Significance
The T-cell receptor (TCR) does not signal on its own. Instead, it is constitutively associated with the CD3 coreceptors, which contain intracellular signaling motifs. Although antigen (Ag) recognition by the TCR and the activation of T cells after CD3 activation have been extensively studied, there is far less known about how the TCR relays the binding of Ag to initiate intracellular signaling. Detailed study of the eight-chain TCR–CD3 complex is hampered by limited structural information for how the components of the complex interact. We use complementary structural approaches to examine where the TCR–CD3 extracellular domains are placed relative to each other and the overall organization of the complex.
Keywords: T-cell receptor, electron microscopy, small-angle X-ray scattering
Abstract
αβ T-cell receptor (TCR) activation plays a crucial role for T-cell function. However, the TCR itself does not possess signaling domains. Instead, the TCR is noncovalently coupled to a conserved multisubunit signaling apparatus, the CD3 complex, that comprises the CD3εγ, CD3εδ, and CD3ζζ dimers. How antigen ligation by the TCR triggers CD3 activation and what structural role the CD3 extracellular domains (ECDs) play in the assembled TCR–CD3 complex remain unclear. Here, we use two complementary structural approaches to gain insight into the overall organization of the TCR–CD3 complex. Small-angle X-ray scattering of the soluble TCR–CD3εδ complex reveals the CD3εδ ECDs to sit underneath the TCR α-chain. The observed arrangement is consistent with EM images of the entire TCR–CD3 integral membrane complex, in which the CD3εδ and CD3εγ subunits were situated underneath the TCR α-chain and TCR β-chain, respectively. Interestingly, the TCR–CD3 transmembrane complex bound to peptide–MHC is a dimer in which two TCRs project outward from a central core composed of the CD3 ECDs and the TCR and CD3 transmembrane domains. This arrangement suggests a potential ligand-dependent dimerization mechanism for TCR signaling. Collectively, our data advance our understanding of the molecular organization of the TCR–CD3 complex, and provides a conceptual framework for the TCR activation mechanism.
T cells are key mediators of the adaptive immune response. Each αβ T cell contains a unique αβ T-cell receptor (TCR), which binds antigens (Ags) displayed by major histocompatibility complexes (MHCs) and MHC-like molecules (1). The TCR serves as a remarkably sensitive driver of cellular function: although TCR ligands typically bind quite weakly (1–200 μM), even a handful of TCR ligands are sufficient to fully activate a T cell (2, 3). The TCR does not possess intracellular signaling domains, uncoupling Ag recognition from T-cell signaling. The TCR is instead noncovalently associated with a multisubunit signaling apparatus, consisting of the CD3εγ and CD3εδ heterodimers and the CD3ζζ homodimer, which collectively form the TCR–CD3 complex (4, 5). The CD3γ/δ/ε subunits each consist of a single extracellular Ig domain and a single immunoreceptor tyrosine-based activation motif (ITAM), whereas CD3ζ has a short extracellular domain (ECD) and three ITAMs (6–11). The TCR–CD3 complex exists in 1:1:1:1 stoichiometry for the αβTCR:CD3εγ:CD3εδ:CD3ζζ dimers (12). Phosphorylation of the intracellular CD3 ITAMs and recruitment of the adaptor Nck lead to T-cell activation, proliferation, and survival (13, 14). Understanding the underlying principles of TCR–CD3 architecture and T-cell signaling is of therapeutic interest. For example, TCR–CD3 is the target of therapeutic antibodies such as the immunosuppressant OKT3 (15), and there is increasing interest in manipulating T cells in an Ag-dependent manner by using naturally occurring and engineered TCRs (16).
Assembly of the TCR–CD3 complex is primarily driven by each protein’s transmembrane (TM) region, enforced through the interaction of evolutionarily conserved, charged, residues in each TM region (4, 5, 12). What, if any, role interactions between TCR and CD3 ECDs play in the assembly and function of the complex remains controversial (5): there are several plausible proposed models of activation, which are not necessarily mutually exclusive (5, 17–19). Although structures of TCR–peptide–MHC (pMHC) complexes (2), TCR–MHC-I–like complexes (1), and the CD3 dimers (6–10) have been separately determined, how the αβ TCR associates with the CD3 complex is largely unknown. Here, we use two independent structural approaches to gain an understanding of the TCR–CD3 complex organization and structure.
Results
Placement of the αβ TCR and CD3εδ ECDs via Small-Angle X-ray Scattering.
We first set out to determine the relative positions of the TCR and CD3 ECDs. In the absence of the interaction between TCR and CD3 TM helices (Fig. 1A) (12, 20), there is no detectable interaction between αβ TCR and CD3 ECDs (9), suggesting any TCR–CD3 ECD intermolecular interactions are very weak. To generate a stable complex of soluble TCR and CD3εδ suitable for structural analysis, we developed a system in which the hydrophobic TM domains were replaced by a parallel heterotrimeric coiled coil (21), providing an analogous structure to the proposed TCRα–CD3εδ TM arrangement (Fig. 1B) (10, 12, 20). Our constructs included the membrane-proximal stalks of the LC13 TCR and CD3εδ to ensure the correct relative juxtapositioning of subunits. The complex was coexpressed in insect cells and, after purification, interacted with the expected affinity to both its cognate pMHC (Fig. S1) and the anti-CD3ε antibody OKT3 (as detailed later) (22). The LC13 TCR–CD3εδ complex molecular mass was 102 kDa as determined by multiangle laser light scattering (MALLS; Fig. S1), similar to that predicted for the glycosylated LC13 TCR–CD3εδ complex (88.1 kDa).
