Significance
FAT10, a ubiquitin-like modifier, is an oncogene that interacts with mitotic arrest-deficient 2 (MAD2) and confers cellular malignancy. Here we identified the MAD2-binding residues of FAT10 and determined the first solution structure, to our knowledge, of the first FAT10 ubiquitin-like domain. Importantly, we demonstrated the proof-of-mechanism for a novel and specific drug-targeting strategy that entails the specific inhibition of the pathological activity of a therapeutic target but not its reported physiological function, thus minimizing undesirable side effects: Abrogation of the FAT10–MAD2 interaction curtailed tumor progression without affecting FAT10’s interaction with its other known physiological binding partners. This study presents a paradigm for drug targeting and paves the way for the development of a novel small-molecule anticancer inhibitor targeting the MAD2-binding interface of FAT10.
Keywords: chromosomal instability, ubiquitin, aneuploidy, cancer progression, FAT10
Abstract
FAT10 (HLA-F-adjacent transcript 10) is a ubiquitin-like modifier that is commonly overexpressed in various tumors. It was found to play a role in mitotic regulation through its interaction with mitotic arrest-deficient 2 (MAD2). Overexpression of FAT10 promotes tumor growth and malignancy. Here, we identified the MAD2-binding interface of FAT10 to be located on its first ubiquitin-like domain whose NMR structure thus was determined. We further proceeded to demonstrate that disruption of the FAT10–MAD2 interaction through mutation of specific MAD2-binding residues did not interfere with the interaction of FAT10 with its other known interacting partners. Significantly, ablation of the FAT10–MAD2 interaction dramatically limited the promalignant capacity of FAT10, including promoting tumor growth in vivo and inducing aneuploidy, proliferation, migration, invasion, and resistance to apoptosis in vitro. Our results strongly suggest that the interaction of FAT10 with MAD2 is a key mechanism underlying the promalignant property of FAT10 and offer prospects for the development of anticancer strategies.
FAT10 (HLA-F-adjacent transcript 10) is a ubiquitin-like modifier protein that functions as a proteasomal degradation signal (1–3). Recent studies, however, have suggested that FAT10’s functions extend beyond protein degradation. FAT10 is expressed mainly in tissues of the immune system, including the spleen and thymus (4, 5). In immune cells, FAT10 is strongly induced by proinflammatory stimuli and facilitates T-cell activation by enhancing antigen presentation of mature dendritic cells (6). FAT10 also is induced by proinflammatory cytokines in various tissues outside the immune system including the liver and colon (7, 8), although the physiological functions of this response remain unknown. What is clear, however, is that constitutive induction of FAT10 has deleterious consequences in promoting cellular malignancy. Our group recently has reported that ectopic expression of FAT10 induced malignant transformation in nontumorigenic cells and tumor promotion in tumorigenic cells (9), implicating FAT10 in facilitating tumor growth and progression. This finding is consistent with multiple reports that found FAT10 to be up-regulated in several tumor types including tumors of the liver and colon (5, 8, 10, 11).
To date, the mechanism underlying FAT10’s promalignant characteristic remains unclear. One compelling albeit indirect piece of evidence stems from the finding that FAT10 interacts with the spindle checkpoint protein mitotic arrest-deficient 2 (MAD2) during mitosis and reduces MAD2 localization to the kinetochores, resulting in aneuploidy (7, 12), a phenomenon closely associated with tumorigenesis and a hallmark of many solid tumors (13). Given the strong association between aneuploidy, chromosomal instability, and cancer development (reviewed in ref. 14), we hypothesized that FAT10 induces malignant progression through its interaction with MAD2. Therefore, the aims of this study were to elucidate the structure of the MAD2-binding interface of FAT10 and subsequently to examine the effect of disrupted FAT10–MAD2 interaction on FAT10-induced tumor progression.
Because of its poor solubility, no information is available thus far on the structure of FAT10. In this study, using heteronuclear multidimensional NMR spectroscopy, we identified the MAD2-binding interface of FAT10 located on its first ubiquitin-like domain and further determined the solution structure of this first domain. Moreover, we demonstrated that this interface is specific for the interaction of FAT10 with MAD2 and that mutation of the MAD2-binding residues of FAT10 did not interfere with the interaction of FAT10 with its other known key interaction partners including ubiquitin-activating E1 enzyme (UBA6) (15), ubiquitin-like protein NEDD8 μLtimate buster-1 long (NUB1L) (2), histone deacetylase 6 (HDAC6) (16), autophagosome receptor p62 (17), and tumor suppressor p53 (18). Significantly, abrogation of the FAT10–MAD2 interaction by mutating the core MAD2-binding residues on FAT10 limited FAT10-induced tumor growth, as evidenced by the reduced tumor sizes in mice xenografts. Additionally, disruption of the FAT10–MAD2 interaction mitigated the promalignant capacity of FAT10 in vitro, including its ability to modulate aneuploidy, cellular proliferation, migration, invasion, and resistance to apoptosis. We further found that the interaction of FAT10 with MAD2 was essential for inducing widespread gene dysregulation that may promote cellular malignancy.
Taken together, our results unequivocally implicate the interaction of FAT10 with MAD2 as a key mechanism underlying the promalignant property of FAT10. Importantly, we have elucidated the MAD2-binding interface of FAT10 and have demonstrated that it is specific for binding to MAD2, opening up prospects for the development of specific anticancer strategies targeting the promalignant function of FAT10 by inhibiting the pathological FAT10–MAD2 interaction.
Results
Structural Characterization of FAT10.
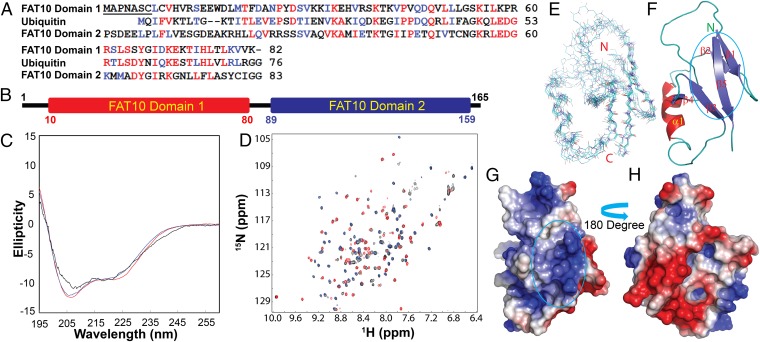
FAT10 is a 165-aa multifunctional protein comprising two ubiquitin-like domains (Fig. 1 A and B). However, the structure of the FAT10 protein remains elusive, largely because of its insoluble nature. Here, by extensively optimizing the buffer conditions, we succeeded in obtaining the full-length FAT10 sample at a protein concentration of 20 µM and collected its circular dichroism (CD) and NMR 1H-15N heteronuclear single-quantum correlation (HSQC) spectra (represented by black in Fig. 1 C and D). The full-length FAT10 protein contains well-formed secondary structures, as is evident from its far-UV CD spectrum comprising two negative signals at 208 and 222 nm as well as a positive signal at 195 nm (Fig. 1C). FAT10’s well-folded structure was evidenced further by its HSQC spectrum with large spectral dispersions at both 1H (∼3.1 ppm) and 15N (∼26 ppm) dimensions (Fig. 1D). However, because the full-length FAT10 protein tended to aggregate, even at 20 µM, we dissected it into its two ubiquitin-like domains: amino acid residues 1–82 (domain 1) and 83–165 (domain 2). Although domain 2 was highly soluble and stable, domain 1 was poorly soluble even at 50-µM concentrations. Hence, we further generated several constructs of domain 1 with sequential deletion of the N-terminal residues and found that the construct with seven N-terminal residues deleted exhibited reasonable solubility and stability for further determination of the NMR structure. The two isolated domains contained well-formed secondary structures and well-folded 3D structures as evident from their far-UV CD spectra (represented by red and blue in Fig. 1C) and HSQC spectra (represented by red and blue in Fig. 1D), respectively. Strikingly, the HSQC peaks of the two isolated domains were almost superimposable on the corresponding peaks of the full-length FAT10 (Fig. 1D). This finding strongly implies that the two ubiquitin-like domains are linked by a flexible loop and that, in the context of the full-length FAT10 protein, there might not be significant packing between the two domains.
