Highlight
An Arabidopsis gene, Dof5.8, is a target of the auxin-responsive MONOPTEROS transcription factor. Mutations in Dof5.8 interfere with embryo development and vein patterning when combined with a monopteros allele.
Key words: Arabidopsis thaliana, auxin response, Dof transcription factor, embryo development, MONOPTEROS, vascular development.
Abstract
MONOPTEROS (MP) is an auxin-responsive transcription factor that is required for primary root formation and vascular development, whereas Dof5.8 is a Dof-class transcription factor whose gene is expressed in embryos as well as the pre- and procambial cells in the leaf primordium in Arabidopsis thaliana. In this study, it is shown that MP directly activates the Dof5.8 promoter. Although no apparent phenotype of the single dof5.8 mutants was found, phenotypic analysis with the mp dof5.8 double mutants revealed that mutations within Dof5.8 enhanced the phenotype of a weak allele of mp, with an increase in the penetrance of the ‘rootless’ phenotype and a reduction in the number of cotyledons. Furthermore, interestingly, although mp mutants showed reduced vascular pattern complexity in cotyledons, the mp dof5.8 double mutants displayed both more simplex and more complex vascular patterns in individual cotyledons. These results imply that the product of Dof5.8 whose expression is regulated by MP at least in part might be involved in multiple processes controlled by MP.
Introduction
MONOPTEROS (MP), which is also known as auxin response factor 5 (ARF5) of the ARF family, is a key regulator that functions in the establishment of vasculature and body patterns in embryonic and post-embryonic development in Arabidopsis thaliana (Hardtke and Berleth, 1998; Ulmasov et al., 1999a, b ). Mutations within the MP gene interfere with the body axis patterning in the early embryo and the formation of vascular strands. Thus, the mp mutants show reduced complexity in vascular patterns in both cotyledons and true leaves, and the mp seedlings are often rootless and have only one cotyledon (Berleth and Jürgens, 1993; Przemeck et al., 1996). Because MP is a transcription factor, direct target genes of MP have been searched for in order to clarify MP-mediated regulations. Consistent with the diverse functions of MP in embryonic root initiation, lateral organ initiation, shoot meristem cell regulation, and vascular patterning in leaves (Berleth and Jürgens, 1993; Przemeck et al., 1996; Hardtke et al., 2004; Schuetz et al., 2008), recent intensive studies revealed that MP activates the expression of DRN involved in cell patterning in embryos (Cole et al., 2009), TMO genes crucial for embryonic root initiation (Schlereth et al., 2010), and LFY for flower initiation (Yamaguchi et al., 2013), and represses the expression of ARR7 and ARR15, negative regulators of cytokinin signalling, in the shoot apical meristem (Zhao et al., 2010). Furthermore, it has been reported that MP regulates the expression of BRX involved in cross-talk between the auxin and brassinosteroid pathways (Bauby et al., 2007; Beuchat et al., 2010; Scacchi et al., 2010), Athb8 associated with the formation of vascular strands in leaves (Donner et al., 2009), and MP itself (Lau et al., 2011). Target genes of MP probably vary depending on organ, tissue type, or developmental stage, and thus MP functions and MP-induced regulation still remain largely unknown.
Dof transcription factors are a family of transcription factors that harbour a plant-specific Dof DNA-binding domain that recognizes 5ʹ-AAAG-3ʹ or 5ʹ-CTTT-3ʹ motifs (Yanagisawa, 2002, 2004). Although the physiological functions of Dof transcription factors are highly diverse, many Dof genes are expressed in the vasculature or during vascular development (Gualberti et al., 2002; Imaizumi et al., 2005; Skirycz et al., 2006; Konishi and Yanagisawa, 2007; Guo et al., 2009; Gardiner et al., 2010; Schlereth et al., 2010; Le Hir and Bellini, 2013). It was shown previously that the promoter of A. thaliana Dof5.8 is specifically active in embryos during the transition and heart stages and the future vasculature of cotyledons at the walking-stick stage, as well as procambial cells (vascular precursors) and pre-procambial cells (cells in the middle of the first stage of vascular development from the ground meristem cells to the procambial cells) in the leaf primordium (Konishi and Yanagisawa, 2007).
As the initial steps of vascular development in leaves in dicots are triggered by auxin flow, and then auxin-induced MP activity modulates gene expression for formation of the vascular network (Donner et al., 2009; Ckurshumova et al., 2011), it is known that pre-procambial and procambial cells (hereafter collectively termed ‘provascular cells’) are characterized by expression of the auxin-responsive marker gene, DR5:GUS (Mattsson et al., 2003), or the auxin efflux carrier protein, PIN1 (Scarpella et al., 2006; Wenzel et al., 2007). Based on the expression pattern of Dof5.8 in embryos and provascular cells in the leaf primordium, we speculated that Dof5.8 might be a target of MP and associated with MP-regulated processes. To examine this hypothesis, molecular genetic and biological analyses were performed in this study. The results indicate that MP directly activates the Dof5.8 promoter whereas mutations within Dof5.8 influence multiple phenotypes of the mp mutant, arf5-2.
Materials and methods
Plant materials
Arabidopsis thaliana ecotype Columbia (Col) was used as the wild-type strain in all experiments. Seeds of the mp mutants, arf5-1, SALK_001058 and arf5-2 (also called mp-S319 or SALK_021319), and SALK T-DNA lines of Dof5.8 were obtained from the Arabidopsis Resource Center (Alonso et al., 2003; Okushima et al., 2005; Donner et al., 2009). DR5-GUS (β-glucuronidase) seeds (Ulmasov et al., 1997b ) were a gift from Dr Tom J. Guilfoyle. For the analysis of transcript levels of Dof5.8 in mp alleles, selfed seeds from heterozygous mp plants were sown. Seedlings exhibiting the rootless phenotype were collected for quantitative reverse transcription–polymerase chain reaction (qRT–PCR) analysis. To generate the double mutants of arf5-2 and dof5.8-1 or dof5.8-2, the dof5.8 plants that are homozygous for a T-DNA insertion were crossed to heterozygous arf5-2 plants, and F2 plants homozygous for the dof5.8 T-DNA allele and heterozygous for arf5-2 allele were selected by PCR-based genotyping. For phenotypic analysis, rootless F3 seedlings, which are homozygous for the arf5-2 allele (Table 1), were picked for analysis of cotyledon numbers and vascular patterns. For the analysis of the Dof5.8 promoter activity in the arf5-2 background, the Dof5.8pro-GUS line harbouring the GUS reporter gene under the control of the Dof5.8 promoter (Konishi and Yanagisawa, 2007) was crossed to the arf5-2 heterozygous plant. The F3 population that was homozygous for the Dof5.8pro-GUS transgene linked to the glufosinate ammonium resistance gene and heterozygous for the arf5-2 allele was selected by phenotypic analysis of the glufosinate ammonium resistance and rootless phenotype or genotyping using a cotyledon of F3 seedlings.