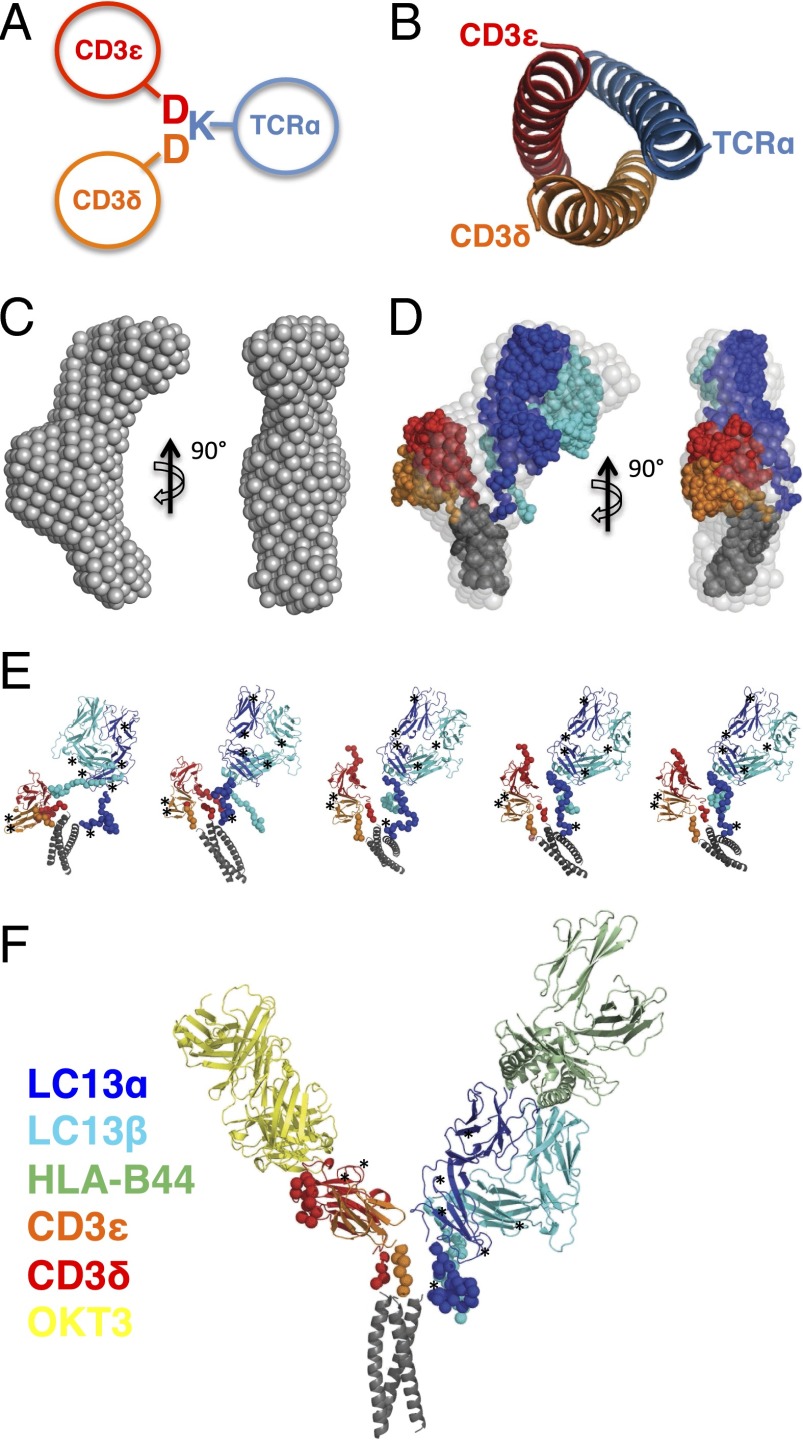
Fig. 1.
Generation of a stable TCR–CD3εδ complex and analysis via SAXS. (A) Cartoon depicting the TM domain-based assembly of TCRα with CD3εδ. The polar Asp (marked as “D”) and Lys (marked as “K”) residues that mediate assembly are depicted. Transmembrane α-helices are represented by circles. (B) The parallel heterotrimeric coiled coil used to link TCR and CD3εδ viewed down the long axis of the helices. (C) The ab initio model of LC13 TCR–CD3εδ shown in two orientations. (D) Overlay of the LC13 TCR–CD3εδ ab initio model (light gray spheres) with a representative CORAL model in which the TCR (α, blue; β, cyan), CD3 (ε, red; δ, orange) and coiled coil (black) are represented as dots. (E) Independent LC13 TCR–CD3εδ models predicted via CORAL. (F) CORAL-constrained model for the LC13 TCR–CD3εδ complex bound to OKT3 Fab and HLA-B44 pMHC. In E and F, potential N-linked glycosylation sites in the TCR and CD3 subunits are indicated by asterisks.
We deduced the structure of the LC13 TCR–CD3εδ by using a series of small-angle X-ray scattering (SAXS) experiments. As controls, we analyzed the ECDs of the LC13 TCR and CD3εδ alone as well as the cognate LC13 TCR ligand, HLA-B8, and a Fab fragment of the anti-CD3ε antibody OKT3. Each of these samples was free of protein aggregation as evidenced by the linearity of Guinier plots (Fig. S2B), and was judged to be monomeric in solution based on the calculated molecular mass, radius of gyration (Rg), and maximal particle dimension (Table S1). Moreover, we observed a good agreement between the calculated scattering curve of each component's previously reported crystal structure and our experimentally derived SAXS data (Fig. S2). Next, we performed SAXS measurements on the isolated TCR–CD3εδ construct, which had a mass of 105 kDa, indicative of a monomeric species. Reconstruction of the particle shape ab initio using the DAMMIF program revealed TCR–CD3εδ to possess an elongated bow shape with approximate dimensions of 150 × 45 × 55 Å and contain a central bulged region (Fig. 1C). To understand how the TCR and CD3 subunits are organized within this complex, we used the program CORAL (complexes with random loops) (23) to find the optimal position and orientation of available high-resolution domain structures (LC13 TCR, CD3εδ, and the coiled-coil subunit), as well as determining the approximate conformation of missing portions of the polypeptide chain, namely the N terminus of CD3ε and the membrane-proximal stalks of TCR and CD3. The independent CORAL models each fitted well to the SAXS data for values of q < 0.2 Å and overlaid very well with each other and the ab initio model (Fig. 1 D and E and Fig. S2). Taken together, our data indicate that the CD3εδ heterodimer occupies the central bulged region whereas the TCR and coiled coil are located at the extremities of the particle. The CD3εδ ECDs are poised to make contact with the ECD and stalks of TCRα, with the CD3εδ ECDs placed below rather than alongside the TCR ECDs (Fig. 1 C–E).
