Graphical abstract
Abbreviations: Top, DNA topoisomerase; CL, cutaneous leishmaniasis; VL, visceral leishmaniasis; DALYs, disability-adjusted life years; kDNA, kinetoplast DNA; NTD, neglected tropical diseases; NGO, non-governmental organization; lk, linking number; NLS, Nuclear Localization Signal
Keywords: Kinetoplastids, Tritryps, Topoisomerases, DNA topology, Target-based drug discovery, Chemotherapy
Highlights
-
•
There is an urgent need of new treatments against trypanosomatids-borne diseases.
-
•
DNA topoisomerases are pointed as potential drug targets against unicellular parasites.
-
•
Trypanosomatids have a full set of DNA topoisomerases in both nucleus and kinetoplast.
-
•
TopII and TopIII are located in the kinetoplast and fully involved in kDNA replication.
-
•
Tritryps TopIB differ in structure from mammalian’s pointing to an attractive target.
Abstract
The Trypanosomatidae family, composed of unicellular parasites, causes severe vector-borne diseases that afflict human populations worldwide. Chagas disease, sleeping sickness, as well as different sorts of leishmaniases are amongst the most important infectious diseases produced by Trypanosoma cruzi, Trypanosoma brucei and Leishmania spp., respectively. All these infections are closely related to weak health care services in low-income populations of less developed and least economically developed countries. Search for new therapeutic targets in order to hit these pathogens is of paramount priority, as no effective vaccine is currently in use against any of these parasites. Furthermore, present-day chemotherapy comprises old-fashioned drugs full of important side effects. Besides, they are prone to produce tolerance and resistance as a consequence of their continuous use for decades. DNA topoisomerases (Top) are ubiquitous enzymes responsible for solving the torsional tensions caused during replication and transcription processes, as well as in maintaining genomic stability during DNA recombination. As the inhibition of these enzymes produces cell arrest and triggers cell death, Top inhibitors are among the most effective and most widely used drugs in both cancer and antibacterial therapies. Top relaxation and decatenation activities, which are based on a common nicking–closing cycle involving one or both DNA strands, have been pointed as a promising drug target. Specific inhibitors that bind to the interface of DNA-Top complexes can stabilize Top-mediated transient DNA breaks. In addition, important structural differences have been found between Tops from the Trypanosomatidae family members and Tops from the host. Such dissimilarities make these proteins very interesting for drug design and molecular intervention. The present review is a critical update of the last findings regarding trypanosomatid’s Tops, their new structural features, their involvement both in the physiology and virulence of these parasites, as well as their use as promising targets for drug discovery.
1. Introduction
The family Trypanosomatidae (order Kinetoplastida) comprises several unicellular flagellated-protozoan parasites responsible for many human infectious diseases in low-income populations of less developed and least economically developed countries (WHO, 2008). Two species of the genus Trypanosoma, Trypanosoma brucei (subspecies gambiense and rhodesiense) and Trypanosoma cruzi, as well as the different species of the genus Leishmania, are collectively known as the “tritryps”. They are responsible for some of the most harmful neglected diseases that were tackled by WHO in the 2003 meeting held in Berlin. Human African trypanosomiasis, sleeping sickness or Congo trypanosomiasis is a lethal infection caused by T. brucei, which is transmitted by tse–tse flies (Glossina) in Sub-Saharan African countries. Sleeping sickness is a biphasic disease. The first phase is clinically characterized by the presence of fever, headache, joint pain and itching. Whilst in the second one, which can be lethal if left untreated, the parasite causes anaemia and neurological symptoms owing to the aggressive pathogen-invasion of the host central nervous system (Lejon et al., 2013). American trypanosomiasis (Chagas disease), caused by T. cruzi in Central and South American countries, is transmitted by the faeces of blood-sucking “kissing bugs” of the subfamily Triatominae. In addition, indirect transmission through blood transfusions or organ transplantation from infected patients has also been reported. Chagas disease symptoms may vary during the course of infection. Local swelling can be noticed in early stages, but disease chronification leads to a cardiomyopathy, which may result in sudden death. The occurrence of damage in the gastrointestinal tract is also very common, with the presence of megacolon and megaesophagus, thereby leading to an inevitable weight loss (Teixeira et al., 2011). Leishmaniasis is transmitted by sandflies of the genera Phlebotomus and Lutzomyia. Cutaneous leishmaniasis (CL), whose etiological agent is Leishmania major in the Old World – among other species – and Leishmania panamensis and Leishmania braziliensis in the New World, should be considered the mildest clinical presentation of this disease. CL presents with erythema, edema and permanent ulcers at the bite site. Although CL is not fatal, it is extremely disfiguring when located on the host’s face. American tegumentary leishmaniasis (also known as mucocutaneous leishmaniasis) arises as a consequence of an inadequately treated New-World CL (WHO, 2007). This clinical form presents with severe facial disfiguration, including nose and lips, which can derive to cancer processes. Last, there is a very aggressive visceral form of leishmaniasis (VL) produced by Leishmania donovani in Asia and Leishmania infantum in the countries of the Mediterranean basin. VL produces organ swelling (specifically targeting liver and spleen) and may be deadly if left untreated (Ashford, 2000; Murray et al., 2005; Antinori et al., 2012).
The impact of these diseases can be measured in terms of prevalence, fatal outcomes, as well as disability-adjusted life years (DALYs) lost as a direct consequence of both pathologic effect and treatments (Goto and Lindoso, 2010). Sleeping sickness is an infection exclusively concentrated in South-Saharan countries. Nowadays, this scourge is prevalent in 36 countries, whose endemic infection involves around 60-million people at risk (Aksoy et al., 2003). The number of reported cases was estimated to be ca. 10,000 but it represented only one-third of the real infections occurred in 2009. In addition, an estimated annual DALYs lost of 1.7-million has a profound impact in the economy of these countries (Simarro et al., 2011). For its part, Chagas disease has presence in 18 Central and South American countries with 17 million individuals currently infected and 100 million people at risk. Statistical annual records show the emergence of 41,200 new cases every year, as well as 12,500 related deaths. Chagas calculated DALYs reach annual values over 570,000 (Moncayo and Silveira, 2009). Lastly, leishmaniases are prevalent in 98 countries with presence in four continents (Oceania is the only one free of these maladies) with approximately 360-million people at risk. Chagas disease has resurged in USA and Europe due to the incoming tourism and migratory fluxes from prevalent countries of South America (Schmunis, 2007; Gascon et al., 2010). The annual estimation of leishmaniasis is close to 1.6 new million cases each year, 20,000–40,000 of which become fatal. Over 50% of the 2-million worldwide-appraised DALYs is mainly focused in South-East Asia, with Bihar (India) as the epicenter of Leishmania-related deaths (Alvar et al., 2012). VL is a re-emerging opportunistic disease in immunosuppressed patients owing to three main facts: (i) the immunosuppressive effect of drugs used in organ transplantation; (ii) as a consequence of HIV infections in South-European countries or (iii) climate change due to global warming (Alvar et al., 2008; van Griensven et al., 2014; Koltas et al., 2014).
Although prevention is considered the best approach to fight against these diseases, any possible vaccine is in a very early stage of development. The main reason is that these infections cause a deregulation of the host immune system. In fact, it has been shown that both human patients, as well as experimental animals, become susceptible to infections caused by these pathogens when Th1/Th2 balance shifts toward Th2-mediated humoral immunity (Alexander and Bryson, 2005). However, when there is a shift toward Th1-dominant immune responses the host becomes resistant and develops (life-span) natural immunity against re-infection. Nowadays, some new therapies take advantage of this circumstance to treat these infectious diseases by means of Th1 stimulants or auto vaccines developed from infected hosts. However, these promising approaches are being only developed for Leishmania-infected dogs but not for humans, making chemotherapy the most feasible alternative against these neglected tropical diseases (NTDs) at present (for a comprehensive review see: Palatnik-de-Sousa, 2012).
Current chemotherapy against these diseases was developed many years ago, is plenty of toxic side effects and it often requires parenteral treatments. These inconveniences make the use of this chemotherapy in field conditions difficult, and since the administration of repeated doses is needed, the appearing of relapses is extremely frequent due to a poor adherence to treatments (Leprohon et al., 2011). African trypanosomiasis is a good example of these statements. Pentamidine (an aromatic diamidine) and suramin are the chosen drugs against the first blood-stage infection (Burri, 2010). However, when the neurological phase is established, only arsenicals (melarsoprol) or the ornithine decarboxylase suicide inhibitor eflornithine can ameliorate the symptoms (Lutje et al., 2013). Recently, the combination of nifurtimox and eflornithine has prompted the change from the old-fashioned melarsoprol to the best treatment available (Simarro et al., 2012). For the Chagas disease, the current pharmacopoeia is reduced to two drugs, nifurtimox and benznidazole, with a 60–90 days long treatment and with noticeable side effects (Rassi et al., 2010).
First choice drugs against leishmaniases are old-fashioned pentavalent antimonium-based drugs (Glucantime and Pentostam), which are the current therapy against these diseases since the 50s all over the World (Balaña-Fouce et al., 1998). These drugs have several flaws, including the need of parenteral administration, the occurrence of many undesirable side effects, as well as the appearing of resistances due to the over-usage (Croft and Olliaro, 2011). Drug combinations of antimonials with allopurinol (a xanthine oxidase inhibitor) very much improve the curative outcomes (Manna et al., 2009). In a second instance, macrolide antibiotics such as Amphotericin B, are a good alternative to the former compounds (Monge-Maillo and López-Vélez, 2013). This antibiotic formulated in liposomes (AmBisome) has resulted in a good therapy against VL, but it is extremely expensive for developing countries and requires intravenous administration (Mohamed-Ahmed et al., 2012). Lipid-ether compounds – miltefosine and edelfosine – are promising oral drugs first synthesized as anti-tumourals but with potent anti-leishmanial effect (Bhattacharya et al., 2007; Berman, 2008). For its part, paromomycin – an aminoglycoside antibiotic – has been approved in 2006 in India against VL (Sundar et al., 2007). These compounds require repeated dosing during 20 days or more, and with the exception of miltefosine, they must be parenterally administered to the patients.
At this point, the need of new drugs against these diseases is clearly justifiable (Barrett and Croft, 2012). However, since NTDs mostly affect low-income countries, the expectable economic profits of the big pharmaceutical trusts are negligible compared to those obtained with other pathologies of the more developed World. Therefore, basic academic research groups have assumed new drug target discovery and drug design research. Interestingly, drug discovery initiatives (i.e., DNDi), promoted by world-wide NGOs and supported by public and private funds are aimed to eliminate NTDs during the current decade. Meanwhile, Glaxo Smith Kline has done the economic effort of offering thousands of compounds from different drug libraries to be tested by basic research groups in order to find new hits against NTDs pathogens.
Within the novel targets for drug intervention, DNA topoisomerases (Top) have attracted attention since their discovery. The role played by Top in cell physiology is of paramount importance and since the beginning many research groups pointed to these enzymes as putative targets against proliferative processes. This is the reason why some Top inhibitors are being marketed against cancer (Pommier et al., 2010) or as promising antiparasitic drugs (Das et al., 2008; García-Estrada et al., 2010).
In the present review we update novel information about Top enzymes in trypanosomatids, making special emphasis in the novel findings on their peculiar structure as well as in the molecules used to target them.
2. DNA topoisomerases
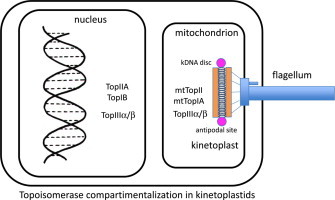
Since the length of the DNA helix containing the genetic information of an organism is several millions times longer than the size of the cellular nucleus, it is a physical pre-requisite to fold it thousands of times in order to fit it inside this organelle (Annunziato, 2008). DNA packaging can be solved by simple folding of the circular chromosome in prokaryotic organisms, or can be condensed around nucleosomes, where histones play a key role in gene expression and structure of eukaryotic chromosomes. Trypanosomatids are ancient eukaryotes that contain a unique disc-shaped extranuclear DNA called kinetoplast inside their single mitochondrion. Adjacent to the flagellar basal body there is an intricate mesh of thousands of interlocked DNA rings called mini- and maxicircles that represents 10 to 20% of the total DNA in the cell (Shapiro and Englund, 1995; Chen et al., 1995a). Minicircles are the most abundant form in kDNA (5.000–10.000 molecules per cell). They are circular DNA molecules with a length perimeter of 100–2500 bp encoding guide RNAs involved in editing mitochondrial nascent RNAs (Stuart and Panigrahi, 2002). Maxicircles are bigger in perimeter – 30,000–50,000 bp – but less abundant than minicircles – 50 copies per cell. Maxicircles are the equivalent of mitochondrial DNA from other eukaryotes, as they codify mitochondrial proteins, ribosomal RNA and tRNAs (Shlomai, 2004). Replication of kDNA occurs synchronously with genomic DNA during cell division. kDNA components must be decatenated from the network before their DNA replication and then catenated again to the nascent mesh when this process is finished (Liu et al., 2005). DNA replication from both mini- and maxicircles takes place in theta (θ)-shaped intermediaries, where Top play a key role (Jensen and Englund, 2012). Since kinetoplast is unique to trypanosomatids, it has been pointed as an important target for drug discovery.
