Summary
Definitive hematopoietic stem cells (HSCs) develop in the aorta gonad mesonephros (AGM) region in a stepwise manner. Type I pre-HSCs express CD41 but lack CD45 expression, which is subsequently upregulated in type II pre-HSCs prior to their maturation into definitive HSCs. Here, using ex vivo modeling of HSC development, we identify precursors of definitive HSCs in the trunk of the embryonic day 9.5 (E9.5) mouse embryo. These precursors, termed here pro-HSCs, are less mature than type I and II pre-HSCs. Although pro-HSCs are CD41+, they lack the CD43 marker, which is gradually upregulated in the developing HSC lineage. We show that stem cell factor (SCF), but not interleukin-3 (IL-3), is a major effector of HSC maturation during E9–E10. This study extends further the previously established hierarchical organization of the developing HSC lineage and presents it as a differentially regulated four-step process and identifies additional targets that could facilitate the generation of transplantable HSCs from pluripotent cells for clinical needs.
Highlights
-
•
Pro-HSCs are early embryonic precursors of definitive HSCs in E9.5 dorsal aorta
-
•
In contrast to CFU-C, pro-HSCs lack CD43 marker and have low cKIT expression
-
•
SCF, but not IL-3, promotes progression of pro-HSCs into definitive HSCs
-
•
HSCs develop hierarchically in a four-step process
In this article, Medvinsky and colleagues show that the E9.5 mouse embryo contains precursors of definitive hematopoietic stem cells (HSCs) that are distinguishable from those at later developmental stages. This CD41+ precursor, termed here “pro-HSC,” does not express CD43 marker, which is upregulated later on in development. Pro-HSCs can mature into definitive HSCs in response to SCF, but not to IL-3.
Introduction
The embryonic aorta gonad mesonephros (AGM) region is an important site of hematopoietic stem cell (HSC) development prior to colonization of the fetal liver (Dzierzak and Speck, 2008; Medvinsky et al., 2011). At the embryonic day 11 (E11) preliver stage, developing HSCs have also been identified in the placenta, extraembryonic arteries, and head (Dzierzak and Speck, 2008; Gekas et al., 2010; Gordon-Keylock et al., 2013; Li et al., 2012). In the AGM region, definitive HSCs (dHSCs) are localized to the endothelial lining of the dorsal aorta (de Bruijn et al., 2002; Rybtsov et al., 2011). The first dHSCs emerging in the AGM region express endothelial-specific markers such as vascular endothelial cadherin (VE-cadherin) (Ivanovs et al., 2014; North et al., 2002; Taoudi et al., 2005). Cell-fate experiments using permanent genetic labeling of embryonic endothelial cells and their progeny provided strong evidence in favor of the endothelial origin of HSCs (Chen et al., 2009; Zovein et al., 2008). Live imaging in zebrafish revealed the formation of Runx1+ HSC/multipotent progenitors from the endothelial lining of the dorsal aorta (Bertrand et al., 2010; Kissa and Herbomel, 2010). Despite the strong indication that HSCs develop from a specialized hematogenic endothelial lining of the dorsal aorta, it is becoming increasingly apparent that this is not a single-step process (Rybtsov et al., 2011; Taoudi et al., 2008). Furthermore, some evidence suggests that divergence of the HSC lineage from the embryonic endothelium may occur prior to E10.5 before extensive formation of intra-aortic clusters (Rybtsov et al., 2011; Swiers et al., 2013a; Yoder et al., 1997).
Ex vivo modeling is a powerful tool for dissecting the mechanisms of embryonic development of HSCs. During a few days ex vivo, the AGM region and extraembryonic arteries are capable of generating a large pool of dHSCs comparable to that developing in the fetal liver (Gordon-Keylock et al., 2013; Taoudi et al., 2008). While AGM region explant cultures initially enabled HSC maturation to be replicated in vitro (Medvinsky and Dzierzak, 1996), a dissociation-reaggregation culture system that was subsequently developed enabled analysis of individual cell populations in the AGM region (Taoudi et al., 2008). Two types of embryonic precursors sequentially developing into dHSCs have been described in the E10.5–E11.5 AGM region: type I pre-HSCs (VE-cad+CD45−CD41lo) and type II pre-HSCs (VE-cad+CD45+) (Rybtsov et al., 2011). Although E11.5 dHSCs are localized to the endothelial layer of the dorsal aorta (de Bruijn et al., 2002), type I and II pre-HSCs are distributed more broadly both in the endothelial and subendothelial cell layers (Rybtsov et al., 2011). At E11, pre-HSCs have also been identified in extraembryonic arteries (Gordon-Keylock et al., 2013). Meanwhile, cells of the yolk sac cleanly severed from extraembryonic arteries lack or show poor capacity to mature into dHSCs in similar culture conditions (Gordon-Keylock et al., 2013; Rybtsov et al., 2011).
Previous studies showed that the caudal part of the E8.5 embryo contains precursors that can give rise to low-level repopulating hematopoietic cells (Cumano et al., 1996). However, the development of true adult-type high-level repopulating HSCs in culture has been achieved to date only from E10.5 AGM region cells (Robin et al., 2006; Rybtsov et al., 2011; Taoudi and Medvinsky, 2007). Understanding the mechanics of HSC development requires tracking and characterization of the entire embryonic pathway leading to dHSC formation, covering earlier stages of development. Here, we aimed to characterize the cell type immediately preceding the emergence of type I pre-HSCs by focusing on the E9.5 stage. We have been able to mature dHSCs from the E9.5 caudal part of the embryo body. As expected, the maturation of these early precursors into dHSCs requires a longer time than of E11.5 AGM region-derived type I and II pre-HSCs.
Embryonic precursors of dHSCs are known to express CD41 (Bertrand et al., 2005; Ferkowicz et al., 2003; McKinney-Freeman et al., 2009; Rybtsov et al., 2011). We show that E9.5 HSC precursors also express CD41 and RUNX1 but in contrast to later (including adult) stages lack expression of CD43 (Moore et al., 1994). E9.5 HSC precursors and type I pre-HSCs can mature into dHSCs in response to stem cell factor (SCF), but not interleukin-3 (IL3), although for type II pre-HSCs, SCF and IL-3 are equally active. Such strong early dependency on SCF, but not IL-3, identifies SCF as a major HSC maturation factor, which is consistent with the dramatic HSC deficiency in SCF knockouts, but not in IL-3 knockouts (Ding et al., 2012). Taken together, these features identify the E9.5 precursor as a distinct hematopoietically committed cell type termed here “pro-HSC.”
In summary, using a modified ex vivo protocol enabling replication of HSC development from the E9.5 mouse embryo, we characterize here an early hematopoietic precursor in the HSC hierarchy (pro-HSC) localized to the aortic region of the E9.5 embryo. Pro-HSCs are VE-cad+CD45−CD41loCD43− and SCF dependent and precede the appearance of type I pre-HSCs in the AGM region. Taken together with previous studies, HSC development between E9.5 and E11.5 can be characterized as a four-step process driven primarily by SCF.
Results
E9.5 Precursors Require a Longer Time than Type I and II Pre-HSCs to Mature into dHSCs
Previous studies showed that 3–4 days in culture is sufficient for dHSCs maturation from E11.5 AGM cells (Medvinsky and Dzierzak, 1996; Taoudi et al., 2008). Our attempts to obtain dHSCs by culturing the caudal part of E9.5 embryo using similar conditions have failed, and we reasoned that precursors of HSCs from this early stage may require a longer culture period. Therefore, cell suspensions obtained from caudal parts of E9.5 embryos were coaggregated with OP9 cells and cultured for 7 days on floating membranes at the gas-liquid interphase in presence of three growth factors (termed further 3GF): IL-3, SCF, and Fms-related tyrosine kinase 3 ligand (FLT3L). After 7 days in culture, cells were transplanted intravenously into adult irradiated recipients, which resulted in a long-term multilineage high-level hematopoietic repopulation (Figure 1A). Systematic analysis demonstrated a significant gap in maturation kinetics between HSC precursors in E9.5 and E10.5/E11.5 embryos. While late E10.5 and E11.5 pre-HSCs can mature into dHSCs during 4 days culture, E9.5 precursors produce dHSC only after 7 days of culture (Figure 1A; Taoudi et al., 2008), and coaggregation with OP9 cells facilitated dHSCs maturation from E9.5 cells (Figure S1A available online). Primary and secondary transplantations of bone marrow from primary recipients repopulated with E9.5-derived cells showed balanced long-term multilineage engraftment detectable in the peripheral blood and lymphohematopoietic organs (Figures S1B–S1E) (Muller-Sieburg et al., 2012).
