metabolites. Glandular trichomes are epidermal protuberances which serve as sites of secretion and accumulation of different compounds such as Ca, Na, Mn and Pb ions, defensive proteins and secondary metabolites such as essential oils, monoterpenoids and phenylpropanoids. Larger numbers of bacteria can also be found on the lower than upper leaf surface, possibly due to lower radiation exposure, or because of higher density of stomata or trichomes and a thinner cuticular layer (Karamanoli et al., 2012). Consequently, bacteria attached on the lower leaf surface find better conditions for survival and growth, which increase the probability of their survival compared with bacteria attached to other parts of the leaf.
Evidence indicates that human enteric pathogens demonstrate similar behaviour on leaves, with a few differences. Salmonella enterica serovar Thompson was shown to attach around stomata of spinach leaves and in cell margins, similar to where native bacteria are detected (Warner et al., 2008). Use of confocal microscopy to visualize cells of E. coli attached at stomata and trichomes of cut lettuce plants concluded that attachment sites for E. coli are similar to those reported for plant pathogens (Seo and Frank, 1999). The stomata provide protective niches for the bacteria, and also can serve as a source of nutrients. Golberg and colleagues confirmed that Salmonella cells prefer this niche by showing that the bacteria are mostly located near and within the stomata of lettuce leaves. However, the ability of Salmonella to colonize the surface around the stomata was observed only with certain serovars on specific plants (Golberg et al., 2011). On the other hand, while E. coli better attached to cut surfaces of lettuce, Pseudomonas fluorescens preferentially attached to the intact areas, and S. Typhimurium attached to both, cut and intact surfaces in a similar manner (Takeuchi et al., 2000). Whether the ability of enteric pathogens to localize to similar adhesion sites on the leaves like plant pathogens or the natural microflora contributes to long-term survival in the plant environment is an issue that should be addressed.
Enteric bacteria penetrating into the soil through water, fertilizers or directly exposed to the roots during hydroponic growth, are able to attach to the rhizosphere of the plant host. Following attachment, these bacteria can invade or move to the upper parts of the plant (Lapidot and Yaron, 2009). In the case of attachment to the root surface, in contrast to leaves and fruits, more significant differences were observed between the location of the natural microflora and enteric pathogens. The natural plant microflora and plant pathogens tend to attach to the epidermis and to the root hairs formed by trichomes. Plant pathogens bind rapidly and particularly well to cut ends of roots and wound sites and bind poorly to the root tips (Matthysse and Kijne, 1998). In contrast, E. coli strains prefer to attach to the root tips of alfalfa sprouts, but attach to the roots very slowly. Further, not all investigated E. coli strains are able to bind to the root hairs (Jeter and Matthysse, 2005).
Bacterial properties
It is mostly believed that attachment of Salmonella and E. coli is an active process, but not all observations support this assumption. Only viable Salmonella cells were able to attach to vegetable tissues such as slices of potatoes (Saggers et al., 2008). On the other hand, similar levels of attachment to lettuce were observed with live E. coli O157:H7, killed E. coli O157:H7 and fluorescent polystyrene microspheres (Solomon and Matthews, 2006). The difference was attributed to the method used for bacterial inactivation. Escherichia coli cells were inactivated with glutaraldehyde, which is known to alter the adhesive properties of the bacterial envelope, while Salmonella cells were inactivated by different methods including formalin, ethanol, kanamycin and thermal treatment (Saggers et al., 2008).
A number of authors investigated the role of cell surface charge, presence of divalent cations, hydrophobicity and capsule production in passive or active attachment of E. coli to lettuce tissue (Hassan and Frank, 2003; 2004; Boyer et al., 2007). Collectively, these studies have shown very little correlation between the presence of cell surface appendages, charge or hydrophobicity and the ability of the bacteria to attach to lettuce tissue. Subsequently, treatment with the hydrophobic surfactant Span85 detached only 80% of attached E. coli O157:H7 from intact lettuce leaves, and this surfactant was ineffective in detaching the pathogen from cut edges, indicating that the nature of surface is heterogeneous (Hassan and Frank, 2003). Alternatively, for Salmonella, a linear correlation was reported between bacterial cell surface hydrophobicity and the strength of attachment to melon fruits (Ukuku and Fett, 2002; 2006).
On the molecular level, studies investigating the role of specific bacterial factors in adhesion to plants have shown contradictory results, and until now, very few genetic elements have been definitively identified as essential for attachment or survival of human pathogens on leaves, roots, fruits or sprouts. The specific bacterial factors that contribute to attachment to plant tissue were identified by different experimental approaches like differential expression analysis upon contact with plants or plants extracts and assessment of the ability of mutants and overexpression strains to attach to plant tissues. Table 2 presents major genes frequently identified and investigated. Interestingly, many genes that have a role in adhesion to plants tissues have also been identified as attachment or virulence factors of E. coli and S. Typhimurium when infecting animals. This phenomenon occurs despite of the fact that many studies not only focused on genes with known function in the host, but also screened for functional genes using whole genome transcription analysis or transposons libraries.
Table 2.
Bacterial genes involved in attachment of pathogens on plants
| Gene | Function | Pathogen | Plant/part | Method | Maximal effect of mutant | Reference |
|---|---|---|---|---|---|---|
| adrA | Regulation of cellulose biosynthesis | S. Newport and S. Enteritidis | Alfalfa sprouts | Directed deletion | expressed on the sprouts, but no effect in attachment | (Barak et al., 2007) |
| bcsA | Cellulose biosynthesis | S. Enteritidis | Alfalfa sprouts | Directed deletion | 1 log reduction in attachment | (Barak et al., 2007) |
| bcsC | Cellulose biosynthesis | S. Typhimurium | Tomato fruits | Directed deletion | 1 log reduction in attachment | (Shaw et al., 2011) |
| csgB/csgD | Curli subunit/regulatory protein | S. Newport | Alfalfa sprouts | Transposon insertion | Eightfold reduction in attachment | (Barak et al., 2005) |
| csgA (agfA) | Curli subunit | E. coli O157:H7 | Alfalfa sprouts | Directed deletion | Fourfold reduction in attachment | (Torres et al., 2005) |
| csgA (agfA) | Curli subunit | E. coli K-12 and E. coli O157:H7 | Lettuce leaves | Directed deletion | 1 log reduction in attachment in the first 30 min | (Fink et al., 2012) |
| csgA/bcsA | Curli subunit/Cellulose biosynthesis | S. Typhimurium | Parsley leaves | Transposon insertion | No effect on attachment | (Lapidot et al., 2006) |
| csgB/bcsA | Curli subunit/Cellulose biosynthesis | S. Enteritidis | Alfalfa sprouts | Directed deletion | 1.5 log reduction in attachment | (Barak et al., 2007) |
| csgD (agfD) | Regulatory protein | E. coli O157:H7 | Alfalfa sprouts | Directed deletion | 12-fold increase in attachment | (Torres et al., 2005) |
| fliC | Flagella | E. coli O157:H7 | Spinach and lettuce leaves | Directed deletion | reduction in attachment | (Xicohtencatl-Cortes et al., 2009) |
| fliC | Flagella | S. Typhimurium and Senftenberg | Basil leaves | Directed deletion | Fivefold reduction in attachment in Senftenberg but not in Typhimurium | (Berger et al., 2009) |
| fliC | Flagella | S. Senftenberg | Tomato fruits | Directed deletion | No effect on attachment | (Shaw et al., 2011) |
| fliC/fliB | Flagella | S. Typhimurium | Tomato fruits | Directed deletion | No effect on attachment | (Shaw et al., 2011) |
| escN | T3SS | E. coli O157:H7 | Arugula leaves | Directed deletion | No attachment | (Shaw et al., 2008) |
| espB | T3SS | E. coli O157:H7 | Arugula leaves | Directed deletion | Twofold reduction in attachment | (Shaw et al., 2008) |
| ompA | Membrane protein | E. coli O157:H7 | Alfalfa sprouts | Directed deletion | 31-fold reduction in attachment | (Torres et al., 2005) |
| pgaC | poly-β-1,6-N-acetylglucosamine production | E. coli O157:H7 | Alfalfa sprouts | Transposon insertion | 3.7 log reduction in attachment | (Matthysse et al., 2008) |
| rpoS | Stationary-phase Sigma factor | S. Newport | Alfalfa sprouts | Transposon insertion | Eightfold reduction in attachment | (Barak et al., 2005) |
| waaI | LPS production | E. coli O157:H7 | Alfalfa sprouts | Transposon insertion | 1 log increment in attachment | (Matthysse et al., 2008) |
| wcaD | Colanic acid production | E. coli O157:H7 | Alfalfa sprouts | Transposon insertion | 2.9 log reduction in attachment | (Matthysse et al., 2008) |
| wcaJ | Colanic acid production | S. Enteritidis | Alfalfa sprouts | Directed deletion | No effect | (Barak et al., 2007) |
| ycfR | Putative membrane protein involved in biofilm | E. coli K-12 and E. coli O157:H7 | Lettuce leaves | Transposon insertion | No effect | (Fink et al., 2012) |
| ycfR | Putative membrane protein involved in biofilm | S. Typhimurium and Saintpaul | Spinach leaves Grape tomatoes | Directed deletion | 4 log reduction in attachment | (Salazar et al., 2013) |
| yhjN | Cellulose production | E. coli O157:H7 | Alfalfa sprouts | Transposon insertion | 1.8 log reduction in attachment | (Matthysse et al., 2008) |
| yidE | Adherence mediator | E. coli O157:H7 | Alfalfa sprouts | Directed deletion | Fivefold increase in attachment | (Torres et al., 2005) |
| yigG | Putative inner membrane protein | S. Typhimurium and Saintpaul | Spinach leaves Grape tomatoes | Directed deletion | 2 log reduction in attachment | (Salazar et al., 2013) |
| yihO | O-antigen capsule assembly | S. Enteritidis | Alfalfa sprouts | Directed deletion | 1 log reduction in attachment | (Barak et al., 2007) |
Strains of E. coli and S. enterica produce a diversity of pili and fimbriae and non-fimbrial adhesins that function as ‘professional’ adhesion systems as well as surface structures such as type III secretion systems (T3SSs) and flagella with alternative major functions (Hernandez-Reyes and Schikora, 2013; Yaron, 2014). Pili and fimbriae are hair-like appendages on the surface of the bacterial cells that often contain adhesins on their tips with affinity to different carbohydrates. Examples are the type 1, P, S and F1C fimbriae in E. coli. The adhesins interact with mammals' components, either non-specifically via hydrophobic or electrostatic interactions, or by binding to specific host cell receptor moieties, and are responsible to the tropism in adhesion to a specific host or tissue (Wagner and Hensel, 2011). Several adhesins and fimbriae of E. coli and Salmonella like amyloid curli fimbriae have widely been investigated in relation to adhesion to plants (Table 2). The studies demonstrated that curli usually have a role in attachment of E. coli and Salmonella to sprouts and leaves, but the effect of their inactivation is low. For example, deletion of csg genes resulted in no more than 1-log reduction in binding (Table 2). Furthermore, these results point out the complexity of adhesion and show that very little is known about the role of these adhesins in adherence of human pathogens to plant tissue. For example, mutations in the csgA gene had a very low effect on the ability of E. coli O157:H7 to bind to sprouts, but increased the binding of the same strain to Caco-2 human cells. On the other hand, insertion of csgA into the E. coli K-12 laboratory strain enabled the bacteria to bind to sprouts, indicating that E. coli O157:H7 possesses several redundant protein adhesins and that overexpression of each adhesin alone is sufficient to promote binding to alfalfa sprouts (Torres et al., 2005).
