Summary
We established an interaction network among three Arabidopsis PERK receptor-like kinases (8, 9, and 10), two AGC VIII kinases (AGC1-9 and KIPK) and KCBP with a role in negatively regulating root growth.
Key words: Proline-rich extensin-like receptor-like kinase, KCBP-interacting protein kinase, kinesin-like calmodulin-binding protein, root growth, signalling, sucrose.
Abstract
The Arabidopsis proline-rich, extensin-like receptor-like kinases (PERKs) are a small group of receptor-like kinases that are thought to act as sensors at the cell wall through their predicted proline-rich extracellular domains. In this study, we focused on the characterization of a subclade of three Arabidopsis predicted PERK genes, PERK8, -9, and -10, for which no functions were known. Yeast two-hybrid interaction studies were conducted with the PERK8,- 9, and -10 cytosolic kinase domains, and two members of the Arabidopsis AGC VIII kinase family were identified as interacting proteins: AGC1-9 and the closely related kinesin-like calmodulin-binding protein (KCBP)-interacting protein kinase (KIPK). As KIPK has been identified previously as an interactor of KCBP, these interactions were also examined further and confirmed in this study. Finally, T-DNA mutants for each gene were screened for altered phenotypes under different conditions, and from these screens, a role for the PERK, KIPK, and KCBP genes in negatively regulating root growth was uncovered.
Introduction
Plant receptor-like kinases make up the largest class of kinases in plant genomes, with over 600 predicted members in Arabidopsis, and have a large range of functions from plant development to plant–microbe interactions and abiotic stress responses (reviewed by Gish and Clark, 2011; Antolin-Llovera et al., 2012; Osakabe et al., 2013). One predicted subclass of the plant receptor-like kinases is the proline-rich, extensin-like receptor-like kinases (PERKs) (Silva and Goring, 2002). With their predicted proline-rich, extensin-like extracellular domains, PERKs are thought to be part of a group of proteins that act as sensors/receptors at the cell wall. These sensors/receptors, for example, may monitor changes to the cell wall during cell expansion in plant growth, or during plant exposure to abiotic/biotic stresses, and activate associated cellular responses (reviewed by Humphrey et al., 2007; Steinwand and Kieber, 2010; Doblin et al., 2014).
The first PERK member to be characterized was Brassica napus PERK1, which was found to be expressed in a number of different tissues as well as rapidly induced by wounding (Silva and Goring, 2002). The expression patterns of the 15-member Arabidopsis PERK gene family were subsequently characterized, and several were found to be broadly expressed (PERK1, -2, -3, -8, -9, -10, -14, and -15), while others showed high expression in specific tissues such as the root (PERK13) and pollen (PERK4, -5, -6, -7, -11, and -12) (Nakhamchik et al., 2004). PERKs are predicted to be membrane localized with a cluster of positively charged amino acids next to the transmembrane domain orienting the proline-rich domains on the external face of the plasma membrane and the kinase domain internally (Silva and Goring, 2002; Nakhamchik et al., 2004). Consistent with this, BnPERK1 was found to have a serine/threonine kinase activity in vitro, and when fused to green fluorescent protein (BnPERK1:GFP) was localized to the plasma membrane when transiently expressed in onion cells (Silva and Goring, 2002). In addition, a BnPERK1:haemagglutinin fusion expressed in Arabidopsis was enriched in the plasma membrane fraction, following aqueous two-phase partitioning (Haffani et al., 2006). Arabidopsis PERK4 has also been shown to have kinase activity, and a GFP:PERK4 fusion was localized to the plasma membrane and appeared to be associated with the cell wall (Bai et al., 2009).
Defining clear roles for PERK genes during plant growth and development has been challenging, and the ectopic expression or suppression of PERK genes in Arabidopsis can lead to complex changes in growth and organ development. When BnPERK1 was expressed ectopically in Arabidopsis, there were several altered traits including an increased number of lateral shoots, and an increased number of ovules per pistil and seeds per silique (Haffani et al., 2006). Ectopic callose and cellulose deposits were also detected in these transgenic plants suggesting cell wall alterations. The antisense expression of BnPERK1 in Arabidopsis led to the suppression of tandemly linked Arabidopsis PERK1 and -3 genes, and resulted in reduced apical dominance and abnormal floral organs (Haffani et al., 2006). In addition, hypocotyls from dark-grown, PERK1- and -3-supressed seedlings were longer relative to the wild-type Arabidopsis Columbia ecotype (Col-0), while seedlings with ectopic BnPERK1 expression displayed shorter hypocotyls (Haffani et al., 2006). Complex traits were similarly found with the ectopic expression of Arabidopsis PERK12 in the activation-tagged mutant inflorescence growth inhibitor1 (igi1). Hwang et al. (2010) identified igi1 in a screen for increased shoot-branching mutants, and igi1 displayed a dwarf phenotype with increased axillary branching and abnormal floral organs. This phenotype resulted from the T-DNA carrying four enhancers inserting in the 5′ end of the PERK12 gene and driving strong ectopic expression of this normally pollen-specific gene (Hwang et al., 2010).
More specific functions have been uncovered for the Arabidopsis PERK4 and -13 genes. PERK13 (also called RHS10) was identified in a screen for Arabidopsis genes displaying root hair cell-specific expression (Won et al., 2009). PERK13/RHS10 Promoter:GFP plants displayed GFP fluorescence in the root hairs, and a perk13/rhs10 T-DNA insertion mutant had longer root hairs compared with wild-type. Using a strong root hair promoter, Won et al. (2009) also overexpressed PERK13/RHS10 in root hairs, and this resulted in an inhibition of root hair elongation and shorter root hairs. In the case of PERK4, perk4 T-DNA mutants were found to be less sensitive to the inhibitory effects of abscisic acid (ABA) and had increased germination rates and root length in the presence of ABA compared with wild-type Col-0 roots (Bai et al., 2009). Under these conditions, the increased root length appeared to be due to increased cell elongation relative to wild-type Col-0 seedlings. Finally, there was reduced ABA-mediated calcium channel activity in the perk4 mutant roots relative to Col-0, suggesting a role for calcium fluxes in the ABA-mediated inhibition of root elongation (Bai et al., 2009).
While nothing is known about downstream interacting/signalling proteins for PERKs, the kinase domain from Arabidopsis PERK1 was isolated previously as an interactor for the nuclear shuttle protein (NSP) from cabbage leaf curl virus (Florentino et al., 2006). NSP is needed to transport the viral DNA from the nucleus to the cytoplasm, and NSP functions with the movement protein in spreading the viral DNA from cell to cell. Arabidopsis PERK1 (also called NSP-associated kinase or NsAK) was found to phosphorylate NSP, and a perk1/nsak T-DNA insertion mutant had increased tolerance to cabbage leaf curl virus infection, with a delay in symptom development, suggesting that PERK1/NsAK positively regulates NSP function, perhaps playing a role in cell-to-cell movement (Florentino et al., 2006).
In this study, we focused on a subclade of three Arabidopsis PERK genes, PERK8, -9, -10, which show widespread expression in Arabidopsis (Silva and Goring, 2002; Nakhamchik et al., 2004). Two different approaches were taken simultaneously to characterize these genes: (i) a yeast two-hybrid screen was conducted with the PERK10 cytosolic domain to identify interacting proteins; and (ii) T-DNA knockout mutants were screened for altered root growth under different conditions. Interactors of PERK8, -9, and -10 were identified as AGC1-9 and the closely related kinesin-like calmodulin-binding protein (KCBP)-interacting protein kinase (KIPK), members of the Arabidopsis AGC VIII kinase family (Zegzouti et al., 2006; Rademacher and Offringa, 2012). KIPK was previously identified as an interactor of KCBP (Day et al., 2000), and this interaction was examined further in this study. Finally, using T-DNA mutants, we uncovered a role for the PERK, KIPK, and KCBP genes in negatively regulating root growth.
Materials and methods
Yeast two-hybrid screen
For the yeast two-hybrid library, random-primed cDNA was synthesized from flower bud mRNA and inserted into pGADT7-Rec (GAL4 activation domain) using the Matchmaker library construction and screening kit (BD Biosciences Clontech). For the PERK constructs, the entire cytosolic kinase domains (just after the transmembrane domains) were PCR amplified and cloned as fusions to the GAL4 DNA-binding domain into pGBKT7 (BD Biosciences Clontech). PERK10 was used to screen the yeast two-hybrid library according to the kit instructions. Following the screening of positive colonies on quadruple dropout plates (SD–Ade–His–Leu–Trp), plate assays were conducted on the remaining positive colonies to test for α-galactosidase activity (MEL1 reporter) in the presence of 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-Gal), or filter lifts were performed to test β-galactosidase activity (lacZ reporter) in the presence of 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal). The plasmids from these positive yeast colonies were isolated directly (Ding et al., 2003), retested for interactions with PERK10, and sequenced (Genome Quebec sequencing facility). Further screening was conducted to remove non-specific interactors, and KIPK2 was chosen for further analysis. The KIPK2 clones from the library screen contained the region of aa 85–324 or 85–355.
