Abstract
Assessing the level of genetic diversity within a germplasm collection contributes to evaluating the potential for its utilization as a gene pool to improve the performance of cultivars. In this study, 45 high-quality simple sequence repeat (SSR) markers were screened and used to estimate the genetic base of a world-wide collection of 248 rapeseed (Brassica napus) inbred lines. For the whole collection, the genetic diversity of A genome was higher than that of C genome. The genetic diversity of C genome for the semi-winter type was the lowest among the different germplasm types. Because B. oleracea is usually used to broaden the genetic diversity of C genome in rapeseed, we evaluated the potential of 25 wild B. oleracea lines. More allelic variations and a higher genetic diversity were observed in B. oleracea than in rapeseed. One B. oleracea line and one oilseed B. rapa line were used to generate a resynthesized Brassica napus line, which was then crossed with six semi-winter rapeseed cultivars to produce 7 F1 hybrids. Not only the allele introgression but also mutations were observed in the hybrids, resulting in significant improvement of the genetic base.
Keywords: genetic diversity, allelic variation, Brassica oleracea, resynthesized Brassica napus, SSR mutation
Introduction
Rapeseed (Brassica napus L., AACC, 2n = 38), the second most important oilseed crop in the world after soybean, is a recent amphidiploid species originated from a spontaneous interspecific hybridization of B. rapa (AA, 2n = 20) and B. oleracea (CC, 2n = 18) during medieval times (Iñiguez-Luy and Federico 2011, U 1935). This species includes rapeseed, rutabaga or swede, vegetable types and fodder crops (Snowdon et al. 2007). Rapeseed came into cultivation 400 years ago in Europe and initially formed a winter oilseed rape (OSR) and a spring OSR, which were grown in distinct environments (Gómez-Campo and Prakash 1999, Prakash et al. 2011). Rapeseed was introduced from Europe and indirectly from Japan to China in the 1930–1940s, which was followed by intensive crossing with the Chinese traditional oilseed crop B. rapa for adaption to local environments (Liu 1985, Qian et al. 2006, Xiao et al. 2012). The rapeseed cultivated in China is primarily semi-winter, which requires a shorter vernalization period to flower than the winter type.
Although both B. oleracea and B. rapa have a great diversity of morphotypes with various origins, B. napus most likely originated from a few interspecific hybridizations between B. rapa and B. oleracea (Iñiguez Luy and Federico 2011), which led to a relatively narrow genetic diversity. Furthermore, under the strong breeding selection for zero seed erucic acid and low seed glucosinolate content, current oilseed rape cultivars have a narrower genetic basis (Bus et al. 2011, Delourme et al. 2013). The genetic diversity of semi-winter OSR has been estimated to be lower than those of winter and spring OSR (Bus et al. 2011, Qian et al. 2006), however, only a few accessions of semi-winter OSR were used in these studies. Recently, the exchanges of germplasm between China and Europe, Australia and Canada are more frequent than ever before; therefore it is important to understand the genetic bases of these three different types.
Because genetically diverse germplasm is a potentially valuable source for the improvement of pathogen and pest resistance, tolerance to abiotic stresses, and heterosis, significant effort has been made to broaden the genetic base of rapeseed germplasm using different strategies. One strategy is to use genetically diverse germplasm from other geographical regions to cross with the adapted cultivars (Qian et al. 2007, Quijada et al. 2004, Udall et al. 2004). Another strategy is utilization of the diploid progenitor species of B. napus, i.e., B. rapa and B. oleracea, both of which have enormous variability in morphology, agronomic characteristics and stress tolerance (Gómez-Campo 1980, Prakash et al. 2011). Introgression of the genomic components of B. rapa into B. napus has been used in the breeding of numerous Chinese cultivars, as mentioned earlier. Recently, a large-scale and highly efficient method was established to develop new types of B. napus carrying genomic components of B. rapa via the bridge of a hexaploid (AACCCC) derived from hybridization between B. napus and B. oleracea (Li, Q. et al. 2013). Resynthesis of B. napus using B. rapa and B. oleracea might be the most extensively adopted method to develop novel B. napus germplasm (Becker et al. 1995, Girke et al. 2012a, Jesske et al. 2013, Seyis et al. 2003). The wild Brassica species that have the same chromosome number (n = 9) as B. oleracea, including B. incana, B. bourgaei, etc. have also successfully been used to generate resynthesized B. napus lines (Jesske et al. 2013). DNA molecular marker analysis has revealed that these resynthesized B. napus lines are genetically highly divergent from B. napus inbred lines/cultivars (Becker et al. 1995, Girke et al. 2012b, Jesske et al. 2013, Seyis et al. 2003). Furthermore, surprisingly high yields were observed in some hybrids resulted from crosses between resynthesized B. napus lines and adapted cultivars (Girke et al. 2012a, Jesske et al. 2013). However, so far, the genetic base of these hybrids has rarely been studied.
The objectives of the present study were to (a) compare the genetic diversities of different germplasm types of rapeseed, (b) appraise the genetic basis of a wild B. oleracea germplasm collection, and (c) broaden the genetic diversity of rapeseed by crossing with resynthesized B. napus and assess their allele variation.