DNA appears in nature as underwound form, acquiring a compact negatively supercoiled organization that is not accessible to most physiological cell processes when needed. For this reason, during DNA replication or transcription to RNA, as well as during DNA repair, DNA supercoils should be resolved to a more relaxed form that permits access to polymerases, transcriptional factors or DNA repair proteins. Top are the proteins responsible for relaxing supercoiled DNA introducing breaks in single or both strands of the double helix (Champoux and Dulbecco, 1972). At this moment, DNA can rotate freely alleviating the tension accumulated by supercoiling, powering the rotation of one strand over the other. This task can be fuelled by ATP hydrolysis or not. Since these enzymes are molecular machines, a support of energy should be necessary to exert the mechanical function. According to these characteristics; the number of DNA strand breaks introduced and the requirement of metabolic energy, Top are divided into two Families. Type I Top (TopI) are ATP-independent enzymes that break one DNA strand, whereas Type II Top (TopII) need ATP hydrolysis to break both DNA strands (Champoux, 2001). Differences in the mode of action suppose differences in changes introduced in DNA topology. TopI are solely involved in the relaxation of supercoiled DNA (Cretaio et al., 2007). TopII, however, are not only involved in supercoiled DNA relaxation, but they are also implicated in the catenation/decatenation and knotting/unknotting of complex DNA topological networks (for reviews see: Wang, 1998, 2002, 2009; Champoux, 2001; Forterre et al., 2007; Pommier et al., 2010).
TopIA was considered, since long time ago, as a prokaryotic-borne enzyme that resolves positive supercoils through the transient covalent binding to the 5′-end of the broken DNA strand followed by passage and rejoining (Corbett and Berger, 2004). Regardless TopIA, TopIB is a true Mg2+-independent eukaryotic enzyme that introduces both positive and negative supercoils, establishing a temporary covalent bond to the 3′-end of cut DNA releasing a 5′-OH free end (Cheng et al., 1998). The catalytic Tyr at the active site of TopIB introduces a nucleophilic attack on a phosphate group of the nucleotide chain, creating a transient phosphodiester bond with the broken DNA strand (Stewart et al., 1998; Pommier, 2009). This allows TopIB to change the linking number (Lk) in multiples of 1 (n), unlike TopIA, which introduces changes strictly in steps of one in the Lk of a single-stranded circular DNA (Lindsley, 2005).
TopII are multimeric proteins that break both DNA strands fuelled by the hydrolysis of ATP. TopII bind transiently to the 5′-ends of both DNA strands relaxing positive and negative supercoils, changing the Lk in steps of +2 (Watt and Hickson, 1994). The transient break at both DNA strands permits the passage of another duplex through the gap and subsequent rejoining. TopII enzymes relax positively supercoiled DNA, but also have additional catenation/decatenation and knotting/unknotting activities that make them needed for chromosomal segregation in the nucleus and for kDNA disassembly in individual minicircles in the kinetoplast during cell division (Chen et al., 1995b).
A common feature to all Top enzymes is the presence of a Tyr residue in the catalytic site of the enzyme (Champoux, 2001). Tyr is the amino acid that produces a nucleophilic attack to the phosphodiester DNA backbone introducing a transient covalent bond with the enzyme. This transient bonding has been exploited to design compounds able to target it. Since the time required to religate cleaved DNA is extremely short, many compounds act stabilizing this enzymatic step introducing an enzyme-DNA adduct that undergoes irreversible damage (Hsiang et al., 1985). When the replication fork collides with one of these stabilized complexes, a double-strand break can be produced, starting the irreversible fragmentation (Hsiang et al., 1989). Since the stabilization of DNA-Top complexes can have a natural origin, eukaryotic cells have developed a DNA repair systems that act removing these tyrosyl-DNA bonds. Tyrosyl-DNA-phosphodiesterase-1 (TDP1) has been regarded as a potential therapeutic co-target of TopIB (Dexheimer et al., 2008; Banerjee et al., 2010).
A full set of Top enzymes is currently available in the TriTryp Genome database http://tritrypdb.org/tritrypdb/; (i) nuclear TopIB and TopIIA enzymes involved in DNA replication and repair that have been fully described as putative drug targets; (ii) mitochondrial TopIA and TopII that have been described closely linked to kDNA replication; (iii) two putative members of the TopIA subfamily TopIII (α and β) involved in resolving DNA entanglements that can arise during DNA recombination (Table 1).
Table 1.
Tritryps Top-encoding genes annotated into GeneDB database (http://www.genedb.org/Homepage). TopIA, TopIB (large and small subunits), TopII and TopIII genes from different species: L. major Friendlin, T. brucei 927 and T.cruzi.
| Protein | L. major Friendlin | T. brucei 927 | T. cruzi |
|---|---|---|---|
| GeneDB database | |||
| TopIA | LmjF21.0125 | Tb927.10.1900 | TcCLB.510121.160 |
| TopIB (L) | LmjF34.3440 | Tb927.4.1330 | TcCLB.508693.20 |
| TopIB (S) | LmjF04.0060 | Tb927.9.6940 | TcCLB.506625.110 |
| TopII | LmjF28.2280 | Tb927.11.11550 Tb927.11.11560 |
TcCLB.508699.10 TcCLB.509203.70 |
| mtTopII | LmjF.15.1290 | Tb927.9.5590 | TcCLB.508277.370 |
| TopIII | LmjF28.1780 LmjF36.3200 |
Tb927.11.9170 Tb927.11.9520 |
TcCLB.510901.100 TcCLB.511589.120 TcCLB.508851.170 |
2.1. Type IB topoisomerases
Unlike other enzymes characterized to date, kinetoplastid TopIB is a heterodimer (Reguera et al., 2008). The first description was done in L. infantum where the functional bisubunit enzyme was composed of a large (L) subunit of 635 amino acids with a MW of 73 kDa (LiTopIL), which contains the N-terminal and core domains, and a small (S) subunit of 262 amino acids with a MW of 28 kDa (LiTopIS), which retains the Tyr catalytic residue. Genes encoding these proteins are located on different chromosomes in the leishmanial genome (Villa et al., 2003). In this very year, similar results were published in African trypanosomes using a classical platform of protein purification. However, a controversy about the oligomeric status (L2S2) is still under debate (Bodley et al., 2003). It seems that this unique genomic organization, in which the genes encoding both TopIB protomers are located on different chromosomes, occurs solely in trypanosomatids (Reguera et al., 2006).
Multiple alignments of DNA sequences and X-ray analysis of a co-crystal containing a truncated LdTopIB bound to nick double stranded DNA at a resolution of 2.27 Å, permitted the identification of specific protein domains in the leishmanial enzyme (Davies et al., 2006). The large LdTopIL subunit contains a short and poorly conserved N-terminal domain, whose function has not yet been clarified so far. The central core domain conserves a high homology with other TopIB proteins (Stewart et al., 1996; Cretaio et al., 2007). It contains several amino acids interacting with DNA; i.e., Arg314 (Arg488 in the human enzyme), Lys352 (Lys532 in the human enzyme), Arg410 (Arg590 in the human enzyme) and His453 (His632 in the human enzyme) (Diaz-González et al., 2008). Finally, this subunit displays a C-terminal extension, which lacks homology with other ortholog proteins (Diaz-Gonzalez et al., 2007a). The small LdTopIS subunit contains a long and non-conserved N-terminal extension enriched in Ser residues, which is susceptible of post-translational phosphorylation. However, the C-terminal domain conserves high homology with the SKINY signature, where Tyr222 (Tyr723 in human enzyme) is the fifth amino acid of the catalytic pentad that plays a role in the nucleophilic attack to DNA. The functionality of these amino acids in the leishmanial enzyme has been confirmed by single site mutation analysis and expression in a TopIB-null yeast mutant (Diaz-Gonzalez et al., 2007b) (Fig. 1A).
Fig. 1.
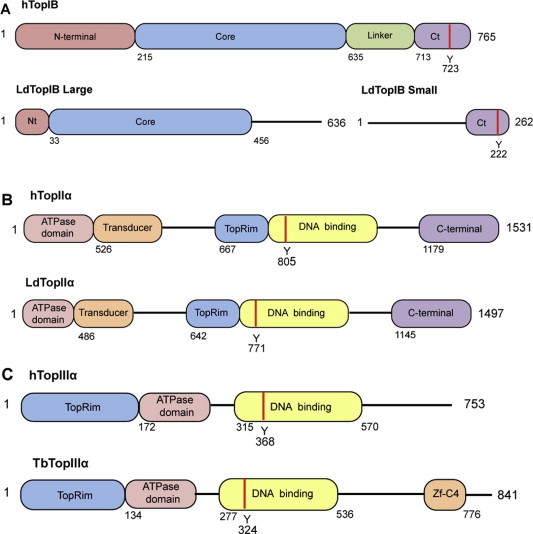
Schematic lineal representations of TopIB, TopII and TopIII from tritryps compared to human counterparts. (A) Human (top) and bisubunit L. donovani (down) TopIB, structural domains: Non-conserved hydrophilic N-terminal domain (Nt); central “core” domain with the representative active site amino acids (“tetrad”); non-conserved “linker” subdomain that connects “core” and the C-terminal domain and C-terminal domain (Ct) containing the DNA cleaving Tyr (red strip). (B) Human (top) and L. donovani TopII α (down) structural domains: N-terminal domain containing the ATPase and transducer domains; the Mg2+ binding site at the TopRim motif; a DNA binding that contains the DNA cleaving Tyr (red strip); C-terminal domain. (C) Human (top) and T. brucei TopIII α (down) structural domains: N-terminal domain containing the Mg2+ binding site at the TopRim motif; the ATPase domain and C-terminal domain; DNA binding site that contains the DNA cleaving Tyr (red strip); Znf-C4 (zinc-finger domain). Distances represented in the drawings are not in scale. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
However, the 3D model obtained from the X-ray analysis fails in describing the linker domain, a poorly conserved region that connects the core and C-terminus end, because it was removed to prepare the crystal (Davies et al., 2006). The linker domain is not merely a connector but it contributes to both DNA binding and sensitivity to TopIB poisons, probably increasing the rigidity of the structure and slowing down the rotation process prior to the religation of the cleaved DNA strand (Stewart et al., 1997). A recent study has disclosed the regions of both subunits that interact to each other to constitute a functional linker in Leishmania (Prada et al., 2012). Using a predictive 3D model combined with a sequential set of internal and lineal truncations on both monomers, the authors were able to identify within the N-extension domain of LdTopIS a conserved heptapeptide motif (RPPVVRS), which is spatially close enough to interact with a region (amino acids 525–581) of the large LdTopIL subunit. Remarkably, the pentapeptide RPPVV introduces a loop in the secondary structure of LdTopIS that permits the interaction with the large subunit. These results agree well with those reported by Stewart and co-workers (1999), who reconstituted enzyme species from hTopIB fragments that lack the linker domain and observed that any alteration on this region produces modifications in the religation step and an impairment to be poisoned by Top inhibitors (Ireton et al., 2000; Fiorani et al., 2003).
TopIB are nucleus-operating enzymes that need to display recognizable Nuclear Localization Signals (NLS) for nucleo-cytoplasmic trafficking. Three NLS motifs have been recently identified in LdTopIB by sequential deletion analyses of the genes encoding the two subunits fused to the GFP-encoding ORF (Prada et al., 2013b). NLS1 is located at the C-terminal extension of LdTopIL and consists of a 44-amino acids region enriched in Lys residues (easily identifiable by nuclear importins). NLS2 and NLS3 are located at the N-terminal and C-terminal domains of the small LdTopIL subunit, respectively. NLS2 is a well-identified 10-amino acids signature preceding a well-conserved cluster of Ser in trypanosomatids. This fact is important since this NLS requires at least one Ser residue to be functional; suggesting that hypothetical phosphorylation of Ser is necessary for nuclear localization (Teng et al., 2011). Finally, NLS3 is a 28-amino acid region that includes the catalytic Tyr222. The amino acid sequence of NLS3 bears no resemblance at all to other canonical NLS signals described so far. However, it looks like the PY-NLSs signals proposed by Lee and co-workers (2006) in human proteins, where proximal Pro and Tyr residues are recognized by a β-importin to drive the protein inside the nucleus. Furthermore, regardless previous results (BoseDasgupta et al., 2008), it was demonstrated that both subunits can reach the nuclear compartment independently (Prada et al., 2013b). Although some authors pointed to dual nucleus/kinetoplast LdTopIB localization, no specific mitochondrial localization signals have been identified in any of the subunits of the dimer. In addition, none of the chimeras fused to GFP co-localize with kDNA, thus proving that none of LdTopIB subunits are bound to DNA minicircles, which in fact rules out the hypothesis of a TopIB operating in the kinetoplast (Das et al., 2006a,b).