Table 1.
Segregation of the arf5-2 allele among populations derived from plants heterozygous for the arf5-2 allele in the wild-type, dof5.8-1 or dof5.8-2 background
| Genotype of parental plant | No. of seedlings with the indicated genotype at the MP locus (% of total) | No. of rootless seedlingsa | |||
|---|---|---|---|---|---|
| MP/MP | MP/arf5-2 | arf5-2/arf5-2 | Total | ||
| MP/arf5-2 | 31 (35.2%) | 38 (43.2%) | 19 (21.6%) | 88 | 2 |
| dof5.8-1/dof5.8-1; MP/arf5-2 | 26 (26.8%) | 51 (52.6%) | 20 (20.6%) | 97 | 14 |
| dof5.8-2/dof5.8-2; MP/arf5-2 | 23 (19.3%) | 69 (58.0%) | 27 (22.7%) | 119 | 15 |
a All rootless seedlings were homozygous for the arf5-2 allele.
Plant growth conditions
Seeds were sterilized and sown on half-strength Murashige and Skoog (1/2MS) agar plates containing 1% sucrose, as described previously (Konishi and Yanagisawa, 2008). After 3–4 d of stratification, plates were transferred to a chamber set at 23 °C with continuous illumination (60 μE m–2 s–1). For 2,4-dichlorophenoxyacetic acid (2,4-D) treatment, seedlings were grown in liquid 1/2MS medium for 3 d and treated or not with 10 μM 2,4-D for 16h. For the analysis of the vascular pattern, seeds were plated on 1/2MS agar medium containing 1% sucrose, solidified with 0.3% agar. For protoplast transient assays, ecotype Col plants were grown on peat containing nutrients (Sakatanotane Co., Yokohama, Kanagawa, Japan) at 23 °C for 3 weeks under continuous light.
Genotyping
DNA extraction was performed according to Konishi and Sugiyama (2003). Primers used in PCR are listed in Supplementary Table S1 available at JXB online.
Protoplast transient assays
The DNA fragment from the Dof5.8 promoter was amplified by PCR (Konishi and Yanagisawa, 2007), and used to replace the Cauliflower mosaic virus 35S RNA promoter in pJD301 (Luehrsen et al., 1992) to produce reporter plasmids containing the luciferase (LUC) gene. The deleted versions of the Dof5.8 promoter were generated by digestion of the full-length promoter fragment with SphI for truncation at position –1301 (relative to the translation start site) and EcoRI for truncation at position –1077, whereas mutated Dof5.8 promoters were generated by PCR-based mutagenesis, as described previously (Konishi and Yanagisawa, 2010). Primers used for the mutagenesis are listed in Supplementary Table S1 at JXB online. For the construction of effector plasmids, MP or the BODENLOS (BDL) cDNA insert was amplified by RT–PCR and inserted in place of EIN3 cDNA in the 35SC4PPDK-EIN3-MYC plasmid (Yanagisawa et al., 2003). All constructs were verified by DNA sequencing. Co-transfection of reporter and effector plasmids and an internal control plasmid (UBQ10-GUS) into A. thaliana mesophyll protoplasts was carried out according to the method of Yoo et al. (2007). Measurement of LUC and GUS activities and calculations of relative LUC activity levels were performed as described previously (Yanagisawa et al., 2003). For auxin treatment, protoplasts were incubated in the absence or presence of 1 μM indole-3-acetic acid (IAA) after co-transfection.
qRT–PCR analysis
RNA preparation and qRT–PCR were performed as described previously (Konishi and Yanagisawa, 2010). The primers used are listed in Supplementary Table S1 at JXB online.
Construction of binary plasmids and generation of transgenic A. thaliana plants
To construct binary vectors for GUS staining, the DNA fragments for the truncated or mutated Dof5.8 promoters were excised from the respective reporter plasmids used in protoplast transient assays and then inserted in place of the Dof5.8 promoter in the pCB-Dof5.8pro-GUS construct. The transformations of A. thaliana were carried out using these binary vectors, as described previously (Konishi and Yanagisawa, 2007).
GUS staining and histological analysis
Histochemical GUS staining was essentially performed as described previously (Konishi and Yanagisawa, 2007). Samples were fixed in 90% acetone at –20 °C, rinsed four times with 0.1M sodium phosphate buffer (pH 7.4), and then incubated in X-Gluc solution [0.1M sodium phosphate (pH 7.4), 3mM potassium ferricyanide, 0.5mM potassium ferrocyanide, 0.5g l–1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide cyclohexilammonium salt] at 37 °C. When GUS activity was weak, the concentration of potassium ferricyanide was reduced according to Donnelly et al. (1999). Potassium ferricyanide at 1.75mM was thus used for the Dof5.8 promoter truncated at –1301 and the promoter containing a 486bp fragment (from –1558 to –1073) of the Dof5.8 promoter upstream of the 35S minimal promoter, and 0.5mM potassium ferricyanide was used for the Dof5.8 promoter truncated at –1077. After staining, samples were incubated in methanol to remove chlorophyll and then mounted in the clearing solution (a mixture of chloral hydrate, water, and glycerol in a ratio of 8:2:1). Observation was performed using a stereomicroscope (MZ16F, Leica Microsystems, Germany) or a microscope equipped with Nomarski optics (BX51, Olympus Co., Tokyo, Japan). For the observation of vascular patterns, cotyledons were fixed in a mixture of ethanol and acetic acid in a ratio of 9:1, hydrated through a graded series of ethanol, and then mounted with the clearing solution (Konishi and Sugiyama, 2003).
Chromatin immunoprecipitation analysis
Chromatin immunoprecipitation (ChIP) analysis was carried out to examine the binding of MP to the Dof5.8 promoter, as described previously (Donner et al., 2009).
Results
Regulation of the provascular expression of Dof5.8 by MP
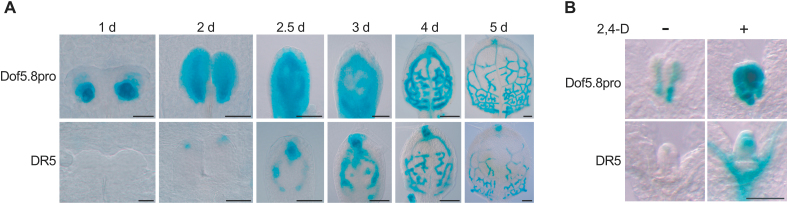
It was shown previously that the Dof5.8 promoter is active in provascular cells in leaf primordia (Konishi and Yanagisawa, 2007). Because the synthetic auxin response element, DR5, induces a similar expression pattern in leaves (Ulmasov et al., 1997b ; Mattsson et al., 2003; Scarpella et al., 2006), the expression patterns produced by a GUS reporter gene under the control of either the Dof5.8 promoter or DR5 during the development of the first leaf primordia were compared (Fig. 1A). The Dof5.8 promoter initially directed strong GUS expression deep inside the bulges of the leaf primordia (Fig. 1A, ‘1d’). This expression then expanded vertically with the upward extension of the primordia. From day 2.5 onwards, reporter expression was localized to the provascular network in a pattern similar to that produced by DR5. This suggested that the activity of the Dof5.8 promoter is regulated at least in part by auxin that accumulated during pre-procambium formation. Consistently, a treatment with the synthetic auxin 2,4-D significantly strengthened expression of the GUS reporter gene under the control of the Dof5.8 promoter (Fig. 1B).