Taking adhesins of plant-associated bacteria into consideration indicates that additional factors that have hardly been investigated may have a role in attachment of enteric pathogens to plants. The type 1 fimbriae are widespread among members of the Enterobacteriaceae and are specified by their binding to mannosides in glycoproteins on the surface of mammalian cells. Type 1 fimbriae were also suggested to mediate adhesion of nitrogen fixating Klebsiella as well as Klebsiella pneumoniae to specific sites on the root hairs of bluegrass (Haahtela et al., 1985). Moreover, E. coli type V-secreted adhesins, such as the antigen 43, bind to integrins in the animal tissue via conserved RGD motifs (Henderson and Owen, 1999). Interestingly, plant pathogens also secrete RGD-containing proteins which might bind to specific receptor proteins of plants, but their involvement in adhesion has not been examined (Senchou et al., 2004).
Contradictory results have been obtained concerning the role of flagella in adherence of Salmonella and E. coli to plants. Deletion of the flagella subunit encoding gene fliC of E. coli O157:H7 or S. Senftenberg rendered the bacteria significantly less adherent to baby spinach and lettuce leaves (Xicohtencatl-Cortes et al., 2009), and leaf epidermis of basil (Berger et al., 2009), respectively, but deletion of the same gene in S. Typhimurium did not affect leaf attachment (Berger et al., 2009). It was suggested that this contradiction relates to the fact that the alternative flagellar subunit protein FliB is functional in this serovar, when FliC is not expressed. However, flagella were also not involved in attachment of S. Typhimurium to tomato fruits, even when both genes, fliB and fliC, were deleted (Shaw et al., 2011). In a further study, two genes of unknown function essential for swarming were found to be important factors for infection of alfalfa sprouts (Barak et al., 2009).
The role of biofilm formation in survival on the plant will be further discussed, but several genes products involved in production of biofilm components like bcs and rpoS have also a significant role in the initial attachment of E. coli and Salmonella. As can be seen in Table 2, the genes that have the most significant effect on the levels of attachment are genes involved in production of extracellular carbohydrates on the bacterial surface such as pgaC [synthesis of poly-β-1,6-N-acetylglucosamine (PGA)], wcaD (synthesis of colanic acid) in E. coli O157:H7 and ycfR (putative membrane protein involved in biofilm formation) in Salmonella. Mutants deficient in cellulose production reduced the ability of E. coli O157:H7 to attach to alfalfa sprouts (Matthysse et al., 2008), or the ability of S. Typhimurium to attach to tomato fruits (Shaw et al., 2011), but the influence of these mutations on the ability of the mutants to attach to other plants was much less noteworthy, with up to 1-log reduction. On the other hand, a plasmid carrying the cellulose gene allowed E. coli K-12 to bind to sprouts (Matthysse et al., 2008). Interestingly, the impact of each of these polysaccharides in attachment to mammalian cells and sprouts was different (Matthysse et al., 2008).
Collectively, these studies demonstrate that enteric pathogens specifically attach rapidly and irreversibly to produce surfaces. Attachment depends on plant and bacterial factors as well as on environmental conditions, but no single factor was found to be essential for attachment, possibly because bacteria use several parallel mechanisms to ensure tight attachment to different plants or to different plant cells under a wide variety of conditions. Moreover, despite the high numbers of publications in this topic, the exact contribution of each identified factor is not clear yet, probably due to redundancy in adhesion factors, diversity of adhesion factors in each pathogen and plant receptors, as well as the differences in cell surface composition.
Biofilm of S. enterica and E. coli and its regulation
After or in parallel to attachment, the bacteria start to produce the biofilm matrix. The extracellular matrix produced by many Enterobacteriaceae is composed of proteinaceous components and exopolysaccharides. A major protein component is curli (amyloid fimbriae also known as thin aggregative fibres), which are encoded by at least seven genes organized in the csgBAC and csgDEFG operons (also termed agf genes). The csgBA genes encode the curli structural genes (Hammar et al., 1996), and the csgDEFG operon encodes, besides the major transcriptional regulator, CsgD, required for curli expression and biofilm formation, three accessory proteins required for the assembly of curli on the cell surface (Hammar et al., 1996; Loferer et al., 1997; Robinson et al., 2006). In Salmonella, the secreted large surface protein BapA (biofilm-associated protein) is also a major component of the biofilm matrix (Barnhart and Chapman, 2006; Latasa et al., 2006). Like fimbriae, this protein mediates the interactions between different cells leading to aggregation (Latasa et al., 2006).
A major exopolysaccharide in S. enterica and E. coli biofilms is cellulose (Zogaj et al., 2001), while some E. coli and S. enterica serovars also secrete capsular polysaccharides or other exopolysaccharides like colanic acid (Gibson et al., 2006). Many E. coli strains also have the potential to secrete PGA (Itoh et al., 2008). Cellulose consists of linear chains of glucose monomers connected by β-1,4-glycosidic bonds, which assemble into macromolecular fibrillar structures. These crystalline fibres are water insoluble and have a rigid structure (Ross et al., 1991). The two operons, bcsABZC and bcsEFG, encode the structural genes required for cellulose biosynthesis (Nobles et al., 2001; Zogaj et al., 2001; Solano et al., 2002), whereby BcsA and BcsB form the cellulose synthase complex (Romling, 2002; Omadjela et al., 2013).
Escherichia coli and Salmonella strains produce more than 80 distinct capsules which can also be a constituent of the biofilm matrix, and are classified into several groups. Group 4 capsules comprise of O-polysaccharides, structurally similar to the O-polysaccharides of the LPS, termed the O-antigen capsules. In E. coli, they are polymerized by the Wxy polymerase, and transferred across the membrane by the Wzx complex (Whitfield and Roberts, 1999). In Salmonella, two yih operons were found to be important for capsule assembly and translocation (Gibson et al., 2006).
A regulatory scheme for the main biofilm components is illustrated in Fig. 1. For clarity, only major regulators and regulators that have been investigated in relation to association of the bacteria with plants are shown. As outlined above, the extracellular matrix components, cellulose, curli fimbriae and in S. enterica BapA are positively regulated by CsgD, the major hub of biofilm formation (Romling et al., 2000; Uhlich et al., 2001; Latasa et al., 2006; Simm et al., 2014). Synthesis of the Salmonella O-antigen capsule coregulates with the cellulose synthesis. CsgD also regulates the yih genes in coordination with cellulose and curli (Gibson et al., 2006). Regulation of CsgD and subsequently the expression of the matrix components is highly responsive to many environmental signals such as growth phase, nutrients, oxygen tension, ethanol, temperature, osmolarity and a number of regulatory proteins (Gerstel and Romling, 2003). For most strains of Salmonella and also a fraction of E. coli strains, csgD expression is optimal at temperatures below 30°C in media with low salt (Romling et al., 1998; Bokranz et al., 2005). Maximal expression is observed during stationary phase upon limitation of nutrients such as nitrogen, phosphate and iron (Gerstel and Romling, 2003), which, directly and indirectly, requires the stationary-phase sigma factor RpoS (Arnqvist et al., 1994). The response regulator OmpR, a component of the two-component regulatory system OmpR/EnvZ that responds to changes in osmolarity (Pratt et al., 1996), is absolutely required for CsgD expression. Oxygen tension also plays a major determinative role in CsgD expression (Romling et al., 1998; Gerstel and Romling, 2001). Investigating 51 S. Typhimurium strains from different origins indeed demonstrated that most strains form optimal biofilm at acidic pH (∼ 5.5), 0.5% NaCl and 25°C (Lianou and Koutsoumanis, 2012). In the same line, the genes involved in curli and cellulose production were highly induced in a panel of S. enterica strains at 25°C and low nutrient availability (Castelijn et al., 2012), and also colanic acid is generally not produced at temperatures above 30°C (Whitfield and Roberts, 1999). PGA is synthesized at 37°C, but it is also implicated in attachment to surfaces during growth at lower temperatures (Wang et al., 2004).
Fig 1.
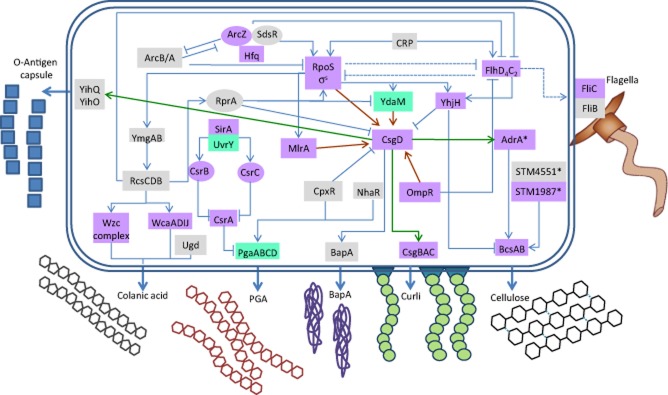
Regulation of components of the biofilm matrix in Salmonella typhimurium and Escherichia coli.Proteins and sRNAs controlling the synthesis of biofilm components are shown. Straight arrows: direct activation. Straight lines with blunt ends: direct inhibition. Dotted lines: indirect effects. Proteins are in boxes (green, E. coli only; grey, S. Typhimurium only; violet, in both species), and regulatory sRNAs are in circle. Cyclic di-GMP binding proteins are marked with an asterisk. Additional regulatory elements were not included for clarity. The figure is mainly based on data from the following references: (Gibson et al., 2006; Mika and Hengge, 2013; Anwar et al., 2014).
Regulation of cellulose biosynthesis by CsgD is indirect. Once CsgD is expressed in the stationary phase of growth, it activates the transcription of adrA encoding a diguanylate cyclase that subsequently leads to cellulose biosynthesis through the production of the allosteric activator cyclic di-GMP (Romling et al., 1998; 2000). However, cellulose biosynthesis can be uncoupled from CsgD expression, as alternative diguanylate cyclases encoded by the S. enterica chromosome like STM4551 and STM1987 can activate cellulose biosynthesis under alternative growth conditions (Anwar et al., 2014; Simm et al., 2014). Cyclic di-GMP has emerged as a major regulatory secondary messenger signalling system activating csgD expression in S. enterica and E. coli (Römling et al., 2013), and is usually with oppositely regulated with the flagella synthesis and function (Mika and Hengge, 2013). Regulation of csgD expression by cyclic di-GMP is complex and involves at least four di-guanylate cyclases and four phosphodiesterases in S. Typhimurium.