2.2. Type II topoisomerases
Type II topoisomerases (Top II) are ubiquitous and essential enzymes required for the regulation of DNA topology (Buck and Zechiedrich, 2004). Top II are multimeric enzymes that hydrolyze ATP to generate transient phosphotyrosine bonds at the 5′ end of both broken strands of the DNA helix. After cutting, the ends of the DNA are separated and another DNA duplex is passed through this break, which is finally religated (reviewed in Berger et al., 1996; Wang, 1996; Champoux, 2001).
Type II enzymes are organized into two families, Top IIA and IIB. The former includes, among others, the homodimeric (A2) eukaryotic Top II, which relaxes positively supercoiled DNA. Type IIB topoisomerases are heterotetrameric (A2B2) enzymes that only include one family member, topoisomerase VI. Although its role is less understood, it shares several features with the bacterial type IIA topoisomerase (reviewed in Nitiss, 2009).
Eukaryotic Top II enzymes participate in the segregation and maintenance of the chromosome structure (Gasser et al., 1986; Rose et al., 1990) and are involved in chromosome condensation and decondensation events (Adachi et al., 1991). In trypanosomatids, besides the essential role in DNA nuclear metabolism Top II are also important for the segregation, replication, remodeling and maintenance of kDNA (Shapiro, 1994; Shapiro and Showalter, 1994; Lindsay et al., 2008). Mitochondrial TopII (mtTopII) is of paramount importance to disaggregate kDNA network during cell division in kinetoplastids previous to cell division (Jensen and Englund, 2012). Early studies carried out with the antitumour drug etoposide in T. brucei, showed that replication of free minicircles was disrupted by the drug, giving rise to multiple interlocked minicircle-dimers that contained newly synthesized DNA (Shapiro et al., 1989). In fact, a loss in kDNA network (diskinetoplasty) and subsequent cell death was reported upon the induction of an RNAi-mediated silencing event in mtTop II, a phenomenon that was preceded by a gradual shrinkage of the network and accumulation of gapped free minicircle replication intermediates (Wang and Englund, 2001). There are other likely functions for mtTopII, such as the mending of kDNA after releasing minicircles for replication and the reattachment of nicked or gapped minicircles to the periphery of the replicating network (Lindsay et al., 2008), where a type I Top enzyme could also play a critical role (Scocca and Shapiro, 2008).
Only one mtTop II, localized primarily to the antipodal sites of the kinetoplast (Melendy et al., 1988), is predicted after the analysis of the genome sequence of several trypanosomatids (Strauss and Wang, 1990; Fragoso and Goldenberg, 1992; Das et al., 2001; Hanke et al., 2003; De Sousa et al., 2003). In addition, two other Top II enzymes, which are encoded by the tandemly-linked single-copy genes Top II α and Top II β, were found in the chromosome 11 of T. brucei (Kulikowicz and Shapiro, 2006). These Top II (74% identical and differing only in the C-terminal region) share a common node with other nuclear topoisomerases. The latter are clearly different from the clade that includes the mtTop II, which is positioned near the DNA gyrase branch that is more closely related to the prokaryotic type II enzymes. RNAi-mediated silencing of the ATP-dependent nuclear Top II α in T. brucei caused abnormalities in nuclear DNA and cell growth arrest at early stages of cell cycle, pointing to this protein as an attractive therapeutic target (Wang and Englund, 2001). Although the localization and function of the putative Top II β protein is unknown, it is phylogenetically related to other eukaryotic nuclear Top II (Kulikowicz and Shapiro, 2006). These researchers produced knock-down mutants in the gene encoding TbTopII β showing that there was no detectable effect on growth rate, remaining its function in trypanosomatids still unclear.
Regarding other trypanosomatids, a single ortholog of Top II α β (but not of TopII β) is present in the L. major chromosome 28. Although two additional ORFs were found in T. cruzi, they appeared to be alleles of the same gene rather than α and β isoforms of Top II (Kulikowicz and Shapiro, 2006).
Top IIA activity has been purified from several trypanosomatids and some of the corresponding coding genes have been cloned from Crithidia fasciculata (Shlomai and Zadok, 1983; Melendy et al., 1988; Melendy and Ray, 1989; Pasion et al., 1992; Hines and Ray, 1997), T. brucei (Strauss and Wang, 1990; Wang and Englund, 2001; Kulikowicz and Shapiro, 2006), T. cruzi (Douc-Rasy et al., 1986; Fragoso et al., 1998), Trypanosoma equiperdum (Shapiro, 1994), L. donovani (Chakraborty and Majumder, 1987, 1991; Das et al., 2001), Leishmania chagasi (De Sousa et al., 2003) and L. infantum (Hanke et al., 2003).
Basically, trypanosomatids TopII α are homodimeric proteins, each subunit ranging in size from 137 to 160 kDa. Similarly to other eukaryotes, each subunit is constituted by the fusion of domains from bacterial DNA gyrase (GyrB and GyrA) and TopIV (ParE and ParC) building a GyrB/ParE fused-domain at the N-terminal-end and a GyrA/ParC fused-domain at the C-terminal-end, respectively (Champoux, 2001).
Three structural domains, namely ATPase, central DNA binding (B′ and A′), and C-terminal domain, which are present in a single polypeptide chain, constitute eukaryotic Top II. These domains are connected to each other by a flexible hinge region (Sengupta et al., 2003). The ATPase domain is found at the enzyme N-terminal end, which spans the first 489 amino acids in T. brucei and the first 384 amino acids in L. donovani (Sengupta et al., 2005a,b). ATP binding to the nucleotide-binding cassette promotes the dimerization of the N-terminal ATPase domains, which control the open/close movements of the protein that are involved in DNA releasing (Wei et al., 2005). Following the ATPase domain the core region comprises the B′ domain (Toprim or unusual Rossman fold that contains the constant Asp-X-Asp motif and coordinates two Mg2+ ions per each subunit) (Aravind et al., 1998) and the N-terminal region of A′ (a central heart-shaped DNA-binding core), the latter containing the catalytic Tyr residue involved in the trans-esterification reaction of DNA (Bjergbaek et al., 2000). The transducer helical domain, which links the ATPase domain to B′, is believed to communicate the nucleotide state of the ATPase domain to the rest of the protein (Bendsen et al., 2009). Finally, after the A′ domain, eukaryotic Top II have an unconserved C-terminal hydrophilic domain that is dispensable for topoisomerization, although it plays in L. donovani an important role in the in vitro TopII cleavage reaction (Das et al., 2006b). In addition, this domain may also include signals for nuclear localization and interaction with other proteins (Reece and Maxwell, 1991). In fact, a potent nuclear localization signal was found within the C-terminus of the L. donovani Top II (Sengupta et al., 2003) (Fig. 1B).
In general, Top II are ATP-dependent enzymes that require the presence of Mg2+ ions to fulfill their catalytic cycle (Bates and Maxwell, 2007). Mg2+ ions seem to facilitate the interaction between the enzyme and substrate in at least two ways: (i) facilitating the formation of TopII/DNA cleavage complexes; (ii) forming the ATP-Mg2+ substrate required for ATPase activity (Sissi and Palumbo, 2009). As it was indicated above, Top II contain the conserved Mg2+-interaction domain called Toprim. Since Top II perform two sequential phosphoryl transfer reactions that promote profound conformational enzymatic changes, the high affinity of Toprim for Mg2+ may be related to the need to keep the free 3′-OH end produced by the cleavage reaction strongly connected to the Toprim acidic sequence, hence in place to perform religation (Zhu et al., 1997).
2.3. Type IA topoisomerases
Subfamily Top IA differs in structure, sequence and catalytic mechanism from TopIB. TopIA enzymes are essentially DNA relaxing enzymes (relaxases) introducing changes in negatively supercolied DNA in discrete numbers (Lk in multiples of 1) (Byrd and Matson, 1997). However, TopIII enzymes appear to be more active catenating/decatenating DNA (decatenases) due to a specific 17-amino insertion located close to the central cavity within the enzyme (Li et al., 2000; Forterre et al., 2007; Viard and Bouthier, 2007). Trypanosomatids have a full set of TopIA subfamily enzymes compartmentalized both in the nucleus and kinetoplast. The T. brucei genome contains three type IA topoisomerase-encoding genes: one in chromosome 10 (annotated as true TopIA) and two in chromosome 11, both annotated as TopIII (http://www.genedb.org/genedb/tryp). For its part, L. major Friedlin has a putative TopIA-encoding gene in chromosome 21 and two putative TopIII ORFs at chromosomes 28 and 36, respectively. A similar composition of putative TopIA orthologous genes is deposited in the T. cruzi genome database.
Trypanosomatid TopIA along with mtTopII are mitochondrial enzymes that play a role in kDNA minicircles segregation during cell division. Unlike TopIB, the catalytic tyrosine establishes the transient phosphotyrosine intermediate bond at the 5′-end of the scissile DNA strand (Champoux, 2001). Structurally T. brucei TopIA (TbTopIA) was characterized as an 88.9 kDa-monomeric protein encoded by a gene of 2418 bp with a high degree of phylogenetic conservation. The L. donovani ortholog consists of a protein of 90 kDa encoded by an ORF of 2453 bp. TbTopIA contains an N-terminal domain, which has a mitochondrial localization signal that drives the protein to kDNA, followed by a phylogenetically conserved region similar to prokaryotic TopIA called core (Scocca and Shapiro, 2008). This region contains all the conserved domains of the protein with two DNA binding topofolds (amino acids 186–660) (Duguet et al., 2006) and a C-terminal end variable in size and sequence. Four structural domains constitute the core region that give rise to a toroidal-shaped protein with a central motif filled of basic amino acids that allows the passage of a single- or double-stranded DNA molecule (Duguet et al., 2006). Between domains II and IV a hinge motif allows two spatial open and close configurations. TbTopIA has the catalytic Tyr-362 residue within domain III near a Mg2+ binding domain enriched in acidic amino acids named toprim (amino acids 10–170), which is involved in the conformational changes of the protein during the relaxation reaction. Unlike prokaryotic TopIA proteins, TbTopIA lack Zn-binding domains (Scocca and Shapiro, 2008).
Immunolocalization assays clearly demonstrated that mtTbTopIA operates inside the single mitochondrion of T. brucei linked to flagellar basal body and associated to the enzymes in charge of DNA replication (SSE-1, PIF5, Pol β, ligase β and mtTopII) at the antipodal sites of kDNA disc (Jensen and Englund, 2012). These studies showed that the position of mtTbTopIA is changing from one pole to the other pole of the kDNA disk as replication is taking place. Silencing studies of mtTbTopIA by RNAi under tetracycline control, showed that after induction with the antibiotic, growth arrest and almost a complete disappearance of the kDNA occurred with no changes in nuclear DNA (Scocca and Shapiro, 2008). A deeper analysis of minicircle network replication after RNA silencing showed an accumulation of late theta forms but not of daughter catenates, suggesting that mtTbTopIA plays a role in the resolution on the entwined parental strands during the resolution step of minicircle replication (Scocca and Shapiro, 2008). Finally, late free minicircles containing gaps are used by mitochondrial TopII enzymes to reattach them to the replicating kDNA network (Wang, 2002). Since mitochondrial trypanosomatid TopIA has no orthologs in the definitive host, and as a result of the deadly effects that its abolition has (as described above), there is no doubt that this enzyme is an ideal target to be explored for the development of novel drugs (Tang and Shapiro, 2010).
