SUMMARY
Just as reference genome sequences revolutionized human genetics, reference maps of interactome networks will be critical to fully understand genotype-phenotype relationships. Here, we describe a systematic map of ~14,000 high-quality human binary protein-protein interactions. At equal quality, this map is ~30% larger than what is available from small-scale studies published in the literature in the last few decades. While currently available information is highly biased and only covers a relatively small portion of the proteome, our systematic map appears strikingly more homogeneous, revealing a “broader” human interactome network than currently appreciated. The map also uncovers significant inter-connectivity between known and candidate cancer gene products, providing unbiased evidence for an expanded functional cancer landscape, while demonstrating how high quality interactome models will help “connect the dots” of the genomic revolution.
INTRODUCTION
Since the release of a high-quality human genome sequence a decade ago (International Human Genome Sequencing Consortium, 2004), our ability to assign genotypes to phenotypes has exploded. Genes have been identified for most Mendelian disorders (Hamosh et al., 2005) and over one hundred thousand alleles have been implicated in at least one disorder (Stenson et al., 2014). Hundreds of susceptibility loci have been uncovered for numerous complex traits (Hindorff et al., 2009) and the genomes of a few thousand human tumors have been nearly fully sequenced (Chin et al., 2011). This genomic revolution is poised to generate a complete description of all relevant genotypic variations in the human population.
Genomic sequencing will however, if performed in isolation, leave fundamental questions pertaining to genotype-phenotype relationships unresolved (Vidal et al., 2011). The causal changes that connect genotype to phenotype remain generally unknown, especially for complex trait loci and cancer-associated mutations. Even when identified, it is often unclear how a causal mutation perturbs the function of the corresponding gene or gene product. To “connect the dots” of the genomic revolution, functions and context must be assigned to large numbers of genotypic changes.
Complex cellular systems formed by interactions among genes and gene products, or interactome networks, appear to underlie most cellular functions (Vidal et al., 2011). Thus, a full understanding of genotype-phenotype relationships in human will require mechanistic descriptions of how interactome networks are perturbed as a result of inherited and somatic disease susceptibilities. This in turn will require high quality and extensive genome and proteome-scale maps of macromolecular interactions such as protein-protein interactions (PPIs), protein-nucleic acid interactions, and post-translational modifiers and their targets.
First-generation binary PPI interactome maps (Rual et al., 2005; Stelzl et al., 2005) have already provided network-based explanations for some genotype-phenotype relationships, but they remain incomplete and of insufficient quality to derive accurate global interpretations (Figure S1A). There is a dire need for empirically-controlled (Venkatesan et al., 2009) high-quality proteome-scale interactome reference maps, reminiscent of the high-quality reference genome sequence that revolutionized human genetics.
The challenges are manifold. Even considering only one splice variant per gene, approximately 20,000 protein-coding genes (Kim et al., 2014; Wilhelm et al., 2014) must be handled and ~200 million protein pairs tested to generate a comprehensive binary reference PPI map. Whether such a comprehensive network could ever be mapped by the collective efforts of small-scale studies remains uncertain. Computational predictions of protein interactions can generate information at proteome scale (Zhang et al., 2012) but are inherently limited by biases in currently available knowledge used to infer such interactome models. Should interactome maps be generated for all individual human tissues using biochemical co-complex association data, or would ‘context-free’ information on direct binary biophysical interaction for all possible PPIs be preferable? To what extent would these approaches be complementary? Even with nearly complete, high-quality reference interactome maps of biophysical interactions, how can the biological relevance of each interaction be evaluated under physiological conditions? Here, we begin to address these questions by generating a proteome-scale map of the human binary interactome and comparing it to alternative network maps.
RESULTS
Vast uncharted interactome zone in literature
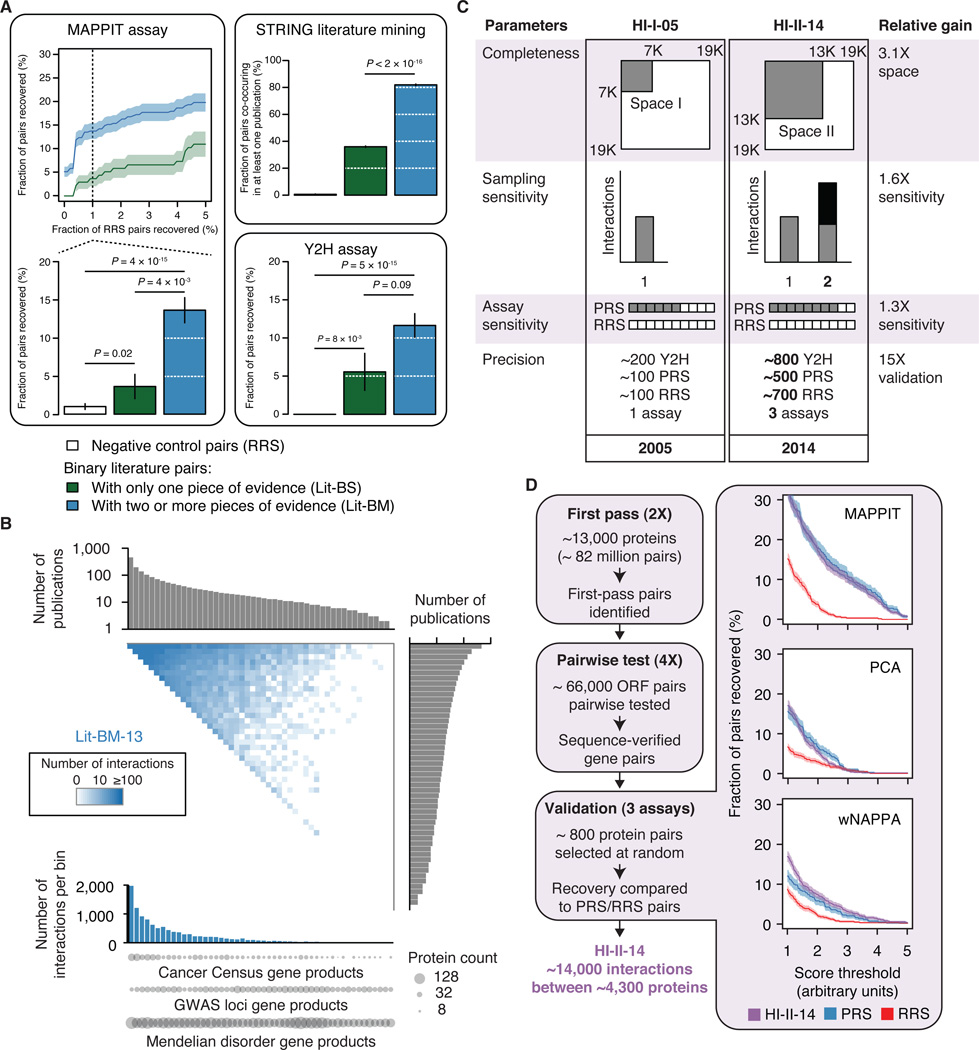
To investigate whether small-scale studies described in the literature are adequate to qualitatively and comprehensively map the human binary PPI network, we assembled all binary pairs identified in such studies and available as of 2013 from seven public databases (Figure S1B, see Extended Experimental Procedures, Section 1). Out of the 33,000 literature binary pairs extracted, two thirds were reported in only a single publication and detected by only a single method (Lit-BS pairs), thus potentially presenting higher rates of curation errors than binary pairs supported by multiple pieces of evidence (Lit-BM pairs; Tables S1A, S1B and S1C) (Cusick et al., 2009). Testing representative samples from both of these sets using the mammalian protein-protein interaction trap (MAPPIT) (Eyckerman et al., 2001) and yeast two-hybrid (Y2H) (Dreze et al., 2010) assays, we observed that Lit-BS pairs were recovered at rates that were only slightly higher than the randomly selected protein pairs used as negative control (random reference set; RRS) and significantly lower than Lit-BM pairs (Figure 1A and Table S2A; see Extended Experimental Procedures, Section 2). Lit-BS pairs co-occurred in the literature significantly less often than Lit-BM pairs as indicated by STRING literature mining scores (Figure 1A and Figure S1C; see Extended Experimental Procedures, Section 2) (von Mering et al., 2003), suggesting that these pairs were less thoroughly studied. Therefore, use of binary PPI information from public databases should be restricted to interactions with multiple pieces of evidence in the literature. In 2013 this corresponded to 11,045 high-quality protein pairs (Lit-BM-13), more than an order of magnitude below current estimates of the number of PPIs in the full human interactome (Stumpf et al., 2008; Venkatesan et al., 2009).
Figure 1. Vast uncharted interactome zone in literature and generation of a systematic binary dataset.
