SUMMARY
The retinal anterior homeobox (rax) gene encodes a transcription factor necessary for vertebrate eye development. rax transcription is initiated at the end of gastrulation in Xenopus, and is a key part of the regulatory network specifying anterior neural plate and retina. We describe here a Xenopus tropicalis rax mutant, the first mutant analyzed in detail from a reverse genetic screen. As in other vertebrates, this nonsense mutation results in eyeless animals, and is lethal peri-metamorphosis. Tissue normally fated to form retina in these mutants instead forms tissue with characteristics of diencephalon and telencephalon. This implies that a key role of rax, in addition to defining the eye field, is in preventing alternative forebrain identities. Our data highlight that brain and retina regions are not determined by the mid-gastrula stage but are by the neural plate stage. An RNA-Seq analysis and in situ hybridization assays for early gene expression in the mutant revealed that several key eye field transcription factors (e.g. pax6, lhx2 and six6) are not dependent on rax activity through neurulation. However, these analyses identified other genes either up- or down-regulated in mutant presumptive retinal tissue. Two neural patterning genes of particular interest that appear up-regulated in the rax mutant RNA-seq analysis are hesx1 and fezf2. These genes were not previously known to be regulated by rax. The normal function of rax is to partially repress their expression by an indirect mechanism in the presumptive retina region in wildtype embryos, thus accounting for the apparent up-regulation in the rax mutant. Knock-down experiments using antisense morpholino oligonucleotides directed against hesx1 and fezf2 show that failure to repress these two genes contributes to transformation of presumptive retinal tissue into non-retinal forebrain identities in the rax mutant.
Keywords: Xenopus tropicalis, forebrain patterning, retina formation
INTRODUCTION
A complex network of transcription factors activated in the early neurula contributes to forebrain and early eye patterning (reviewed by Ogino et al., 2012; Papalopulu, 1995; Shimizu and Hibi, 2009; Sinn and Wittbrodt, 2013; Zuber, 2010). The retinal anterior homeobox (rax) gene is one key factor in this network. rax is expressed in the anterior neural plate starting during late gastrula stages (St. 12; stages according to Nieukoop and Faber, 1994) in regions fated to form retina and hypothalamus and later appears in the pineal anlagen (Furukawa et al., 1997; Mathers et al., 1997). Mutations in the DNA-binding homeodomain of the zebrafish rax homolog rx3 result in eyeless fish (Kennedy et al., 2004; Loosli et al., 2003), and the mouse Rax null mutant is also eyeless (Mathers et al., 1997; Voronina et al., 2005). Human RAX mutations result in anophthalmia in compound heterozygotes (Voronina et al., 2004). Morpholino (MO) knockdown experiments targeting rax in Xenopus also resulted in eyeless phenotypes (Andreazzoli et al., 2003).
To utilize the many advantages of Xenopus for studying early embryogenesis together with genetic manipulations (Harland and Grainger, 2011), we developed the N-ethyl-N-nitrosourea (ENU) mutagenesis protocol described here, which entails spermatogonial mutagenesis of Xenopus tropicalis, and the use of F1 mutagenized animals in a Targeting Induced Local Lesions IN Genomes (TILLING) screen to identify mutations in genes of interest. This study describes a new nonsense mutation in the Xenopus tropicalis rax gene, which prematurely terminates protein translation upstream of the rax homeodomain, consistent with this acting as a null mutation.
We performed an RNA-Seq analysis targeting just the affected tissue, the anterior neural plate, in rax mutants at the time of eye specification (stage 15, Saha and Grainger, 1992), which allowed us to greatly reduce background from a whole embryo analysis. This analysis identified differentially expressed genes in the rax mutant, revealing genes of interest not previously known to be regulated by rax.
In rax mutants the tissue that would normally be fated to become retina is transformed into both telencephalic and diencephalic tissue. This implies that a key role of rax is not only to help define the retina in a positive sense, but also to inhibit adjacent factors from specifying this region towards other forebrain fates, and highlights that regional determination of the brain and retina are not fixed at the time of onset of rax expression.
MATERIALS AND METHODS
ENU mutagenesis and TILLING
We have utilized spermatogonial mutagenesis, a widely used approach for vertebrate ENU mutagenesis, e.g. in mouse (Weber et al., 2000). Adult males were injected with ENU (100 mg/kg body weight) at weekly intervals for two or three weeks. Higher doses, or treatments at 100 mg/kg for more than three weeks, resulted in higher lethality and lower doses or single week treatments at 100 mg/kg were not effective. After recovering for several months when males were effectively sterile, F1 populations were generated for evaluation of recessive phenotypes or for TILLING studies.
A small-scale TILLING screen was performed by Sanger sequencing of a limited number of gene targets in an F1 population. The calculated mutation rate in the loci examined was 1 in 200 to 300 kb, varying among offspring of different mutagenized males, but in the range expected for efficient ENU mutagenesis in other vertebrates (Beier, 2000; Kettleborough et al., 2011), and similar to the initial X. tropicalis TILLING screen using direct mutagenesis of mature sperm (Goda et al., 2006). The rax mutation described here was the first nonsense mutation recovered during our screen that affected a key eye regulatory gene.
rax, rax-EnR mRNA and BAC injections
rax or rax-EnR (rax-engrailed) capped mRNA was synthesized from pCS2-Xrx1 or pCS2-Xrx1-EnR (kind gifts from Massimiliano Andreazzoli, Andreazzoli et al., 1999, 2003) using the mMESSAGE mMACHINE® SP6 Kit (Invitrogen). 5 pg (or 5 pg and 10 pg for rax-EnR experiments) mRNA was injected into one dorsal blastomere at the 4-cell stage of X. tropicalis embryos along with 20 ng fluorescein-labeled dextran (FLDx) as tracer, where un-injected sides served as controls. The rax-containing X. tropicalis BAC clone ISB1-349A23 was recombineered and injected into Xenopus embryos as described (Fish et al., 2012).
RNA-Seq Analysis
Anterior neural plates (ANPs) from neural plate stage (St. 15) embryos were dissected from a rax mutant heterozygote cross, collected individually and frozen in multi-well dishes. The remaining embryonic tissue was processed for genotyping as described in Goda et al., (2006), using RaxGenoF and RaxGenoR primers for PCR amplification and subsequent Sanger sequencing: RaxGenoF: 5’-ACCAGGCACCTCTTTTTGTG-3’
RaxGenoR: 5’-CTGACCTCGGGCAAGTTTAC-3’
Homozygous wildtype and mutant ANPs were collected into three pools per genotype. Pooled tissues were lysed in Trizol (Invitrogen) and processed according to manufacturer's instructions. cDNA libraries were prepared using the Illumina mRNA kit, and sequenced using Illumina Solexa sequencing. RNAseq raw data from two experiments is available on the European Nucleotide Archive (ENA), study number ERP002095, (http://www.ebi.ac.uk/ena/data/view/ERP002095). The analysis used existing gene models in Ensembl and those generated by TopHat (Trapnell et al., 2009) and Cufflinks (Trapnell et al., 2010), to run a differential gene expression analysis, using Cuffdiff. Cuffdiff was run with a GTF file, produced from Cuffcompare. RNAseq analysis data is published on Array Express, experiment number E-ERAD-130 (http://www.ebi.ac.uk/arrayexpress/experiments/E-ERAD-130/). A simplified table of statistically significant differentially expressed genes and their calculated read values, as generated by the Cuffdiff analysis, is available in the Supplementary Materials (Table S4).
Genotyping of rax mutant and wildtype embyros
Since we can easily distinguish between rax mutant and wildtype phenotypes after St. 24 due to differential morphologies of the eye region, we do not necessarily perform genotyping on embryos older than St. 24. In this case, the genotype of wildtype embryos is shown as (+/?). All embryos shown at earlier stages were genotyped. Embryos were lysed as described (Fish et al., 2012; Nakayama et al., 2013) followed by genomic PCR of the rax region (PCR primers are listed above). The PCR amplicon was purified and sequenced. In most experiments, phenotypes are indistinguishable between +/+ and +/−, and thus one representative embryo of either genotype has been chosen as a “wild-type” exemplar to image and labeled as (+/?).
Histology
For analysis of the mutant phenotype, embryos were fixed overnight in Bouin's fixative and embedded in JB-4 Plus® resin (Polysciences). 3 μm sections were cut and stained with toluidine blue.
For assessing expression domains of certain transcription factors following in situ hybridization, embryos were embedded in Paraplast® Plus and sectioned at 10 μm. Other embryos were embedded in 2% low melt agarose in 0.1 × MBS and vibratome sectioned at 30 μm.
DiI Labeling
Presumptive retina rudiments were labeled at St. 15 using DiI (Molecular Probes® Invitrogen). A 10mg/ml stock of DiI in DMSO was diluted 1:20 in 250 mM sucrose, loaded into a microinjection needle and applied to the surface of embryos. Injection sites were standardized using an ocular grid, measuring along the line of the inner margin of the anterior neural fold (x-axis) and a line running through the embryonic midline of the embryo (y-axis).
Tissue Transplantations
Small pieces of anterior neural plate containing the left eye rudiment (roughly equivalent to the rax expression domain) were removed at St. 15 from rax +/? and rax −/−embryos and transplanted to the posterior flank of same stage hosts. Hosts were allowed to develop to approximately St. 37 and fixed with MEMFA (100 mM MOPS (pH 7.4), 2 mM EGTA, 1 mM MgSO4, 3.7% (v/v) formaldehyde) for in situ hybridization. Host and donor embryo pairs were raised in parallel so that the genotype of donor tissue could be determined by the phenotype of the eye on the un-operated side of the donor embryo.
Fluorescence microscopy and photomicroscopy
Fluorescent lineage tracer (FLDx) in embryos was observed using a Zeiss Discovery V12 microscope with fluorescent illumination and 470nm GFP filter set. Photos were taken with a Zeiss AxioCam Mrc5 CCD camera assembled on the microscope, using Axiovision 4.8 software.