Top III are TopIA subfamily proteins, which in turn are subdivided into TopIII α and TopIII β isoforms (Viard and Bouthier, 2007). Trypanosomatids contain phylogenetically well-conserved genes encoding both TopIII α and TopIII β (Kim and Cross, 2010). Genetic analysis revealed the presence of a 2754-bp ORF encoding the 102.5-kDa T. brucei TopIII α (TbTopIII α) and a putative 2523-bp ORF encoding the 937-kDa T. brucei TopIII β (TbTopIII β) (Kim and Cross, 2010). Similarly, the putative gene encoding L. donovani TopIII α (LdTopIII α) is 2844 bp long and encodes a 104-kDa monomer, whereas the 2601-bp gene encoding TopIIIb (LdTopIII β) produces a 95-kDa protein (http://www.genedb.org/Homepage/Ldonovani_BPK282A1). The deduced amino acid sequence of LdTopIII β has the active Tyr at position 327, which is located within a highly conserved GYISYPRTES signature. This protein contains seven CXXC sequences instead of the eight motifs found in other TopIIIβ proteins (Banerjee et al., 2011). In a similar way, TbTopIII α has been characterized and contains a Zn-binding domain at the C-terminus. The sequences of tritryps TopIIIα are very well conserved in T. brucei, T. cruzi and L. major and contain Zn-binding motif(s) at their C-terminal regions. A LdTopIII β fused to GFP chimera was used to identify the localization of this protein, revealing that LdTopIIIβ is targeted inside both the parasite nucleus and kinetoplast (Banerjee et al., 2011) (Fig. 1C).
TopIII proteins show decatenase rather than relaxase activity by their own. TopIII α proteins play a role in concert with RecQ helicases maintaining genomic stability and integrity by controlling recombination events (Kim and Wang, 1992). TbTopIII α acts as a single-stranded DNA decatenase and release non-crossover products resulting from the dissolution of recombinogenic structures, such as double Holliday junctions. Regarding other eukarya organisms, TbTopIII α may be part of a putative RecQ-TopIII-Rmi1 (RTR) complex that is involved in the recombination-mediated antigenic switching in T. brucei, removing undesirable recombination intermediates (Yang et al., 2010; Knoll et al., 2014). Null-TbTopIII α mutants have been shown to play a critical role in homologous recombination processes and antigenic switching. Double replacement of the two alleles encoding the TbTopIII α produces no changes in the proliferation rate of the parasites, but generates an elevated frequency of recombinant events and hyper-switcher phenotype always linked to RAD51. These experiments concluded that TopIII α plays a crucial role in homologous recombination processes and is closely linked to adaptive VSG switching that allows blood forms of T. brucei to evade from the host immune response (Kim and Cross, 2010).
Very few studies have been carried out with TopIII β enzymes in trypanosomatids. Although TopIII β is not essential for cell survival, null-mutants display unique phenotypes in eukarya. Knock-out mice lacking the TopIII β -encoded gene develop normally up to maturity, but have a shorter lifespan and exhibit decreased fertility (Kwan and Wang, 2001; Kwan et al., 2003). In addition, these mutants have a deficient activation of p53, pointing to an important role of TopIIIβ in apoptosis induced by DNA damage (Mohanty et al., 2008). Deficient β top3 mutant yeasts are viable but display a slow-growth phenotype (Gangloff et al., 1999). Experiments with the LdTopIII β gene, showed a partial recovery of phenotype (Banerjee et al., 2011). In addition, this property is lost when rescuing is produced by a partially deleted ORF lacking the Zn-binding domain of the protein. However, no experiments regarding genome stability and programmed cell death have been performed in trypanosomatids, thus the role of this enzyme remaining a mystery in these parasites.
3. DNA topoisomerases as potential drug targets
Both TopIB and TopII are validated targets for antineoplastic drugs in human medicine. TopIB inhibitors, such as topotecan (a water soluble derivative of CPT) and irinotecan (Camptosar) – a prodrug that must be processed hydrolytically by intracellular esterases – are currently in clinical use against ovarian carcinoma (Wethington et al., 2008) and colorectal cancer, respectively (Kuppens et al., 2004). On the other hand, TopII inhibitors are also part of the antitumour drug Vademecum, such as the epipodophyllotoxin derivatives (etoposide and teniposide) (Hande, 1998) or acridine analogues (amsacrine, doxorubicin, etc.) (Louie and Issell, 1985). Last but not least, the bacterial DNA gyrase has been used for years as a target for a number of antibiotics of the fluoroquinolone family (Drlica and Malik, 2003).
Top inhibitors are classified into two categories: (i) Class I or enzyme poisons are those compounds that displace the cleaving/ligation balance toward cleavage, thus stabilizing the cleavage complexes with DNA. This selectively kills the cells found in S phase by collision with the advancing replication fork during DNA replication (Tsao et al., 1993); (ii) Class II or “true inhibitors” are those compounds that interfere with the catalytic functions of the enzyme without trapping the covalent complexes (Pommier, 2006).
With this favourable background, the opportunity to explore these enzymes as potential drug targets in trypanosomatids was early underlined, even when their molecular structures had not yet been described (Burri et al., 1996; Cheesman, 2000). The presence of decatenase and relaxase activities in the single mitochondria of these parasites and their role on the structure of the kinetoplast minicircle network, pointed to these enzymes as extremely attractive targets to design specific drugs to deconstruct kDNA. In 1995 the group led by Shapiro showed that CPT and etoposide triggered kDNA linearization and further disappearance in T. brucei, a phenomenon called diskinetoplasticity, which led to parasite death within hours (Bodley and Shapiro, 1995).
With the exception of yeast, TopIBs are essential for eukaryotic organisms (Uemura et al., 1987). Double gene replacement of the gene encoding LdTopIS, originates a non-viable phenotype in L. major (Balaña-Fouce et al., 2008). In addition, gene silencing of each of the T. brucei TopIB subunits, results in a drastic reduction in DNA and RNA synthesis in vitro (Bakshi and Shapiro, 2004). These findings and the structural differences from host orthologs pointed to these enzymes as putative targets for drug discovery (Balaña-Fouce et al., 2006). First, the uncompetitive inhibitor CPT and its derivatives have been used as inhibitors against T. brucei and Leishmania spp. TopIB with promising results (Das et al., 2004). The primary effect of the formation of single strand breaks (SSBs) can lead to the induction of the TDP1 repair system enzyme studied by Pommier in mammalian cells (Dexheimer et al., 2008) and recently described in trypanosomatids (Banerjee et al., 2010). The collision of the replication fork during DNA replication transforms SSBs into irreversible double-stranded DNA breaks (DSBs), which can be repaired only by homologous DNA recombination mechanisms. The persistence of no repaired DSBs may cause a process of programmed cell death both in amastigotes and promastigotes, which has been described in detail as a result of the exposure of L. donovani to Top poisons (Sen et al., 2004).
In addition to CPT, topotecan and Camptosar were experimentally tested against ex vivo cultures of experimentally infected murine splenocytes with L. infantum amastigotes, showing that gimatecan, an orphan drug approved by the European Medicines Agency for the treatment of glioblastoma (Hu et al., 2013), has a strong leishmanicidal activity and good selective index (Prada et al., 2013a). An interesting group of non-CPT TopIB poisons are indenoisoquinolines, which were initially developed as antitumour compounds with an improved ability to stabilize cleavage complexes (Antony et al., 2005). Some of these compounds have shown interesting therapeutic profiles against L. infantum and T. brucei in experimental infections of mice (Bakshi et al., 2009; Balaña-Fouce et al., 2012). Indotecan, administered intraperitoneally every 2 days for 15 days (a total of eight doses) had a similar cure rate that paromomycin alone or in combination with 200-mg/kg-body-weight Glucantime in experimental infections of VL in BALB/c mice (Gangneux et al., 1997). Furthermore, indotecan was more effective than the same dose schedule of CPT either free or liposome encapsulated, in a murine model of L. donovani leishmaniasis (Proulx et al., 2001). Some naturally occurring molecules have shown to be TopIB poisons and have anti-leishmanial effects. Amongst others, the bisnapthaquinone dyospirin (Ray et al., 1998), the pentacyclic triterpenoid dihydrobetulinic acid (Chowdhury et al., 2003) or the naturally occurring flavones (baicalein, luteolin and quercetin) are potent LdTopIB poisons in vitro (Mittra et al., 2000; Das et al., 2006a,b). For their part, unsaturated fatty acids from marine organisms have shown to be selective Class II inhibitors of recombinant L. infantum TopIB (Carballeira, 2013) (Table 2).
Table 2.
TopIB inhibitors assayed in trypanosomatids.
| Class | TopIB inhibitors | Trypanosomatid | References |
|---|---|---|---|
| Camptothecins | CPT, SN38, topotecan, gimatecan | L. infantum | Prada et al. (2013a) |
| CPT, irinotecan, topotecan | T. brucei |
Bodley and Shapiro (1995) Deterding et al. (2005) |
|
| Indocarbazoles | Rebeccamycin | T. brucei | Deterding et al. (2005) |
| Napthoquinones | Diospyrin diospyrin, naphthalene derivatives |
L. donovani T. brucei, T.cruzi |
Hazra et al. (1987) Ray et al. (1998) Ganapaty et al. (2006) |
| Flavonoids | Baicalein, luteolin, quercetin | L. donovani | Das et al. (2006a,b) |
| Poliheterocyclis | Berberin |
L. braziliensis L. donovani T. brucei, T. congolense |
Vennerstrom et al. (1990) Marquis et al. (2003) Merschjohann et al. (2001) |
| Pentavalent antimony | Pentostam, glucantime | L. donovani | Chakraborty and Majumder (1988) |
| Indenoisoquinolines | Indotecan, AM 13-55 alkylamine N-6, N-6 3-aminopropyl, N-6 3-imidazolylprolyl |
L. infantum T. brucei |
Balaña-Fouce et al. (2012) Bakshi et al. (2009) |
| Bis-benzimidazoles | Hoechst-33258, Hoechst-33342 | L. donovani | Walker and Saravia (2004) |
| Triterpenoids | Betulinic acid and derivatives dihydrobetulinic acid |
L. donovani, L. amazonensis, L. braziliensis, T. cruzi L. donovani |
Domínguez-Carmona et al. (2010) Chowdhury et al. (2003) |
| Insaturated fatty acids | 6-Heptadecynoic acid and 6-icosynoic acid (±)-17-methyltrans-4,5-methyleneoctadecanoic acid 2-Alkynoic fatty acids |
L. donovani |
Carballeira et al. (2009) Carballeira et al. (2010) Carballeira et al. (2012) |
| Lignan glycosides | Lyoniside, sacaroside | L. donovani | Saha et al. (2013) |
Similarly to TopIB, TopII silencing in T. brucei, leads to the progressive degradation of kDNA network and parasite death (see above). Eukaryotic TopII was proposed as a putative target for anti-parasitic drugs since the 1990s (Burri et al., 1996; Cheesman, 2000). One of the first groups of compounds that showed therapeutic potential against trypanosomatids was derived from the 9-anilinoacridine backbone, and had potential anti-tumour activity owing to TopII inhibition (Werbovetz et al., 1994). Linearization of kDNA minicircles has also been reported in parasites treated with these compounds because of TopII inhibition, preventing the growth of L. major promastigotes and amastigotes (Gamage et al., 1997). Epipodophyllotoxins are semisynthetic derivatives of non-alkaloid toxins from the root extracts of plants from the genus Podophyllum (Table 3). The two leading compounds, etoposide and teniposide, stabilize the double-stranded cleavage mediated by TopII and inhibit the religation of DNA breaks (Yang et al., 2009). These compounds are clinically in use against different types of cancer (Anonymous, 2000), and have been proven effective against different parasites (García-Estrada et al., 2010), inducing apoptotic-like cell death (Meslin et al., 2007). However, etoposide have not shown any effect on trypanosomatids, pointing to important structural differences between this enzyme and that from the host counterpart (Sengupta et al., 2005a,b).
Table 3.