(A) Validation of binary literature pairs extracted from public databases (Bader et al., 2003; Berman et al., 2000; Chatr-aryamontri et al., 2013; Kerrien et al., 2012; Licata et al., 2012; Prasad et al., 2009; Salwinski et al., 2004). Fraction of pairs recovered by MAPPIT at increasing RRS recovery rates (top left) and at 1% RRS recovery rate (bottom left), found to co-occur in the literature as reported in the STRING database (upper right), and recovered by Y2H (lower right). Shading and error bars indicate standard error of the proportion. P values, two-sided Fisher’s exact tests. For n values, see Table S6.
(B) Adjacency matrix showing Lit-BM-13 interactions, with proteins in bins of ~350 and ordered by number of publications along both axes. Upper and right histograms show the median number of publications per bin. The color intensity of each square reflects the total number of interactions between proteins for the corresponding bins. Total number of interactions per bin (lower histogram). Number of gene products from GWAS loci (Hindorff et al., 2009), Mendelian disease genes (Hamosh et al., 2005) and Sanger Cancer Gene Census (Cancer Census) (Futreal et al., 2004) genes per bin (circles).
(C) Improvements from first-generation to second-generation interactome mapping based on an empirically-controlled framework (Venkatesan et al., 2009). Completeness: fraction of all pairwise protein combinations tested; Assay sensitivity: fraction of all true biophysical interactions that are identifiable by a given assay; Sampling sensitivity, fraction of identifiable interactions that are detected in the experiment; Precision: fraction of reported pairs that are true positives.
(D) Experimental pipeline for identifying high-quality binary protein-protein interactions (left). ORF: Open Reading Frame. Fraction of HI-II-14, PRS and RRS pairs (right) recovered by MAPPIT, PCA and wNAPPA at increasing assay stringency. Shading indicates standard error of the proportion. P > 0.05 for all assays when comparing PRS and HI-II-14 at 1% RRS, twosided Fisher’s exact tests. For n values, see Table S6.
See also Figures S1 and S2 and Tables S1 and S2.
The relatively low number of high-quality binary literature PPIs may reflect inspection biases inherent to small-scale studies. Some genes such as RB1 are described in hundreds of publications while most have been mentioned only in a few (e.g. the un-annotated C11orf21 gene). To investigate the effect of such biases on the current coverage of the human interactome network, we organized the interactome search space by ranking proteins according to the number of publications in which they are mentioned (Figure 1B). Interactions between highly studied proteins formed a striking “dense zone” in contrast to a large sparsely populated zone, or “sparse zone”, involving poorly studied proteins. Candidate gene products identified in genome-wide association studies (GWAS) or associated with Mendelian disorders distribute homogeneously across the publication-ranked interactome space (Figure 1B and Figure S1D), demonstrating a need for unbiased systematic PPI mapping to cover this uncharted territory.
A proteome-wide binary interactome map
Based on literature-curated information, the human interactome appears to be restricted to a narrow dense zone, suggesting that half of the human proteome participates only rarely in the interactome network. Alternatively, the zone that appears sparse in the literature could actually be homogeneously populated by PPIs that have been overlooked due to sociological or experimental biases.
To distinguish between these possibilities and address other fundamental questions outlined above, we generated a new proteome-scale binary interaction map. By acting on all four parameters of our empirically-controlled framework (Venkatesan et al., 2009), we increased the coverage of the human binary interactome with respect to our previous human interactome dataset obtained by investigating a search space defined by ~7,000 protein-coding genes (“Space I”) and published in 2005 (HI-I-05) (Rual et al., 2005) (Figures 1C and 1D; see Extended Experimental Procedures, Section 3). A search space consisting of all pairwise combinations of proteins encoded by ~13,000 genes (“Space II”; Table S2B) was systematically probed, representing a 3.1-fold increase with respect to the HI-I-05 search space. To gain in sensitivity, we performed the Y2H assay in different strain backgrounds that showed increased detection of pairs of a positive reference set (PRS) composed of high-quality pairs from the literature without increasing the detection rate of RRS pairs. To increase our sampling, the entire search space was screened twice independently. Pairs identified in this first pass were subsequently tested pairwise in quadruplicate starting from fresh yeast colonies. To ensure reproducibility, only pairs testing positive at least three times out of the four attempts and with confirmed identity were considered interacting pairs, resulting in ~14,000 distinct interacting protein pairs.
We validated these binary interactions using three binary protein interaction assays that rely on different sets of conditions than the Y2H assay: i) reconstituting a membrane-bound receptor complex in mammalian cells using MAPPIT, ii) in vitro using the well-based nucleic acid programmable protein array (wNAPPA) assay (Braun et al., 2009; Ramachandran et al., 2008), and iii) reconstituting a fluorescent protein in Chinese hamster ovary cells using a protein-fragment complementation assay (PCA) (Nyfeler et al., 2005) (see Extended Experimental Procedures, Section 4). The Y2H pairs exhibited validation rates that were statistically indistinguishable from a PRS of ~500 Lit-BM interactions while significantly different from an RRS of ~700 pairs with all three orthogonal assays and over a large range of score thresholds (Figure 1D, Tables S2A and S2C), demonstrating the quality of the entire dataset. Using three-dimensional co-crystal structures available for protein complexes in the Protein Data Bank (Berman et al., 2000) and for domain-domain interactions (Stein et al., 2011) (Figure S2, Tables S2D, S2E and S2F; see Extended Experimental Procedures, Sections 5 and 6), we also demonstrated that our binary interactions reflect direct biophysical contacts, a conclusion in stark contrast to a previous report suggesting that Y2H interactions are inconsistent with structural data (Edwards et al., 2002). Our results also suggested that Y2H sensitivity correlates with the number of residue-residue contacts and thus presumably with interaction affinity. The corresponding human interactome dataset covering Space II and reported in 2014 (HI-II-14; Table S2G) is the largest experimentally-determined binary interaction map yet reported, with 13,944 interactions amongst 4,303 distinct proteins.
Overall biological significance
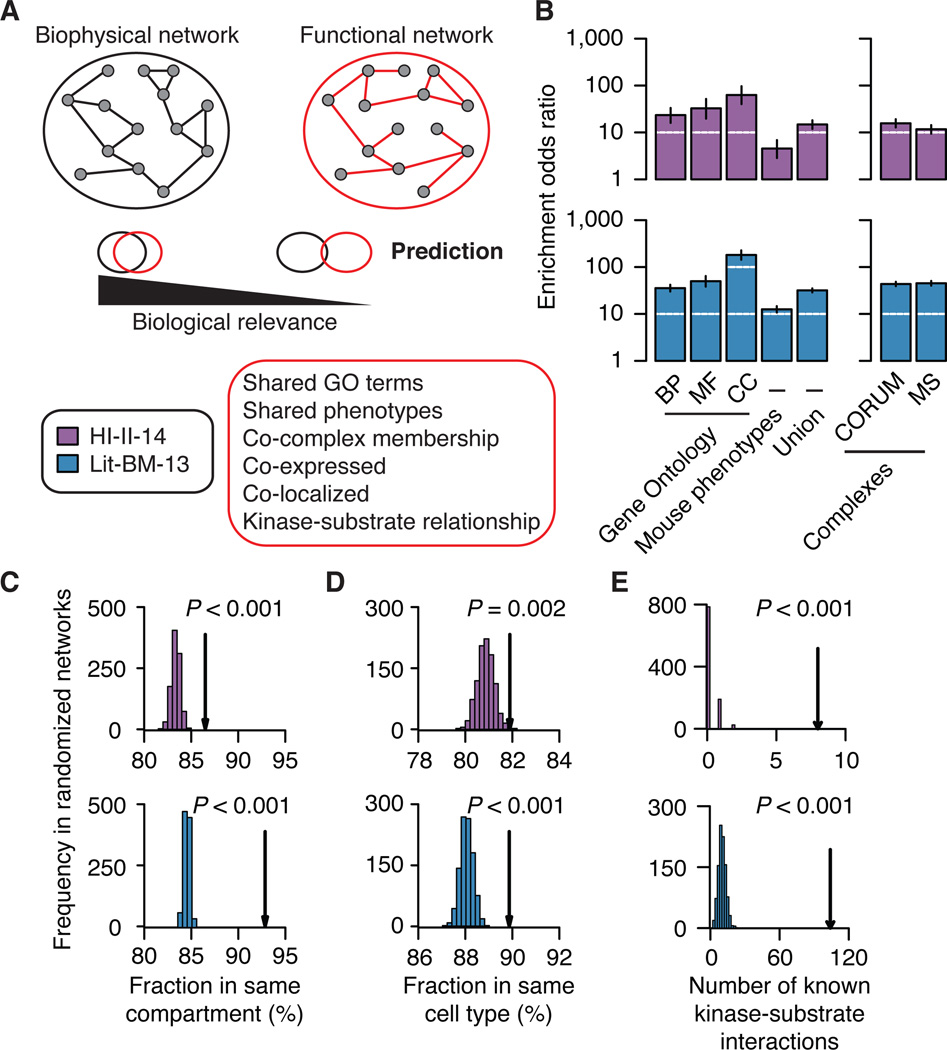
To assess the overall functional relevance of HI-II-14, we combined computational analyses with a large-scale experimental approach. We first measured enrichment for shared Gene Ontology (GO) terms and phenotypic annotations and observed that HI-II-14 shows significant enrichments that are similar to those of Lit-BM-13 (Figures 2A and 2B; see Extended Experimental Procedures, Section 7). Second, we measured how much binary interactions from HI-II-14 reflect membership in larger protein complexes as annotated in CORUM (Ruepp et al., 2010) or reported in a co-complex association map (Woodsmith and Stelzl, 2014). In both cases, we observed a significant enrichment for binary interactions between protein pairs that belong to a common complex (P < 0.001; Figure 2B). Third, we performed a similar analysis using tissue-specific mRNA expression data across the 16 human tissues of the Illumina Human Body Map 2.0 project as well as cellular compartment localization annotations from the GO Slim terms. Again, HI-II-14 was enriched for interactions mediated by protein pairs present in at least one common compartment or cell type (Figures 2C and 2D). Finally, we measured the overlap of HI-II-14 with specific biochemical relationships, as represented by kinase-substrate interactions. Both HI-II-14 and Lit-BM-13 contained significantly more PPIs reflecting known kinase-substrate relationships (Hornbeck et al., 2012) than the corresponding degree-controlled randomized networks (Figure 2E). In addition, HI-II-14 tends to connect tyrosine and serine/threonine kinases (Manning et al., 2002) to proteins with tyrosine or serine/threonine phospho-sites (Hornbeck et al., 2012; Olsen et al., 2010), respectively (Figure S3A), pointing to the corresponding interactions being genuine kinase-substrate interactions. In short, our systematic interactome map, which was generated independently from any pre-existing biological information, reveals functional relationships at levels comparable to those seen for the literature-based interaction map.