In situ hybridization
Whole-mount in situ hybridization was carried out essentially according to the procedure in Sive et al.,(2000), though modified when genotyping was required. In these cases the acetic anhydride step and lengthy overnight fixation of post-stained embryos were omitted. Both steps interfere with the ability to accurately sequence DNA. After photographing embryos following this procedure, embryos were genotyped as described above. All antisense probes were labeled with digoxigenin (Roche) and detected by BCIP/NBT (Roche). See Table S1 for details of probe construction.
Morpholino antisense oligonucleotides and validation mRNA injections
See Tables S2 and S3 for injected-morpholino sequences and mRNA constructs used to assay morpholino efficiency.
Gfp Immunostaining
Embryos were fixed in MEMFA for 1 hour, then washed and stored in phosphate buffered saline (PBS) at 4°C. Vibratome sections were blocked in PBS with 0.1% Tween 20, 2 mg/ml BSA and 10% normal goat serum (PBST/NGS) and incubated overnight with primary anti-Gfp antibody (Invitrogen A11122- rabbit, 1:500). Sections were washed and blocked again, and then incubated overnight with secondary antibody (GARGG-AlexaFluor488, 1:500). Embryos were washed in PBST and then mounted on subbed slides in 70% glycerol/PBS.
RESULTS
A Xenopus tropicalis rax mutant results in an eyeless phenotype
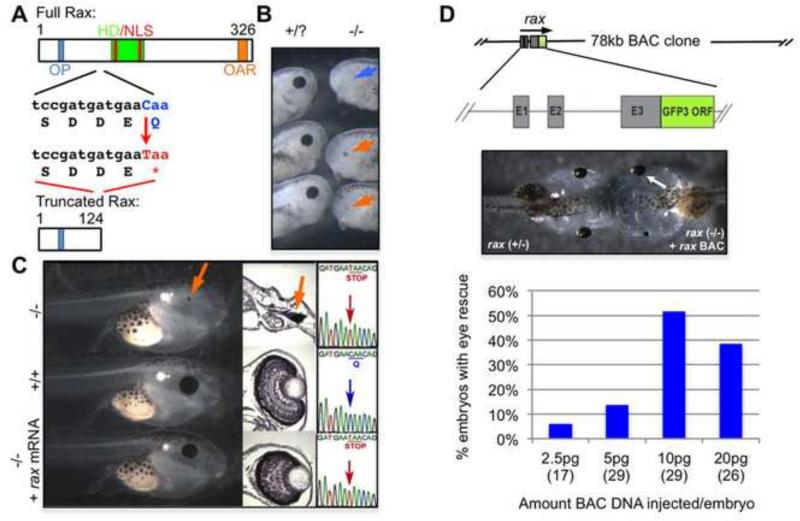
A reverse genetic screen (as described in the Materials and Methods) performed in Xenopus tropicalis identified a nonsense mutation in the rax gene. The X. tropicalis Rax protein consists of 326 amino acids containing an octapeptide (OP) domain, homeodomain (which also contains 2 nuclear localization signals (NLSs), Voronina et al., 2004) and OAR (otp, al, and rax) domain (Fig. 1A). The OP domain may be involved in repression while the C-terminus OAR domain is thought to function as a transactivation domain (Furukawa et al., 1997; Pan et al., 2006). The nonsense mutation identified in our spermatogonial mutagenesis/TILLING screen occurs in the second of three exons, changing a glutamine codon into a stop codon, halting the translation of Rax protein after the OP domain and upstream of the homeodomain and OAR domain (Fig. 1A). Notably, a similar truncation previously described in human patients (Q147X) results in a protein product that cannot translocate to the nucleus and has no DNA-binding activity, as it does not contain an NLS and eliminates helices 2 and 3 of the homeodomain (Voronina et al., 2004). Since our mutant locus could potentially encode only a shorter protein (Q125X) lacking the entire homeodomain (including two NLSs), the truncated protein, if it is present, we expect has no ability to interact with DNA or to localize properly to the nucleus and thus most likely behaves as a null. Animals homozygous for the mutant allele are eyeless (Fig. 1B, right and 1C, top). A small amount of retinal pigmented epithelium (RPE) is sometimes observed in mutant embryos (Fig. 1B and 1C, orange arrows). Embryos illustrating the range of observed RPE were chosen from one clutch in Fig. 1B; it should be noted that this range does not represent the observed ratios of RPE phenotypes, which is variable depending on the parental mating.
Figure 1. A nonsense mutation in the retinal anterior homeobox gene (rax) results in an eyeless phenotype.
(A) The Rax protein structure includes an octapeptide domain (OA, blue), homeodomain (HD, green) and OAR domain (orange). Nuclear localization signals (NLSs, red) are found in the homeodomain. A nonsense mutation results in a truncation prior to the homeodomain and NLSs. (B-C) Tadpoles homozygous for the rax mutant allele fail to form eyes (B, right; C, top). A small amount of retinal pigmented epithelium (RPE) is sometimes seen in mutant animals (B, C, orange arrows) whereas some mutant embryos have no sign of RPE (B, blue arrow); this variation is observed within the same clutch of embryos. The eyeless phenotype can be rescued with the injection of 5 pg of rax mRNA (C, bottom, 10 out of 24 injected). Sections through the mutant, wildtype, and mRNA-rescued mutant tadpole eye regions are shown in the middle panel, illustrating the loss of eye tissue, except small residual RPE in some mutant embryos, and rescue of the retinal and lens structures in mRNA-injected mutant. Far right of (C), insets of Sanger-sequencing showing point mutation in exon 2, which results in a premature stop codon. (D) A Xenopus tropicalis BAC clone containing a rax-gfp fusion gene can rescue eye formation when transiently expressed in mutant embryos. Note that BAC injection tends to be effective dominantly (or only) in one side as described before (Fish et al. 2012). E1-E3 indicate rax exons; the gfp3 open reading frame (ORF) has been fused to the 3’ end of the final rax exon (E3) to produce a rax-gfp3 fusion gene. Quality of rescue is highly dose-sensitive, with the best rescue observed with 5 pg mRNA (C, data not shown) and 10 pg of BAC DNA injected (D).
The eyeless phenotype can be rescued in approximately 40% of embryos by injecting homozygous mutants with low levels (5 pg) of mRNA encoding the wildtype Rax protein (Fig 1C, bottom, while another 40% showed partial rescue (i.e. microphthalmic eyes, not shown) as well as by the injection of a Bacterial Artificial Chromosome (BAC) clone that expresses a Rax-Gfp3 fusion protein (Fish et al., 2012, Fig. 1D). Both rescue experiments support the conclusion that the eyeless phenotype is indeed caused by the rax nonsense mutation and not by other background mutations induced by ENU mutagenesis.
The expression of key eye field transcription factors is unaffected in neurula stage rax mutant embryos
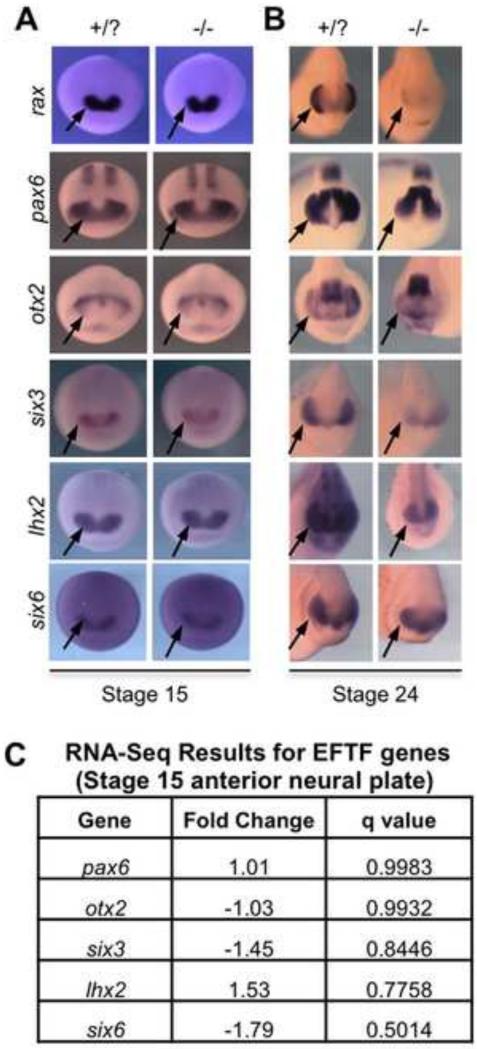
Originally, experiments performed in a mouse Rax mutant supported a regulatory network model placing key eye field transcription factors (EFTFs) (Pax6, Six3, Lhx2, and Six6) downstream of Rax because these genes are down-regulated in the mutant at early stages (Zhang et al., 2000; and reviewed in Bailey et al., 2004). We examined the activation of these genes by in situ hybridization in neural plate stage Xenopus embryos and did not detect any differences in expression of pax6, otx2, six3, lhx2 or six6 (Fig. 2A). This is consistent with previous examinations of the early expression patterns of pax6, otx2 and six3 in zebrafish rx3 mutants (Loosli et al., 2003, Kennedy et al., 2004). By early tailbud stages, however, all of these genes are down-regulated in rax mutants in the presumptive retinal region (Fig. 2B). In the case of genes that are normally expressed both in the retina and other brain regions, such as pax6, otx2, and lhx2, only the retinal expression is affected in the rax mutant (Fig. 2B).
Figure 2. Key eye field transcription factor (EFTF) expression is unaffected in neural plate stage embryos.
(A) The expression patterns and levels of rax, pax6, otx2, six3, lhx2, and six6 assayed via in situ hybridization in neural plate stage (St. 15) embryos with at least one wildtype allele versus homozygous mutant embryos reveal that key EFTF expression levels are unaffected in the rax mutant. Presumptive retinal regions indicated (arrows). (B) By early tailbud stage (St. 24), down-regulation of EFTFs can be detected in regions where the retina normally forms (arrows). (C) RNA-Seq analysis comparing wildtype and mutant St. 15 anterior neural plates confirms that EFTFs are not significantly affected in early stage rax mutant embryos. Fold change calculated using normalized read counts averaged from triplicate pooled samples. Q-values calculated using analysis utility Cuffdiff, part of the Cufflinks software package (Trapnell et al., 2010), with a False Discovery Rate (FDR) of 0.05. High q-values indicate that listed genes are not significantly affected in neural plate stage rax mutant embryos.