TopII inhibitors assayed in trypanosomatids.
| Class | TopII inhibitors | Trypanosomatid | References |
|---|---|---|---|
| Fluoroquinolones | Norfloxacin, ciprofloxacin, ofloxacin | T. brucei | Nenortas et al. (1999, 2003) |
| Ciprofloxacin, enoxacin | L. panamensis | Cortazar et al. (2007) | |
| Norfloxacin, ofloxacin | T. cruzi | Gonzales-Perdomo et al. (1990) | |
| Podophylotoxins | Etoposide, teniposide | L. donovani | Sengupta et al. (2005a,b) |
| T. equiperdum | Shapiro and Showalter (1994) | ||
| Ellipticines | 2-Methyl-9-OH-ellipticin | T. equiperdum | Shapiro et al. (1989) |
| Flavonoids | Baicalein, luteolin, quercetin |
L. donovani L. panamensis |
Tasdemir et al. (2006) Mittra et al. (2000) Cortazar et al. (2007) |
| Anthracyclins | Doxorrubicin | L. donovani | Singh and Dey (2007) |
| Diamidines | Pentamidine |
L. donovani L. panamensis |
Singh and Dey (2007) Cortazar et al. (2007) |
| Aminocoumarins | Novobiocin |
T. cruzi L. donovani |
Gonzales-Perdomo et al. (1990) Singh et al. (2005) |
| Acridines | Anilinoacridines |
L. chagasi T. brucei |
Werbovetz et al. (1992) Gamage et al. (1997) |
| Triterpenoids | Dihydrobetulinic acid | L. donovani | Chowdhury et al. (2003) |
4. Concluding remarks
Trypanosomatids have a complete set of six classes of Top enzymes encoded in their respective genomes. Each class, defined by structure and mechanism of action, has distinct but sometimes overlapping cellular localizations and functions. TopIB enzymes are structurally dissimilar to the rest of proteins described so far, but they conserve the ability to pass a single DNA strand in a processive manner to relief torsional stress. TopIA, including TopIII α and TopIII β enzymes have decatenase rather than relaxase activity and work in concert with helicases to resolve noncrossover products by dissolution of recombination intermediates. TopII enzymes, both nuclear and mitochondrial, carry out strand passage through a double-strand break, including decatenating and unknotting activities and are key enzymes in kDNA replication. TopIB and TopII are important targets for drug intervention in cancer. Many of the drugs used as antitumourals have been used in a “piggy backing” strategy against tritryps with different success. A full structural knowledge of tritryps Top proteins based on 3D X-ray analysis of Top crystals, will allow in silico high-throughput virtual screening of potential inhibitors in a structure-based drug discovery strategy.
Acknowledgements
This research was supported by Ministerio de Economía y Competitividad (AGL2010-16078/GAN and CYTED 214RT0482), Instituto de Salud Carlos III (PI12/00104) and Junta de Castilla y León (grants Gr238 and LE182U13).
References
- Adachi Y., Luke M., Laemmli U.K. Chromosome assembly in vitro: topoisomerase II is required for condensation. Cell. 1991;64:137–148. doi: 10.1016/0092-8674(91)90215-k. [DOI] [PubMed] [Google Scholar]
- Aksoy S., Gibson W.C., Lehane M.J. Interactions between tsetse and trypanosomes with implications for the control of trypanosomiasis. Adv. Parasitol. 2003;53:1–83. doi: 10.1016/s0065-308x(03)53002-0. [DOI] [PubMed] [Google Scholar]
- Alexander J., Bryson K. T helper (h)1/Th2 and Leishmania: paradox rather than paradigm. Immunol. Lett. 2005;99:17–23. doi: 10.1016/j.imlet.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Alvar J., Aparicio P., Aseffa A., Den Boer M., Cañavate C., Dedet J.P., Gradoni L., Ter Horst R., López-Vélez R., Moreno J. The relationship between leishmaniasis and AIDS: the second 10 years. Clin. Microbiol. Rev. 2008;21:334–359. doi: 10.1128/CMR.00061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvar J., Vélez I.D., Bern C., Herrero M., Desjeux P., Cano J., Jannin J., den Boer M., Leishmaniasis Control Team W.H.O. Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato A. DNA packaging: nucleosomes and chromatin. Nat. Educ. 2008;1:26. [Google Scholar]
- Anonymous DNA topoisomerase II inhibitors. IARC Monogr. Eval. Carcinog. Risks Hum. 2000;76:175–344. [PMC free article] [PubMed] [Google Scholar]
- Antinori S., Schifanella L., Corbellino M. Leishmaniasis: new insights from an old and neglected disease. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:109–111. doi: 10.1007/s10096-011-1276-0. [DOI] [PubMed] [Google Scholar]
- Antony S., Kohlhagen G., Agama K., Jayaraman M., Cao S., Durrani F.A., Rustum Y.M., Cushman M., Pommier Y. Cellular topoisomerase I inhibition and antiproliferative activity by MJ-III-65 (NSC 706744), an indenoisoquinoline topoisomerase I poison. Mol. Pharmacol. 2005;67:523–530. doi: 10.1124/mol.104.003889. [DOI] [PubMed] [Google Scholar]
- Aravind L., Leipe D.D., Koonin E.V. Toprim: a conserved catalytic domain in type IA and II topoisomerases, DNAG-type primases, OLD family nucleases and RecR proteins. Nucl. Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashford R.W. The leishmaniasis as emerging and reemerging zoonoses. Int. J. Parasitol. 2000;30:1269–1281. doi: 10.1016/s0020-7519(00)00136-3. [DOI] [PubMed] [Google Scholar]
- Bakshi R.P., Shapiro T.A. RNA interference of Trypanosoma brucei topoisomerase IB: both subunits are essential. Mol. Biochem. Parasitol. 2004;136:249–255. doi: 10.1016/j.molbiopara.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Bakshi R.P., Sang D., Morrell A., Cushman M., Shapiro T.A. Activity of indenoisoquinolines against African trypanosomes. Antimicrob. Agents Chemother. 2009;53:123–128. doi: 10.1128/AAC.00650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaña-Fouce R., Cubría J.C., Reguera R.M., Ordóñez D. The pharmacology of leishmaniasis. Gen. Pharmacol. 1998;30:435–443. doi: 10.1016/s0306-3623(97)00268-1. [DOI] [PubMed] [Google Scholar]
- Balaña-Fouce R., Redondo C.M., Pérez-Pertejo Y., Diaz-González R., Reguera R.M. Targeting atypical trypanosomatid DNA topoisomerase. Drug Discov. Today. 2006;11:733–740. doi: 10.1016/j.drudis.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Balaña-Fouce R., García-Estrada C., Pérez-Pertejo Y., Reguera R.M. Gene disruption of the DNA topoisomerase IB small subunit induces a non-viable phenotype in the hemoflagellate Leishmania major. BMC Microbiol. 2008;8:113. doi: 10.1186/1471-2180-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaña-Fouce R., Prada C.F., Requena J.M., Cushman M., Pommier Y., Álvarez-Velilla R., Prada C.F., Pérez-Pertejo Y., Reguera R.M. Indotecan (LMP400) and AM13-55: two novel indenoisoquinolines show potential for treating visceral leishmaniasis. Antimicrob. Agents Chemother. 2012;56:5264–5270. doi: 10.1128/AAC.00499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee B., Roy A., Sen N., Majumder H.K. A tyrosyl DNA phosphodiesterase 1 from kinetoplastid parasite Leishmania donovani (LdTdp1) capable of removing topo I-DNA covalent complexes. Mol. Microbiol. 2010;78:119–137. doi: 10.1111/j.1365-2958.2010.07318.x. [DOI] [PubMed] [Google Scholar]
- Banerjee B., Sen N., Majumder H.K. Identification of a functional type IA topoisomerase, LdTopIIIb, from kinetoplastid parasite Leishmania donovani. Enz. Res. 2011:230542. doi: 10.4061/2011/230542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett M.P., Croft S.L. Management of trypanosomiasis and leishmaniasis. Br. Med. Bull. 2012;104:175–196. doi: 10.1093/bmb/lds031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates A.D., Maxwell A. Energy coupling in type II topoisomerases: why do they hydrolyze ATP? Biochemistry. 2007;46:7929–7941. doi: 10.1021/bi700789g. [DOI] [PubMed] [Google Scholar]
- Bendsen S., Oestergaard V.H., Skouboe C., Brinch M., Knudsen B.R., Andersen A.H. The QTK loop is essential for the communication between the N-terminal atpase domain and the central cleavage–ligation region in human topoisomerase IIalpha. Biochemistry. 2009;48:6508–6515. doi: 10.1021/bi9005978. [DOI] [PubMed] [Google Scholar]
- Berger J.M., Gamblin S.J., Harrison S.C., Wang J.C. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- Berman J.J. Treatment of leishmaniasis with miltefosine: 2008 status. Expert Opin. Drug Metab. Toxicol. 2008;4:1209–1216. doi: 10.1517/17425255.4.9.1209. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S.K., Sinha P.K., Sundar S., Thakur C.P., Jha T.K., Pandey K., Das V.R., Kumar N., Lal C., Verma N., Singh V.P., Ranjan A., Verma R.B., Anders G., Sindermann H., Ganguly N.K. Phase 4 trial of miltefosine for the treatment of Indian visceral leishmaniasis. J. Infect. Dis. 2007;196:591–598. doi: 10.1086/519690. [DOI] [PubMed] [Google Scholar]
- Bjergbaek L., Kingma P., Nielsen I.S., Wang I., Westergaard O., Osheroff N., Andersen A.M. Communication between the ATPase and cleavage/religation domains of human topoisomerase IIα. J. Biol. Chem. 2000;275:13041–13048. doi: 10.1074/jbc.275.17.13041. [DOI] [PubMed] [Google Scholar]
- Bodley A.L., Shapiro T.A. Molecular and cytotoxic effects of camptothecin, a topoisomerase I inhibitor, on trypanosomes and Leishmania. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3726–3730. doi: 10.1073/pnas.92.9.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodley A., Chakraborty A.K., Xie S., Burri C., Shapiro T.A. An unusual type IB topoisomerase from African trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7539–7544. doi: 10.1073/pnas.1330762100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BoseDasgupta S., Ganguly A., Das B.B., Roy A., Khalkho N.V., Majumder H.K. The large subunit of Leishmania topoisomerase I functions as the ‘molecular steer’ in type IB topoisomerase. Mol. Microbiol. 2008;67:31–46. doi: 10.1111/j.1365-2958.2007.06002.x. [DOI] [PubMed] [Google Scholar]
- Buck G.R., Zechiedrich E.L. DNA disentangling by type-2 topoisomerases. J. Mol. Biol. 2004;340:933–939. doi: 10.1016/j.jmb.2004.05.034. [DOI] [PubMed] [Google Scholar]
- Burri C. Chemotherapy against human African trypanosomiasis: is there a road to success? Parasitology. 2010;137:1987–1994. doi: 10.1017/S0031182010001137. [DOI] [PubMed] [Google Scholar]
- Burri C., Bodley A.L., Shapiro T.A. Topoisomerases in kinetoplastids. Parasitol. Today. 1996;12:226–231. doi: 10.1016/0169-4758(96)10017-x. [DOI] [PubMed] [Google Scholar]
- Byrd D.R., Matson S.W. Nicking by transesterification: the reaction catalysed by a relaxase. Mol. Microbiol. 1997;25:1011–1022. doi: 10.1046/j.1365-2958.1997.5241885.x. [DOI] [PubMed] [Google Scholar]
- Carballeira N.M. Recent developments in the antiprotozoal and anticancer activities of the 2-alkynoic fatty acids. Chem. Phys. Lipids. 2013;172–173:58–66. doi: 10.1016/j.chemphyslip.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballeira N.M., Cartagena M.M., Prada C.F., Rubio C.F., Balaña-Fouce R. Total synthesis and antileishmanial activity of the natural occurring acetylenic fatty acids 6-heptadecynoic acid and 6-icosynoic acid. Lipids. 2009;44:953–961. doi: 10.1007/s11745-009-3345-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballeira N.M., Montano N., Reguera R.M., Balaña-Fouce R. The first total synthesis of the (±)-17-methyl-trans-4,5-methyleneoctadecanoic acid and related analogs with antileishmanial activity. Tetrahedron Lett. 2010;51:6153–6155. doi: 10.1016/j.tetlet.2010.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballeira N.M., Cartagena M., Sanabria D., Tasdemir D., Prada C.F., Reguera R.M., Balaña-Fouce R. 2-Alkynoic fatty acids inhibit topoisomerase IB from Leishmania donovani. Bioorg. Med. Chem. Lett. 2012;22:6185–6189. doi: 10.1016/j.bmcl.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A.K., Majumder H.K. Decatenation of kinetoplast DNA by an ATP-dependent DNA topoisomerase from the kinetoplast hemoflagellate Leishmania donovani. Mol. Biochem. Parasitol. 1987;26:215–224. doi: 10.1016/0166-6851(87)90074-0. [DOI] [PubMed] [Google Scholar]
- Chakraborty A.K., Majumder H.K. Mode of action of pentavalent antimonials: specific inhibition of type I DNA topoisomerase of Leishmania donovani. Biochem. Biophys. Res. Commun. 1988;152:605–611. doi: 10.1016/s0006-291x(88)80081-0. [DOI] [PubMed] [Google Scholar]
- Chakraborty A.K., Majumder H.K. An ATP-independent catenating enzyme from the kinetoplast hemoflagellate Leishmania donovani. Biochem. Biophys. Res. Commun. 1991;180:279–285. doi: 10.1016/s0006-291x(05)81289-6. [DOI] [PubMed] [Google Scholar]
- Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Champoux J.J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA – a possible swivel for DNA replication. Proc. Natl. Acad. Sci. U.S.A. 1972;69:143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesman S.J. The topoisomerases of protozoan parasites. Parasitol. Today. 2000;16:277–281. doi: 10.1016/s0169-4758(00)01697-5. [DOI] [PubMed] [Google Scholar]
- Chen J., Rauch C.A., White J.H., Englund P.T., Cozzarelli N.R. The topology of the kinetoplast DNA network. Cell. 1995;80:61–69. doi: 10.1016/0092-8674(95)90451-4. [DOI] [PubMed] [Google Scholar]
- Chen J., Englund P.T., Cozzarelli N.R. Changes in network topology during the replication of kinetoplast DNA. EMBO J. 1995;14:6339–6347. doi: 10.1002/j.1460-2075.1995.tb00325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Kussie P., Pavletich N., Shuman S. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- Chowdhury A.R., Mandal S., Goswami A., Ghosh M., Mandal L., Chakraborty D., Ganguly A., Tripathi G., Mukhopadhyay S., Bandyopadhyay S., Majumder H.K. Dihydrobetulinic acid induces apoptosis in Leishmania donovani by targeting DNA topoisomerase I and II: implications in antileishmanial therapy. Mol. Med. 2003;9:26–36. [PMC free article] [PubMed] [Google Scholar]