Fig. 1.
Structural characterization of FAT10 and its two isolated domains. (A) Sequence alignment of two ubiquitin-like domains of FAT10 with ubiquitin. FAT10 residues that are identical or homologous to ubiquitin are shown in red and blue, respectively. (B) Domain organization of the 165-residue FAT10 containing two ubiquitin-like domains designated FAT10 domain 1 and FAT10 domain 2, respectively. The boundaries of the two domains are based on the 3D structure we determined. (C) Far-UV CD spectra of the full-length FAT10 (black trace), domain 1 (red trace), and domain 2 (blue trace) at 20-µM protein concentration. (D) Superimposition of 2D 1H-15N HSQC spectra of full-length FAT10 (black), domain 1 (red), and domain 2 (blue). (E) Superimposition of the eight selected backbone structures of domain 1 at the secondary structure regions. N refers to the amino-terminus and C refers to the carboxyl-terminus of FAT10 domain 1. (F) Three-dimensional structure of FAT10 domain 1 in ribbon format comprising a typical ubiquitin-like fold composed of five β-strands and one α-helix. (G and H) The electrostatic potential surface of FAT10 domain 1 oriented as in F (G) and with the orientation rotated 180° along the z axis (H). Positively charged, negatively charged, and nonpolar/neutral residues are represented by blue, red, and gray respectively. The main MAD2-binding regions in F and G are outlined in cyan.
NMR Structures and Identification of the MAD2-Binding Surface of FAT10.
Because FAT10 domain 1 started to unfold at pH below 7.2 and showed a detectable coexistence of the folded and unfolded states at pH 7.0, we subsequently conducted all biophysical studies at pH 7.4. By analyzing 15N-edited HSQC-total correlation spectroscopy (TOCSY) and HSQC-NOESY, we achieved both backbone and side-chain assignments of most nonproline residues except for 10 residues within the long loop from Pro59 to Thr73 for which HSQC peaks were not detected at pH 7.4. Next, the 3D structure was determined for FAT10 domain 1. Briefly, NOE-derived distance and TALOS-based dihedral angle restraints (19) were used in the CYANA software package (20) to calculate 50 NMR structures. The eight structures with the lowest target functions were selected for further refinement with AMBER force field (21). The eight superimposed NMR structures of FAT10 domain 1 are shown in Fig. 1E, and the calculation statistics and structure quality are summarized in Table S1. The eight NMR structures consist of well-defined secondary structures comprising five β-strands and one α-helix, with rms deviations over the secondary structure regions of 0.45 Å for backbone atoms and 1.15 Å for all atoms as compared with the mean structure. The loop regions were less defined, with distinctive conformations in different structures. In particular, the loop residues Arg60 and Glu61 were highly variable because of the lack of NOE connectivities, and their corresponding HSQC peaks were very broad, implying possible conformational changes (22). As a consequence, rms deviations of the whole FAT10 domain 1 became 1.08 Å for backbone atoms and 2.24 Å for all atoms. Furthermore, no slowly exchanged amide protons could be identified by the NMR H–D exchange experiments at pH 7.4, suggesting that FAT10 domain 1 also undergoes a global folding–unfolding exchange that usually occurs on the microsecond-to-second time scale. The presence of the conformational exchanges on the microsecond-to-second time scale thus rationalizes the relatively low number of long-range NOEs, which contributes to the relatively high rms deviations for FAT10 domain 1. Although FAT10 domain 1 possesses the characteristic ubiquitin-like fold comprising five β-strands and one α-helix, the electrostatic potential surface of this domain differs from that of ubiquitin (Fig. S1) (23). For FAT10 domain 1, the surface constituted by some of the residues from β-strands 1, 3, and 5 is highly positive (Fig. 1G), whereas the opposite side is largely negative (Fig. 1H).
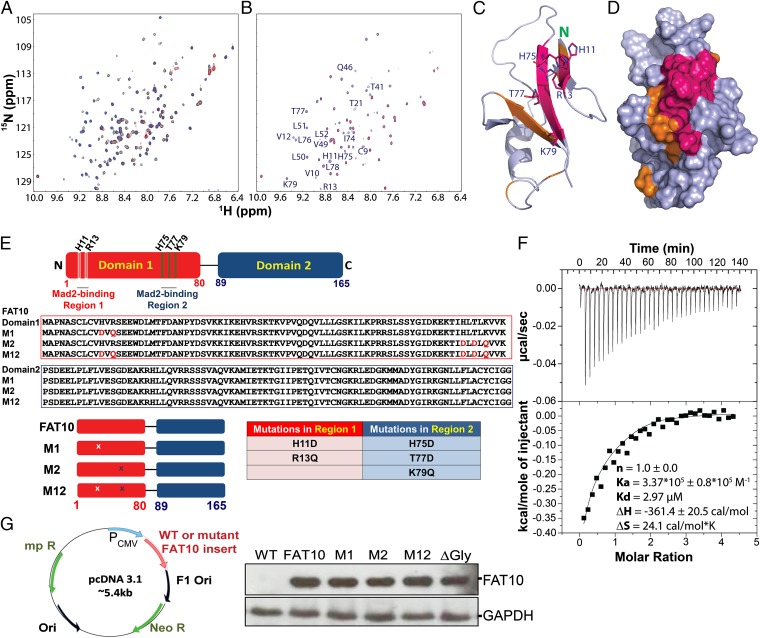
We also conducted extensive investigations on the binding interaction between FAT10 and MAD2. FAT10 proteins including FAT10 (aa 1–165), FAT10 (aa 8–165), FAT10 domain 1 (aa 1–82), FAT10 domain 1 (aa 8–82), and FAT10 domain 2 (aa 82–165) were subjected to NMR HSQC titration by adding MAD2. The HSQC titration allowed the detection of weak binding events and mapping of the binding interface (22, 24, 25). Titration of the 15N-labeled full-length FAT10 sample by the gradual addition of unlabeled MAD2 triggered a significant reduction in intensity or even the disappearance of a portion of FAT10 HSQC peaks (Fig. 2A). Upon closer inspection, we found that these peaks were primarily from domain 1 (Fig. 2B), suggesting that FAT10 domain 1 is responsible for binding to MAD2. Furthermore, HSQC peaks of residues 1–7 were not perturbed significantly by the addition of MAD2. To confirm our finding, 15N-labeled samples of the two isolated domains of FAT10 were titrated with MAD2. As expected, the gradual addition of MAD2 induced significant reduction in intensity or even the disappearance of HSQC peaks of domain 1, but no chemical shift or intensity change was detected in HSQC peaks of domain 2 even at a FAT10 domain 2:MAD2 molar ratio of 1:4, despite FAT10 domain 2 assuming the same ubiquitin-like fold (Fig. S2). These data strongly suggest that only domain 1 of FAT10 interacts with MAD2. Subsequently, we mapped the MAD2-binding interface of FAT10 domain 1 by examining residues with HSQC peaks that either disappeared or significantly broadened at FAT10 domain 1:MAD2 molar ratios of 1:0.5 and 1:1 (represented by red and brown, respectively, in Fig. 2 C and D). Interestingly, the MAD2-binding surface, which is positively charged, is composed mainly of β-strands 1, 2, 3, and 5 with the core residues located on the β1 (Val9–Arg12) and β5 (Ile74–Lys79) strands (Fig. 2 C and D). Of the core residues, only the side chains of His11 and Arg13 on β1 and His75, Thr77, and Lys79 on β5 were exposed to the solvent; the other core residues were buried within the protein. Because buried hydrophobic residues usually are involved in folding and stabilization, and exposed residues often play a central role in binding, we focused our investigations on the five exposed residues, namely His11, Arg13, His75, Thr77, and Lys79, and their possible contributions to the FAT10–MAD2 interaction.