An RNA-Seq analysis was performed to compare the gene expression profiles of St.15 anterior neural plates of rax mutant and wildtype embryos. Although rax expression is initially detected in late gastrula Xenopus embryos, the neural plate (St.15) was chosen to capture genes that are directly affected by loss of functional Rax protein or indirectly regulated but part of the immediate network of genes associated with Rax activity. At this stage the retina is also determined, though it is not determined when rax is first activated (Saha and Grainger, 1992). In addition, the neural plate provides landmarks that make precise dissections possible and detailed fate maps exist for this stage (Eagleson and Harris, 1990; Eagleson et al., 1995). Our RNA-Seq analysis (Fig. 2C) supports the in situ hybridization results showing no significant expression changes in pax6, otx2, six6, or lhx2. The gene six6 may be slightly, though not significantly, reduced, though no differences between wildtype and mutant are seen by in situ hybridization (e.g. Fig. 2A).
Morphological changes in the rax mutant can be detected as early as St. 21, prior to the down-regulation of key EFTFs
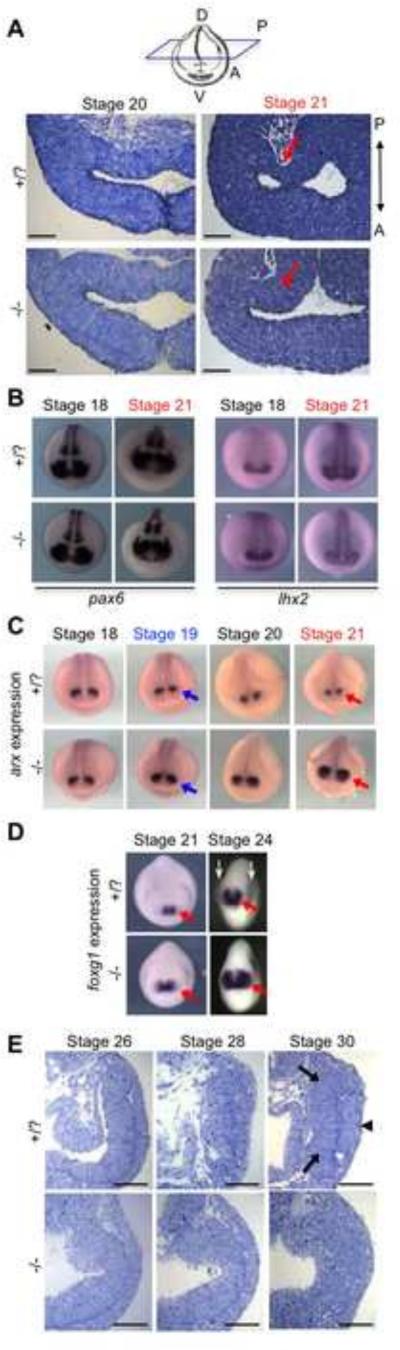
Histological sections were examined to look for the earliest morphological changes in mutant embryos. Although embryos of any genotype look similar at St. 20, by St. 21 a thinning of the posterior optic vesicle can be observed in wildtype embryos that is not observed in mutant embryos (Fig. 3A, red arrows). Interestingly, this morphological change is detectable before the down-regulation of key EFTFs, e.g. pax6 and lhx2, begins to occur (Fig. 3B). One transcript of interest found to be significantly increased in mutant embryos before the morphological change at st. 21 is the aristaless-related homeobox (arx) gene. This transcription factor is expressed early in the developing diencephalon (posterior to the retina, El-Hodiri et al., 2003; Miura et al., 1997) and is important for forebrain patterning in Xenopus, mouse and zebrafish (Filippi et al., 2012; Kitamura et al., 2002; Rodríguez-Seguel et al., 2009). The expression of the transcriptional repressor arx in rax mutant embryos begins to expand from the initial diencephalic territory into the presumptive retina at St. 19 (Fig. 3C, blue arrows). By the stage where morphological changes are first detected (St. 21, Fig. 3C, red arrows), arx expression has further expanded from its posterior domain into the region where the optic vesicle normally forms in wildtype embryos.
Figure 3. Morphological changes in rax mutant embryos can be detected by St. 21, prior to differential expression of key EFTFs, but following up-regulation of arx. Up-regulation of telencephalic marker foxg1 is also observed by St. 21.
(A) Histological sections through St. 20 and St. 21 embryos reveal no detectable change in optic vesicle morphology at St. 20, but a lack of thinning of the posterior optic vesicle wall in mutant embryos at St. 21 (red arrows). The plane of sectioning is indicated above, and is the same plane for all stages shown throughout (serial sections were carefully examined to correctly match the plane of sectioning). The anterior and posterior ends of sections are marked A and P, respectively. (B) Examination of key EFTFs (pax6 and lhx2 shown) via in situ hybridization at St. 18 and St. 21 reveals no detectable change in expression. (C) By St. 19, anterior expansion of the arx expression domain is observed in mutant embryos (blue arrows). This expansion becomes more pronounced at St. 21(red arrows). (D) Examination of telencephalic marker foxg1 at stages 21 and 24 reveals lateral expansion in rax mutant embryos (red arrows). At stage 24, lateral bulging of the optic vesicles can be observed in whole embryos from an anterior view (white arrows); these bulges are not observed in rax mutant embryos, and foxg1 expression is expanded into a region of the territory the optic vesicle would occupy in wildtype embryos. (E) Histological sections at St. 26, St. 28 and St. 30 show the increasing divergence of wildtype and mutant morphologies. By St. 30, the optic cup and lens tissue are forming in wildtype embryos (black arrows and arrowhead, respectively), but typically fail to form in rax mutant embryos. In panels A and E, a minimum of three embryos for each stage and genotype were sectioned and consistently displayed the shown morphology. Scale bars in panels A and E measure 75 micrometers.
We also examined the expression of the primarily telencephalic gene foxg1, both because expansion of the telencephalon is observed in zebrafish rx3 mutants (Stigloher et al., 2006), and since it is a target of hesx1 (previously xanf), which is also up-regulated in the rax mutant (see below), though hesx1 is no longer active at stages we examine here. The transcription factor foxg1 is an important integrator of signaling pathways patterning the forebrain and regulating telencephalon formation (Andoniadou et al., 2011; Danesin et al., 2009). We observe expansion of foxg1 expression in mutant embryos as early as St. 21 (Fig. 3D, red arrows), with significant lateral expansion into the presumptive optic vesicle territory (white arrows) observed by St. 24. Divergence in eye morphology continues at later stages, and by St. 30 a well-formed optic cup is discernable in wildtype embryos (Fig. 3E, black arrows) and the lens is beginning to form (Fig. 3E, black arrowhead). These structures typically fail to form in rax mutants (Fig. 3E).
Retinal tissue in the rax mutant is transformed into tissue with diencephalic and telencephalic properties
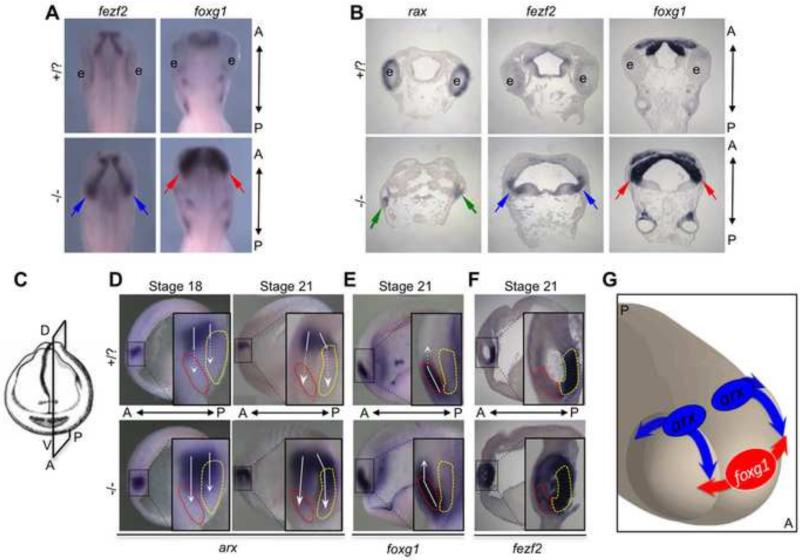
Since the region fated to become retina appears to persist in some form, as illustrated in Fig. 3 (and substantiated below), we wanted to establish what the fate of the presumptive retina might be in rax mutant embryos, i.e., is it transformed, for example, into adjacent brain tissue types? The data mentioned above showing that the diencephalic gene arx is expanded into the retina region supports this proposal. Toward that end we examined expression of the gene fezf2 in rax mutant embryos because it has significant diencephalic expression posterior to the retina, it is up-regulated in the mutant (as discussed further below), and it regulates arx activity (Rodriguez-Seguel et al., 2009). fezf2 is a zinc-finger transcription factor that plays a key role in diencephalic and telencephalic fates (Jeong et al., 2007). In mouse knockout studies, it functions similarly and partially redundantly with closely-related fezf1 to localize the zona limitans intrathalamica (ZLI), the signaling center that marks the boundary between the prethalamus and thalamus subdivisions of the diencephalon (Hirata et al., 2006; Lavado et al., 2008). In Xenopus, fezf2, together with fezf1, suppresses formation of the caudal diencephalon as part of its role in ZLI localization (Shimizu and Hibi, 2009). Examining the fezf2 expression pattern in rax mutant embryos at tadpole stages (St. 32 and 35) in both whole-mount (Fig. 4A) and sectioned embryos (Fig, 4B), we observed that it was greatly expanded in mutant embryos, extending laterally into the region where the retina would normally form (Fig. 4A-B, blue arrows).
Figure 4. Diencephalic and telencephalic tissue is expanded in the rax mutant.