Hfq, a RNA chaperone, is another central positive regulator of biofilm formation in S. Typhimurium and E. coli (Holmqvist et al., 2010; Monteiro et al., 2012). In S. Typhimurium, Hfq together with its associated small RNAs ArcZ and SdsR positively control the expression of CsgD. ArcZ also regulates the transition between sessility and motility, and the timing of type 1 fimbriae versus curli fimbriae surface attachment at ambient temperatures (Monteiro et al., 2012).
Recently, it has been shown that hyper-expression of the T3SS-1 of Salmonella facilitates a cell aggregation phenotype that results in biofilm formation (Jennings et al., 2012). A similar observation was also described in the plant pathogen Erwinia chrysanthemi (Yap et al., 2005). This biofilm is not dependent on cellulose, curli or flagella production, and its extracellular matrix contains different virulence proteins secreted by the T3SS-1 system like SipA and SopB (Jennings et al., 2012). Other types of biofilms of S. Typhimurium are mainly dependent on type 1 fimbriae (Monteiro et al., 2012) and flagella (Crawford et al., 2010).
Biofilm formation by S. enterica and E. coli on produce surfaces and its role in survival on plants
The formation of biofilms by plant epiphytic or pathogenic bacteria has long been known (Danhorn and Fuqua, 2007); however, the discovery that human enteric pathogens are able to establish biofilms on plant surfaces was unexpected. Escherichia coli, Salmonella, Campylobacter, Listeria and Shigella have been found to form distinct biofilms on surfaces of produce such as tomatoes, melons and parsley (Agle, 2003; Annous et al., 2005; Iturriaga et al., 2007). Consequently, the number of studies on the formation of biofilms by foodborne pathogens on produce surfaces has expanded in the last decade only. A correlation between the ability to form biofilms and the attachment to fresh produce and/or survival was shown as strains with pronounced biofilm formation in vitro also attached to plants in significantly higher numbers, or survived better after disinfection (Lapidot et al., 2006; Patel and Sharma, 2010). This correlation extended to strains isolated from produce-related outbreaks. Isolates of Salmonella collected during tomato outbreaks produced biofilms and better adhered and attached to tomato leaflets compared with non-biofilm-producing strains (Cevallos-Cevallos et al., 2012). The E. coli O104:H4 strain that caused the devastating fenugreek seed outbreak in 2011 possesses a unique composition of multiple virulence factors, as well as the ability to produce rapidly a more stable and thicker biofilm compared with E. coli O157:H7. The ability of this strain to form a biofilm is probably aided by the production of high levels of exopolysaccharides and fimbriae due to overexpression of pgaA and aggR genes products, involved in the production of PGA, and regulation of type 1 aggregative adherence fimbriae respectively (Al Safadi et al., 2012). Also, E. coli strains isolated from plants formed significantly more biofilms and produced more cellulose and curli compared with E. coli strains isolated from human and other animals (Meric et al., 2013). Similarly, Salmonella isolates from produce commodities formed significantly thicker biofilms and persisted on spinach plants at higher numbers than poultry Salmonella isolates. Increasing persistence of the isolates was attributed to curli expression in several isolates. However, S. Tennessee formed a greater biofilm, but produced less curli, suggesting that other extracellular appendages contribute to attachment and biofilm formation (Patel et al., 2013). In contrast to the observations indicating that the ability to produce a biofilm improves the survival of the pathogens on the plant, recent evidence has shown that during adaptation of S. Typhimurium in green tomato fruits, the bacteria lose the ability to produce biofilms (Salazar et al., 2013). In line with this finding, a survey of the Salmonella strains recovered from produce-associated outbreaks revealed that many of them do not synthesize cellulose and/or curli (Zaragoza et al., 2012). Similarly, the majority of E. coli O157:H7 strains isolated from various outbreaks do not express curli and show only weak or non-biofilm formation in vivo (Liu et al., 2014). The reasons for these opposing observations are not clear.
Bacteria embedded within biofilms on plant tissue are more difficult to remove and more resistant to inactivation than their planktonic counterparts (Chmielewski and Frank, 2003). Biofilm-associated cells as well as capsule producing cells are also significantly more tolerant to desiccation, as deletion of CsgD, the major biofilm activator in Salmonella, resulted in increased susceptibility to desiccation stress (Gibson et al., 2006). Generally, in recent years, the role of biofilm-related genes in survival on sprouts, leaves or fruits of different plants was investigated (Table 3). The most investigated genes demonstrated various effects under different experimental conditions. For example, deletion of the cellulose synthase gene bcsA in Salmonella serovars resulted in a significant reduction in survival on alfalfa sprouts (Barak et al., 2007), but caused no effect on tomatoes (Noel et al., 2010). The genes with the most significant effect in survival on both alfalfa sprouts and tomatoes were the yih genes, involved in the synthesis of O-antigen capsule in Salmonella (Table 3).
Table 3.
Bacterial genes involved in biofilm formation and/or survival on plants
| Gene | Function | Pathogen | Plant/part | Method | Comments | Reference |
|---|---|---|---|---|---|---|
| adrA | Regulation of cellulose synthesis | S. Newport | Alfalfa sprouts | Directed deletion | No effect on survival 24–48 hpi | (Barak et al., 2007) |
| aroA | 5-enolpyruvylshikimate 3-phosphate synthase | S. Typhimurium | Red tomatoes | Directed deletion | reduction in survival | (Noel et al., 2010) |
| bcsA | Cellulose synthase | S. Enteritidis | Alfalfa sprouts | Directed deletion | 2 log reduction in survival | (Barak et al., 2007) |
| bcsA | Cellulose synthase | S. Typhimurium | Red tomatoes | Directed deletion | No effect on survival | (Noel et al., 2010) |
| csgB/bcsA | Curli/Cellulose | S. Enteritidis | Alfalfa sprouts | Directed deletion | 1.5 log reduction in survival | (Barak et al., 2007) |
| csgA | Curli subunit | E. coli K-12 and E. coli O157:H7 | lettuce leaves | Transposon insertion | 0.5–3 log reduction in survival | (Fink et al., 2012) |
| csgA | Curli subunit | E. coli O157:H7 | Spinach grown on hydroponics and in soil | Directed deletion and overexpression | No effect on root uptake and internalization | (Macarisin et al., 2014) |
| csgB/csgC | Curli subunit and curli assembly | S. Typhimurium | Red tomatoes | Directed deletion | No effect on survival | (Noel et al., 2010) |
| csgD | Regulation of biofilm and rdar morphotype | S. Typhimurium | Tomato leaves and fruits | Directed deletion and point mutation | rdar morphotype better survived in the plant | (Gu et al., 2011) |
| csrB/csrC | sRNAs, Global regulation | S. Typhimurium | Red tomatoes | Directed deletion | No effect on survival | (Noel et al., 2010) |
| cysB | Regulation of cysteine operon and antibiotic resistance | S. Typhimurium | Red tomatoes | Directed deletion | reduction in survival | (Noel et al., 2010) |
| flhDC | Flagella regulators | S. Typhimurium | Red tomatoes | Transposon insertion | No effect on survival | (Noel et al., 2010) |
| flhF | Flagella | S. Typhimurium | Red tomatoes | Directed deletion | No effect on survival | (Noel et al., 2010) |
| hilA | Regulator of virulence genes | S. Typhimurium | Red tomatoes | Directed deletion | Trend of improved survival | (Noel et al., 2010) |
| lpfA | Long polar fimbriae | S. Typhimurium | Red tomatoes | Directed deletion | No effect on survival | (Noel et al., 2010) |
| motA | Flagella motor | S. Typhimurium | Red tomatoes | Transposon insertion | Improved survival | (Noel et al., 2010) |
| pgaC | Poly-β-1,6-N-acetylglucosamine production | E. coli | Alfalfa sprouts | Transposon insertion | Decreased biofilm formation | (Matthysse et al., 2008) |
| sirA | Type VI secretion protein | S. Typhimurium | Red tomatoes | Directed deletion | Improved survival | (Noel et al., 2010) |
| sirA | Type VI secretion protein | S. Typhimurium and Saintpaul | Spinach leaves Grape tomatoes | Directed deletion | 2 log reduction in survival | (Salazar et al., 2013) |
| wcaD | Colanic acid production | E. coli | Alfalfa sprouts | Transposon insertion | Lowers biofilm formation | (Matthysse et al., 2008) |
| wcaJ | Colanic acid production | S. Enteritidis | Alfalfa sprouts | Directed deletion | No effect on survival | (Barak et al., 2007) |
| ycfR | Putative membrane protein involved in stress response and biofilm | E. coli K-12 and E. coli O157:H7 | Lettuce leaves | Transposon insertion | 1.5–3 log reduction in survival | (Fink et al., 2012) |
| yhjN | Cellulose production | E. coli | Alfalfa sprouts | Transposon insertion | Lowers biofilm formation | (Matthysse et al., 2008) |
| yihO | O-antigen capsule assembly | S. Enteritidis | Alfalfa sprouts | Directed deletion | 2.5 log reduction in survival 24 hpi | (Barak et al., 2007) |
| yihT | O-antigen capsule assembly | S. Typhimurium | Green and red tomatoes | Directed deletion | 2–3 log reduction in survival 34 dpa on green tomatoes and in fruits defective in ethylene perception | (Marvasi et al., 2013) |
Presumably, these and other mechanisms aid long-term survival of biofilm-associated cells on the plant surface not only in the field but also during harvest, transport, sanitation and storage. On parsley plants, for instance, resistance to disinfection treatments (i.e. chlorination) was improved in biofilm producing S. Typhimurium compared with non-producing mutants, when washing was conducted a week after inoculation, but no differences were observed when the treatment was carried out a few hours after inoculation (Lapidot et al., 2006).
Regulation of biofilm formation on plants
As discussed above, several environmental conditions were shown to have an impact on biofilm production. For example, biofilm formation of Salmonella has been reported to be maximal under reduced nutrient availability, aerobic conditions, low osmolarity and mid temperatures (25–28°C) (Gerstel and Romling, 2003). All these conditions exist on a plant surface rather than in the gut environment. Indeed, several studies reported about induction of biofilm-associated genes in the plant environment. Assessment of the transcriptional profile in E. coli O157:H7 attached to intact lettuce leaves showed that 10% of the genes have at least a twofold expression change between day 0 and day 1 and/or day 3 (Fink et al., 2012). Among them, csgA and csgB showed 34.6-fold and 13.9-fold induction, respectively, and rpoS 2.1-fold induction. Other genes probably involve in biofilm regulation, such as ybiM and yceP, also show a very strong induction (Fink et al., 2012). In line with these findings, E. coli attached to the lettuce rhizosphere upregulates the csg genes (Hou et al., 2012), and S. Weltevreden upregulated csg and TTSS-2 genes during alfalfa sprout colonization as compared with M9 minimal medium (Brankatschk et al., 2013). Screening for Salmonella genes differentially regulated in tomatoes relative to growth in Luria–Bertani soft agar identified several genes, including genes associated with attachment, stress response, biofilm formation and capsule formation (Noel et al., 2010). Furthermore, deletion of SirA, involved in regulation of type 1 fimbriae and biofilm formation (Teplitski et al., 2006), and MotA, involved in motility, modestly affected fitness, while YihT, involved in synthesis of the capsule, affected the fitness in green, but not ripe tomatoes. In these studies, known Salmonella genes associated with regulation of motility and virulence in animals, such as hilA, flhDC and fliF, did not contribute to the fitness of the bacteria (Noel et al., 2010; Marvasi et al., 2013).
