Abstract
Although rising ocean temperatures threaten scleractinian corals and the reefs they construct, certain reef corals can acclimate to elevated temperatures to which they are rarely exposed in situ. Specimens of the model Indo-Pacific reef coral Pocillopora damicornis collected from upwelling reefs of Southern Taiwan were previously found to have survived a 36-week exposure to 30°C, a temperature they encounter infrequently and one that can elicit the breakdown of the coral–dinoflagellate (genus Symbiodinium) endosymbiosis in many corals of the Pacific Ocean. To gain insight into the subcellular pathways utilized by both the coral hosts and their mutualistic Symbiodinium populations to acclimate to this temperature, mRNAs from both control (27°C) and high (30°C)-temperature samples were sequenced on an Illumina platform and assembled into a 236 435-contig transcriptome. These P. damicornis specimens were found to be ∼60% anthozoan and 40% microbe (Symbiodinium, other eukaryotic microbes, and bacteria), from an mRNA-perspective. Furthermore, a significantly higher proportion of genes from the Symbiodinium compartment were differentially expressed after two weeks of exposure. Specifically, at elevated temperatures, Symbiodinium populations residing within the coral gastrodermal tissues were more likely to up-regulate the expression of genes encoding proteins involved in metabolism than their coral hosts. Collectively, these transcriptome-scale data suggest that the two members of this endosymbiosis have distinct strategies for acclimating to elevated temperatures that are expected to characterize many of Earth's coral reefs in the coming decades.
Keywords: acclimation, coral reef, dinoflagellate, endosymbiosis, gene expression, Symbiodinium, transcriptome
Introduction
Coral reefs have been hypothesized to undergo extensive degradation in the coming decades due to both local (Fabricius 2005) and global-scale (Hoegh-Guldberg et al. 2007) anthropogenic impacts. Specifically, increasing seawater temperature and acidity associated with global climate change (GCC) could potentially lead to more frequent episodes of coral bleaching (Veron 2011), whereby the endosymbiotic association between the anthozoan host and its gastrodermal endosymbionts of the genus Symbiodinium breaks down (Gates 1990). However, most GCC models do not account for acclimation, which has recently shown to be notable in reef-building corals exposed to increases in temperature (e.g., Mayfield et al. 2011, 2012a, 2013a; Barshis et al. 2013; Palumbi et al. 2014). For instance, populations of the model reef-building coral Pocillopora damicornis from upwelling reefs in Southern Taiwan were found to acclimate for nine months to a temperature (30°C) they encounter in situ for only several hours on an annual timescale (Mayfield et al. 2013b), although the physiological mechanisms by which such acclimation occurred are still poorly understood.
In recent years, molecular biology-driven techniques have advanced the collective knowledge of both the cell biology of coral–dinoflagellate endosymbioses (e.g., Mayfield et al. 2009, 2010, 2012b, 2014b; Peng et al. 2011; Chen et al. 2012; Wang et al. 2013) and their responses to environmental changes (e.g., Mayfield et al. 2013c, d; Putnam et al. 2013). Microarrays (e.g., DeSalvo et al. 2008) and next-generation sequencing (NGS; e.g., Vidal-Dupiol et al. 2013) are particularly promising approaches for addressing an array of functional questions in this globally important mutualism; however, only one work to date (Shinzato et al. 2014) has considered both endosymbiotic compartments in its transcriptomic characterization of a reef coral, and this study was not set within an experimental framework. Indeed, it has been posed that both members of the coral–Symbiodinium endosymbiosis should be acknowledged when attempting to gauge the response of this mutualism to environmental shifts (Fitt et al. 2009). Inhibition of the photosynthetic pathways of Symbiodinium in response to high light and temperature is known to elicit reactive oxygen species (ROS) production, a phenomenon that can ultimately lead to bleaching (Lesser 1997). These findings suggest to many that the Symbiodinium populations represent the more sensitive compartment to environmental change, although the host coral surely plays an important role in the stress (or acclimation) response of the holobiont, as well (Mayfield & Gates 2007).
NGS-based mRNA sequencing (i.e., ‘RNA-Seq’) could not only aid in uncovering the genetic basis of acclimation to 30°C in P. damicornis, but it could also directly test the hypothesis that one compartment of the endosymbiosis is more responsive to temperature change at the subcellular level (sensu Leggat et al. 2011). In fact, a series of recent studies have reported that Symbiodinium does not show a marked response at the mRNA level to elevated temperatures (e.g., Boldt et al. 2010; Rosic et al. 2011; Mayfield et al. 2014a), although transcriptome-scale approaches may better clarify whether or not this is a ubiquitous property of in hospite Symbiodinium populations. Herein, mRNAs from P. damicornis samples that had been exposed to a control (27°C) or high (30°C) temperature for 2 or 36 weeks (n = 3 replicates for each treatment–time) were sequenced with an Illumina Tru-Seq™ kit (ver. 2) on an Illumina Genome Analyzer IIx (GAIIx), and the resulting sequences were assembled into a meta-transcriptome against which mRNA expression of the 12 individual samples was assessed. It was hypothesized that the transcriptome-scale data set generated herein could aid in the development of a subcellular mechanism by which P. damicornis specimens from upwelling reefs of Southern Taiwan were able to acclimate to high temperature over a multi-season timescale.
Methods
Elevated temperature experiment, cDNA library preparation, and cDNA sequencing
The experiment was first described in Mayfield et al. (2013b). Briefly, P. damicornis nubbins originally collected from Houbihu, Nanwan Bay, Taiwan (21°56′18.01″N, 120°44′45.54″E) were exposed to either a control (27°C, n = 3 coral reef mesocosms) or elevated (30°C, n = 3 mesocosms) temperature for 0, 2, 4, 8, 24, or 36 weeks (n = 3 biological replicates for each treatment–sampling time) under natural, partially shaded light (average photosynthetically active radiation [PAR] at the time of sampling = ∼100 μmol/m2/s; maximum PAR = ∼800–900 μmol/m2/s; see Mayfield et al. 2013b for details.). The temperature was not reduced at night as in previous studies conducted with conspecifics from shallow (0.5–1.5 m) tide pools of the same site (e.g., Mayfield et al. 2013a); briefly, the coral specimens utilized herein were from approximately 3 m depth, one at which seawater temperature does not differ significantly between day and night except during upwelling events (see Mayfield et al. 2012a for a treatise on the temperature environment of Houbihu.). It was hypothesized that corals would bleach after ∼2 weeks of treatment exposure given that they only experience temperatures above 30 °C for several hours annually. However, corals exposed to 30 °C continued to grow at similar rates as experimental controls, as well as demonstrated similar phenotypes with regard to all response variables documented (Mayfield et al. 2013b). Instead, rather than a ‘stress’ experiment, the study became one seeking to document the mRNA-level basis of the acclimation response.
Only mRNAs from the samples sacrificed after 2 and 36 weeks of exposure were sequenced (1 nubbin/mesocosm/sampling time × 3 mesocosms/treatment × 2 treatments × 2 sampling times = 12 samples), representing boreal winter and summer sampling times, respectively. These samples represented a mix of six genotypes, as the original 72 nubbins used in the experiments (see Mayfield et al. 2013b for details.) originated from six colonies (12 nubbins/colony) that were mixed haphazardly in a seawater table prior to random assignment to a specific mesocosm/experimental treatment. It was hypothesized that mechanisms of coral acclimation might differ across seasons; corals were anticipated to face more severe stress in the summer months, at which point higher light levels are typically documented in subtropical seas. Further purification of the 12 RNAs from Mayfield et al. (2013b), preparation of the 12 barcoded Tru-Seq cDNA libraries, and sequencing (paired end, 100 bp) on one flow cell of the Illumina GAIIx were described by Mayfield et al. (2013d).