For further yeast two-hybrid interaction testing, the full-length KIPK2 cDNA was PCR amplified and cloned into pGEM-T Easy (Promega) while the full-length KIPK1 cDNA was obtained as a pUni clone (U22072) (Yamada et al., 2003) from the Arabidopsis Biological Resource Center (ABRC). Different segments from the KIPK1 and KIPK2 cDNA were then transferred to create an in-frame fusion with the GAL4 activation domain in pGADT7-Rec. For KIPK1, the KIPK184–312 segment was PCR amplified, while internal restriction sites were used for the other constructs: KIPK184–582 (Cfr10I–SmaI), KIPK1303–582 (SspI–SmaI), and KIPK1303–934 (SspI–SalI in vector). For KIPK2, one of the original yeast two-hybrid clones was KIPK285–324, and internal restriction sites were used for the other constructs: KIPK285–624 (start of KIPK285–324–FspI), KIPK2277–624 (NdeI–FspI), and KIPK2277–948 (NdeI–EcoR1 in vector). For KCBP, the pAS1CYH2/N-terminal KCBP plasmid containing aa 12–923 was obtained from Dr Anireddy Reddy. KCBP12–923 was then transferred from this plasmid as an NcoI fragment to construct an in-frame fusion to the GAL4 DNA-binding domain in pGBKT7. For the smaller KCBP segments, internal restriction sites were used: KCBP12–294 (NcoI–NdelI), KCBP12–496 (NcoI–EcoRI), KCBP12–622 (NcoI–Psp1406I), and KCBP465–923 (EcoRVNcoI). Pairwise combinations of the pGBKT7- and pGADT7-based plasmids were transformed into the yeast Y187 strain and selected for on SD–Leu–Trp plates. Filter lifts were performed on the transformed colonies to test for β-galactosidase activity in the presence of X-Gal (producing a blue colour for positive interactions).
Plant material and growth conditions
The Arabidopsis SALK T-DNA insertion lines (Alonso et al., 2003) were obtained from the ABRC at Ohio State University. The Arabidopsis GABI-Kat T-DNA insertion line for KIPK2 was obtained from the GABI-Kat collection at the Max Planck Institute of Plant Breeding Research (Rosso et al., 2003). Wild-type Arabidopsis thaliana (Col-0) and transgenic seeds were surface sterilized with a 1:1 (95% ethanol:3% H2O2) solution. Seeds were rinsed three to five times with sterile distilled water. All seeds were placed at 4 °C in the dark in either a 0.01% agar solution or on the medium plates themselves to synchronize germination. Seeds were germinated on ½ Murashige and Skoog salts (½ MS) (Cassion Laboratories) and 0.8% (w/v) agar (Phytoblend; Cassion Laboratories) at pH 5.8, unless otherwise stated. Long-day growth chamber conditions consisted of a 16h light/8h dark photoperiod at 22 °C. Constant light conditions were a 24h light cycle at 22 °C. Plants were grown on autoclaved potting soil (Premiere ProMix) and fertilized with a 20-20-20 mixture (Plant Pro Fertilizer).
T-DNA insertion line screens and reverse transcription (RT)-PCR analyses
Combinations of T-DNA and gene-specific PCR-based screens on genomic DNA were used to identify homozygous T-DNA insertion lines. These lines were then crossed to generate the multiple perk, kipk, and kcbp homozygous T-DNA insertion mutants. The homozygous plants were confirmed as knockouts by conducting RT-PCR using RNA extracted from flower buds (except for KCBP where seedling RNA was used). For expression profiling of PERK8, PERK9, PERK10, KIPK1, KIPK2, and KCBP genes, RNA was extracted from 2-week-old seedling tissue (whole seedlings, rosettes, roots, or hypocotyls). The PCR primers used are listed in Supplementary Table S1 at JXB online.
Sucrose response assays
Seeds were sterilized as described above and plated on Phytoblend medium plates. After 48h at 4 °C in the dark, the plates were placed vertically in a 16h light/8h dark photoperiod at 22 °C for 7 d. The 7-d-old seedlings were then transferred to sucrose treatment plates (Phytoblend) containing 0, 0.5, or 4.5% sucrose, lining up the root tips, and placed at 22 °C under 24h light (under approx. 80–110 µmol m–2 sec–1). After 7 d, digital photographs were taken, and the new root growth (since transfer) was measured from the digital images using ImageJ software (Abramoff et al., 2004).
DEX::PERK10 transgenic Arabidopsis
The full-length PERK10 cDNA was inserted next to the dexamethasone (DEX)-inducible promoter in the pTA7002 plant transformation vector (Aoyama and Chua, 1997) and transformed into Col-0, the perk8-1,9-1,10-1 triple mutant, the kipk1-1,2-1 double mutant, and the kcbp-1 mutant using the floral dip method (Clough and Bent, 1998). Primary transformants were identified by hygromycin B selection (25mg l–1) on ½ MS agar plates and then transferred to soil. Seeds were then collected from each individual primary transformant for further testing. For the DEX plate germination assays, seeds were sterilized, sown on ½ MS agar plates with 10 µM DEX, and stratified at 4 °C for 3 d in the dark. The plates were then transferred to a 16h light/8h dark photoperiod at 22 °C, and after 5 d, the phenotypes of the germinated seedlings were scored. To confirm PERK10 expression with DEX treatment, RNA was extracted from untreated and treated seedlings for RT-PCR analysis. For phloroglucinol staining of lignin, samples were placed in a 1% phloroglucinol/HCl solution, shaken gently for 10min, and then mounted in a 50% glycerol/6M HCl solution and observed under bright-field illumination (Rogers et al., 2005). For detection of callose, roots were placed in a decolorized aniline blue solution (Sigma; resuspended at 0.1mg ml–1 in phosphate buffer, pH 9.0).
Results
Yeast two-hybrid screen for PERK10-interacting proteins
Both BnPERK1 and AtPERK4 have been shown to localize to the plasma membrane (Silva and Goring, 2002; Haffani et al., 2006; Bai et al., 2009), and similarly, PERK8, -9, and -10 are predicted to be transmembrane proteins (Fig. 1A; Nakhamchik et al., 2004). Thus, to search for potential downstream signalling proteins for PERK8, -9, and -10, only the cytosolic domains were used in the yeast two-hybrid system. These constructs were first tested for background activity, and from this, PERK10 was chosen for the full-scale screen of a yeast two-hybrid library constructed of random-primed cDNA from Arabidopsis flower bud mRNA. Among the positive clones sequenced from this screen was a predicted member of the Arabidopsis AGC VIII kinase family (AGC1-9; Zegzouti et al., 2006; Rademacher and Offringa, 2012), which was most closely related to the previously characterized KIPK (Day et al., 2000; Zegzouti et al., 2006). KIPK was renamed KIPK1, and AGC1-9 was renamed KIPK2 (Fig. 1A). Two different KIPK2 clones were identified in this yeast two-hybrid screen and both were partial cDNAs covering aa 85–324 and 85–355, respectively.
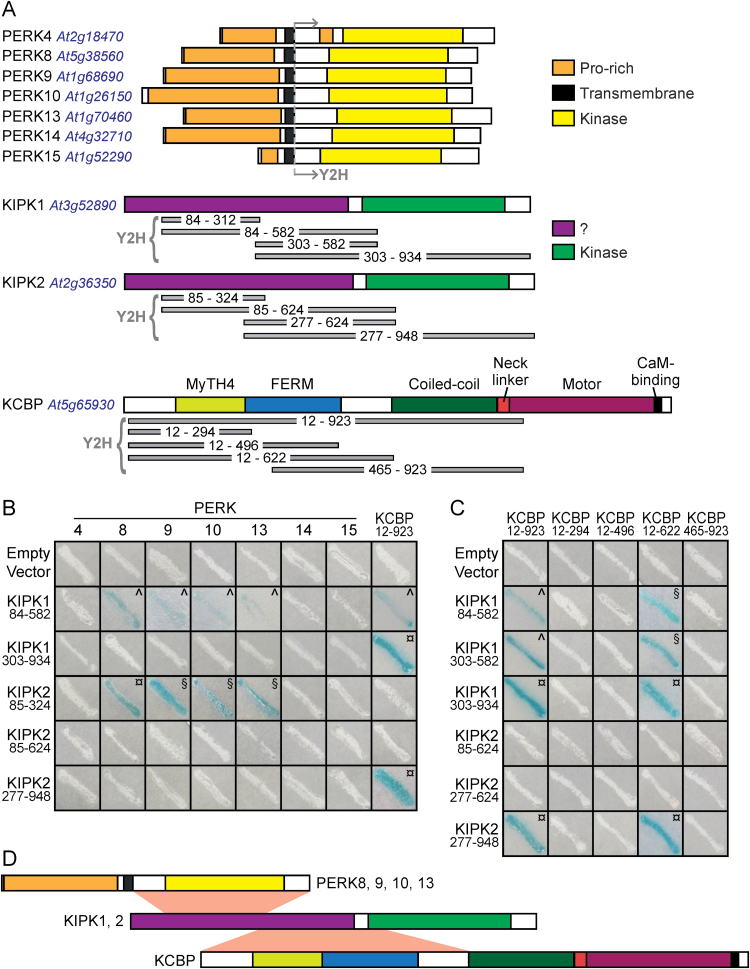
Fig. 1.