Fig. 2.
NMR identification and mutation of MAD2-binding residues of FAT10. (A) Superimposition of HSQC spectra of 15N-labeled full-length FAT10 in the absence (black) and presence (red) of unlabeled MAD2 at a FAT10:MAD2 molar ratio of 1:1.5 and FAT10 domain 2 (blue). (B) Superimposition of HSQC spectra of 15N-labeled FAT10 domain 1 in the absence (blue) and presence (red) of the unlabeled MAD2 at a molar ratio of 1:1. Corresponding residues of HSQC peaks that disappeared or displayed significantly reduced intensity are labeled. (C and D) Three-dimensional structure of FAT10 domain 1 in ribbon (C) and surface (D) format. Residues with significantly altered HSQC intensities at the FAT10 domain 1:MAD2 molar ratios of 1:0.5 and 1:1 are shown in red and brown, respectively. (E) Site-directed mutagenesis to introduce various mutations into human FAT10 cDNA. (Top) Schematic showing mutations at MAD2-binding region 1 (M1), region 2 (M2), or at both regions (M12). The specific mutations are shown by alignment of primary amino acid sequences (Middle), schematically (Bottom Left), and in a table (Bottom Right). (F, Upper) ITC profiles of the binding reaction of FAT10 domain 1 with MAD2 and (Lower) integrated values for reaction heats with subtraction of the corresponding blank results normalized against the amount of ligand injected versus the molar ratio of FAT10 domain 1:MAD2. The thermodynamic binding parameters obtained from data fitting are shown. (G, Left) Wild-type and the various FAT10 mutant cell lines were generated following cloning of the respective FAT10 cDNA into the pcDNA3.1 vector and subsequent transfection into HCT116 cells. (Right) Western blot analysis confirmed comparable expression of wild-type and mutant FAT10 in HCT116 cells. FAT10 mutants containing mutated di-glycine residues (∆Gly: G164A, and G165A) were generated also.
We therefore generated various FAT10 domain 1 mutants: M1 with H11D and R13Q mutations in region 1; M2 with H75D, T77D, and K79Q mutations in region 2; and M12 with all the aforementioned mutations (Fig. 2E). The respective amino acid substitutions were chosen to disrupt potential amino acid charge-dependent interactions while retaining their molecular weight. Importantly, the amino acid substitutions did not result in significant denaturation or misfolding of the respective mutant proteins, as indicated by their CD spectra (Fig. S3A). To quantify the binding event, the thermodynamic parameters of the binding between MAD2 and wild-type FAT10 domain 1 as well as its mutants were measured by isothermal titration calorimetry (ITC). The binding affinity obtained for the wild-type FAT10 domain 1 (dissociation constant, Kd, of 2.97 µM) (Fig. 2F) was consistent with the observed disappearance of HSQC peaks of the FAT10 domain 1 upon binding to MAD2 (22, 25). On the other hand, M1 showed significantly reduced binding affinity (Fig. S3B) and thus could not be fitted to obtain the binding parameters, and M12 showed almost no binding to MAD2 (Fig. S3C). The M2–MAD2 interaction could not be determined because of its low solubility.
Mutation of MAD2-Binding Residues on FAT10 Does Not Affect FAT10’s Binding to Other Interaction Partners.
We next examined the involvement of the aforementioned FAT10 residues in the interaction of FAT10 with MAD2. Mutations of the putative MAD2-binding regions of FAT10 (Fig. 2E) were introduced into full-length human FAT10 cDNA using site-directed mutagenesis and were cloned into a pcDNA3.1 vector (Invitrogen) for stable expression of the wild-type and mutant FAT10 proteins in HCT116 parental cells (Fig. 2G). Importantly, the expression levels of wild-type and mutant FAT10 proteins used in the subsequent experiments of this study were within the range of FAT10 expression detected in hepatocellular carcinoma (HCC) tumors (Fig. S4).
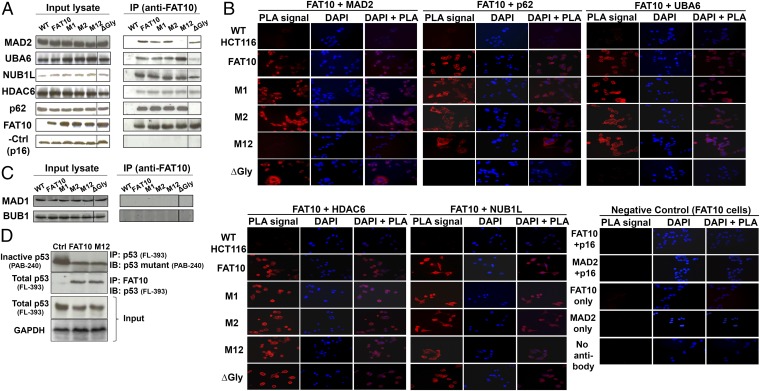
To identify the FAT10 residues that are critical for its interaction with MAD2, we examined the interaction of wild-type and mutant FAT10 with endogenous MAD2 using coimmunoprecipitation (co-IP) and in situ proximity ligation assay (PLA), a sensitive technique that detects sites of protein–protein interaction through the visualization of red fluorescent signals. Strikingly, both methods of detecting protein–protein interaction showed abrogation of the FAT10–MAD2 interaction only upon mutation of both MAD2-binding regions (i.e., mutant M12) (Fig. 3 A and B, Upper Left and Fig. S5). FAT10 mutated at either binding region (M1 and M2) retained the ability to interact with MAD2, albeit to a lesser extent for M1 (Fig. 3 A and B, Upper Left). Furthermore, mutation of both MAD2-binding regions did not affect the binding of FAT10 to its known substrates namely UBA6, NUB1L, HDAC6, and p62 (Fig. 3 A and B), suggesting that the MAD2-binding interface of FAT10 is specific to MAD2. Separately, mutation of the C terminus di-glycine motif of FAT10 abolished binding to UBA6 and p62 (Fig. 3 A and B, Upper Middle and Upper Right), consistent with reports that FAT10 binds to these proteins through its di-glycine residues (15, 17). An irrelevant antibody, anti-p16, was used as a negative control in the interaction experiments. We further ascertained that FAT10 binds to free MAD2 and not to MAD2 bound to the spindle checkpoint complex, because members of the MAD2 complex, MAD1 and Bub1 (budding uninhibited by benzimidazoles 1), could not be detected following immunoprecipitation using FAT10-specific antibodies (Fig. 3C and Fig. S5). Because Li et al. (18) previously found that FAT10 overexpression increased the population of transcriptionally active p53, we next tested if mutation of the MAD2-binding regions of FAT10 interfered with this function. To this end, transcriptionally inactive p53 levels were determined using PAB240 antibodies following immunoprecipitation of full-length p53 (FL-393 antibody) in wild-type FAT10, M12, and wild-type parental cells. Consistently, a significant reduction in transcriptionally inactive p53 levels was detected in wild-type FAT10 cells as compared with parental cells, although total p53 levels were similar in both cell lines (Fig. 3D). A similar reduction in transcriptionally inactive p53 levels also was observed in M12 cells (Fig. 3D), suggesting that the abrogation of the FAT10–MAD2 interaction did not interfere with the ability of FAT10 to increase the population of transcriptionally active p53. Furthermore, mutation of the MAD2-binding residues of FAT10 did not interfere with the interaction of FAT10 with p53 (Fig. 3D), supporting the specificity of the MAD2-binding interface of FAT10 for MAD2.