Read quality control and assembly
SolexaQA (Cox et al. 2010) was used to check the global quality of the ∼100-bp transcript reads and trim each to its longest contiguous read segment. Only sequences with a Phred score >20 (‘h-20’ setting in SolexaQA; representing a call accuracy of 99%) were assembled. Short reads (<25 bp) were discarded, and the remaining 42 117 250 reads were assembled with two additional, Taiwan-based RNA-seq data sets derived from adult (12 samples sequenced across one flow cell of a GAIIx [100 bp, paired end]) and larval (6 samples from Putnam et al. [2013] sequenced across one flow cell of a GAIIx [100 bp, paired end]) P. damicornis specimens. The 123 806 185 read pairs from the three experiments were assembled using Trinity (Grabherr et al. 2011; paired end mode) on a supercomputer at the Institute of Information Sciences (Academia Sinica, Taiwan) using the following script:../../trinityrnaseq_r2013-02-25/Trinity.pl –seqType fa –JM 200G –left 3PD_trim_q20_r1_re.fa –right 3PD_trim_q20_r2_re.fa –output 3PD_trinity_paired –CPU 40 –bflyCalculateCPU –bflyHeapSpaceMax 300G &; this yielded 315 133 contigs and 19 855 110 unassembled reads. Roche 454-dervied reads from ‘PocilloporaBase’ (Traylor-Knowles et al. 2011) were used to create a hybrid assembly with the unassembled reads with MIRA (Chevreux et al. 2004). Specifically, MIRA's default arguments were used under ‘454_SETTINGS’ and ‘Solexa_SETTINGS.’ Afterwards, 87% of the unassembled reads were ‘recovered’ and grouped into 39 019 MIRA contigs. Cap3 (Huang & Madan 1999; default settings) was then used for assembly of the Trinity- and MIRA-derived contigs, as well as 454 singlets from MIRA, and this resulted in 350 045 sequences (fasta output). Short contigs (<200 bp) were discarded, and the remaining 334 284 contigs were clustered using cd-hit-est (Huang et al. 2010; ‘-c 0.95 -M 0 -T 40 -r 0;’ i.e., a 95% similarity threshold) into 279 533 unique transcripts.
Protein identification and functional annotation
‘Transdecoder’ (from Trinity) was used to identify and translate open reading frames (ORFs), and protein sequences <30 amino acids (AA) were discarded. Only the longest continuous AA sequence generated from a contig was used in the BLASTp analysis (e-value cut-off of 10−10) of NCBI's nr database (Pruitt et al. 2007). KEGG was used to gain more insight into the pathways and signal transduction interactions represented in the assembly, and 52 768 of the conceptually translated proteins could be functionally annotated further via the KEGG database. The HMM (Söding 2005) algorithm was used to search for Pfam domains in the AA sequences, and HMMER (Zhang & Wood 2003) was used to acquire functional modules of AA sequence domains (e-value < 10−2). Gene ontology (GO; Ashburner et al. 2000) annotation from the Pfam-GO mapping table was used to assign GO tags to the conceptually translated proteins.
Taxonomy distribution
The taxonomy classification structure of the NCBI database was used in conjunction with blast to determine the likely taxonomic origin of each contig. Contigs demonstrating significant (e-value < 10−5) alignments to published metazoan (NCBI Taxonomy ID: 33208) sequences, particularly those from the model anemone Nematostella vectensis (NCBI Taxonomy ID: 45351) or the reef coral Acropora digitifera (NCBI Taxonomy ID: 70779), were assumed to be of host coral origin. Those aligning significant to conceptually translated alveolate (NCBI Taxonomy ID: 33630), stramenopile (NCBI Taxonomy ID: 33634), or green plant (NCBI Taxonomy ID: 33090) proteins were assumed to be of Symbiodinium origin. A repeated-measures anova was used to determine whether the host:Symbiodinium contig ratio was similar across the two treatments and two sampling times. jmp® (SAS Institute, ver. 11) was used to conduct this and all other statistical tests described below.
Read quantification and statistical analyses
RSEM (Li & Dewey 2011; default settings) was used to map 42 117 250 high-quality reads onto 279 533 unique transcripts. This resulted in 36 779 588 paired alignments that were converted into 236 435 unique transcripts. Statistical analyses were conducted on the fragments per kilobases mapped (fpkm) data obtained from mapping these paired alignments onto the reference transcriptome. Repeated-measures anovas were performed to determine the effects of temperature and time, as well as the temperature × time interaction, on expression of these 236 435 contigs. Non-adjusted P-values for all comparisons have been presented on the website, described below. Tukey's honestly significant difference (HSD) tests (P < 0.05) were used to detect individual mean differences when a treatment effect was documented in the model.
GOs were assigned to ∼89 000 contigs from the overall assembly, as well to both host coral and Symbiodinium contigs found to be expressed at significantly different (P < 0.01) levels between treatments. Two-sample proportion tests were used to determine whether the proportions of the major functional categories differed significantly (P < 0.01) between the respective reference assembly of each compartment and the DEG pool for that compartment. Such proportion tests were also conducted to determine whether the relative proportions of the different functional groups differed significantly between the DEG pools of these two major eukaryotic compartments of this coral holobiont: host coral and Symbiodinium.
To combat the issue of generating false-positive results from having conducted nearly two million statistical tests (∼236 000 contigs × 9 treatment-time comparisons), a Bonferroni-adjusted α level was considered, although this resulted in an α level of 10−7, at which only one coral and six Symbiodinium contigs were differentially expressed between treatments (Table S1, Supporting information). Instead, comparisons were conducted at α level cut-offs from 1 to typically 10−5–10−10 for each of four comparisons: control (C) vs. high (H) temperature (data pooled across sampling times), C vs. H comparisons at the 2-week sampling time (‘C2 vs. H2’), C vs. H comparisons at the 36-week sampling time (‘C36 vs. H36’), and 2 vs. 36 weeks (data pooled across treatments) at each of four different comparison models: 10-fold difference, 2-fold difference, H = 0/C > 0, and H > 0/C = 0 (Table S1, Supporting information). The latter two comparisons were conducted because of the inability to calculate a fold change when a gene was not expressed by samples of one treatment group. The comparative framework developed in Table S1 (Supporting information), Fig.4, and the website (http://ips.iis.sinica.edu.tw/coral/) aimed to accommodate the needs of a greater cadre of researchers by considering a range of statistical stringency for each biological comparison of interest.
Fig 4.
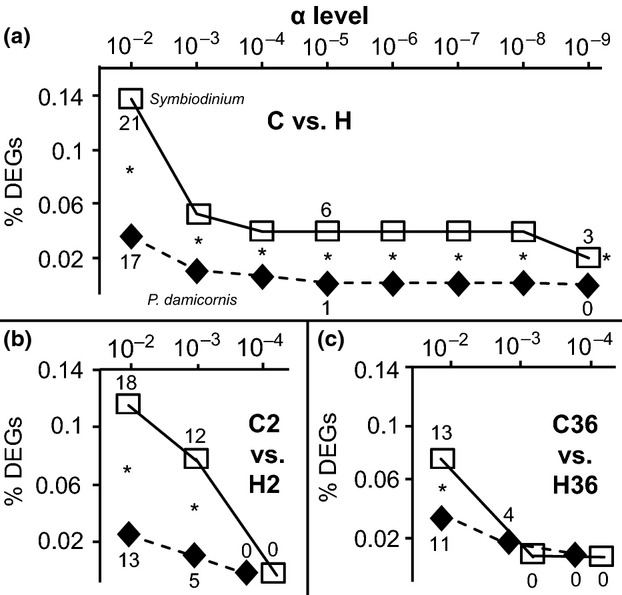
The percentage of genes differentially expressed at various α levels for each compartment of the Pocillopora damicornis–Symbiodinium endosymbiosis. When looking at the expression of genes that were transcribed only by the high-temperature samples and expressed at significantly different levels between temperatures (repeated-measures anova, P < 0.01), the percentage of differentially expressed genes (‘% DEGs’) has been calculated for the Symbiodinium (hollow squares; #Symbiodinium DEGs/total Symbiodinium contig count × 100) and host coral (black diamonds; host coral DEGs/total host coral contig count × 100) compartments at a variety of α levels. Two-sample proportion tests were used to determine whether there were differences between the respective % DEGs of each compartment at each α level across the two temperature treatments (a) (data pooled across sampling times), as well as between treatments in the 2-week sampling period only (b), and between treatments in the 36-week sampling time only (c). The actual numbers of DEGs have been placed next to to certain icons, and asterisks (‘*’) denote significant differences (two-sample proportion test, P < 0.01) in % DEGs between compartments within each α level of each panel.