Yeast two-hybrid analyses for protein–protein interactions between the PERK, KIPK, and KCBP proteins. (A) Predicted protein domains and segments tested in the yeast two-hybrid system. The predicted PERK domains are from Nakhamchik et al. (2004), the KIPK1 and -2 kinase domains were predicted from The Arabidopsis Information Resource (TAIR) and SMART (Schultz et al., 1998), and the KCBP domain organization is from Abdel-Ghany et al. (2005). The regions tested in the yeast two-hybrid system are shown in grey beneath each gene. (B) Yeast two-hybrid interactions between the PERK and KIPK proteins. The cytosolic domains from PERK4, -8, -9, -10, -13, -14, and -15 were tested for interactions against different subdomains of KIPK1 and KIPK2. Both PERK1 and KIPK184–312 were initially included in the screen but may have been toxic, as yeast colonies could not be recovered for either construct. (C) Yeast two-hybrid interactions between the KIPK and KCBP proteins. Different subdomains of KIPK1 and KIPK2 were tested for interactions with different subdomains of KCBP. For both (B) and (C), yeast Y187 cells were transformed with the indicated constructs, and positive interactions were scored by the development of a blue colour (from activation of the lacZ reporter). Symbols represent time for the blue colour to develop: ¤,Less than 1h; §, 1–3h; ^, more than 3h. The negative interactions (white streaks) were monitored for up to 24h (with the exception of PERK8 and -9 transformants, which were left for shorter incubation times due to some background activity). (D) Schematic of proposed minimal interaction domains between the PERK, KIPK, and KCBP proteins.
The full-length KIPK2 protein was predicted to be 948 aa with the catalytic domain of a serine/threonine protein kinase predicted to be from aa 559 to 898 (Fig. 1A). KIPK2 and KIPK1 shared 76% amino acid sequence homology (alignment shown in Supplementary Fig. S1 at JXB online.). In addition to the kinase domains, KIPK1 and -2 shared a highly similar N-terminal domain that was enriched for serine (Fig. 1A and Supplementary Fig. S1). The specificity of the KIPK2 interaction was tested against a series of PERKs (PERK4, -8, -9, -10, -13, -14, and -15), and only PERK8, -9, -10, and -13 were found to interact with the library clone, KIPK284–324 (Fig. 1B). Of the PERKs tested, PERK8, -9, -10, and -13 were the most closely related to each other (Fig. 2A; Nakhamchik et al., 2004). Despite the high sequence similarity of KIPK1 to KIPK2, it was not isolated in the yeast two-hybrid screen, and so a similarly small KIPK1 construct was designed and tested for interactions with PERKs. However, the KIPK184–312 construct, which covered the same interacting region in KIPK284–324, appeared to be toxic in yeast, as colonies could not be recovered. When a larger segment covering the N-terminal region, KIPK184–582, was tested, it interacted with PERK8, -9, -10, and -13 (Fig. 1B). Interestingly, when a KIPK2 fragment encompassing the same region was tested, KIPK284–624, no interaction could be observed, which may explain why only small partial cDNAs (encoding KIPK284–324 and KIPK284–355) were isolated from the original yeast two-hybrid screen. It is possible that the larger KIPK2 fragment had some type of inhibitory activity associated with it. Finally, constructs lacking a portion of the N-terminal region, KIPK1303–934 and KIPK2277–948, did not show any interactions with the PERKs (Fig. 1A, B).
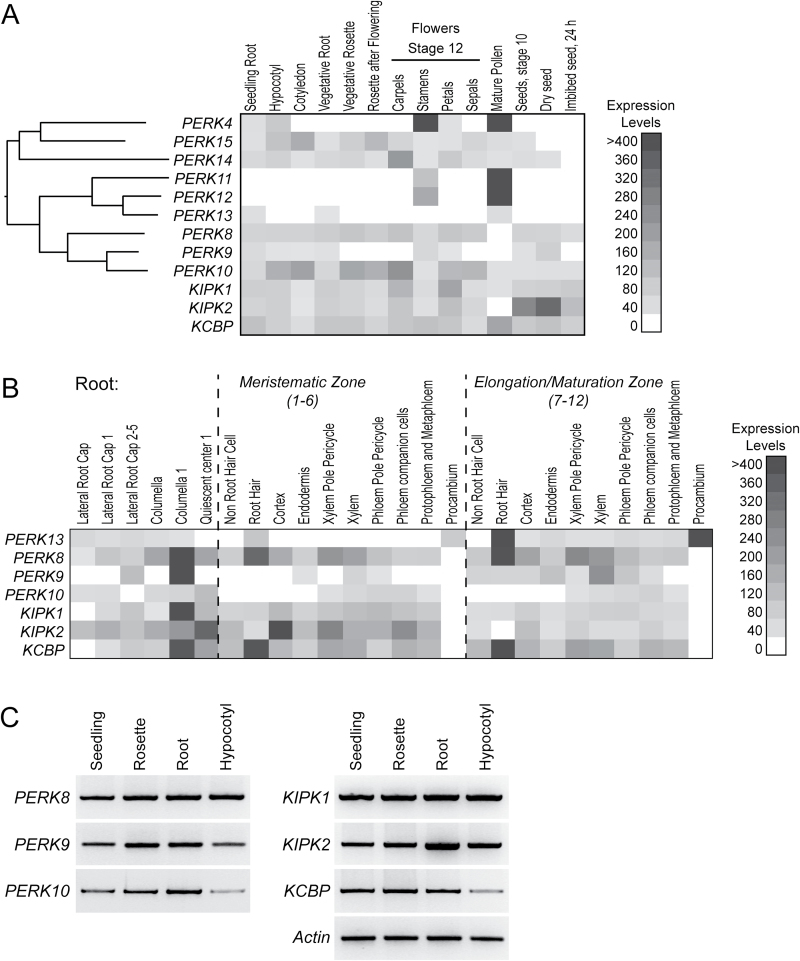
Fig. 2.
Expression profiles for the PERK, KIPK, and KCBP genes. (A) Heatmap of the microarray expression profiles for the PERK, KIPK, and KCBP genes in datasets for different tissues (Schmid et al., 2005) and seeds (Nakabayashi et al., 2005). Shown next to the PERKs is a protein distance phylogeny built using the cytosolic protein domains (Mobyle, http://mobyle.pasteur.fr) to illustrate the relationships among these family members (Nakhamchik et al., 2004). (B) Heatmap of the microarray expression profiles for the PERK, KIPK, and KCBP genes in the different root tissues dataset (Brady et al., 2007). E-northern analyses of the public microarray datasets were performed through the BioArray Resource (Winter et al., 2007). (C) RT-PCR analysis of PERK, KIPK, and KCBP expression in 2-week-old seedlings. RNA was extracted from whole seedlings, rosettes, roots, and hypocotyls. Actin was used as a positive control for expression in all tissues.