Fig. 1.
Provascular activity of the Dof5.8 promoter and its enhancement by auxin. (A) Time course analysis of GUS expression under the control of the Dof5.8 promoter (Dof5.8pro) or the DR5 element (DR5) during development of the primordia of the first true leaves. Two primordia flanking the shoot apical meristem (1 d and 2 d) and individual leaf primordia (2.5–5 d) are shown. Scale bars=20 μm in 1 d and 2 d, 50 μm in 2.5 d and 3 d, 100 μm in 4 d and 5 d. (B) Effect of auxin treatment on the activity of the Dof5.8 promoter. Seedlings were treated or not with 10 μM 2,4-D for 16h. Scale bar=100 μm.
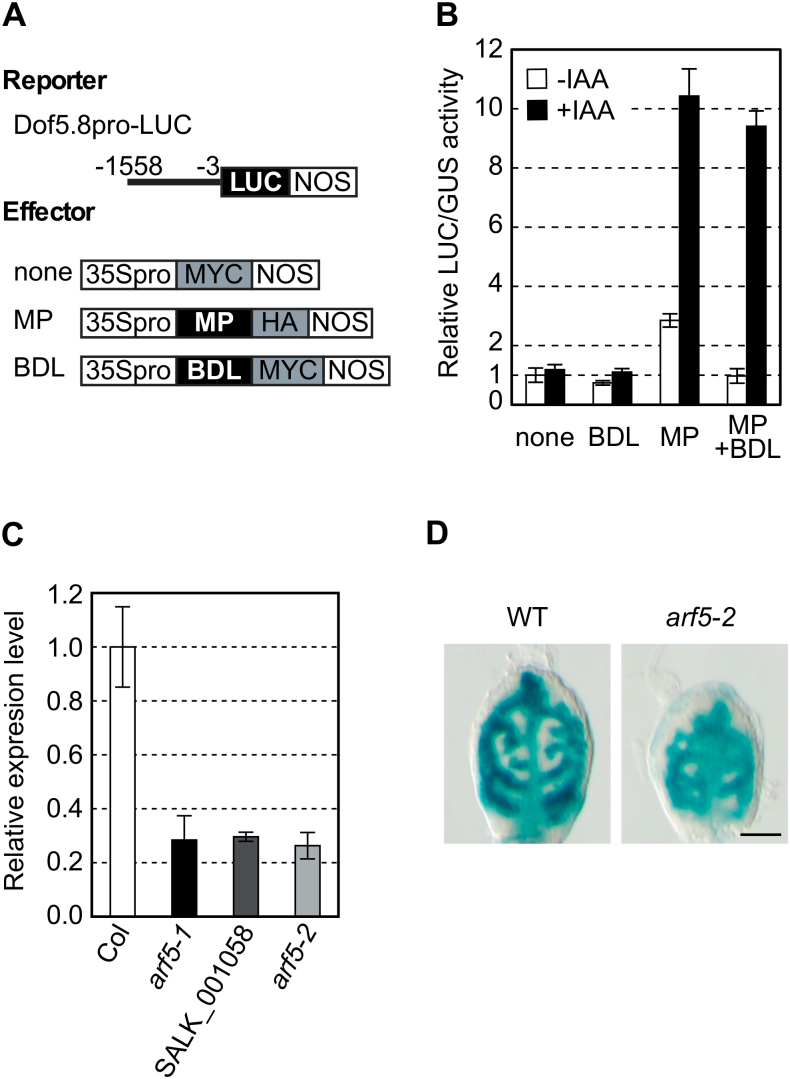
Among the ARFs, MP is known to be involved in vascular development. mp mutants show reduced vascular pattern complexity and sometimes form a disconnected vasculature (Berleth and Jürgens, 1993; Przemeck et al., 1996; Hardtke and Berleth, 1998). Thus, it was hypothesized that the Dof5.8 promoter is activated by MP. To examine this possibility, transactivation assays were performed in A. thaliana protoplasts using a reporter plasmid containing the LUC gene under the control of the Dof5.8 promoter (Dof5.8pro-LUC) and an effector plasmid directing the constitutive high level expression of MP (Fig. 2A). The results indicated that MP increases Dof5.8 promoter activity by ~3-fold and that the MP-induced activation was hampered by the co-expression of BDL/IAA12 (Fig. 2B), a cognate repressor protein of MP (Hamann et al., 2002). Furthermore, the addition of a natural auxin, IAA, into the incubation buffer of the transfected protoplasts enhanced the effect of MP. This could be due to the activation of MP through the degradation of repressor proteins of MP, namely endogenous Aux/IAA proteins or co-expressed BDL, by auxin (Mockaitis and Estelle, 2008). These findings suggested that MP could transactivate the Dof5.8 promoter in response to auxin.
Fig. 2.
Activation of the Dof5.8 promoter by MP. (A) Schematic diagrams of the reporter and effector constructs used in the protoplast transactivation assay described in (B). Numbers indicate nucleotide positions relative to the translation start codon. ‘MYC’ and ‘HA’ are MYC- and haemagglutinin-tag peptides. (B) Effects of MP and auxin on the activity of the Dof5.8 promoter in protoplasts. The Dof5.8pro-LUC reporter construct was co-transfected with the expression plasmid of BDL, MP, or both, or an empty vector (none) and then protoplasts were incubated in the presence (+ IAA) or absence (– IAA) of 1 μM IAA. An internal control plasmid (UBQ10-GUS) was also co-transfected to normalize LUC reporter activity levels. Relative levels of LUC activity are shown as the means ±SD (n=3). (C) Relative Dof5.8 transcript levels in three mp alleles. Total RNA was extracted from the shoots of 4-day-old seedlings and used for qRT–PCR analysis. The transcript levels in wild-type A. thaliana (Col) were set to 1, as all three mp alleles (arf5-1, SALK_001058, and arf5-2) were in the Colombia background. Data are shown as the means ±SD (n=3). (D) GUS staining of the first true leaves of 4-day-old wild-type and arf5-2 seedlings that harbour the GUS gene under the control of the Dof5.8 promoter. The arf5-2 homozygous seedlings obtained from a segregating population of the plant homozygous for the Dof5.8pro-GUS transgene and heterozygous for the arf5-2 allele were used. Scale bar=50 μm.