Genes that were differentially expressed at an α level of 0.01 were used to generate Figs.2 and3a, as well as target DEGs for real-time PCR analysis, described below. To uncover differences in metabolism-targeted gene expression across treatments and compartments (Fig.3b,c), only genes that were differentially expressed at an α level of 0.01 and whose expression levels differed by 2-fold or more between treatments (at either or both sampling times) were included in the analysis. When comparing the total number of DEGs for each compartment (Fig.3a) with a student's t-test, a correction was made to reflect the fact that 3.2-fold more contigs were of host coral origin; the Symbiodinium DEG contig counts were multiplied by 3.2 prior to the statistical tests but are presented as their uncorrected values in the figure itself. Such a correction was unnecessary when calculating the percentage of genes that were differentially expressed (i.e., ‘% DEGs;’ #DEGs/total contig count) in each compartment (sensu Figs.3b and4).
Fig 2.
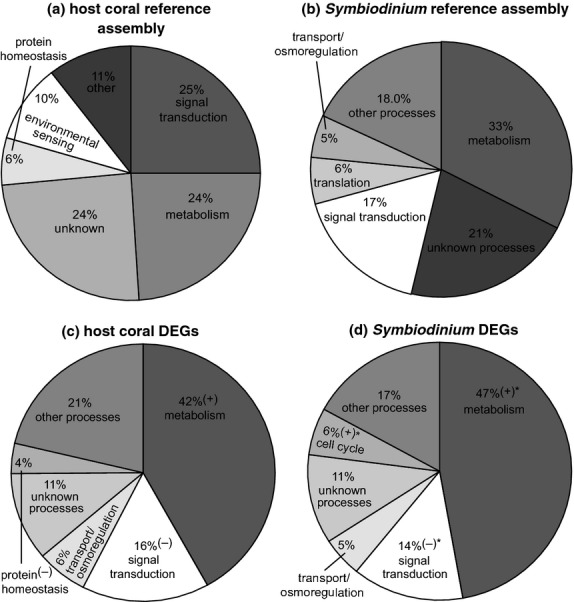
Functional distribution of differentially expressed genes (DEGs). A gene ontology (GO) was assigned to a high percentage of the 48 288-contig host coral (a) and 15 374-contig Symbiodinium (b) reference assemblies. A subset of 970 host coral (c) and 879 Symbiodinium (d) DEGs (repeated-measures anova, P < 0.01) was then analysed separately and assigned GO functional category tags. In (c-d), GO categories that were down- or up-regulated relative to the respective reference assembly for each compartment (two-sample proportion test, P < 0.01) are marked with a ‘(−)’ or a ‘(+),’ respectively. When neither icon has been placed next to a GO category in (c–d), the respective GO category was represented at a similar proportion as in the respective reference assembly for that compartment (two-sample proportion test, P > 0.01). GO category percentages that differed significantly between host coral and Symbiodinium DEG pools are denoted by asterisks (‘*’) in (d). In all panels, ‘unknown processes’ refer to contigs for which GO tags could be assigned, although the GOs did not correspond to a particular cellular process (e.g., ‘diabetes’).
Fig 3.
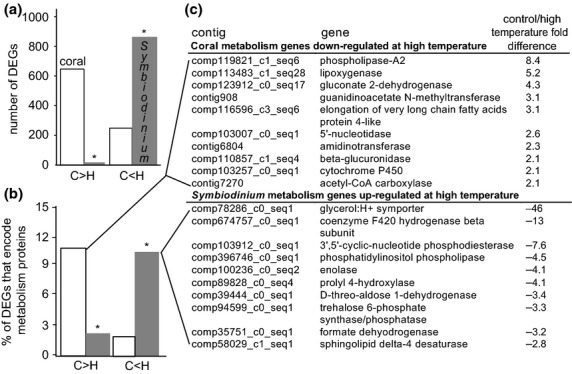
Differentially expressed genes (DEGs) across temperature treatments for each compartment of the Pocillopora damicornis–Symbiodinium endosymbiosis, with an emphasis on metabolism-targeted genes. Differentially expressed genes (DEGs P < 0.01) were sorted into those that were expressed at higher levels in controls (‘C>H’) and those expressed at higher levels in the high-temperature samples (‘C<H’) for both coral (white column) and Symbiodinium (grey column) compartmentss (a). Then, the fraction of the DEG pool from each compartment that was comprised of metabolism genes was calculated (#metabolism DEGs/total number of DEGs × 100) for each compartment for genes differing by twofold or greater between treatments (at either or both sampling times) (b). A random selection of the metabolism-targeted genes that were down-regulated at high temperature at twofold or greater levels in the host coral and up-regulated at twofold or greater levels at high temperature in the Symbiodinium compartment can be found in the embedded table (c). When differences between compartments within each comparison group were determined to be statistically significant by student's t-tests and two-sample proportion tests (P < 0.01) for (a) and (b), respectively, an asterisk (‘*’) has been placed above one of the two columns.
Real-time PCR verification of select genes
Real-time PCR assays were designed for either genes hypothesized a priori to be thermosensitive or those unveiled in a non-Bonferroni-adjusted assessment of significant differences identified from analysis of the fpkm data (see Table1 for distinctions.). The latter group included genes that were expressed at significantly different levels between treatments in the repeated-measures anova model, as well as those found to be expressed at significantly different levels at either one or both sampling times by Tukey's HSD tests (P < 0.05). There was a prerequisite that all three biological replicates of the treatment exhibiting higher expression levels expressed the gene. Genes whose expression was hypothesized a priori to be responsive to temperature (Table1) predominantly featured those encoding proteins involved in the restoration of homeostasis in temperature-stressed cells (Hochachka & Somero 2002), and these included genes encoding heat-shock proteins and superoxide dismutases. a priori-selected target genes also included those involved in photosynthesis (e.g., photosystem I) and metabolite flux (e.g., carbonic anhydrase) within the coral holobiont, as these two processes have been suggested to be thermosensitive in reef corals (Jones et al. 2000 and Mayfield & Gates 2007, respectively).
Table 1.