Figure 1.
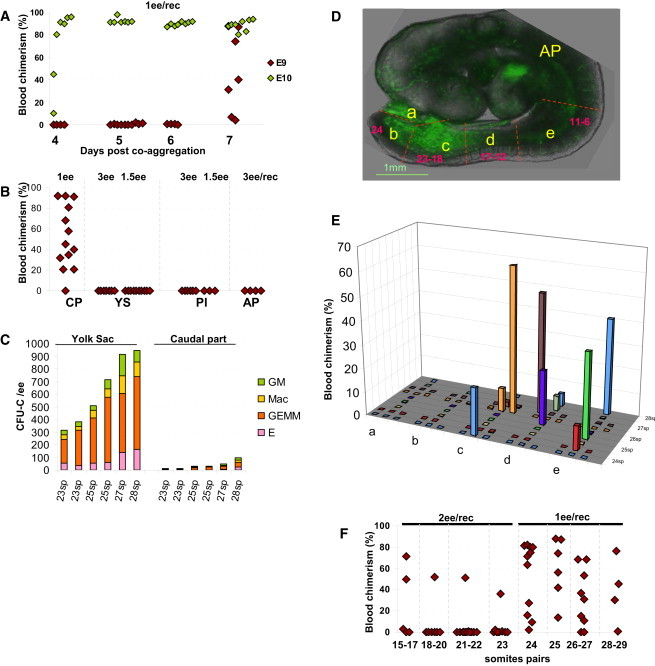
Pro-HSC Maturation into dHSCs
(A) While dHSCs from E10.5 precursors appeared after 4 days in culture (green diamonds), E9.5 caudal part precursors matured into dHSCs only by day 7 in culture (red diamonds) (two independent experiments).
(B) Pro-HSCs are localized to the caudal part of the embryo body, but not in other intra- or extraembryonic tissues (three independent experiments).
(C) The yolk sac, but not the caudal part, contains large numbers of CFU-C (comparisons between yolk sac and caudal parts of individually staged embryos are shown; two independent experiments).
(D) Dissection strategy for E9.5 embryo (24 sp): a, umbilical cord; b, tip of the tail; c, posteriormost one-third of the caudal part excluding the tail tip; d, intermediate one-third of the caudal part; and e, rostralmost one-third of the caudal part immediately below the heart level; anterior part (AP) including the heart, neck, and head.
(E) Subdissected regions from individual embryos were separately cocultured with OP9 cells. Presence of pro-HSCs in different regions (x axis) and level of donor-derived engraftment (y axis) is indicated. Age of individual embryos (24–28 sp) is indicated (z axis). Each color depicts one embryo (four independent experiments).
(F) Pro-HSCs stably emerge in the embryonic caudal part from 24 sp stage. On rare occasions, dHSCs developed in embryos between 15 and 23 sp (three independent experiments).
Diamond symbols represent individual recipients. Number of e.e. transplanted per recipient (ee/rec) is shown. Dissection scheme shown on CD41Cre::sGFP embryo for better orientation (Rybtsov et al., 2011). For multilineage analyses, see Figure S1. Each plot represents two to four independent experiments.
Pro-HSCs Are Distributed along the Aortic Region between the Heart and the Base of the Umbilical Cord
To investigate the localization of pro-HSCs in the E9.5 embryo, cell suspensions obtained from the E9.5 caudal part (trunk below heart level), anterior part (AP; including the rest of the body), yolk sac, and placenta were coaggregated with OP9 cells, cultured for 7 days with 3GF, and then transplanted into irradiated recipients (Figure 1B). Only the caudal part was able to generate dHSCs. None of other tissues, including those that harbor HSCs at later stages (placenta, yolk sac, and anterior part above the heart, including the head), contained precursors capable of maturing into dHSCs. Of note, the caudal part generates HSCs regardless of low content of colony-forming units in culture (CFU-C), whereas the yolk sac lacks HSC potential, despite the presence of large numbers of CFU-C (Figures 1B and 1C).
To determine more accurately the localization of pro-HSCs, E9.5 caudal parts were sectioned into thick transverse slices along the longitudinal axis (Figure 1D) that were tested for their ability to generate dHSCs. The following subregions were tested: umbilical cord (a), tail (b), posterior caudal part (one-third of the caudal part excluding tail harboring the region of ∼5 somite pairs (sp) around the base of the umbilical cord) (c); intermediate caudal part (intermediate one-third of the caudal part ∼5 sp) (d); rostral caudal part (one-third of the caudal part adjacent to the heart, including the base of the vitelline vein, but not the vitelline artery) (e); and anterior part including the heart, neck, and head (AP). We found that all trunk sections (except umbilical cord and tip of tail) below the heart level were able to generate dHSCs following 7-day coaggregation culture (Figure 1E).
Effective Emergence of Pro-HSCs Occurs from 23/24 Sp Stage
To determine the exact stage at which pro-HSCs appear, embryos were grouped in accordance with their age (sp) and coaggregated with OP9 cells. A few embryos in E9.5 litters were found to be retarded and had only 15–17 sp, normally detected in E8.5 litters. Each adult recipient was transplanted with 2–1–2 e.e. (embryo equivalents) of cultured cells from each stage. Although on rare occasions, embryos from 15–17 sp to 23 sp were able to generate dHSCs, only 24 sp and older embryos showed consistent presence of pro-HSCs (Figure 1F). Indeed, only 5 out of 33 recipients of younger than 24 sp caudal parts showed hematopoietic repopulation (of total 66 e.e. transplanted), whereas 26 of 30 recipients transplanted with cultured 24–29 sp caudal parts (in total 30 e.e.) achieved high-level long-term hematopoietic engraftment.
Pro-HSCs are CD41+CD43− and Upregulate CD43 during HSC Lineage Development
To identify the phenotype of pro-HSCs, the Lin-VC+CD45− population isolated from the E9.5 (24–29 sp) caudal part was sorted into four fractions based on CD41 and CD43 staining. After coaggregation with OP9 cells and 7-day culture, cells were transplanted into irradiated recipients (Figure S2A). Only the VC+CD45−CD41loCD43− fraction gave rise to dHSCs (Figures 2Aa and 2Ba′). By the beginning of E10 (30–34 sp), not only the CD41+CD43− but also the CD41+CD43+ subsets within the VC+CD45− population were capable of maturing into dHSCs (Figures 2Ab and 2Bb′). By E11.5, both type I and type II pre-HSCs were exclusively CD43+ (Figures 2Ac, 2Bc′, 2Ad, and 2Bd′, respectively). Of note, CD43 expression increases during transition from type I pre-HSCs into type II pre-HSCs and upregulates further in adult bone marrow HSCs (Figures S2B and S2C). Of note, E11.5 VC+CD43+ populations enriched for both types of pre-HSCs are very efficient at generating CFU-C (Figure 2H).
Figure 2.