KIPK1 was first identified as an interactor for KCBP by Day et al. (2000), who used KCBP12–923 to screen an Arabidopsis yeast two-hybrid library and pulled out KIPK1148–934 as an interactor. When subclones of KIPK1 were further tested, KIPK1513–934 was found to interact with KCBP12–923 while KIPK1190–537 did not (Day et al., 2000). KCBP, which belongs to the kinesin-14 family, is unique in its domain organization and is encoded by a single-copy gene in Arabidopsis (Fig. 1A) (Reddy et al., 1996; Narasimhulu et al., 1997; Abdel-Ghany et al., 2005). For testing KCBP interactions in our system, a series of KIPK1 and -2 constructs were tested against KCBP12–923, and interactions were observed for both KIPK1303–934 and KIPK2277–948 (Fig. 1A, B). KIPK1303–934 and KIPK2277–948 interactions were specific to KCBP, confirming that the N-terminal region is needed for interactions with PERKs (e.g. KIPK284–324; Fig. 1A, B). A series of smaller constructs was made to further define the regions of interactions. Both KIPK184–582 and KIPK1303–582 interacted with KCBP12–923 and the smaller KCBP12–622 construct (Fig. 1A, C). KCBP12–923 and KCBP12–622 only interacted with KIPK2 when the kinase domain was present (KIPK2277–948; Fig. 1C). Similar to what was seen for the PERK kinases, the smaller KIPK2 constructs (KIPK285–624 and KIPK2277–624) failed to show any detectable interaction with KCBP. In an attempt to further delineate which region of KCBP12–622 was interacting with KIPK1 and -2, three other constructs were tested, KCBP12–294, KCBP12–496, and KCBP496–923, but no interactions were detected (Fig. 1C). Altogether from these results, we propose a model where the proximal N-terminal region in KIPK1 and -2 mediates interactions with the PERK kinase domains while the distal N-terminal residues in KIPK1 and -2 are required for interactions with the N-terminal half of KCBP (Fig. 1D).
Phenotypic screening of perk, kipk, and kcbp T-DNA insertion mutants
With the discovery of the PERK–KIPK–KCBP interactions, the next step was to uncover where this potential signalling pathway might be functioning in planta. We focused on PERK8, -9, and -10 as they were the most closely related and were more broadly expressed in Arabidopsis (Fig. 2) (Nakhamchik et al., 2004). While PERK13 also interacted with KIPK1 and -2, it had a more specific expression profile (Fig. 2A, B) (Nakhamchik et al., 2004) and mutant phenotype (root hairs; Won et al., 2009), and was most closely related to the pollen-specific PERK11 and -12 (Fig. 2A) (Nakhamchik et al., 2004). Similar to PERK8, -9, and -10, a survey of the public microarray datasets shows that KIPK1, KIPK2, and KCBP were expressed across a range of Arabidopsis tissues (Fig. 2A). Overlapping expression profiles for the PERK(8,9,10)–KIPK(1,2)–KCBP set were detected in the microarray datasets for the different seedling tissues, the vegetative root, some of the floral tissues, and seeds (Fig. 2A). RT-PCR also confirmed expression of the PERK(8,9,10), KIPK(1,2), and KCBP genes in whole seedlings, rosettes, roots and hypocotyls (Fig. 2C).
KCBP is the only gene in the PERK(8,9,10)–KIPK(1,2)–KCBP set with a known function in Arabidopsis; that is, in trichome morphogenesis. The zwichel (zwi) mutant, identified in a screen for trichome mutants, was determined to be a loss-of-function kcbp mutant and displayed defects in trichome cell expansion resulting in shorter stalks and reduced branching (Oppenheimer et al., 1997). To search for overlapping in planta functions for the PERK(8,9,10)–KIPK(1,2)–KCBP set, loss-of-function homozygous T-DNA mutants were identified for all six genes (Fig. 3). The mutants were also crossed to form multiple gene knockout mutants and screened for altered phenotypes. As expected, kcbp-1 displayed the reduced trichome branching phenotype reported previously for the zwi/kcbp mutant (Oppenheimer et al., 1997). Any mutant combinations with kcbp-1 also showed the reduced trichome branching: the perk8-1,9-1,10-1,kcbp-1 quadruple mutant and kipk1-1,2-1,kcbp-1 triple mutant (Supplementary Fig. S2 at JXB online.). However, the trichomes were normal in appearance for the perk8-1,9-1,10-1 triple mutant and the kipk1-1,2-1 double mutant (Supplementary Fig. S2), suggesting that these proteins did not participate in the regulation of KCBP during trichome formation. In addition, plants from all these mutant combinations did not display any detectable growth or floral defects when grown on soil (Supplementary Fig. S2).
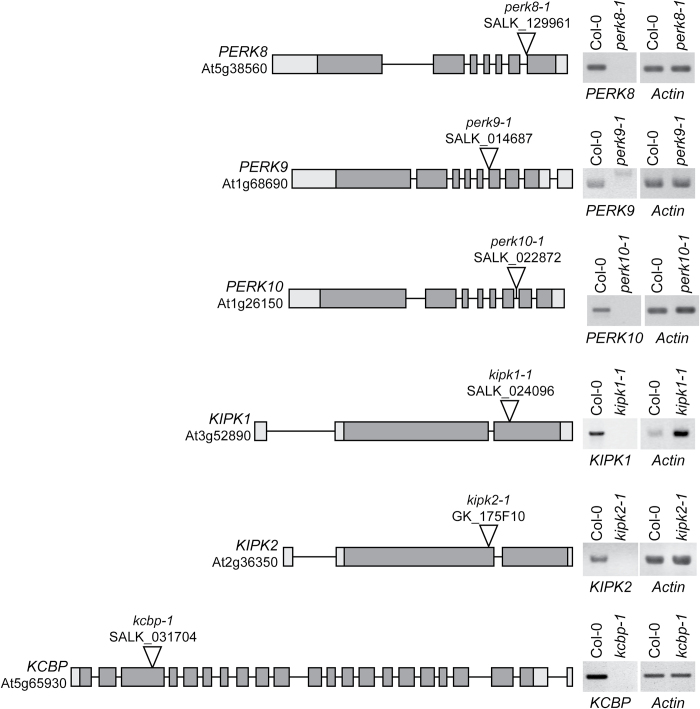
Fig. 3.
T-DNA insertion sites for the PERK, KIPK, and KCBP genes. On the left, schematics display the Arabidopsis SALK T-DNA insertion lines for the PERK8, PERK9, PERK10, KIPK1, and KCBP genes (Alonso et al., 2003) and the GABI-Kat T-DNA insertion line for KIPK2 (Rosso et al., 2003). On the right, RT-PCR analyses using gene-specific primers show the loss of gene-specific transcripts in each homozygous mutant, indicating that each T-DNA insertion is a knockout line. Actin primers were used as positive controls for each sample. T-DNA insertion sites were obtained from the SIGnAL website (http://signal.salk.edu), and gene models were obtained from The Arabidopsis Information Resource on 1 June, 2011 (http://www.arabidopsis.org) (Rhee et al., 2003).
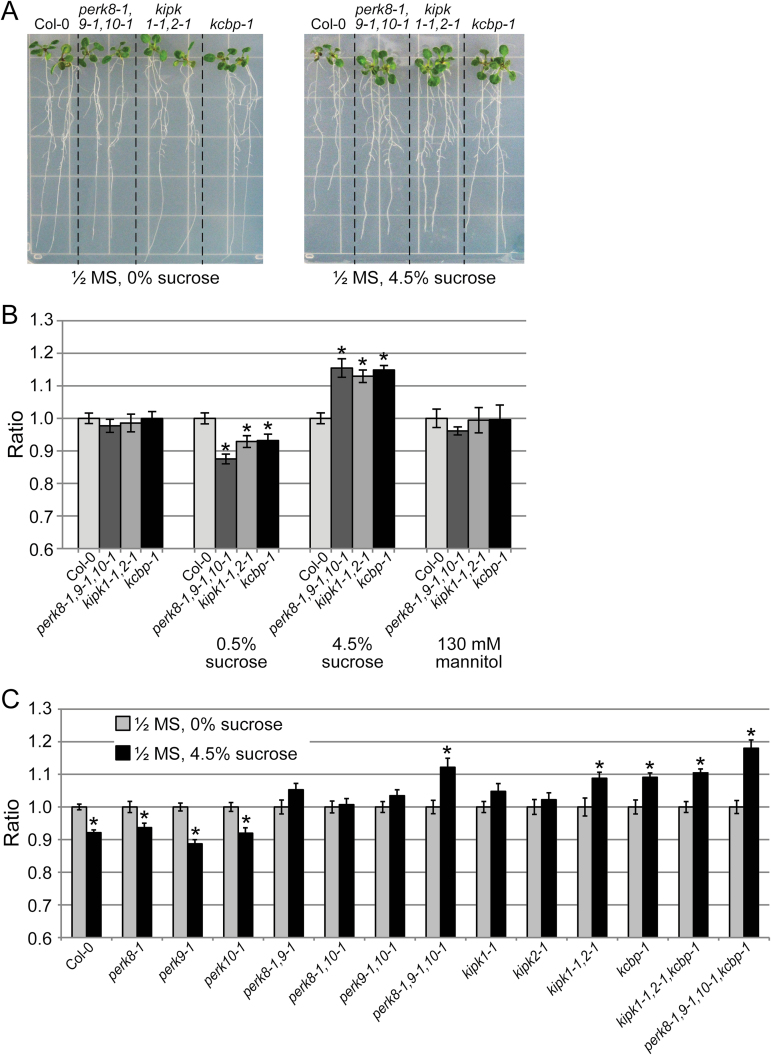
With the root being one of the tissues expressing the PERK(8,9,10)-KIPK(1,2)-KCBP set (Fig. 2), more focused root growth assays were conducted to search for shared mutant phenotypes. One of the root growth conditions tested was growing 7-d-old seedlings on different concentrations of sucrose (Hauser et al., 1995) under a 16h light/8h dark cycle or a 24h light cycle (Supplementary Fig. S3 at JXB online.). The most significant changes were seen when the primary root lengths on plates without sucrose were compared with plates with 4.5% sucrose after 7 d of growth under a 24h light cycle (Fig. 4A, B, Supplementary Fig. S3, and Supplementary Table S2 at JXB online.). The perk8-1,9-1,10-1 triple mutant, kipk1-1,2-1 double mutant, and the kcbp-1 mutant all displayed a significant increase in primary root length on 4.5% sucrose under 24h lighting when compared with Col-0 roots. This was not an osmotic effect, as the comparable concentration of mannitol produced no change in the root lengths of the mutants compared with Col-0. The increased primary root length of the mutants was also not seen on 0.5% sucrose, where the mutant roots tended to be a bit shorter than Col-0 (Fig. 4B).