Incorporation of pathogens in multi-species biofilms
Bacterial cells introduced to the leaf surface, if not producing their own aggregates or biofilms, have a better chance of surviving when they are deposited on or in aggregates of existing bacteria (Monier and Lindow, 2005). Salmonella enterica serovar Thompson and the plant pathogen Pantoea agglomerans, for instance, were shown to form co-aggregates on cilantro leaves (Brandl and Mandrell, 2002), and the epiphytes Wausteria paucula supported the survival of E. coli O157:H7 on lettuce leaves (Cooley et al., 2006). Also, the fungal plant pathogens Cladosporium cladosporioides and Penicillium expansum promoted the colonization of Salmonella in cantaloupe (Richards and Beuchat, 2005). A recent study has shown that E. coli O157:H7 strains promote biofilm formation of some strong biofilm-producing species isolated from fresh produce processing facilities, such as Burkholderia caryophylli and Ralstonia insidiosa. Likewise, the population of E. coli increases by 1 log in the dual-species biofilms (Liu et al., 2014). These studies demonstrate that indigenous microorganisms may aid attachment and long-term survival of foodborne pathogens on the plant surface and during washing with antimicrobials. However, it is not clear whether this positive effect results from embedding of foodborne pathogens in the already existing biofilms or from other interactions like lesion and release of nutrients.
The plant response and its effect on bacterial biofilms
Discussion of plant-associated biofilms is not completed without referring to the plant defence response. Plants apply a range of mechanisms for protection against microorganisms. The plant protection includes local or systemic production of defence enzymes and antimicrobial molecules. Some mechanisms, like the production of essential oils with a broad-range antimicrobial activity, are constitutively active, while others, like secretion of reactive oxygen species (ROS), are induced after exposure to pathogens. Evidence indicates that plants apply similar mechanisms of defence in response to internalization of human enteric pathogens as against plant pathogens. Microorganisms encounter surface receptors on host cells, termed pattern recognition receptors (PRRs), that recognize conserved pathogen-associated molecular patterns (PAMPs) and trigger downstream defence signalling pathways. PAMPs include conserved microbial surface molecules such as flagellin, LPS and glycoproteins (Nurnberger et al., 2004). Recognition of PAMPs by PRRs initiates PAMP-triggered immunity (PTI), which usually prevents microbial growth and halts infection before the microorganism gains a hold in the plant (Chisholm et al., 2006). Many surface components of E. coli and Salmonella such as LPS, flagella, peptidoglycans, curli and pili are homologous to PAMPs of plant pathogens. Deletion of these PAMPs from E. coli or Salmonella usually resulted in better colonization of the interior of the plants than the wild-type strains (Iniguez et al., 2005; Seo and Matthews, 2012). Some of these molecules are also components of the biofilm matrix. Curli can serve as a PAMP (Seo and Matthews, 2012), and the S. Typhimurium LPS being a strong PAMP in tobacco plants (Shirron and Yaron, 2011). The flagellin subunit FliC, but not FliB, is recognized by Nicotiana benthamiana and tomato plants as a PAMP and activates their PTI (Meng et al., 2013). These observations indicate that components of the bacteria and the biofilm matrix may induce the plant response. On the other hand, biofilm production may mask the underlying bacterial surface, providing further protection. Indeed, an E. coli O157:H7 mutant that produces a great amount of exopolysaccharides and a thick capsule exhibits a better survival pattern on Arabidopsis compared with the wild-type strain (Meng et al., 2013).
The response of the plant cells results in production of ROS, nitric oxide, different ions as well as molecular signals like jasmonate, salicylic acid and ethylene (Jones and Dangl, 2006). Some of these compounds were found to have a role not only against persistence of enteric pathogens in plants but also against biofilm formation. Ethylene and salicylic acid decreased endophytic colonization of Salmonella in alfalfa (Iniguez et al., 2005). Interestingly, an inverse correlation was observed between in vitro biofilm formation by uropathogenic E. coli and the salicylate concentration (Vila and Soto, 2012). Moreover, the synthesis of fimbriae such as P fimbriae and type 1 fimbriae was reduced following growth in the presence of salicylate (Kunin et al., 1994). In the case of type 1 fimbriae, salicylate decreased the expression of fimA coding for its major structural subunit (Vila and Soto, 2012). Similarly, jasmonic acid reduced the synthesis of the O-antigen capsule by Salmonella in tomatoes. In mammals, the yih operon of Salmonella is induced by bile and enhances biofilm formation on gallbladder by triggering the O-antigen capsule production. In tomatoes, on the other hand, jasmonic acid and its precursors, produced during the plant response, strongly reduce the expression of YihT, and thus inhibit biofilm formation (Marvasi et al., 2013).
Other microorganisms in the plant environment can also affect, directly or indirectly, through induction of the plant defence response, the biofilm formation by human enteric pathogens. The soil bacterium B. subtilis colonizes roots of different plants and is currently used as a biocontrol agent against plant pathogens. During colonization of the roots, it produces surfactin, an antimicrobial lipopeptide that not only inhibits the growth of common plant pathogens, but also induces the plant defence response (reviewed in Vlamakis et al., 2013). Interestingly, surfactin molecules also delay adhesion of Salmonella and Listeria strains to solid surfaces (Nitschke et al., 2009), and inhibit biofilm formation by Salmonella (Mireles et al., 2001). However, their influence on colonization of enteric pathogens on plants has not been investigated yet.
In addition to the plant immune response triggered upon exposure to microorganisms, many plants produce antimicrobial compounds constitutively. Interestingly, some of these compounds do not only inhibit bacterial growth, but also inhibit biofilm formation or even efficiently kill the pathogens specifically in biofilms. For example, 3% of 498 investigated plant extracts inhibited biofilm formation of E. coli O157:H7. The most active extract, Carex dimorpholepis, inhibits curli formation and decreases swimming and swarming motility, probably by repression of quorum sensing genes (Lee et al., 2013). In another study, Carex plant extracts inhibited E. coli O157:H7 and Pseudomonas aeruginosa biofilm formation without affecting planktonic cells growth. One of the active antibiofilm compounds in these extracts was ε-viniferin (Cho et al., 2013). Plant sterols such as the β-sitosterol glucoside from citrus are potent inhibitors of E. coli O157:H7 biofilm formation and motility, without affecting the cell viability. This inhibition probably occurs through repression of rssAB and hns, regulatory elements of the flagellar operon (Vikram et al., 2013). Essential oils from cassia (Cinnamomum aromaticum) and Peru balsam (Myroxylon balsamum) kill plant and human pathogens within biofilms in similar efficacy as the planktonic cells, and the oil of red thyme (Thymus vulgaris) is even more effective against biofilms cells than planktonic bacteria (Kavanaugh and Ribbeck, 2012). Helichrysum italicum produces several metabolites such as acylated styrylpyrones derivatives with anti-biofilm properties against P. aeruginosa (D'Abrosca et al., 2013).
In conclusion, regulation of biofilm formation on the plant is a complex process affected by many factors. There are several levels of interactions of the plant response with bacteria and their biofilms. Current study indicates a role of biofilm formation in triggering or evading the plant response, while molecules produced by the plant either constituently or in response to the bacteria may trigger or inhibit the process of biofilm formation.
Conclusion
Most likely, plants do not provide optimal conditions for growth of human enteric pathogens, but the pathogens use different mechanisms to tightly attach to favourable sites on the plant organs and to fit the harsh conditions in order to survive in the plants long enough and in sufficient numbers for successful infection of new mammalian hosts with the consequence of a significant number of outbreaks. The studies detailed in this review indicate that production of biofilms is a common strategy employed by pathogenic Salmonella and E. coli strains to survive in different niches of the plant. Following a tight adhesion on favourable sites, the process of biofilm formation on the plant is affected by many environmental factors, as well as properties of the plant and characters of the pathogenic strain or the ingenious plant microflora. The synthesis of the biofilm matrix components such as cellulose and curli occurs under environmental conditions similar to the ones existing on the plants and may be further induced or inhibited by compounds produced by the plants or neighbouring microorganisms (Fig. 2). Although intensive research activity is ongoing, our current understanding of the factors that affect biofilm formation and its role in survival of foodborne pathogens on the plants is rudimentary. The different strains, serovars and pathovars and a wide variety of host plants in combination with diverse experimental methods of inoculation and plant growth, limit the ability to compare the results of different studies and to unambiguously conclude the dominant factors for adhesion and biofilm formation.
Fig 2.
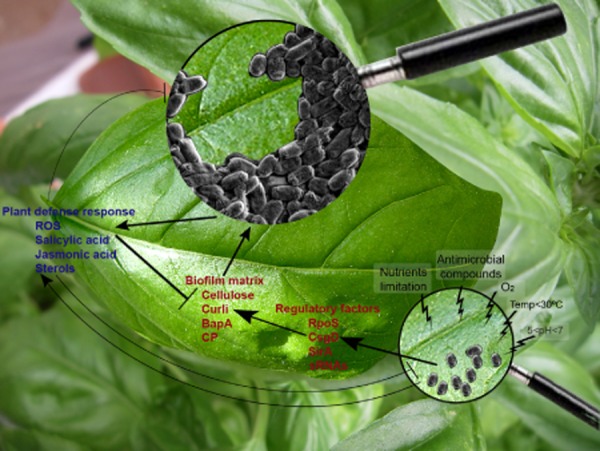
Illustration of biofilm formation by Salmonella cells on a leaf.Upon attachment of Salmonella cells to the leaf, the bacteria are exposed to environmental conditions (temperature below 30°C, atmospheric oxygen, etc.) that trigger expression of regulatory sRNAs and proteins such as RpoS, CsgD and SirA. Expression of these proteins and sRNAs is enhanced by stress signals existing on the leaf surface such as low availability of nutrients, and activity of antimicrobial compounds produced by the plant or indigenous microorganisms. The induced regulatory proteins activate the genes involved in production of components of the biofilm matrix such as cellulose, curli, BapA and capsules (CP), leading to the development of biofilms on the leaf surface. While the biofilm structure stabilizes the colonization on the plant and provides protection from different stresses, its components also contribute to the induction of the local and systemic plant defence response. As part of the plant response, triggered by both, single bacteria and biofilms, the plant produces and secretes different signal and antimicrobial compounds such as ROS compounds, salicylic acid, jasmonic acid and sterols. Some of these compounds kill free and biofilm associated bacteria and/or inhibit the process of biofilm formation.