Fig. 3.
Disruption of the FAT10–MAD2 interaction does not affect the binding of FAT10 with its known interaction partners and its function in protein degradation. (A and B) Mutation of both MAD2-binding regions of FAT10 (M12) abrogates the FAT10–MAD2 interaction but not FAT10’s interaction with other known interaction partners including UBA6, NUB1L, HDAC6, and p62 as shown by co-IP (A, Right) and PLA (B). Antibody against p16 was included as a negative control. Loading controls for co-IP (input lysate) are shown (A, Left). (B) Right columns show red fluorescence signals representing sites of protein–protein interaction within cells. Center columns show nuclear staining with DAPI. Right columns show overlay of PLA and DAPI signals. Single antibody (anti-FAT10 or anti-MAD2) and no primary antibody were included as negative controls (B, Lower Right). (C) FAT10 binds to free MAD2. Members of the MAD2 mitotic spindle checkpoint complex MAD1 and Bub1 were probed on FAT10-immunoprecipitated lysates obtained in A. (D) Mutation of both MAD2-binding regions of FAT10 (M12) does not affect the modulation of p53 by FAT10. Inactive p53 was probed using p53 mutant-specific antibody (PAB-240) in immunoblotting (IB) following immunoprecipitation (IP) of full-length p53 (FL-393) of HCT116 cell lysates transiently overexpressing wild-type or mutant FAT10 (M12) for 48 h. To probe for total p53 interacting with FAT10, p53 (FL393) was used for immunoblotting following immunoprecipitation with FAT10-specific antibodies.
Abrogation of the FAT10–MAD2 Interaction Ameliorates FAT10-Induced Chromosomal Instability and Mitotic Checkpoint Dysregulation.
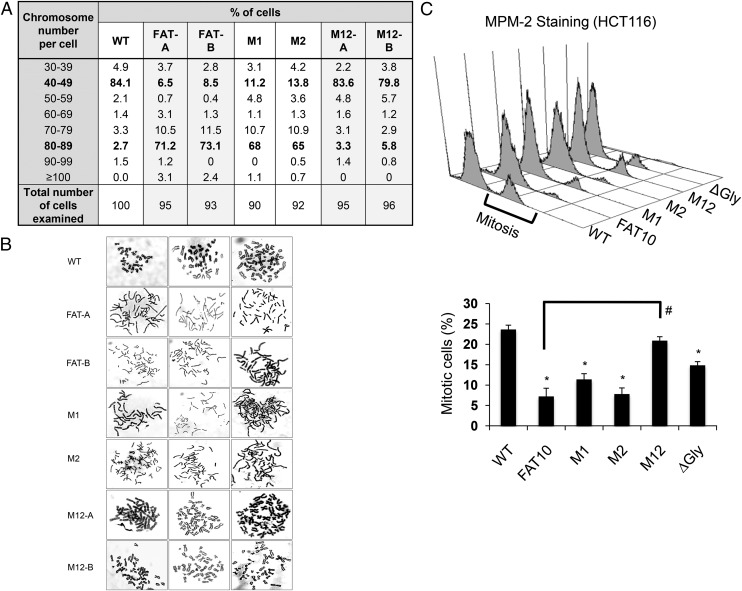
FAT10 overexpression has been shown to promote chromosomal instability (7, 12), tumor initiation, and malignant features in both nontumorigenic and tumorigenic cells (9). To examine the effect of disrupted FAT10–MAD2 interaction on FAT10-induced chromosomal instability, chromosomes from parental wild-type HCT116 cells and from wild-type FAT10 (FAT-A and FAT-B), M1, M2, and M12 (M12-A and M12-B) cells were karyotyped. Consistent with our previous findings (7, 12), overexpression of wild-type FAT10 showed a markedly higher proportion of aneuploid cells compared with parental cells (Fig. 4A). Specifically, the majority (>70%) of wild-type FAT10-expressing cells displayed more than the usually observed number of chromosomes (80–89), whereas the majority (>80%) of the parental cells retained the modal number of chromosomes (40–49) (Fig. 4A) (12). Further, a majority (>68%) of MAD2-deficient cells (Fig. S6A) similarly showed more than the usually observed number of chromosomes (80–89) (Fig. S6B). Although similarly high proportions of aneuploid cells were observed in M1 and M2 cells and in wild-type FAT10-expressing cells, the majority (>79%) of M12 cells deficient for FAT10–MAD2 interaction retained modal chromosome numbers (Fig. 4A), suggesting that the FAT10–MAD2 interaction may be critical for FAT10-induced aberrations in chromosome number.
Fig. 4.
Disruption of the FAT10–MAD2 interaction inhibits FAT10-induced chromosomal instability. (A) Tabulation of the karyotype analysis of wild-type parental cells, FAT10-expressing cells (FAT-A, FAT-B), and the various stable FAT10 mutant cells (M1, M2, M12-A, M12-B). (B) Three representative metaphase spreads of parental wild-type, stable wild-type, and mutant FAT10-expressing cells. (C) Schematic (Upper) and graphical (Lower) representation of mitotic profiles of parental wild-type, stable wild-type, and mutant FAT10-expressing cells. Cells were synchronized at G1/S phase by single thymidine block followed by treatment with the M-phase inhibitor nocodazole for 8 h. Cells were stained with the mitosis marker MPM-2 (27) before FACS analysis. All data shown are mean ± SE. *P < 0.05 compared with parental wild type; #P < 0.05 compared with FAT10-expressing cells.
A closer examination of the morphology of the metaphase chromosomes in the various cell lines revealed that wild-type FAT10-expressing cells contained incompletely condensed, long “thread-like” chromosomes as opposed to the condensed, short “ribbon-like” chromosomes in parental cells (Fig. 4B). Strikingly, the morphology of the metaphase chromosomes in M1 and M2 cells resembled that of wild-type FAT10-expressing cells, whereas the morphology of metaphase chromosomes in M12 cells resembled that of parental cells (Fig. 4B). Notably, MAD2 depletion has been reported to alter chromosome morphology similarly (26). Taken together, these data suggest that the interaction of FAT10 with MAD2 contributes to FAT10-induced abnormalities in chromosome structure and number.