Materials and Methods
Plant materials
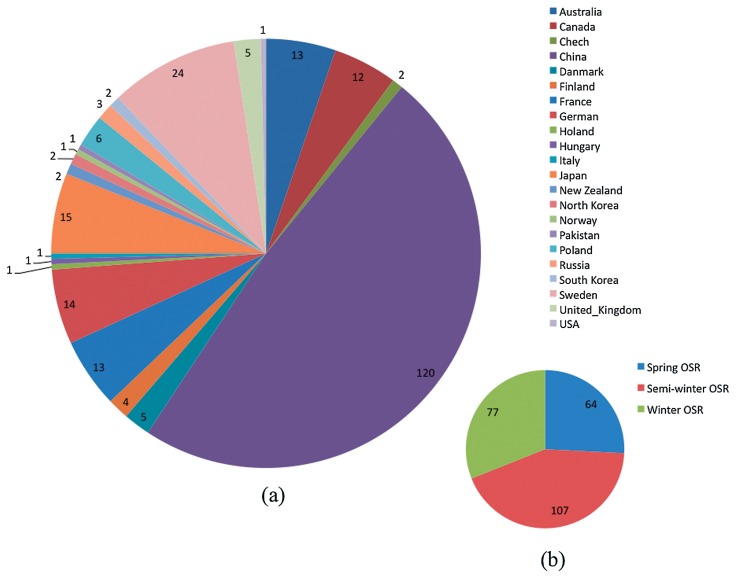
Eight double haploid (DH) lines derived from geographically different inbred lines of B. napus (Supplemental Table 1) were employed to screen SSR primers. To investigate the genetic diversity of the A and C genomes of B. napus, a total of 248 rapeseed inbred lines collected in the National Mid-term Gene Bank for Oil Crops of China, including 77 winter OSR lines, 107 semi-winter OSR lines and 64 spring OSR lines (Fig. 1, Supplemental Table 2), were used in this study. Based on their origins, 140 lines originated from Asia, 80 from Europe, 15 from Oceania, and 13 from North America. Considering broadening the genetic diversity of rapeseed, the genetic diversity of a collection of 25 wild B. oleracea (B. oleracea ssp. oleracea) lines was evaluated (Supplemental Table 3). These wild B. oleracea lines were collected from rocky Atlantic coasts of Spain (Bay of Biscay) and cultivated perennially in a glass house of Oil Crops Research Institute, CAAS. A line ‘C3-8’ of wild B. oleracea was randomly selected to cross with a cultivar of B. rapa, ‘Jingxuanhuangxinwu’, to develop a resynthesized B. napus line ‘Res-1’. Subsequently, Res-1 was crossed as a female parent with five modern cultivars of semi-winter OSR (male parents), and reciprocally crossed with another one modern cultivar, ‘Zhongshuang12’, of semi-winter OSR to produce F1 lines (Supplemental Table 3).
Fig. 1.
A summarized description of 248 rapeseed lines. a. Based on regional origins; b. Based on germplasm types.
DNA isolation and SSR genotyping
Genomic DNA was extracted from young leaves using the CTAB method (Saghai-Maroof et al. 1984). SSR primer pairs from different resources were used for genotyping. Primers with Ol, Na, BRAS, and CB prefixes were from http://www.brassica.info/resource/markers.php, and those prefixed with BnGMS, BnEMS, BrGMS, and BoGMS were developed by Huazhong Agriculture University (Cheng et al. 2009, Fan et al. 2010, Li, H. et al. 2011, 2013, Wang et al. 2011, Xu et al. 2010).
The genomic DNA of the eight DH lines was used to screen SSR primers. A polymerase chain reaction (PCR) was performed in a reaction mixture of 10 μl containing 10 ng genomic DNA as a PCR template, 0.5 μM of primers, 0.5 U Qiagen Taq polymerase (Applied Biosystems, USA), 1× PCR buffer (Applied Biosystems), and 200 μM of dNTPs. PCR amplification was conducted using touchdown methodology with 3 minutes of initial denaturation followed by 10 cycles of denaturing at 94°C for 30 s, annealing at 60°C for 30 s (temperature reduced by 0.5°C for each cycle) and extension at 72°C for 30 s, then by 30 cycles of 94°C for 30 seconds, 57°C for 30 seconds, and 72°C for 30 seconds with a final extension of 20 min at 72°C. The PCR products were denatured and visualized on 6% denaturing polyacrylamide gel, and only those that could produce less than two amplicons, i.e, two loci, were selected.
The selected SSR primers were labeled by fluorescence-dye 6-FAM and used to amplify the genomic DNA of the eight DH lines again. The PCR products with internal size standard (GeneScan™ 500 LIZ® from Applied Biosystems) were denatured, and capillary electrophoresis was performed on a 3730xl DNA Analyzer (Applied Biosystems). Subsequently, GeneMapper v4.0 (Applied Biosystems) was used for the allele size estimation. The SSR markers whose allelic variations could easily be ascertained were selected and used to analyze the genetic diversity of B. napus and B. oleracea. Multiplexed analysis by mixing two or three PCR products with significantly different sizes was employed in this study to reduce cost.
The positions of the selected SSR markers on the chromosomes were determined based on previous genetic maps (Cheng et al. 2009, Fan et al. 2010, Li, H. et al. 2011, 2013, Wang et al. 2011, Xu et al. 2010). For markers that were not located on the existing genetic maps, their chromosome information was inferred by aligning their sequences with the genome sequences of B. rapa and B. oleracea by using BLASTn (http://brassicadb.org/brad/index.php) (Cheng et al. 2011).
Data analysis
For each SSR locus, allelic variations were scored as 1 for presence or 0 for absence of an allele. The number of alleles (Na), gene diversity (D), polymorphism information content (PIC), and Nei’s distance (Nei et al. 1983) were estimated using PowerMarker version 3.51 (Liu et al. 2005). The differences in genetic diversity indices between different subgenomes or groups were assessed using one-way analysis of variance (ANOVA) implemented in SPSS 9.0. The expected heterozygosity (He) and observed heterozygosity (Ho) were calculated using Genepop 4.2.2 (Rousset 2008). Genepop 4.2.2 was also used to estimate the inbreeding coefficients (FIS) (Weir and Cockerham 1984) and perform Hardy-Weinberg equilibrium exact test.
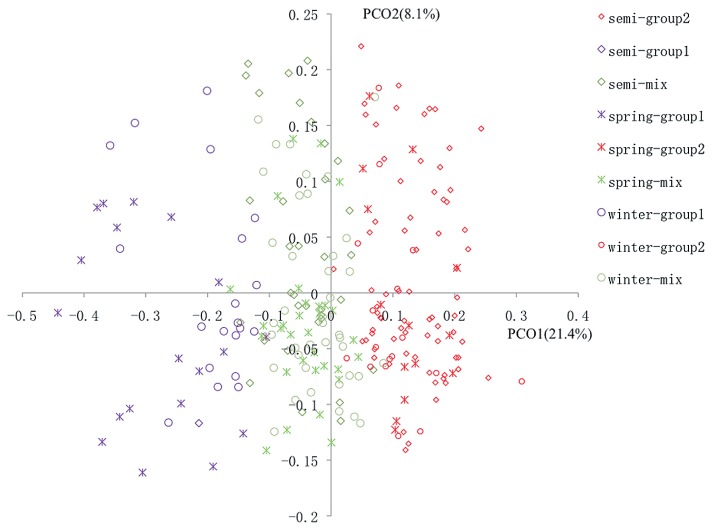
Nei’s genetic distance matrix among individuals was used to construct dendrograms using the UPGMA (unweighted pair-group method with arithmetic means) method implemented in PowerMarker, and the dendrograms were drawn using the program MEGA5 (http://www.megasoftware.net/mega.html) (Tamura et al. 2011). Nei’s genetic distance matrix was also imported into the software NTSYS-pc 2.1 (Rohlf 1992) to perform principal coordinate (PCO) analysis. The 2-D plots were made from the first two eigenvectors.