SAXS data of LC13 TCR–CD3εδ in complex with the OKT3 Fab fragment or the high-affinity ligand pHLA-B44 (22) further improved our understanding of the TCR–CD3 architecture. The availability of cocrystal structures of CD3εγ–OKT3 Fab (8) and LC13 TCR–HLA-B44 (22) combined with the capability of CORAL to simultaneously fit multiple scattering curves from subsets of the entire system allowed us to generate a highly constrained pseudo–high-resolution model. LC13 TCR–CD3εδ formed stable complexes with OKT3 Fab and HLA-B44, as judged by the SAXS-derived measurements (Table S1) and the size and shape of the corresponding ab initio models, which displayed an additional region of density that corresponded to the bound ligand or OKT3 Fab fragment (Fig. S3). The generated CORAL models fitted well to the SAXS data and exhibited a V-shaped structure in which one of the arms of the V is occupied by the bound antibody (Fig. 1F and Fig. S4). Based on our model, the regions that are consistently in close proximity in independent CORAL models, and thus well positioned to make potential intersubunit interactions, are the membrane-proximal stalks of TCR and CD3.
EM of Membrane-Associated TCR–CD3 Complex.
We sought to place the TCR–CD3εδ ECDs in the larger, membrane-associated TCR–CD3 complex. We first examined full-length versions of the human TCRs LC13 and 1G4 complexed to CD3 by negative-stain electron microscopy (EM) (Fig. S5). EM of membrane-bound TCR–CD3 has been previously reported, but produced particles whose composition and relative arrangement were difficult to interpret (24). Although we obtained monodisperse particles that suggested monomeric TCR–CD3 complexes, the averages were heterogeneous and did not show sufficient features to definitively determine the TCR–CD3 oligomerization state or relative placement of TCR and CD3 domains (Fig. S5).
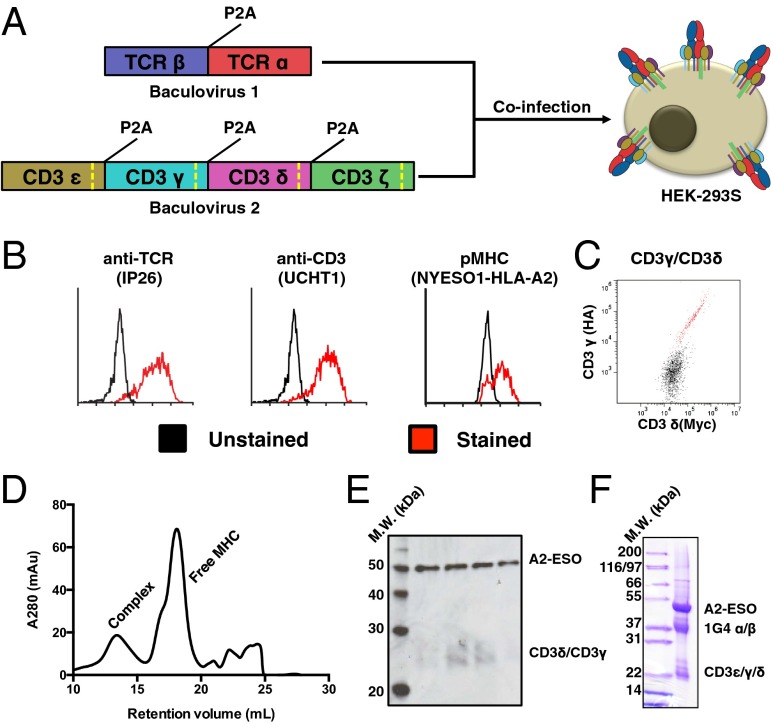
To increase the molecular weight of the TCR–CD3 complex for study via EM, we set out to express a stable TCR–CD3 transmembrane complex bound to pMHC. As pMHC–TCR interactions are generally low affinity, we used a version of the human 1G4 TCR that was affinity-matured to 20-pM affinity for its cognate ligand, HLA-A2 presenting the NY-ESO1 peptide (A2-ESO1) (25). 1G4 TCR has also recently been functionally reconstituted in HEK-293 cells (26). We created one baculovirus each for TCR and CD3 expression in mammalian cells (27), with individual TCR/CD3 chains cleaved into individual polypeptides through use of viral 2A peptides (Fig. 2A) (28). Cells transduced with both viruses stained robustly with antibodies against TCR and CD3, as well as with fluorescently labeled A2-ESO1 (Fig. 2B). When we stained for epitope tags fused to the N termini of CD3γ and CD3δ, we observed excellent correlation between the two chains, indicating the expected 1:1 ratio (Fig. 2C). To solubilize and purify the TCR–CD3 complex, we tandem-affinity purified the complex via histidine and Fc tags on CD3γ/δ and A2-ESO, respectively (Fig. S6). The protein was then cleaved from the Fc via engineered protease sites recognized by Rhinovirus 3C protease. To reduce sample heterogeneity, intracellular domains (ICDs) had engineered protease sites and were concurrently removed via 3C protease (Fig. S6). The purified protein contained CD3 and pMHC, indicative of a correctly assembled complex (Fig. 2 D–F).
Fig. 2.