Figure 2. Overall biological significance.
(A) Schematic of the method to assess biological relevance of binary maps.
(B) Enrichment of binary interactome maps for functional relationships (left) and co-complex memberships (right). Error bars indicate 95% confidence intervals. BP: Biological process, MF: Molecular function, CC: Cellular component. Mouse phenotypes: Shared phenotypes in mouse models by orthology mapping. MS: Mass-spectrometry based map. Enrichments: P ≤ 0.05 for all annotations and maps, two-sided Fisher’s exact tests. For n values, see Table S6.
(C) and (D) Fraction of binary interactions between proteins localized in a common cellular compartment and proteins co-present in at least one cell type (arrows) compared to those in 1,000 degree-controlled randomized networks. Empirical P values. For n values, see Table S6.
(E) Number of known kinase-substrate interactions found in binary maps (arrows) compared to those in 1,000 randomized networks. Empirical P values are shown. See also Figure S3.
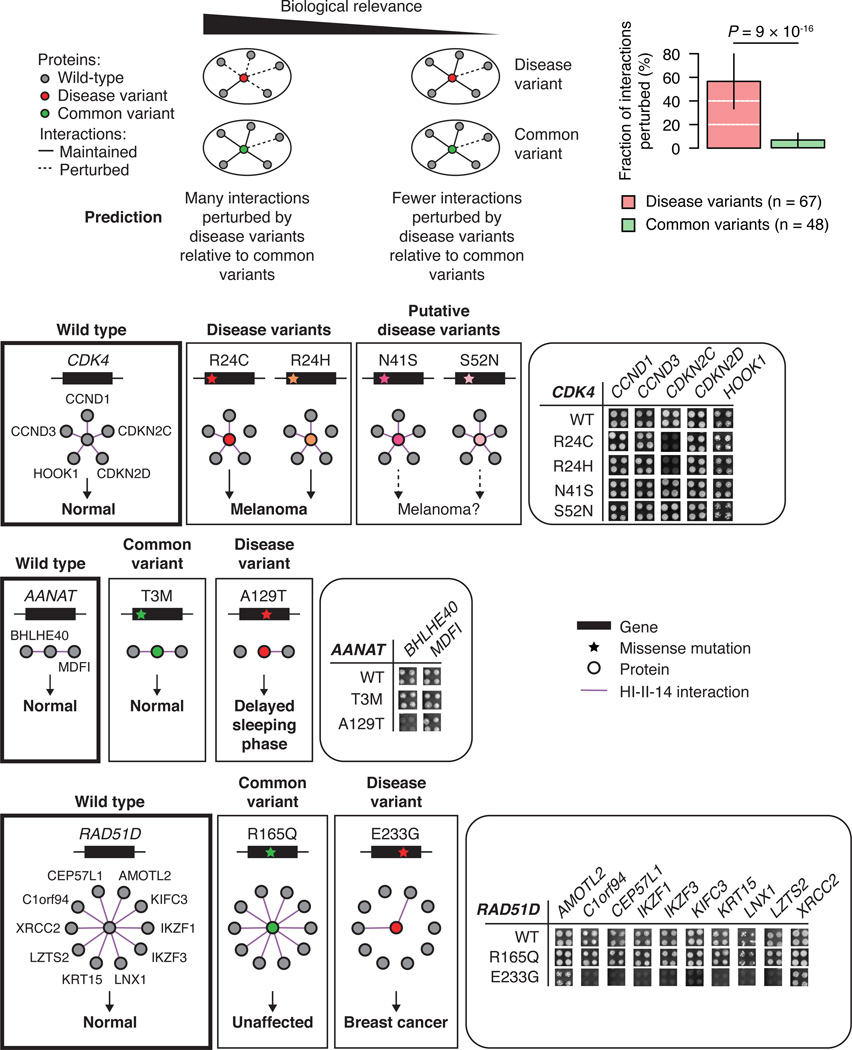
To further investigate the overall biological relevance of HI-II-14, we used an experimental approach that compares the impact of mutations associated with human disorders to that of common variants with no reported phenotypic consequences on biophysical interactions (Figure 3). Our rationale is that a set of interactions corresponding to genuine functional relationships should more likely be perturbed by disease-associated mutations than by common variants. The following example will illustrate this concept. Mutations R24C and R24H in CDK4 are clearly associated with melanoma by conferring resistance to CDKN2A inhibition (Wölfel et al., 1995), whereas N41S and S52N mutations are of less clear clinical significance (Zhong et al., 2009) and have remained functionally uncharacterized. HI-II-14 contains five CDK4 interactors: two inhibitors (CDKN2C and CDKN2D), two cyclins (CCND1 and CCND3), and HOOK1, a novel interacting partner and a potential phosphorylation target of CDK4 (Figure S3B). In agreement with previous reports the comparative interaction profile shows that R24C and R24H, but not N41S and S52N, specifically perturb CDK4 binding to CDKN2C (Figure 3).
Figure 3. Perturbations of protein interactions by disease and common variants.
Fraction of interactions of the wild-type gene product lost by mutants bearing the disease-associated or common variants (top right, error bars indicate standard error of the proportion). P value, two-sided Fisher’s exact test. Comparison of interaction profile of wild-type CDK4, AANAT, and RAD51D to the interaction profiles of mutant bearing disease or common variants (bottom). Yeast growth phenotypes on SC-Leu-Trp-His+3AT media in quadruplicate experiments are shown.
In total, we identified 32 human genes for which: (i) the corresponding gene product is reported to have binary interactors in HI-II-14, (ii) germline disease-associated mutations have been reported, and (iii) common coding variants unlikely to be involved in any disease have been identified in the 1,000 genomes project (1000 Genomes Project Consortium, 2012). To avoid over-representation of certain genes, we selected a total of 115 variants, testing up to 4 disease and 4 common variants per disease gene for their impact on the ability of the corresponding proteins to interact with known interaction partners (see Extended Experimental Procedures, Section 8). Disease variants were 10-fold more likely to perturb interactions than non-disease variants (Figure 3 and Table S3). Strikingly, more than 55% of the 107 HI-II-14 interactions tested were perturbed by at least one disease-associated variant, and the same trend was observed when considering only mutants with evidence of expression in yeast as indicated by their ability to mediate at least one interaction (Figure S3C). Examples of novel specifically-perturbed interactions include AANAT-BHLHE40 and RAD51D-IKZF1 (Figure 3). In the first case, the A129T mutation in AANAT is known to be associated with a delayed sleeping phase syndrome, and specifically perturbs an interaction between AANAT and BHLHE40, the product of a gene reported to function in circadian rhythm regulation (Nakashima et al., 2008). In the second case, the breast cancer associated RAD51D E233G mutation perturbs interactions with a number of partners, including the known cancer gene product IKZF1 (Futreal et al., 2004).