(A) Dorsal views of St. 32 embryo heads assayed for diencephalic marker fezf2 (left) or telencephalic marker foxg1 (right) via in situ hybridization. Both fezf2 (blue arrows) and foxg1 (red arrows) expression is expanded into regions where the eyes would normally form in wildtype. (B) Frontal sections through St. 35wildtype and mutant embryos detecting rax, fezf2, foxg1 or foxd1 expression by in situ hybridization. In rax mutant embryos (bottom row), green arrows indicate small remaining rudiment still expressing rax. In the absence of functional rax, expression of fezf2 (blue arrows) and foxg1 (red arrows) is expanded into the tissue where the retina would normally form. (C) Visualization of the parasagittal plane of sectioning performed on embryos in D-F. (D) Lateral views of parasagittally-bisected embryos at indicated stages stained via in situ hybridization for early diencephalic marker arx. In panels D-F, red dashed outlines indicate developing telencephalic tissue, and yellow dashed outlines indicate developing posterior secondary prosencephalon. White arrows cover the expression range of each marker, and dashed white arrows indicate into which region expression expands in rax mutant embryos (bottom row). arx expression is expanded ventrally in mutant embryos, and expands both anteriorly into the telencephalon and posteriorly into the secondary prosencephalon, with significant expansion observed by St. 21. (E) Parasagittally-bisected embryos at St. 21 show ventral expansion of telencephalic marker foxg1, which expands beyond the wildtype telencephalic domain in rax mutants. (F) fezf2 expression in the wildtype marks the posterior secondary prosencephalon. In rax mutant embryos this expression expands into the telencephalon and optic vesicle. (Note: for clarification of dense staining, this embryo was thinly parasagittally-sectioned via vibratome, instead of simply bisected as in panels D-E. The plane of sectioning is the same in panels D-F). (G) A cartoon illustrating the 3-dimensional movements of arx and foxg1 expansion into the presumptive optic vesicle regions of rax mutant embryos. Observed expansion of arx is moving ventrally, both anteriorly and posteriorly. Expansion of foxg1 is primarily lateral. Movement of these marker boundaries (and others) likely results in the expansion of diencephalic and telencephalic markers observed in the later mutant embryos shown in A-B.
In both whole-mount and sectioned embryos we also observed posterior and lateral expansion of the telencephalon (defined by foxg1 expression) into the region where the retina would normally form (Fig. 4A-B, red arrows). In rax mutant tadpoles we also observed the persistence of low levels of rax expression in a small rudiment that remains (Fig. 4B and 5A, green arrows), which may be what remains of the optic stalk.
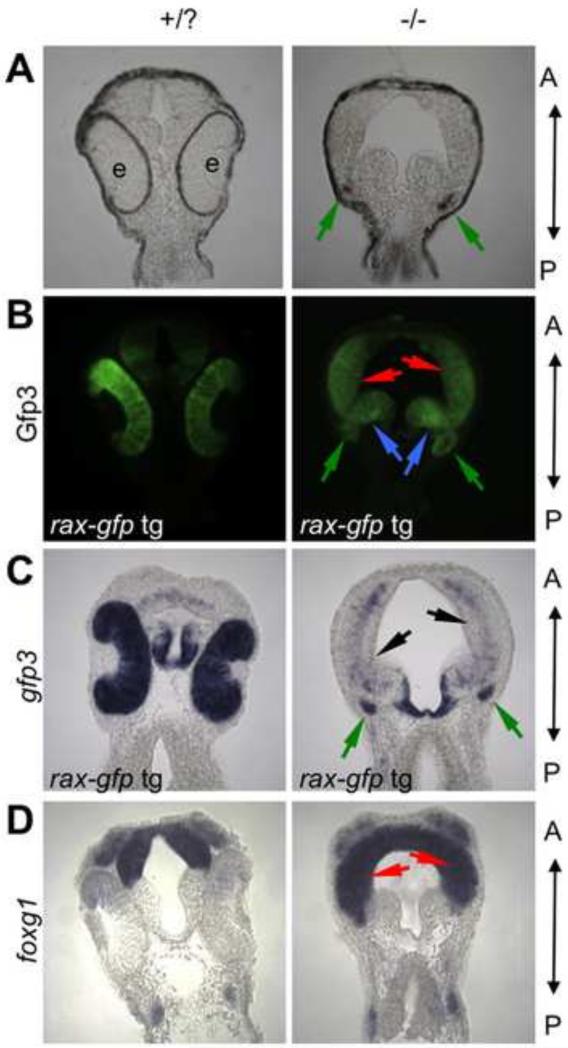
Figure 5. Presumptive retinal tissue is transformed into diencephalic and telencephalic tissue in the rax mutant.
(A-C) Genetic labeling of retinal fated tissue in wildtype and mutant embryos. A transgenic line expressing gfp3 under control of the rax-promoter was crossed into the rax mutant background. (A) Brightfield image of frontal sections from stage 32 embryos. (B) Gfp3 protein expression detected via immunostaining. (C) Transgene (gfp3) mRNA expression visualized via in situ hybridization. (D) Telencephalic marker foxg1 mRNA expression visualized via in situ hybridization. In all panels, A marks the anterior and P the posterior. Red arrows mark regions of telencephalic expansion, and blue arrows mark diencephalic expansion into eye region. Green arrows mark the small rudiment that is pigmented (A) in this specific section and continues to express Gfp3 (B-C). Although Gfp3 protein can still be readily discerned, the mRNA is mostly absent in the expanded telencephalic region (black arrows). Five embryos from each phenotype (wild-type or mutant eye morphology) and treatment were scored for these analyses; expression patterns were highly stereotyped within each phenotype. A minimum of two embryos for each phenotype and treatment were then chosen for serial sectioning and imaging.
Embryos were either cut or sectioned in a parasagittal plane in the brain rather than in the more lateral optic vesicle in order to better visualize the early changes in the expression domains of these key genes in mutant embryos (Fig. 4C-F). We observed that arx expression expands ventrally toward and into the presumptive telencephalon (red outlines), which, more laterally, would be the anterior face of the optic vesicle. In addition, arx is expanded posteriorly into part of what is the secondary prosencephalon (yellow outlines) in mutant embryos (Fig. 4D), a region which is contiguous to the posterior face of the optic vesicle. Telencephalic marker foxg1 expands less significantly dorsally (Fig. 4E), but as discussed above (Fig. 3D) expands laterally into the optic vesicle instead. In this plane of section the predominantly diencephalic and posterior secondary prosencephalic marker fezf2 has expanded expression into the telencephalic territory (Fig. 4F) and, as discussed below, is in fact expanded throughout the optic vesicle. A three-dimensional model summarizes the proposed directions of expansion of arx and foxg1 in the rax mutant. The expression of arx expands ventrally, both anteriorly and posteriorly in the presumptive optic vesicle in the mutant, while the telencephalic gene foxg1 has a primarily lateral expansion into the anterior face of the optic vesicle (Fig. 4G).
To distinguish whether these forebrain tissues expand via proliferation into the space occupied by the eye field, or if the eye field tissue is transformed into these alternative fates, we utilized a transgenic line to label eye field cells. This line, which expresses gfp3 under the control of the rax upstream promoter (rax pro-gfp3) in the same domain as endogenous rax (Hirsch et al., 2002), was crossed to our rax mutant line to create rax mutant heterozygotes with the rax pro-gfp3 transgene. When intercrossed, animals from this line produce rax homozygous mutant embryos in which both endogenous rax and rax pro-gfp3 transcription is eventually down-regulated and the eyes fail to form, whereas Gfp3 protein accumulates, remaining for days in cells where rax was initially transcribed because of its longevity compared to mRNA (Verkhusha et al., 2003). St. 32 embryos from such a cross were collected, sectioned and immunostained to detect Gfp3 protein, which labels tissues originally fated to contribute to the retina (Fig. 5B). In wildtype embryos, most of the labeling is observed in the retina, and the gfp3 RNA expression also is high in the retina (Fig 5C, left panel). By contrast, very little gfp3 mRNA is seen in rax mutant embryos (Fig. 5C, black arrows). However, Gfp3protein is still detected by immunochemistry (Fig. 5B). By comparing the mutant Gfp3- labeled brain regions with mutant embryos showing the expansion of telencephalic marker foxg1, we can observe the overlap in expression domains, supporting the conclusion that at least some of the presumptive retina has been transformed into telencephalic tissue in the mutant (Fig. 5B and D). We also observe some Gfp3-labeled regions that are not coincident with foxg1 expression (Fig. 5B, blue arrows) but include the expanded fezf2 expression zone (Fig. 4B) indicating an enlarged region of diencephalic character within the original eye field.
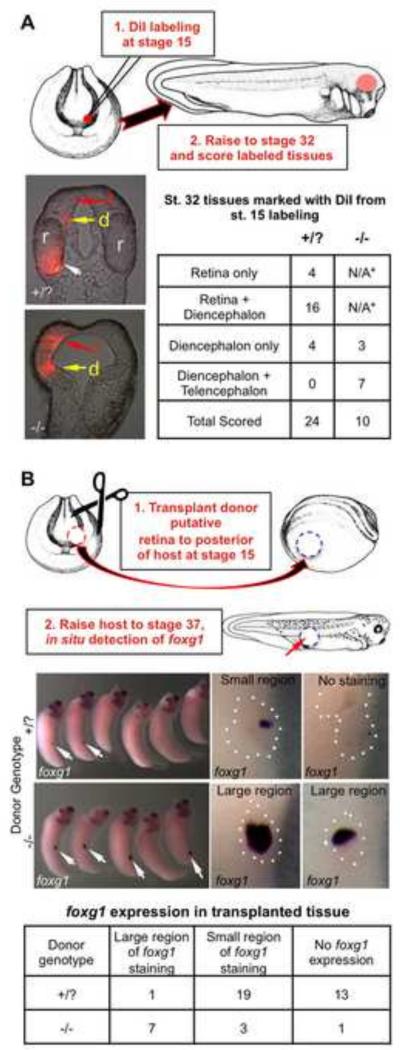
In another experiment testing this transformation hypothesis, we labeled presumptive retinal tissue with DiI marker at St. 15, and observed where the marked cells were found in St. 32 embryos (Fig. 6A). In wildtype embryos labeled cells were usually seen in retinal and small patches of diencephalic tissue, while in mutant embryos we found labeled cells in tissues that overlap with the expanded diencephalic and telencephalic marker-expressing tissues (i.e. fezf2 and foxg1, Fig. 4B).
Figure 6. Presumptive retinal tissue is transformed into both diencephalic and telencephalic fates in rax mutant embryos.