Next, because wild-type FAT10 overexpression was reported to facilitate escape from nocodazole-induced mitotic arrest (12), we examined if this escape was mediated by the interaction of FAT10 with MAD2. Mitotic cell populations were obtained by synchronization at G1/S phase followed by nocodazole-induced arrest for 8 h. Cells then were assessed by FACS analysis following staining with the mitosis marker mitotic protein monoclonal 2 (MPM-2) (27). Consistent with our earlier observations, a significantly reduced proportion of mitotic cells was observed in wild-type FAT10-expressing cells (7.2%) compared with parental cells (23.7%) following mitotic arrest (Fig. 4C). Similarly, depletion of MAD2 resulted in a significantly reduced proportion of mitotic cells (Fig. S6C), phenocopying the reduction seen with FAT10 overexpression. Notably, abrogation of the FAT10–MAD2 interaction (i.e., in the M12 mutant) limited the cell’s ability to escape mitotic arrest to levels comparable to that of parental cells (Fig. 4C). Mutation of the di-glycine motif of FAT10 (∆Gly) moderately restored the mitotic cell population, albeit to a lesser extent (Fig. 4C). These data support a role for the FAT10–MAD2 interaction in disrupting the mitotic checkpoint and in inducing aneuploidy.
Abrogation of the FAT10–MAD2 Interaction Attenuates FAT10-Induced Malignancy in Vitro.
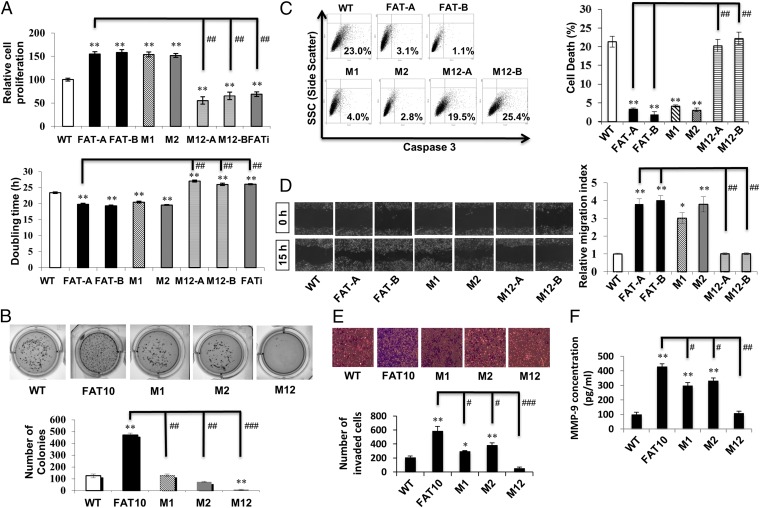
We next investigated the role of the FAT10–MAD2 interaction on FAT10-induced malignancy because FAT10 overexpression was previously found to enhance the cell’s anchorage-independent growth, resistance to cell death, and its invasion and migration abilities (9).
Examination of the cellular proliferation profiles of wild-type parental, wild-type FAT10-expressing (FAT-A and FAT-B), FAT10 knock-down (FATi), M1, M2, and M12 (M12-A and M12-B) cells revealed enhanced proliferation of cells that retained the FAT10–MAD2 interaction (FAT-A, FAT-B, M1, and M2) compared with parental cells (Fig. 5A). However, disruption of the FAT10–MAD2 interaction (M12) significantly attenuated cellular proliferation to levels observed in FATi cells, which were notably lower than those of parental cells (Fig. 5A). Next, investigation of the anchorage-independent growth profiles of the various cell lines showed that wild-type FAT10 overexpression enhanced anchorage-independent growth of cells relative to parental cells (Fig. 5B). Mutation of either MAD2-binding region of FAT10 (M1 and M2) reduced colony formation on soft agar to that observed in parental cells, whereas abrogation of the FAT10–MAD2 interaction (M12) almost obliterated anchorage-independent growth of the cells (Fig. 5B).
Fig. 5.
Abrogated FAT10–MAD2 interaction attenuates FAT10-induced malignancy in vitro. In vitro malignant characteristics of parental wild-type, stable wild-type, and mutant FAT10-expressing cells were examined. (A, Upper) Cell-proliferation profiles were assessed by the colorimetric WST assay in which the number of viable cells is measured by the reduction of the tetrazolium compound WST in the colored formazan product. (Lower) The corresponding cell-doubling time in hours. (B) Anchorage-independent growth profiles on soft agar were assessed. (Upper) Representative pictures of stained colonies. (Lower) Corresponding graphical representation. (C, Left) Cellular apoptosis profiles following camptothecin-induced cell death were assessed by FACS detection of activated caspase-3. (Right) Corresponding graphical representation. (D, Left) Cell migration profiles were assessed by scratch-wound assay over 15 h. (Right) Subsequent quantification. (E) Cellular invasion profiles were assessed by Matrigel invasion assay for 24 h. (Upper) Representative pictures of the invaded cells following staining. (Lower) Quantification. (F) Cellular MMP-9 secretion profiles were detected by ELISA and are represented graphically. All data are shown as mean ± SE. *P < 0.05 and **P < 0.01 compared with parental wild-type; #P < 0.05 and ##P < 0.01 compared with FAT10-expressing cells.
Examination of the cell’s ability to resist camptothecin-induced cell death revealed that ablation of the FAT10–MAD2 interaction (M12) increased susceptibility to cell death, whereas cells harboring mutation of either MAD2-binding region of FAT10 (M1 and M2) retained an ability to resist cell death similar to that of wild-type FAT10-expressing cells (Fig. 5C). Additionally, abrogation of the FAT10–MAD2 interaction (M12) attenuated FAT10-induced cellular migration and invasion to levels comparable to or below those of parental cells (Fig. 5 D and E, respectively). Notably, mutation of either MAD2-binding region of FAT10, particularly M1, only partially impaired the invasive and migratory ability of the cells. Further, analysis of a protein that degrades extracellular matrix and is associated with colon cancer progression (28), matrix metalloproteinase 9 (MMP-9), showed that wild-type FAT10 induced cells to secrete significantly higher amounts of MMP-9 compared with parental cells (Fig. 5F). Abolishment of the FAT10–MAD2 interaction inhibited FAT10-induced MMP-9 secretion to levels observed in parental cells, whereas mutation of either MAD2-binding region of FAT10 only moderately attenuated the increased MMP-9 secretion (Fig. 5F).
Collectively, disruption of the interaction of FAT10 with MAD2 curtailed various FAT10-induced malignant characteristics in vitro, and these findings were observed consistently in transiently-expressing FAT10 SNU449-transformed liver cells (Fig. S7) suggesting a role for the FAT10–MAD2 interaction in tumor progression.