Altogether these computational and experimental results provide strong evidence that HI-II-14 pairs correspond to biologically relevant interactions and represent a valuable resource to further our understanding of the human interactome and its perturbations in human disease.
A “broader” interactome
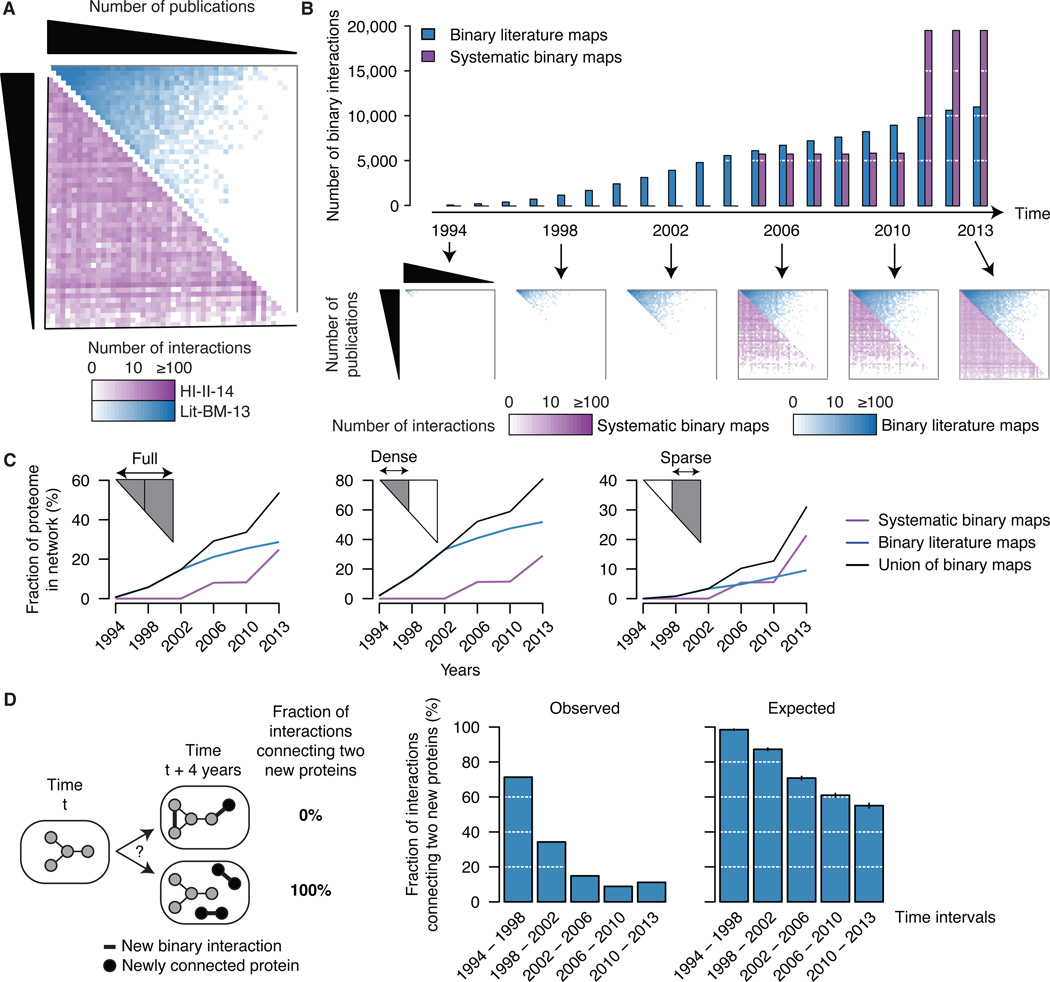
Unlike literature-curated interactions, HI-II-14 protein pairs are distributed homogeneously across the interactome space (Figure 4A), indicating that sociological biases, and not fundamental biological properties, underlie the existence of a densely populated zone in the literature. Since 1994, the number of high-quality binary literature PPIs has grown roughly linearly to reach ~10,000 interactions in 2013 (Figure 4B), while systematic datasets are punctuated by a few large-scale releases. Although the sparse territory of the literature map gradually gets populated, interaction density in this zone continues to lag behind that of the dense zone (Figure 4B). In terms of proteome coverage, the expansion rate is faster for systematic maps than for literature maps, especially in the sparse territory (Figure 4C and Figure S4A; see Extended Experimental Procedures, Section 9). While Lit-BM-13 provides more information in the dense zone, HI-II-14 reveals interactions for more than 2,000 proteins absent from Lit-BM-13. These observations are likely due to a tendency of the literature map to expand from already connected proteins (Figure 4D).
Figure 4. A “broader” interactome.
(A) Adjacency matrices showing Lit-BM-13 (blue) and HI-II-14 (purple) interactions, with proteins in bins of ~350 and ordered by number of publications along both axes. The color intensity of each square reflects the total number of interactions for the corresponding bins.
(B) Total number of binary interactions in literature and systematic interactome maps over the past 20 years (top), with years reflecting either date of public release of systematic binary datasets or date of publication that resulted in inclusion of interactions in Lit-BM-13. Adjacency matrices (bottom) as in Figure 4A.
(C) Fraction of the human proteome present in binary interactome maps at selected time points since 1994, considering the full interactome space (left) or only dense (middle) and sparse (right) zones of Lit-BM-13 with respect to number of publications.
(D) Fraction of new interactions connecting two proteins that were both absent from the map at the previous time point (four years interval; middle) compared to 1,000 randomized networks (right).
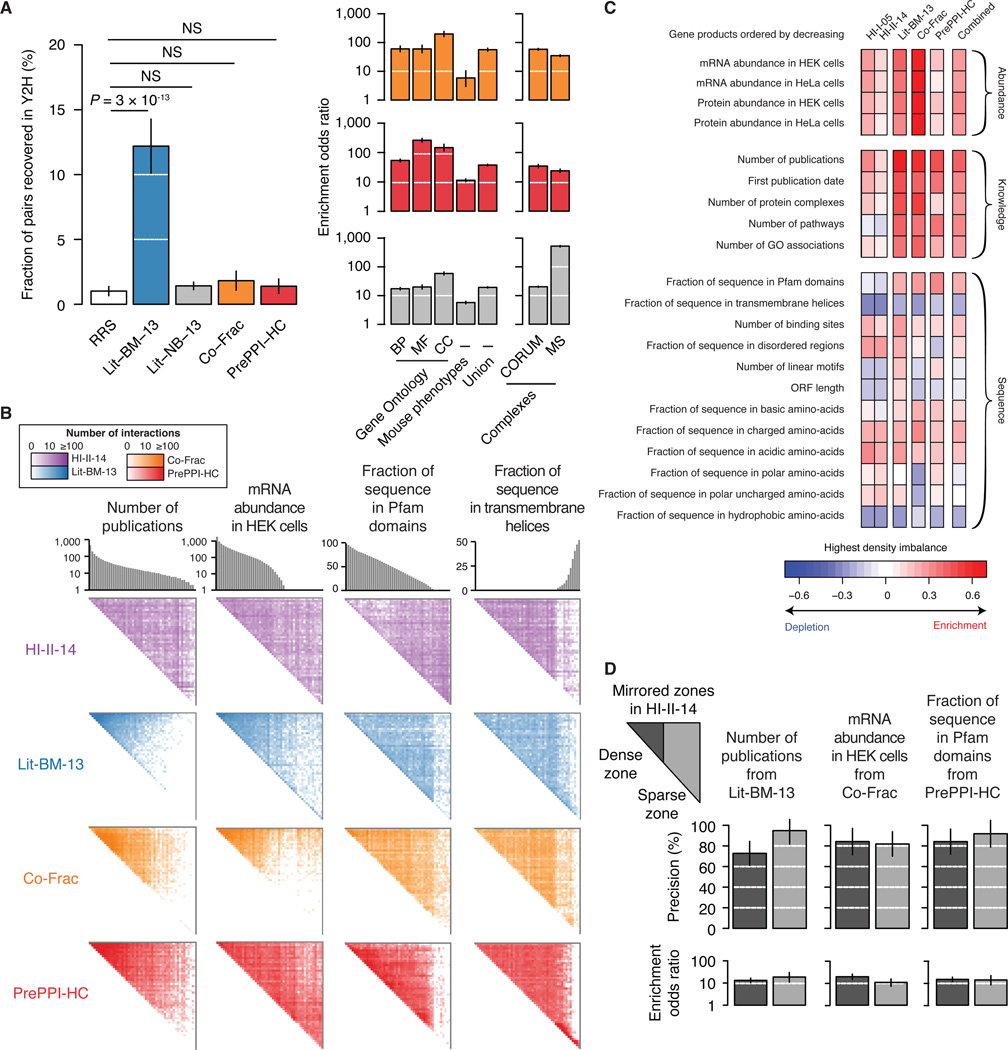
To more deeply explore the heterogeneous coverage of the human interactome, we compared HI-II-14 and Lit-BM-13 to a collection of ~25,000 predicted binary PPIs of high-confidence (PrePPI-HC) (Zhang et al., 2012) and a co-fractionation map of ~14,000 potentially binary interactions (Co-Frac) (Havugimana et al., 2012). We tested the extent to which these two datasets contain binary interactions (see Extended Experimental Procedures, Section 10). Representative samples from both Co-Frac and PrePPI-HC were recovered by Y2H at a much lower rate than a sample of Lit-BM-13 and appeared statistically indistinguishable from random pairs (Figure 5A and Table S4A). A literature non-binary dataset (Lit-NB-13) performed similarly. However, Co-Frac and PrePPI-HC, like Lit-NB-13, were both significantly enriched for functionally relevant relationships. Thus, although these datasets represent potentially valuable resources, both Co-Frac and PrePPI-HC appear to be more comparable to non-binary than to binary datasets. Surprisingly, even though PrePPI-HC and Co-Frac systematically surveyed the full human proteome and map different portions of the interactome (Figures S4B), both exhibit a strong tendency to report interactions amongst well-studied proteins (Figure 5B). This bias is likely due to the integration of functional annotations in the generation of both datasets.