To substantiate that the expression of Dof5.8 is under the control of MP in planta, Dof5.8 transcript levels were first analysed in three mp alleles by qRT–PCR. The expression levels of Dof5.8 in all mp mutants, including arf5-1 (Alonso et al., 2003; Okushima et al., 2005) and arf5-2 (Alonso et al., 2003; Donner et al., 2009), were reduced to ~30% of those in the wild-type A. thaliana (Fig. 2C). Furthermore, when the GUS gene fused to the Dof5.8 promoter was introduced into the arf5-2 mutant by crossing, the GUS expression was found to be lower in leaf primordia of the arf5-2 seedlings than in those of the wild-type seedlings (Fig. 2D). These results indicate that Dof5.8 expression is regulated by MP activity, at least in part.
Identification of the MP-binding sites required for provascular activation of the Dof5.8 promoter
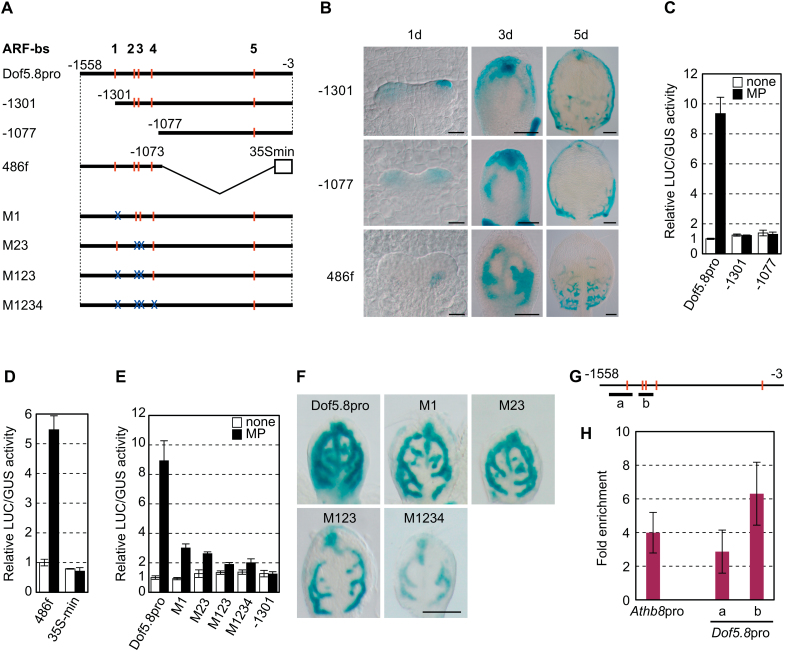
To determine the region within the Dof5.8 promoter that is required for provascular expression, increasing segments of this promoter were deleted from its 5’ end (Fig. 3A). Deletion of the region from –1558 to –1301 in the Dof5.8 promoter diminished GUS expression in provascular cells (Fig. 3B). In addition, a synthetic promoter in which the 486bp fragment from –1558 to –1073 was fused to the 35S minimal promoter (the 486f promoter) was able to direct provascular GUS reporter expression (Fig. 3A, B), indicating that this 486bp region is sufficient to confer Dof5.8 expression in provascular cells.
Fig. 3.
Identification of MP-binding sites and the promoter region required for Dof5.8 expression in provascular cells. (A) Schematic representation of the promoter fragments used for deletion and mutational analyses of the Dof5.8 promoter. The sequences matching the ARF-binding consensus sequence (ARF-bs), and disruptions in these sequences are indicated by red bars and blue ‘X’s, respectively. Numbers indicate nucleotide positions relative to the translation start codons. A 486bp fragment from –1558 to –1073 was fused to the 35S minimal promoter truncated at –72 (35S min) to generate a fusion promoter (the 486f promoter). The mutated sequences are 5’-ACAGAG-3’ in ARF-bs 1–3 and 5’-ACAGTG-3’ in ARF-bs 4. (B) The activity of the truncated Dof5.8 promoters and the 486f promoter in the primordia of the first leaves. GUS staining of the first leaves of the transgenic seedlings carrying the GUS gene under the control of the deletion promoters described in (A). Scale bars=20 μm (1 d), 50 μm (3 d), and 100 μm (5 d). (C–E) MP-mediated transactivation of truncated Dof5.8 promoters (C), the 486f synthetic promoter (D), and mutated Dof5.8 promoters (E) in protoplasts. Promoters fused to LUC were co-transfected with the 35S-MP-HA plasmid (black bars) or an empty vector (white bars). Data are shown as the means ±SD (n=3). (F) GUS staining of the first leaves of the 4-day-old transgenic seedlings carrying the GUS gene under the control of the mutated promoters described in (A). Scale bar=100 μm. (G) Schematic representation of the Dof5.8 promoter showing ARF-binding sites (red bars) and the positions of the amplified DNA fragments (bars a and b) used in the ChIP-qPCR analysis shown in (H). (H) ChIP analysis of the binding of MP to the Dof5.8 promoter. Four-day-old transgenic seedlings expressing CFP-tagged MP were used. A DNA fragment from the Athb8 promoter was amplified as a positive control (Donner et al., 2009). The values were normalized using amplified DNA from a promoter unrelated to MP (the UBQ10 promoter).
The relationship between MP-mediated activation and expression of Dof5.8 in provascular cells was examined using the series of truncated Dof5.8 promoters. The results revealed that MP transactivated only the 486f synthetic promoter (Fig. 3C, D), indicating that the region from –1558 to –1301 is required, whilst the region from –1558 and –1073 is sufficient for both provascular cell expression and activation by MP (Fig. 3B–D). These data further indicate that an intimate relationship exists between MP-mediated activation and expression of Dof5.8 in provascular cells.
ARFs are known to recognize and bind to 5ʹ-TGTCNC-3ʹ sequences (Ulmasov et al.,1997a, 1999b). Four putative ARF-binding sequences were identified in the region between positions –1558 and –1073 of the Dof5.8 promoter (Fig. 3A). A mutation in site 1 (M1) or simultaneous mutations in sites 2 and 3 (M23) reduced the magnitude of activation by MP (Fig. 3A, E). Combination of these mutations (M123) led to an enhanced reduction in reporter enzyme activity, whereas disruption of the fourth site (M1234) had no apparent additional effect. The effects of these mutations on provascular expression in leaf primordia were also assessed (Fig. 3F). The mutated Dof5.8 promoters still directed provascular expression in leaf primordia, but mutations significantly decreased the GUS expression levels. These results suggest that MP recognizes multiple sites in the Dof5.8 promoter that are involved in provascular expression of Dof5.8, although other sites in addition to these putative ARF-binding sites analysed are probably involved in provascular expression of Dof5.8, as is argued in detail in the Discussion.