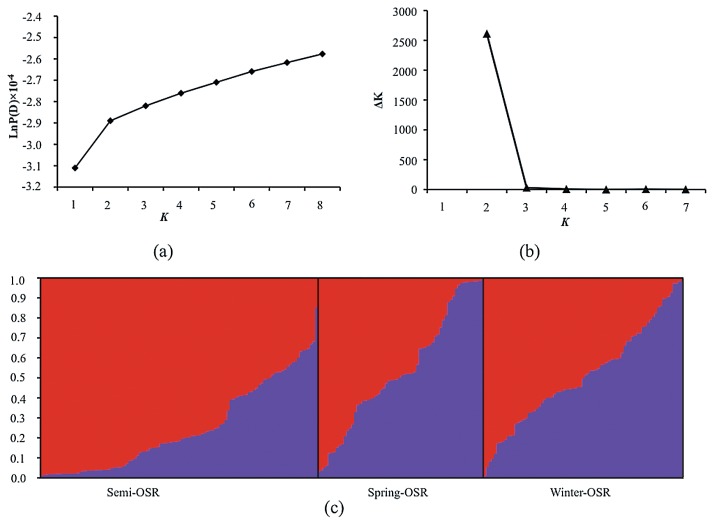
The software package STRUCTURE v2.3.4 (Pritchard et al. 2000) was used to infer population structure using a burn-in of 10,000, run length of 100,000, and a model allowing for admixture and correlated allele frequencies. Five independent runs were performed with K value (the putative number of genetic groups) varying from 1 to 8. The most likely K value was determined by log probability of data (LnP(D)) and an ad hoc statistic ΔK based on the rate of change of LnP(D) between successive K value, following Evanno et al. (2005). Lines with probability of membership ≥0.7 were assigned to corresponding clusters, and those <0.7 were assigned to a mixed group.
SSR mutation rate was calculated as the number of the mutated alleles divided by the total number of observed alleles. For estimating the SSR mutation rates in resynthesized B. napus and the F1 lines, respectively, only the alleles mutated in each generation were considered.
Results
Identification of genome-specific SSR markers
According to the results of denaturing polyacrylamide gel electrophoresis and capillary electrophoresis, 43 SSR primer pairs that could produce less than two loci and whose allelic variations could be clearly discriminated were selected (Supplemental Table 4). The sizes of the PCR amplicons of 20 primer pairs for the SSR markers located in the C genome were compared in B. napus and B. oleracea. The primer pairs that could detect a single locus in both B. oleracea and B. napus were considered to be C-genome-specific. As a result, 18 pairs except for BnEMS0628 and BoGMS0949 were identified (Supplemental Table 4). The primer pair BnEMS0628 was reported to detect two loci, BnEMS0628a and BnEMS0628b, on C9 of B. napus (Wang et al. 2011), but could be detected only one locus with allele sizes of 267, 271, and 274 bp in B. oleracea. The allele sizes of BnEMS0628a included 267 and 271 bp, and those of BnEMS0628b included 258 and 261 bp in 248 inbred lines of B. napus (Supplemental Fig. 1, Supplemental Table 4). Aligning the sequence of BnEMS0628 against the genome of B. rapa by BLASTn showed that BnEMS0628 was located on A9 with an allele size of 260 bp. Therefore, we inferred that BnEMS0628a might derive from progenitor B. oleracea and BnEMS0628b from B. rapa. Using the same method, we inferred that BoGMS0949a is located on C5 and BoGMS0949b on A10 in B. napus. Among the 23 primer pairs of SSR markers that were reported to be located in the A genome, 22 pairs with the exception of CB10524 could be detected only one locus (Supplemental Table 4), indicating they were A-genome-specific. CB10524 could detect two loci in B. napus, including one locus, CB10524a, with allelic variations of 216, 222, and 224 bp and the other no polymorphism locus, CB10524b, with only one allelic variation of 237 bp in 248 inbred lines of B. napus. CB10524a was reported to be on A10 of B. napus, and because no polymorphism was detected for CB10524b, CB10524b was not used in the present study. Hence, a total of 45 SSR markers, of which 24 markers were A-genome-specific and 21 markers were C-genome-specific, were employed for assessing the genetic diversity of rapeseed in this study (Supplemental Table 4).
Genetic structure analysis of the rapeseed inbred lines
Clustering inference performed with possible clusters (K) from 1 to 8 showed that the most significant change of likelihood occurred when K increased from 2 to 3, and the highest ΔK value was observed at K = 2 (Fig. 2). Both parameters suggested that the 248 genotypes could be assigned into two groups. Using a probability of membership threshold of 70%, 40 and 103 lines were assigned into the two groups, respectively. The remaining 105 lines were classified into a mixed group. The results of clustering and PCO analysis were incorporated and shown in Fig. 3. Almost all of the lines in group1 were spring and winter OSR except of one line of semi-winter OSR. The group2 included 72 semi-winter lines, 14 spring lines, and 17 winter OSR lines. The UPGMA dendrogram also displayed two clear clades, one of which was mostly comprised of winter and spring OSR, another of which contained three OSR types (Supplemental Fig. 2a). A subtree, consisted of four cultivars developed by Oil Crops Research Institute, CAAS, was consistent with their pedigree (Supplemental Fig. 2b), indicating the reliability of the results.
Fig. 2.
Analysis of the population structure of 248 rapeseed accessions by STRUCTURE. a. Estimated LnP(D) of possible clusters (K) from 1 to 8; b. ΔK based on the rate of change of LnP(D) between successive K; c. Population structure based on K = 2. Each individual is represented by a thin vertical line, which is partitioned into red and violet segments that represent the individual’s estimated membership fractions in two clusters. Black lines separate individuals of different germplasm types, which are labeled below the figure.
Fig. 3.
Principal coordinate (PCO) analysis of 248 rapeseed lines based on 45 SSR marekers. PCO1 and PCO2 are the two first principal coordinates and the proportion of variance explained by these coordinates is indicated in parentheses. Colors represent groups defined by STRCTURE and symbols represent germplasm types.