Real-time PCR assays. Twenty-nine real-time PCR assays developed for the model reef coral Pocillopora damicornis and its endosymbiotic Symbiodinium populations were conducted with the triplicate biological replicates sampled at each of two temperature treatments [control (27 °C) and high (30 °C)] at each of two sampling times (2 and 36 weeks; n = 12 samples in total). When a significant correlation was documented between Illumina- and real-time PCR-derived data (P < 0.05), a ‘Y’ has been placed in ‘Correlation between techniques?’ column. When Illumina-derived expression of a gene was found to be up-regulated at high temperature, the sampling time(s) at which such a change was documented has/have been included in the right-most column
| Biological processGene (abbreviation) | Contig name/# | Compartment | Reference | Results-Table | Results – Fig. | Correlation between techniques? | Up-regulated at high temperature? |
|---|---|---|---|---|---|---|---|
| Metabolism | |||||||
| carbonic anhydrase (ca)† | comp301310_c0_seq1 | Host coral | Mayfield et al. (2013d) | S2 | S1a–c | Y | 36 weeks only |
| amylase | comp378549_c0_seq1 | Host coral | Mayfield et al. (2014c) | S2 | S1d–f | Y | 2 weeks only |
| transketolase | comp47495_c0_seq1 | Symbiodinium | Mayfield et al. (2013d) | S3 | S3a–c | Y | |
| zinc-induced facilitator-like 1-like (zifl1-l) | comp47495_c0_seq1 | Symbiodinium | Mayfield et al. (2013d) | S3 | S3d–f | Y | |
| nitrate transporter 2 (nrt2)† | Contig6284 | Symbiodinium | Mayfield et al. (2013a) | S3 | S3g–i | Y | |
| Molecular transport/redox balance/osmoregulation | |||||||
| ion transporter (iontrans) | comp1089337_c0_seq1 | Host coral | Mayfield et al. (2013d) | S2 | S1g–i | 36 weeks only | |
| sulfotransferase (sftase) | comp1050556_c0_seq1 | Host coral | Mayfield et al. (2013d) | S2 | S1j–l | ||
| mitochondrial carrier protein (mcp) | comp298112_c0_seq1 | Host coral | Mayfield et al. (2014c) | S2 | S1m–o | Y | 2 weeks only |
| voltage-dependent ion channel (vdic) | comp1050556_c0_seq1 | Symbiodinium | Mayfield et al. (2013d) | S3 | S3j–l | Y | 2 weeks only |
| potassium channel (K+ channel) | comp517306_c0_seq1 | Symbiodinium | Mayfield et al. (2013d) | S3 | S3m–o | Y | 2 weeks only |
| calcium channel (Ca2+ channel) | comp1535924_c0_seq1 | Symbiodinium | Mayfield et al. (2013d) | S3 | S3p–r | Y | |
| Cell adhesion (host coral genes only) | |||||||
| lectin | comp19963_c0_seq1 | Host coral | Mayfield et al. (2013d) | S2 | S1p–r | Y | 36 weeks only |
| selectin | comp1535924_c0_seq1 | Host coral | Mayfield et al. (2013d) | S2 | S1s–u | Y | 2 weeks only |
| Photosynthesis (Symbiodinium genes only) | |||||||
| RuBisCO (rbcL)† | Contig12235 | Symbiodinium | Mayfield et al. (2012a) | S3 | S3s–u | Y | |
| photosystem I subunit III (psI)† | comp114066_c1_seq2 | Symbiodinium | Mayfield et al. (2012a) | S3 | S3v–x | Y | |
| phosphoglycolate phosphatase (pgpase)† | comp101919_c0_seq2 | Symbiodinium | Crawley et al. (2010) | S3 | S3y–aa | Y | |
| Stress response | |||||||
| heat shock protein 70 (hsp70)† | contig2062 | Host coral | Putnam et al. (2013) | S2 | S2a–c | ||
| cu-Zn superoxide dismutase (cu-zn-sod)† | comp2083_c0_seq1 | Host coral | Mayfield et al. (2014c) | S2 | S2d–f | ||
| heat shock protein 40 (hsp40)† | comp17042_c0_seq1 | Symbiodinium | Mayfield et al. (2014c) | S3 | S4a–c | Y | |
| heat shock protein 70 (hsp70)† | Contig 7114 | Symbiodinium | Mayfield et al. (2009) | S3 | S4d–f | Y | |
| heat shock protein 90 (hsp90)† | comp2083_c0_seq1 | Symbiodinium | Mayfield et al. (2014c) | S3 | S4g–i | Y | Both times |
| ascorbate peroxidase (apx1)† | comp90407_c0_seq1 | Symbiodinium | Mayfield et al. (2012a) | S3 | S4j–l | Y | |
| ubiquitin ligase (ubiqlig)† | comp784314_c0_seq1 | Symbiodinium | Mayfield et al. (2014c) | S3 | S4m–o | Y | 2 weeks only‡ |
| Glycoprotein | |||||||
| von Willebrand factor α (vwfα) | comp880232_c0_seq1 | Host coral | Mayfield et al. (2014c) | S2 | S2g–i | 36 weeks only | |
| DNA replication | |||||||
| DNA polymerase (DNAp) | comp144001_c0_seq1 | Host coral | Mayfield et al. (2014c) | S2 | S2j–l | ||
| chromosome segregation protein (smc-csp) | comp880232_c0_seq1 | Host coral | Mayfield et al. (2013d) | S2 | S2m–o | ||
| Other processes | |||||||
| green fluorescent protein-like chromoprotein (gfp-like cp) | comp818_c0_seq1 | Host coral | Mayfield et al. (2013d) | S2 | S2p–r | Y | 36 weeks only‡ |
| ciliary dynein (cildyn) | comp144001_c0_seq1 | Symbiodinium | Mayfield et al. (2014c) | S3 | S4p–r | Y | 2 weeks only |
| RNA helicase | comp324200_c0_seq1 | Symbiodinium | Mayfield et al. (2013d) | S3 | S4s–u | Y | 2 weeks only |
a-priori-selected (prior to assembly) target gene.
up-regulation corroborated by real-time PCR.
RNAs from the 12 samples from which Illumina libraries were created were diluted to 20 ng/μL, and 10 μL (200 ng) were reverse transcribed to cDNA alongside an exogenous Solaris™ RNA spike (Thermo-Scientific) with the High Capacity™ cDNA synthesis kit (Life Technologies) according to the manufacturers’ instructions. Three-fold-diluted cDNAs (2 μL/reaction) were amplified in real time with 10 μL (1×) EZ-TIME™ SYBR® Green mastermix with ROX® passive reference dye (Yeastern, Biotech., LTD.) and the primer concentrations mentioned in the respective references of Table1 on either an Applied Biosystems 7500 real-time PCR machine or a StepOnePlus™ real-time PCR system (Life Technologies) at the thermocycling conditions described in the respective references of Table1. Recovery of the Solaris spike was quantified with the proprietary Taqman™ FAM probe-based assay, instead of the SYBR Green-based chemistry used for the 29 target genes. Expression of the target genes was normalized to recovery of the RNA spike as described in Putnam et al. (2013).
A biological composition control, which is the normalization method of choice for coral samples demonstrating spatio-temporal (e.g., Mayfield et al. 2010) or treatment-induced changes in the biological material ratio of the holobiont (e.g., loss of Symbiodinium due to bleaching), was not utilized, as an assessment of a breakdown of the contigs in each sample (Fig.1e) found that a similar coral:Symbiodinium contig ratio was characteristic of all four treatment–time groups (repeated-measures anova, P > 0.05). Furthermore, the Symbiodinium densities were previously found to be similar across samples (Mayfield et al. 2013b). However, the biological composition of the coral holobiont with respect to the ratio of the host coral:Symbiodinium must be considered and used as a normalizing factor in studies in which this parameter changes, such as those eliciting the bleaching response, and a variety of previous works have developed protocols for the quantification of the Symbiodinium contribution to total holobiont nucleic acid extracts (e.g., Mieog et al. 2009).
Fig 1.
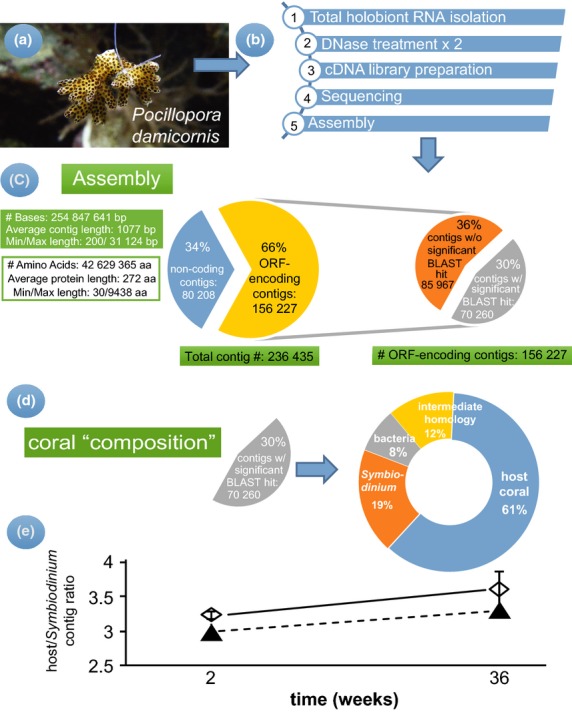
Schematic of the protocol and overall assessment of the Pocillopora damicornis transcriptome. mRNAs from the model reef coral P. damicornis (a) were converted to barcoded cDNA libraries, and assembled (b). A breakdown of the transcriptome assembly (c) and the taxonomy distribution of the assembled contigs (d) have been presented. The host coral/Symbiodinium contig ratio (e) was calculated in both control (hollow diamonds) and high-temperature (black triangles) samples collected after 2 or 36 weeks, and error bars represent standard deviation of the mean.
Database construction and content
A website was created to serve as a perpetual repository of the 236 435 (84.6%) expressed contigs, as well as the 43 098 (15.4%) non-expressed contigs that were generated upon the meta-assembly of the multiple P. damicornis transcriptomes mentioned above. Coral-trans-dbase (http://ips.iis.sinica.edu.tw/coral/), which was constructed in SQLite (ver. 3.4.2), was generated with the Python programming language (ver. 2.5.2), and the flask framework and sqlalchemy ORM are used to access the database. The database is hosted on a computer running Ubuntu Linux 8.04.4 LTS using the Apache http server (ver. 2.2.8) and contains all contig nucleic acid sequences, as well as the AA sequences of the conceptually translated ORFs. The website also includes extensive annotation (e.g., top blast hits, Pfam, KEGG, GO)-related information, as well as plots of the gene expression results (including error bars) across the two treatments and two sampling times. The raw expression data can be downloaded, as well.