Fig. 4.
Root growth of the different perk, kipk, and kcbp mutants under high sucrose. (A) Images showing seedling roots for Col-0, the perk8-1,9-1,10-1 triple mutant, the kipk1-1,2-1 double mutant, and the kcbp-1 mutant, grown on ½ MS plates with 0 or 4.5% sucrose under 24h light. (B) Graph showing the ratios of root length at d 7 on different medium conditions, under 24h light, for Col-0, the perk8-1,9-1,10-1 triple mutant, the kipk1-1,2-1 double mutant, and the kcbp-1 mutant. Ratios are relative to Col-0 growth under each condition. Asterisks represent statistically significant differences (P<0.05) when compared with Col-0 under each condition (n≥25). (C) Graph showing the ratios of root length at d 7 in the presence or absence of 4.5% sucrose, for the different perk, kipk, and kcbp mutant combinations. Ratios are root growth on ½ MS with 4.5% sucrose compared with ½ MS alone for each genotype. Asterisks represent statistically significant differences (P<0.05) when compared with ½ MS alone conditions for each genotype (n≥25). Col-0 and the single perk mutants (perk8-1, perk-1, and perk10-1) showed significant decreases in root growth on 4.5% sucrose plates. In contrast, significant increases in root growth on 4.5% sucrose plates were observed for the perk8-1,9-1,10-1 triple mutant, the kipk1-1,2-1 double mutant, the kcbp-1 mutant, the kipk1-1,2-1,kcbp-1 triple mutant, and the perk8-1,9-1,10-1,kcbp-1 quadruple mutant. Seedlings for the other genotypes did not show any significant changes.
When the primary root lengths from 4.5% sucrose plates were compared with plates without sucrose, a further trend was observed: whereas Col-0 displayed reduced growth on 4.5% sucrose, the multiple mutants had increased primary root lengths on 4.5% sucrose under 24h lighting (Supplementary Fig. S3, Supplementary Table S2). Thus, root growth in the absence or presence of 4.5% sucrose, under a 24h light cycle, was further tested across a number of different mutant combinations (Fig. 4C, Supplementary Table S2). The single perk mutants (perk8-1, perk9-1, and perk10-1) had a similar profile to Col-0 where there was decreased root length in the presence of 4.5% sucrose. However, the perk double mutants (perk8-1,9-1, perk8-1,10-1, and perk9-1,10-1) and the kipk single mutants (kipk1-1 and kipk1-2) showed an intermediate phenotype where there was no significant change in primary root length under the two conditions. Finally, increased primary root lengths on 4.5% sucrose were observed for the perk8-1,9-1,10-1 triple mutant, the kipk1-1,2-1 double mutant, the kcbp-1 mutant, the kipk1-1,2-1,kcbp-1 triple mutant, and the perk8-1,9-1,10-1,kcbp-1 quadruple mutant. Thus, while 4.5% sucrose had a somewhat inhibitory effect on Col-0 and the single perk mutant roots, the multiple perk, kipk, and kcbp mutants displayed increased root length on 4.5% sucrose under a 24h light cycle (Fig 4C and Supplementary Table S2).
Overexpression of PERK10 in Col-0 and the perk, kipk, and kcbp mutants
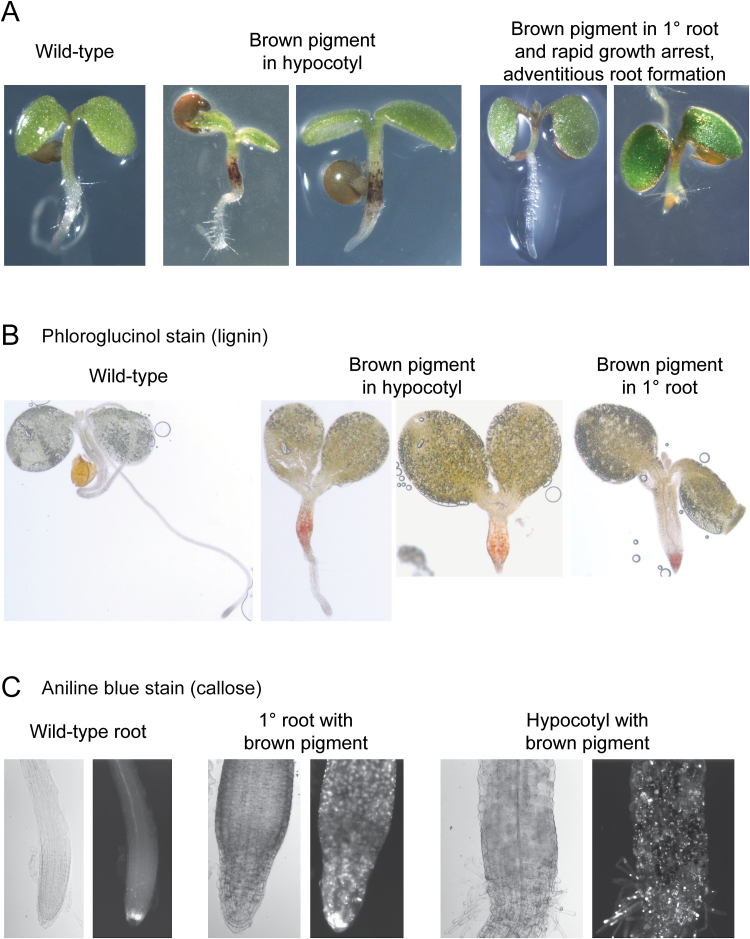
With the multiple perk, kipk, and kcbp loss-of-function mutants displaying increased root growth (under high sucrose and 24h lighting conditions), we also tested the opposite effect of overexpressing PERK10. We first attempted to overexpress PERK10 using the constitutive cauliflower mosaic virus 35S promoter but failed to recover any transgenic Arabidopsis seedlings. As a result, the PERK10 cDNA was then expressed under the control of a DEX-inducible promoter in Arabidopsis, following treatment with DEX. Seeds from primary Col-0/DEX::PERK10 transformants were germinated in the present of 10 µM DEX, and three different groups of phenotypes were observed: (i) seedlings that were wild type in appearance; (ii) seedlings with visible brown pigments in the hypocotyl; and (iii) seedlings with visible brown pigments in the root and rapid growth arrest of the primary root (these seedlings also produced adventitious roots) (Fig. 5A). When seeds from 20 independent Col-0/DEX::PERK10 primary transformants were germinated in the presence of DEX, there was a range in the frequency of seedlings displaying brown pigments, with the strongest line showing 77% of the seedlings with visible brown pigments in the hypocotyl or the root (Supplementary Table S3 at JXB online.). These mutant phenotypes were never observed when transgenic Col-0 seeds with the DEX-only construct were tested (Supplementary Table S3). To test whether the brown pigmented tissues contained ectopic lignin (Rogers et al., 2005), phloroglucinol staining was performed and produced the pink staining expected for lignin (Fig. 5B). The brown pigmented primary root and hypocotyl were also found to have ectopic callose deposits when stained with aniline blue (Fig. 5C). Thus, the overexpression of PERK10 in Col-0 seedlings resulted in some seedlings displaying root growth arrest with ectopic deposits of lignin and callose.
Fig. 5.
Phenotypes for the Col-0/DEX::PERK10 seedlings. (A) Images of 5-d-old germinated seedlings. Seeds from primary Col-0/DEX::PERK10 transformants were germinated in the presence of 10 µM DEX, and three different phenotypes were observed as shown. (B) Phloroglucinal staining for lignin in 5-d-old Col-0/DEX::PERK10 seedlings. (C) Aniline blue staining for callose in 5-d-old Col-0/DEX::PERK10 seedlings.