(A) Retinal rudiment was labeled in St. 15 embryos using DiI marking. Embryos were then raised to St. 32 and scored for the fate of labeled tissues. Frontal sections of St. 32 embryos show DiI marking the retina (r, white arrow) and small amount of diencephalon (d, yellow arrow) in wildtype embryos, but not the telencephalon (t, red arrow). In mutant embryos, labeled tissues are found extensively in both the diencephalon and expanded telencephalon. Results shown in the table. *Since eyes fail to form in rax mutants, DiI labeling in the retina is not applicable (N/A). (B) Presumptive retinal tissue from St. 15 rax mutant or wildtype donor embryos was transplanted to the posterior of host embryos, which were raised to St. 37 and fixed. Donor embryos were allowed to develop in order to determine donor phenotype. In situ hybridization to detect telencephalic marker foxg1 was performed on hosts with donor tissue to determine the extent of telencephalic character in donor transplants. Left panels show lateral views of whole embryos, white arrows indicate regions positive for foxg1 expression; right panels show higher magnification view of transplanted region (outlined with white dots). Representative examples of each category (large or small region of transplant that is positive for foxg1expression domain relative to entire transplant size) are shown in right panels. Results from scoring are tallied in the bottom table.
Finally we tested the extent to which this fate switch was autonomously determined by St. 15 through a tissue transplantation assay. Putative retinal tissue from St. 15 homozygous rax mutant or wildtype embryos was transplanted to the posterior of St. 15 host embryos (Fig. 6B). These embryos were raised to St. 37, fixed and the telencephalic marker foxg1 was assayed by in situ hybridization. Consistently, in wildtype transplants only small regions or no regions with telencephalic properties were detected (Fig. 6B) based on the low levels of foxg1 expression. In rax mutant transplants, however, we saw large domains of foxg1 expression in transplanted tissue, indicating that this tissue had at least in part already been transformed to an autonomously determined telencephalic character by St. 15.
Examination of genes differentially expressed in the rax mutant at St. 15 reveals the up-regulation of forebrain patterning genes and down-regulation of key eye genes
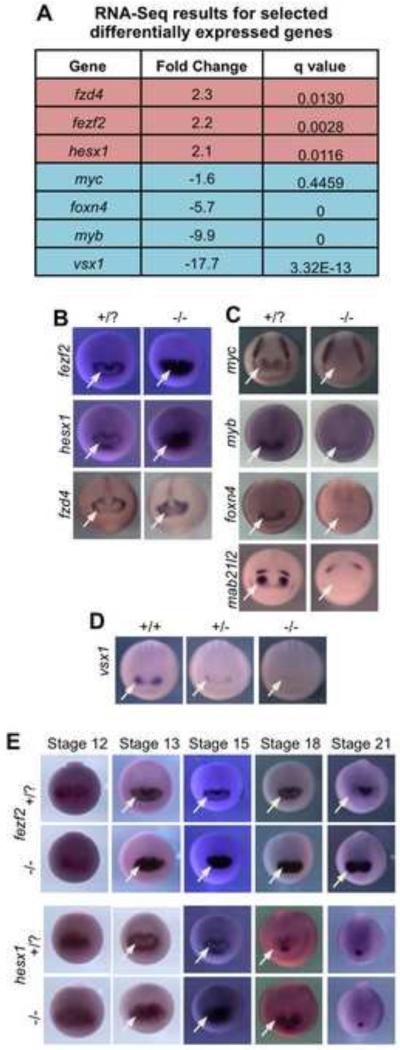
Our transplant experiment from neural plate stage embryos (St. 15) indicated that a fate transformation to non-retinal forebrain tissue has occurred by this stage, the same stage as the RNA-Seq analysis that was performed here. To identify which genes may be causing transformation of retina to brain in the mutant, genes identified as highly differentially expressed in the RNA-Seq analysis were examined by in situ hybridization (Fig. 7A-D; see Table S4 for all differentially expressed genes). Three transcripts identified as significantly increased in rax mutants were fezf2, hesx1, and fzd4. The wild-type expression pattern of all three genes is similar at St. 15, where transcripts are absent from the central eye field and detectable only in the periphery; the pattern forms a central domain lacking expression within the eye field (hereafter, we refer to this as an expression “hole”), which is not observed in rax mutants (Fig. 7B, arrows). Down-regulated genes were also examined, and in the cases of myc, myb and foxn4, among others, expression in the presumptive retina region is lost in rax mutant embryos (Fig. 7C, arrows). Although the mab21l2 gene was not identified in the RNA-Seq data, we assayed its expression since it has been shown to be down-regulated in zebrafish rx3 mutant lines and necessary for eye development (Kennedy et al., 2004; Yamada et al., 2004). Indeed, mab21l2 expression is lost in the retina region of rax mutants (Fig. 7C, arrows). Finally, although we do not observe any heterozygote phenotype morphologically, or in the expression of most genes assayed via in situ hybridization, in the case of vsx1 we do observe reduced expression in heterozygous embryos and a loss of expression in homozygous mutants (Fig. 7D, arrows).
Figure 7. Genes with altered expression in rax mutants include those necessary for retina formation and forebrain patterning.
(A) Table listing key up-regulated (red) and down-regulated (blue) genes from RNA-Seq analysis. (All significantly affected genes identified in this analysis can be found in Table S4) (B) In situ hybridization for up-regulated genes in St. 15 embryos. White arrows indicate regions where up-regulation is observed in mutants in the presumptive retinal field. (C) In situ hybridization for down-regulated genes in St.15 embryos. White arrows indicate regions in the eye field where expression is not detected in the mutant. (D) vsx1 expression (arrows) at neural plate stage is reduced in the heterozygote compared to wildtype and not detected in rax homozygous mutant embryos. (E) A time course of fezf2 and hesx1 expression from late gastrula (St. 12) through neural tube stage (St. 21) in rax mutant and wildtype embryos. White arrows indicate regions where repression is seen in wildtype embryos and not observed in rax mutants.
The genes fezf2 and hesx1 were chosen for further analysis because of their known roles in forebrain patterning (Ermakova et al., 1999; Shimizu and Hibi, 2009; Wang et al., 2011). A time-course of expression patterns for both genes reveals that they are initially activated more broadly in the region where the anterior neural plate will form at St.12, and at this stage their expression patterns look the same in wildtype and rax mutant embryos (Fig. 7E). By St, 13, however, the apparent repression of both transcripts in the central eye field can be detected resulting in the distinctive expression pattern ring; this repression does not occur in mutant embryos and therefore the expression “hole” is not observed (Fig. 7E). In the case of fezf2 expression, the expanded expression domain in the mutant is observed through late neurula stages (St. 21) and beyond, and is strongly expressed in the putative retina region in rax mutants (Fig. 7E, Fig 4A, B). Increased expression of hesx1 persists through St. 18, although by St. 21 the expression domain in wildtype and mutant embryos has receded to just the presumptive anterior pituitary gland (Fig. 7E). It is important to highlight then that the higher expression levels of fezf2 and hesx1 in the rax mutant, that were initially detected by both RNA-Seq of St. 15 ANPs, are a consequence of the failure in the mutant of earlier repressive interactions. The expression patterns of these two genes imply a localized transcriptional repression in the early central eye field that must occur after an initial, broad activation. It does not appear that Rax is directly acting as this repressor, however, based on two observations: 1) the expression domain of rax is larger than the repression region; and 2) the results of experiments where an obligate repressor form of Rax was expressed in Xenopus embryos (Fig. S1). This fused repressor, RaxEnR (containing the Rax DNA-binding domain fused to the Drosophila Engrailed repressor domain), was shown to function as a dominant repressor in Xenopus (Andreazzoli et al., 1999). When overexpressed in wildtype embryos, RaxEnR increased the expression of fezf2 and hesx1, mimicking the rax mutant phenotype. RaxEnR mimics loss of function of Rax, and thus, wildtype Rax must function as an activator, not a repressor, in this context.
Knockdown of forebrain patterning genes fezf2 and hesx1 can reduce the expansion of telencephalic marker foxg1 and diencephalic marker arx in rax mutant embryos
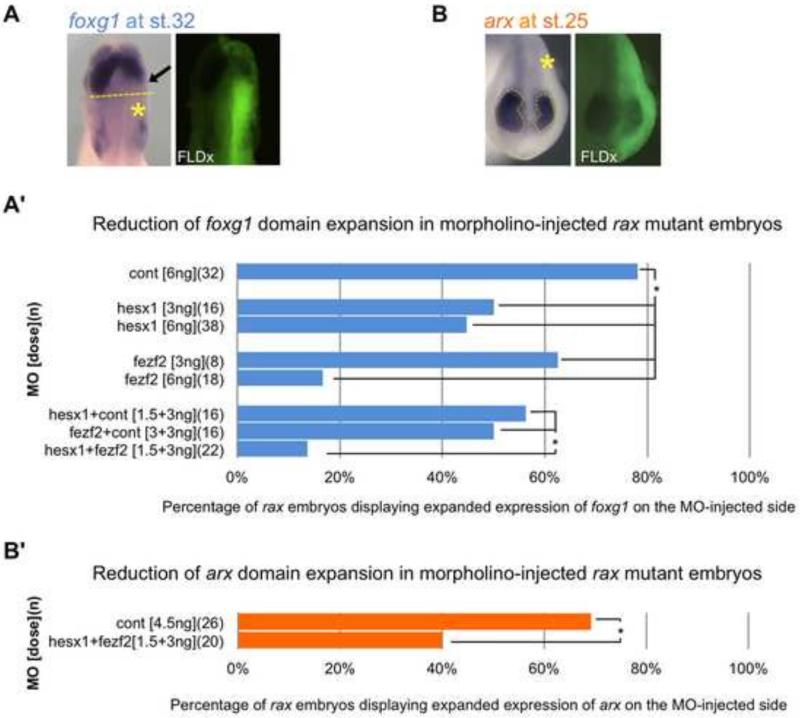
To examine the effects of the lack of regional repression of fezf2 and hesx1 in rax mutants on telencephalic and diencephalic expansion, we performed translation-blocking antisense MO microinjections to discern effects on foxg1 and arx domain expansion. MOs were first tested for their efficiency (Fig. S2) and both individually and in combination were injected into one dorsal blastomere of 4-cell stage embryos to knockdown the expression of Fezf2 and/or Hesx1 in one lateral half of the embryo. To assay for foxg1 expansion, in situ hybridization was performed to assess the effect of MO injection on expanded foxg1 expression on the injected side, using the un-injected side as control for identifying mutant embryos (Fig. 8A). Under our experimental conditions, a control MO injection had little effect and close to 80% of injected mutant embryos showed the typical expansion of foxg1 expression, whereas moderate doses of either fezf2 or hesx1-targeted MOs reduced the expanded foxg1-expressing region significantly (Fig. 8A’). Furthermore, less than 20% of mutant embryos showed the typical expansion of the foxg1 domain when injected with low doses of both MOs together, whereas comparable doses of each individual MO injection still showed typical expanded foxg1 expression in up to 60% of mutant embryos.