Figure 5. Comparison of interaction mapping approaches.
(A) Evaluation of the quality of Co-Frac, PrePPI-HC and pairs from small-scale experiments in the literature with no binary evidence (Lit-NB-13). Fraction of pairs recovered by Y2H as compared to pairs from Lit-BM-13 and pairs of randomly selected proteins (RRS) (left). Enrichment in functional interactions and co-complex memberships (right). Legend as in Figure 2B. For n values, see Table S6.
(B) Adjacency matrices for HI-II-14, Lit-BM-13, Co-Frac and PrePPI-HC maps, with proteins per bins of ~350 and ordered by number of publications, mRNA abundance in HEK cells, fraction of protein sequence covered by Pfam domains, or fraction of protein sequence in transmembrane helices. Figure legend as in Figure 1B.
(C) Highest interaction density imbalances (observed minus expected) are shown in the four maps, the union of all four maps, and our previous binary map (HI-I-05) for 21 protein properties.
(D) Precision at 1% RRS recovery in the MAPPIT assay (top, error bars indicate standard error of the proportion) and functional enrichment (bottom, union of Gene Ontology and mouse phenotypes based annotations, error bars indicate 95% confidence intervals) of HI-II-14 pairs found in dense and sparse zones mirrored from Lit-BM-13, Co-Frac and PrePPI-HC. P > 0.05 for all pairwise comparisons of dense and sparse zones, two-sided Fisher’s exact tests. For n values, see Table S6.
Because coverage might depend on gene expression levels, we also examined interactome maps for expression-related sparse versus dense zones. Co-Frac shows a strong bias towards interactions involving proteins encoded by genes highly expressed in the cell lines used (Figure 5B). This expression-dependent bias is echoed in the literature map, perhaps reflecting a general tendency to study highly expressed proteins. In contrast, both HI-II-14 and PrePPI-HC exhibit a uniform interaction density across the full spectrum of expression levels, likely explained by the standardized expression of proteins tested in Y2H and by the independence of homology-based predictions from expression levels.
We more broadly explored the intrinsic biases that might influence the appearance of sparsely populated zones by examining 21 protein or gene properties, roughly classified as expression-, sequence-, or knowledge-based (Figures 5B and 5C, Tables S4B and S4C; see Extended Experimental Procedures, Section 9). For example, PrePPI-HC is virtually devoid of interactions between proteins lacking Pfam domains, consistent with conserved domains forming the basis of the prediction method. HI-II-14 appears depleted of interactions amongst proteins containing predicted transmembrane helices, consistent with expected limitations of the Y2H assay (Stagljar and Fields, 2002). Co-Frac is similarly depleted in interactions involving proteins with transmembrane helices, which may result from membrane-bound proteins being filtered out during biochemical fractionations. Compared to HI-II-14, HI-I-05 presented a less homogenous coverage of the space with respect to abundance and knowledge properties, likely reflecting the content of early versions of the hORFeome (Figure S4C). Importantly, no single map appeared unbiased in all 21 examined properties. A combined map presented a slightly increased homogeneity although intrinsic knowledge biases of the three maps using literature-derived evidence were still predominant.
To confirm that HI-II-14 interactions found in the sparse zones of the three other maps are of as high quality as those found in dense zones, we compared MAPPIT validation rates and functional enrichment across these zones for all protein properties examined. MAPPIT validation rates of dense and sparse zone pairs were consistent for nearly all properties (Figure 5D and Figure S4D), indicating that HI-II-14 interactions are of similar biophysical quality throughout the full interactome space. Functional enrichment within the sparse zone was statistically indistinguishable from that of the dense zone (Figure 5D and Figure S4E), demonstrating the functional importance of HI-II-14 biophysical interactions in zones covered sparsely by other types of interactome maps.
Considering all current maps, more than half of the proteome is now known to participate in the interactome network. Our systematic exploration of previously uncharted territories dramatically expands the interactome landscape, suggesting that the human interactome network is broader in scope than previously observed, and that the entire proteome may be represented within a fully mapped interactome.
Interactome network and cancer landscape
Genes associated with the same disease are believed to be preferentially inter-connected in interactome networks (Barabási et al., 2011; Vidal et al., 2011). However, in many cases, these observations were made with interactome maps that are composites of diverse evidence, e.g. binary PPIs, co-complex memberships and functional associations, a situation further complicated by the uneven quality and sociological biases described above. Using HI-II-14, we revisited this concept for cancer gene products. Our goal was to investigate whether the cancer genomic landscape is limited to the known cancer genes curated in the Sanger Cancer Gene Census (Futreal et al., 2004) (“Cancer Census”), or if, alternatively, it might extend to some of the hundreds of additional candidate genes enriched in somatic mutations uncovered by systematic cancer genome sequencing (“SM genes”) (Chin et al., 2011) and/or identified by functional genomic strategies such as Sleeping Beauty transposon-based screens in mice (“SB genes”) (Copeland and Jenkins, 2010) or global investigations on DNA tumor virus targets (“VT genes”) (Rozenblatt-Rosen et al., 2012).
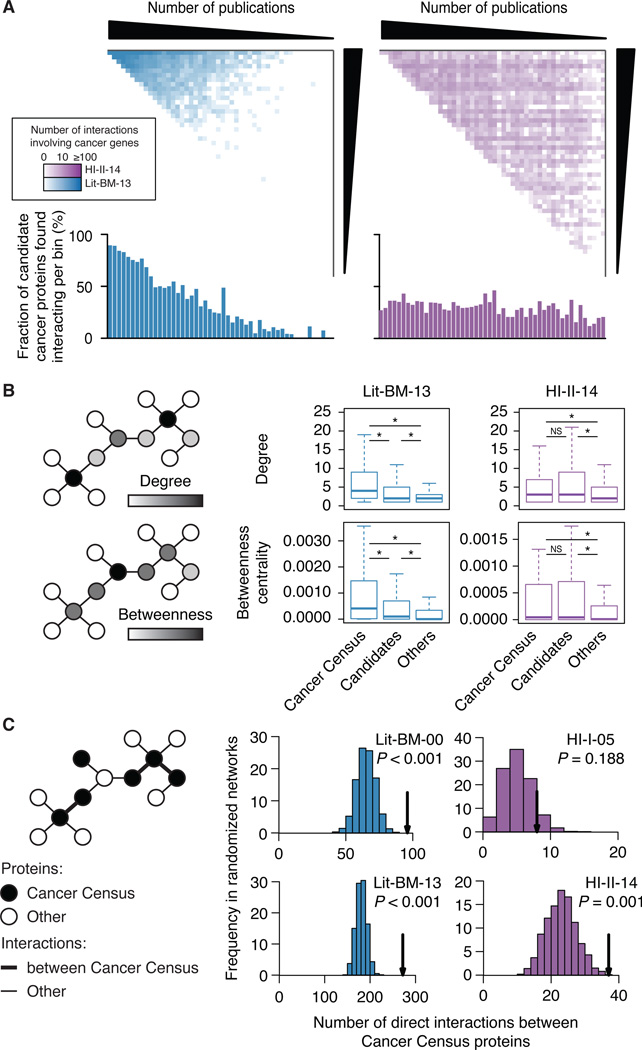
Given our homogeneous coverage of the space for known (Cancer Census genes) and candidate (SB, SM and VT genes) cancer genes (Figure 6A), we first tested the postulated central role of cancer gene products in biological networks (Barabási et al., 2011) and verified that both sets tend to have more interactions and to be more central in the systematic map than proteins not associated with cancer (Figure 6B). We then examined the inter-connectivity of known cancer proteins and showed that Cancer Census gene products interact with each other more frequently than expected by chance, a trend not apparent in HI-I-05 (Figure 6C). We sought to use this topological property as the basis for novel cancer gene discovery in the large lists of cancer candidates from genomic and functional genomic screens.