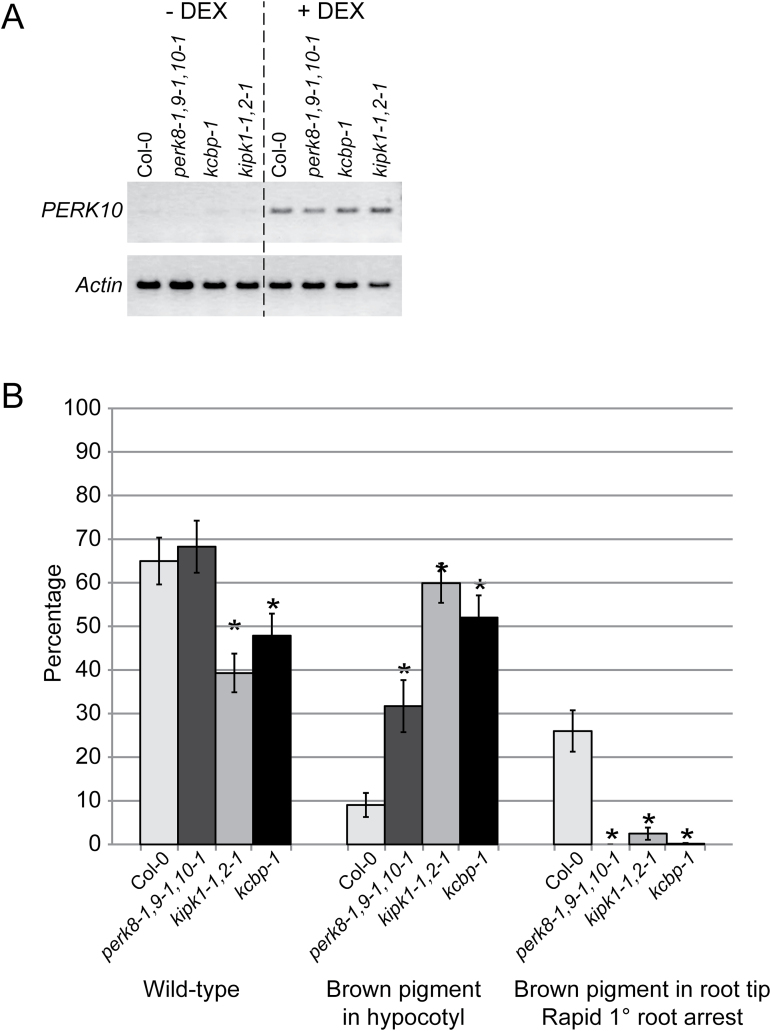
The effects of overexpressing PERK10 was also tested in the different mutant backgrounds with the DEX::PERK10 construct introduced into the perk8-1,9-1,10-1 triple mutant, the kipk1-1,2-1 double mutant, and the kcbp-1 single mutant. RT-PCR analysis showed that expression of the DEX::PERK10 construct was induced in all backgrounds with DEX treatment (Fig. 6A). For each background, seeds were collected from 20 independent primary transformants, germinated in the presence of DEX, and scored for the phenotypes resulting from PERK10 overexpression (Supplementary Table S3). Interestingly, while wild-type seedlings and seedlings with the brown pigmented hypocotyls were observed, very few to no seedlings were found to have the brown pigmented primary root with the rapid growth arrest (Supplementary Table S3). Figure 6B shows the percentage of seedlings in each category averaged over the 20 independent lines for Col-0 and the different mutant backgrounds. While Col-0/DEX::PERK10 seedlings had a frequency of 26.01% with the brown pigmented primary root, 0% of the perk8-1,9-1,10-1/DEX::PERK10 seedlings displayed this phenotype, and very low levels of the brown pigmented primary roots were detected in the kipk1-1,2-1/DEX::PERK10 seedlings (2.45%) and the kcbp-1/DEX::PERK10 seedlings (0.15%) (Fig. 6B and Supplementary Table S3). Interestingly, in all three mutant backgrounds, there was an increase in seedlings with brown pigmented hypocotyls in comparison with Col-0/DEX::PERK10. Nevertheless, in terms of root growth, the overexpressing PERK10 in the three different mutant backgrounds resulted in an attenuation of the brown pigmented primary root arrest phenotype observed in Col-0 (Fig. 6B).
Fig. 6.
DEX::PERK10 expression in the different mutant backgrounds. (A) RT-PCR analysis of PERK10 expression in 5-d-old DEX::PERK10 seedlings from the different backgrounds, following no treatment (– DEX) or treatment with 10 µM DEX (+ DEX). Actin was used as a positive control for expression in all tissues. (B) Graph of the different phenotypes observed in 5-d-old DEX::PERK10 seedlings. Seeds from primary transformants for Col-0/DEX::PERK10, perk8-1,9-1,10-1/DEX::PERK10, kipk1-2,2-1/DEX::PERK10, and kcbp-1/DEX::PERK10 were germinated in the presence of 10 µM DEX and scored for the different phenotypes at d 5. For each genotype, seeds from 20 independent primary transformants were scored, and the mean was determined across the 20 lines (see Supplementary Table S3). Up to 25% of the seeds may be untransformed wild type, depending on the number of integrated transgenes, since the seeds were from primary transformants, and no transgene selection was applied.
Discussion
In this study, we set out to identify in planta functions for the predicted PERK8, -9, and -10 receptor-like kinases and to identify their potential downstream signalling proteins. KIPK1 and -2 were found to be interactors of the cytosolic kinase domains from PERK8, -9, and -10, and as such, they represented candidate downstream signalling proteins. A GFP:KIPK1 was reported to be targeted to both the nucleus and cytoplasm in yeast (Zegzouti et al., 2006), which would place KIPK1 in an overlapping compartment with the PERK8, -9, and -10 cytosolic domains for interactions. KIPK1 was identified previously as an interactor of KCBP (Day et al., 2000), and we demonstrated here that this interaction is conserved for KIPK2. Future research will need to investigate the nature of these interactions in vivo, and the role of phosphorylation by the PERK(8,9,10) and KIPK(1,2) kinase domains during these interactions. To determine if the PERK(8,9,10)–KIPK(1,2) –KCBP interactions were biologically relevant and to determine if they were functioning as positive or negative regulators, T-DNA insertion mutants were screened for altered growth phenotypes. No obvious differences were apparent under standard growth conditions, and while kcbp mutants displayed a trichome phenotype (Oppenheimer et al., 1997), this was not seen in the perk or kipk mutants. However, changes were observed on root growth assays under high sucrose and 24h lighting where the perk8-1,9-1,10-1 triple mutant, the kipk1-1,2-1 double mutant, and the kcbp-1 mutant all displayed significant increases in primary root length compared with Col-0. Since all three mutant combinations displayed the same phenotype of increased root growth, this suggests that PERK(8,9,10) –KIPK(1,2)–KCBP are functioning in the same direction to negatively regulate root growth.
KIPK1 and -2 are members of the Arabidopsis AGC VIII kinases, which belong to the eukaryotic AGC serine/threonine protein kinase family, named after cAMP-dependent protein kinase, cGMP-dependent protein kinase, and protein kinase C (Bogre et al., 2003; Zegzouti et al., 2006). The Arabidopsis AGC VIII kinases are further divided into four subgroups, and KIPK1 and -2 belong to the AGC1 subgroup (reviewed by Rademacher and Offringa, 2012). Well-characterized Arabidopsis AGC VIII kinases include the AGC4 subgroup members, the PHOTOTROPIN1 and PHOTOTROPIN2 blue-light photoreceptors (Huala et al., 1997; Jarillo et al., 2001; Takemiya et al., 2005). There are also the AGC3 subgroup members, PINOID, WAG1, and WAG2, which are proposed to regulate auxin fluxes by regulating the localization of PIN-FORMED (PIN) auxin efflux carriers (Christensen et al., 2000; Santner and Watson, 2006; Dhonukshe et al., 2010). Interestingly, the AGC1 subgroup includes D6 PROTEIN KINASE members that have also been implicated in regulating PIN proteins in relation to phototropic responses such as hypocotyl bending (Willige et al., 2013). Finally, the AGC2 subgroup includes UNICORN functioning in planar growth regulation (Enugutti et al., 2012) and OXIDATIVE SIGNAL INDUCIBLE1 required for signalling following oxidative bursts during root hair growth and basal pathogen resistance (Rentel et al., 2004). One of the defining features of the AGC VIII kinases is the variable N-terminal domains that are thought to confer regulatory functions (Rademacher and Offringa, 2012). For KIPK1 and -2, this region is approximate 500 aa and displays 63% amino acid identity between the two proteins. The conservation of this region appears to be restricted to KIPK1 and -2 and not found in the other Arabidopsis AGC VIII kinases. There are no known predicted protein–protein interaction domains in this conserved N-terminal domain, and the only noticeable feature is an enrichment of serines. Our yeast two-hybrid data showed that the proximal N-terminal residues are required for KIPK1 and -2 to interact with the PERK cytosolic kinase domains while the distal N-terminal residues along with the C-terminal kinase domain are involved in the interaction with KCBP.