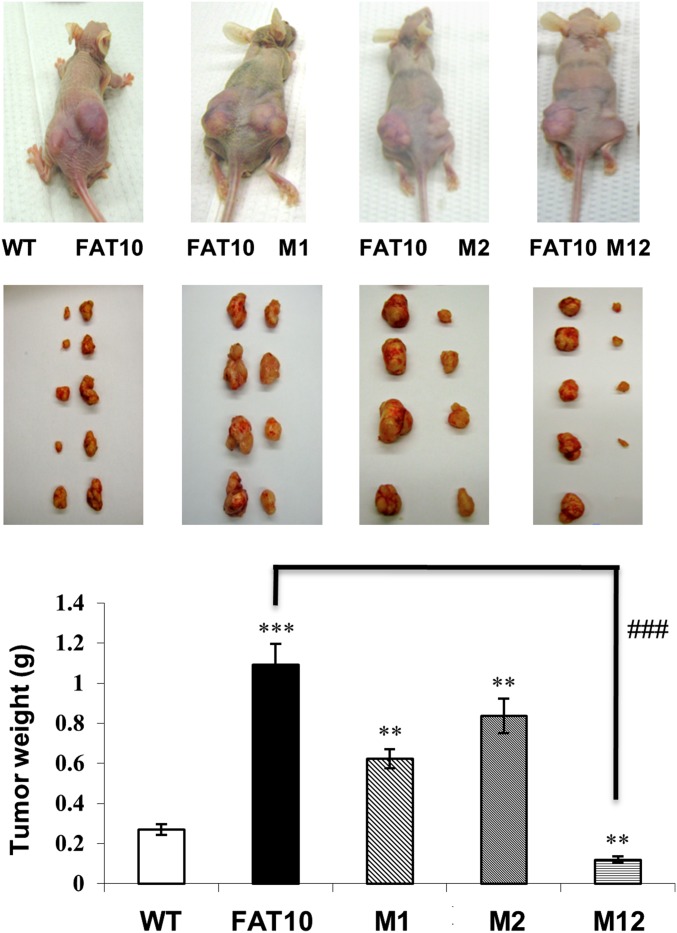
Abrogation of the FAT10–MAD2 Interaction Attenuates FAT10-Induced Tumor Formation in Vivo.
We previously had demonstrated that FAT10 overexpression promotes tumor formation in nude mice, recapitulating the malignant phenotype observed in vitro (9). To investigate if the tumor-promoting ability of FAT10 is mediated through its interaction with MAD2, parental and various FAT10-expressing HCT116 stable cells were injected s.c. into opposite flanks of nude mice. Consistent with our previous report, wild-type FAT10-expressing cells induced significantly larger tumor formation at the injected sites than did parental cells (Fig. 6). Strikingly, MAD2 binding-deficient FAT10-expressing cells (M12 cells) formed dramatically smaller tumors, even smaller than those arising from parental cells, whereas mutation of either MAD2-binding region of FAT10 (M1 and M2) moderately retarded FAT10-induced tumor growth (Fig. 6). These data highlight the tumor-promoting ability of FAT10 and further suggest that its promalignant characteristic is mediated by its interaction with MAD2.
Fig. 6.
Abrogated FAT10–Mad2 interaction attenuates FAT10-induced tumor formation in vivo. Representative pictures of tumors in vivo (Top) and excised tumors (Middle) following s.c. injection of parental wild-type, stable wild-type (FAT10), and mutant (M1, M2, and M12) FAT10-expressing HCT116 cells. (Bottom) Mean tumor weights of six nude mice per group, 3 wk post s.c. injection, are represented graphically. All data are shown as mean ± SE. **P < 0.01 and ***P < 0.001 compared with parental wild-type; ###P < 0.001 compared with FAT10-expressing cells.
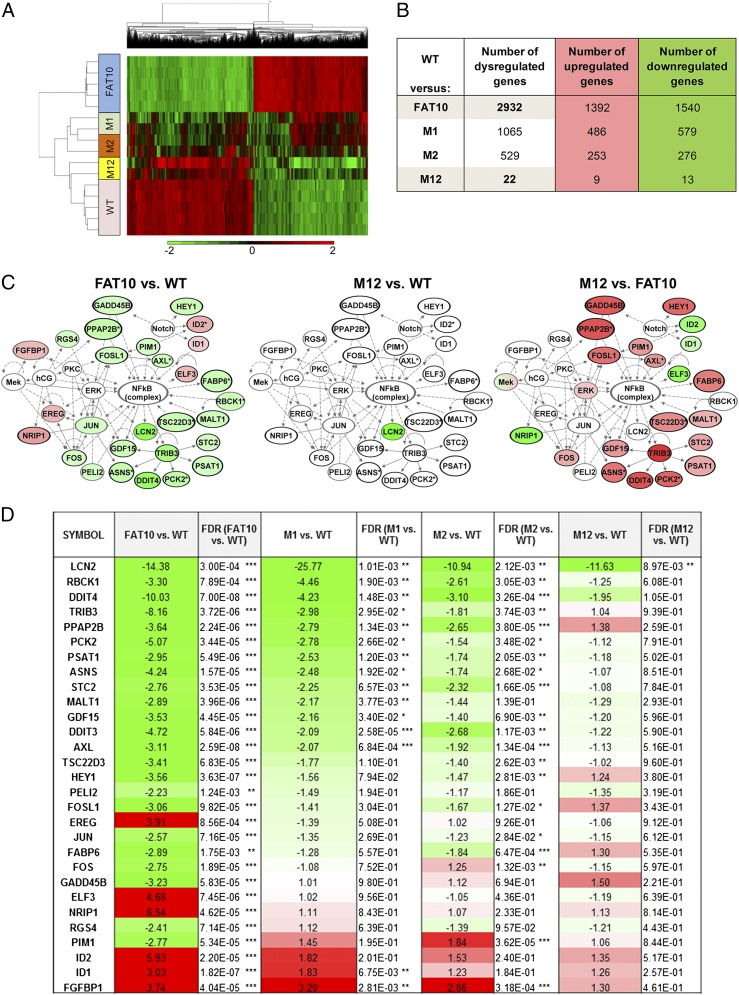
Mutation of MAD2-Binding Residues of FAT10 Inhibited FAT10-Induced Global Gene-Expression Changes.
To assess the effect of mutating the MAD2-binding regions of FAT10 on global gene expression, expression profiling was performed on cells stably expressing wild-type FAT10 and on cells expressing the various FAT10 mutants M1, M2, and M12. All cells were synchronized using double thymidine treatment before expression profiling to exclude potential cell cycle-dependent confounding effects on gene expression. Although wild-type FAT10 induced significant changes in global gene-expression profiles, the gene-expression profiles of the MAD2 binding-deficient FAT10 mutant (M12) resembled those of the parental cells (Fig. 7A). Specifically, the number of dysregulated genes in M12 cells was markedly reduced compared with the wild-type counterpart (Fig. 7B). In addition, deregulated gene-expression profiles of the top associated network function deregulated by wild-type FAT10, namely, Cellular Growth and Proliferation, Cellular Development, and Cell Death (Tables S2 and S3) (9), could be restored by abrogating the interaction of FAT10 with MAD2 (Fig. 7C). As evident from Fig. 7D, a majority of the significantly deregulated genes in the top associated network of wild-type FAT10-expressing cells were not significantly altered in M12 cells. These data suggest that the interaction between FAT10 and MAD2 is important for FAT10-induced deregulation of genes of key pathways associated with tumor progression such as cellular growth and proliferation, cellular development, and cell death. Taken together, our data demonstrate that FAT10 promotes tumor progression through its interaction with MAD2 and that disruption of the FAT10–MAD2 interaction may curtail cellular malignancy.
Fig. 7.
Abrogated FAT10–MAD2 interaction mitigated FAT10-induced global gene-expression changes. (A) Hierarchical clustering of genes dysregulated in wild-type FAT10-expressing cells (FAT10) compared with parental wild-type cells and mutant FAT10-expressing cells (M1, M2, and M12). Down-regulated genes (relative to parental wild-type cells) are represented in green; up-regulated genes are represented in red. (B) Table showing the number of dysregulated genes in the indicated microarray analysis. (C and D) The top associated network “Cellular Growth and Proliferation, Cellular Development, and Cell Death” of genes differentially expressed in wild-type FAT10 and parental HCT116 cells as identified by IPA. The genes derived from this top associated network and their expression profiles in FAT10, M1, M2, and M12 cells relative to parental wild-type cells are presented graphically (C) and in a table that includes their fold change and False Discovery Rate (FDR)-adjusted P value (D). Green and red represent down- and up-regulation of gene expression, respectively, relative to the parental wild-type cells; white ovals in C represent genes with no significant change of expression between the indicated comparisons. ***P < 0.001, **P < 0.01, *P < 0.05.