Sequence data can be accessed by a variety of methods, including via contig name, keyword, functional category, GO tag, statistical parameters (e.g., α level or fold change) or input sequences (blast). Within each search category, contigs can be sorted by compartment (coral, Symbiodinium, bacteria, or intermediate homology), as well as by GO tag, enabling users to determine which compartment and functional pathway, respectively, contributed more significantly to the contig list presented by the search engine (sensu Figs.4). On the blast page, an alignment viewer based on Mviewer (Brown et al. 1998) has been implemented. A detailed description of the components of the website can be found on the homepage.
Results
Assembly and taxonomy distribution
The paired end, 100-bp reads from the 12 coral nubbins (Fig.1a) generated ∼255 Mbp of sequencing data (Fig.1b) that were assembled into 236 435 expressed contigs (N50 length = 1936 bp). Approximately 66% (156 227) of these contigs possessed an ORF, and 70 260 (30%) of these ORF-encoding contigs were conceptually translated into a protein product that aligned significantly (e < 10−10) to a protein in the NCBI nr database (Fig.1c). Of these 70 260 protein-coding contigs, 61% (48 228) were found to be of metazoan (e < 10−5), and thus presumably coral, origin (Fig.1d). A partial portion of the mitochondrial genome aligned most closely (e = 0) to published mitogenomes of the α genotype of P. damicornis (Schmidt-Roach et al. 2014).
Only 19% of the reference assembly aligned significantly (e < 10−5) to conceptually translated proteins of alveolates, other algae, or green plants; these 15 374 contigs were assumed to be of Symbiodinium (clade C; Mayfield et al. 2013b) origin. Approximately 8% (5600) of the contigs aligned significantly (e < 10−5) to bacterial genes. Given that a poly-A selection step was conducted, it could not be determined whether these sequences were actually from coral-associated bacteria or whether they represented contamination; therefore, these data are not discussed further. Finally, ∼12% of the protein-coding contigs were found to align significantly (e < 10−5) to conceptually translated proteins from a mix of both metazoans and alveolates or other eukaryotic microbes; hence, their compartment of origin could not be confidently ascribed, and these contigs have been labelled as ‘intermediate homology’ in Fig.1 and on the website.
Reference assemblies vs. DEG pools
When looking at the host coral (Fig.2a) and Symbiodinium (Fig.2b) reference assemblies, it is clear that the two most represented GO functional categories were metabolism and signal transduction, which made up about 50% of the transcriptome of each compartment when combined. Of the 970 coral DEGs (P < 0.01; Fig.2c), the most dominant functional categories were also metabolism and signal transduction; only the former process was at a significantly higher proportion than in the host coral reference assembly. The latter process was actually significantly under-represented in the host coral DEG pool, as was protein homeostasis (i.e., protein processing, turnover, and regulation). Genes involved in metabolism and signal transduction were also over- and under-represented, respectively, in the Symbiodinium DEG pool (n = 879; Fig.2d).
Coral vs. Symbiodinium response
For genes that were expressed at significantly higher levels (P < 0.01) in the controls relative to the high-temperature samples (‘C>H’), a significantly higher quantity (Fig.3a) and proportion (data not shown) of DEGs were detected in the host compartment. However, when looking at genes that were expressed at higher levels in samples of the high-temperature treatment (‘C<H’), there were nearly four-fold more Symbiodinium DEGs (Fig.3a). These differences are paralleled by changes in the number of metabolism-targeted genes in each compartment (Fig.3b); nearly 11% of the genes that were down-regulated at two-fold or greater levels in the host coral at high temperatures encoded proteins involved in metabolism. In contrast, over 10% of the genes that were significantly up-regulated at two-fold or higher levels at high temperature in the Symbiodinium compartment encoded metabolism proteins (Fig.3b), and this is a significantly higher percentage than that of the host coral metabolism-targeted genes for this comparison (∼2%). A selection of the metabolism genes that were down-regulated at high temperature in the coral hosts and up-regulated at high temperature in their resident Symbiodinium populations is shown in Fig.3c.
When only fold changes between treatments were considered, and the α levels were set to >0.1 (Table S1, Supporting information), a significantly higher percentage of the host coral transcriptome was found to be differentially expressed (host coral % DEGs [#coral DEGs/total coral contig count × 100] > Symbiodinium % DEGs [#Symbiodinium DEGs/total Symbiodinium contig count × 100]). As the α levels became more stringent, a relatively higher percentage of Symbiodinium genes was found to be differentially expressed (Table S1, Supporting information; also see Fig.4a for genes only expressed by high-temperature samples.). For instance, for the C vs. H comparisons, approximately three-fold more Symbiodinium genes (5.8% of the transcriptome) were differentially expressed between temperature treatments at an α level of 0.01 relative to host coral genes (2.0% of the transcriptome; two-sample proportion test, P < 0.0001) when fold changes were not considered. Furthermore, a significantly higher proportion of the Symbiodinium transcriptome was differentially expressed relative to their host corals when the α level was set to 10−6 or lower for a variety of the comparison modules (Table S1, Supporting information); in fact, this proportion was significantly higher than that of the host corals at α levels of 10−2 and lower for genes only expressed by the high-temperature samples (Fig.4a).
A similar, albeit less dramatic, trend was revealed when looking at the C2 vs. H2 (Fig.4b) and C36 vs. H36 (Fig.4c) comparisons for genes only expressed by high-temperature samples. In the former, there were significantly higher proportions (and absolute values) of Symbiodinium DEGs over host coral DEGs at both 10−2 and 10−3 α levels. There was also a statistically higher proportion of Symbiodinium DEGs over host coral DEGs at the 10−2 α level for the 36-week samples. However, at an α level of 10−3, a similar proportion of host and Symbiodinium DEGs was documented at the 36-week sampling time.
Real-time PCR validation
Genes that were either differentially expressed or hypothesized a priori to be temperature sensitive, such as those encoding proteins involved in metabolism (Figs. S1a–f and S3a–i [Supporting information] for the coral and Symbiodinium, respectively), osmoregulation/transport (Figs S1g–o and S3j–r [Supporting information] for the coral and Symbiodinium, respectively), cell adhesion (Fig. S1p–u, Supporting information; host coral only), photosynthesis (Fig. S3s–aa, Supporting information; Symbiodinium only) and the stress response (Figs S2a–f and S4a–o [Supporting information] for the coral and Symbiodinium, respectively), were targeted by real-time PCR. Genes encoding proteins involved in several additional processes were targeted in the host coral (Fig. S2g–r, Supporting information) and Symbiodinium (Fig. S4p–u, Supporting information) compartments, as well. Of the 13 host coral genes targeted (Figs S1 and S2, Supporting information), the expression of 7 (58.3%) was found to correlate significantly between techniques. In contrast, Illumina- and real-time PCR-derived mRNA expression values correlated significantly for all 16 Symbiodinium target genes, and this proportion was significantly higher than that of the host (χ2 proportion test, Z = 3.69, P < 0.05).
One and two genes in the coral and Symbiodinium compartments, respectively, both yielded congruent results between techniques and exhibited treatment differences within a sampling time by Tukey's HSD tests. These were the Symbiodinium metabolism gene zinc-induced facilitator-like-1-like (zifl1-l; Fig. S3e, Supporting information) and the proteasome gene ubiquitin ligase (ubiqlig; Fig. S4n, Supporting information), as well as host coral green fluorescent protein-like chromoprotein (gfp-like cp; Fig. S2q, Supporting information). zifl1-l was expressed at ∼10-fold higher levels in the control samples over the high-temperature samples after 2 weeks of treatment exposure, while ubiqlig was expressed at 2.5-fold higher levels in the high-temperature samples at this sampling time. The expression of both genes was similar at the 36-week sampling time. In contrast, host coral gfp-like cp was expressed at similar levels between treatments at the 2-week sampling time but was expressed at 90-fold higher levels in samples of the high-temperature treatment relative to experimental controls after 36 weeks; this represents one of the most differentially expressed genes in the entire transcriptome.