Recent evidence has shown that several plant hosts and environmental plant-associated bacteria such as B. subtilis are able to synthesize and secrete compounds that not only target planktonic cells, but also inhibit attachment and biofilm formation or even specifically kill bacteria in biofilms. Future studies on plant-related compounds or plant symbionts that affect bacterial biofilm formation are required and might discover novel substances involved in biofilm formation. Subsequently, understanding of the mode of action of such compounds will aid in development of novel strategies to decrease the fitness of human enteric pathogens on fresh produce and prevent outbreaks, and also in inhibition of biofilm formation not only on such foodstuff, but also on many other types of solid surfaces of medical, industrial and agricultural importance.
Acknowledgments
The authors expresses thanks to Dr. Natali Shirron, Lee Shavit, Eden Eran and Noga Cohen who contributed to the preparation of the figures in this review.
Conflict of interest
None declared.
References
- Agle ME. 2003. Shigella boydii18: Characterization and biofilm formation. PhD thesis. Champaign, IL: University of Illinois.
- Ahmad I, Wigren E, Le Guyon S, Vekkeli S, Blanka A, El Mouali Y, et al. The EAL-like protein STM1697 regulates virulence phenotypes, motility and biofilm formation in Salmonella Typhimurium. Mol Microbiol. 2013;90:1216–1232. doi: 10.1111/mmi.12428. [DOI] [PubMed] [Google Scholar]
- Al Safadi R, Abu-Ali GS, Sloup RE, Rudrik JT, Waters CM, Eaton KA. Manning SD. Correlation between in vivo biofilm formation and virulence gene expression in Escherichia coli O104:H4. PLoS ONE. 2012;7:e41628. doi: 10.1371/journal.pone.0041628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allende A, Martinez B, Selma V, Gil MI, Suarez JE. Rodriguez A. Growth and bacteriocin production by lactic acid bacteria in vegetable broth and their effectiveness at reducing Listeria monocytogenes in vitro and in fresh-cut lettuce. Food Microbiol. 2007;24:759–766. doi: 10.1016/j.fm.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Annous BA, Burke A. Sites JE. Surface pasteurization of whole fresh cantaloupes inoculated with Salmonella Poona or Escherichia coli. J Food Prot. 2004;67:1876–1885. doi: 10.4315/0362-028x-67.9.1876. [DOI] [PubMed] [Google Scholar]
- Annous BA, Solomon EB, Cooke PH. Burke A. Biofilm formation by Salmonella spp. on cantaloupe melons. J Food Saf. 2005;25:276–287. [Google Scholar]
- Anonymous 2008. Outbreak alert! Closing the gaps in our federal food-safety net. Center for Science in the Public Interest: Washington, DC. URL http://cspinet.org/new/pdf/outbreak_alert_2008_report_final.pdf.
- Anriany YA, Weiner RM, Johnson JA, De Rezende CE. Joseph SW. Salmonella enterica serovar Typhimurium DT104 displays a rugose phenotype. Appl Environ Microbiol. 2001;67:4048–4056. doi: 10.1128/AEM.67.9.4048-4056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar N, Rouf SF, Romling U. Rhen M. Modulation of biofilm-formation in Salmonella enterica serovar Typhimurium by the periplasmic DsbA/DsbB oxidoreductase system requires the GGDEF-EAL domain protein STM3615. PLoS ONE. 2014;9:e106095. doi: 10.1371/journal.pone.0106095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist A, Olsen A. Normark S. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol. 1994;13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x. [DOI] [PubMed] [Google Scholar]
- Barak JD, Whitehand LC. Charkowski AO. Differences in attachment of Salmonella enterica serovars and Escherichia coli O157:H7 to alfalfa sprouts. Appl Environ Microbiol. 2002;68:4758–4763. doi: 10.1128/AEM.68.10.4758-4763.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak JD, Gorski L, Naraghi-Arani P. Charkowski AO. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl Environ Microbiol. 2005;71:5685–5691. doi: 10.1128/AEM.71.10.5685-5691.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak JD, Jahn CE, Gibson DL. Charkowski AO. The role of cellulose and O-antigen capsule in the colonization of plants by Salmonella enterica. Mol Plant Microbe Interact. 2007;20:1083–1091. doi: 10.1094/MPMI-20-9-1083. [DOI] [PubMed] [Google Scholar]
- Barak JD, Gorski L, Liang AS. Narm KE. Previously uncharacterized Salmonella enterica genes required for swarming play a role in seedling colonization. Microbiology. 2009;155:3701–3709. doi: 10.1099/mic.0.032029-0. [DOI] [PubMed] [Google Scholar]
- Barnhart MM. Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie GA. Lindow SE. Bacterial colonization of leaves: a spectrum of strategies. Phytopathology. 1999;89:353–359. doi: 10.1094/PHYTO.1999.89.5.353. [DOI] [PubMed] [Google Scholar]
- Berger CN, Shaw RK, Brown DJ, Mather H, Clare S, Dougan G, et al. Interaction of Salmonella enterica with basil and other salad leaves. ISME J. 2009;3:261–265. doi: 10.1038/ismej.2008.95. [DOI] [PubMed] [Google Scholar]
- Beuchat LR. Comparison of chemical treatments to kill Salmonella on alfalfa seeds destined for sprout production. Int J Food Microbiol. 1997;34:329–333. doi: 10.1016/s0168-1605(96)01202-0. [DOI] [PubMed] [Google Scholar]
- Beuchat LR, Adler BB. Lang MM. Efficacy of chlorine and a peroxyacetic acid sanitizer in killing Listeria monocytogenes on iceberg and Romaine lettuce using simulated commercial processing conditions. J Food Prot. 2004;67:1238–1242. doi: 10.4315/0362-028x-67.6.1238. [DOI] [PubMed] [Google Scholar]
- Blair KM, Turner L, Winkelman JT, Berg HC. Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- Bokranz W, Wang X, Tschape H. Romling U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J Med Microbiol. 2005;54:1171–1182. doi: 10.1099/jmm.0.46064-0. [DOI] [PubMed] [Google Scholar]
- Boyer RR, Sumner SS, Williams RC, Pierson MD, Popham DL. Kniel KE. Influence of curli expression by Escherichia coli 0157:H7 on the cell's overall hydrophobicity, charge, and ability to attach to lettuce. J Food Prot. 2007;70:1339–1345. doi: 10.4315/0362-028x-70.6.1339. [DOI] [PubMed] [Google Scholar]
- Brandl MT. Fitness of human enteric pathogens on plants and implications for food safety. Annu Rev Phytopathol. 2006;44:367–392. doi: 10.1146/annurev.phyto.44.070505.143359. [DOI] [PubMed] [Google Scholar]
- Brandl MT. Mandrell RE. Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl Environ Microbiol. 2002;68:3614–3621. doi: 10.1128/AEM.68.7.3614-3621.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brankatschk K, Kamber T, Pothier JF, Duffy B. Smits TH. Transcriptional profile of Salmonella enterica subsp. enterica serovar Weltevreden during alfalfa sprout colonization. Microb Biotechnol. 2013 doi: 10.1111/1751-7915.12104. doi: 10.1111/1751-7915.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JV, Mohle-Boetani J, Reporter R, Abbott S, Farrar J, Brandl M, et al. An outbreak of Salmonella serotype Thompson associated with fresh cilantro. J Infect Dis. 2001;183:984–987. doi: 10.1086/319254. [DOI] [PubMed] [Google Scholar]
- Castelijn GA, van der Veen S, Zwietering MH, Moezelaar R. Abee T. Diversity in biofilm formation and production of curli fimbriae and cellulose of Salmonella Typhimurium strains of different origin in high and low nutrient medium. Biofouling. 2012;28:51–63. doi: 10.1080/08927014.2011.648927. [DOI] [PubMed] [Google Scholar]
- CDC. 2014. List of selected multistate foodborne outbreak investigations. URL http://www.cdc.gov/foodsafety/outbreaks/multistate-outbreaks/outbreaks-list.html.