Production and characterization of full-length 1G4 TCR–CD3 complex. (A) Schematic for expression of 1G4 TCR–CD3 complex in HEK-293 cells via coinfection of baculoviruses. The baculoviruses respectively encode for TCRα/β and CD3ε/γ/δ/ζ, with each polypeptide chain separated by a viral 2A peptide (P2A). Each CD3 subunit contains a Rhinovirus 3C protease cleavage site (dashed yellow line) to remove intracellular domains after protein expression. (B) Expression of folded TCR–CD3 complex on HEK-293 cells as demonstrated by an anti-TCR antibody (Left), anti-CD3ε antibody (Center), and cognate pMHC (Right). A high-affinity TCR allows for staining of monomeric pMHC. (C) Equal staining for orthogonal epitope tags on the N termini of CD3γ and CD3γδ indicate 1:1 incorporation of CD3εγ and CD3εδ into the TCR–CD3 complex. (D) Size-exclusion chromatography for the TCR–CD3 complex bound by pMHC. (E) Western blot of size-exclusion chromatography fractions of the pMHC–TCR–CD3 complex peak shows staining for pMHC (via anti-β2m antibody) and CD3γ/δ (via anti-His antibody). The blot was simultaneously treated with both primary antibodies. (F) SDS-PAGE gel of final 1G4 TCR–CD3–ESO–A2 material showing presence of TCR, CD3, and MHC. CD3ζ is not visible because of its small size after protease cleavage (5 kDa).
To observe the overall molecular architecture of the pMHC–TCR–CD3 complex, we analyzed soluble pMHC–TCR (Fig. 3A), membrane-bound pMHC–TCR–CD3 (Figs. 2 and 3B), and membrane-bound pMHC–TCR–CD3 decorated with an anti-CD3 Fab (7) (Fig. 3C and Fig. S7) by negative-stain EM (Fig. S8). Class averages showed the soluble pMHC–TCR complex to be monomeric (Fig. 3A) and to be consistent with the previously solved X-ray crystal structure of 1G4 TCR bound to A2-ESO1 (Fig. S9) (29, 30).
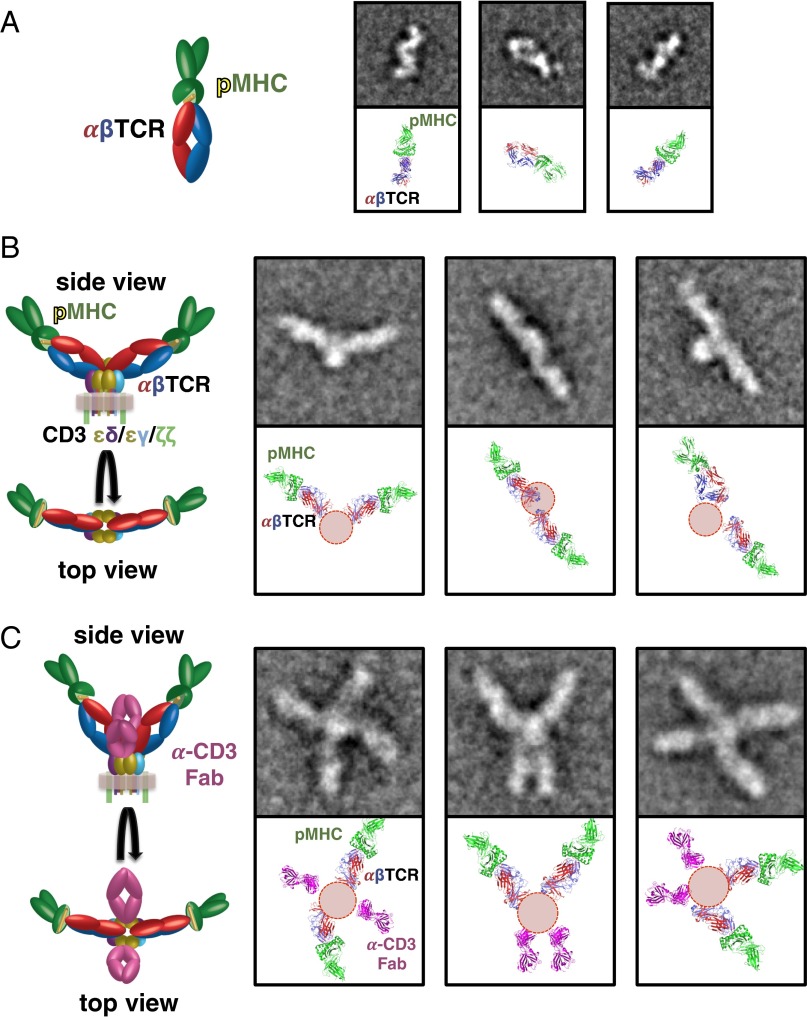
Fig. 3.
Negative-stain EM of soluble pMHC–1G4–TCR complex (A), membrane-bound pMHC–1G4–TCR–CD3 complex (B), and pMHC-1G4–TCR–CD3 complex decorated with anti-CD3 Fab (C) indicates a dimeric membrane-bound TCR–CD3 complex. (Left) Cartoon representation of the domain structure of each complex. Representative 2D class averages for each complex (Top Right) are oriented to a model based on the solved crystal structure of the pMHC–TCR and CD3εδ–UCHT1 complexes (Bottom Right). Red circles (B and C) are density noted in the class averages not accounted for by the TCR or pMHC ECDs, likely consisting of the CD3 ECDs and the TCR/CD3 transmembrane helices. The side length of the class averages in A is 25.6 nm, and the side length of the class averages in B and C is 42.6 nm.
EM images of pMHC–TCR–CD3 clearly revealed largely dimeric complexes (Fig. 3B and Fig. S8), and class averages revealed them to consist of two elongated “wings” projecting out from a central region of additional density (Fig. 3B). The observed wings in the 2D class averages were consistent with the observed size and shape for the pMHC–TCR ECDs (Fig. 3 A and B). Notably, the particles did not show any observable bulging outward at the base of the wings. Instead, the density in between the two wings likely consists of the CD3 ECDs and the TCR–CD3 TM regions (Fig. 3B). The EM class averages are therefore consistent with what we observed for the soluble TCR–CD3εδ via SAXS (Fig. 1), and suggest that the CD3εγ heterodimer is placed as CD3εδ, but on the other side of the TCR, proximal to the TCR β-chain (Fig. 3B). The relative orientation of the dimer arms exhibited significant variability and flexibility relative to the central CD3/TM region (Movie S1), but were generally oriented at obtuse angle relative to one another, with many class averages displaying an essentially antiparallel arrangement (Fig. S8 and Movie S1).