Discussion
What is Pocillopora damicornis?
This Illumina-based P. damicornis transcriptome is larger than a previous, 454-based one (Traylor-Knowles et al. 2011); an additional ∼175 000 contigs were produced herein, and only ∼3000 contigs in the Traylor-Knowles et al. (2011) transcriptome were not identified. This discrepancy is partially due to the previous assembly containing only a small proportion (<5%) of Symbiodinium genes, but could also be due to the sequencing of a large number of splice variants or even non-coding RNAs (despite the employment of a poly-A-selection step) in the present work, of which only ∼30% of the contigs could be confidently assigned a protein identity.
The low percentage of Symbiodinium genes in the previous P. damicornis transcriptome (Traylor-Knowles et al. 2011) is in stark contrast to not only the results obtained herein (∼20%), but also those of Shinzato et al. (2014; ∼35%). The differences in the host:Symbiodinium contig ratio between this study and Shinzato et al. (2014), which was conducted with the massive coral Porites australiensis, may highlight the fact that not all corals have a similar biological composition. It should be mentioned, however, that the in vivo biomass ratio may not necessarily approximate the mRNA ratio; it is likely that the former is closer to 50/50% for many anthozoan–dinoflagellate endosymbioses based on microscopic images of Symbiodinium in hospite (Chen et al. 2012; Mayfield et al. 2013b), in which the majority of the gastrodermal volume is occupied by these dinoflagellates. It is also possible that methodological differences accounted, in part, for the differences in host/Symbiodinium ratio between this study and that of Shinzato et al. (2014); both the host coral and Symbiodinium transcriptomes were assembled de novo herein, and the origin of the assembled contigs was hypothesized based on alignments to published sequences queried across the entire NCBI database. In contrast, Shinzato et al. (2014) aligned their host and Symbiodinium transcriptomes directly to published host coral (A. digitifera; Shinzato et al. 2011) and Symbiodinium (Shoguchi et al. 2013) genomes, respectively; doing so may have allowed for the assignment of a higher percentage of Symbiodinium contigs.
Symbiodinium vs. host coral mRNA-level response to high-temperature exposure
The clade C Symbiodinium populations of the samples sequenced herein demonstrated a more pronounced mRNA-level response than their coral hosts after 2 weeks of exposure to 30°C; specifically, a higher number and percentage of DEGs were detected in the Symbiodinium compartment at the 2-week, but not the 36-week, sampling time at a 10−3 α level. Whether the Symbiodinium transcriptome always responds more strongly to elevated temperature than that of their hosts remains to be determined; for instance, it could be that the host coral transcriptome changed dramatically after several hours of exposure to elevated temperature, with gene expression levels quickly returning to baseline before the first sampling time. Future work should seek to assess transcriptome-wide changes in this coral holobiont over a more fine-tuned timescale to determine whether this is indeed the case.
High-temperature effects on coral metabolism and osmoregulation
Although genes related to metabolism compromised ∼30% of the host coral and Symbiodinium reference assemblies, ∼45% of the Symbiodinium and host DEGs were involved in metabolism (Fig.2), suggesting that this process may have been altered in corals exposed to high temperatures. Whereas their coral hosts were found to down-regulate the expression of metabolism genes at high temperature, Symbiodinium were more likely to up-regulate genes involved in metabolism (although zifl1-l represents an exception). This could mean that the Symbiodinium populations incubated at high temperatures were exhibiting elevated metabolic rates, a hallmark of many organisms exposed to abnormally elevated temperatures (Hochachka & Somero 2002).
Such elevated metabolic rates in the Symbiodinium populations of the high-temperature samples could theoretically have driven the metabolism gene expression changes documented in their hosts. Symbiodinium populations within corals exposed to elevated temperatures typically become photoinhibited (Jones et al. 2000), at which point flux of the food source and compatible osmolyte glycerol may be diminished (Gates & Edmunds 1999). As most cnidarians rely on glycerol and similar small organic compounds to establish their osmotic pressure (Shick 1991), coral respiration of the remaining glycerol pools may cause a collapse of the host cytoskeleton over the Symbiodinium cell(s) in the coral gastroderm (Mayfield et al. 2010). In contrast, the increases in Symbiodinium metabolism inferred herein from the gene expression data may have driven increases in osmolyte flux into the host gastroderm, causing both the down-regulation of many metabolism-targeted genes and the gastrodermal tissue swelling revealed previously by scanning electron microscopy (Mayfield et al. 2013b).
The role of chromoproteins in coral acclimation to high temperature
This increase in gastrodermal thickness was only observed in samples analysed after 36 weeks of high-temperature exposure (Mayfield et al. 2013b), which was during the boreal summer. Given the higher light levels experienced by all corals at this time (Mayfield et al. 2013b), the thicker gastroderms of the high-temperature samples could also have been necessary to provide a greater space in which GFP-like CPs could absorb high light levels that might otherwise have caused Symbiodinium photoinhibition (Mayfield et al. 2014c). As both high light and temperature are typically needed to elicit bleaching (Hoegh-Guldberg 1999), such shading by gastrodermal GFP-like CPs (sensu Smith et al. 2013), whose respective gene mRNA was expressed at 90-fold higher levels by high-temperature corals, might have allowed for a greater degree of self-shading that ultimately allowed these corals to acclimate to high temperatures during the summer months. Future work will attempt to verify these chromoprotein expression differences at the protein level, as well as to determine their capacity to buffer the intra-gastrodermal Symbiodinium populations from excessive irradiation. Such studies may also elucidate whether photoprotection or osmoregulatory/metabolic influences were more important in driving the increases in gastrodermal thickness witnessed in these high-temperature samples (Mayfield et al. 2013b).
Ubiquitin ligase, protein turnover, and coral acclimation to elevated temperatures
In addition to the inferred increases in Symbiodinium metabolism and enhanced shading by GFP-like CP under high light levels, ubiquitin ligase proteins may also have aided in the ability of these corals to acclimate to high temperatures given the dramatic increases in expression of the respective gene, ubiqlig, in Symbiodinium populations exposed to 30°C for 2 weeks. Ubiquitin ligases are involved in tagging proteins to be degraded by the proteasome (Welchman et al. 2005), and elevated levels of ubiquitin-conjugated proteins were associated with corals inhabiting thermally extreme backreefs in Samoa (Barshis et al. 2010). High levels of ubiquitin ligase protein expression are likely to be associated with high protein turnover rates, and while Symbiodinium protein turnover was not measured herein, the elevated levels of Symbiodinium ubiglig expression at high temperature may provide evidence for this phenomenon.
Gates & Edmunds (1999) suggested that corals characterized by high levels of protein turnover should have a consequently greater capacity for acclimatization, as is the case for a plethora of other organisms (Hochachka & Somero 2002); as such, the high protein turnover rates of the Symbiodinium compartment in corals exposed to high temperatures for 2 weeks might have, for instance, aided in their ability to readily catabolize and process temperature-denatured proteins in an efficient manner and hence allowed for their ultimate acclimation. Although host coral protein turnover was not measured herein, genes involved in protein homeostasis were actually under-represented in the host coral DEG pool; this may mean that protein processing/turnover may be less important in the acclimation response of the coral host. Furthermore, P. damicornis has been hypothesized, but not directly shown, to demonstrate low rates of protein turnover given its high growth rates and low metabolic rates relative to those of massive poritids (Loya et al. 2001), which have repeatedly been found to be among the most resilient corals to environmental change (Brown 1997). Therefore, the low protein turnover rates hypothesized by a combined assessment of conjectures put forth by Gates & Edmunds (1999) and Loya et al. (2001), in conjunction with the metabolic suppression inferred from the down-regulation of a multitude of metabolism-targeted genes in high-temperature samples herein, may have allowed such high-temperature nubbins to continue to grow at comparable rates to controls (Mayfield et al. 2013b) over the duration of the experiment by, for instance, conserving metabolic energy for growth-related processes.