- Cevallos-Cevallos JM, Gu G, Danyluk MD. van Bruggen AH. Adhesion and splash dispersal of Salmonella enterica Typhimurium on tomato leaflets: effects of rdar morphotype and trichome density. Int J Food Microbiol. 2012;160:58–64. doi: 10.1016/j.ijfoodmicro.2012.09.021. [DOI] [PubMed] [Google Scholar]
- Chia TW, Goulter RM, McMeekin T, Dykes GA. Fegan N. Attachment of different Salmonella serovars to materials commonly used in a poultry processing plant. Food Microbiol. 2009;26:853–859. doi: 10.1016/j.fm.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B. Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Chmielewski RAN. Frank JF. Biofilm formation and control in food processing facilities. Comp Rev Food Sci Food Safety. 2003;2:22–32. doi: 10.1111/j.1541-4337.2003.tb00012.x. [DOI] [PubMed] [Google Scholar]
- Cho HS, Lee JH, Ryu SY, Joo SW, Cho MH. Lee J. Inhibition of Pseudomonas aeruginosa and Escherichia coli O157:H7 biofilm formation by plant metabolite epsilon-viniferin. J Agric Food Chem. 2013;61:7120–7126. doi: 10.1021/jf4009313. [DOI] [PubMed] [Google Scholar]
- Collignon S. Korsten L. Attachment and colonization by Escherichia coli O157:H7, Listeria monocytogenesSalmonella enterica subsp. enterica serovar Typhimurium, and Staphylococcus aureus on stone fruit surfaces and survival through a simulated commercial export chain. J Food Prot. 2010;73:1247–1256. doi: 10.4315/0362-028x-73.7.1247. [DOI] [PubMed] [Google Scholar]
- Cooley MB, Chao D. Mandrell RE. Escherichia coli O157:H7 survival and growth on lettuce is altered by the presence of epiphytic bacteria. J Food Prot. 2006;69:2329–2335. doi: 10.4315/0362-028x-69.10.2329. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR. Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS. Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Craven SE. Williams DD. In vitro attachment of Salmonella Typhimurium to chicken cecal mucus: effect of cations and pretreatment with Lactobacillus spp. isolated from the intestinal tracts of chickens. J Food Prot. 1998;61:265–271. doi: 10.4315/0362-028x-61.3.265. [DOI] [PubMed] [Google Scholar]
- Crawford RW, Reeve KE. Gunn JS. Flagellated but not hyperfimbriated Salmonella enterica serovar Typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J Bacteriol. 2010;192:2981–2990. doi: 10.1128/JB.01620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crull K, Rohde M, Westphal K, Loessner H, Wolf K, Felipe-Lopez A, et al. Biofilm formation by Salmonella enterica serovar Typhimurium colonizing solid tumours. Cell Microbiol. 2011;13:1223–1233. doi: 10.1111/j.1462-5822.2011.01612.x. [DOI] [PubMed] [Google Scholar]
- Danhorn T. Fuqua C. Biofilm formation by plant associated bacteria. Annu Rev Microbiol. 2007;61:401–422. doi: 10.1146/annurev.micro.61.080706.093316. [DOI] [PubMed] [Google Scholar]
- Davey ME. O'Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Abrosca B, Buommino E, D'Angelo G, Coretti L, Scognamiglio M, Severino V, et al. Spectroscopic identification and anti-biofilm properties of polar metabolites from the medicinal plant Helichrysum italicum against Pseudomonas aeruginosa. Bioorg Med Chem. 2013;21:7038–7046. doi: 10.1016/j.bmc.2013.09.019. [DOI] [PubMed] [Google Scholar]
- Dinu LD. Bach S. Induction of viable but nonculturable Escherichia coli O157:H7 in the phyllosphere of lettuce: a food safety risk factor. Appl Environ Microbiol. 2011;77:8295–8302. doi: 10.1128/AEM.05020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MP. Erickson MC. Summer meeting 2007 – the problems with fresh produce: an overview. J Appl Microbiol. 2008;105:317–330. doi: 10.1111/j.1365-2672.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- Dunne WM., Jr Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev. 2002;15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves CL, Jones BD. Clegg S. Biofilm formation by Salmonella enterica serovar Typhimurium and Escherichia coli on epithelial cells following mixed inoculations. Infect Immun. 2005;73:5198–5203. doi: 10.1128/IAI.73.8.5198-5203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink RC, Black EP, Hou Z, Sugawara M, Sadowsky MJ. Diez-Gonzalez F. Transcriptional responses of Escherichia coli K-12 and O157:H7 associated with lettuce leaves. Appl Environ Microbiol. 2012;78:1752–1764. doi: 10.1128/AEM.07454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz E, Visser AA, Van Diepeningen AD, Klerks MM, Termorshuizen AJ. van Bruggen AH. Quantification of contamination of lettuce by GFP-expressing Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium. Food Microbiol. 2007;24:106–112. doi: 10.1016/j.fm.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Gandhi M, Golding S, Yaron S. Matthews KR. Use of green fluorescent protein expressing Salmonella Stanley to investigate survival, spatial location, and control on alfalfa sprouts. J Food Prot. 2001;64:1891–1898. doi: 10.4315/0362-028x-64.12.1891. [DOI] [PubMed] [Google Scholar]
- Gerstel U. Romling U. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella Typhimurium. Environ Microbiol. 2001;3:638–648. doi: 10.1046/j.1462-2920.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- Gerstel U. Romling U. The csgD promoter, a control unit for biofilm formation in Salmonella Typhimurium. Res Microbiol. 2003;154:659–667. doi: 10.1016/j.resmic.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Gibson DL, White AP, Snyder SD, Martin S, Heiss C, Azadi P, et al. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J Bacteriol. 2006;188:7722–7730. doi: 10.1128/JB.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golberg D, Kroupitski Y, Belausov E, Pinto R. Sela S. Salmonella Typhimurium internalization is variable in leafy vegetables and fresh herbs. Int J Food Microbiol. 2011;145:250–257. doi: 10.1016/j.ijfoodmicro.2010.12.031. [DOI] [PubMed] [Google Scholar]
- Gonzalez RJ, Luo Y, Ruiz-Cruz S. McEvoy JL. Efficacy of sanitizers to inactivate Escherichia coli O157:H7 on fresh-cut carrot shreds under simulated process water conditions. J Food Prot. 2004;67:2375–2380. doi: 10.4315/0362-028x-67.11.2375. [DOI] [PubMed] [Google Scholar]
- Gu G, Hu J, Cevallos-Cevallos JM, Richardson SM, Bartz JA. van Bruggen AH. Internal colonization of Salmonella enterica serovar Typhimurium in tomato plants. PLoS ONE. 2011;6:e27340. doi: 10.1371/journal.pone.0027340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haahtela K, Tarkka E. Korhonen TK. Type 1 fimbria-mediated adhesion of enteric bacteria to grass roots. Appl Environ Microbiol. 1985;49:1182–1185. doi: 10.1128/aem.49.5.1182-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann J, Quadt-Hallmann A, Miller WG, Sikora RA. Lindow SE. Endophytic colonization of plants by the biocontrol agent Rhizobium etli G12 in relation to Meloidogyne incognita infection. Phytopathology. 2001;91:415–422. doi: 10.1094/PHYTO.2001.91.4.415. [DOI] [PubMed] [Google Scholar]
- Hammar M, Bian Z. Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Sherman DM, Linton RH, Nielsen SS. Nelson PE. The effects of washing and chlorine dioxide gas on survival and attachment of Escherichia coli O157: H7 to green pepper surfaces. Food Microbiol. 2000;17:521–533. [Google Scholar]
- Hassan AN. Frank JF. Influence of surfactant hydrophobicity on the detachment of Escherichia coli O157:H7 from lettuce. Int J Food Microbiol. 2003;87:145–152. doi: 10.1016/s0168-1605(03)00062-x. [DOI] [PubMed] [Google Scholar]
- Hassan AN. Frank JF. Attachment of Escherichia coli O157:H7 grown in tryptic soy broth and nutrient broth to apple and lettuce surfaces as related to cell hydrophobicity, surface charge, and capsule production. Int J Food Microbiol. 2004;96:103–109. doi: 10.1016/S0168-1605(03)00160-0. [DOI] [PubMed] [Google Scholar]
- Henderson IR. Owen P. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and oxyR. J Bacteriol. 1999;181:2132–2141. doi: 10.1128/jb.181.7.2132-2141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Reyes C. Schikora A. Salmonella, a cross-kingdom pathogen infecting humans and plants. FEMS Microbiol Lett. 2013;343:1–7. doi: 10.1111/1574-6968.12127. [DOI] [PubMed] [Google Scholar]
- Holmqvist E, Reimegard J, Sterk M, Grantcharova N, Romling U. Wagner EG. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010;29:1840–1850. doi: 10.1038/emboj.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Fink RC, Black EP, Sugawara M, Zhang Z, Diez-Gonzalez F. Sadowsky MJ. Gene expression profiling of Escherichia coli in response to interactions with the lettuce rhizosphere. J Appl Microbiol. 2012;113:1076–1086. doi: 10.1111/j.1365-2672.2012.05412.x. [DOI] [PubMed] [Google Scholar]
- Iniguez AL, Dong Y, Carter HD, Ahmer BM, Stone JM. Triplett EW. Regulation of enteric endophytic bacterial colonization by plant defenses. Mol Plant Microbe Interact. 2005;18:169–178. doi: 10.1094/MPMI-18-0169. [DOI] [PubMed] [Google Scholar]
- Islam M, Doyle MP, Phatak SC, Millner P. Jiang X. Persistence of enterohemorrhagic Escherichia coli O157:H7 in soil and on leaf lettuce and parsley grown in fields treated with contaminated manure composts or irrigation water. J Food Prot. 2004;67:1365–1370. doi: 10.4315/0362-028x-67.7.1365. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, et al. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2008;190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriaga MH, Escartin EF, Beuchat LR. Martínez-Peniche R. Effect of inoculum size, relative humidity, storage temperature, and ripening stage on the attachment of Salmonella Montevideo to tomatoes and tomatillos. J Food Prot. 2003;66:1756–1761. doi: 10.4315/0362-028x-66.10.1756. [DOI] [PubMed] [Google Scholar]
- Iturriaga MH, Tamplin ML. Escartin EF. Colonization of tomatoes by Salmonella Montevideo is affected by relative humidity and storage temperature. J Food Prot. 2007;70:30–34. doi: 10.4315/0362-028x-70.1.30. [DOI] [PubMed] [Google Scholar]
- Jennings ME, Quick LN, Ubol N, Shrom S, Dollahon N. Wilson JW. Characterization of Salmonella type III secretion hyper-activity which results in biofilm-like cell aggregation. PLoS ONE. 2012;7:e33080. doi: 10.1371/journal.pone.0033080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeter C. Matthysse AG. Characterization of the binding of diarrheagenic strains of E. coli to plant surfaces and the role of curli in the interaction of the bacteria with alfalfa sprouts. Mol Plant Microbe Interact. 2005;18:1235–1242. doi: 10.1094/MPMI-18-1235. [DOI] [PubMed] [Google Scholar]
- Jones JD. Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Joseph B, Otta SK. Karunasagar I. Biofilm formation by Salmonella spp. on food contact surfaces and their sensitivity to sanitizers. Int J Food Microbiol. 2001;64:367–372. doi: 10.1016/s0168-1605(00)00466-9. [DOI] [PubMed] [Google Scholar]
- Karamanoli K, Thalassinos G, Karpouzas D, Bosabalidis AM, Vokou D. Constantinidou HI. Are leaf glandular trichomes of oregano hospitable habitats for bacterial growth? J Chem Ecol. 2012;38:476–485. doi: 10.1007/s10886-012-0117-7. [DOI] [PubMed] [Google Scholar]