Arabidopsis KCBP was first isolated in a screen for proteins that bound calmodulin (CaM). The CaM-binding domain was mapped to the C-terminal end of KCBP and required Ca2+ for the interaction with CaM (Reddy et al., 1996). The novel KCBP-interacting Ca2+-binding (KIC) protein was also demonstrated to bind to the C-terminal end of KCBP (Reddy et al., 2004). KCBP is part of the kinesin-14 family, and kinesin-14 members are known to play roles in the organization of spindle poles and transporting cargo, and have C-terminal motor domains that drive minus-end-directed movement along microtubules (reviewed by Verhey and Hammond, 2009). As predicted, the KCBP motor domain binds microtubules, but this binding could be inhibited by both CaM and KIC in the presence of Ca2+ (Song et al., 1997; Reddy et al., 2004). Thus, CaM and KIC were proposed to be negative regulators of KCBP, and in keeping with this role, the overexpression of KIC resulted in trichomes with reduced branches, a phenotype found in the kcbp/zwi loss-of-function mutant (Oppenheimer et al., 1997; Reddy et al., 2004). With the possibility of KCBP also binding actin through the MyTH4 and the FERM domains (Reddy and Day 2000), Smith and Oppenheimer (2005) proposed a speculative model of KCBP interacting with microtubules and actin filaments to facilitate the transport of cell wall material from the Golgi to the plasma membrane for trichome branching (Smith and Oppenheimer 2005).
Thus, one possible cellular role of the PERK(8,9,10)–KIPK(1,2)–KCBP interactions in root growth may be to regulate cell expansion through the transport of cell wall material. We did not observe any differences in the lengths of expanded root cells (Supplementary Fig. S3B), and so the potential role of this pathway may be to regulate the rate of root elongation. Wuyts et al. (2011) found that when Col-0 seedlings were grown on 1% sucrose, the seedlings had longer roots and a faster rate of root elongation when compared with seedling roots grown with no sucrose. Therefore, the increased growth of the perk8-1,9-1,10-1 triple mutant, the kipk1-1,2-1 double mutant, and the kcbp-1 mutant may be related to faster root elongation rates in the presence of 4.5% sucrose and 24h lighting. In contrast to the mutants, these conditions had an inhibitory effect on Col-0 roots (when compared with Col-0 seedling roots grown in the absence of sucrose), as reported previously (Hauser et al., 1995). KCBP has also been found to be localized to microtubules in dividing tobacco BY-2 cells, suggesting a role during the cell cycle (Bowser and Reddy, 1997). This was further supported when antibodies to KCBP were injected into dividing stamen hair cells causing a disrupted phragmoplast formation and delayed cell division (Vos et al., 2000). Thus, it is quite possible that the PERK(8,9,10)–KIPK(1,2)–KCBP interactions may also be influencing cell division rates during root growth.
In contrast to the mutant root phenotypes, the overexpression of PERK10 led to a rapid arrest in primary root growth in wild-type Col-0 seedlings. The frequency of this phenotype was significantly reduced or absent in the transgenic DEX::PERK10 perk8-1,9-1,10-1 triple mutant, the kipk1-1,2-1 double mutant, and the kcbp-1 mutant compared with transgenic Col-0/DEX::PERK10 seedlings. This is what would be expected as the overactivation of this signalling pathway would be attenuated in these mutants compared with Col-0; that is, one would predict impairment of the signalling pathway downstream of PERK8, -9, and -10 for the kipk1-1,2-1 double mutant and the kcbp-1 mutant. For the transgenic DEX::PERK10 perk8-1,9-1,10-1 triple mutant results, the simplest explanation is that the loss of PERK8, -9, and -10 expression in the perk8-1,9-1,10-1 triple mutant would decrease the total PERK activity following DEX::PERK10 induction with DEX treatment, and as a result, primary root growth arrest is not observed. Thus, the occurrence of the primary root growth arrest phenotype in Col-0/DEX::PERK10 seedlings, and reduced frequency of this phenotype in the transgenic DEX::PERK10 perk8-1,9-1,10-1 triple mutant, the kipk1-1,2-1 double mutant, and the kcbp-1 mutant is consistent with the proposed role of PERK(8,9,10)–KIPK(1,2)–KCBP as negatively regulators of root growth. The second distinct phenotype observed with PERK10 overexpression was germinating DEX::PERK10 transgenic seedlings with brown pigment in the hypocotyl. This phenotype was enhanced in the transgenic DEX::PERK10 perk8-1,9-1,10-1 triple mutant, the kipk1-1,2-1 double mutant, and the kcbp-1 mutant compared with transgenic Col-0/DEX::PERK10 seedlings. It is unclear why there was an increased frequency of this hypocotyl brown pigment phenotype in the mutant backgrounds, but perhaps there are other unknown signalling components functioning in the hypocotyl to cause this enhanced phenotype with PERK10 overexpression.
There is a general trend observed with other perk mutants of these genes acting as negative regulators of growth. A perk13/rhs10 mutant had longer root hairs compared with wild-type, while overexpression of PERK13/RHS10 in root hairs caused reduced root hair elongation and shorter root hairs (Won et al., 2009). In addition, in the presence of ABA, perk4 mutants displayed increased root elongation relative to wild-type Col-0 (Bai et al., 2009). Among the complex traits present in the ectopic and antisense expression of BnPERK1 in Arabidopsis were changes in hypocotyl length in dark-grown seedlings (Haffani et al., 2006). Hypocotyls were significantly shorter with ectopic BnPERK1 expression, while the antisense expression of BnPERK1 leading to the suppression of Arabidopsis PERK1 and -3 resulted in longer hypocotyls (Haffani et al., 2006). Thus, the overall theme suggests that PERKs act as negative regulators of plant growth, and under specific conditions or tissues, the loss of PERK function is associated with increased growth while increased PERK activity is associated with reduced growth. In the case of PERK8, -9, and -10, we propose that KIPK1 and -2 and KCBP function downstream to mediate their signalling responses.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Amino acid sequence alignment of KIPK1 and KIPK2.
Supplementary Fig. S2. Phenotypes of perk, kipk and kcbp mutant plants.
Supplementary Fig. 3. Root growth of wild-type Col-0 and perk8-1,9-1,10-1 triple mutant seedlings under different conditions.
Supplementary Table S1. Primers for RT-PCR analyses
Supplementary Table S2. Root lengths for the perk, kipk, and kcbp mutants under different conditions.
Supplementary Table S3. DEX::PERK10 seedling phenotypes in Col-0 and the perk, kipk, and kcbp mutants.
Acknowledgements
We thank the Salk Institute Genomic Analysis Laboratory, GABI-Kat, and ABRC for providing the sequence-indexed Arabidopsis T-DNA insertion lines and the pUNI-KIPK1 cDNA, and thank Dr Anireddy Reddy for the KCBP clone. We thank Dr Nancy Silva and Sarah Keatley for assistance with the PERK T-DNA insertion line screens and Sneha Mehta for assistance with the yeast two-hybrid screen. We are very grateful to Dr Malcolm Campbell and Dr Dario Bonetta for helpful discussions. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) and a Canada Research Chair to DRG.
Glossary
Abbreviations:
- ABA
abscisic acid
- CaM
calmodulin
- DEX
dexamethasone
- GFP
green fluorescent protein
- KCBP
kinesin-like calmodulin-binding protein
- KIC
KCBP-interacting Ca2+-binding
- KIPK
KCBP-interacting protein kinase
- NSP
nuclear shuttle protein
- MS
Murashige and Skoog
- PERK
proline-rich, extensin-like receptor-like kinase
- RT-PCR
reverse transcription-PCR.