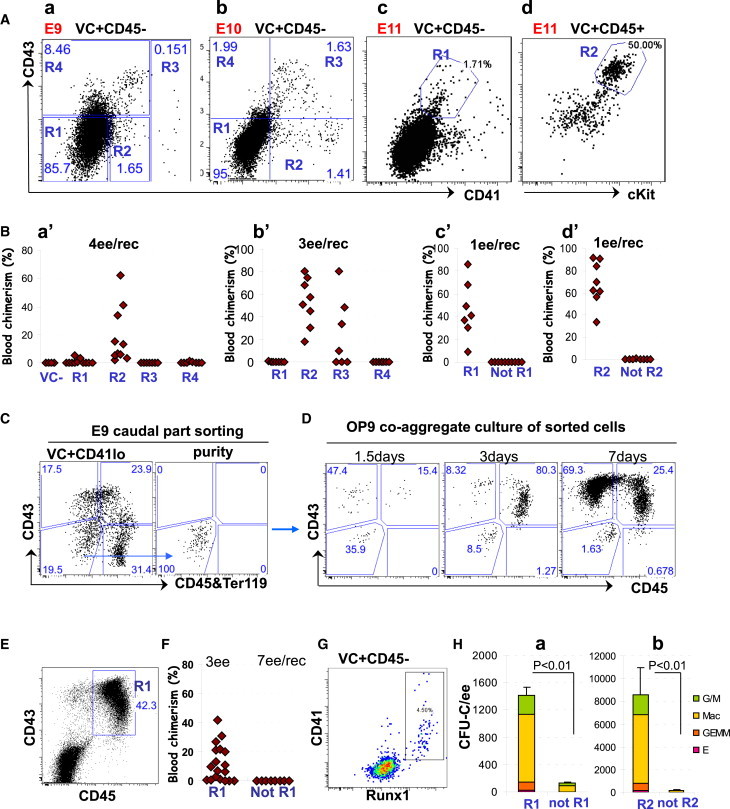
Stepwise Phenotypic Progression of the HSC Lineage during E9.5 to E11.5
(A) Sorting strategies for the developing HSC lineage (plots show 7AAD−Lin− cells).
(B) Functional identification of members of the developing HSC hierarchy. Sorting strategy (see also Figure S2) and functional validation for:
(a and a′) E9 VC+CD45−: repopulation achieved with R2 cells (note that the VC-negative population was also tested and gave no repopulation), four independent experiments;
(b and b′) E10 VC+CD45−: repopulation achieved with R2 and R3 cells, two independent experiments;
(c and c′) E11 VC+CD45−: repopulation achieved only with R1 cells, three independent experiments;
(d and d′) E11 VC+CD45+: repopulation achieved only with R2 cells, three independent experiments;
(C and D) Tracking CD43 and CD45 upregulation in cultured pro-HSCs. Sorting strategy of pro-HSCs (Lin−VC+CD41loCD43−CD45−) from fresh E9 caudal parts (C, left). Sorting purity of this population (C, right). Pure, sorted, Lin−VC+ CD41loCD43−CD45− pro-HSCs show gradual upregulation of CD43 in cultured during 7 days ex vivo (note that CD45 upregulation occurs with a slight delay) (D) (two independent experiments).
(E and F) By the end of culture, E9 caudal parts produce definitive HSCs with the CD43+CD45+ phenotype (E) (located in R1), as demonstrated by transplantation of sorted live cells (F) (two independent experiments).
(G) Demonstration of coexpression of CD41 and RUNX1 in E9 live VC+CD45− population shown by intracellular RUNX1 antibody staining.
(H) CFU-C potential of sorted E11 populations. (a) CFU-C are generated predominantly by R1 cells sorted as in (Ac) (three independent experiments). Error bars represent SD. (b) CFU-C are generated predominantly by R2 cells sorted as in (Ad) (three independent experiments). Error bars represent SD.
We then monitored phenotypic progression of pro-HSCs maturing ex vivo. Stepwise transition of pro-HSCs (VC+CD45−CD41loCD43−Lin−) into the CD45−CD43+ (type I pre-HSCs) phenotype and subsequently into the CD45+CD43+ (dHSCs) phenotype could be followed over time in culture (Figures 2C and 2D). The CD45+CD43+ phenotype of dHSCs emerging in culture was further confirmed by the functional transplantation analysis (Figures 2E and 2F).
Pro-HSCs Are Present in the Dorsal Aorta, but Not in Other Large Vessels
In E9.5 embryos, the VC+CD45−CD41+ population enriched for pro-HSCs coexpresses RUNX1, as shown by intracellular antibody staining (Figure 2G). The E9.5 VC+ aortic lining almost completely lacks CD43 expression but shows RUNX1 expression (predominantly in its ventral domain), suggesting the presence of pro-HSCs in this location (Figures 3A and 3A′, yellow arrow). Small vessels branching from the ventral domain of the aorta also contain individual VC+CD43−RUNX1+ cells (Figures 3A′ and 3B, yellow arrowheads). In contrast, the omphalomesenteric (OA) and vitelline and umbilical arteries (UA) contain strings of large prominent clusters composed of VC+CD43+RUNX1+ cells (Figures 3B, 3B′, and S3A–S3C). Rare VC−CD43+RUNX1+ cells can also be detected in the E9.5 circulation (Figure S3D).
Figure 3.
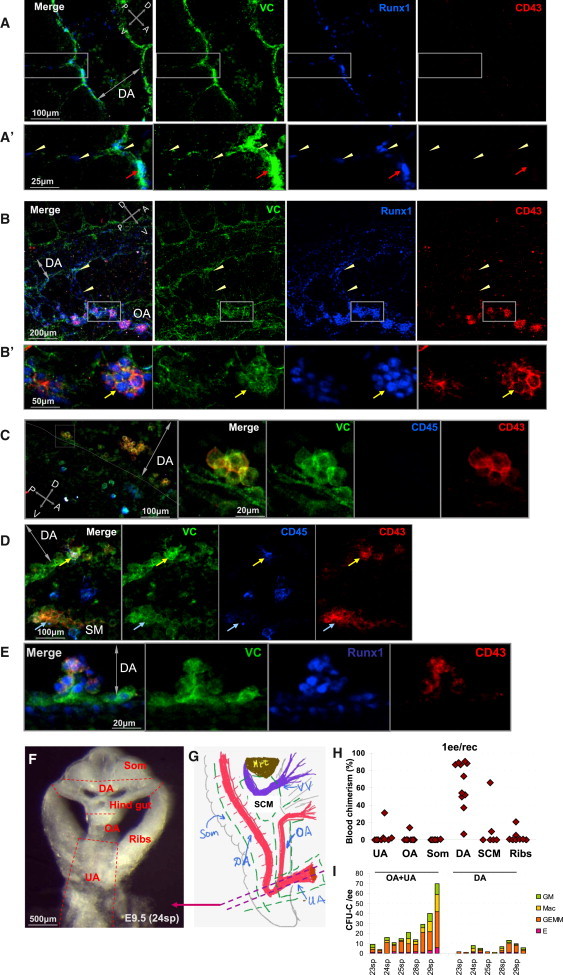
Phenotypic Mapping of Pro-HSC Localization
(A) VC+CD43+RUNX1+ cells localize to the ventral floor of the E9 dorsal aorta. Zoomed boxed area is shown in (A′). Yellow arrowheads indicate cells with pro-HSC phenotype. Red arrows show RUNX1+VC+ pro-HSCs localized to the ventral endothelium.
(B) Hematopoietic development in the E9 omphalomesenteric artery (OA). The OA, but not the dorsal aorta (DA), contains strings of large VC+CD43+RUNX1+ clusters. Note the presence of VC+CD43−RUNX1+cells in small vessels connecting OA and DA (shown by yellow arrowheads). Zoomed image of the boxed area is shown in (B′). Yellow arrow indicates a large cluster inside the OA composed of VC+CD43+RUNX1+ cells.
(C) Small ventrally located intra-aortic clusters appearing at E10 contain cells expressing CD43, but not CD45.
(D) E10 rare VC+CD43+ cells in small intra-aortic clusters show expression of CD45 (yellow arrow). Subendothelial (aortic) mesenchyme (SM) contains large clusters of VE-cad+CD45−CD43+ (blue arrow) not associated with vessels.
(E) E10 intra-aortic clusters are VE-cad+CD43+RUNX1+.