- Kavanaugh NL. Ribbeck K. Selected antimicrobial essential oils eradicate Pseudomonas spp. and Staphylococcus aureus biofilms. Appl Environ Microbiol. 2012;78:4057–4061. doi: 10.1128/AEM.07499-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH. Wei CI. Biofilm formation by multidrug-resistant Salmonella enterica serotype Typhimurium phage type DT104 and other pathogens. J Food Prot. 2007;70:22–29. doi: 10.4315/0362-028x-70.1.22. [DOI] [PubMed] [Google Scholar]
- Kisluk G. Yaron S. Presence and persistence of Salmonella enterica serotype Typhimurium in the phyllosphere and rhizosphere of spray-irrigated parsley. Appl Environ Microbiol. 2012;78:4030–4036. doi: 10.1128/AEM.00087-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisluk G, Hoover D, Kniel K. Yaron S. Quantification of low and high levels of Salmonella enterica serovar Typhimurium on leaves. LWT – Food Sci Technol. 2012;45:36–42. [Google Scholar]
- Kisluk G, Kalily E. Yaron S. Resistance to essential oils affects survival of Salmonella enterica serovars in growing and harvested basil. Environ Microbiol. 2013;15:2787–2798. doi: 10.1111/1462-2920.12139. [DOI] [PubMed] [Google Scholar]
- Klerks MM, van Gent-Pelzer M, Franz E, Zijlstra C. van Bruggen AH. Physiological and molecular responses of Lactuca sativa to colonization by Salmonella enterica serovar Dublin. Appl Environ Microbiol. 2007;73:4905–4914. doi: 10.1128/AEM.02522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N, Murata M. Isshiki K. Efficiency of sodium hypochlorite, fumaric acid, and mild heat in killing native microflora and Escherichia coli O157:H7, Salmonella Typhimurium DT104, and Staphylococcus aureus attached to fresh-cut lettuce. J Food Prot. 2006;69:323–329. doi: 10.4315/0362-028x-69.2.323. [DOI] [PubMed] [Google Scholar]
- Kroupitski Y, Pinto R, Belausov E. Sela S. Distribution of Salmonella Typhimurium in romaine lettuce leaves. Food Microbiol. 2011;28:990–997. doi: 10.1016/j.fm.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Kunin CM, Hua TH, Guerrant RL. Bakaletz LO. Effect of salicylate, bismuth, osmolytes, and tetracycline resistance on expression of fimbriae by Escherichia coli. Infect Immun. 1994;62:2178–2186. doi: 10.1128/iai.62.6.2178-2186.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter S, Hartmann A. Schmid M. Colonization of barley (Hordeum vulgare) with Salmonella enterica and Listeria spp. FEMS Microbiol Ecol. 2006;56:262–271. doi: 10.1111/j.1574-6941.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Lapidot A. Yaron S. Transfer of Salmonella enterica serovar Typhimurium from contaminated irrigation water to parsley is dependent on curli and cellulose, the biofilm matrix components. J Food Prot. 2009;72:618–623. doi: 10.4315/0362-028x-72.3.618. [DOI] [PubMed] [Google Scholar]
- Lapidot A, Romling U. Yaron S. Biofilm formation and the survival of Salmonella Typhimurium on parsley. Int J Food Microbiol. 2006;109:229–233. doi: 10.1016/j.ijfoodmicro.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Latasa C, Solano C, Penades JR. Lasa I. Biofilm-associated proteins. C R Biol. 2006;329:849–857. doi: 10.1016/j.crvi.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Laus MC, van Brussel AA. Kijne JW. Role of cellulose fibrils and exopolysaccharides of Rhizobium leguminosarum in attachment to and infection of Vicia sativa root hairs. Mol Plant Microbe Interact. 2005;18:533–538. doi: 10.1094/MPMI-18-0533. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cho HS, Joo SW, Chandra Regmi S, Kim JA, Ryu CM, et al. Diverse plant extracts and trans-resveratrol inhibit biofilm formation and swarming of Escherichia coli O157:H7. Biofouling. 2013;29:1189–1203. doi: 10.1080/08927014.2013.832223. [DOI] [PubMed] [Google Scholar]
- Lehmacher A, Bockemuhl J. Aleksic S. Nationwide outbreak of human salmonellosis in Germany due to contaminated paprika and paprika-powdered potato chips. Epidemiol Infect. 1995;115:501–511. doi: 10.1017/s0950268800058660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianou A. Koutsoumanis KP. Strain variability of the biofilm-forming ability of Salmonella enterica under various environmental conditions. Int J Food Microbiol. 2012;160:171–178. doi: 10.1016/j.ijfoodmicro.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Liao CH. Cooke PH. Response to trisodium phosphate treatment of Salmonella Chester attached to fresh-cut green pepper slices. Can J Microbiol. 2001;47:25–32. doi: 10.1139/w00-116. [DOI] [PubMed] [Google Scholar]
- Liu NT, Nou X, Lefcourt AM, Shelton DR. Lo YM. Dual-species biofilm formation by Escherichia coli O157:H7 and environmental bacteria isolated from fresh-cut processing facilities. Int J Food Microbiol. 2014;171:15–20. doi: 10.1016/j.ijfoodmicro.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Loferer H, Hammar M. Normark S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol. 1997;26:11–23. doi: 10.1046/j.1365-2958.1997.5231883.x. [DOI] [PubMed] [Google Scholar]
- Macarisin D, Patel J, Bauchan G, Giron JA. Sharma VK. Role of curli and cellulose expression in adherence of Escherichia coli O157:H7 to spinach leaves. Foodborne Pathog Dis. 2012;9:160–167. doi: 10.1089/fpd.2011.1020. [DOI] [PubMed] [Google Scholar]
- Macarisin D, Patel J, Bauchan G, Giron JA. Ravishankar S. Effect of spinach cultivar and bacterial adherence factors on survival of Escherichia coli O157:H7 on spinach leaves. J Food Prot. 2013;76:1829–1837. doi: 10.4315/0362-028X.JFP-12-556. [DOI] [PubMed] [Google Scholar]
- Macarisin D, Patel J. Sharma VK. Role of curli and plant cultivation conditions on Escherichia coli O157:H7 internalization into spinach grown on hydroponics and in soil. Int J Food Microbiol. 2014;173:48–53. doi: 10.1016/j.ijfoodmicro.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Mahon BE, Ponka A, Hall WN, Komatsu K, Dietrich SE, Siitonen A, et al. An international outbreak of Salmonella infections caused by alfalfa sprouts grown from contaminated seeds. J Infect Dis. 1997;175:876–882. doi: 10.1086/513985. [DOI] [PubMed] [Google Scholar]
- Mariani-Kurkdjian P. Bingen E. Escherichia coli 0104:H4: a hybrid pathogen. Arch Pediatr. 2012;19(Suppl. 3):S97–S100. doi: 10.1016/S0929-693X(12)71281-2. [DOI] [PubMed] [Google Scholar]
- Markland SM, Shortlidge KL, Hoover DG, Yaron S, Patel J, Singh A, et al. Survival of pathogenic Escherichia Coli on basil, lettuce, and spinach. Zoonoses Public Health. 2012;60:563–571. doi: 10.1111/zph.12033. [DOI] [PubMed] [Google Scholar]
- Marvasi M, Cox CE, Xu Y, Noel JT, Giovannoni JJ. Teplitski M. Differential regulation of Salmonella Typhimurium genes involved in O-antigen capsule production and their role in persistence within tomato fruit. Mol Plant Microbe Interact. 2013;26:793–800. doi: 10.1094/MPMI-09-12-0208-R. [DOI] [PubMed] [Google Scholar]
- Matthysse AG, Kijne JW. Attachment of Rhizobiaceae to plant cells. In: Spaink HP, Kondorosi A, Hooykaas PJJ, editors. The Rhizobiaceae: Dordrecht, the Netherlands: Kluwer; 1998. pp. 235–249. Molecular Biology of Model Plant Associated Bacteria. [Google Scholar]
- Matthysse AG, Deora R, Mishra M. Torres AG. Polysaccharides cellulose, poly-B-1,6-N-acetyl-D-glucosamine, and colanic acid are required for optimal binding of Escherichia coli O157:H7 strains to alfalfa sprouts and K-12 strains to plastic but not for binding to epithelial cells. Appl Environ Microbiol. 2008;74:2384–2390. doi: 10.1128/AEM.01854-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Altier C. Martin GB. Salmonella colonization activates the plant immune system and benefits from association with plant pathogenic bacteria. Environ Microbiol. 2013;15:2418–2430. doi: 10.1111/1462-2920.12113. [DOI] [PubMed] [Google Scholar]
- Meric G, Kemsley EK, Falush D, Saggers EJ. Lucchini S. Phylogenetic distribution of traits associated with plant colonization in Escherichia coli. Environ Microbiol. 2013;15:487–501. doi: 10.1111/j.1462-2920.2012.02852.x. [DOI] [PubMed] [Google Scholar]
- Mika F. Hengge R. Small regulatory RNAs in the control of motility and biofilm formation in E. coli and Salmonella. Int J Mol Sci. 2013;14:4560–4579. doi: 10.3390/ijms14034560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mireles JR, 2nd, Toguchi A. Harshey RM. Salmonella enterica serovar Typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J Bacteriol. 2001;183:5848–5854. doi: 10.1128/JB.183.20.5848-5854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier JM. Lindow SE. Aggregates of resident bacteria facilitate survival of immigrant bacteria on leaf surfaces. Microb Ecol. 2005;49:343–352. doi: 10.1007/s00248-004-0007-9. [DOI] [PubMed] [Google Scholar]
- Monteiro C, Papenfort K, Hentrich K, Ahmad I, Le Guyon S, Reimann R, et al. Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol. 2012;9:489–502. doi: 10.4161/rna.19682. [DOI] [PubMed] [Google Scholar]
- Neff NT, Binns AN. Brandt C. Inhibitory effects of a pectin-enriched tomato cell wall fraction on Agrobacterium tumefaciens binding and tumor formation. Plant Physiol. 1987;83:525–528. doi: 10.1104/pp.83.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke M, Araujo LV, Costa SG, Pires RC, Zeraik AE, Fernandes AC, et al. Surfactin reduces the adhesion of food-borne pathogenic bacteria to solid surfaces. Lett Appl Microbiol. 2009;49:241–247. doi: 10.1111/j.1472-765X.2009.02646.x. [DOI] [PubMed] [Google Scholar]
- Nobles DR, Romanovicz DK. Brown RM., Jr Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiol. 2001;127:529–542. [PMC free article] [PubMed] [Google Scholar]
- Noel JT, Arrach N, Alagely A, McClelland M. Teplitski M. Specific responses of Salmonella enterica to tomato varieties and fruit ripeness identified by in vivo expression technology. PLoS ONE. 2010;5:e12406. doi: 10.1371/journal.pone.0012406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger T, Brunner F, Kemmerling B. Piater L. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev. 2004;198:249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- Ochman H. Groisman EA. The evolution of invasion by enteric bacteria. Can J Microbiol. 1995;41:555–561. doi: 10.1139/m95-074. [DOI] [PubMed] [Google Scholar]
- Olaimat AN. Holley RA. Factors influencing the microbial safety of fresh produce: a review. Food Microbiol. 2012;32:1–19. doi: 10.1016/j.fm.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Omadjela O, Narahari A, Strumillo J, Melida H, Mazur O, Bulone V. Zimmer J. BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc Natl Acad Sci USA. 2013;110:17856–17861. doi: 10.1073/pnas.1314063110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W. Schaffner DW. Modeling the growth of Salmonella in cut red round tomatoes as a function of temperature. J Food Prot. 2010;73:1502–1505. doi: 10.4315/0362-028x-73.8.1502. [DOI] [PubMed] [Google Scholar]
- Patel J. Sharma M. Differences in attachment of Salmonella enterica serovars to cabbage and lettuce leaves. Int J Food Microbiol. 2010;139:41–47. doi: 10.1016/j.ijfoodmicro.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Patel J, Singh M, Macarisin D, Sharma M. Shelton D. Differences in biofilm formation of produce and poultry Salmonella enterica isolates and their persistence on spinach plants. Food Microbiol. 2013;36:388–394. doi: 10.1016/j.fm.2013.06.019. [DOI] [PubMed] [Google Scholar]