Figure 8. Knockdown of fezf2 and/or hesx1 function reduces expanded foxg1- and arx- expressing region(s) in rax mutant embryos.
(A) Representative example of the dorsal view of St. 32 rax mutant embryos injected with fezf2 and/or hesx1 MO(s), and assayed for foxg1 expression by in situ hybridization (left panel). Note that reduced signal is seen in the ectopically expanded foxg1-expressing region (arrow, left panel) on the injected side of mutant embryos (yellow asterisk, left panel), as visualized by FLDx tracer (right panel). (A’) Percentage of embryos with mutant phenotype (expanded foxg1 expression) on injected side after receiving MO(s) injection. Embryos were injected into one dorsal blastomere at the 4-cell stage with either control, fezf2 or hesx1 morpholinos, or a combination of two morpholinos, as indicated in the y-axis. (B) Representative example of the frontal view of a St. 25 rax mutant embryo injected with combined fezf2 and hesx1 MOs, and assayed for arx expression by in situ hybridization (left panel). Dashed yellow outlines mark the region of arx expression, which is reduced on the injected side (marked with an asterisk, and visualized by FLDx tracer in right panel). (B’) Percentage of embryos with mutant phenotype (expanded arx expression) on injected side after receiving MO(s) injection. Embryos were injected into one dorsal blastomere at the 4-cell stage with either control or combined fezf2 or hesx1 morpholinos. (A’-B’) Injected doses of morpholinos is indicated in brackets, and numbers of embryos scored in parentheses. Asterisks indicate p-values of <0.05 by chi-squared test.
To examine the effects of increased fezf2 and hesx1 expression on diencephalic expansion, we performed a similar experiment, knocking down expression of fezf2 and hesx1, and assaying instead for diencephalic marker arx expansion in rax mutant embryos (Fig. 8B). We similarly observed a reduction of the severity of the mutant phenotype (expanded arx expression) in a majority (60%) of MO-injected embryos (compared to 30% of control MO-injected, Fig. 8B’).
These results suggest that the telencephalic and diencephalic expansions observed in rax mutant animals are likely mediated by many of the same genes, and that the inability to repress fezf2 and hesx1 in the central eye field, (most likely resulting from failure to express some repressive factor(s) downstream of rax), is a key mediator of the expansion of forebrain identities into the region where the retina would normally form.
DISCUSSION
Using the Xenopus tropicalis rax mutant line, we have gained significant insights regarding the role of rax in the gene regulatory network forming the retina. In addition, we have further elucidated the role of rax in preventing encroachment of non-retinal forebrain fates into the eye field, utilizing the synergy of working with a mutant line together with the many other experimental advantages of the Xenopus system for studying development.
EFTF genes are activated normally in the absence of functional Rax
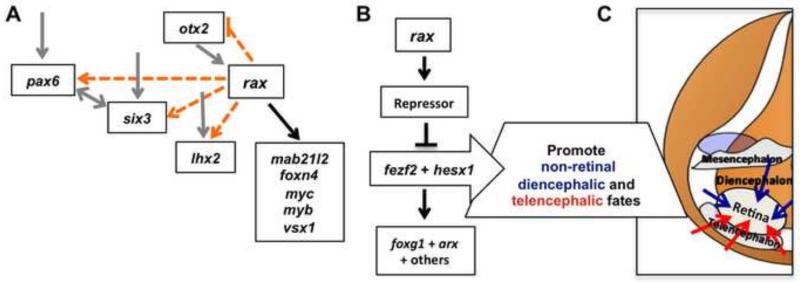
RNA-Seq and in situ hybridization analyses support the conclusion that rax is not necessary to activate key eye field factors pax6, lhx2, or six3 (Fig. 9A). In fact, these genes maintain comparable levels of expression to wildtype embryos in the rax mutant through St. 21, after morphological divergence in optic vesicle formation is first observed along with the increased expression domain of forebrain patterning gene arx, strengthening the argument that there are independent pathways controlling retina formation and highlighting the key role of rax in defining the eye field. Reduced expression of these EFTF genes can be observed by St. 24, and could be due to the reassignment of the putative retinal tissue to different identities that either fail to maintain EFTF expression or actively repress these factors. It is important to note that even with the normal expression of pax6, lhx2, otx2, six6 and six3, morphogenesis of the optic rudiment is halted prior to full optic vesicle formation in the absence of rax, suggesting a master role of rax in retina formation.
Figure 9. Models of rax function.
(A) Analysis of the rax mutant indicates that rax function is not necessary for activation of key EFTFs pax6, otx2, lhx2, or six3. rax expression is necessary for the correct expression of other key eye genes mab21l2, foxn4, myc, myb, vsx1, in addition to other factors. Dotted orange lines indicate putative, early interactions within the eye field gene regulatory network not supported by these analyses, although later, indirect effects of rax inactivation does affect their expression. Solid grey lines indicate previously described interactions not addressed in this work, but consistent with our data, and solid black lines indicate interactions supported by these analyses. (B) A key forebrain patterning event necessary for correct retina formation is the activation of a repressor(s) by rax, whose role is to down-regulate fezf2 and hesx1 in the anterior neural plate, creating a hole within their expression domains starting by late gastrula stages (St. 13). As shown in (C), when this repression of fezf2 and hesx1 fails to occur we propose that they contribute to establishment of non-retinal diencephalic (blue arrows) and telencephalic (red arrows) properties within presumptive retinal tissue, resulting in the expansion of these forebrain identities into the retinal region. Fate map in (C) adapted with permission from (Eagleson and Harris, 1990).
Genes downstream of rax
We have confirmed key retinal factors previously identified as downstream of rax, such as mab21l2 (Kennedy et al., 2004), as well as identifying new factors such as myc, myb, foxn4, and vsx1 (Fig. 9A). The placement of myc expression downstream of rax has also been inferred from experiments using MO knockdown assays in Xenopus, although a direct interaction has not been shown (Terada et al., 2006). Instead, an intermediate factor, xhmgb3, was proposed to mediate the activation of myc by rax. However, xhmgb3 is actually not differentially expressed in our RNA-Seq analysis; this is one of a handful of discrepancies in genetic interactions observed between the Xenopus rax mutant phenotype and those reported in MO-based knockdown or overexpression studies (discussed in more detail below). Nevertheless, the role of the oncogene myc will be a focus for further study, as its dual roles in promoting proliferation and in determining cell fate are likely important but complex and not well understood (Bellmeyer et al., 2003; Rodrigues et al., 2008). Interestingly, vsx1 expression is one example of the few instances where a heterozygote phenotype can be detected, and its transcription may therefore be highly sensitive to Rax levels.
More recent work on the zebrafish rx3 mutant has implicated Eph/Ephrin signaling downstream of rx3 as key to proper morphogenesis of the eye (Cavodeassi et al., 2013). Future analysis of our Xenopus rax mutant is planned to examine the fate of this signaling pathway as well.
Mechanism of creating the expression “hole” within fezf2, hesx1, and fzd4 domains
Novel repressive interactions downstream of rax were identified over the course of these experiments: forebrain patterning genes fezf2 and hesx1 and Wnt-signaling pathway component fzd4 each display a distinctive expression domain “hole” in the anterior neural plate of wildtype embryos that is not observed in the rax mutant. RNASeq and in situ hybridization of the expression patterns of many genes expressed in the eye region at early stages rule out the possibility that increased apoptosis or reduced proliferation resulting in elimination of tissues corresponding to the “hole” is involved in the process of losing the expression “hole” in rax mutants (if this were the case, all genes expressed in the retina region should be affected and at least several, e.g. pax6 and lhx2, are not). Instead, our experiments support the conclusion that fezf2 and hesx1 are initially activated normally in the mutant, but are not properly repressed in the central eye field at neural plate stages, leading to improper expression of these factors and their downstream targets in the presumptive retinal region and causing expansion of alternative forebrain identities into the eye field (Fig. 9B). Experiments designed to clarify which repressor(s) downstream of rax are acting to prevent transcription of these genes in the repressed region (after their early, broad activation) have yet to identify convincing candidates, although several putative repressors down-regulated in the rax mutant have been tested, including myc, foxn4 and tbx3 (data not shown). Identification of the repressor or repressors active in this role will be an important future step in elucidating this portion of the eye field regulatory network. The possible role of fzd4 in this network is also an intriguing line of inquiry, as discussed further below. Like fezf2, its expression, normally restricted to a ring in early forebrain, fills the presumptive eye field in the rax mutant (data not shown), thus potentially having a role in the observed phenotype.
Another role for rax gene in brain patterning
A second key role for rax revealed by these experiments is the prevention of alternative forebrain fates via repression of diencephalic and telencephalic patterning genes, in addition to the activation of eye field-promoting genes. Fig. 9C illustrates the proposed expansion of alternative brain fates. As noted above, expansion of the telencephalon is also seen in a rx3 mutation in zebrafish (Stigloher et al., 2006). However, the expansion of the diencephalon displayed in the Xenopus mutant is not observed in the zebrafish rx3 mutant, and similarly arx gene expression is not expanded. One reason for this may be the duplication and subfunctionalization of rax genes rx1, rx2, and rx3 in zebrafish (Chuang and Raymond, 2001; Chuang et al., 1999). Although rx3 is the earliest expressed of the three genes, rx1 is expressed soon after and appears initially unaffected in the zebrafish rx3 mutant (Stigloher et al., 2006) and may impact the nature of the patterning changes seen in zebrafish. Like human (Wang et al., 2004), but unlike mouse, Xenopus also has a second rax gene, rax2, which is expressed later, after neurulation (Pan et al., 2006, data not shown), and could not have any possible redundant function with rax at early stages unlike the multi-gene family in zebrafish.