Genetic diversity in the rapeseed inbred lines
In the panel of 248 inbred lines of B. napus, a total of 83 alleles were detected at 24 SSR loci with 3.46 alleles per locus in the A genome and 61 alleles at 21 SSR loci with 2.9 alleles per locus in the C genome (Table 1). The mean D and PIC of the SSR markers in the A genome were 0.50 and 0.44, respectively, whereas those of the C genome was 0.43 and 0.36, respectively. For the individual germplasm type, the mean D and PIC in the A genome showed no significant difference, whereas the mean D of semi-winter OSR in the C genome were 0.36, significantly lower than those of the other types in the C genome and those of all types in the A genome (Table 1).
Table 1.
Number of alleles (Na), genetic diversity (D), polymorphism information content (PIC) between A and C genomes for 248 B.napus inbred lines based on 45 SSR markers
| Genome | Genetic group | Na | D | PIC | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||
| Total | Range | Meana | Range | Meana | Range | Meana | ||
| A Genome | Semi-winter OSR | 80 | 2–6 | 3.33a | 0.12–0.69 | 0.50a | 0.11–0.64 | 0.43a |
| Spring OSR | 79 | 2–6 | 3.29a | 0.21–0.76 | 0.49a | 0.19–0.73 | 0.43a | |
| Winter OSR | 80 | 2–6 | 3.33a | 0.22–0.77 | 0.48a | 0.19–0.73 | 0.42a | |
| Total | 83 | 2–6 | 3.46a | 0.17–0.74 | 0.50a | 0.16–0.70 | 0.44a | |
| C Genome | Semi-winter OSR | 54 | 2–4 | 2.57b | 0.08–0.70 | 0.36b | 0.07–0.65 | 0.30b |
| Spring OSR | 59 | 2–5 | 2.80ab | 0.08–0.67 | 0.45a | 0.08–0.63 | 0.37ab | |
| Winter OSR | 59 | 2–5 | 2.81ab | 0.19–0.67 | 0.46a | 0.18–0.61 | 0.38ab | |
| Total | 61 | 2–5 | 2.90ab | 0.17–0.69 | 0.43a | 0.16–0.63 | 0.36ab | |
Mean values marked with the same letter are not significantly different (P = 0.05).
Based on the SSR markers on the A genome, the average genetic distances of pairs of inbred lines within semi-winter OSR (intra-semi-winter OSR) was 0.465, which was significantly higher than those of other intra-groups, whereas, based on the SSR markers on the C genome or all of the 45 SSR markers, the average genetic distance of intra-spring OSR was the highest (Table 2). Among the average genetic distances between pairs of inbred lines from different groups, those of semi-winter OSR vs. spring OSR were the highest, i.e, 0.477 for the A genome, 0.411 for the C genome and 0.447 for the whole genome.
Table 2.
The genetic distance in intra-group and inter-group calculated by Nei’s method (Nei 1983) based on 45 SSR markers
| Genetic group | A genome | C genome | A and C genome | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Mean ± SDa | Maximum | Minimum | Mean ± SDa | Maximum | Minimum | Mean ± SDa | Maximum | Minimum | |
| Intra-group | |||||||||
| Semi-winter OSR | 0.465 ± 0.100e | 0.804 | 0.066 | 0.343 ± 0.115a | 0.799 | 0.015 | 0.409 ± 0.083a | 0.722 | 0.071 |
| Spring OSR | 0.447 ± 0.108c | 0.882 | 0.100 | 0.407 ± 0.119c | 0.815 | 0.083 | 0.428 ± 0.090bc | 0.752 | 0.118 |
| Winter OSR | 0.420 ± 0.100a | 0.790 | 0.155 | 0.400 ± 0.114b | 0.805 | 0.069 | 0.411 ± 0.083a | 0.760 | 0.144 |
| Inter-group | |||||||||
| Semi-winter OSR VS. Spring OSR | 0.477 ± 0.108f | 0.894 | 0.123 | 0.411 ± 0.131c | 0.865 | 0.031 | 0.447 ± 0.096d | 0.826 | 0.157 |
| Semi-winter OSR VS. Winter OSR | 0.460 ± 0.097d | 0.845 | 0.142 | 0.394 ± 0.117b | 0.815 | 0.017 | 0.430 ± 0.081c | 0.745 | 0.125 |
| Winter OSR VS. Spring OSR | 0.441 ± 0.105b | 0.871 | 0.115 | 0.407 ± 0.114c | 0.823 | 0.081 | 0.425 ± 0.085b | 0.784 | 0.163 |
Mean ± SD values marked with the same letter are not significantly different (P = 0.05).
Variance of genetic diversity of the C genome in wild B. oleracea and rapeseed
The C-genome-specific SSR markers with the exception of BnEMS0628b were used to analyze the 25 wild B. oleracea lines. A total of 75 alleles were detected at 20 SSR loci with 3.75 alleles per locus in the C genome, which was significantly higher than the number found in 248 B. napus lines (Table 3). Among all of the allelic variations (85) detected in the C genomes of B. napus and B. oleracea, 10 (10/85 = 11.8%) were unique to B. napus and 28 (28/85 = 32.9%) to B. oleracea. The mean genetic diversity and PIC of the SSR markers in the 25 B. oleracea lines were 0.50 and 0.45, respectively, slightly higher than those in the 248 B. napus inbred lines (0.44 and 0.37, respectively). The expected heterozygosities of the B. oleracea and B. napus inbred lines/culitvars were higher than their observed heterozygosities. Large heterozygote deficit was observed in the B. napus inbred lines with Fis of 0.72, indicating a serious inbreeding depression. The Fis of the wild B. oleracea was also high as 0.47, showing significant departure from Hardy–Weinberg equilibrium, which might be caused by the small population. The cross of synthesized B. napus and rapeseed cultivars significantly increased the genetic diversity, causing the observed heterozygosity higher than expected heterozygosity and FIS less than 0.
Table 3.
Number of alleles per locus (Na), genetic diversity (D), polymorphism information content (PIC) of the 20 SSR markers on C genome in B. oleracea and B. napus
| Genetic groups | Na | D | PIC | Heterozygosity | FISb | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||
| Total | Range | Meana | Range | Meana | Range | Meana | Expected (He) | Observed (Ho) | ||
| B. oleracea | 75 | 1–9 | 3.75c | 0–0.76 | 0.50b | 0–0.74 | 0.45b | 0.52 | 0.27 | 0.47* |
| Inbred lines of B. napus | 59 | 2–5 | 2.95b | 0.16–0.66 | 0.44ab | 0.15–0.60 | 0.37ab | 0.44 | 0.13 | 0.72* |
| Modern cultivars of B. napus | 42 | 1–3 | 2.10a | 0–0.63 | 0.38a | 0–0.55 | 0.31a | 0.41 | 0.13 | 0.69* |
| F1 of B. napus | 49 | 2–4 | 2.45ab | 0.13–0.66 | 0.44ab | 0.12–0.59 | 0.37ab | 0.48 | 0.68 | −0.46 |
The values marked with the same letter are not significantly different (P = 0.05).