(F) Fine spatial localization of pro-HSCs in the caudal part of the body. Dissection strategy shown on a thick transverse slice in the area of UA as indicated schematically by double dotted line in (G) (green dotted line indicates subdissections). DA, OA, and UA are indicated by red color. Violet color depicts vitelline veins (VV).
(H) Dorsal aorta is the main site of pro-HSC residence. Dissected subregions of E9.5 caudal part (as shown in (F) and (G) were cocultured with OP9 cells and transplanted into irradiated recipients.
(I) CFU-C contents in UA + OA and DA. UA + OA contains significantly more CFU-C than dorsal aorta (comparisons between UA + OA and DA of individually staged embryos are shown).
Abbreviations: OA, omphalomesenteric artery; DA, dorsal aorta; UA, umbilical artery; VV, vitelline vein; SCM, subcardiac mesenchyme; Som, somites; Hrt, heart. Crossed double-headed arrows indicate dorsoventral (D-V) and posterior-arterial (P-A) orientations. Double-headed arrow indicates the lumen of dorsal aorta (DA). Each plot represents two independent experiments.
Cell clusters emerge in the dorsal aorta only by E10 contain mainly VC+CD43+CD45− cells and some VC+CD43+CD45+ cells (Figures 3C and 3D). Some cells in clusters also coexpress RUNX1 (Figure 3 E). Similar to intra-aortic clusters, intra-UA and intra-OA clusters by E10–E10.5 are mainly composed of CD43+ cells, some of which start upregulating CD45 (Figures S3A–S3C). Most of the CD45+ cells scattered in the embryo body lack CD43 expression and are likely to be mature macrophages (Figures 3C and 3D).
To more precisely determine the location of pro-HSCs, we subdissected the E9.5 caudal part (Figures 3F and 3G). After separation of the yolk sac, somites, and lateral body walls (“ribs”), we isolated and cultured the OA, the UA, the vitelline vein with surrounding mesenchyme (including liver rudiment), and the dorsal aorta. We found that dHSCs predominantly developed from the aortic region, which has smooth endothelial lining, and practically none were generated by the OA (Figure 3H), despite the presence of large CD43+ clusters.
Pro-HSCs and CFU-C in the E9.5 Embryo Are Distinguishable by CD43 and cKIT Expression Levels
The above experiments determined that pro-HSCs reside within the VC+CD45−CD41+CD43− population. To test whether CFU-C also reside in this fraction, VC+CD45− populations were sorted from the E9.5 caudal parts on the basis of CD41 and CD43 staining and plated in a methylcellulose assay (Figure 4A). The absolute majority of CFU-C (20 CFU-C/e.e.) were detected in the CD41+CD43+ fraction, whereas the CD41+CD43− population, which is enriched for pro-HSCs, contained only 1 CFU-C/e.e. (Figure 4B). This suggests that CFU-C are localized mainly to UA and OA. To test this, we dissected and separately examined the dorsal aorta and UA + OA in the methylcellulose assay and found approximately 2- to 5-fold more CFU-C in UA + OA than in the dorsal aorta (depending on the exact age of the embryo) (Figure 3I). These data suggest that large CD41+CD43+ clusters within the OA and UA are the sources of CFU-C.
Figure 4.
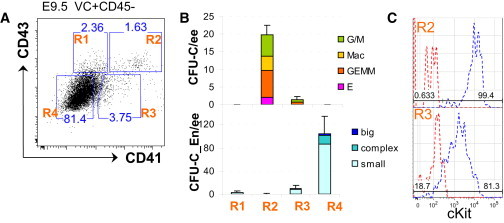
CFU-C in the E9.5 Embryo Are Marked by High cKIT and CD43 Expression
(A) Gating strategy for identification of CFU-C and endothelial progenitors in E9.5 caudal part; live Lin−VC+CD45− cells were sorted on the basis of CD41 and CD43 expression.
(B) Four sorted populations (as indicated in A) were tested for hematopoietic CFU-C and endothelial progenitors CFU-En (frequencies per e.e. are shown). While CFU-C resided in R2 (note the large numbers of multilineage colonies, CFU-GEMM generated), CFU-En resided in R4. Error bars represent SD.
(C) CFU-C population (R2) shows high level of cKIT expression, whereas pro-HSCs (R3) express cKIT at intermediate levels (cKIT expression and control FMO are depicted by blue and red colors, respectively). Each plot represents two independent experiments.
We then investigated cKIT (SCF receptor) expression on CFU-C and found that the entire VC+CD45−CD41+CD43+ population expresses cKIT at a high level. Meanwhile, the VC+CD45−CD41loCD43− population enriched for pro-HSCs express cKIT at lower levels (Figure 4C). Thus, pro-HSCs and CFU-C are phenotypically distinct and tend to be spatially segregated in the embryo.
Pro-HSCs Are Devoid of Endothelial Potential
We have previously shown that type I and II pre-HSCs do not produce endothelial colonies in the clonogenic endothelial assay (Rybtsov et al., 2011; Taoudi et al., 2005). As expected, the main endothelial potential in E9.5 embryos is associated with the VC+CD45−CD41−CD43− population as shown by clonogenic endothelial assays where approximately 100 endothelial colonies (CFU-C_En) were detected per 1 e.e. (Figure 4B). Even though 6 e.e. were taken for each experiment, the VC+CD45−CD41+CD43+ populations enriched for CFU-C produced no CFU-C_En. We then tested the endothelial potential of pro-HSCs (VC+CD45−CD41+CD43−) and found that this population produces very few endothelial colonies (∼10 CFU-C_En/e.e.) represented mainly by single tubules that were spatially segregated from rare CFU-C hematopoietic colonies (Figure 4B).
SCF Alone Is Sufficient to Induce Maturation of Pro-HSCs and Type I Pre-HSCs into dHSCs
Previous studies showed that IL-3, on its own or in combination with other factors (SCF and FLT3L), is capable of inducing maturation of dHSCs from the E10.5–E11.5 AGM region (Robin et al., 2006; Taoudi et al., 2008). We have shown above that 3GF combination is also effective for induction of E9.5 pro-HSCs (Figures 1A–1C.) and further tested which of these factors are essential for this process. In contrast to Il3, Scf is expressed at a substantial level in the E9.5 dorsal aorta and increases 2.5-fold by E10.5 (Figure 5C). Given the expression of cKIT in pro-HSCs (Figure 4C) and the increase in Scf expression in vivo (Figure 5C), we tested whether SCF can induce maturation of pro-HSCs in vitro. Indeed, the addition of soluble SCF induced efficient production of dHSCs (in seven out of nine recipients repopulated) with high engraftment capacity (range of blood chimerism, 45%–84%) (Figure 5A). In contrast, the effect of IL-3 was much weaker. Only 3 of 11 recipients transplanted with IL-3-treated cultures of pro-HSC showed donor-derived hematopoietic engraftment at relatively modest levels (7%–25% blood chimerism) (Figure 5A). Incubation with SCF and IL-3 together showed a moderate potentiating effect (13 out of 13 recipients with 8%–86% donor-derived engraftment in blood) (Figures 5A and 5D). FLT3L on its own showed little or practically no effect and in combination with SCF or/and IL-3 only slightly improved engraftment, if at all (data not shown). Of note, combined incubation with SCF and IL-3 tends to increase myeloid donor-derived contribution (Figure 5D).
Figure 5.
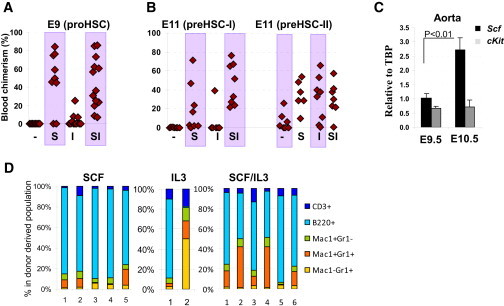
Growth Factor Requirements for Pro-/Pre-HSC Development
(A) E9 pro-HSCs mature into dHSCs in presence of SCF (S), but not IL-3 (I) (1 e.e./recipient; four independent experiments).