Figure 6. Network properties of cancer gene products.
(A) Adjacency matrices for Lit-BM-13 and HI-II-14 only showing interactions involving the product of a Cancer Census (Futreal et al., 2004) or of a candidate cancer gene. Figure legend as in Figure 1B. Lower histograms show for each bin, the fraction of cancer candidates having at least one interaction.
(B) Distribution of the number of interactions (degree) and normalized number of shortest paths between proteins (betweenness centrality) for products of Cancer Census and of candidate cancer genes in Lit-BM-13 and in HI-II-14 maps as compared to other proteins (right; * for P < 0.05, NS for P > 0.05, two-sided Wilcoxon rank sum tests). For n values, see Table S6.
(C) Number of interactions between products of Cancer Census genes (arrows) in HI-I-05, HI-II-14, Lit-BM as of 2000 (Lit-BM-00) and as of 2013 (Lit-BM-13), as compared to 1,000 degree-controlled randomized networks. Empirical P values. For n values, see Table S6.
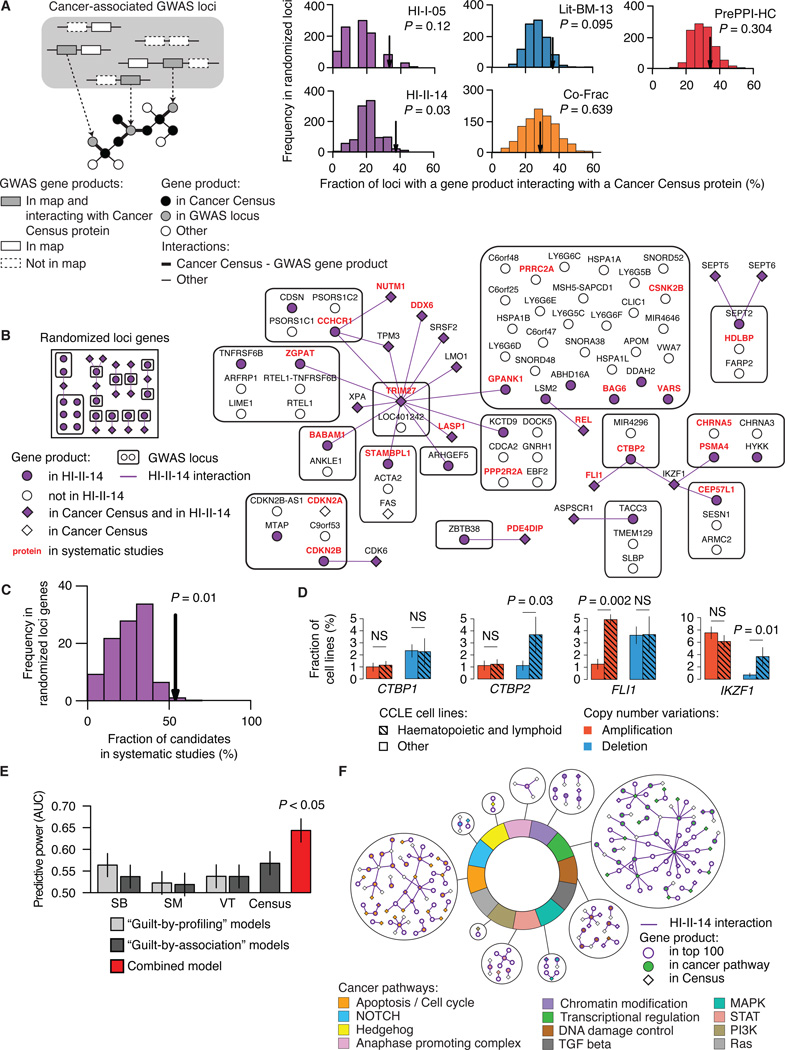
We examined whether products of candidate cancer genes identified in GWAS (Table S5A) tend to be connected to Cancer Census proteins, and observed significant connectivity in all four maps (Figure S5A; see Extended Experimental Procedures, Section 11). When loci containing a known cancer gene were excluded, only HI-II-14 showed such connectivity, supporting its unique value to identify cancer candidate genes beyond those already well demonstrated (Figure 7A and Figure S5A). In further support of their association with cancer, genes in cancer GWAS loci prioritized by “guilt-by-association” in HI-II-14 tend to correspond to cancer candidates from systematic cancer studies (Figures 7B and 7C). These results suggest that cancer-associated proteins tend to form subnetworks perturbed in tumorigenesis, and that HI-II-14 provides new context to prioritize cancer genes from genome-wide studies.
Figure 7. Interactome network and cancer landscape.
(A) Fraction of cancer-related GWAS loci containing at least one gene encoding a protein that interacts with the product of a Cancer Census gene in HI-I-05, HI-II-14, Lit-BM-13, Co-Frac and PrePPI-HC (arrows) as compared to randomly selected loci genes. GWAS loci already containing a Cancer Census gene are excluded. Empirical P values. For n values, see Table S6.
(B) Network representing products of genes in cancer-associated GWAS loci and their interactions with Cancer Census proteins in HI-II-14 (right), and a representative example of the network obtained for randomized loci genes (left).
(C) Fraction of GWAS loci gene products interacting with a Cancer Census protein also identified in systematic genomic and functional genomic studies (arrow) as compared to the fraction obtained for randomized loci genes (bottom right). Empirical P value.
(D) CTBP2 and IKZF1 are deleted in significantly more haematopoietic and lymphoid cancer cell lines than in other cancer cell lines. CCLE, Cancer Cell Line Encyclopedia. Each barplot compares the fraction of cell lines from the 163 haematopoietic and lymphoid (hatched bars) or 717 other (empty bars) cell types where CTBP1, CTBP2, FLI1 or IKZF1 were found amplified (red) or deleted (blue). P values, two-sided Fisher’s exact tests (NS for P > 0.05).
(E) Predictive power of guilt-by-profiling and guilt-by-association models compared to the combined model (Figure S6; see Extended Experimental Procedures, Section 11). AUC: Area under the curve in Figure S6C. P value, two-sided Wilcoxon rank sum test. SB, Sleeping Beauty transposon-based mouse cancer screen; SM, Somatic mutation screen in cancer tissues; VT, Virus targets.
(F) Binary interactions from HI-II-14 involving the top candidates and Cancer Census gene products in the twelve pathways associated to cancer development and progression. See also Figures S5, S6 and S7 and Table S5.
The following example illustrates the power of our combined approach. C-terminal Binding Protein 2 (CTBP2) is encoded at a locus associated with prostate cancer susceptibility (Thomas et al., 2008) and belongs to both SB and VT gene lists (Mann et al., 2012; Rozenblatt-Rosen et al., 2012). Two Cancer Census genes, IKZF1 and FLI1, encode interacting partners of CTBP2 in HI-II-14. These are transcription factors with tumor suppressor (Payne and Dovat, 2011) and proto-oncogene (Kornblau et al., 2011) roles, respectively, in lymphoid tumors. Given its interactions with IKZF1 and FLI1, we investigated the potential role of CTBP2 in lymphoid tumorigenesis. In the Cancer Cell Line Encyclopedia (Barretina et al., 2012), FLI1 was significantly more often amplified in lymphoid than in other cell lines (Figure 7D), consistent with its proposed proto-oncogenic role in these tumors. In contrast, both CTBP2 and IKZF1, but not CTBP1, were deleted significantly more often in lymphoid cancer cell lines. Notably, deletion of CTBP2 or IKZF1 and amplification of FLI1 were mostly non-overlapping in the different cell lines, suggesting that either event may be sufficient to affect tumorigenesis (Figure S5B). Altogether, these results suggest a role for CTBP2 in suppressing lymphoid tumors by direct repression of FLI1 function, potentially involving IKZF1.
Finally, we assessed how HI-II-14 interactions can be integrated with genomic and functional genomic datasets. Going beyond the “guilt-by-profiling” concept, we also used these gene sets in “guilt-by-association” predictions in a combined model (Figure S6A), which leads to substantially improved cancer gene rankings over those found using either predictive strategy alone (Figure 7E, Figures S6B and S6C, Table S5B; see Extended Experimental Procedures, Section 12). In contrast, a similar analysis using HI-I-05 interactions showed that its limited size prevented inclusion of any guilt-by-association terms (Figure S6D). Genes significantly mutated in cancer patients from recent TCGA pan-cancer mutation screens (Table S5C) (Lawrence et al., 2014) were enriched amongst highly ranked predictions from the combined model (P = 6 × 10−3, one-sided Wilcoxon rank test), supporting the validity of our integrated cancer gene predictions. Our top-ranked prediction was the cyclin-dependent kinase 4 (CDK4), a well-known cancer gene product. Four other genes from the Cancer Census list appeared among the top 25 ranked genes. Strikingly, STAT3, which ranked third, was added to the Cancer Census after our training set was established, highlighting the ability of this approach to identify novel cancer gene products.