Asterisks indicate significant departure from Hardy–Weinberg equilibrium (P = 0.05).
Allele mutation in the resynthesized B. napus and its offsprings
Compared with the alleles in the resynthesized B. napus line Res-1and its C genome donor ‘C3-8’, 4 (10%) mutant SSR alleles were observed in Res-1 (Table 4). Seven F1 hybrids were obtained by crossing six modern cultivars of semi-winter OSR and Res-1. Comparing the alleles in the parental Res-1 and modern cultivars with those in F1 lines revealed a total of 26 (9.3%) mutant SSR alleles in the generation of F1 lines. Different combinations had different rates of SSR mutation. The highest rate, 20%, was observed in the F1 lines of ‘Zhongshuang12 × Res-1’ and ‘Res-1 × Zheyou17’, whereas there were no SSR mutations in two F1 lines of ‘Res-1 × Huyou21’ and ‘Res-1 × G142’. Different rates of SSR mutation were also observed in the F1 lines of the reciprocal cross between ‘Zhongshuang12’ and ‘Res-1’. F1 of ‘Res-1 × Zhongshuang12’ exhibited a mutation rate of 15.0%. Random mutations, including insertions or deletions of different repeats of motifs, occurred in each SSR marker in the F1 lines (Table 5). The locus CB10369 with motif of GAT had the highest mutation rate of 29.4%. Noticeably, an insertion and a deletion of 20 motifs were observed at the locus of BnGMS631 in two F1 lines.
Table 4.
SSR mutations rate occurred in resynthesized B. napus and each F1 line derived from resynthesized B. napus and modern rapeseed cultivar
| Materials | Number of locia | Number of mutations | Number of alleles | Mutation rate (%) |
|---|---|---|---|---|
| Res-1 | 20 | 4 | 40 | 10.0 |
| Res-1 (♀) × Zhongshuang12 (♂) | 20 | 6 | 40 | 15.0 |
| Zhongshuang12 (♀) × Res-1 (♂) | 20 | 8 | 40 | 20.0 |
| Res-1 (♀) × Gaoyouzhongshuang11 | 20 | 1 | 40 | 2.5 |
| Res-1 (♀) × Huyou21 (♂) | 20 | 0 | 40 | 0.0 |
| Res-1 (♀) × H40 (♂) | 20 | 3 | 40 | 7.5 |
| Res-1 (♀) × G142 (♂) | 20 | 0 | 40 | 0.0 |
| Res-1 (♀) × Zheyou17 (♂) | 20 | 8 | 40 | 20.0 |
| Total of F1 lines | 140 | 26 | 280 | 9.3 |
The 20 C-genome specific markers were used in this study.
Table 5.
Mutation rate of each microsatellite locus in 7 F1 lines derived from resynthesized B. napus and modern rapeseed cultivars
| Locusa | Total number of observed alleles | Motif | Mutationsb | Total number of mutations | Mutation rate (%) |
|---|---|---|---|---|---|
| BnEMS0158 | 14 | AGA | −3 | 1 | 7.1 |
| BnEMS0628 | 14 | AG | −1, +1 | 2 | 14.3 |
| BnGMS3 | 14 | CTT | −2, +10 | 2 | 14.3 |
| BnGMS509 | 14 | AG | +1 | 1 | 7.1 |
| BnGMS631 | 14 | TA | −20, +20 | 2 | 14.3 |
| BnGMS633 | 14 | AT | −1 | 1 | 7.1 |
| BoGMS0486 | 14 | ATT | +3 | 1 | 14.3 |
| BoGMS0738 | 14 | TCT | +2 | 1 | 7.1 |
| BoGMS0949 | 14 | TC | +1 | 1 | 7.1 |
| BoGMS1909 | 14 | AT | +2, +7 | 2 | 14.3 |
| BoGMS2095 | 14 | AT | +4 | 2 | 14.3 |
| BoGMS2499 | 14 | AG | −9 | 1 | 7.1 |
| CB10258 | 14 | CAT | −1, +2 | 2 | 14.3 |
| CB10320 | 14 | CT | −3, +3 | 2 | 14.3 |
| CB10369 | 14 | GAT | −3c, +3, +6, +9 | 5 | 29.4 |
The loci that no mutations were observed were not listed.
The positive value indicates motif insertion and the negative value indicates motif deletion.
The same motif deletions were observed in two F1 lines.
Discussion
Because the amphidiploid B. napus contains two homologous but divergent A and C genomes that had been triplicated relative to A. thaliana (Cheung et al. 2009, Town et al. 2006), multiple loci are usually detected by a pair of SSR primers (Cheng et al. 2009, Li, H. et al. 2013, Piquemal et al. 2005). In the process of analyzing multi-locus SSRs, it is usually difficult to assign alleles to specific loci, which easily results in ambiguous genotyping. In the present study, we screened 41 high-quality primer pairs of single-locus SSRs using both denaturing polyacrylamide gel electrophoresis and capillary electrophoresis. According to the previous genetic maps and allele sizes of the SSRs detected in B. napus and B. oleracea and those estimated in B. rapa, the alleles were precisely assigned to specific SSR loci. Due to the limited number of primer pairs of single-locus SSRs, those of double-locus SSRs were also considered. It was estimated that approximately 35.9% SSR primers could produce two amplicons (Li, H. et al. 2013). Using this method, the abovementioned shortcoming of double-locus SSRs could be overcome and more SSR primers could be used for detection of genome-specific allelic variation and population genetics studies.
The 248 inbred lines were classified into two groups using the STRUCTURE, which was consistent with the results of Xiao et al. (2012). One group mainly consisted of semi-winter OSR, and partial of winter and spring OSR as well, which is reasonable since the semi-winter OSR was genetically derived from winter and spring OSR. However, in another group only one line of semi-winter OSR was comprised and other lines were winter or spring OSR, indicating that these germplasm resources were nearly not utilized in the breeding of semi-winter OSR cultivars. We also observed that some spring OSR lines and winter OSR lines did not show distinct clustering which might be due to relatively short history of domestication of rapeseed (Gómez-Campo and Prakash 1999, Prakash et al. 2011) or the mutual introgression between different gene pools (Butruille et al. 1999, Rahman 2013). These results also suggested that population structure had not strong contribution to the genetic differentiation between the three germplasm types in our rapeseed collection. However, these three germplasm types were usually regarded to be distinctly different types according to their growth habit and different geographic distribution, and in previous studies, comparisons of the genetic diversities in these three types were often performed (Bus et al. 2011, Qian et al. 2006). Therefore, the genetic diversities of the three germplasm types were compared in our study.