Discussion
In recent years, increasing evidence has pointed toward a pathological role for FAT10 in promoting cellular malignancy through mechanisms beyond its proteasome-targeting function. Liu et al. (4) first reported the interaction of FAT10 with the mitotic spindle assembly checkpoint protein MAD2, and we further showed that this interaction induced chromosomal instability (7, 12), a hallmark of many solid tumors. Recently, a causal role for FAT10 in tumor initiation and progression was established (9, 29). However, the role of the FAT10–MAD2 interaction in promoting malignancy remained unclear, largely because the insoluble nature of the FAT10 protein in vitro hampered the identification of the FAT10 protein structure and its interaction interface with MAD2.
In this study, we achieved a breakthrough to this conundrum, and with the FAT10 domain 1 structure presented here we report the successful determination of the first (to our knowledge) solution structures of the FAT10 domains through extensive dissection of the protein. We show through NMR studies that FAT10 contains two domains that adopt the same ubiquitin-like fold and that they may not possess tight packing between each other. Importantly, we mapped the MAD2-binding interface of FAT10 to domain 1 of FAT10 with a Kd of 2.97 µM. The very different electrostatic potential surface of domain 2 may be largely responsible for its inability to bind MAD2 (Fig. S2). We further identified and characterized five core residues within this interface that are crucial for the interaction of FAT10 with MAD2. Mutation of these five key residues abolished the FAT10–MAD2 interaction both in vitro and in vivo and mitigated the promalignant features of FAT10, strongly implicating the interaction of FAT10 with MAD2 in mediating FAT10-induced malignancy. Importantly, disruption of the FAT10–MAD2 interaction significantly curtailed FAT10-induced tumor growth in vivo, suggesting that the role of FAT10 in tumor progression is critically dependent on its interaction with MAD2.
In agreement with literature reports implicating FAT10 in mitotic regulation (7, 12), a recent study revealed FATylation of mitosis-related cell cycle regulators, including MAD2, during mitosis (30). Here, we provide further mechanistic insight into FAT10-induced mitotic dysregulation through its interaction with the mitotic spindle assembly checkpoint protein MAD2. We demonstrated that the capacity of FAT10 to induce numerical and structural chromosomal aberrations is critically dependent on its interaction with MAD2. Perhaps this finding is not surprising, given that MAD2 serves as an important checkpoint during mitosis, ensuring proper attachment of chromosomes to the spindle microtubules during prometaphase. During this phase, unattached kinetochores prevent metaphase-to-anaphase transition by inducing the conversion of mitotic checkpoint proteins such as MAD2 into diffusible inhibitors of anaphase-promoting complex-cdc20 (APCcdc20) (31). Interestingly, the MAD2-dependent FAT10-induced malignant phenotype strikingly resembles the high prevalence of aneuploidy and spontaneous tumors observed in mice heterozygous for MAD2 and overexpressing MAD2 (32, 33), implicating a weakened mitotic checkpoint in promoting cellular malignancy. The significance of checkpoint aberration and aneuploidy in promoting neoplastic transformation also has been highlighted by several studies that used mice with genetically altered levels of checkpoint components (reviewed in ref. 13). Notably, we found that other FAT10-induced malignant characteristics such as enhanced proliferation, migration, and invasion and resistance to apoptosis also were dependent on its interaction with MAD2, consistent with the reported promalignant phenotype of MAD2-deficient cells (34–36). These findings suggest that MAD2 may have other roles that are independent of its role in mitosis, although the underlying mechanisms at present remain unclear. Additionally, our data suggest that the interaction of FAT10 with MAD2 induces widespread gene deregulation that may promote cellular malignancy. Although it is not immediately clear whether the aberrant gene-expression patterns observed were a result of FAT10-induced chromosomal instability or a consequence of the FAT10–MAD2 interaction, our data undoubtedly show that abrogating the FAT10–MAD2 interaction protects on cells against global gene deregulation induced by FAT10.
In this study, we have demonstrated that the MAD2-binding interface of FAT10 is specific to MAD2, thus presenting a previously unidentified and specific strategy for targeting the MAD2-related tumor-promoting activities of FAT10. Mutation of the five core residues of FAT10 that are crucial for the FAT10–MAD2 interaction did not affect the covalent binding of FAT10 with its E1 enzyme UBA6 or with other known binding partners through the FAT10 di-glycine residues, consistent with the notion that the two FAT10 domains are structurally independent. Noncovalent interactions between FAT10 and proteins NUB1L and HDAC6 also were unaffected by the disruption of the MAD2-binding interface of FAT10. Specific targeting of the MAD2-related pathological activity of FAT10 offers the advantage of minimizing potential undesirable side effects for future therapy and is of paramount importance, because our current understanding of the physiological roles of FAT10 is incomplete. Specific targeting of FAT10 also appears more favorable than targeting MAD2 because MAD2 is ubiquitously expressed and plays a critical physiological role in cell cycle regulation. In contrast, the frequent selective overexpression of FAT10 in tumors (5) makes extremely attractive the development of small-molecule inhibitors targeting the MAD2-binding interface of FAT10 for use as a cytostatic agent in anticancer therapies for patients with premalignant neoplasms or tumors.
In summary, this study has shed light on the mechanism underlying FAT10-induced promotion of cellular malignancy through its interaction with MAD2. Importantly, our findings present a paradigm for drug targeting as well as the foundation for the development of novel, small-molecule anticancer drugs that specifically target the MAD2-related promalignant functions of FAT10.
Materials and Methods
Detailed materials and methods are included in SI Materials and Methods. Briefly, HCT116 cell lines were purchased from American Type Culture Collection (ATCC). The MAD2-binding interface of FAT10 was obtained by NMR studies. FAT10 mutants were generated using site-directed mutagenesis. Protein–protein interactions in cells were verified using co-IP and PLA. Chromosome numbers and mitotic index analyses were performed as previously described (12). Cell proliferation was measured using the water-soluble tetrazolium salt (WST-1) assay. Anchorage-independent growth was investigated using soft agar colony formation assay. Apoptotic cells were measured using FACS analysis following FITC-caspase3 staining. Cell migration was assessed using the scratch-wound assay. Invasion assays were performed using the Matrigel Invasion Assay. MMP-9 secretion was measured using ELISA. Details of the expression microarray analyses to identify genes affected by the FAT10–MAD2 interaction are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by National Medical Research Council (NMRC) Grant NMRC/1306/2011 and block funding from National Cancer Centre Singapore (to C.G.L.) and by NMRC Grant R154-000-454-213 and Ministry of Education of Singapore (MOE) Tier 2 Grant MOE 2011-T2-1-096 (to J.S.). W.W. is a recipient of an MOE graduate scholarship under MOE Grant R-154-000-388-112 (to J.S.). S.S.T. and W.-C.M. are recipients of National University of Singapore Graduate School for Integrative Sciences and Engineering Scholarships.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: NMR, atomic coordinates, chemical shifts, and restraints reported in this paper have been deposited in the Protein Data Bank (PDB), www.pdb.org (PDB ID code 2MBE), and the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE54167).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403383111/-/DCSupplemental.