To characterize the biological processes in which the candidate cancer genes predicted by the combined model are likely to be involved, we identified binary interactions linking them to each other or to Cancer Census proteins in the twelve ‘pathways of cancer’ relevant to cancer development and progression (Table S5D) (Vogelstein et al., 2013). Of our top 100 candidates, 60 mapped to at least one cancer pathway (Figure 7F and Figure S7), twice as many as would be expected from predictions using either the guilt-by-profiling or guilt-by-association approach alone. We propose that many novel cancer candidates can be annotated to specific processes based on their interactions with Cancer Census gene products and known participation in cellular pathways. For example, the candidate protein ID3, a DNA-binding inhibitor, interacts with the two Cancer Census transcription factors TCF12 and TCF3, suggesting a role for ID3 in the regulation of transcription by inhibiting binding of specific transcription factors to DNA (Loveys et al., 1996; Richter et al., 2012). CTBP2, which we identified as a potential suppressor in lymphoid tumors, represents another example (Figure 5E and Figure S7).
In summary, the increased and uniform coverage of HI-II-14 demonstrates that known and candidate cancer gene products are highly connected in the interactome network, which in turn provides unbiased evidence for an expanded functional cancer landscape.
DISCUSSION
By systematically screening half of the interactome space with minimal inspection bias, we more than doubled the number of high-quality binary PPIs available from the literature. Covering zones of the human interactome landscape that have been weakly charted by other approaches, our systematic binary map provides deeper functional context to thousands of proteins, as demonstrated for candidates identified in unbiased cancer genomic screens. Systematic binary mapping therefore stands as a powerful approach to “connect the dots” of the genomic revolution.
Combining high-quality binary pairs from the literature with systematic binary maps, 30,000 high-confidence interactions are now available. It is likely that a large proportion of the human interactome can soon be mapped by taking advantage of the emergence of reference proteome maps (Kim et al., 2014; Wilhelm et al., 2014), a combination of nearly complete clone collections (Yang et al., 2011), rapid improvements in Y2H assay sensitivity and emerging interaction-mapping technologies that drastically reduce cost (Caufield et al., 2012; Stagljar and Fields, 2002; Yu et al., 2011).
Reference binary interactome maps of increased coverage and quality will be required to interpret condition-specific interactions and to characterize the effects of splicing and genetic variation on interactions (Zhong et al., 2009). While protein-protein interactions represent an important class of interactions between macromolecules, future efforts integrating this information with protein-DNA, protein-RNA, RNA-RNA or protein-metabolite interactions will provide a unified view of the molecular interactions governing cell behaviour. Just as a reference genome enabled detailed maps of human genetic variation (1000 Genomes Project Consortium, 2012), completion of a reference interactome network map will enable deeper insight into genotype-phenotype relationships in human.
EXPERIMENTAL PROCEDURES
Extraction of the literature-based datasets
Human PPIs annotated with tractable publication records were extracted from seven databases through August 2013. Large-scale systematic datasets and pairs involving products of UBC, SUMO1, SUMO2, SUMO3, SUMO4 or NEDD8, were excluded. The remaining pairs were divided into those having no piece of binary evidence (Lit-NB) and those with at least one piece of binary evidence based on PSI-MI experimental method codes. Binary ones were divided between pairs with one and two or more pieces of evidence (Lit-BS and Lit-BM, respectively). For benchmark experiments in Y2H and MAPPIT, equivalent datasets were extracted similarly in December 2010.
Generation of the binary protein-protein interaction map
HI-II-14 was generated by screening all pairwise combinations of 15,517 ORFs from hORFeome v5.1 (Space II) as described previously (Dreze et al., 2010). ORFs encoding first pass pairs were identified either by Sanger or by Stitch-seq (Yu et al., 2011). HI-II-14 was validated by comparing a subset of 809 interactions to a positive and a random reference set of 460 and 698 protein pairs, respectively, using MAPPIT, PCA and wNAPPA assays.
Interaction perturbation by disease and common variants
Disease variants were obtained from the Human Gene Mutation Database (HGMD 2009 V2) (Stenson et al., 2014) and common variants were derived from the 1,000 genomes project (1000 Genomes Project Consortium, 2012). Only variants with a minor allele frequency above 1% were considered common. All successfully cloned disease and common variants were systematically tested for interaction with all interactors of their wild-type counterpart.
Interaction density imbalance
For each protein property, we ranked all proteins and, for any property threshold, partitioned the interactome space into a first region containing pairs of proteins both above (or below) the threshold, and a second region containing all remaining pairs. Interaction density imbalance of a given PPI map for a given threshold was calculated as the fraction of interactions observed in the first region minus the fraction of PPIs expected assuming a uniform distribution in the space. Dense and sparse zones were defined by identifying the threshold for which the deviation from expectation is maximal.
Measure of functional enrichment
For each pairwise comparison, PPI and functional maps were trimmed to interactions where both proteins were present in both maps and restricted to Space II to allow comparison between PPI maps. Functional enrichment odds ratios were calculated using Fisher’s exact tests.
GWAS analysis
307 distinct cancer-associated SNPs were identified from 75 GWAS publications covering 10 types of cancer and 142 distinct loci were identified at a linkage disequilibrium threshold of 0.9. For each map, we calculated the number of loci encoding an interactor of a Cancer Census protein over the number of loci encoding a protein in the PPI map. To assess significance, we measured the corresponding fraction when randomly selecting for each locus the same number of proteins than genes with products in the PPI map.
Cancer association scoring system
For each gene, 7 features were measured. Three features represent membership in the SB, SM and VT lists of candidate cancer genes (“guilt-by-profiling” features). The four other features represent its number of interactors in HI-II-14 that are present in these three lists and in the Cancer Census list, normalized by the expected numbers in degree-controlled randomized networks (“guilt-by-association” features). We measured the ability of each feature to prioritize known Cancer Census genes with separate logistic regression models. We combined all seven features in a forward stepwise logistic regression model using the Akaike information criterion to determine the stepwise halting. The final set of features selected was: the SB, SM and VT guilt-by-profiling and the Cancer Census and SB guilt-by-association features. “Receiver Operating Characteristic” curves were obtained by measuring at decreasing score threshold the fraction of known Cancer Census genes recovered and the corresponding fraction of proteins predicted as candidate cancer genes.
Datasets
For reference datasets used in this study, see Extended Experimental Procedures, Section 13. All high-quality binary PPIs described in this paper can be accessed on this website: http://interactome.dfci.harvard.edu/H_sapiens/
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to acknowledge past and present members of the Center for Cancer Systems Biology (CCSB) and particularly H. Yu for helpful discussions. This work was supported primarily by NHGRI grant R01/U01HG001715 awarded to M.V., D.E.H., F.P.R. and J.T. and in part by the following grants and agencies: NHGRI P50HG004233 to M.V., F.P.R. and A.-L.B.; NHLBI U01HL098166 subaward to M.V.; NHLBI U01HL108630 subaward to A.-L.B.; NCI U54CA112962 subaward to M.V.; NCI R33CA132073 to M.V.; NIH RC4HG006066 to M.V., D.E.H. and T.H.; NICHD ARRA R01HD065288, R21MH104766 and R01MH105524 to L.M.I.; NIMH R01MH091350 to L.M.I. and T.H.; NSF CCF-1219007 and NSERC RGPIN-2014-03892 to Y.X.; Canada Excellence Research Chair, Krembil Foundation, Ontario Research Fund–Research Excellence Award, Avon Foundation, Canadian Institute for Advanced Research Fellowship to F.P.R.; grant CSI07A09 from Junta de Castilla y Leon (Valladolid, Spain), grant PI12/00624 from Ministerio de Economia y Competitividad (AES 2012, ISCiii, Madrid, Spain,) and grant i-Link0398 from Consejo Superior de Investigaciones Científicas (CSIC, Madrid, Spain) to J.D.L.R.; Spanish Ministerio de Ciencia e Innovación (BIO2010-22073) and the European Commission through the FP7 project SyStemAge grant agreement n:306240 to P.A.; Group-ID Multidisciplinary Research Partnerships of Ghent University, grant FWO-V G.0864.10 from the Fund for Scientific Research-Flanders and ERC Advanced Grant N° 340941 to J.T.; EMBO long-term fellowship to A.K.; Institute Sponsored Research funds from the Dana-Farber Cancer Institute Strategic Initiative to M.V. I.L. is a postdoctoral fellow with the FWO-V. M.V. is a "Chercheur Qualifié Honoraire" from the Fonds de la Recherche Scientifique (FRS-FNRS, Wallonia-Brussels Federation, Belgium). Since performing the work described, Ce.F. has become an employee of Celgene Research SL, part of the Celgene Corporation.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information includes Extended Experimental Procedures, 7 figures, 6 tables.