The binding of MP to the Dof5.8 promoter in vivo was investigated using ChIP analysis of transgenic A. thaliana plants expressing a functional cyan fluorescent protein (CFP)-tagged MP under the control of its own promoter (Donner et al., 2009). As shown in Fig. 3G and H, the results indicated that the binding of MP to the Dof5.8 promoter is comparable with (amplicon ‘a’) and even stronger than (amplicon ‘b’) that to the Athb8 promoter, a known target of MP during vascular development (Donner et al., 2009). Collectively, these results suggest that the Dof5.8 promoter is a direct target of MP in provascular cells and that the entire region from –1558 to –1077 contributes to provascular expression of Dof5.8.
Loss of Dof5.8 affects the root and cotyledon phenotypes of the arf5-2 mutant
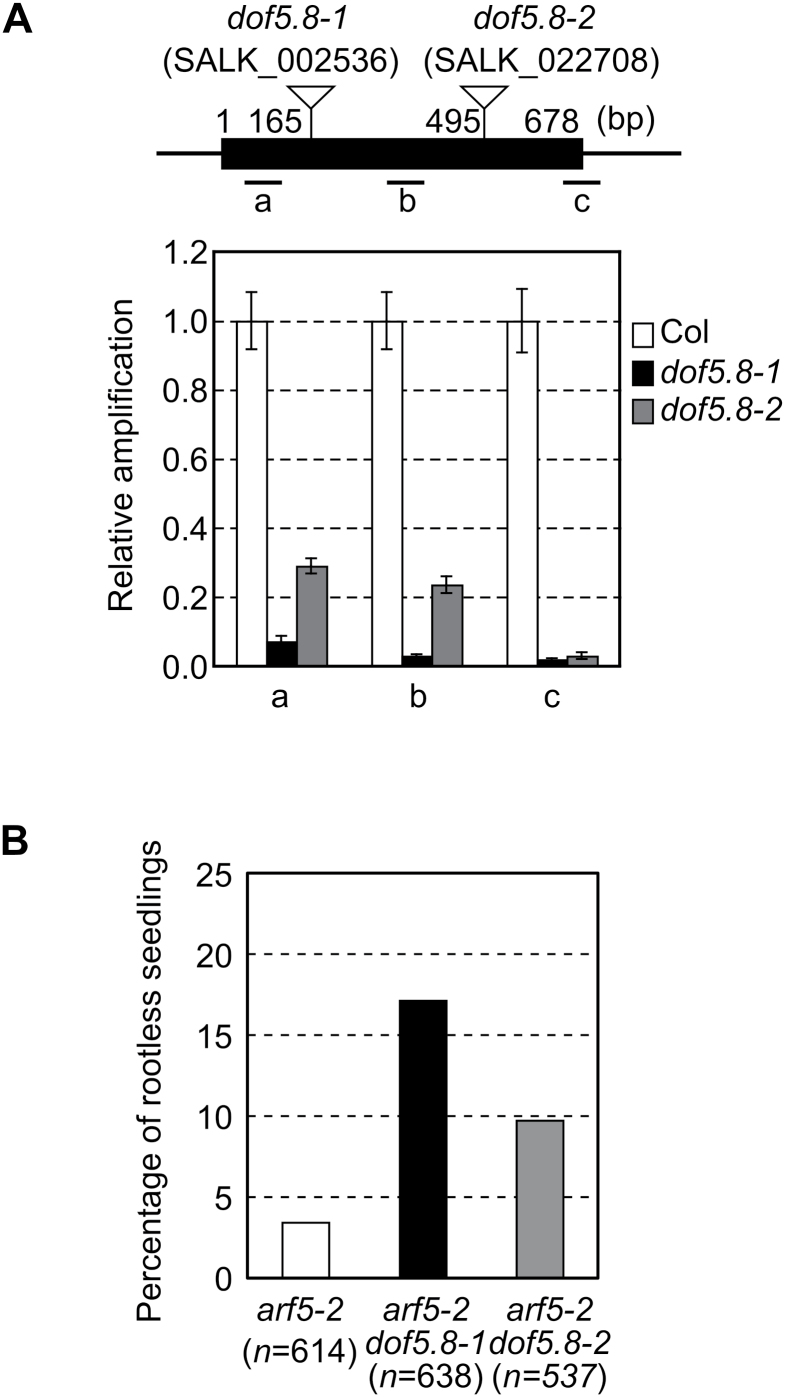
To explore the role of Dof5.8, two T-DNA insertion lines, dof5.8-1 (SALK_002536) and dof5.8-2 (SALK_022708), were analysed. Because T-DNA was inserted into the region encoding the Dof DNA-binding domain in the dof5.8-1 allele, this allele is a null allele. In dof5.8-2, T-DNA was inserted into the C-terminal region flanking the N-terminal Dof domain, suggesting that the product of this allele may retain its DNA-binding activity (Fig. 4A). However, because of the reduced transcript level in dof5.8-2, it is probably a loss-of-function allele (Fig. 4A). Although neither allele exhibited an apparent phenotype, it was found that they enhanced the phenotypes of a weak allele of mp, arf5-2, including abnormal root and cotyledon development. Seedlings of the mp mutants are rootless and often have only one cotyledon (Berleth and Jürgens, 1993). The penetrance of both phenotypes is low in the arf5-2 allele (Donner et al., 2009; Rademacher et al., 2011). In an experiment using a population from the MP/arf5-2 parent plant, 3.4% of the arf5-2 seedlings were rootless, as 21 seedlings out of 614 showed the phenotype (Fig. 4B). Considerably larger numbers of seedlings were found to be rootless when populations from the parent plants that are homozygous for the dof5.8 mutation and heterozygous for the arf5-2 allele were investigated: 17.1% of the dof5.8-1 arf5-2 population and 9.7% of the dof5.8-2 arf5-2 population were rootless. Since the result of genotyping indicated that the percentage of arf5-2 homozygotes was mostly the same (~20%) in these three populations (Table 1; Supplementary Fig. 1 at JXB online), it was concluded that dof5.8 mutations increased the penetrance of the rootless phenotype of arf5-2.
Fig. 4.
Enhancement of the rootless phenotype of the arf5-2 mutant by dof5.8 mutations. (A) The positions of T-DNA insertions and transcript levels of Dof5.8 in the dof5.8-1 and dof5.8-2 mutant lines. The black box indicates the exon, and the positions of the three amplicons (a–c) are shown below. Nucleotide numbers are given relative to the translation start codon. The transcript levels in wild-type A. thaliana (Col) were set to 1. Data are shown as the means ±SD (n=3). (B) The percentages of the seedlings with the rootless phenotype from segregating populations of arf5-2 single and arf5-2 dof5.8 double mutants. Populations derived from parental plants heterozygous for the arf5-2 allele in the wild-type, dof5.8-1 homozygous or dof5.8-2 homozygous background were analysed.