(B) E11 type I pre-HSCs respond by maturing into dHSCs in the presence of SCF, but not IL-3. Meanwhile, type II pre-HSCs respond equally well to SCF and IL-3 (left and right, respectively). Transplantations: type I pre-HSCs: 0.2 ee/recipient (left) and type II pre-HSCs: 0.3 ee/recipient (right) (three independent experiments).
(C) Scf expression in the dorsal aorta, as measured by quantitative PCR, increases 2.5-fold between E9.5 and E10.5. Note that cKit expression remains at the same level during this period of time. Expression levels are normalized to Tbp housekeeping gene expression (three independent RNA preparations). Error bars represent SD.
(D) Long-term donor-derived lymphoid and myeloid contribution in recipient blood derived from E9.5 caudal parts. While SCF on its own generates dHSCs with balanced lymphomyeloid differentiation (∼15%–20% of lymphoid cells), IL-3 on its own or in combination with SCF tends to shift balance more toward myeloid differentiation. Each bar represents an individual recipient (two independent experiments).
We then tested whether the next stage (type I pre-HSCs) retains the predominant responsiveness to SCF. Sorted E11.5 AGM type I pre-HSCs were cocultured with OP9 cells in the presence of either SCF or IL-3. While cultures supplemented with exogenous SCF repopulated seven out of eight recipients (three low and four high) (Figure 5B, left), cultures supplemented with IL-3 gave repopulation in only one of seven recipients. Again, SCF and IL-3 in combination showed a potentiating effect on dHSC development (Figures 5A and 5B).
We then tested effects of IL-3 and SCF on type II pre-HSCs isolated from the E11.5 AGM region and coaggregated with OP9 cells. Control cultures (no exogenous growth factors) generated a few dHSCs resulting in repopulation of four out of seven recipients with modest engraftment levels (3%–27%) (Figure 5B, right). However, addition of exogenous SCF resulted in engraftment of all six recipient mice with high-level donor-derived chimerism (10%–54%). In line with previous reports, incubation with IL-3 was also effective, with seven out of eight recipients being repopulated at various levels (Figure 5B, right). Culturing with SCF and IL-3 together showed no improvement over the individual effects of IL-3 or SCF (Figure 5B). Thus, pro-HSCs and type I pre-HSCs show strong responsiveness to SCF, whereas type II pre-HSCs can mature equally well in response to SCF or IL-3 alone.
Discussion
Although the first hematopoietic differentiation occurs in the yolk sac of the E7.5 mouse embryo, definitive HSCs giving rise to the entire adult hematopoietic system emerge only by late E10.5–E11.0 (Dzierzak and Speck, 2008; Medvinsky et al., 2011; Swiers et al., 2013b). There is a strong line of evidence to suggest an endothelial origin for the adult hematopoietic hierarchy (Bertrand et al., 2010; Chen et al., 2009; de Bruijn et al., 2002; Kissa and Herbomel, 2010; Zovein et al., 2008). From early developmental stages, the HSC lineage shares VE-cadherin expression with endothelial cells, which is subsequently downregulated during fetal life (Kim et al., 2005; Taoudi et al., 2005). Hematopoietic progenitors (CFU-C) in the early embryo express CD41 and only later become CD45+ (Mikkola et al., 2003). The HSC lineage evolves rapidly through sequential developmental stages marked by upregulation of CD41 (type I pre-HSCs: VC+CD41loCD45−) followed by upregulation of CD45 in type II pre-HSCs (VC+CD45+) before becoming competent dHSCs (Rybtsov et al., 2011; Taoudi et al., 2008). The analysis of earlier precursors of HSCs has to date been hampered by the lack of an adequate in vitro system that would support HSC development from very early developmental stages.
Here, we developed an in vitro system which allowed us to characterize E9.5 dHSCs precursors, termed here pro-HSCs, preceding the appearance of type I pre-HSCs in E10.5 embryos (Rybtsov et al., 2011). Pro-HSCs differ from subsequent stages by two important characteristics: (1) in agreement with a more immature status, pro-HSCs require a longer culture period (7 days) to mature into dHSCs compared to pre-HSCs; and (2) pro-HSCs are VC+CD41+RUNX1+CD45− but lack expression of the CD43 marker, which is upregulated at later stages beginning from type I pre-HSCs.
HSC development is regulated by various transcription factors and signaling pathways (Bigas et al., 2013; Chanda et al., 2013; Chen et al., 2009; Guiu et al., 2013; Leung et al., 2013; Nimmo et al., 2013; Richard et al., 2013). Previous studies showed that IL-3 is a potent inducer of dHSC maturation in the E10.5–11.5 AGM region (Robin et al., 2006); however, we demonstrate that IL-3 has very little effect on pro-HSCs. By contrast, SCF has a strong maturation effect on pro-HSCs. Furthermore, SCF is a strong trigger of dHSC maturation from both type I and type II pre-HSCs. Scf is clearly expressed in the E9.5 aorta and is significantly upregulated by E10.5, thus correlating with pro-HSC to pre-HSC transition. Of note, weak and inconsistent Il3 expression becomes detectable only in the E11.5 dorsal aorta in parallel with the emergence of type II pre-HSCs (Gordon-Keylock et al., 2013). These findings indicate that SCF is the major early factor initiating HSC development, in agreement with a critical decline in HSC activity in SCF mutants, which die perinatally (Ding et al., 2012), while IL3 knockouts remain viable into adulthood (Lantz et al., 1998; Robin et al., 2006). Concurrent SCF and IL-3 action has only an additive or weakly potentiating effect on pro-HSC/pre-HSC maturation, and a similar interaction between SCF and IL-3 has been described in mast cells (Dvorak et al., 1994).
The HSC lineage and CFU-C develop in parallel in the embryo, and their exact relationship is poorly understood (Medvinsky et al., 2011). Some data indicate that they emerge from distinct endothelial compartments (Chen et al., 2011). However, the analysis of mechanistic differences between these two cell types is hampered by our inability to reliably distinguish developing HSCs and CFU-C by phenotype. Here, we show that CD43 marker clearly segregates these populations in the E9.5 embryo as pro-HSCs, in contrast to CFU-C, do not express CD43. In addition, pro-HSCs express cKIT at significantly lower levels than E9.5 CFU-C. Low cKIT expression is also characteristic of quiescent adult bone marrow HSCs (Grinenko et al., 2014; Matsuoka et al., 2011; Shin et al., 2014).
Confocal analysis showed that CD43+ cells reside mainly within large cell clusters in the OA, but not in the dorsal aorta, suggesting association of CFU-C activity with intra-OA clusters. The VE-cad+CD43−RUNX1+ population, which is enriched for pro-HSCs, is localized to the ventral endothelial lining of the dorsal aorta and small vessels branching from the aorta.
Subdissection of E9.5 caudal parts followed by functional analysis confirmed that pro-HSCs are closely associated with the dorsal aorta and could be found at different levels between the heart and the base of the umbilical cord. Thus, pro-HSCs develop prior to formation of intra-aortic clusters. Contrary to a previous supposition (Zovein et al., 2010), strings of large VC+CD43+RUNX1+ clusters inside OA and in extraembryonic vessels at E9.5 are not enriched for pro-HSCs. Only later do extraembryonic vessels develop pre-HSCs in fairly low numbers (Gordon-Keylock et al., 2013). Pre-HSCs can be also found at later stages in subendothelial layers of the dorsal aorta (Rybtsov et al., 2011). The majority of cells with pro-HSC phenotype (E9.5) are localized to the endothelium of the dorsal aorta and we confirmed their nature functionally. However, similar rare cells can be found at E9.5 both in subaortic capillaries and in larger vitelline/umbilical arteries. It needs to be elucidated further whether pre-HSCs in these sites develop locally with delay or are colonized later by pre-HSCs from the dorsal aorta.