When we observed the Fab-decorated pMHC–TCR–CD3 complex, the two-winged particles were replaced by particles that had four arms (Fig. 3C). Class averages of the particles showed that the anti-CD3 Fab (appearing as the shorter arms in the complex) was not appended to the wings of the previous pMHC–TCR–CD3 particles (Fig. 3B), but rather to the central region between the wings (Fig. 3C). Given that the Fab-binding epitope of CD3ε is composed of the membrane-distal Ig loops (7), the CD3 ECDs are likely “bent” at an angle relative to the TCR. Indeed, the class averages of Fab-decorated membrane-bound particles bore a close resemblance to one half of the CORAL-generated SAXS model of the soluble TCR–CD3εδ (Figs. 1E and 3C), thereby supporting our use of the trimeric coiled-coil domain as a TM mimetic. Interestingly, we observed only two OKT3 Fab fragments per dimeric TCR–CD3 particle despite the presumed presence of four CD3ε chains, suggesting that only one of the CD3 heterodimers is accessible in the fully assembled TCR–CD3 complex (Fig. 3C). However, we cannot discount the possibility that the dimeric TCR–CD3 complex retains only two CD3 heterodimers, as has been previously suggested (31). Class averages of the Fab-decorated particles still retained considerable flexibility, illustrated by the longer arms in the particles being located in cis as well as in trans of the central CD3/TM region mass (Fig. 3C) and by the relative mobility of each wing relative to the central CD3/TM region (Movie S2).
Discussion
Our analyses have determined that the CD3 ECDs are situated underneath, rather than alongside, the TCR constant domains (Figs. 1 D–F and 3C). The TCR and CD3 ECD–TM domain “connecting peptides” are highly conserved and affect TCR–CD3 function when altered (32–37). Given the longer length of the TCR connecting peptides relative to the CD3 connecting peptides, there is sufficient space for the TCR to be placed over the CD3 ECDs, consistent with what was observed for our pMHC–TCR–CD3 complexes (Fig. 3 B and C), the SAXS TCR–CD3 ECD structure (Fig. 1 D and E), and previously discovered TCR ECD mutations that affect TCR–CD3 assembly and function, which are primarily located toward the “bottom” of the TCR Cα/Cβ domains (5, 38, 39).
There is evidence pointing to the existence of monomeric (12, 40, 41) and multimeric (41–44) TCR–CD3 complexes. These oligomeric states may arise from differences in preparation of the TCR–CD3 complexes (41), developmental TCR–CD3 association with the pre-Tα molecule (45), or cell-intrinsic differences such as membrane composition (41). Our data provide additional evidence that the TCR–CD3 complex can exist as a stable dimer, even upon solubilization in detergent (41, 44). The consistent site of dimerization and lack of higher-order species (Fig. 3 and Figs. S5 and S8) suggest the observed particles dimerize through a specific interaction site, likely mediated through the TCR–CD3 TMs and/or membrane-proximal portions of the ECDs. Although our data do not definitively speak to the oligomeric state of the TCR–CD3 complex on the surface of T cells, it leaves open the possibility that the observed dimerization may be brought about by binding to Ags, which could place the TCR–CD3 complex into a signaling-permissive state (46) (Fig. 4). However, TCR–CD3 likely requires molecular reorientation beyond simple dimerization to initiate signaling, as seen from previous observations that not all bivalent antibodies can induce TCR–CD3 signaling (47) and that relatively high-affinity pMHC ligands bound in certain topologies cannot activate T cells, even when oligomerized (48).
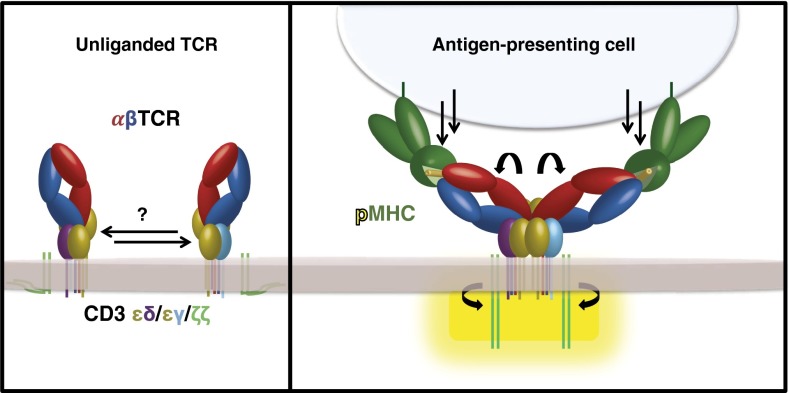
Fig. 4.
Proposed mechanism for TCR–CD3 signaling. TCR binding of pMHC ligand may induce a reorientation of the TCR–CD3 subunits, which would propagate through the TCR–CD3 ECDs and TM domains, exposing the CD3 ICDs for activation. It is also possible that pMHC (or other Ag) itself induces dimerization of the TCR–CD3 complex.