References
- 1.Schmidtke G, Kalveram B, Groettrup M. Degradation of FAT10 by the 26S proteasome is independent of ubiquitylation but relies on NUB1L. FEBS Lett. 2009;583(3):591–594. doi: 10.1016/j.febslet.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Rani N, Aichem A, Schmidtke G, Kreft SG, Groettrup M. FAT10 and NUB1L bind to the VWA domain of Rpn10 and Rpn1 to enable proteasome-mediated proteolysis. Nat Commun. 2012;3:749. doi: 10.1038/ncomms1752. [DOI] [PubMed] [Google Scholar]
- 3.Hipp MS, Kalveram B, Raasi S, Groettrup M, Schmidtke G. FAT10, a ubiquitin-independent signal for proteasomal degradation. Mol Cell Biol. 2005;25(9):3483–3491. doi: 10.1128/MCB.25.9.3483-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu YC, et al. A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2. Proc Natl Acad Sci USA. 1999;96(8):4313–4318. doi: 10.1073/pnas.96.8.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CG, et al. Expression of the FAT10 gene is highly upregulated in hepatocellular carcinoma and other gastrointestinal and gynecological cancers. Oncogene. 2003;22(17):2592–2603. doi: 10.1038/sj.onc.1206337. [DOI] [PubMed] [Google Scholar]
- 6.Ebstein F, Lehmann A, Kloetzel PM. The FAT10- and ubiquitin-dependent degradation machineries exhibit common and distinct requirements for MHC class I antigen presentation. Cell Mol Life Sci. 2012;69(14):2443–2454. doi: 10.1007/s00018-012-0933-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren J, Wang Y, Gao Y, Mehta SB, Lee CG. FAT10 mediates the effect of TNF-α in inducing chromosomal instability. J Cell Sci. 2011;124(Pt 21):3665–3675. doi: 10.1242/jcs.087403. [DOI] [PubMed] [Google Scholar]
- 8.Lukasiak S, et al. Proinflammatory cytokines cause FAT10 upregulation in cancers of liver and colon. Oncogene. 2008;27(46):6068–6074. doi: 10.1038/onc.2008.201. [DOI] [PubMed] [Google Scholar]
- 9.Gao Y, et al. FAT10, an Ubiquitin-like Protein, Confers Malignant Properties in Non-tumorigenic and Tumorigenic Cells. Carcinogenesis. 2013;35(4):923–34. doi: 10.1093/carcin/bgt407. [DOI] [PubMed] [Google Scholar]
- 10.Qing X, French BA, Oliva J, French SW. Increased expression of FAT10 in colon benign, premalignant and malignant epithelial neoplasms. Exp Mol Pathol. 2011;90(1):51–54. doi: 10.1016/j.yexmp.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan DW, et al. Ubiquitin D is correlated with colon cancer progression and predicts recurrence for stage II-III disease after curative surgery. Br J Cancer. 2010;103(7):961–969. doi: 10.1038/sj.bjc.6605870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren J, et al. FAT10 plays a role in the regulation of chromosomal stability. J Biol Chem. 2006;281(16):11413–11421. doi: 10.1074/jbc.M507218200. [DOI] [PubMed] [Google Scholar]
- 13.Holland AJ, Cleveland DW. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10(7):478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: Aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5(10):773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 15.Gavin JM, et al. Mechanistic studies on activation of ubiquitin and di-ubiquitin-like protein, FAT10, by ubiquitin-like modifier activating enzyme 6, Uba6. J Biol Chem. 2012;287(19):15512–15522. doi: 10.1074/jbc.M111.336198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalveram B, Schmidtke G, Groettrup M. The ubiquitin-like modifier FAT10 interacts with HDAC6 and localizes to aggresomes under proteasome inhibition. J Cell Sci. 2008;121(Pt 24):4079–4088. doi: 10.1242/jcs.035006. [DOI] [PubMed] [Google Scholar]
- 17.Aichem A, et al. The proteomic analysis of endogenous FAT10 substrates identifies p62/SQSTM1 as a substrate of FAT10ylation. J Cell Sci. 2012;125(Pt 19):4576–4585. doi: 10.1242/jcs.107789. [DOI] [PubMed] [Google Scholar]
- 18.Li T, et al. FAT10 modifies p53 and upregulates its transcriptional activity. Arch Biochem Biophys. 2011;509(2):164–169. doi: 10.1016/j.abb.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13(3):289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 20.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 21.Christen M, et al. The GROMOS software for biomolecular simulation: GROMOS05. J Comput Chem. 2005;26(16):1719–1751. doi: 10.1002/jcc.20303. [DOI] [PubMed] [Google Scholar]
- 22.Qin H, et al. Structural characterization of the EphA4-Ephrin-B2 complex reveals new features enabling Eph-ephrin binding promiscuity. J Biol Chem. 2010;285(1):644–654. doi: 10.1074/jbc.M109.064824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komander D, et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10(5):466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin H, Shi J, Noberini R, Pasquale EB, Song J. Crystal structure and NMR binding reveal that two small molecule antagonists target the high affinity ephrin-binding channel of the EphA4 receptor. J Biol Chem. 2008;283(43):29473–29484. doi: 10.1074/jbc.M804114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williamson MP. Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Michel L, et al. Complete loss of the tumor suppressor MAD2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc Natl Acad Sci USA. 2004;101(13):4459–4464. doi: 10.1073/pnas.0306069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tapia C, et al. Two mitosis-specific antibodies, MPM-2 and phospho-histone H3 (Ser28), allow rapid and precise determination of mitotic activity. Am J Surg Pathol. 2006;30(1):83–89. doi: 10.1097/01.pas.0000183572.94140.43. [DOI] [PubMed] [Google Scholar]
- 28.Wagenaar-Miller RA, Gorden L, Matrisian LM. Matrix metalloproteinases in colorectal cancer: Is it worth talking about? Cancer Metastasis Rev. 2004;23(1-2):119–135. doi: 10.1023/a:1025819214508. [DOI] [PubMed] [Google Scholar]
- 29.Liu L, et al. As an independent prognostic factor, FAT10 promotes hepatitis B virus-related hepatocellular carcinoma progression via Akt/GSK3beta pathway. Oncogene. 2014;33(7):909–920. doi: 10.1038/onc.2013.236. [DOI] [PubMed] [Google Scholar]
- 30.Merbl Y, Refour P, Patel H, Springer M, Kirschner MW. Profiling of ubiquitin-like modifications reveals features of mitotic control. Cell. 2013;152(5):1160–1172. doi: 10.1016/j.cell.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu H. Structural activation of Mad2 in the mitotic spindle checkpoint: The two-state Mad2 model versus the Mad2 template model. J Cell Biol. 2006;173(2):153–157. doi: 10.1083/jcb.200601172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michel LS, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409(6818):355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 33.Sotillo R, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11(1):9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, et al. Depression of MAD2 inhibits apoptosis and increases proliferation and multidrug resistance in gastric cancer cells by regulating the activation of phosphorylated survivin. Tumour biol. 2010;31(3):225–232. doi: 10.1007/s13277-010-0036-6. [DOI] [PubMed] [Google Scholar]
- 35.Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell. 2011;19(6):701–714. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prencipe M, et al. Cellular senescence induced by aberrant MAD2 levels impacts on paclitaxel responsiveness in vitro. Br J Cancer. 2009;101(11):1900–1908. doi: 10.1038/sj.bjc.6605419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.