References
- Abdel-Ghany SE, Day IS, Simmons MP, Kugrens P, Reddy AS. 2005. Origin and evolution of Kinesin-like calmodulin-binding protein. Plant Physiology 138, 1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image Processing with ImageJ. Biophotonics International 11, 36–42. [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana . Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Antolin-Llovera M, Ried MK, Binder A, Parniske M. 2012. Receptor kinase signaling pathways in plant–microbe interactions. Annual Review of Phytopathology 50, 451–473. [DOI] [PubMed] [Google Scholar]
- Aoyama T, Chua NH. 1997. A glucocorticoid-mediated transcriptional induction system in transgenic plants. The Plant Journal 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Bai L, Zhang G, Zhou Y, Zhang Z, Wang W, Du Y, Wu Z, Song CP. 2009. Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca signalling, is required for abscisic acid responses in Arabidopsis thaliana . The Plant Journal 60, 314–327. [DOI] [PubMed] [Google Scholar]
- Bogre L, Okresz L, Henriques R, Anthony RG. 2003. Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends in Plant Science 8, 424–431. [DOI] [PubMed] [Google Scholar]
- Bowser J, Reddy AS. 1997. Localization of a kinesin-like calmodulin-binding protein in dividing cells of Arabidopsis and tobacco. The Plant Journal 12, 1429–1437. [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Dagenais N, Chory J, Weigel D. 2000. Regulation of auxin response by the protein kinase PINOID. Cell 100, 469–478. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Day IS, Miller C, Golovkin M, Reddy AS. 2000. Interaction of a kinesin-like calmodulin-binding protein with a protein kinase. Journal of Biological Chemistry 275, 13737–13745. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Huang F, Galvan-Ampudia CS, et al. 2010. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137, 3245–3255. [DOI] [PubMed] [Google Scholar]
- Ding X, Snyder AK, Shaw R, Farmerie WG, Song WY. 2003. Direct retransformation of yeast with plasmid DNA isolated from single yeast colonies using rolling circle amplification. Biotechniques 35, 774–776, 778–779. [DOI] [PubMed] [Google Scholar]
- Doblin MS, Johnson KL, Humphries J, Newbigin EJ, Bacic AT. 2014. Are designer plant cell walls a realistic aspiration or will the plasticity of the plant’s metabolism win out? Current Opinion in Biotechnologynol 26C, 108–114. [DOI] [PubMed] [Google Scholar]
- Enugutti B, Kirchhelle C, Oelschner M, Torres Ruiz RA, Schliebner I, Leister D, Schneitz K. 2012. Regulation of planar growth by the Arabidopsis AGC protein kinase UNICORN. Proceedings of the National Academy of Sciences, USA 109, 15060–15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentino LH, Santos AA, Fontenelle MR, Pinheiro GL, Zerbini FM, Baracat-Pereira MC, Fontes EP. 2006. A PERK-like receptor kinase interacts with the geminivirus nuclear shuttle protein and potentiates viral infection. Journal of Virology 80, 6648–6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish LA, Clark SE. 2011. The RLK/Pelle family of kinases. The Plant Journal 66, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffani YZ, Silva-Gagliardi NF, Sewter SK, Grace Aldea M, Zhao Z, Nakhamchik A, Cameron RK, Goring DR. 2006. Altered expression of PERK receptor kinases in Arabidopsis leads to changes in growth and floral organ formation. Plant Signaling & Behavior 1, 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MT, Morikami A, Benfey PN. 1995. Conditional root expansion mutants of Arabidopsis. Development 121, 1237–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, Schulze WX. 2008. PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Research 36, D1015–D1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. 1997. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278, 2120–2123. [DOI] [PubMed] [Google Scholar]
- Humphrey TV, Bonetta DT, Goring DR. 2007. Sentinels at the wall: cell wall receptors and sensors. New Phytologist 176, 7–21. [DOI] [PubMed] [Google Scholar]
- Hwang I, Kim SY, Kim CS, Park Y, Tripathi GR, Kim SK, Cheong H. 2010. Over-expression of the IGI1 leading to altered shoot-branching development related to MAX pathway in Arabidopsis. Plant Molecular Biology 73, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Gabrys H, Capel J, Alonso JM, Ecker JR, Cashmore AR. 2001. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 410, 952–954. [DOI] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Engineering Design & Selection 17, 527–536. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. 2005. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. The Plant Journal 41, 697–709. [DOI] [PubMed] [Google Scholar]
- Nakhamchik A, Zhao Z, Provart NJ, Shiu SH, Keatley SK, Cameron RK, Goring DR. 2004. A comprehensive expression analysis of the Arabidopsis proline-rich extensin-like receptor kinase gene family using bioinformatic and experimental approaches. Plant and Cell Physiology 45, 1875–1881. [DOI] [PubMed] [Google Scholar]
- Narasimhulu SB, Kao YL, Reddy AS. 1997. Interaction of Arabidopsis kinesin-like calmodulin-binding protein with tubulin subunits: modulation by Ca2+-calmodulin. The Plant Journal 12, 1139–1149. [DOI] [PubMed] [Google Scholar]
- Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. 2009. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 10, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DG, Pollock MA, Vacik J, Szymanski DB, Ericson B, Feldmann K, Marks MD. 1997. Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proceedings of the National Academy of Sciences, USA 94, 6261–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. 2013. Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. Journal of Experimental Botany 64, 445–458. [DOI] [PubMed] [Google Scholar]
- Rademacher EH, Offringa R. 2012. Evolutionary adaptations of plant AGC kinases: from light signaling to cell polarity regulation. Frontiers in Plant Science 3, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Day IS. 2000. The role of the cytoskeleton and a molecular motor in trichome morphogenesis. Trends in Plant Science 5, 503–505. [DOI] [PubMed] [Google Scholar]
- Reddy AS, Safadi F, Narasimhulu SB, Golovkin M, Hu X. 1996. A novel plant calmodulin-binding protein with a kinesin heavy chain motor domain. Journal of Biological Chemistry 271, 7052–7060. [DOI] [PubMed] [Google Scholar]
- Reddy VS, Day IS, Thomas T, Reddy AS. 2004. KIC, a novel Ca2+ binding protein with one EF-hand motif, interacts with a microtubule motor protein and regulates trichome morphogenesis. Plant Cell 16, 185–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel MC, Lecourieux D, Ouaked F, et al. 2004. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427, 858–861. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, et al. 2003. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Research 31, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers LA, Dubos C, Surman C, Willment J, Cullis IF, Mansfield SD, Campbell MM. 2005. Comparison of lignin deposition in three ectopic lignification mutants. New Phytologist 168, 123–140. [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. 2003. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Molecular Biology 53, 247–259. [DOI] [PubMed] [Google Scholar]
- Santner AA, Watson JC. 2006. The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis. The Plant Journal 45, 752–764. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. 2005. A gene expression map of Arabidopsis thaliana development. Nature Genetics 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proceedings of the National Academy of Sciences, USA 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva NF, Goring DR. 2002. The proline-rich, extensin-like receptor kinase-1 (PERK1) gene is rapidly induced by wounding. Plant Molecular Biology 50, 667–685. [DOI] [PubMed] [Google Scholar]
- Smith LG, Oppenheimer DG. 2005. Spatial control of cell expansion by the plant cytoskeleton. Annual Review of Cell and Developmental Biology 21, 271–295. [DOI] [PubMed] [Google Scholar]
- Song H, Golovkin M, Reddy AS, Endow SA. 1997. In vitro motility of AtKCBP, a calmodulin-binding kinesin protein of Arabidopsis. Proceedings of the National Academy of Sciences, USA 94, 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinwand BJ, Kieber JJ. 2010. The role of receptor-like kinases in regulating cell wall function. Plant Physiology 153, 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemiya A, Inoue S, Doi M, Kinoshita T, Shimazaki K. 2005. Phototropins promote plant growth in response to blue light in low light environments. Plant Cell 17, 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, Hammond JW. 2009. Traffic control: regulation of kinesin motors. Nature Reviews Molecular Cell Biology 10, 765–777. [DOI] [PubMed] [Google Scholar]
- Vos JW, Safadi F, Reddy ASN, Hepler PK. 2000. The kinesin-like calmodulin binding protein is differentially involved in cell division. Plant Cell 12, 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Ahlers S, Zourelidou M, et al. 2013. D6PK AGCVIII kinases are required for auxin transport and phototropic hypocotyl bending in Arabidopsis. Plant Cell 25, 1674–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won SK, Lee YJ, Lee HY, Heo YK, Cho M, Cho HT. 2009. Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiology 150, 1459–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts N, Bengough AG, Roberts TJ, Du C, Bransby MF, McKenna SJ, Valentine TA. 2011. Automated motion estimation of root responses to sucrose in two Arabidopsis thaliana genotypes using confocal microscopy. Planta 234, 769–784. [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, et al. 2003. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302, 842–846. [DOI] [PubMed] [Google Scholar]
- Zegzouti H, Li W, Lorenz TC, Xie M, Payne CT, Smith K, Glenny S, Payne GS, Christensen SK. 2006. Structural and functional insights into the regulation of Arabidopsis AGC VIIIa kinases. Journal of Biological Chemistry 281, 35520–35530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.