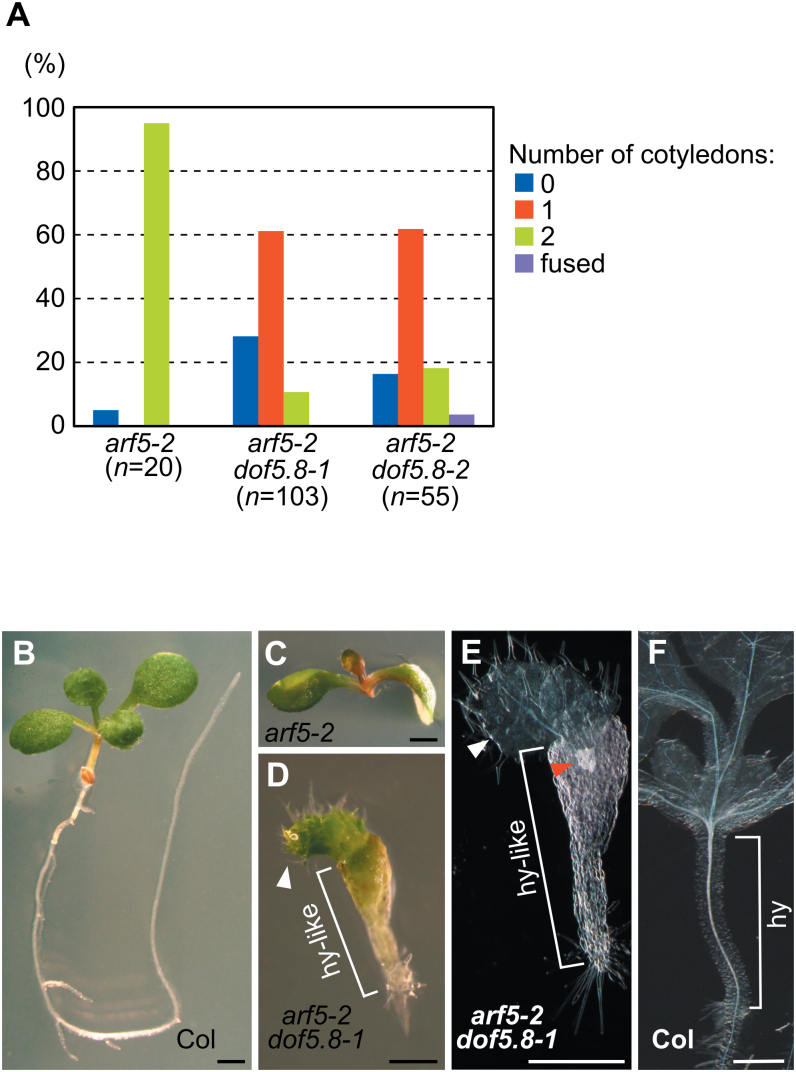
Another effect of dof5.8 mutations in the arf5-2 mutant was also found. Most of the rootless arf5-2 seedlings (95%) possessed two cotyledons, while far fewer seedlings of rootless arf5-2 dof5.8-1 (10.7%) and arf5-2 dof5.8–2 (18.2%) seedlings possessed two cotyledons (Fig. 5A). Furthermore, considerable numbers of the double mutant seedlings had no cotyledons, although such a phenotype was rarely seen in the single mp mutants. The cotyledon-less seedlings of the double mutants always had a fat hypocotyl-like structure, which was topped with true leaves with trichomes but did not include developed vascular elements (Fig. 5D, E). This result suggests that an interaction between arf5-2 and dof5.8 mutations influenced embryonic development and thus formation of cotyledons.
Fig. 5.
The synergistic effects caused by dof5.8 and arf5-2 mutations. (A) The number of cotyledons in rootless seedlings of arf5-2 and arf5-2 dof5.8 mutants. (B–D) Images of 7-day-old wild-type (B), arf5-2 (C), and arf5-2 dof5.8–1 (D) seedlings. (E, F) Cleared images of arf5-2 dof5.8-1 (E) and Col (F) seedlings. White and red arrowheads in (D, E) indicate true leaves and vascular elements, respectively. ‘hy-like’ and ‘hy’ in (D–F) indicate a hypocotyl-like structure and hypocotyl, respectively. Scale bars=1mm in (B–D) and 0.5mm in (D–F).
Effects of dof5.8 mutations on vascular patterning in the arf5-2 mutant
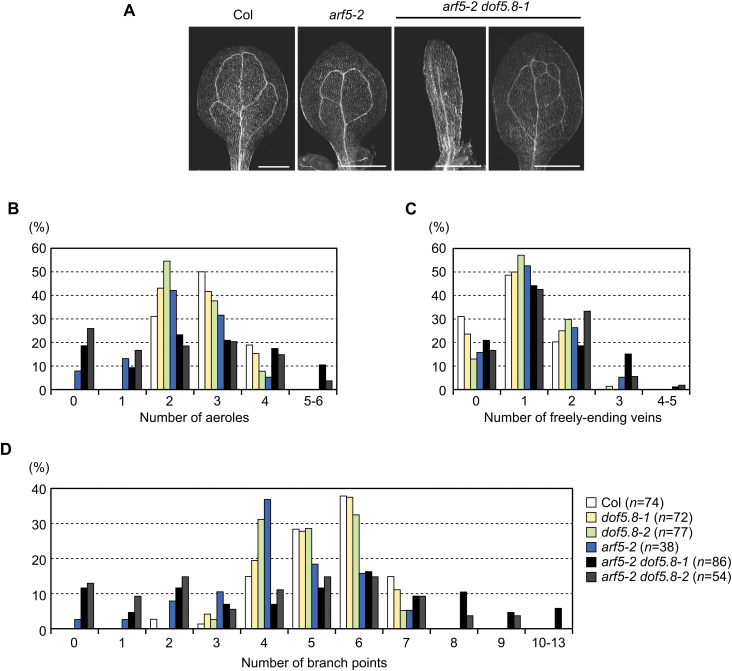
Vein patterns of the double mutants were also analysed, and it was found that the dof5.8 mutation modulates the vascular pattern of arf5-2 cotyledons. Vascular patterns in the cotyledons of wild-type A. thaliana are relatively simple and invariant, with the secondary veins delimiting the two upper aeroles and the zero to two lower aeroles (Fig. 6A). The majority of arf5-2 cotyledons showed vascular patterns similar to those of the wild type (Fig. 6A). However, a small portion (7.9%) had no aeroles due to incomplete formation of the secondary veins (Fig. 6B). The dof5.8 mutations increased the ratio of cotyledons lacking aeroles (18.6% in arf5-2 dof5.8-1 and 25.9% in arf5-2 dof5.8-2; Fig. 6B). However, at the same time, some arf5-2 dof5.8 cotyledons showed more complex vascular patterns with extra aeroles (10.4% in arf5-2 dof5.8-1 and 3.7% in arf5-2 dof5.8-2; Fig. 6A, B). The number of branch points in arf5-2 dof5.8 cotyledons also a showed similar, broader distribution (Fig. 6D). The formation of extra aeroles and branch points was not observed in the wild type, or in arf5-2 or dof5.8 single mutants.