Using the selected 45 SSR markers, a total of 144 alleles with an average of 3.16 alleles per locus were detected in 248 rapeseed inbred lines. The number of alleles per locus was similar to that detected in 192 rapeseed inbred lines by Xiao et al. (2012) but lower than that detected in 509 B. napus species-wide inbred lines (Bus et al. 2011), which might be due to more germplasm types, including fodder, swede, and vegetable types, being included in the materials of that study. The availability of genome-specific primer pairs makes it possible to assess genetic diversity separately for the A and C genomes. The average number of alleles per locus, genetic diversity, and PIC of the markers on the A genome were slightly higher than those on the C genome for the entire germplasm set as well as three germplasm types. This result is consistent with previous studies (Bus et al. 2011, Li et al. 2014), which might be explained by more natural or artificial introgression to the B. napus from B. rapa than from B. oleracea. The interspecific crosses between B. napus canola and B. rapa occur readily, while those between B. napus and B. oleracea more difficultly (Myers 2006). The genetic diversity indices of different germplasm types vary significantly (Bus et al. 2011, Delourme et al. 2013, Hasan et al. 2006); however, that of the semi-winter type has seldom been studied. Although 139 lines from China were used in the materials of Xiao et al. (2012), as the pedigree information of many inbred lines was unknown, the genetic diversity values of the semi-winter type were not compared with other types. The present study showed that the genetic diversity of the C genome for the semi-winter type was the lowest, which might be due to its short cultivation history of only approximately eighty years (Liu 1985). These results suggest that the genetic diversity of the C genome should be broadened, especially for the semi-winter type. Interestingly, the genetic diversity of the A genome in the semi-winter type was significantly higher than those in both spring and winter OSR and might be explained by the fact that the Chinese B. rapa was intensively introgressed into B. napus as mentioned earlier (Liu 1985, Qian et al. 2006, Xiao et al. 2012). Since the Chinese traditional oil crop B. rapa has much longer evolution and domestication history than that of B. napus, the semi-winter OSR might have some unique characters which might be introgressed into the breeding gene pools in other counties. For example, a cultivar Zhongyou821 was regarded to have strong resistance to Sclerotinia sclerotiorum, and this cultivar was extensively studied (Zhao et al. 2009, http://www.canolacouncil.org/media/504998/randy_kutcher.pdf).
The C genome of B. napus is closely related to that of B. oleracea and its related wild types, such as B. incana, B. bourgeaui, B. montana, etc. (Mei et al. 2011). All of them have the same chromosome number and show a wide range of morphological and genetic diversity (Gómez-Campo 1980, Mei et al. 2011, Prakash et al. 2011). The present study revealed that the genetic diversity of only 25 wild lines of B. oleracea was higher than that of 248 lines of B. napus, indicating the enormous potential of B. oleracea and its related wild types for broadening the genetic base of the C genome of B. napus. The resynthesized B. napus lines derived from an artificial cross between the parental species B. rapa and B. oleracea and its closely related wild types have been used for many years to broaden the genetic diversity of B. napus (Becker et al. 1995, Girke et al. 2012b, Jesske et al. 2013). However, only the genetic diversity of the resynthesized B. napus was studied in those works. In the present study, we noticed that not only the introgression of allelic variations in B. oleracea through resynthesized B. napus but also the SSR mutations that occurred in the process of development of resynthesized B. napus and its successive generations obviously widened the genetic base of B. napus. The SSR mutations appeared to be random, and 0–20% mutation rates were observed in resynthesized B. napus and its F1 lines derived from crossing with other cultivars. No SSR mutations were observed in two F1 lines, which might be due to the nature of random SSR mutation and the limited number of SSR markers used in this study. SSRs are tandem repeats of a basic motif (1–6 bp) and exist throughout the genome. Such genomic regions are intrinsically instable and prone to mutation by mechanisms of replication slippage or unequal crossing over (Ellegren 2004). Mutation rates have been estimated for a variety of crop plants, ranging from 0 to 5 × 10−3 per locus per generation (Marriage et al. 2009). Exposed to external stresses such as irradiation, oxidative damage, in vitro culture etc., will increase SSR mutation rates. For example, in a DH population of B. rapa, a 1.9% mutation rate was observed (Yu et al. 2013); However, 31.3% novel SSR mutations were revealed in a recombinant inbred line (RIL) population of B. napus with substantial A genome introgressions from B. rapa (Zou et al. 2011), indicating that interspecific hybridization significantly increases the SSR mutation rate. In present study, the highest mutation rate was 29.4% per locus per generation. These high-rate SSR mutations are obviously different from those occurred in generations derived from self-cross. It was revealed that the number of genomic rearrangement events and the number of novel SSR variants were positively correlated, and the novel genomic variations were associated with a higher number of QTLs for agronomic traits in the RIL population than the alleles alone that were introgressed from the A genome of B. rapa (Zou et al. 2011). A high frequency of structural genomic changes has been observed in early generations of the resynthesized B. napus (Song et al. 1995, Xiong et al. 2011), which can lead to altered gene expression (Chen and Ni 2006, Osborn et al. 2003) and proteomic changes (Kong et al. 2011). To our knowledge, the present study is the first to detect the averagely high rate of allele mutation in the resynthesized B. napus and its F1 hybrids. These allele mutations might be associated with genomic rearrangement, which, along with the allele introgression and effects of heterosis, might collaboratively contribute to the surprisingly high yield of the hybrids derived from the resynthesized B. napus lines and the adapted cultivars which was observed in previously studies (Jesske et al. 2013, Udall et al. 2004).
Supplementary Information
Acknowledgments
The authors thank anonymous reviewers for their comments on the manuscript and constructive suggestions. This work was supported by National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (2011BAD35B09), Chengguang Program for Young Scientists of Wuhan Municipal Government (2013070104010031), the National Natural Science Foundation of China (31301360, 31101124), and the Strategic Japanese-Chinese Cooperative Program on “Climate Change” (2012DFG90290).