CD43 expression allows clear morphological tracking of intra-aortic cluster formation in E10.5 embryos. During HSC lineage development, CD43 is progressively upregulated, reaching high expression levels by E11.5 in pre- type II HSCs. Whether CD43, known to have antiadherent function (Drew et al., 2005), is involved in release of dHSCs from the AGM region niche prior to colonization of the fetal liver needs to be elucidated in future. In the adult CD43 is implicated in transendothelial migration of T-lymphocytes and possibly HSCs through interaction with E-selectin (Manjunath et al., 1995; Winkler et al., 2012; Woodman et al., 1998).
The prevailing dogma in the field describes emergence of HSCs directly from hematogenic endothelium. All described precursors of the HSC lineage express VE-cadherin. However, while pro-HSCs are hematopoietically committed (CD41+RUNX1+) and readily generate hematopoietic colonies, they lack endothelial potential. It has recently been proposed that the hematogenic endothelium expresses only Runx1 mRNA, but not the protein (Swiers et al., 2013a); however, pro-HSCs express RUNX1 at the protein level. Taken together, this places pro-HSCs in an advanced developmental position and indicates that they have already segregated from the hematogenic endothelium.
Based on the level of immaturity, surface immunophenotype and responsiveness to SCF/IL-3, E9.5 precursors identified here are termed pro-HSCs. Based on this understanding, we propose a model that includes four consecutive stages of HSC development (Figure 6).
Figure 6.
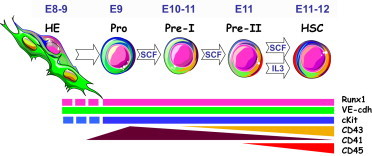
Schematic Development of the HSC Lineage in the Context of Growth Factor—SCF and IL-3—Dependency
Proposed stepwise development of the HSC lineage in the dorsal aorta beginning from E9.5 stage based on cumulative data obtained from current research and previous publications (Rybtsov et al., 2011; Taoudi et al., 2008). Color coding shows a transient increase in CD41 expression at E9.5 followed by stable gradual upregulation of CD43 and subsequent elevation of CD45 expression. Modulation of VE-cadherin, cKIT, and RUNX1 expression was not studied in detail, and these are presented schematically at steady levels throughout E9–E11 stages. Stage-specific dependency on SCF and IL-3 is indicated within arrows showing transition of one stage to another. Abbreviations: HE, hematogenic endothelium; Pro, pro-HSC; pre- I, type I pre-HSCs; pre- II, type II pre-HSCs; HSC, definitive HSCs. Images of cells were downloaded from Servier Medical Art (http://www.servier.com/) with the owner’s permission.
In summary, this study highlights extended hierarchical organization of the developing HSC lineage. We identify an E9.5 pro-HSC as an upstream precursor distinct from subsequent members of the developing HSC lineage based on CD43− phenotype, dependence on SCF, but not on IL-3, and the extended time required for maturation into dHSCs in culture. We show that SCF is a major effector of HSC maturation during E9–E10. Interestingly, soluble SCF has a dramatic enhancing effect on maturation of HSCs, although OP9 cells express high levels of membrane-bound SCF. This study defines both a key cellular intermediate and appropriate culture conditions that may improve protocols for generating transplantable HSCs from pluripotent cells for clinical needs, which remains a challenge in the field. Further studies will need to elucidate molecular mechanisms underlying stepwise HSC development.
Experimental Procedures
Animals
Staged embryos were obtained by mating C57BL/6 (CD45.2/2) or C57BL/6 enhanced GFP mice. The morning of discovery of the vaginal plug was designated as day 0.5. More accurate embryo staging was performed by counting somite pairs (sp) and grouped as E9.0 (15–24 sp), E9.5 (25–29 sp), E10.0 (31–34 sp), E10.5 (35–39 sp), and E11.0–E11.5 (>40 sp). All experiments with animals were performed under Project License granted by the Home Office (UK), University of Edinburgh Ethical Review Committee, and conducted in accordance with local guidelines.
Long-Term Repopulation Assay
CD45.2/2 (or GFP+) cells were injected into adult irradiated CD45.1/2 heterozygous recipients along with 100,000 CD45.1/1 nucleated bone marrow carrier cells per recipient. Recipients were Ɣ-irradiated by split dose (600 + 550 rad) with a 3 hr interval. Numbers of cells injected are expressed in embryo equivalents (e.e.), where 1 e.e. corresponds to a cell population sorted from one embryo after adjustment for dead cells. Percentage donor-derived chimerism was evaluated in peripheral blood at 6.5 weeks, 14 weeks, and 12 months posttransplantation using FACSCalibur or Fortessa (BD Biosciences). Erythrocytes were depleted using PharM Lyse (BD Bioscience), and cells were stained with anti-CD16/32 (Fc-block) followed by anti-CD45.1-APC (cloneA20) and anti-CD45.2-PE (clone 104) monoclonal antibodies (eBioscience). HSC numbers were assessed using ELDA analysis (Hu and Smyth, 2009). Multilineage donor-derived hematopoietic contribution in recipient blood and organs was determined by staining with anti-CD45.1-V450, anti-CD45.2-V500 and lineage-specific anti-Mac1 fluorescein isothiocyanate (FITC), Gr1-PE CD3e-APC, B220-PE-Cy7 monoclonal antibodies (BD Pharmingen).
E9.5 Embryo Dissection
Fine needles suitable for microdissection of E9 embryos were prepared by electrolysis of tungsten wire in 2 M NaOH with one or two drops of Decon 60 (Ekvall et al., 1999), using an engineered automated programmable machine built in-house. Embryo isolation and dissection were performed in Dulbecco’s PBS (+Ca+Mg) (GIBCO) supplemented with 7% heat-inactivated fetal bovine serum (FBS) (PAA Laboratories), 100 U/ml penicillin (Life Technologies), and 100 μg/ml streptomycin (Life Technologies). Tissues were incubated with collagenase/dispase (1 mg/ml collagenase/dispase, Roche) at 37° and washed with fluorescence-activated cell sorting (FACS) buffer (Ca2+ and Mg2+-free PBS, Sigma) supplemented with 5% FBS and dissociated in FACS buffer. Yolk sacs were carefully separated from the vitelline/omphalomesenteric arteries and umbilical cords.
HSC Ex Vivo Maturation
Cells from E9.5 caudal parts dissociated by collagenase/dispase were sorted and either reaggregated or coaggregated with OP9 cells. To this end, cell suspension containing 1 e.e. caudal part or sorted cells and 105 OP9 cells in 30 μl volume of media (Iscove’s modified Dulbecco’s medium [IMDM], Invitrogen-GIBCO, 20% of preselected, heat-inactivated FBS, PAA Laboratories, L-glutamine, penicillin/streptomycin) were centrifuged at 450 × g/12 min in 200 μl pipette tips sealed with parafilm (Taoudi et al., 2008). When appropriate, the IMDM was supplemented with murine recombinant cytokines in concentration 100 ng/ml (all from PeproTech). Coaggregates were cultured on floating 0.8 μm AAWP 25 mm nitrocellulose membranes (Millipore) for 24 hr followed by complete replacement of medium followed by additional 4–6 days culture. E11.5 pre-HSCs were assayed as previously described (Rybtsov et al., 2011). Cultured aggregates were dissociated using collagenase/dispase. All experiments were performed at least twice independently.
CFU-C and Endothelial Assays
Dissociated cells were plated in methylcellulose culture (MethoCult3434 medium; STEMCELL Technologies) according to the manufacturer’s instructions. For endothelial hematopoietic clonogenic assays, we used an OP9-based assay (Taoudi et al., 2005). Briefly, a cell population was plated on OP9 cells in the methylcellulose medium supplemented with 100 ng/ml SCF and 50 ng/ml vascular endothelial growth factor (PeproTech). After 11 days, cultures were stained with anti-CD31 antibodies to assess formation of hematopoietic and endothelial colonies.