Fig. 6.
The vascular patterns of arf5-2 dof5.8 cotyledons. (A) Representative images of the vascular pattern of cotyledons of wild-type (Col), arf5-2 mutant, and arf5-2 dof5.8-1 double mutant seedlings. Scale bars=1mm. (B–D) The percentage of cotyledons with the indicated number of aeroles (B), freely ending veins (C), and branch points (D). Seeds from plants heterozygous for arf5-2 in the wild-type, dof5.8-1 homozygous or dof5.8-2 homozygous background were sown, and rootless seedlings were used in this analysis. (This figure is available in colour at JXB online.)
Discussion
In this study, it was shown that MP regulates the expression of the Dof5.8 transcription factor gene through its direct binding to the Dof5.8 promoter sequence. It was also shown that two Dof5.8 mutations (dof5.8-1 and dof5.8-2) influence abnormal root and cotyledon development and vascular patterning in the arf5-2 mutant, although the effects of dof5.8 mutations alone were not recognizable in the wild-type genetic background. These phenotypes, together with the evidence that the expression of Dof5.8 is regulated by MP, suggest a genetic interaction between MP and Dof5.8 in the developmental programme in A. thaliana.
Provascular expression of Dof5.8
The results of the deletion and mutation analyses of the Dof5.8 promoter and a ChIP analysis indicated the contribution of MP to the activity of the Dof5.8 promoter in provascular cells (Figs 2, 3). Nevertheless, there are still questions as to the observed phenomenon. The early activity of the Dof5.8 promoter was observed inside leaf primordia where DR5 activity is absent. Because MP mRNA is present within primordia at this stage (Wenzel et al., 2007), the lack of DR5 activity may reflect the lack of the activity of MP protein and the regulation of the Dof5.8 promoter by other transcription factors, although there is another possibility that this phenomena is due to the limitation of using this synthetic reporter. Furthermore, the results of deletion and point mutational analyses in protoplasts and in planta were not perfectly consistent with each another: the effect of deletion of the region from –1558 to –1301 was stronger than that of the M1 mutation (Fig. 3). On the other hand, the result of a ChIP analysis suggests that binding to the region from –1558 to –1301 is weaker than that to the region from –1301 to –1077. Therefore, cis-elements for other transcription factors that act co-operatively with or independently of MP may be present in this region. Alternatively, MP might bind to two 5’-TGTC-3’ sequences in this region in addition to the first putative MP-binding site (site 1 in Fig. 3A), since the TGTC sequence, a part of the consensus sequence for MP binding (5’-TGTCNC-3’), functioned as an MP-binding site in the Athb8 and TMO7 promoters (Donner et al., 2009; Schlereth et al., 2010). Moreover, the mutations on four putative MP-binding sites (M1–M4) affected the Dof promoter activity differently in mesophyll protoplasts and leaf primordia. For instance, the M1 mutation appeared to reduce MP-dependent activation in mesophyll protoplasts and provascular cells to different extents (Fig. 3E, F). This fact also implies the possibility that other transcription factors besides MP are involved in the regulation of the expression of the Dof5.8 promoter in provascular cells. Taken together, although the results indicated that the activity of the Dof5.8 promoter in provascular cells is modulated by MP through the interaction with the region from –1558 to –1077, further analysis focused on this region would be necessary for complete understanding of the molecular mechanisms underlying the expression of Dof5.8 in provascular cells.
The role of Dof5.8 in cotyledon and root formation, and vein patterning
The dof5.8 mutations enhanced the effect of the mp mutation on embryonic root and cotyledon development, consistent with the expression of Dof5.8 in embryos as well as in the immature veins of young leaves (Konishi and Yanagisawa, 2007). The dof5.8 mutations increased the penetrance of the rootless phenotype of arf5-2, suggesting that, when the activity of MP is compromised, Dof5.8 becomes critical for embryonic root formation. Such an auxiliary role to that of MP was also reported for ARF6 (Rademacher et al., 2011). The dof5.8 mutations also produced a synergistic effect with the mp mutation on cotyledon development, which resulted in cotyledon-less seedlings (Fig. 5). Although such a severe phenotype has rarely been reported for any single mp alleles, the combination of mp with a gain-of-function allele of BDL, or with the nph4 mutation produces cotyledon-less seedlings (Hamann et al., 1999; Hardtke et al., 2004). BDL is an inhibitor of ARFs including MP, and NPH4 encodes ARF7. Therefore, a more severe defect in auxin response during embryonic development could cause such cotyledon-less seedlings. The fact that dof5.8 mp double mutants showed such a severe phenotype implies that Dof5.8 is associated with the auxin- and MP-induced developmental programme during embryogenesis.
In contrast to the effects in embryonic development, dof5.8 mutations exert both positive and negative effects on vascular formation in leaf primordia of the arf5-2 mutant. The vein patterns of cotyledons in both the wild type and the arf5-2 mutant are relatively invariant, whereas the vein patterns of the arf5-2 dof5.8 mutants exhibited larger variation, with both reduced and increased complexity in vein pattern (Fig. 6). Although this phenomenon is interesting, it is difficult to explain it if Dof5.8 merely plays an auxiliary role to MP in vascular formation in leaf primordia. Thus, the molecular basis of this is currently unclear. More detailed analysis of dof5.8 mutants in combination with mutations within other Dof genes that are expressed in provasuclar cells or other genes downstream of MP, as well as the identification of target genes of Dof5.8 would be necessary to reveal of the role of Dof5.8 in vascular formation in leaves.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Genotyping of segregating populations of the arf5-2 mutant harbouring the heterozygous arf5-2 allele and the mutants that are heterozygous for the arf5-2 allele and homozygous for the dof5.8 allele.
Table S1. Primer list.