Our data provide a structural framework that can potentially begin to reconcile disparate observations about how the TCR relays a ligation event to signaling. Such observations include studies showing that monomeric pMHC possesses little to no ability to induce signaling regardless of affinity (49), there exist signaling-permissive and -nonpermissive pMHC–TCR docking geometries (48), conformational changes occur in the TCR upon pMHC ligation (46, 50–52), the CD4 and CD8 coreceptors are required to place Lck relative to the CD3 ITAMs (53, 54), phosphatases are occluded from the T-cell Ag-presenting cell synapse (18, 26, 55), and mechanotransduction appears to be important for TCR signaling (19, 36, 56–59) (Fig. 4). In the dimeric model we have visualized by EM, the combined interactions of the TCR–CD3 TMs, connecting peptides, and ECDs appear to create a centrally disposed interacting unit that could act as a fulcrum to transmit information of Ag binding to the CD3 ICDs upon engaging pMHC on an opposing cell (Figs. 1 D–F, 3, and 4). The TCR–CD3 complex may rely upon dimerization and/or reorientation of the observed TCR wings by a vectorial force from the opposing cell (Fig. 4) to trigger the transmission of a signal, consistent with the notion that two TCRs need to be engaged in a signaling-permissive orientation beyond simple dimerization (60, 61) and explaining why monomeric Ags in solution and certain multimeric Ags are not signaling competent.
Collectively, our structural models provide several important pieces of structural information about the TCR–CD3–pMHC complex, namely that the assembly is dimerized and the TCR is situated over top, rather than alongside, the CD3 ectodomains. With this snapshot in hand, mechanistic models of TCR activation can be probed experimentally with much higher precision and clarity.
Methods
Expression and Purification of Soluble TCR–CD3δε Complex.
To generate stably associated protein complexes, the C terminus of the human LC13 TCRα, CD3ε, and CD3δ ECDs (residues 1–227, 1–105, and 1–80, respectively) were fused to the N terminus of an engineered heterotrimeric coiled-coil domain (21) by using splice-by-overlap PCR. LC13 TCR and CD3 chains were then separately cloned into modified versions of the bicistronic baculovirus expression vector pFastbac dual (Invitrogen), downstream of a GP67 signal sequence. The final vector sequences were as follows: vector 1, LC13 TCRα-CoilA (AEIAAIEYEQAAIKEEIAAIKDKIAAIKEYIAAI) and LC13 TCRβ-Thrombin-His6); vector 2, CD3ε-coilB (EKIAAIKEEQAAIEEEIQAIKEEIAAIKYLIAQI), and CD3δ-coilC (AEIAAIKYKQAAIKNEIAAIKQEIAAIEQMIAAI). Viral stocks derived from both vectors were separately amplified in Sf9 cells and used to coinfect High Five cells (Invitrogen) according to the manufacturer’s instructions. Insect cell supernatant was concentrated, and buffer exchanged into 10 mM Tris, pH 8, containing 0.5 M NaCl and 10 mM imidazole before purification via nickel-affinity chromatography. Protein was further purified via size-exclusion and anion-exchange chromatography by using Superdex 200 16/60 and HiTrapQ HP columns (GE Healthcare).
Expression and Purification of Full-Length 1G4 TCR–CD3 Complex.
TCR and CD3 constructs, each codon-optimized for human expression, were synthesized as single ORFs (Genscript), with each gene separated by a short GSG linker followed by a 2A sequence from porcine teschovirus (P2A; ATNFSLLKQAGDVEENPGP). The construct designs were as follows: for TCR, the previously reported affinity-matured version of 1G4 was constructed as 1G4β-2A-1G4α; for CD3, CD3ε-2A-CD3γ-2A-CD3δ-2A-CD3ζ. Native leader sequences for each protein were used. Initial characterization experiments and EM characterization of 1G4–CD3 were conducted with each CD3 chain having an orthogonal epitope tag to ensure that all chains were present (CD3ε Flag, CD3γ HA, and CD3δ Myc, all on the N terminus; CD3ζ 1D4 on the C terminus). TCRα/β were each tagged with His6 and EE tags on the N terminus. For purification of pMHC-bound 1G4–CD3, CD3γ, and CD3δ were expressed with N-terminal His6 tags and the TCR chains were untagged. Additionally, 3C protease sites (LVELFQGP) were installed 9 aa after the predicted end of the transmembrane region for each CD3 chain. The constructs were cloned into the BacMam expression vector pVLAD6 and transfected to create baculovirus as previously described (27). Viruses were amplified to P2 before protein expression.
TCR–CD3 complexes were expressed in HEK-293S GnTi− cells, which were maintained as previously described (27). SI Methods provides further details.
SAXS.
SAXS data were collected at the Australian Synchrotron by using a 1M Pilatus detector. For individual components, buffers/samples were loaded into 1-mm quartz capillaries and continuously flowed through the beam during data collection. For multicomponent protein complexes, samples were loaded onto an in-line Superdex 200 (10/300) size-exclusion column (GE Healthcare). In both cases, multiple 1-s exposures were collected, checked for radiation damage, and averaged where appropriate. SI Methods provides further details.
EM.
Purified 1G4–MHC, 1G4–CD3, 1G4–CD3–MHC, and 1G4–CD3–MHC–anti-CD3 Fab complexes were prepared by conventional negative staining with 0.75% (wt/vol) uranyl formate (62). Images were collected with a Tecnai T12 electron microscope (FEI) equipped with an LaB6 filament and operated at an acceleration voltage of 120 kV. Images were recorded by using low-dose procedures on an UltraScan 895 4K × 4K CCD camera (Gatan) using a defocus of −1.5 μm and a nominal magnification of 52,000×. The calibrated magnification was 70,527×, yielding a pixel size of 2.13 Å on the specimen level. Purified LC13-CD3 was stained with 2% (wt/vol) uranyl acetate and imaged on a Tecnai TF30 transmission electron microscope operated at 300 kV. Images were recorded on a 2K × 2K CCD camera using an underfocus range of 0.4–2.6 μm and a nominal magnification of 52,000×, yielding a pixel size of 1.8 Å on the specimen level. The use of two electron microscopes results from different sample preparation and data collection locations. SI Methods provides details of data processing.