Fig. 9C and nomenclature in this paper follow the widely used fate map for Xenopus (Eagleson and Harris, 1990) suggesting that tissue posterior to the retina is diencephalic, though recent work argues that tissue immediately posterior to neural plate stage presumptive retina (and the retina) derives from secondary prosencephalon (Puelles, 1995; Puelles et al., 2013). After global neural patterning during gastrulation, fine-tuning of forebrain patterning occurs through later gradients created by local patterning centers in the anterior neural ridge (ANR), and, from the posterior, in the ZLI (Echevarría et al., 2003; Kiecker and Lumsden, 2012). These secondary organizers, along with roofplate and floorplate, create local gradients of Wnt, Shh, Bmp, and Fgf8 that define the boundaries of the major prosencephalic regions, the telencephalon, secondary prosencephalon (including retina and hypothalamus), and diencephalon (Bielen and Houart, 2012; Puelles et al., 2013). However, in the absence of Rax, the existing patterning gradients result in expansion of the forebrain tissues neighboring the presumptive retina. The effects of the loss of rax function on Wnt signaling may be of particular interest for further study, as both canonical and non-canonical Wnt signaling play key roles in patterning the eye field and diencephalon (Cavodeassi et al., 2005; Wilson and Houart, 2004). Studies in zebrafish have demonstrated that non-canonical Wnt signaling via Wnt11 and Fz5 can antagonize canonical Wnt signaling in the eye field; when non-canonical Wnt signaling is reduced, the eye field is lost and re-specified into caudal diencephalon (Cavodeassi et al., 2005). Our observation that fzd4 expression is up-regulated in the rax mutant is intriguing in this regard and may reveal complex interactions, since the fzd4 receptor has been shown to mediate both canonical and non-canonical Wnt signaling, and can act as both an activator or repressor of signaling (Swain et al., 2005).
Returning to the basic finding that properties of the brain both posterior to and anterior to the retina are expanded in the rax mutant, it is important to note that the retina region is likely not completely respecified toward telencephalic and diencephalic fates. We note that, as shown in Fig. 4, the expanded regions of foxg1 and arx, indicators of telencephalic and diencephalic fate, respectively, overlap in the rax mutant, indicating that the respecified retinal tissue likely has a blended or incompletely transformed regional identity.
Timing of rax gene function
It is also important to note that the fate shift resulting from the rax mutation implies that when rax activation occurs in the late gastrula (St. 12), forebrain regional boundaries including the presumptive retina are not yet determined and that the default fate of the retina-forming region would be to assume alternative forebrain identities. This lability is also supported by work showing that brain and eye regions in Xenopus are not determined within the anteroposterior neuroaxis at St. 11.5 (Saha and Grainger, 1992) though they are by the neural plate stage (St. 14). In addition, recent work (S. Louie, M. Fisher and R.M. Grainger, unpublished) argues that the Xenopus retina becomes determined by St. 12.5, suggesting a key role for rax activation in commitment. Consistent with this time frame for regional determination, the transplant experiment presented here shows that the fate of the presumptive retinal field in St 15 rax mutant embryos has already been stably transformed to non-retinal forebrain. Determination of diencephalic regions in zebrafish occurs at a similar stage (Staudt and Houart, 2007). Resolving the determination mechanisms here, both for the retina and brain regions, will be greatly aided by the many approaches the Xenopus affords for examining early development.
Comparison of knockdown strategies and genetic lesions for untangling gene networks during Xenopus embryogenesis
The studies performed with the rax mutant illustrate some clear advantages over transient methods for interfering with gene expression (MOs or dominant-negative constructs). The stability of using embryonic tissue resulting from a consistent genetic lesion allows one to perform manipulations, like the RNA-Seq or transplants performed here, that would be much more uncontrollable because of the genetic variation from embryo to embryo were an injection-based strategy (e.g. using MOs) used instead.
Schulte-Merker and Stainer (2014) report that there accumulated evidence in zebrafish that, for many genes, MO-mediated gene-knockdown phenotypes are not always the same as genetic mutants and in some cases may result from artifacts not accurately assessing the in vivo function of a gene. While Xenopus researchers have used MOs to reveal a multitude of important biological functions, the utility of direct gene mutations as described here allows not only an accurate assessment of gene function but also permits one to screen phenotypes throughout the life of an animal, i.e. with the limitations of MOs to examining early development. Our own observations also at least raise questions about the differences observed between these two methodologies and highlight potential advantages of using mutations for studying development. For example, although we have identified new targets downstream of rax in addition to confirming previously known targets as described above, our observations are not consistent with some interactions implied by experiments utilizing strategies to assay Rax function through MO-knockdown or expression of transcriptionally repressive forms of Rax (Andreazzoli et al., 1999, 2003; Giannaccini et al., 2013; Terada et al., 2006). Specific downstream genetic interactions inferred from such studies, such as placing xhmgb3 downstream of rax (Terada et al., 2006) or transcriptional repressors crx, tle2 and hes4 (Giannaccini et al., 2013), are not consistent with the RNA-Seq analyses presented here where we see that the expression levels of these transcripts are unaffected in rax mutant tissue at neural plate stages. Although it is possible that some of these previously implied downstream effects occur later in development, (i.e. after our RNA-Seq analysis was performed), these differences may also reflect some of the questions raised by Schulte-Merker and Stanier about inferring genetic connections from experiments using MOs (which may also apply to overexpression of dominant-negative constructs). Thus, although MOs or dominant-negative constructs can be and have been very useful reagents for rapid loss-of-function studies, one needs to be aware that they may not always reveal native genetic interactions or at least need to be reconciled with genetic data.
This work highlights the value of generating mutations in Xenopus for studies of early embryonic patterning and organogenesis events that are less tractable in many other vertebrate model systems. While the TILLING technology used to generate the mutation studied here requires significant labor to produce mutations, new genome editing techniques developed for many model organisms (TALENs and CRISPR/Cas9) are now available in the Xenopus system (Blitz et al., 2013; Guo et al., 2014; Lei et al., 2012; Nakajima and Yaoita, 2013; Nakayama et al., 2013; Suzuki et al., 2013). These advances will allow researchers to combine the advantages of Xenopus with facile genetic approaches that will allow rapid generation of null mutations, providing the experimental framework for gaining a more accurate understanding of the gene networks involved in normal developmental processes along with the genetic alterations underlying many human diseases.
Supplementary Material
Highlights.
We identified an eyeless X. tropicalis rax mutant during a mutagenesis screen.
RNA-Seq analysis identified novel genes downstream of rax.
Key eye field transcription factors are expressed normally in this eyeless mutant.
Eye tissue is transformed to non-retinal forebrain identities in rax mutants.
Failure to locally repress fezf2 and hesx1 in mutants leads to forebrain expansion.
Acknowledgements
We thank past Grainger lab members Lyle Zimmerman, Selina Noramly, Hajime Ogino, Hui Wang and Mary Burns for help establishing the ENU-mutagenesis methodology. We also thank Ruben Bautista in the Stemple lab for contributions to RNAseq experiments, and Massimiliano Andreazzoli and Ken Cho for sharing plasmids. The authors gratefully acknowledge support of the National Xenopus Resource and its staff, from which rax mutant frogs described here are available.
Funding
This work was funded by the National Institutes of Health [NIH R01 EY017400, EY018000 and EY022954] and a research award from the Sharon Stewart Aniridia Trust to RMG. Fellowship funding that contributed to this work was awarded by the University of Virginia Society of Fellows to MBF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
M.B.F., T.N., M.F., N.H., A.C., R.R., S.C. and A.H. performed experiments. M.B.F., T.N., M.F., S.C., A.H., D.L.S. and R.M.G analyzed data. M.B.F., T.N., M.F. and R.M.G. conceived the study and wrote the manuscript. M.B.F. and T.N. contributed equally as joint first authors.
Competing interests
The authors declare no competing financial interests.