Acknowledgements
We thank Dr Tom J. Guilfoyle (University of Missouri) for generously providing DR5-GUS seeds. We also thank the Arabidopsis Biological Resource Center for providing seeds for the SALK_002536, SALK_022708, arf5-1, SALK_001058, and SALK_021319 lines, and the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed A. thalaina T-DNA insertion mutants (the SIGnAL indexed insertion mutant collection). This research was supported by a Grant-in-Aid for Scientific Research on Innovative Areas (no. 21114004) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) (to SY), grants from the Japan Society for the Promotion of Science (JSPS; nos 22380043 and 25252014 to SY and no. 25840099 to MK), and by a Discovery Grant of the Natural Sciences and Engineering Research Council of Canada (to ES). TJD was supported by an NSERC CGS-D Scholarship and an Alberta Ingenuity Student Scholarship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Glossary
Abbreviations:
- ARF
auxin response factor
- BDL
BODENLOS
- ChIP
chromatin immunoprecipitation
- 2,4-D
2,4-dichlorophenoxyacetic acid
- GUS
β-glucuronidase
- IAA
indole-3-acetic acid
- LUC
luciferase
- MP
MONOPTEROS
- MS
Murashige and Skoog
- qRT–PCR
quantitative reverse transcription–polymerase chain reaction.
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana . Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Bauby H, Divol F, Truernit E, Grandjean O, Palauqui JC. 2007. Protophloem differentiation in early Arabidopsis thaliana development. Plant and Cell Physiology 48, 97–109. [DOI] [PubMed] [Google Scholar]
- Berleth T, Jürgens G. 1993. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118, 575–587. [Google Scholar]
- Beuchat J, Scacchi E, Tarkowska D, Ragni L, Strnad M, Hardtke CS. 2010. BRX promotes Arabidopsis shoot growth. New Phytologist 188, 23–29. [DOI] [PubMed] [Google Scholar]
- Ckurshumova W, Scarpella E, Goldstein RS, Berleth T. 2011. Double-filter identification of vascular-expressed genes using Arabidopsis plants with vascular hypertrophy and hypotrophy. Plant Science 181, 96–104. [DOI] [PubMed] [Google Scholar]
- Cole M, Chandler J, Weijers D, Jacobs B, Comelli P, Werr W. 2009. DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136, 1643–1651. [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. 1999. Cell cycling and cell enlargement in developing leaves of Arabidopsis . Developmental Biology 215, 407–419. [DOI] [PubMed] [Google Scholar]
- Donner TJ, Sherr I, Scarpella E. 2009. Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 136, 3235–3246. [DOI] [PubMed] [Google Scholar]
- Gardiner J, Sherr I, Scarpella E. 2010. Expression of DOF genes identifies early stages of vascular development in Arabidopsis leaves. International Journal of Developmental Biology 54, 1389–1396. [DOI] [PubMed] [Google Scholar]
- Gualberti G, Papi M, Bellucci L, Ricci I, Bouchez D, Camilleri C, Costantino P, Vittorioso P. 2002. Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. The Plant Cell 14, 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Qin G, Gu H, Qu LJ. 2009. Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis . The Plant Cell 21, 3518–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Baurle I, Kientz M, Jürgens G. 2002. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes and Development 16, 1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G. 1999. The auxin-insensitive bodenlos mutation affects primary root formation and apical–basal patterning in the Arabidopsis embryo. Development 126, 1387–1395. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. 1998. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO Journal 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T. 2004. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4 . Development 131, 1089–1100. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. 2005. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis . Science 309, 293–297. [DOI] [PubMed] [Google Scholar]
- Konishi M, Sugiyama M. 2003. Genetic analysis of adventitious root formation with a novel series of temperature-sensitive mutants of Arabidopsis thaliana . Development 130, 5637–5647. [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S. 2007. Sequential activation of two Dof transcription factor gene promoters during vascular development in Arabidopsis thaliana . Plant Physiology and Biochemistry 45, 623–629. [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S. 2008. Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. The Plant Journal 55, 821–831. [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S. 2010. Identification of a nitrate-responsive cis-element in the Arabidopsis NIR1 promoter defines the presence of multiple cis-regulatory elements for nitrogen response. The Plant Journal 63, 269–282. [DOI] [PubMed] [Google Scholar]
- Lau S, De Smet I, Kolb M, Meinhardt H, Jürgens G. 2011. Auxin triggers a genetic switch. Nature Cell Biology 13, 611–615. [DOI] [PubMed] [Google Scholar]
- Le Hir R, Bellini C. 2013. The plant-specific dof transcription factors family: new players involved in vascular system development and functioning in Arabidopsis . Frontiers in Plant Science 4, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen KR, de Wet JR, Walbot V. 1992. Transient expression analysis in plants using firefly luciferase reporter gene. Methods in Enzymology 216, 397–414. [DOI] [PubMed] [Google Scholar]
- Mattsson J, Ckurshumova W, Berleth T. 2003. Auxin signaling in Arabidopsis leaf vascular development. Plant Physiology 131, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. 2008. Auxin receptors and plant development: a new signaling paradigm. Annual Review of Cell and Developmental Biology 24, 55–80. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, et al. 2005. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. The Plant Cell 17, 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przemeck GK, Mattsson J, Hardtke CS, Sung ZR, Berleth T. 1996. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200, 229–237. [DOI] [PubMed] [Google Scholar]
- Rademacher EH, Moller B, Lokerse AS, Llavata-Peris CI, van den Berg W, Weijers D. 2011. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. The Plant Journal 68, 597–606. [DOI] [PubMed] [Google Scholar]
- Scacchi E, Salinas P, Gujas B, Santuari L, Krogan N, Ragni L, Berleth T, Hardtke CS. 2010. Spatio-temporal sequence of cross-regulatory events in root meristem growth. Proceedings of the National Academy of Sciences, USA 107, 22734–22739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E, Marcos D, Friml J, Berleth T. 2006. Control of leaf vascular patterning by polar auxin transport. Genes and Development 20, 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Moller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D. 2010. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464, 913–916. [DOI] [PubMed] [Google Scholar]
- Schuetz M, Berleth T, Mattsson J. 2008. Multiple MONOPTEROS-dependent pathways are involved in leaf initiation. Plant Physiology 148, 870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Reichelt M, Burow M, et al. 2006. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. The Plant Journal 47, 10–24. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1997a. ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1999a. Activation and repression of transcription by auxin-response factors. Proceedings of the National Academy of Sciences, USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1999b. Dimerization and DNA binding of auxin response factors. The Plant Journal 19, 309–319. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997b. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel CL, Schuetz M, Yu Q, Mattsson J. 2007. Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana . The Plant Journal 49, 387–398. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Wu MF, Winter CM, Berns MC, Nole-Wilson S, Yamaguchi A, Coupland G, Krizek BA, Wagner D. 2013. A molecular framework for auxin-mediated initiation of flower primordia. Developmental Cell 24, 271–282. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. 2002. The Dof family of plant transcription factors. Trends in Plant Science 7, 555–560. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. 2004. Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant and Cell Physiology 45, 386–391. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J. 2003. Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425, 521–525. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU. 2010. Hormonal control of the shoot stem-cell niche. Nature 465, 1089–1092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.