- Corbett K.D., Berger J.M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- Cortázar T.M., Coombs G.H., Walker J. Leishmania panamensis: comparative inhibition of nuclear DNA topoisomerase II enzymes from promastigotes and human macrophages reveals anti-parasite selectivity of fluoroquinolones, flavonoids and pentamidine. Exp. Parasitol. 2007;116:475–482. doi: 10.1016/j.exppara.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Cretaio E., Pattarello L., Fontebasso Y., Benedetti P., Losasso C. Human DNA topoisomerase IB: structure and functions. Ital. J. Biochem. 2007;56:91–102. [PubMed] [Google Scholar]
- Croft S.L., Olliaro P. Leishmaniasis chemotherapy–challenges and opportunities. Clin. Microbiol. Infect. 2011;17:1478–1483. doi: 10.1111/j.1469-0691.2011.03630.x. [DOI] [PubMed] [Google Scholar]
- Das A., Dasgupta A., Sharma S., Ghosh M., Sengupta T., Bandopadhyay S., Majumder H.K. Characterisation of the gene encoding type II DNA topoisomerase from Leishmania donovani: a key molecular target in antileishmanial therapy. Nucl. Acids Res. 2001;29:1844–1851. doi: 10.1093/nar/29.9.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Dasgupta A., Sengupta T., Majumder H.K. Topoisomerases of kinetoplastid parasites as potential chemotherapeutic targets. Trends Parasitol. 2004;20:381–387. doi: 10.1016/j.pt.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Das B.B., Sen N., Roy A., Dasgupta S.B., Ganguly A., Mohanta B.C., Dinda B., Majumder H.K. Differential induction of Leishmania donovani bi-subunit topoisomerase I-DNA cleavage complex by selected flavones and camptothecin: activity of flavones against camptothecin-resistant topoisomerase I. Nucl. Acids Res. 2006;34:1121–1132. doi: 10.1093/nar/gkj502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B.B., Sengupta T., Ganguly A., Majumder H.K. Topoisomerases of kinetoplastid parasites: why so fascinating? Mol. Microbiol. 2006;62:917–927. doi: 10.1111/j.1365-2958.2006.05428.x. [DOI] [PubMed] [Google Scholar]
- Das B.B., Ganguly A., Majumder H.K. DNA topoisomerases of Leishmania: the potential targets for anti-leishmanial therapy. Adv. Exp. Med. Biol. 2008;625:103–115. doi: 10.1007/978-0-387-77570-8_9. [DOI] [PubMed] [Google Scholar]
- Davies D.R., Mushtaq A., Interthal H., Champoux J.J., Hol W.G. The structure of the transition state of the heterodimeric topoisomerase I of Leishmania donovani as a vanadate complex with nicked DNA. J. Mol. Biol. 2006;357:1202–1210. doi: 10.1016/j.jmb.2006.01.022. [DOI] [PubMed] [Google Scholar]
- De Sousa J.M., Lareau S.M., Pearson R.D., Carvalho E.M., Mann B.J., Jeronimo S.M. Characterization of Leishmania chagasi DNA topoisomerase II: a potential chemotherapeutic target. Scand. J. Infect. Dis. 2003;35:826–829. doi: 10.1080/00365540310017023. [DOI] [PubMed] [Google Scholar]
- Deterding A., Dungey F.A., Thompson K.A., Steverding D. Antitrypanosomal activities of DNA topoisomerase inhibitors. Acta Trop. 2005;93:311–316. doi: 10.1016/j.actatropica.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Dexheimer T.S., Antony S., Marchand C., Pommier Y. Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer Agents Med. Chem. 2008;8:381–389. doi: 10.2174/187152008784220357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz González R., Pérez Pertejo Y., Ordóñez D., Balaña-Fouce R., Reguera R.M. Deletion study of DNA topoisomerase IB from Leishmania donovani: searching for a minimal functional heterodimer. PLoS One. 2007;2:e1177. doi: 10.1371/journal.pone.0001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz González R., Pérez Pertejo Y., Redondo C.M., Pommier Y., Balaña-Fouce R., Reguera R.M. Structural insights on the small subunit of DNA topoisomerase I from the unicellular parasite Leishmania donovani. Biochimie. 2007;89:1517–1527. doi: 10.1016/j.biochi.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Díaz-González R., Pérez-Pertejo Y., Pommier Y., Balaña-Fouce R., Reguera R.M. Mutational study of the “catalytic tetrad” of DNA topoisomerase IB from the hemoflagellate Leishmania donovani: role of Asp-353 and Asn-221 in camptothecin resistance. Biochem. Pharmacol. 2008;76:608–619. doi: 10.1016/j.bcp.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Carmona D.B., Escalante-Erosa F., García-Sosa K., Ruiz-Pinell G., Gutierrez-Yapu D., Chan-Bacab M.J., Giménez-Turba A., Peña-Rodríguez L.M. Antiprotozoal activity of betulinic acid derivatives. Phytomedicine. 2010;17:379–382. doi: 10.1016/j.phymed.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Douc-Rasy S., Kayser A., Riou J.F., Riou G. ATP independent type II topoisomerase from trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 1986;83:7152–7156. doi: 10.1073/pnas.83.19.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Malik M. Fluoroquinolones: action and resistance. Curr. Top. Med. Chem. 2003;3:249–282. doi: 10.2174/1568026033452537. [DOI] [PubMed] [Google Scholar]
- Duguet M., Serre M.C., Bouthier de la Tour C. A universal type IA topoisomerase fold. J. Mol. Biol. 2006;359:805–812. doi: 10.1016/j.jmb.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Fiorani P., Bruselles A., Falconi M., Chillemi G., Desideri A., Benedetti P. Single mutation in the linker domain confers protein flexibility and camptothecin resistance to human topoisomerase I. J. Biol. Chem. 2003;278:43268–43275. doi: 10.1074/jbc.M303899200. [DOI] [PubMed] [Google Scholar]
- Forterre P., Gribaldo S., Gadelle D., Serre M.C. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Fragoso S.P., Goldenberg S. Cloning and characterization of the gene encoding Trypanosoma cruzi DNA topoisomerase II. Mol. Biochem. Parasitol. 1992;55:127–134. doi: 10.1016/0166-6851(92)90133-5. [DOI] [PubMed] [Google Scholar]
- Fragoso S.P., Mattei D., Hines J.C., Ray D., Goldenberg S. Expression and cellular localization of Trypanosoma cruzi type II DNA topoisomerase. Mol. Biochem. Parasitol. 1998;94:197–204. doi: 10.1016/s0166-6851(98)00066-8. [DOI] [PubMed] [Google Scholar]
- Ganapaty S., Steve Thomas P., Karagianis G., Waterman P.G., Brun R. Antiprotozoal and cytotoxic naphthalene derivatives from Diospyros assimilis. Phytochemistry. 2006;67:1950–1956. doi: 10.1016/j.phytochem.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Gamage S.A., Figgitt D.P., Wojcik S.J., Ralph R.K., Ransijn A., Mauel J., Yardley V., Snowdon D., Croft S.L., Denny W.A. Structure–activity relationships for the antileishmanial and antitrypanosomal activities of 1′-substituted 9-anilinoacridines. J. Med. Chem. 1997;40:2634–2642. doi: 10.1021/jm970232h. [DOI] [PubMed] [Google Scholar]
- Gangloff S., de Massy B., Arthur L., Rothstein R., Fabre F. The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 1999;18:1701–1711. doi: 10.1093/emboj/18.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangneux J.P., Sulahian A., Garin Y.J., Derouin F. Efficacy of aminosidine administered alone or in combination with meglumine antimoniate for the treatment of experimental visceral leishmaniasis caused by Leishmania infantum. J. Antimicrob. Chemother. 1997;40:287–289. doi: 10.1093/jac/40.2.287. [DOI] [PubMed] [Google Scholar]
- García-Estrada C., Prada C.F., Fernández-Rubio C., Rojo-Vázquez F., Balaña-Fouce R. DNA topoisomerases in apicomplexan parasites: promising targets for drug discovery. Proc. Biol. Sci. 2010;277:1777–1787. doi: 10.1098/rspb.2009.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon J., Bern C., Pinazo M.J. Chagas disease in Spain, the United States and other non-endemic countries. Acta Trop. 2010;115:22–27. doi: 10.1016/j.actatropica.2009.07.019. [DOI] [PubMed] [Google Scholar]
- Gasser S.M., Laroche T., Falzaet J., Tour E.B.D., Laemmli U.K. Metaphase chromosome structure involvement of topoisomerase II. J. Mol. Biol. 1986;188:613–624. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- Gonzales-Perdomo M., Lisboa de Castro S., Meirelles M.N.S.L., Goldenberg S. Trypanosoma cruzi proliferation and differentiation are blocked by topoisomerase II inhibitors. Antimicrob. Agents Chemother. 1990;34:1707–1714. doi: 10.1128/aac.34.9.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H., Lindoso J.A. Current diagnosis and treatment of cutaneous and mucocutaneous leishmaniasis. Expert Rev. Anti. Infect. Ther. 2010;8:419–433. doi: 10.1586/eri.10.19. [DOI] [PubMed] [Google Scholar]
- Hande K.R. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur. J. Cancer. 1998;34:1514–1521. doi: 10.1016/s0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- Hanke T., Ramiro M.J., Trigueros S., Roca J., Larraga V. Cloning, functional analysis and post-transcriptional regulation of a type II DNA topoisomerase from Leishmania infantum. A new potential target for anti-parasite drugs. Nucl. Acids Res. 2003;31:4917–4928. doi: 10.1093/nar/gkg671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra B., Saha A.K., Ray R., Roy D.K., Sur P., Banerjee A. Antiprotozoal activity of diospyrin towards Leishmania donovani promastigotes in vitro. Trans. R. Soc. Trop. Med. Hyg. 1987;81:738–741. doi: 10.1016/0035-9203(87)90013-7. [DOI] [PubMed] [Google Scholar]
- Hines J.C., Ray D.S. Periodic synthesis of kinetoplast DNA type II topoisomerases during the cell cycle. Mol. Biochem. Parasitol. 1997;88:249–252. doi: 10.1016/s0166-6851(97)00076-5. [DOI] [PubMed] [Google Scholar]
- Hsiang Y.H., Hertzberg R., Hecht S., Liu L.F. Camptothecin induces protein-linked DNA break via mammalian DNA topoisomerase I. J. Biol. Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- Hsiang Y.H., Lihou M.G., Liu L.F. Arrest of replication forks by drugs stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- Hu J., Wen P.Y., Abrey L.E., Fadul C.E., Drappatz J., Salem N., Supko J.G., Hochberg F.A. Phase II trial of oral gimatecan for recurrent glioblastoma. J. Neurooncol. 2013;111:347–353. doi: 10.1007/s11060-012-1023-0. [DOI] [PubMed] [Google Scholar]
- Ireton G.C., Stewart L., Parker L.H., Champoux J.J. Expression of human topoisomerase I with a partial deletion of the linker region yields monomeric and dimeric enzymes that respond differently to camptothecin. J. Biol. Chem. 2000;275:25820–25830. doi: 10.1074/jbc.M002144200. [DOI] [PubMed] [Google Scholar]
- Jensen R.E., Englund P.T. Network news: the replication of kinetoplast DNA. Annu. Rev. Microbiol. 2012;66:473–491. doi: 10.1146/annurev-micro-092611-150057. [DOI] [PubMed] [Google Scholar]
- Kim H.S., Cross G.A. TOPO3alpha influences antigenic variation by monitoring expression-site-associated VSG switching in Trypanosoma brucei. PLoS Pathog. 2010;6:e1000992. doi: 10.1371/journal.ppat.1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R.A., Wang J.C. Identification of the yeast TOP3 gene product as a single strand-specific DNA topoisomerase. J. Biol. Chem. 1992;267:17178–17185. [PubMed] [Google Scholar]
- Knoll A., Schröpfer S., Puchta H. The RTR complex as caretaker of genome stability and its unique meiotic function in plants. Front. Plant Sci. 2014;5:33. doi: 10.3389/fpls.2014.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltas I.S., Eroglu F., Alabaz D., Uzun S. The emergence of Leishmania major and Leishmania donovani in southern Turkey. Trans. R. Soc. Trop. Med. Hyg. 2014;108:154–158. doi: 10.1093/trstmh/trt119. [DOI] [PubMed] [Google Scholar]
- Kulikowicz T., Shapiro T.A. Trypanosoma brucei mitochondrion in the protozoan parasite topoisomerases for the nucleus and distinct genes encode type II. J. Biol. Chem. 2006;281:3048–3056. doi: 10.1074/jbc.M505977200. [DOI] [PubMed] [Google Scholar]
- Kuppens I.E., Beijnen J., Schellens J.H. Topoisomerase I inhibitors in the treatment of gastrointestinal cancer: from intravenous to oral administration. Clin. Colorectal Cancer. 2004;4:163–180. doi: 10.3816/ccc.2004.n.017. [DOI] [PubMed] [Google Scholar]
- Kwan K.Y., Wang J.C. Mice lacking DNA topoisomerase IIIβ develop to maturity but show a reduced mean lifespan. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5717–5721. doi: 10.1073/pnas.101132498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K.Y., Moens P.B., Wang J.C. Infertility and aneuploidy in mice lacking a type IA DNA topoisomerase IIIβ. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2526–2531. doi: 10.1073/pnas.0437998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.J., Cansizoglu A.E., Süel K.E., Louis T.H., Zhang Z., Chook Y.M. Rules for nuclear localisation sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejon V., Bentivoglio M., Franco J.R. Human African trypanosomiasis. Handb. Clin. Neurol. 2013;114:169–181. doi: 10.1016/B978-0-444-53490-3.00011-X. [DOI] [PubMed] [Google Scholar]
- Leprohon P., Légaré D., Ouellette M. ABC transporters involved in drug resistance in human parasites. Essays Biochem. 2011;50:121–144. doi: 10.1042/bse0500121. [DOI] [PubMed] [Google Scholar]
- Li Z., Mondragón A., Hiasa H., Marians K.J., DiGate R.J. Identification of a unique domain essential for Escherichia coli DNA topoisomerase III-catalysed decatenation of replication intermediates. Mol. Microbiol. 2000;35:4888–4895. doi: 10.1046/j.1365-2958.2000.01763.x. [DOI] [PubMed] [Google Scholar]
- Lindsay M.E., Gluenz E., Gull K., Englund P.T. A new function of Trypanosoma brucei mitochondrial topoisomerase II is to maintain kinetoplast DNA network topology. Mol. Microbiol. 2008;70:1465–1476. doi: 10.1111/j.1365-2958.2008.06493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, J.E., 2005. DNA topology: supercoiling and linking. In: Encyclopedia of Life Sciences. John Wiley and Sons: Hoboken, NJ, pp. 1–7.