Conclusions
This represents the first study to simultaneously measure global expression patterns of genes derived from two compartments of an endosymbiotic organism within an experimental framework. From a comprehensive assessment of the DEGs, it is evident that the Symbiodinium compartment responds more strongly at the mRNA level to a 2-week, but not a 36-week, elevated temperature exposure, relative to its host coral, the model reef-building coral P. damicornis. Furthermore, it was found that Symbiodinium were more likely than the host corals in which they resided to up-regulate metabolism-targeted genes at high temperature; this observation highlights the fact that the two members of this endosymbiosis have different sub-cellular strategies for acclimating to high temperatures. Host coral gfp-like cp molecules may have contributed to this acclimation capacity via the light-absorbing capacity of the proteins they encode, while the high expression of ubiqlig by the Symbiodinium populations may have allowed them to rapidly metabolize proteins via the proteasome that had become denatured by high temperatures. Collectively, then, it appears that host control of the light environment, Symbiodinium protein turnover, and both host coral and Symbiodinium metabolism are modulated by elevated temperature, and future work will attempt to decipher which of these processes is most critical for coral acclimatization to temperatures they are likely to face in the coming decades as part of GCC.
Acknowledgments
Dr. Tung-Yung Fan is thanked for establishing the elevated temperature mesocosm facility. A.B.M. was funded by a postdoctoral research fellowship from the National Science Foundation (NSF) of the United States of America (OCE-0852960), as well as a postdoctoral research fellowship from the Khaled bin Sultan Living Oceans Foundation. Research grants from Taiwan's National Science Council (NSC 101-2311-B-291-002-MY3 and 102-2923-B-291-001-MY2 to C.-S.C.) and Ministry of Science and Technology (103-2311-B-001-033-MY3 to C.-Y.L. and 102-2811-B-001 -046 to S.-H.C.) also supported this work. The Illumina-based sequencing work was funded in part by the University of Hawaii (UH) NSF EPSCoR program (Investing in Multidisciplinary University Activities, EPS-0903833 to J. Gaines) in support of the Core Functional Genomics Facility at the Hawaii Institute of Marine Biology, SOEST, UH-Manoa.
A.B.M. conceived and executed the experiment and performed all laboratory work. A.B.M., Y.-B.W., S.-H.C. and C.-Y.L. analysed the data. Y.-B.W. developed the website. All authors contributed computing and laboratory resources. A.B.M. wrote the manuscript.
Data accessibility
The unassembled sequences reported herein have been deposited in the Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra/SRX378204), which is cross-referenced with the Transcriptome Shotgun Assembly database (www.ncbi.nlm.nih.gov/genbank/tsa; BioProject PRJNA227785). Also, an interactive website (http://ips.iis.sinica.edu.tw/coral) that is described in the manuscript has been designed to serve as a perpetual depository for the sequence data set. Finally, assembled contigs, as well as the associated expression data (in spreadsheet format), have been deposited at Dryad (www.datadryad.org, doi: 10.5061/dryad.rh04 m).
Supporting Information
Additional supporting information may be found in the online version of this article.
Differentially expressed gene quantities compared across the two predominant eukaryotic compartments of the Pocillopora damicornis coral holobiont.
Table S2 Repeated-measures anova results of host coral gene expression.
Table S3 Repeated-measures anova results of Symbiodinium gene expression.
Fig. S1. Host coral metabolism, transport, and cell adhesion gene expression.
Fig. S2. Expression of host coral genes involved in the stress response and other processes.
Fig. S3.Symbiodinium metabolism, ion transport, and photosynthesis gene expression.
Fig. S4. Expression of Symbiodinium genes involved in the stress response and other processes.
References
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barshis DJ, Stillman JH, Gates RD, et al. Protein expression and genetic structure of the coral Porites lobata in an environmentally extreme Samoan back reef: does host genotype limit phenotypic plasticity? Molecular Ecology. 2010;19:1705–1720. doi: 10.1111/j.1365-294X.2010.04574.x. [DOI] [PubMed] [Google Scholar]
- Barshis DJ, Ladner JT, Oliver TA, et al. Genomic basis for coral resilience to climate change. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1387–1392. doi: 10.1073/pnas.1210224110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt L, Yellowlees D, Leggat W. Measuring Symbiodinium sp. gene expression patterns with quantitative real-time PCR. Ft Lauderdale, Florida: Proceedings of the 11th ICRS; 2010. pp. 118–122. 7–11 July 2008. [Google Scholar]
- Brown BE. Coral bleaching: causes and consequences. Coral Reefs. 1997;16:s129–s138. [Google Scholar]
- Brown NP, Leroy C, Sander C. MView: a web-compatible database search or multiple alignment viewer. Bioinformatics. 1998;14:380–381. doi: 10.1093/bioinformatics/14.4.380. [DOI] [PubMed] [Google Scholar]
- Chen WNU, Kang HJ, Weis VM, et al. Diel rhythmicity of lipid body formation in a coral-Symbiodinium endosymbiosis. Coral Reefs. 2012;31:521–534. [Google Scholar]
- Chevreux B, Pfisterer T, Drescher B, et al. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Research. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MP, Peterson DA, Biggs PJ. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley A, Kline D, Dunn S, Anthony K, Dove S. The effect of ocean acidification on symbiont photorespiration and productivity in Acropora formosa. Global Change Biology. 2010;16:851–863. [Google Scholar]
- DeSalvo MK, Voolstra CA, Sunagawa S, et al. Differential gene expression during thermal stress and bleaching in the Caribbean coral Montastraea faveolata. Molecular Ecology. 2008;17:3952–3971. doi: 10.1111/j.1365-294X.2008.03879.x. [DOI] [PubMed] [Google Scholar]
- Fabricius KE. Effects of terrestrial runoff on the ecology of corals and coral reefs: review and synthesis. Marine Pollution Bulletin. 2005;50:125–146. doi: 10.1016/j.marpolbul.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Fitt WK, Gates RD, Hoegh-Guldberg O, et al. Response of two species of Indo-Pacific corals, Porites cylindrica and Stylophora pistillata, to short-term thermal stress: the host does matter in determining the tolerance of corals to bleaching. Journal of Experimental Marine Biology and Ecology. 2009;373:102–110. [Google Scholar]
- Gates RD. Seawater temperature and sublethal coral bleaching in Jamaica. Coral Reefs. 1990;8:193–197. [Google Scholar]
- Gates RD, Edmunds PJ. The physiological mechanisms of acclimatization in tropical reef corals. American Zoologist. 1999;39:30–43. [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochachka PW, Somero GN. Biochemical Adaptation: Mechanism and Process in Physiological Evolution. New York: Oxford University Press; 2002. [Google Scholar]
- Hoegh-Guldberg O. Climate change, coral bleaching and the future of the world's coral reefs. Marine and Freshwater Research. 1999;50:839–866. [Google Scholar]
- Hoegh-Guldberg O, Mumby PH, Hooten AJ, et al. Coral reefs under rapid climate change and ocean acidification. Science. 2007;318:1737–1742. doi: 10.1126/science.1152509. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Research. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RJ, Ward S, Amri AY, Hoegh-Guldberg O. Changes in quantum efficiency of Photosystem II of symbiotic dinoflagellates of corals after heat stress and of bleached corals after the 1998 Great Barrier Reef bleaching event. Marine and Freshwater Research. 2000;51:63–71. [Google Scholar]
- Leggat W, Seneca F, Wasmund K, et al. Differential responses of the coral host and their algal symbiont to thermal stress. PLoS One. 2011:e26687. doi: 10.1371/journal.pone.0026687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser MP. Oxidative stress causes coral bleaching during exposure to elevated temperatures. Coral Reefs. 1997;16:187–192. [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loya Y, Sakai K, Yamazato K, Nakano Y, Sambali H, van Woesik R. Coral bleaching: the winners and the losers. Ecology Letters. 2001;4:122–131. [Google Scholar]
- Mayfield AB, Gates RD. Osmoregulation in anthozoan-dinoflagellate symbiosis. Comparative Biochemistry and Physiology A: Molecular and Integrative Physiology. 2007;147:1–10. doi: 10.1016/j.cbpa.2006.12.042. [DOI] [PubMed] [Google Scholar]