Literature Cited
- Becker, H.C., Engqvist, G.M. and Karlsson, B. (1995) Comparison of rapeseed cultivars and resynthesized lines based on allozyme and RFLP markers. Theor. Appl. Genet. 91: 62–67. [DOI] [PubMed] [Google Scholar]
- Bus, A., Körber, N., Snowdon, R.J. and Stich, B. (2011) Patterns of molecular variation in a species-wide germplasm set of Brassica napus. Theor. Appl. Genet. 123: 1413–1423. [DOI] [PubMed] [Google Scholar]
- Butruille, D.V., Guries, R.P. and Osborn, T.C. (1999) Increasing yield of spring oilseed rape hybrids through introgression of winter germplasm. Crop Sci. 39: 1491–1496. [Google Scholar]
- Chen, Z.J. and Ni, Z. (2006) Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. BioEssays 28: 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, F., Liu, S., Wu, J., Fang, L., Sun, S., Liu, B., Li, P., Hua, W. and Wang, X. (2011) BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 11: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X., Xu, J., Xia, S., Gu, J., Yang, Y., Fu, J., Qian, X., Zhang, S., Wu, J. and Liu, K. (2009) Development and genetic mapping of microsatellite markers from genome survey sequences in Brassica napus. Theor. Appl. Genet. 118: 1121–1131. [DOI] [PubMed] [Google Scholar]
- Cheung, F., Trick, M., Drou, N., Lim, Y.P., Park, J.-Y., Kwon, S.-J., Kim, J.-A., Scott, R., Pires, J.C., Paterson, A.H.et al. (2009) Comparative analysis between homoeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. Plant Cell 21: 1912–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delourme, R., Falentin, C., Fomeju, B., Boillot, M., Lassalle, G., André, I., Duarte, J., Gauthier, V., Lucante, N., Marty, A.et al. (2013) High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napus L. BMC Genomics 14: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren, H. (2004) Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5: 435–445. [DOI] [PubMed] [Google Scholar]
- Evanno, G., Regnaut, S. and Goudet, J. (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Fan, C., Cai, G., Qin, J., Li, Q., Yang, M., Wu, J., Fu, T., Liu, K. and Zhou, Y. (2010) Mapping of quantitative trait loci and development of allele-specific markers for seed weight in Brassica napus. Theor. Appl. Genet. 121: 1289–1301. [DOI] [PubMed] [Google Scholar]
- Girke, A., Schierholt, A. and Becker, H.C. (2012a) Extending the rapeseed gene pool with resynthesized Brassica napus II: Heterosis. Theor. Appl. Genet. 124: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke, A., Schierholt, A. and Becker, H.C. (2012b) Extending the rapeseed genepool with resynthesized Brassica napus L. I: Genetic diversity. Genet. Resour. Crop Evol. 59: 1441–1447. [Google Scholar]
- Gómez-Campo, C. (1980) Morphology and morphotaxonomy of the Tribe Brassiceae. In:Tsunoda, S., Hinata, K. and Gómez-Campo, C. (eds.) Brassica crops and wild allies, Japan Science Societies Press, Tokyo, pp. 3–31. [Google Scholar]
- Gómez-Campo, C. and Prakash, S. (1999) Origin and domestication. In:Gómez-Campo, C. (ed.) Biology of Brassica Coenospecies, Elsevier, Amsterdam, pp. 33–58. [Google Scholar]
- Hasan, M., Seyis, F., Badani, A., Pons-Kuhnemann, J., Friedt, W., Lühs, W. and Snowdon, R. (2006) Analysis of genetic diversity in the Brassica napus L. Gene pool using SSR markers. Genet. Res. Crop Evol. 53: 793–802. [Google Scholar]
- Iñiguez-Luy, F. and Federico, M. (2011) The genetics of Brassica napus L. In:Bancroft, I. and Schmidt, R. (eds.) Genetics and genomics of the Brassicaceae, Springer, New York Dordrecht Heidelberg London, pp. 215–260. [Google Scholar]
- Jesske, T., Olberg, B., Schierholt, A. and Becker, H.C. (2013) Resynthesized lines from domesticated and wild Brassica taxa and their hybrids with B. napus L.: genetic diversity and hybrid yield. Theor. Appl. Genet. 126: 1053–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, F., Mao, S., Jiang, J., Wang, J., Fang, X. and Wang, Y. (2011) Proteomic changes in newly synthesized Brassica napus allotetraploids and their early generations. Plant Mol. Biol. Rep. 29: 927–935. [Google Scholar]
- Li, F., Chen, B., Xu, K., Wu, J., Song, W., Bancroft, I., Harper, A.L., Trick, M., Liu, S., Gao, G.et al. (2014) Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.) DNA Res. 21: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Chen, X., Yang, Y., Xu, J., Gu, J., Fu, J., Qian, X., Zhang, S., Wu, J. and Liu, K. (2011) Development and genetic mapping of microsatellite markers from whole genome shotgun sequences in Brassica oleracea. Mol. Breed. 28: 585–596. [Google Scholar]
- Li, H., Younas, M., Wang, X., Li, X., Chen, L., Zhao, B., Chen, X., Xu, J., Hou, F., Hong, B.et al. (2013) Development of a core set of single-locus SSR markers for allotetraploid rapeseed (Brassica napus L.). Theor. Appl. Genet. 126: 937–947. [DOI] [PubMed] [Google Scholar]
- Li, Q., Mei, J., Zhang, Y., Li, J., Ge, X., Li, Z. and Qian, W. (2013) A large-scale introgression of genomic components of Brassica rapa into B. napus by the bridge of hexaploid derived from hybridization between B. napus and B. oleracea. Theor. Appl. Genet. 126: 2073–2080. [DOI] [PubMed] [Google Scholar]
- Liu, H. (1985) Rapeseed genetics and breeding. Shanghai Science and Technology Press, Shanghai. [Google Scholar]
- Liu, N., Chen, L., Wang, S., Oh, C. and Zhao, H. (2005) Comparison of single-nucleotide polymorphisms and microsatellites in inference of population structure. BMC Genet. 6 (Suppl 1): S26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriage, T.N., Hudman, S., Mort, M.E., Orive, M.E., Shaw, R.G. and Kelly, J.K. (2009) Direct estimation of the mutation rate at dinucleotide microsatellite loci in Arabidopsis thaliana (Brassicaceae). Heredity 103: 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei, J., Li, Q., Qian, L., Fu, Y., Li, J., Frauen, M. and Qian, W. (2011) Genetic investigation of the origination of allopolyploid with virtually synthesized lines: application to the C subgenome of Brassica napus. Heredity 106: 955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, J.R. (2006) Outcrossing potential for Brassica species and implications for vegetable crucifer seed crops of growing oilseed Brassicas in the Willamette Valley. Extension publication, Oregon State University. [Google Scholar]