Fluorescence-Activated Cell Sorting and Analysis
The following antibodies were used: anti-CD41-BV421 (brilliant violet 421) or Alexa Fluor 488 (clone MW30reg) anti-CD45 FITC (clone 30-F11), anti-CD43-PE (clone eBioR2/60), and biotinylated anti-VE-cadherin (clone 11.D4.1) followed by incubation with streptavidin-APC (all purchased from BD Pharmingen or Biolegend). Anti-mouse VE-cadherin antibody was biotinylated in-house using FluoReporter Mini-Biotin-XX Protein labeling kit (Invitrogen). For depletion of differentiated hematopoietic cells (Lineage, Lin), anti-mouse B220, CD3, and Ter119 antibodies conjugated with PerCP-Cy5.5 were used (eBioscience). Cell populations were sorted using a FACSAria-II sorter (BD) followed by purity checks. Gating of negative populations was performed on the basis of fluorescence minus one (FMO) staining where one of the antibodies was replaced with isotype control (IC) (BD Pharmingen). Dead cells were excluded by 7AAD staining. For intracellular staining, cells labeled with antibodies to surface antigens were incubated with 0.5 μg/mL ethidium monoazide bromide (EMA) solution (Sigma-Aldrich) to exclude dead cells. The cells were then washed twice and fixed and stained with BD Cytofix/Cytoperm Kit and anti-RUNX1 rabbit monoclonal antibody (clone EPR3099, Epitomics) followed by anti-rabbit Alexa Fluor 488 staining. Rabbit isotype control was used as FMO control. Data acquisition and data analysis were performed by Fortessa (BD) using FlowJo software (Tree Star).
Confocal Microscopy
Whole-mount immunostaining was performed as previously described (Yokomizo et al., 2012), with slight modifications. Embryos dissected from the yolk sac and amnion were fixed with cold acetone and, following dehydration by increasing concentrations of methanol, the head, limbs, and one body wall were removed. After rehydration in 50% methanol, washing with PBS and blocking in 50% fetal calf serum/0.5% Triton X-100, the samples were incubated overnight with antibodies. For staining with antibodies from the same species, incubations were performed sequentially. Primary antibodies used were unconjugated goat anti-mouse CD43 (clone M19, Santa Cruz), rat anti-mouse VE-cadherin (clone 11D4.1, BD Pharmingen), rat anti-mouse CD45 (clone 30-F11, BD Pharmingen), rabbit anti-mouse RUNX1 (clone EPR3099, Abcam), and these were detected by the secondary antibodies anti-goat NL557 (R&D), anti-rat Alexa Fluor 647 (Invitrogen), or anti-rat Alexa 488 (Invitrogen) and anti-rabbit Alexa Fluor 647 (Abcam). After washing, the embryos were dehydrated with methanol and cleared with BABB (one part benzyl alcohol, two parts benzyl benzoate) solution (Yokomizo and Dzierzak, 2010). Images were acquired with an inverted confocal microscope (Leica SP8) and processed using Volocity software.
Statistics
Data on histograms presented as average ± SD and difference evaluated using t test. Numbers of pro-HSCs or HSCs were validated by single-hit Poisson model using ELDA software (Hu and Smyth, 2009).
Acknowledgments
The authors thank Suling Zhao, J. Verth, C. Manson, R. McInnis, and BRR staff for assistance with mouse maintenance and breeding; C. Watt., C. Flockhart, C. Forrest, J. Agnew, and K. Anderson for irradiations; and S. Monard and O. Rodriguez for cell sorting. We thank Drs. Sabrina Gordon-Keylock, Paul Travers, Val Wilson, Celine Souilhol, and Jennifer Easterbrook for helpful comments and Andrejs Ivanovs for technical assistance. This work was supported by BBSRC, LLR, MRC, and the Wellcome Trust.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information
References
- Bertrand J.Y., Giroux S., Golub R., Klaine M., Jalil A., Boucontet L., Godin I., Cumano A. Characterization of purified intraembryonic hematopoietic stem cells as a tool to define their site of origin. Proc. Natl. Acad. Sci. USA. 2005;102:134–139. doi: 10.1073/pnas.0402270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J.Y., Chi N.C., Santoso B., Teng S., Stainier D.Y., Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigas A., Guiu J., Gama-Norton L. Notch and Wnt signaling in the emergence of hematopoietic stem cells. Blood Cells Mol. Dis. 2013;51:264–270. doi: 10.1016/j.bcmd.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Chanda B., Ditadi A., Iscove N.N., Keller G. Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell. 2013;155:215–227. doi: 10.1016/j.cell.2013.08.055. [DOI] [PubMed] [Google Scholar]
- Chen M.J., Yokomizo T., Zeigler B.M., Dzierzak E., Speck N.A. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.J., Li Y., De Obaldia M.E., Yang Q., Yzaguirre A.D., Yamada-Inagawa T., Vink C.S., Bhandoola A., Dzierzak E., Speck N.A. Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell. 2011;9:541–552. doi: 10.1016/j.stem.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumano A., Dieterlen-Lievre F., Godin I. Lymphoid potential, probed before circulation in mouse, is restricted to caudal intraembryonic splanchnopleura. Cell. 1996;86:907–916. doi: 10.1016/s0092-8674(00)80166-x. [DOI] [PubMed] [Google Scholar]
- de Bruijn M.F., Ma X., Robin C., Ottersbach K., Sanchez M.J., Dzierzak E. Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity. 2002;16:673–683. doi: 10.1016/s1074-7613(02)00313-8. [DOI] [PubMed] [Google Scholar]
- Ding L., Saunders T.L., Enikolopov G., Morrison S.J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew E., Merzaban J.S., Seo W., Ziltener H.J., McNagny K.M. CD34 and CD43 inhibit mast cell adhesion and are required for optimal mast cell reconstitution. Immunity. 2005;22:43–57. doi: 10.1016/j.immuni.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Dvorak A.M., Seder R.A., Paul W.E., Morgan E.S., Galli S.J. Effects of interleukin-3 with or without the c-kit ligand, stem cell factor, on the survival and cytoplasmic granule formation of mouse basophils and mast cells in vitro. Am. J. Pathol. 1994;144:160–170. [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E., Speck N.A. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat. Immunol. 2008;9:129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekvall I., Wahlstrom E., Claesson D., Olin H., Olsson E. Preparation and characterization of electrochemically etched W tips for STM. Meas. Sci. Technol. 1999;10:11–18. [Google Scholar]
- Ferkowicz M.J., Starr M., Xie X., Li W., Johnson S.A., Shelley W.C., Morrison P.R., Yoder M.C. CD41 expression defines the onset of primitive and definitive hematopoiesis in the murine embryo. Development. 2003;130:4393–4403. doi: 10.1242/dev.00632. [DOI] [PubMed] [Google Scholar]
- Gekas C., Rhodes K.E., Van Handel B., Chhabra A., Ueno M., Mikkola H.K. Hematopoietic stem cell development in the placenta. Int. J. Dev. Biol. 2010;54:1089–1098. doi: 10.1387/ijdb.103070cg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Keylock S., Sobiesiak M., Rybtsov S., Moore K., Medvinsky A. Mouse extraembryonic arterial vessels harbor precursors capable of maturing into definitive HSCs. Blood. 2013;122:2338–2345. doi: 10.1182/blood-2012-12-470971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinenko T., Arndt K., Portz M., Mende N., Günther M., Cosgun K.N., Alexopoulou D., Lakshmanaperumal N., Henry I., Dahl A., Waskow C. Clonal expansion capacity defines two consecutive developmental stages of long-term hematopoietic stem cells. J. Exp. Med. 2014;211:209–215. doi: 10.1084/jem.20131115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiu J., Shimizu R., D’Altri T., Fraser S.T., Hatakeyama J., Bresnick E.H., Kageyama R., Dzierzak E., Yamamoto M., Espinosa L., Bigas A. Hes repressors are essential regulators of hematopoietic stem cell development downstream of Notch signaling. J. Exp. Med. 2013;210:71–84. doi: 10.1084/jem.20120993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Ivanovs A., Rybtsov S., Anderson R.A., Turner M.L., Medvinsky A. Identification of the niche and phenotype of the first human hematopoietic stem cells. Stem Cell Reports. 2014;2:449–456. doi: 10.1016/j.stemcr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Yilmaz O.H., Morrison S.J. CD144 (VE-cadherin) is transiently expressed by fetal liver hematopoietic stem cells. Blood. 2005;106:903–905. doi: 10.1182/blood-2004-12-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K., Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- Lantz C.S., Boesiger J., Song C.H., Mach N., Kobayashi T., Mulligan R.C., Nawa Y., Dranoff G., Galli S.J. Role for interleukin-3 in mast-cell and basophil development and in immunity to parasites. Nature. 1998;392:90–93. doi: 10.1038/32190. [DOI] [PubMed] [Google Scholar]