- del Pozo JL. Patel R. The challenge of treating biofilm-associated bacterial infections. Clin Pharmacol Ther. 2007;82:204–209. doi: 10.1038/sj.clpt.6100247. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Hsing W, Gibson KE. Silhavy TJ. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol Microbiol. 1996;20:911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- Prouty AM. Gunn JS. Comparative analysis of Salmonella enterica serovar Typhimurium biofilm formation on gallstones and on glass. Infect Immun. 2003;71:7154–7158. doi: 10.1128/IAI.71.12.7154-7158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty AM, Schwesinger WH. Gunn JS. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect Immun. 2002;70:2640–2649. doi: 10.1128/IAI.70.5.2640-2649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina LD, Fleming HP. Breidt F., Jr Bacterial contamination of cucumber fruit through adhesion. J Food Prot. 2002;65:1881–1887. doi: 10.4315/0362-028x-65.12.1881. [DOI] [PubMed] [Google Scholar]
- Richards GR. Beuchat LR. Metabiotic associations of molds and Salmonella Poona on intact and wounded cantaloupe rind. Int J Food Microbiol. 2005;97:329–339. doi: 10.1016/j.ijfoodmicro.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Robinson LS, Ashman EM, Hultgren SJ. Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U. Molecular biology of cellulose production in bacteria. Res Microbiol. 2002;153:205–212. doi: 10.1016/s0923-2508(02)01316-5. [DOI] [PubMed] [Google Scholar]
- Romling U, Bian Z, Hammar M, Sierralta WD. Normark S. Curli fibers are highly conserved between Salmonella Typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Rohde M, Olsen A, Normark S. Reinkoster J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella Typhimurium regulates at least two independent pathways. Mol Microbiol. 2000;36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- Römling U, Galperin MY. Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Mayer R. Benziman M. Cellulose biosynthesis and function in bacteria. Microbiol Rev. 1991;55:35–58. doi: 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saggers EJ, Waspe CR, Parker ML, Waldron KW. Brocklehurst TF. Salmonella must be viable in order to attach to the surface of prepared vegetable tissues. J Appl Microbiol. 2008;105:1239–1245. doi: 10.1111/j.1365-2672.2008.03795.x. [DOI] [PubMed] [Google Scholar]
- Salazar JK, Deng K, Tortorello ML, Brandl MT, Wang H. Zhang W. Genes ycfRsirA and yigG contribute to the surface attachment of Salmonella enterica Typhimurium and Saintpaul to fresh produce. PLoS ONE. 2013;8:e57272. doi: 10.1371/journal.pone.0057272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher K, Romling U. Yaron S. Effect of heat, acidification, and chlorination on Salmonella enterica serovar Typhimurium cells in a biofilm formed at the air-liquid interface. Appl Environ Microbiol. 2005;71:1163–1168. doi: 10.1128/AEM.71.3.1163-1168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher K, Kesselman E, Shimoni E. Yaron S. Morphological analysis of young and old pellicles of Salmonella Typhimurium. Biofouling. 2007;23:385–394. doi: 10.1080/08927010701648265. [DOI] [PubMed] [Google Scholar]
- Schlisselberg DB. Yaron S. The effects of stainless steel finish on Salmonella Typhimurium attachment, biofilm formation and sensitivity to chlorine. Food Microbiol. 2013;35:65–72. doi: 10.1016/j.fm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Senchou V, Weide R, Carrasco A, Bouyssou H, Pont-Lezica R, Govers F. Canut H. High affinity recognition of a Phytophthora protein by Arabidopsis via an RGD motif. Cell Mol Life Sci. 2004;61:502–509. doi: 10.1007/s00018-003-3394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo KH. Frank JF. Attachment of Escherichia coli O157:H7 to lettuce leaf surface and bacterial viability in response to chlorine treatment as demonstrated using confocal scanning laser microscopy. J Food Prot. 1999;62:3–9. doi: 10.4315/0362-028x-62.1.3. [DOI] [PubMed] [Google Scholar]
- Seo S. Matthews KR. Influence of the plant defense response to Escherichia coli O157:H7 cell surface structures on survival of that enteric pathogen on plant surfaces. Appl Environ Microbiol. 2012;78:5882–5889. doi: 10.1128/AEM.01095-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RK, Berger CN, Feys B, Knutton S, Pallen MJ. Frankel G. Enterohemorrhagic Escherichia coli exploits EspA filaments for attachment to salad leaves. Appl Environ Microbiol. 2008;74:2908–2914. doi: 10.1128/AEM.02704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RK, Lasa I, Garcia BM, Pallen MJ, Hinton JC, Berger CN. Frankel G. Cellulose mediates attachment of Salmonella enterica serovar Typhimurium to tomatoes. Environ Microbiol Rep. 2011;3:569–573. doi: 10.1111/j.1758-2229.2011.00263.x. [DOI] [PubMed] [Google Scholar]
- Shirron N. Yaron S. Active suppression of early immune response in tobacco by the human pathogen Salmonella Typhimurium. PLoS ONE. 2011;6:e18855. doi: 10.1371/journal.pone.0018855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirron N, Kisluk G, Zelikovich Y, Eivin I, Shimoni E. Yaron S. A comparative study assaying commonly used sanitizers for antimicrobial activity against indicator bacteria and a Salmonella Typhimurium strain on fresh produce. J Food Prot. 2009;72:2413–2417. doi: 10.4315/0362-028x-72.11.2413. [DOI] [PubMed] [Google Scholar]
- Simm R, Ahmad I, Rhen M, Le Guyon S. Römling U. Regulation of biofilm formation in Salmonella Typhimurium. Future Microbiology. 2014 doi: 10.2217/fmb.14.88. (in press) [DOI] [PubMed] [Google Scholar]
- Solano C, Garcia B, Valle J, Berasain C, Ghigo JM, Gamazo C. Lasa I. Genetic analysis of Salmonella Enteritidis biofilm formation: critical role of cellulose. Mol Microbiol. 2002;43:793–808. doi: 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
- Solomon EB. Matthews KR. Interaction of live and dead Escherichia coli O157:H7 and fluorescent microspheres with lettuce tissue suggests bacterial processes do not mediate adherence. Lett Appl Microbiol. 2006;42:88–93. doi: 10.1111/j.1472-765X.2005.01816.x. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Matute CM, Hassan AN. Frank JF. Comparison of the attachment of Escherichia coli O157:H7, Listeria monocytogenesSalmonella Typhimurium, and Pseudomonas fluorescens to lettuce leaves. J Food Prot. 2000;63:1433–1437. doi: 10.4315/0362-028x-63.10.1433. [DOI] [PubMed] [Google Scholar]
- Tan MS, Wang Y. Dykes GA. Attachment of bacterial pathogens to a bacterial cellulose-derived plant cell wall model: a proof of concept. Foodborne Pathog Dis. 2013;10:992–994. doi: 10.1089/fpd.2013.1536. [DOI] [PubMed] [Google Scholar]
- Teplitski M, Al-Agely A. Ahmer BM. Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar Typhimurium. Microbiology. 2006;152:3411–3424. doi: 10.1099/mic.0.29118-0. [DOI] [PubMed] [Google Scholar]
- Thomas CJ. McMeekin TA. Attachment of Salmonella spp. to chicken muscle surfaces. Appl Environ Microbiol. 1981;42:130–134. doi: 10.1128/aem.42.1.130-134.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Jeter C, Langley W. Matthysse AG. Differential binding of Escherichia coli O157:H7 to alfalfa, human epithelial cells, and plastic is mediated by a variety of surface structures. Appl Environ Microbiol. 2005;71:8008–8015. doi: 10.1128/AEM.71.12.8008-8015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlich GA, Keen JE. Elder RO. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl Environ Microbiol. 2001;67:2367–2370. doi: 10.1128/AEM.67.5.2367-2370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukuku DO. Fett WF. Relationship of cell surface charge and hydrophobicity to strength of attachment of bacteria to cantaloupe rind. J Food Prot. 2002;65:1093–1099. doi: 10.4315/0362-028x-65.7.1093. [DOI] [PubMed] [Google Scholar]
- Ukuku DO. Fett WF. Effects of cell surface charge and hydrophobicity on attachment of 16 Salmonella serovars to cantaloupe rind and decontamination with sanitizers. J Food Prot. 2006;69:1835–1843. doi: 10.4315/0362-028x-69.8.1835. [DOI] [PubMed] [Google Scholar]
- Ukuku DO. Sapers GM. Effect of sanitizer treatments on Salmonella Stanley attached to the surface of cantaloupes and cell transfer to fresh-cut tissues during cutting practices. J Food Prot. 2001;64:1286–1291. doi: 10.4315/0362-028x-64.9.1286. [DOI] [PubMed] [Google Scholar]
- Vestby LK, Moretro T, Langsrud S, Heir E. Nesse LL. Biofilm forming abilities of Salmonella are correlated with persistence in fish meal- and feed factories. BMC Vet Res. 2009;5:20. doi: 10.1186/1746-6148-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikram A, Jayaprakasha GK, Uckoo RM. Patil BS. Inhibition of Escherichia coli O157:H7 motility and biofilm by beta-sitosterol glucoside. Biochim Biophys Acta. 2013;1830:5219–5228. doi: 10.1016/j.bbagen.2013.07.022. [DOI] [PubMed] [Google Scholar]
- Vila J. Soto SM. Salicylate increases the expression of marA and reduces in vitro biofilm formation in uropathogenic Escherichia coli by decreasing type 1 fimbriae expression. Virulence. 2012;3:280–285. doi: 10.4161/viru.19205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Chai Y, Beauregard P, Losick R. Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 2013;11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C, Hensel M. Adhesive mechanisms of Salmonella enterica. In: Linke D, Goldman A, editors. Bacterial Adhesion, Advances in Experimental Medicine and Biology. Dordrecht, The Netherlands: Springer; 2011. pp. 17–34. [DOI] [PubMed] [Google Scholar]
- Wang X, Preston JF., 3rd Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JC, Rothwell SD. Keevil CW. Use of episcopic differential interference contrast microscopy to identify bacterial biofilms on salad leaves and track colonization by Salmonella Thompson. Environ Microbiol. 2008;10:918–925. doi: 10.1111/j.1462-2920.2007.01511.x. [DOI] [PubMed] [Google Scholar]
- Whitfield C. Roberts IS. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- Winfield MD. Groisman EA. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl Environ Microbiol. 2003;69:3687–3694. doi: 10.1128/AEM.69.7.3687-3694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xicohtencatl-Cortes J, Sanchez Chacon E, Saldana Z, Freer E. Giron JA. Interaction of Escherichia coli O157:H7 with leafy green produce. J Food Prot. 2009;72:1531–1537. doi: 10.4315/0362-028x-72.7.1531. [DOI] [PubMed] [Google Scholar]
- Yap MN, Yang CH, Barak JD, Jahn CE. Charkowski AO. The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J Bacteriol. 2005;187:639–648. doi: 10.1128/JB.187.2.639-648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron S. Microbial attachment and persistence on plants. In: Matthews KR, editor. The Produce Contamination Problem. 2nd edn. San Diego, CA, USA: Elsevier; 2014. pp. 21–58. [Google Scholar]
- Zaragoza WJ, Noel JT. Teplitski M. Spontaneous non-rdar mutations increase fitness of Salmonella in plants. Environ Microbiol Rep. 2012;4:453–458. doi: 10.1111/j.1758-2229.2012.00364.x. [DOI] [PubMed] [Google Scholar]
- Zogaj X, Nimtz M, Rohde M, Bokranz W. Romling U. The multicellular morphotypes of Salmonella Typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]