- Nei, M., Tajima, F. and Tateno, Y. (1983) Accuracy of estimated phylogenetic trees from molecular data. II. gene frequency data. J. Mol. Evol. 19: 153–170. [DOI] [PubMed] [Google Scholar]
- Osborn, T.C., Pires, J.C., Birchler, J.A., Auger, D.L., Chen, Z.J., Lee, H.S., Comai, L., Madlung, A., Doerge, R.W., Colot, V.et al. (2003) Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19: 141–147. [DOI] [PubMed] [Google Scholar]
- Piquemal, J., Cinquin, E., Couton, F., Rondeau, C., Seignoret, E., Doucet, I., Perret, D., Villeger, M.J., Vincourt, P. and Blanchard, P. (2005) Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theor. Appl. Genet. 111: 1514–1523. [DOI] [PubMed] [Google Scholar]
- Prakash, S., Wu, X.-M. and Bhat, S.R. (2011) History, evolution, and domestication of Brassica crops. In: Janick, J. (ed.) Plant Breeding Reviews, vol. 35 John Wiley & Sons, Inc., New Jersey, pp. 19–84. [Google Scholar]
- Pritchard, J.K., Stephens, M. and Donnelly, P. (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, W., Meng, J., Li, M., Frauen, M., Sass, O., Noack, J. and Jung, C. (2006) Introgression of genomic components from Chinese Brassica rapa contributes to widening the genetic diversity in rapeseed (B. napus L.), with emphasis on the evolution of Chinese rapeseed. Theor. Appl. Genet. 113: 49–54. [DOI] [PubMed] [Google Scholar]
- Qian, W., Sass, O., Meng, J., Li, M., Frauen, M. and Jung, C. (2007) Heterotic patterns in rapeseed (Brassica napus L.): I. Crosses between spring and Chinese semi-winter lines. Theor. Appl. Genet. 115: 27–34. [DOI] [PubMed] [Google Scholar]
- Quijada, P.A., Udall, J.A., Polewicz, H., Vogelzang, R.D. and Osborn, T.C. (2004) Phenotypic effects of introgressing French winter germplasm into hybrid spring canola. Crop Sci. 44: 1982–1989. [Google Scholar]
- Rahman, H. (2013) Review: Breeding spring canola (Brassica napus L.) by the use of exotic germplasm. Can. J. Plant Sci. 93: 363–373. [Google Scholar]
- Rohlf, F.J. (1992) NTSYS-pc: numerical taxonomy and multivariate analysis system, version 1.50. Exeter Publications, New York. [Google Scholar]
- Rousset, F. (2008) genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8: 103–106. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof, M.A., Soliman, K.M., Jorgensen, R.A. and Allard, R.W. (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81: 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyis, F., Snowdon, R.J., Luhs, W. and Friedt, W. (2003) Molecular characterization of novel resynthesized rapeseed (Brassica napus) lines and analysis of their genetic diversity in comparison with spring rapeseed cultivars. Plant Breeding 122: 473–478. [Google Scholar]
- Snowdon, R., Lühs, W. and Friedt, W. (2007) Oilseed rape. In:Kole, C. (ed.) Genome mapping and molecular breeding in plants, vol. 2: Oilseeds, Springer, Berlin Heidelberg, pp. 55–114. [Google Scholar]
- Song, K., Lu, P., Tang, K. and Osborn, T.C. (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92: 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town, C.D., Cheung, F., Maiti, R., Crabtree, J., Haas, B.J., Wortman, J.R., Hine, E.E., Althoff, R., Arbogast, T.S., Tallon, L.J.et al. (2006) Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18: 1348–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U, N. (1935) Genome-analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7: 389–452. [Google Scholar]
- Udall, J., Quijada, P.A., Polewicz, H., Vogelzang, R. and Osborn, T. (2004) Phenotypic effects of introgressing Chinese winter and resynthesized Brassica napus L. germplasm into hybrid spring canola. Crop Sci. 44: 1990–1996. [Google Scholar]
- Wang, J., Lydiate, D.J., Parkin, I.A., Falentin, C., Delourme, R., Carion, P.W. and King, G.J. (2011) Integration of linkage maps for the amphidiploid Brassica napus and comparative mapping with Arabidopsis and Brassica rapa. BMC Genomics 12: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, B.S. and Cockerham, C.C. (1984) Estimating F-statistics for the analysis of population-structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Xiao, Y., Cai, D., Yang, W., Ye, W., Younas, M., Wu, J. and Liu, K. (2012) Genetic structure and linkage disequilibrium pattern of a rapeseed (Brassica napus L.) association mapping panel revealed by microsatellites. Theor. Appl. Genet. 125: 437–447. [DOI] [PubMed] [Google Scholar]
- Xiong, Z., Gaeta, R.T. and Pires, J.C. (2011) Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 108: 7908–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Qian, X., Wang, X., Li, R., Cheng, X., Yang, Y., Fu, J., Zhang, S., King, G.J., Wu, J.et al. (2010) Construction of an integrated genetic linkage map for the A genome of Brassica napus using SSR markers derived from sequenced BACs in B. rapa. BMC Genomics 11: 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, S., Zhang, F., Zhao, X., Yu, Y., Zhang, D., Zhao, X. and Wang, W. (2013) An improved Brassica rapa genetic linkage map and locus-specific variations in a doubled haploid population. Plant Mol. Biol. Rep. 31: 558–568. [Google Scholar]
- Zhao, J., Buchwaldt, L., Rimmer, S.R., Sharpe, A., McGregor, L., Bekkaoui, D. and Hegedus, D. (2009) Patterns of differential gene expression in Brassica napus cultivars infected with Sclerotinia sclerotiorum. Mol. Plant Pathol. 10: 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J., Fu, D., Gong, H., Qian, W., Xia, W., Pires, J.C., Li, R., Long, Y., Mason, A.S., Yang, T.J.et al. (2011) De novo genetic variation associated with retrotransposon activation, genomic rearrangements and trait variation in a recombinant inbred line population of Brassica napus derived from interspecific hybridization with Brassica rapa. Plant J. 68: 212–224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.