AUTHOR CONTRIBUTIONS
Computational analyses were performed by T.R., M.T., B.C., S.J.P., Ce.F., R.M., A.K. and S.D.G. with help from A.-R.C., J.C.-H., Ch.F., E.F., M.J., S.K., G.N.L., K.L., J.M., Am.S., and Y.S. Experiments were performed by Q.Z., N.S., S.Y., I.L., X.Y. and L.G. with help from D.B., B.E.B., P.B., M.B., M.P.B., D.C.-Z., R.C., E.D., M.D., A.D., F.G., B.J.G., M.F.H., R.K., A.M., R.R.M., A.P., M.M.P., X.R., J.R., P.R., V.R., E.R., J.M.S., An.S., A.A.S., K.S., S.T., A.O.T., S.A.W., J.-C.T., K.V., and J.W. Structural analyses were done by T.R., M.T. and R.M. Extraction of the literature datasets was performed by Ce.F., A.K., Ch.F., M.E.C, and T.H. MAPPIT validation was done by I.L. The adjacency matrix interactome representation was developed by S.J.P. with M.T. Functional enrichment analysis was done by T.R., M.T. and B.C. Interaction perturbation experiments were performed by N.S. and S.Y. with Q.Z. Interactome and proteome coverage analyses were done by T.R. and B.C. Comparison of alternative maps was done by T.R., M.T., B.C. and S.J.P. The density imbalance measure was conceived by M.T. Topological analyses were done by T.R., M.T., B.C., S.J.P. and S.D.G. Cancer-related analyses were done by T.R., M.T. and B.C. The cancer association scoring system was done by M.T. CTBP2 and cancer landscape analyses were done by T.R. Interactome mapping was supervised by D.E.H. and M.V. Principal investigators overseeing primary data management, structural biology, literature recuration and reference set construction, MAPPIT validation, and other computational analyses were T.H., P.A., J.D.L.R., J.T., and F.P.R., respectively. B.C., Y.X., A.-L.B., L.M.I., P.A., J.D.L.R., J.T., M.A.C., D.E.H., T.H., F.P.R. and M.V. designed and/or advised the overall research effort. T.R., M.T., B.C., Q.Z., M.E.C., J.D.L.R., M.A.C., D.E.H., T.H., F.P.R. and M.V. wrote the manuscript with contributions from other co-authors.
The authors declare no competing financial interest.
REFERENCES
- 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun P, Taşan M, Dreze M, Barrios-Rodiles M, Lemmens I, Yu H, Sahalie JM, Murray RR, Roncari L, de Smet AS, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods. 2009;6:91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caufield JH, Sakhawalkar N, Uetz P. A comparison and optimization of yeast two-hybrid systems. Methods. 2012;58:317–324. doi: 10.1016/j.ymeth.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C, Nixon J, Ramage L, Kolas N, O'Donnell L, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Hahn WC, Getz G, Meyerson M. Making sense of cancer genomic data. Genes Dev. 2011;25:534–555. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA. Harnessing transposons for cancer gene discovery. Nat. Rev. Cancer. 2010;10:696–706. doi: 10.1038/nrc2916. [DOI] [PubMed] [Google Scholar]
- Cusick ME, Yu H, Smolyar A, Venkatesan K, Carvunis A-R, Simonis N, Rual JF, Borick H, Braun P, Dreze M, et al. Literature-curated protein interaction datasets. Nat. Methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreze M, Monachello D, Lurin C, Cusick ME, Hill DE, Vidal M, Braun P. High-quality binary interactome mapping. Methods Enzymol. 2010;470:281–315. doi: 10.1016/S0076-6879(10)70012-4. [DOI] [PubMed] [Google Scholar]
- Edwards AM, Kus B, Jansen R, Greenbaum D, Greenblatt J, Gerstein M. Bridging structural biology and genomics: assessing protein interaction data with known complexes. Trends Genet. 2002;18:529–536. doi: 10.1016/s0168-9525(02)02763-4. [DOI] [PubMed] [Google Scholar]
- Eyckerman S, Verhee A, der Heyden JV, Lemmens I, Ostade XV, Vandekerckhove J, Tavernier J. Design and application of a cytokine-receptor-based interaction trap. Nat. Cell Biol. 2001;3:1114–1119. doi: 10.1038/ncb1201-1114. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblau SM, Qiu YH, Zhang N, Singh N, Faderl S, Ferrajoli A, York H, Qutub AA, Coombes KR, Watson DK. Abnormal expression of FLI1 protein is an adverse prognostic factor in acute myeloid leukemia. Blood. 2011;118:5604–5612. doi: 10.1182/blood-2011-04-348052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveys DA, Streiff MB, Kato GJ. E2A basic-helix-loop-helix transcription factors are negatively regulated by serum growth factors and by the Id3 protein. Nucleic Acids Res. 1996;24:2813–2820. doi: 10.1093/nar/24.14.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann KM, Ward JM, Yew CC, Kovochich A, Dawson DW, Black MA, Brett BT, Sheetz TE, Dupuy AJ, et al. Australian Pancreatic Cancer Genome Initiative. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5934–5941. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Nakashima A, Kawamoto T, Honda KK, Ueshima T, Noshiro M, Iwata T, Fujimoto K, Kubo H, Honma S, Yorioka N, et al. DEC1 modulates the circadian phase of clock gene expression. Mol. Cell. Biol. 2008;28:4080–4092. doi: 10.1128/MCB.02168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyfeler B, Michnick SW, Hauri HP. Capturing protein interactions in the secretory pathway of living cells. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6350–6355. doi: 10.1073/pnas.0501976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Payne KJ, Dovat S. Ikaros and tumor suppression in acute lymphoblastic leukemia. Crit. Rev. Oncog. 2011;16:3–12. doi: 10.1615/critrevoncog.v16.i1-2.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TSK, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran N, Raphael JV, Hainsworth E, Demirkan G, Fuentes MG, Rolfs A, Hu Y, LaBaer J. Next-generation high-density self-assembling functional protein arrays. Nat. Methods. 2008;5:535–538. doi: 10.1038/nmeth.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J, Schlesner M, Hoffmann S, Kreuz M, Leich E, Burkhardt B, Rosolowski M, Ammerpohl O, Wagener R, Bernhart SH, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat. Genet. 2012;44:1316–1320. doi: 10.1038/ng.2469. [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J-F, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes—2009. Nucleic Acids Res. 2010;38:D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagljar I, Fields S. Analysis of membrane protein interactions using yeast-based technologies. Trends Biochem. Sci. 2002;27:559–563. doi: 10.1016/s0968-0004(02)02197-7. [DOI] [PubMed] [Google Scholar]
- Stein A, Ceol A, Aloy P. 3did: identification and classification of domain-based interactions of known three-dimensional structure. Nucleic Acids Res. 2011;39:D718–D723. doi: 10.1093/nar/gkq962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf MP, Thorne T, de Silva E, Stewart R, An HJ, Lappe M, Wiuf C. Estimating the size of the human interactome. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- Venkatesan K, Rual J-F, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, et al. An empirical framework for binary interactome mapping. Nat. Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabási A-L. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- Wölfel T, Hauer M, Schneider J, Serrano M, Wölfel C, Klehmann-Hieb E, De Plaen E, Hankeln T, Meyer zum Büschenfelde KH, Beach D. A p16(INK4a)-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- Woodsmith J, Stelzl U. Studying post-translational modifications with protein interaction networks. Curr. Opin. Struct. Biol. 2014;24:34–44. doi: 10.1016/j.sbi.2013.11.009. [DOI] [PubMed] [Google Scholar]
- Yang X, Boehm JS, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, Green TM, et al. A public genome-scale lentiviral expression library of human ORFs. Nat. Methods. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Tardivo L, Tam S, Weiner E, Gebreab F, Fan C, Svrzikapa N, Hirozane-Kishikawa T, Rietman E, Yang X, et al. Next-generation sequencing to generate interactome datasets. Nat. Methods. 2011;8:478–480. doi: 10.1038/nmeth.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Simonis N, Li QR, Charloteaux B, Heuze F, Klitgord N, Tam S, Yu H, Venkatesan K, Mou D, et al. Edgetic perturbation models of human inherited disorders. Mol. Syst. Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.