Supplementary Material
Acknowledgments
We thank Ignacio Moraga, Michael Kuhns, and Mark Davis for helpful discussions; Dr. Eric Hanssen and the staff at the Australian synchrotron for assistance with data collection; and Lars Kjer-Nielsen for establishing a number of the systems underpinning this work. This work was supported by a Regina Casper Stanford Graduate Fellowship (to M.E.B.), a Gerald J. Lieberman Fellowship (to M.E.B.), a National Science Foundation Graduate Fellowship (to M.E.B.), National Health and Medical Research Council (NHMRC) Peter Doherty Fellowship GNT1035636 (to R.B.), an NHMRC Australia Fellowship (to J.R.), NIH Grant R01 AI03867 (to K.C.G.), and Howard Hughes Medical Institute (T.W. and K.C.G.), NHMRC (J.R. and J.M.), and Australian Research Council (J.R. and J.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420936111/-/DCSupplemental.
References
- 1.Eckle SB, Turner SJ, Rossjohn J, McCluskey J. Predisposed αβ T cell antigen receptor recognition of MHC and MHC-I like molecules? Curr Opin Immunol. 2013;25(5):653–659. doi: 10.1016/j.coi.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 3.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002;419(6909):845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 4.Wucherpfennig KW, Gagnon E, Call MJ, Huseby ES, Call ME. Structural biology of the T-cell receptor: Insights into receptor assembly, ligand recognition, and initiation of signaling. Cold Spring Harb Perspect Biol. 2010;2(4):a005140. doi: 10.1101/cshperspect.a005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhns MS, Badgandi HB. Piecing together the family portrait of TCR-CD3 complexes. Immunol Rev. 2012;250(1):120–143. doi: 10.1111/imr.12000. [DOI] [PubMed] [Google Scholar]
- 6.Sun ZJ, Kim KS, Wagner G, Reinherz EL. Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer. Cell. 2001;105(7):913–923. doi: 10.1016/s0092-8674(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 7.Arnett KL, Harrison SC, Wiley DC. Crystal structure of a human CD3-epsilon/delta dimer in complex with a UCHT1 single-chain antibody fragment. Proc Natl Acad Sci USA. 2004;101(46):16268–16273. doi: 10.1073/pnas.0407359101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kjer-Nielsen L, et al. Crystal structure of the human T cell receptor CD3 epsilon gamma heterodimer complexed to the therapeutic mAb OKT3. Proc Natl Acad Sci USA. 2004;101(20):7675–7680. doi: 10.1073/pnas.0402295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun ZY, et al. Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling. Proc Natl Acad Sci USA. 2004;101(48):16867–16872. doi: 10.1073/pnas.0407576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Call ME, et al. The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127(2):355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry R, et al. Structure of the chicken CD3εδ/γ heterodimer and its assembly with the αβT cell receptor. J Biol Chem. 2014;289(12):8240–8251. doi: 10.1074/jbc.M113.544965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111(7):967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samelson LE. Signal transduction mediated by the T cell antigen receptor: the role of adapter proteins. Annu Rev Immunol. 2002;20:371–394. doi: 10.1146/annurev.immunol.20.092601.111357. [DOI] [PubMed] [Google Scholar]
- 14.Borroto A, Abia D, Alarcón B. Crammed signaling motifs in the T-cell receptor. Immunol Lett. 2014;161(1):113–117. doi: 10.1016/j.imlet.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Chatenoud L, Bluestone JA. CD3-specific antibodies: A portal to the treatment of autoimmunity. Nat Rev Immunol. 2007;7(8):622–632. doi: 10.1038/nri2134. [DOI] [PubMed] [Google Scholar]
- 16.Phan GQ, Rosenberg SA. Adoptive cell transfer for patients with metastatic melanoma: The potential and promise of cancer immunotherapy. Cancer Control. 2013;20(4):289–297. doi: 10.1177/107327481302000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhns MS, Davis MM, Garcia KC. Deconstructing the form and function of the TCR/CD3 complex. Immunity. 2006;24(2):133–139. doi: 10.1016/j.immuni.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 18.van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol. 2011;11(1):47–55. doi: 10.1038/nri2887. [DOI] [PubMed] [Google Scholar]
- 19.Wang JH, Reinherz EL. The structural basis of αβ T-lineage immune recognition: TCR docking topologies, mechanotransduction, and co-receptor function. Immunol Rev. 2012;250(1):102–119. doi: 10.1111/j.1600-065X.2012.01161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Call ME, Wucherpfennig KW, Chou JJ. The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nat Immunol. 2010;11(11):1023–1029. doi: 10.1038/ni.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nautiyal S, Alber T. Crystal structure of a designed, thermostable, heterotrimeric coiled coil. Protein Sci. 1999;8(1):84–90. doi: 10.1110/ps.8.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macdonald WA, et al. T cell allorecognition via molecular mimicry. Immunity. 2009;31(6):897–908. doi: 10.1016/j.immuni.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 23.Petoukhov MV, et al. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Cryst. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arechaga I, et al. Structural characterization of the TCR complex by electron microscopy. Int Immunol. 2010;22(11):897–903. doi: 10.1093/intimm/dxq443. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, et al. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nat Biotechnol. 2005;23(3):349–354. doi: 10.1038/nbt1070. [DOI] [PubMed] [Google Scholar]
- 26.James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature. 2012;487(7405):64–69. doi: 10.1038/nature11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dukkipati A, Park HH, Waghray D, Fischer S, Garcia KC. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr Purif. 2008;62(2):160–170. doi: 10.1016/j.pep.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22(5):589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 29.Chen JL, et al. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J Exp Med. 2005;201(8):1243–1255. doi: 10.1084/jem.20042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sami M, et al. Crystal structures of high affinity human T-cell receptors bound to peptide major histocompatibility complex reveal native diagonal binding geometry. Protein Eng Des Sel. 2007;20(8):397–403. doi: 10.1093/protein/gzm033. [DOI] [PubMed] [Google Scholar]