- Mayfield AB, Hirst MB, Gates RD. Gene expression normalization in a dual-compartment system: a real-time PCR protocol for symbiotic anthozoans. Molecular Ecology Resources. 2009;9:462–470. doi: 10.1111/j.1755-0998.2008.02349.x. [DOI] [PubMed] [Google Scholar]
- Mayfield AB, Hsiao YY, Fan TY, Chen CS, Gates RD. Evaluating the temporal stability of stress-activated protein kinase and cytoskeleton gene expression in the Pacific corals Pocillopora damicornis and Seriatopora hystrix. Journal of Experimental Marine Biology and Ecology. 2010;395:215–222. [Google Scholar]
- Mayfield AB, Wang LH, Tang PC, et al. Assessing the impacts of experimentally elevated temperature on the biological composition and molecular chaperone gene expression of a reef coral. PLoS One. 2011:e26529. doi: 10.1371/journal.pone.0026529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield AB, Chan PS, Putnam HM, Chen CS, Fan TY. The effects of a variable temperature regime on the physiology of the reef-building coral Seriatopora hystrix: results from a laboratory-based reciprocal transplant. The Journal of Experimental Biology. 2012a;215:4183–4195. doi: 10.1242/jeb.071688. [DOI] [PubMed] [Google Scholar]
- Mayfield AB, Hsiao YY, Fan TY, Chen CS. Temporal variation in RNA/DNA and protein/DNA ratios in four anthozoan-dinoflagellate endosymbioses of the Indo-Pacific: implications for molecular diagnostics. Platax. 2012b;9:1–24. [Google Scholar]
- Mayfield AB, Chen M, Meng PJ, et al. The physiological response of the reef coral Pocillopora damicornis to elevated temperature: results from coral reef mesocosm experiments in Southern Taiwan. Marine Environmental Research. 2013a;86:1–11. doi: 10.1016/j.marenvres.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Mayfield AB, Fan TY, Chen CS. Physiological acclimation to elevated temperature in a reef-building coral from an upwelling environment. Coral Reefs. 2013b;32:909–921. [Google Scholar]
- Mayfield AB, Fan TY, Chen CS. The physiological impact of ex situ transplantation on the Taiwanese reef-building coral Seriatopora hystrix. Journal of Marine Biology. 2013c Article ID 569369. [Google Scholar]
- Mayfield AB, Fan TY, Chen CS. Real-time PCR-based gene expression analysis in the model reef-building coral Pocillopora damicornis: insight from a salinity stress study. Platax. 2013d;10:1–29. [Google Scholar]
- Mayfield AB, Chen YH, Dai CF, Chen CS. The effects of temperature on gene expression in the Indo-Pacific reef-building coral Seriatopora hystrix: insight from aquarium studies in Southern Taiwan. International Journal of Marine Science. 2014a;4:1–22. [Google Scholar]
- Mayfield AB, Hsiao YY, Chen HK, Chen CS. Rubisco expression in the dinoflagellate Symbiodinium sp. is influenced by both photoperiod and endosymbiotic lifestyle. Marine Biotechnology. 2014b;16:371–384. doi: 10.1007/s10126-014-9558-z. [DOI] [PubMed] [Google Scholar]
- Mayfield AB, Wang YB, Chen CS, Liu PJ. Decreased green fluorescent protein-like chromoprotein gene expression in specimens of the reef-building coral Pocillopora damicornis undergoing high temperature-induced bleaching. Platax. 2014c;11:1–23. [Google Scholar]
- Mieog JC, van Oppen MJH, Berkelmans R, Stam WT, Olsen JL. Quantification of algal endosymbionts (Symbiodinium) in coral tissue using real-time PCR. Molecular Ecology Resources. 2009;9:74–82. doi: 10.1111/j.1755-0998.2008.02222.x. [DOI] [PubMed] [Google Scholar]
- Palumbi SR, Barshis DJ, Traylor-Knowles N, Bay RA. Mechanisms of reef coral resistance to future climate change. Science. 2014;334:895–898. doi: 10.1126/science.1251336. [DOI] [PubMed] [Google Scholar]
- Peng SE, Chen WNU, Chen HK, et al. Lipid bodies in coral-dinoflagellate endosymbiosis: ultrastructural and proteomic analyses. Proteomics. 2011;17:3540–3555. doi: 10.1002/pmic.201000552. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Research. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam HM, Mayfield AB, Fan TY, Chen CS, Gates RD. The physiological and molecular responses of larvae from the reef-building coral Pocillopora damicornis exposed to near-future increases in temperature and pCO2. Marine Biology. 2013;160:2157–2173. [Google Scholar]
- Rosic NN, Pernice M, Dove S, Dunn S, Hoegh-Guldberg O. Gene expression profiles of cytosolic heat-shock proteins Hsp70 and Hsp90 from symbiotic dinoflagellates in response to thermal stress: possible implications for coral bleaching. Cell Stress and Chaperones. 2011;16:69–80. doi: 10.1007/s12192-010-0222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Roach S, Miller KJ, Lundgren P, Andreakis N. With eyes wide open: a revision of species within and closely related to the Pocillopora damicornis species complex (Scleractinia; Pocilloporidae) using morphology and genetics. Zoological Journal of the Linnean Society. 2014;170:1–33. [Google Scholar]
- Shick JM. Functional Biology of Sea Anemones. London: Chapman and Hall; 1991. [Google Scholar]
- Shinzato C, Shoguci E, Kawashima T, et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476:320–323. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- Shinzato C, Inoue M, Kusakabe M. A snapshot of a coral “holobiont”: a transcriptome assembly of the scleractinian coral, porites, captures a wide variety of genes from both the host and symbiotic zooxanthellae. PLoS One. 2014:e85182. doi: 10.1371/journal.pone.0085182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoguchi E, Shinzato C, Kawashima T, et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Current Biology. 2013;23:1399–1408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- Smith EG, D'Angelo C, Salih A, Wiedenmann J. Screening by coral green fluorescent protein (GFP)-like chromoproteins supports a role in photoprotection of zooxanthellae. Coral Reefs. 2013;32:463–474. [Google Scholar]
- Söding J. Protein homology detection by HMM–HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Traylor-Knowles N, Granger BR, Lubinski TJ, et al. Production of a reference transcriptome and a transcriptomic database (PocilloporaBase) for the cauliflower coral, Pocillopora damicornis. BMC Genomics. 2011;12:585. doi: 10.1186/1471-2164-12-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veron JEN. Ocean acidification and coral reefs: an emerging big picture. Diversity. 2011;3:262–274. [Google Scholar]
- Vidal-Dupiol J, Zoccola D, Tambutte E, et al. Genes related to ion transport and energy production are up-regulated in response to CO2-driven pH decrease in corals: new insights from transcriptomic analyses. PLoS One. 2013:e58652. doi: 10.1371/journal.pone.0058652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Lee HH, Fang LS, Mayfield AB, Chen CS. Normal fatty acid and phospholipid synthesis are prerequisites for the cell cycle of Symbiodinium and their endosymbiosis with sea anemones. PLoS One. 2013:e72486. doi: 10.1371/journal.pone.0072486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nature Reviews: Molecular Cell Biology. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wood WI. A profile hidden Markov model for signal peptides generated by HMMER. Bioinformatics. 2003;19:307–308. doi: 10.1093/bioinformatics/19.2.307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed gene quantities compared across the two predominant eukaryotic compartments of the Pocillopora damicornis coral holobiont.
Table S2 Repeated-measures anova results of host coral gene expression.
Table S3 Repeated-measures anova results of Symbiodinium gene expression.
Fig. S1. Host coral metabolism, transport, and cell adhesion gene expression.
Fig. S2. Expression of host coral genes involved in the stress response and other processes.
Fig. S3.Symbiodinium metabolism, ion transport, and photosynthesis gene expression.
Fig. S4. Expression of Symbiodinium genes involved in the stress response and other processes.
Data Availability Statement
The unassembled sequences reported herein have been deposited in the Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra/SRX378204), which is cross-referenced with the Transcriptome Shotgun Assembly database (www.ncbi.nlm.nih.gov/genbank/tsa; BioProject PRJNA227785). Also, an interactive website (http://ips.iis.sinica.edu.tw/coral) that is described in the manuscript has been designed to serve as a perpetual depository for the sequence data set. Finally, assembled contigs, as well as the associated expression data (in spreadsheet format), have been deposited at Dryad (www.datadryad.org, doi: 10.5061/dryad.rh04 m).