- Liu B., Liu Y., Molyka S.A., Agbo E.E.C., Englund P.T. Felowships of the rings: the replication of kinetoplast DNA. Trends Parasitol. 2005;21:363–369. doi: 10.1016/j.pt.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Louie A.C., Issell B.F. Amsacrine (AMSA): a clinical review. J. Clin. Oncol. 1985;3:562–592. doi: 10.1200/JCO.1985.3.4.562. [DOI] [PubMed] [Google Scholar]
- Lutje V., Seixas J., Kennedy A. Chemotherapy for second-stage human African trypanosomiasis. Cochrane Database Syst. Rev. 2013;6:CD006201. doi: 10.1002/14651858.CD006201.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna L., Vitale F., Reale S., Picillo E., Neglia G., Vescio F., Gravino A.E. Study of efficacy of miltefosine and allopurinol in dogs with leishmaniosis. Vet. J. 2009;182:441–445. doi: 10.1016/j.tvjl.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Marquis J.F., Makhey D., LaVoie E.J., Olivier M. Effects of topoisomerases inhibitors protoberberine on Leishmania donovani growth, macrophage function, and infection. J. Parasitol. 2003;89:1048–1052. doi: 10.1645/GE-3161. [DOI] [PubMed] [Google Scholar]
- Melendy T., Sheline C., Ray D.S. Localization of a type II DNA topoisomerase to two sites at the periphery of the kinetoplast DNA of Crithidia fasciculata. Cell. 1988;55:1083–1088. doi: 10.1016/0092-8674(88)90252-8. [DOI] [PubMed] [Google Scholar]
- Melendy T., Ray D.S. Novobiocin affinity purication of a mitochondrial type II topoisomerase from the trypanosomatid Crithidia fasciculata. J. Biol. Chem. 1989;264:1870–1876. [PubMed] [Google Scholar]
- Merschjohann K., Sporer F., Steverding D., Wink M. In vitro effect of alkaloids on bloodstream forms of Trypanosoma brucei and T. congolense. Planta Med. 2001;67:623–627. doi: 10.1055/s-2001-17351. [DOI] [PubMed] [Google Scholar]
- Meslin B., Barnadas C., Boni V., Latour C., De Monbrison F., Kaiser K., Picot S. Features of apoptosis in Plasmodium falciparum erythrocytic stage through a putative role of PfMCA1 metacaspase-like protein. J. Infect. Dis. 2007;195:1852–1859. doi: 10.1086/518253. [DOI] [PubMed] [Google Scholar]
- Mittra B., Saha A., Chowdhury A.R., Pal C., Mandal S., Mukhopadhyay S., Bandyopadhyay S., Majumder H.K. Luteolin, an abundant dietary component is a potent anti-leishmanial agent that acts by inducing topoisomerase II-mediated kinetoplast DNA cleavage leading to apoptosis. Mol. Med. 2000;6:527–541. [PMC free article] [PubMed] [Google Scholar]
- Monge-Maillo B., López-Vélez R. Therapeutic options for old world cutaneous leishmaniasis and new world cutaneous and mucocutaneous leishmaniasis. Drugs. 2013;73:1889–1920. doi: 10.1007/s40265-013-0132-1. [DOI] [PubMed] [Google Scholar]
- Murray H.W., Berman J.D., Davies C.R., Saravia N.G. Advances in leishmaniasis. Lancet. 2005;366:1561–1577. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- Mohamed-Ahmed A.H., Brocchini S., Croft S.L. Recent advances in development of amphotericin B formulations for the treatment of visceral leishmaniasis. Curr. Opin. Infect. Dis. 2012;25:695–702. doi: 10.1097/QCO.0b013e328359eff2. [DOI] [PubMed] [Google Scholar]
- Mohanty S., Town T., Yagi T., Scheidig C., Kwan K.Y., Allore H.G., Flavell R.A., Shaw A.C. Defective p53 engagement after the induction of DNA damage in cells deficient in topoisomerase 3β. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5063–5068. doi: 10.1073/pnas.0801235105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncayo A., Silveira A.C. Current epidemiological trends for Chagas disease in Latin America and future challenges in epidemiology, surveillance and health policy. Mem. Inst. Oswaldo Cruz. 2009;104(Suppl. 1):17–30. doi: 10.1590/s0074-02762009000900005. [DOI] [PubMed] [Google Scholar]
- Nenortas E., Burri C., Shapiro T.A. Antitrypanosomal activity of fluoroquinolones. Antimicrob. Agents Chemother. 1999;43:2066–2068. doi: 10.1128/aac.43.8.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenortas E., Kulikowicz T., Burri C., Shapiro T.A. Antitrypanosomal activities of fluoroquinolones with pyrrolidinyl substitutions. Antimicrob. Agents Chemother. 2003;47:3015–3017. doi: 10.1128/AAC.47.9.3015-3017.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik-de-Sousa C.B. Vaccines for canine leishmaniasis. Front. Immunol. 2012;3:69. doi: 10.3389/fimmu.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasion S.G., Hines J.C., Aebersold R., Ray D.S. Molecular cloning and expression of the gene encoding the kinetoplast-associated type II DNA topoisomerase of Crithidia fasciculata. Mol. Biochem. Parasitol. 1992;50:57–67. doi: 10.1016/0166-6851(92)90244-e. [DOI] [PubMed] [Google Scholar]
- Pommier Y. Topoisomerase inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y., Leo E., Zhang H., Marchand C. DNA Topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada C.F., Álvarez-Velilla R., Diaz-González R., Prieto C., Pérez-Pertejo Y., Balaña-Fouce R., Reguera R.M. A pentapeptide signature motif plays a pivotal role in Leishmania DNA topoisomerase IB activity and camptothecin sensitivity. Biochim. Biophys. Acta. 2012;1820:2062–2071. doi: 10.1016/j.bbagen.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Prada C.F., Álvarez-Velilla R., Balaña-Fouce R., Prieto C., Calvo-Álvarez E., Escudero-Martínez J.M., Requena J.M., Ordóñez C., Desideri A., Pérez-Pertejo Y., Reguera R.M. Gimatecan and other camptothecin derivatives poison leishmania DNA-topoisomerase IB leading to a strong leishmanicidal effect. Biochem. Pharmacol. 2013;85:1433–1440. doi: 10.1016/j.bcp.2013.02.024. [DOI] [PubMed] [Google Scholar]
- Prada C.F., Álvarez-Velilla R., Diaz-González R., Pérez-Pertejo Y., Balaña-Fouce R., Reguera R.M. Identification and characterization of the regions involved in the nuclear translocation of the heterodimeric leishmanial DNA topoisomerase IB. PLoS One. 2013;8:e73565. doi: 10.1371/journal.pone.0073565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx M.E., Desormeaux A., Marquis J.F., Olivier M., Bergeron M.G. Treatment of visceral leishmaniasis with sterically stabilized liposomes containing camptothecin. Antimicrob. Agents Chemother. 2001;45:2623–2627. doi: 10.1128/AAC.45.9.2623-2627.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassi A., Jr., Rassi A., Marin-Neto J.A. Chagas disease. Lancet. 2010;375:1388–1402. doi: 10.1016/S0140-6736(10)60061-X. [DOI] [PubMed] [Google Scholar]
- Ray S., Hazra B., Mittra B., Das A., Majumder H.K. Diospyrin, a bisnaphthoquinone: a novel inhibitor of type I DNA topoisomerase of Leishmania donovani. Mol. Pharmacol. 1998;54:994–999. doi: 10.1124/mol.54.6.994. [DOI] [PubMed] [Google Scholar]
- Reece R.J., Maxwell A. The C-terminal domain of the Escherichia coli DNA gyrase A subunit is a DNA-binding protein. Nucl. Acids Res. 1991;19:1399–1405. doi: 10.1093/nar/19.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera R.M., Redondo C.M., Gutiérrez de Prado R., Pérez-Pertejo Y., Balaña-Fouce R. DNA topoisomerase I from parasitic protozoa: a potential target for chemotherapy. Biochim. Biophys. Acta. 2006;1759:117–131. doi: 10.1016/j.bbaexp.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Reguera R.M., Díaz-González R., Pérez-Pertejo Y., Balaña-Fouce R. Characterizing the bi-subunit type IB DNA topoisomerase of Leishmania parasites; a novel scenario for drug intervention in trypanosomatids. Curr. Drug Targets. 2008;9:966–978. doi: 10.2174/138945008786786118. [DOI] [PubMed] [Google Scholar]
- Rose D., Thomas W., Holm C. Segregation of recombined chromosomes in meiosis I requires DNA topoisomerase II. Cell. 1990;60:1009–1017. doi: 10.1016/0092-8674(90)90349-j. [DOI] [PubMed] [Google Scholar]
- Saha S., Mukherjee T., Chowdhury S., Mishra A., Chowdhury S.R., Jaisankar P., Mukhopadhyay S., Majumder H.K. The lignan glycosides lyoniside and saracoside poison the unusual type IB topoisomerase of Leishmania donovani and kill the parasite both in vitro and in vivo. Biochem. Pharmacol. 2013;86:1673–1687. doi: 10.1016/j.bcp.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Schmunis G.A. Epidemiology of Chagas disease in non-endemic countries: the role of international migration. Mem. Ins. Oswaldo Cruz. 2007;102(Suppl 1):75–85. doi: 10.1590/s0074-02762007005000093. [DOI] [PubMed] [Google Scholar]
- Scocca J.R., Shapiro T.A. A mitochondrial topoisomerase IA essential for late theta structure resolution in African trypanosomes. Mol. Microbiol. 2008;67:820–829. doi: 10.1111/j.1365-2958.2007.06087.x. [DOI] [PubMed] [Google Scholar]
- Sen N., Das B.B., Ganguly A., Mukherjee T., Tripathi G., Bandyopadhyay S., Rakshit S., Sen T., Majumder H.K. Camptothecin induced mitochondrial dysfunction leading to programmed cell death in unicellular hemoflagellate Leishmania donovani. Cell Death Differ. 2004;11:924–936. doi: 10.1038/sj.cdd.4401435. [DOI] [PubMed] [Google Scholar]
- Sengupta T., Mukherjee M., Mandal C., Das A., Majumder H.K. Functional dissection of the C-terminal domain of type II DNA topoisomerase from the kinetoplastid hemoflagellate Leishmania donovani. Nucl. Acids Res. 2003;31:5305–5306. doi: 10.1093/nar/gkg727. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sengupta T., Mukherjee M., Das A., Mandal C., Das R., Mukherjee T., Majumder H.K. Characterization of the ATPase activity of topoisomerase II from Leishmania donovani and identification of residues conferring resistance to etoposide. Biochem. J. 2005;390:419–426. doi: 10.1042/BJ20042128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta T., Mukherjee M., Das R., Das A., Majumder H.K. Characterization of the DNA-binding domain and identification of the active site residue in the gyr A half of Leishmania donovani topoisomerase II. Nucl. Acids Res. 2005;33:2364–2373. doi: 10.1093/nar/gki527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro T.A. Mitochondrial topoisomerase II activity is essential for kinetoplast DNA minicircle segregation. Mol. Cell. Biol. 1994;14:3660–3667. doi: 10.1128/mcb.14.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro T.A., Klein V.A., Englund P.T. Drug-promoted cleavage of kinetoplast DNA minicircles. Evidence for type II topoisomerase activity in trypanosome mitochondria. J. Biol. Chem. 1989;264:4173–4178. [PubMed] [Google Scholar]