- Leung A., Ciau-Uitz A., Pinheiro P., Monteiro R., Zuo J., Vyas P., Patient R., Porcher C. Uncoupling VEGFA functions in arteriogenesis and hematopoietic stem cell specification. Dev. Cell. 2013;24:144–158. doi: 10.1016/j.devcel.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Lan Y., He W., Chen D., Wang J., Zhou F., Wang Y., Sun H., Chen X., Xu C. Mouse embryonic head as a site for hematopoietic stem cell development. Cell Stem Cell. 2012;11:663–675. doi: 10.1016/j.stem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Manjunath N., Correa M., Ardman M., Ardman B. Negative regulation of T-cell adhesion and activation by CD43. Nature. 1995;377:535–538. doi: 10.1038/377535a0. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y., Sasaki Y., Nakatsuka R., Takahashi M., Iwaki R., Uemura Y., Sonoda Y. Low level of c-kit expression marks deeply quiescent murine hematopoietic stem cells. Stem Cells. 2011;29:1783–1791. doi: 10.1002/stem.721. [DOI] [PubMed] [Google Scholar]
- McKinney-Freeman S.L., Naveiras O., Yates F., Loewer S., Philitas M., Curran M., Park P.J., Daley G.Q. Surface antigen phenotypes of hematopoietic stem cells from embryos and murine embryonic stem cells. Blood. 2009;114:268–278. doi: 10.1182/blood-2008-12-193888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvinsky A., Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- Medvinsky A., Rybtsov S., Taoudi S. Embryonic origin of the adult hematopoietic system: advances and questions. Development. 2011;138:1017–1031. doi: 10.1242/dev.040998. [DOI] [PubMed] [Google Scholar]
- Mikkola H.K., Fujiwara Y., Schlaeger T.M., Traver D., Orkin S.H. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003;101:508–516. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- Moore T., Huang S., Terstappen L.W., Bennett M., Kumar V. Expression of CD43 on murine and human pluripotent hematopoietic stem cells. J. Immunol. 1994;153:4978–4987. [PubMed] [Google Scholar]
- Muller-Sieburg C.E., Sieburg H.B., Bernitz J.M., Cattarossi G. Stem cell heterogeneity: implications for aging and regenerative medicine. Blood. 2012;119:3900–3907. doi: 10.1182/blood-2011-12-376749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo R., Ciau-Uitz A., Ruiz-Herguido C., Soneji S., Bigas A., Patient R., Enver T. MiR-142-3p controls the specification of definitive hemangioblasts during ontogeny. Dev. Cell. 2013;26:237–249. doi: 10.1016/j.devcel.2013.06.023. [DOI] [PubMed] [Google Scholar]
- North T.E., de Bruijn M.F., Stacy T., Talebian L., Lind E., Robin C., Binder M., Dzierzak E., Speck N.A. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- Richard C., Drevon C., Canto P.Y., Villain G., Bollérot K., Lempereur A., Teillet M.A., Vincent C., Rosselló Castillo C., Torres M. Endothelio-mesenchymal interaction controls runx1 expression and modulates the notch pathway to initiate aortic hematopoiesis. Dev. Cell. 2013;24:600–611. doi: 10.1016/j.devcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin C., Ottersbach K., Durand C., Peeters M., Vanes L., Tybulewicz V., Dzierzak E. An unexpected role for IL-3 in the embryonic development of hematopoietic stem cells. Dev. Cell. 2006;11:171–180. doi: 10.1016/j.devcel.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Rybtsov S., Sobiesiak M., Taoudi S., Souilhol C., Senserrich J., Liakhovitskaia A., Ivanovs A., Frampton J., Zhao S., Medvinsky A. Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J. Exp. Med. 2011;208:1305–1315. doi: 10.1084/jem.20102419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.Y., Hu W., Naramura M., Park C.Y. High c-Kit expression identifies hematopoietic stem cells with impaired self-renewal and megakaryocytic bias. J. Exp. Med. 2014;211:217–231. doi: 10.1084/jem.20131128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiers G., Baumann C., O’Rourke J., Giannoulatou E., Taylor S., Joshi A., Moignard V., Pina C., Bee T., Kokkaliaris K.D. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat. Commun. 2013;4:2924. doi: 10.1038/ncomms3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiers G., Rode C., Azzoni E., de Bruijn M.F. A short history of hemogenic endothelium. Blood Cells Mol. Dis. 2013;51:206–212. doi: 10.1016/j.bcmd.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoudi S., Medvinsky A. Functional identification of the hematopoietic stem cell niche in the ventral domain of the embryonic dorsal aorta. Proc. Natl. Acad. Sci. USA. 2007;104:9399–9403. doi: 10.1073/pnas.0700984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoudi S., Morrison A.M., Inoue H., Gribi R., Ure J., Medvinsky A. Progressive divergence of definitive haematopoietic stem cells from the endothelial compartment does not depend on contact with the foetal liver. Development. 2005;132:4179–4191. doi: 10.1242/dev.01974. [DOI] [PubMed] [Google Scholar]
- Taoudi S., Gonneau C., Moore K., Sheridan J.M., Blackburn C.C., Taylor E., Medvinsky A. Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell. 2008;3:99–108. doi: 10.1016/j.stem.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Winkler I.G., Barbier V., Nowlan B., Jacobsen R.N., Forristal C.E., Patton J.T., Magnani J.L., Lévesque J.P. Vascular niche E-selectin regulates hematopoietic stem cell dormancy, self renewal and chemoresistance. Nat. Med. 2012;18:1651–1657. doi: 10.1038/nm.2969. [DOI] [PubMed] [Google Scholar]
- Woodman R.C., Johnston B., Hickey M.J., Teoh D., Reinhardt P., Poon B.Y., Kubes P. The functional paradox of CD43 in leukocyte recruitment: a study using CD43-deficient mice. J. Exp. Med. 1998;188:2181–2186. doi: 10.1084/jem.188.11.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder M.C., Hiatt K., Dutt P., Mukherjee P., Bodine D.M., Orlic D. Characterization of definitive lymphohematopoietic stem cells in the day 9 murine yolk sac. Immunity. 1997;7:335–344. doi: 10.1016/s1074-7613(00)80355-6. [DOI] [PubMed] [Google Scholar]
- Yokomizo T., Dzierzak E. Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development. 2010;137:3651–3661. doi: 10.1242/dev.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomizo T., Yamada-Inagawa T., Yzaguirre A.D., Chen M.J., Speck N.A., Dzierzak E. Whole-mount three-dimensional imaging of internally localized immunostained cells within mouse embryos. Nat. Protoc. 2012;7:421–431. doi: 10.1038/nprot.2011.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein A.C., Hofmann J.J., Lynch M., French W.J., Turlo K.A., Yang Y., Becker M.S., Zanetta L., Dejana E., Gasson J.C. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein A.C., Turlo K.A., Ponec R.M., Lynch M.R., Chen K.C., Hofmann J.J., Cox T.C., Gasson J.C., Iruela-Arispe M.L. Vascular remodeling of the vitelline artery initiates extravascular emergence of hematopoietic clusters. Blood. 2010;116:3435–3444. doi: 10.1182/blood-2010-04-279497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