- 31.Alarcón B, Swamy M, van Santen HM, Schamel WW. T-cell antigen-receptor stoichiometry: Pre-clustering for sensitivity. EMBO Rep. 2006;7(5):490–495. doi: 10.1038/sj.embor.7400682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bäckström BT, et al. A motif within the T cell receptor alpha chain constant region connecting peptide domain controls antigen responsiveness. Immunity. 1996;5(5):437–447. doi: 10.1016/s1074-7613(00)80500-2. [DOI] [PubMed] [Google Scholar]
- 33.Xu C, Call ME, Wucherpfennig KW. A membrane-proximal tetracysteine motif contributes to assembly of CD3deltaepsilon and CD3gammaepsilon dimers with the T cell receptor. J Biol Chem. 2006;281(48):36977–36984. doi: 10.1074/jbc.M607164200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minguet S, Swamy M, Schamel WW. The short length of the extracellular domain of zeta is crucial for T cell antigen receptor function. Immunol Lett. 2008;116(2):195–202. doi: 10.1016/j.imlet.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, et al. A conserved CXXC motif in CD3epsilon is critical for T cell development and TCR signaling. PLoS Biol. 2009;7(12):e1000253. doi: 10.1371/journal.pbio.1000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brazin KN, et al. Constitutively oxidized CXXC motifs within the CD3 heterodimeric ectodomains of the T cell receptor complex enforce the conformation of juxtaposed segments. J Biol Chem. 2014;289(27):18880–18892. doi: 10.1074/jbc.M114.574996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martínez-Martín N, et al. Cooperativity between T cell receptor complexes revealed by conformational mutants of CD3epsilon. Sci Signal. 2009;2(83):ra43. doi: 10.1126/scisignal.2000402. [DOI] [PubMed] [Google Scholar]
- 38.Kuhns MS, Davis MM. Disruption of extracellular interactions impairs T cell receptor-CD3 complex stability and signaling. Immunity. 2007;26(3):357–369. doi: 10.1016/j.immuni.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 39.Fernandes RA, et al. T cell receptors are structures capable of initiating signaling in the absence of large conformational rearrangements. J Biol Chem. 2012;287(16):13324–13335. doi: 10.1074/jbc.M111.332783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Germain RN. T-cell signaling: The importance of receptor clustering. Curr Biol. 1997;7(10):R640–644. doi: 10.1016/s0960-9822(06)00323-x. [DOI] [PubMed] [Google Scholar]
- 41.Schamel WW, et al. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J Exp Med. 2005;202(4):493–503. doi: 10.1084/jem.20042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuhns MS, et al. Evidence for a functional sidedness to the alphabetaTCR. Proc Natl Acad Sci USA. 2010;107(11):5094–5099. doi: 10.1073/pnas.1000925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molnár E, Deswal S, Schamel WW. Pre-clustered TCR complexes. FEBS Lett. 2010;584(24):4832–4837. doi: 10.1016/j.febslet.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Schrum AG, Gil D, Turka LA, Palmer E. Physical and functional bivalency observed among TCR/CD3 complexes isolated from primary T cells. J Immunol. 2011;187(2):870–878. doi: 10.4049/jimmunol.1100538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pang SS, et al. The structural basis for autonomous dimerization of the pre-T-cell antigen receptor. Nature. 2010;467(7317):844–848. doi: 10.1038/nature09448. [DOI] [PubMed] [Google Scholar]
- 46.Beddoe T, et al. Antigen ligation triggers a conformational change within the constant domain of the alphabeta T cell receptor. Immunity. 2009;30(6):777–788. doi: 10.1016/j.immuni.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 47.Yoon ST, Dianzani U, Bottomly K, Janeway CA., Jr Both high and low avidity antibodies to the T cell receptor can have agonist or antagonist activity. Immunity. 1994;1(7):563–569. doi: 10.1016/1074-7613(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 48.Adams JJ, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35(5):681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie J, et al. Photocrosslinkable pMHC monomers stain T cells specifically and cause ligand-bound TCRs to be ‘preferentially’ transported to the cSMAC. Nat Immunol. 2012;13(7):674–680. doi: 10.1038/ni.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gil D, Schamel WW, Montoya M, Sánchez-Madrid F, Alarcón B. Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell. 2002;109(7):901–912. doi: 10.1016/s0092-8674(02)00799-7. [DOI] [PubMed] [Google Scholar]
- 51.van Boxel GI, Holmes S, Fugger L, Jones EY. An alternative conformation of the T-cell receptor alpha constant region. J Mol Biol. 2010;400(4):828–837. doi: 10.1016/j.jmb.2010.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hawse WF, et al. Cutting edge: Evidence for a dynamically driven T cell signaling mechanism. J Immunol. 2012;188(12):5819–5823. doi: 10.4049/jimmunol.1200952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin Y, Wang XX, Mariuzza RA. Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4. Proc Natl Acad Sci USA. 2012;109(14):5405–5410. doi: 10.1073/pnas.1118801109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Yin Y, Mariuzza RA. Structural and biophysical insights into the role of CD4 and CD8 in T cell activation. Front Immunol. 2013;4:206. doi: 10.3389/fimmu.2013.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA. T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature. 2005;436(7050):578–582. doi: 10.1038/nature03843. [DOI] [PubMed] [Google Scholar]
- 56.Kim ST, et al. The alphabeta T cell receptor is an anisotropic mechanosensor. J Biol Chem. 2009;284(45):31028–31037. doi: 10.1074/jbc.M109.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell. 2014;157(2):357–368. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim ST, et al. Distinctive CD3 heterodimeric ectodomain topologies maximize antigen-triggered activation of alpha beta T cell receptors. J Immunol. 2010;185(5):2951–2959. doi: 10.4049/jimmunol.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu C, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell. 2008;135(4):702–713. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minguet S, Swamy M, Alarcón B, Luescher IF, Schamel WW. Full activation of the T cell receptor requires both clustering and conformational changes at CD3. Immunity. 2007;26(1):43–54. doi: 10.1016/j.immuni.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 61.Minguet S, Schamel WW. A permissive geometry model for TCR-CD3 activation. Trends Biochem Sci. 2008;33(2):51–57. doi: 10.1016/j.tibs.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 62.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.