- Shapiro T.A., Showalter A.F. In vivo inhibition of trypanosome mitochondrial topoisomerase II: effects on kinetoplast DNA maxicircles. Mol. Cell. Biol. 1994;14:5891–5897. doi: 10.1128/mcb.14.9.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro T.A., Englund P.T. The structure and replication of kinetoplast DNA. Annu. Rev. Microbiol. 1995;49:117–143. doi: 10.1146/annurev.mi.49.100195.001001. [DOI] [PubMed] [Google Scholar]
- Shlomai J. The structure and replication of kinetoplast DNA. Curr. Mol. Med. 2004;4:623–647. doi: 10.2174/1566524043360096. [DOI] [PubMed] [Google Scholar]
- Shlomai J., Zadok A. Reversible decatenation of kinetoplast DNA by a DNA topoisomerase from trypanosomatids. Nucl. Acids Res. 1983;11:4019–4034. doi: 10.1093/nar/11.12.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simarro P.P., Diarra A., Ruiz Postigo J.A., Franco J.R., Jannin J.G. The human African trypanosomiasis control and surveillance programme of the World Health Organization 2000–2009: the way forward. PLoS Negl. Trop. Dis. 2011;5:e1007. doi: 10.1371/journal.pntd.0001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simarro P.P., Franco J.R., Diarra A., Ruiz Postigo J.A., Jannin J.G. Update on field use of the available drugs for the chemotherapy of human African trypanosomiasis. Parasitology. 2012;139:842–846. doi: 10.1017/S0031182012000169. [DOI] [PubMed] [Google Scholar]
- Singh G., Dey C.S. Induction of apoptosis-like cell death by pentamidine and doxorubicin through differential inhibition of topoisomerase II in arsenite-resistant L. donovani. Acta Trop. 2007;103:172–185. doi: 10.1016/j.actatropica.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Singh G., Jayanarayan K.G., Dey C.S. Novobiocin induces apoptosis-like cell death in topoisomerase II over-expressing arsenite resistant Leishmania donovani. Biochem. Parasitol. 2005;141:57–69. doi: 10.1016/j.molbiopara.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Sissi C., Palumbo M. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucl. Acids Res. 2009;37:702–711. doi: 10.1093/nar/gkp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart L., Ireton G.C., Champoux J.J. The domain organization of human topoisomerase I. J. Biol. Chem. 1996;271:7602–7608. doi: 10.1074/jbc.271.13.7602. [DOI] [PubMed] [Google Scholar]
- Stewart L., Ireton G.C., Champoux J.J. Reconstitution of human topoisomerase I by fragment complementation. J. Biol. Mol. 1997;269:355–372. doi: 10.1006/jmbi.1997.1056. [DOI] [PubMed] [Google Scholar]
- Stewart L., Redinbo M.R., Qiu X., Hol W.G., Champoux J.J. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- Stewart L., Ireton G.C., Champoux J.J. A functional linker in human topoisomerase I is required for maximum sensitivity to camptothecin in a DNA relaxation assay. J. Biol. Chem. 1999;274:32950–32960. doi: 10.1074/jbc.274.46.32950. [DOI] [PubMed] [Google Scholar]
- Strauss P.R., Wang J.C. The TOP2 gene of Trypanosoma brucei: a single-copy gene that shares extensive homology with other TOP2 genes encoding eukaryotic DNA topoisomerase II. Mol. Biochem. Parasitol. 1990;38:141–150. doi: 10.1016/0166-6851(90)90214-7. [DOI] [PubMed] [Google Scholar]
- Stuart K., Panigrahi A.K. RNA editing: complexity and complications. Mol. Microbiol. 2002;45:591–596. doi: 10.1046/j.1365-2958.2002.03028.x. [DOI] [PubMed] [Google Scholar]
- Sundar S., Jha T.K., Thakur C.P., Sinha P.K., Bhattacharya S.K. Injectable paromomycin for visceral leishmaniasis in India. N. Engl. J. Med. 2007;356:2571–2581. doi: 10.1056/NEJMoa066536. [DOI] [PubMed] [Google Scholar]
- Tang S.C., Shapiro T.A. Newly identified antibacterial compounds are topoisomerase poisons in African trypanosomes. Antimicrob. Agents Chemother. 2010;54:620–626. doi: 10.1128/AAC.01025-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir D., Kaiser M., Brun R., Yardley V., Schmidt T.J., Tosun F., Rüedi P. Antitrypanosomal and antileishmanial activities of flavonoids and their analogues: in vitro, in vivo, structure–activity relationship, and quantitative structure–activity relationship studies. Antimicrob. Agents Chemother. 2006;50:1352–1364. doi: 10.1128/AAC.50.4.1352-1364.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira A.R., Hecht M.M., Guimaro M.C., Sousa A.O., Nitz N. Pathogenesis of chagas’ disease: parasite persistence and autoimmunity. Clin. Microbiol. Rev. 2011;24:592–630. doi: 10.1128/CMR.00063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng A.C., Al-Montashiri N.A., Cheng B.L., Lou P., Ozmizrak P., Chen H.H., Stewart A.F. Identification of a phosphorylation-dependent nuclear localization motif in interferon regulatory factor 2 binding protein 2. PLoS One. 2011;6:e24100. doi: 10.1371/journal.pone.0024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao Y.P., Russo A., Nyamuswa G., Silber R., Liu L.F. Interaction between replication forks and topoisomerase I-DNA cleavable complexes: studies in a cell-free SV40 DNA replication system. Cancer Res. 1993;53:5908–5914. [PubMed] [Google Scholar]
- Uemura T., Morino K., Uzawa S., Shiozaki K., Yanagida M. Cloning and sequencing of Schizosaccharomyces pombe DNA topoisomerase I gene, and effect of gene disruption. Nucl. Acids Res. 1987;15:9727–9739. doi: 10.1093/nar/15.23.9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Griensven J., Carrillo E., López-Vélez R., Lynen L., Moreno J. Leishmaniasis in immunosuppressed individuals. Clin. Microbiol. Infect. 2014;20:286–299. doi: 10.1111/1469-0691.12556. [DOI] [PubMed] [Google Scholar]
- Vennerstrom J.L., Lovelace J.K., Waits V.B., Hanson W.L., Klayman D.L. Berberine derivatives as antileishmanial drugs. Antimicrob. Agents Chemother. 1990;34:918–921. doi: 10.1128/aac.34.5.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viard T., Bouthier de la Tour C. Type IA topoisomerases: a simple puzzle? Biochimie. 2007;89:456–467. doi: 10.1016/j.biochi.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Villa H., Otero-Marcos A.R., Reguera R.M., Balaña-Fouce R., García-Estrada C., Pérez-Pertejo Y., Tekwani B.L., Myler P.J., Stuart K.D., Bjornsti M.A., Ordóñez D. A novel active DNA topoisomerase I in Leishmania donovani. J. Biol. Chem. 2003;278:3521–3526. doi: 10.1074/jbc.M203991200. [DOI] [PubMed] [Google Scholar]
- Walker J., Saravia N.G. Inhibition of Leishmania donovani promastigote DNA topoisomerase I and human monocyte DNA topoisomerases I and II by antimonial drugs and classical antitopoisomerase agents. J. Parasitol. 2004;90:1155–1162. doi: 10.1645/GE-3347. [DOI] [PubMed] [Google Scholar]
- Wang J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- Wang J.C. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- Wang J.C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wang J.C. A journey in the world of DNA rings and beyond. Ann. Rev. Biochem. 2009;78:31–54. doi: 10.1146/annurev.biochem.78.030107.090101. [DOI] [PubMed] [Google Scholar]
- Wang Z., Englund P.T. RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J. 2001;20:4674–4683. doi: 10.1093/emboj/20.17.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt P.M., Hickson I.D. Structure and function of type II DNA topoisomerases. Biochem. J. 1994;303:681–695. doi: 10.1042/bj3030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Ruttenburg A.J., Bechis S.K., Verdine G.L. Nucleotide-dependent domain movement in the ATPase domain of human type IIA DNA topoisomerase. J. Biol. Chem. 2005;280:37041–37047. doi: 10.1074/jbc.M506520200. [DOI] [PubMed] [Google Scholar]
- Werbovetz K.A., Lehnert E.K., Macdonald T.L., Pearson R.D. Cytotoxicity of acridine compounds for Leishmania promastigotes in vitro. Antimicrob. Agents Chemother. 1992;36:495–497. doi: 10.1128/aac.36.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werbovetz K.A., Spoors P.G., Pearson R.D., Macdonald T.L. Cleavable complex formation in Leishmania chagasi treated with anilinoacridines. Mol. Biochem. Parasitol. 1994;65:1–10. doi: 10.1016/0166-6851(94)90110-4. [DOI] [PubMed] [Google Scholar]
- Wethington S.L., Wright J.D., Herzog T.J. Key role of topoisomerase I inhibitors in the treatment of recurrent and refractory epithelial ovarian carcinoma. Expert Rev. Anticancer Ther. 2008;8:819–831. doi: 10.1586/14737140.8.5.819. [DOI] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; Geneva: 2008. The Global Burden of Disease: 2004 Update. [Google Scholar]
- World Health Organization . World Health Organization; Geneva: 2007. Global Plan to Combat Neglected Tropical Diseases 2008–2015. [Google Scholar]
- Yang J., Bogni A., Schuetz E.G., Ratain M., Dolan M.E., McLeod H., Gong L., Thorn C., Relling M.V., Klein T.E., Altman R.B. Etoposide pathway. Pharmacogenet. Genomics. 2009;19:552–553. doi: 10.1097/FPC.0b013e32832e0e7f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Bachrati C.Z., Ou J., Hickson I.D., Brown G.W. Human topoisomerase IIIalpha is a single-stranded DNA decatenase that is stimulated by BLM and RMI1. J. Biol. Chem. 2010;285:21426–21436. doi: 10.1074/jbc.M110.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C.X., Roche C.J., Tse-Dinh Y.C. Effect of Mg(II) binding on the structure and activity of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 1997;272:16206–16210. doi: 10.1074/jbc.272.26.16206. [DOI] [PubMed] [Google Scholar]