References
- Andoniadou CL, Signore M, Young RM, Gaston-Massuet C, Wilson SW, Fuchs E, Martinez-Barbera JP. HESX1- and TCF3-mediated repression of Wnt/β-catenin targets is required for normal development of the anterior forebrain. Development. 2011;138:4931–4942. doi: 10.1242/dev.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreazzoli M, Gestri G, Angeloni D, Menna E, Barsacchi G. Role of Xrx1 in Xenopus eye and anterior brain development. Development. 1999;126:2451–2460. doi: 10.1242/dev.126.11.2451. [DOI] [PubMed] [Google Scholar]
- Andreazzoli M, Gestri G, Cremisi F, Casarosa S, Dawid IB, Barsacchi G. Xrx1 controls proliferation and neurogenesis in Xenopus anterior neural plate. Development. 2003;130:5143–5154. doi: 10.1242/dev.00665. [DOI] [PubMed] [Google Scholar]
- Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers EH, Jamrich M. Regulation of vertebrate eye development by Rx genes. Int. J. Dev. Biol. 2004;48:761–770. doi: 10.1387/ijdb.041878tb. [DOI] [PubMed] [Google Scholar]
- Beier DR. Sequence-based analysis of mutagenized mice. Mamm. Genome. 2000;11:594–597. doi: 10.1007/s003350010113. [DOI] [PubMed] [Google Scholar]
- Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c myc is an essential regulator of neural crest formation in xenopus. Dev. Cell. 2003;4:827–839. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- Bielen H, Houart C. BMP signaling protects telencephalic fate by repressing eye identity and its Cxcr4-dependent morphogenesis. Dev. Cell. 2012;23:812–822. doi: 10.1016/j.devcel.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz IL, Cho KW. Anterior neurectoderm is progressively induced during gastrulation: the role of the Xenopus homeobox gene orthodenticle. Development. 1995;121:993–1004. doi: 10.1242/dev.121.4.993. [DOI] [PubMed] [Google Scholar]
- Blitz IL, Biesinger J, Xie X, Cho KWY. Biallelic genome modification in F(0) Xenopus tropicalis embryos using the CRISPR/Cas system. genesis. 2013;51:827–834. doi: 10.1002/dvg.22719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavodeassi F, Carreira-Barbosa F, Young RM, Concha ML, Allende ML, Houart C, Tada M, Wilson SW. Early stages of zebrafish eye formation require the coordinated activity of Wnt11, Fz5, and the Wnt/beta-catenin pathway. Neuron. 2005;47:43–56. doi: 10.1016/j.neuron.2005.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavodeassi F, Ivanovitch K, Wilson SW. Eph/Ephrin signalling maintains eye field segregation from adjacent neural plate territories during forebrain morphogenesis. Development. 2013;140:4193–4202. doi: 10.1242/dev.097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JC, Raymond PA. Zebrafish genes rx1 and rx2 help define the region of forebrain that gives rise to retina. Dev. Biol. 2001;231:13–30. doi: 10.1006/dbio.2000.0125. [DOI] [PubMed] [Google Scholar]
- Chuang JC, Mathers PH, Raymond PA. Expression of three Rx homeobox genes in embryonic and adult zebrafish. Mech. Dev. 1999;84:195–198. doi: 10.1016/s0925-4773(99)00077-5. [DOI] [PubMed] [Google Scholar]
- Danesin C, Peres JN, Johansson M, Snowden V, Cording A, Papalopulu N, Houart C. Integration of telencephalic Wnt and hedgehog signaling center activities by Foxg1. Dev. Cell. 2009;16:576–587. doi: 10.1016/j.devcel.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Eagleson GW, Harris WA. Mapping of the presumptive brain regions in the neural plate of Xenopus laevis. J. Neurobiol. 1990;21:427–440. doi: 10.1002/neu.480210305. [DOI] [PubMed] [Google Scholar]
- Eagleson G, Ferreiro B, Harris WA. Fate of the anterior neural ridge and the morphogenesis of the Xenopus forebrain. J. Neurobiol. 1995;28:146–158. doi: 10.1002/neu.480280203. [DOI] [PubMed] [Google Scholar]
- Echevarría D, Vieira C, Gimeno L, Martínez S. Neuroepithelial secondary organizers and cell fate specification in the developing brain. Brain Res. Rev. 2003;43:179–191. doi: 10.1016/j.brainresrev.2003.08.002. [DOI] [PubMed] [Google Scholar]
- El-Hodiri HM, Qi X-L, Seufert DW. The Xenopus arx gene is expressed in the developing rostral forebrain. Dev. Genes Evol. 2003;212:608–612. doi: 10.1007/s00427-002-0282-8. [DOI] [PubMed] [Google Scholar]
- Ermakova GV, Alexandrova EM, Kazanskaya OV, Vasiliev OL, Smith MW, Zaraisky AG. The homeobox gene, Xanf-1, can control both neural differentiation and patterning in the presumptive anterior neurectoderm of the Xenopus laevis embryo. Development. 1999;126:4513–4523. doi: 10.1242/dev.126.20.4513. [DOI] [PubMed] [Google Scholar]
- Filippi A, Jainok C, Driever W. Analysis of transcriptional codes for zebrafish dopaminergic neurons reveals essential functions of Arx and Isl1 in prethalamic dopaminergic neuron development. Dev. Biol. 2012;369:133–149. doi: 10.1016/j.ydbio.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Fish MB, Nakayama T, Grainger RM. Simple, fast, tissue-specific bacterial artificial chromosome transgenesis in Xenopus. Genesis. 2012;50:307–315. doi: 10.1002/dvg.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann S. Wnt signaling in eye organogenesis. Organogenesis. 2008;4:60–67. doi: 10.4161/org.4.2.5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Kozak CA, Cepko CL. rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc. Natl. Acad. Sci. 1997;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannaccini M, Giudetti G, Biasci D, Mariotti S, Martini D, Barsacchi G, Andreazzoli M. Brief Report: Rx1 Defines Retinal Precursor Identity by Repressing Alternative Fates Through the Activation of TLE2 and Hes4. Stem Cells. 2013;31:2842–2847. doi: 10.1002/stem.1530. [DOI] [PubMed] [Google Scholar]
- Goda T, Abu-Daya A, Carruthers S, Clark MD, Stemple DL, Zimmerman LB. Genetic Screens for Mutations Affecting Development of Xenopus tropicalis. PLoS Genet. 2006;2:e91. doi: 10.1371/journal.pgen.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhang T, Hu Z, Zhang Y, Shi Z, Wang Q, Cui Y, Wang F, Zhao H, Chen Y. Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development. 2014;141:707–714. doi: 10.1242/dev.099853. [DOI] [PubMed] [Google Scholar]
- Harland RM, Grainger RM. Xenopus research: metamorphosed by genetics and genomics. Trends Genet. 2011;27:507–515. doi: 10.1016/j.tig.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T, Nakazawa M, Muraoka O, Nakayama R, Suda Y, Hibi M. Zinc-finger genes Fez and Fez-like function in the establishment of diencephalon subdivisions. Development. 2006;133:3993–4004. doi: 10.1242/dev.02585. [DOI] [PubMed] [Google Scholar]
- Hirsch N, Zimmerman LB, Gray J, Chae J, Curran KL, Fisher M, Ogino H, Grainger RM. Xenopus tropicalis transgenic lines and their use in the study of embryonic induction. Dev. Dyn. 2002;225:522–535. doi: 10.1002/dvdy.10188. [DOI] [PubMed] [Google Scholar]
- Jeong J-Y, Einhorn Z, Mathur P, Chen L, Lee S, Kawakami K, Guo S. Patterning the zebrafish diencephalon by the conserved zinc-finger protein Fezl. Development. 2007;134:127–136. doi: 10.1242/dev.02705. [DOI] [PubMed] [Google Scholar]
- Kennedy BN, Stearns GW, Smyth VA, Ramamurthy V, van Eeden F, Ankoudinova I, Raible D, Hurley JB, Brockerhoff SE. Zebrafish rx3 and mab21l2 are required during eye morphogenesis. Dev. Biol. 2004;270:336–349. doi: 10.1016/j.ydbio.2004.02.026. [DOI] [PubMed] [Google Scholar]
- Kettleborough RNW, Bruijn E, de, Eeden F, van, Cuppen E, Stemple DL. High-throughput target-selected gene inactivation in zebrafish. Methods Cell Biol. 2011;104:121–127. doi: 10.1016/B978-0-12-374814-0.00006-9. [DOI] [PubMed] [Google Scholar]
- Kiecker C, Lumsden A. The role of organizers in patterning the nervous system. Annu. Rev. Neurosci. 2012;35:347–367. doi: 10.1146/annurev-neuro-062111-150543. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kusaka M, Omichi K, Suzuki R, Kato-Fukui Y, Kamiirisa K, et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat. Genet. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- Lavado A, Lagutin OV, Oliver G. Six3 inactivation causes progressive caudalization and aberrant patterning of the mammalian diencephalon. Development. 2008;135:441–450. doi: 10.1242/dev.010082. [DOI] [PubMed] [Google Scholar]
- Lei Y, Guo X, Liu Y, Cao Y, Deng Y, Chen X, Cheng CHK, Dawid IB, Chen Y, Zhao H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc. Natl. Acad. Sci. 2012;109:17484–17489. doi: 10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loosli F, Staub W, Finger-Baier KC, Ober EA, Verkade H, Wittbrodt J, Baier H. Loss of eyes in zebrafish caused by mutation of chokh/rx3. EMBO Rep. 2003;4:894–899. doi: 10.1038/sj.embor.embor919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers PH, Grinberg A, Mahon KA, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- Miura H, Yanazawa M, Kato K, Kitamura K. Expression of a novel aristaless related homeobox gene “Arx” in the vertebrate telencephalon, diencephalon and floor plate. Mech. Dev. 1997;65:99–109. doi: 10.1016/s0925-4773(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Yaoita Y. Comparison of TALEN scaffolds in Xenopus tropicalis. Biol. Open. 2013;2:1364–1370. doi: 10.1242/bio.20136676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis. 2013;51:835–843. doi: 10.1002/dvg.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieukoop PD, Faber J. Normal Table of Xenopus laevis. Garland Publishing Inc; 1994. [Google Scholar]
- Ogino H, Ochi H, Reza HM, Yasuda K. Transcription factors involved in lens development from the preplacodal ectoderm. Dev. Biol. 2012;363:333–347. doi: 10.1016/j.ydbio.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Pan Y, Nekkalapudi S, Kelly LE, El-Hodiri HM. The Rx-like homeobox gene (Rx-L) is necessary for normal photoreceptor development. Invest. Ophthalmol. Vis. Sci. 2006;47:4245–4253. doi: 10.1167/iovs.06-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalopulu N. Regionalization of the forebrain from neural plate to neural tube. Perspect. Dev. Neurobiol. 1995;3:39–52. [PubMed] [Google Scholar]
- Puelles L. A segmental morphological paradigm for understanding vertebrate forebrains. Brain. Behav. Evol. 1995;46:319–337. doi: 10.1159/000113282. [DOI] [PubMed] [Google Scholar]
- Puelles L, Harrison M, Paxinos G, Watson C. A developmental ontology for the mammalian brain based on the prosomeric model. Trends Neurosci. 2013;36:570–578. doi: 10.1016/j.tins.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Rodrigues CO, Nerlick ST, White EL, Cleveland JL, King ML. A Myc-Slug (Snail2)/Twist regulatory circuit directs vascular development. Development. 2008;135:1903–1911. doi: 10.1242/dev.011296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Seguel E, Alarcón P, Gómez-Skarmeta JL. The Xenopus Irx genes are essential for neural patterning and define the border between prethalamus and thalamus through mutual antagonism with the anterior repressors Fezf and Arx. Dev. Biol. 2009;329:258–268. doi: 10.1016/j.ydbio.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Saha MS, Grainger RM. A labile period in the determination of the anterior-posterior axis during early neural development in Xenopus. Neuron. 1992;8:1003–1014. doi: 10.1016/0896-6273(92)90123-u. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, Stainier DYR. Out with the old, in with the new: reassessing morpholino knockdowns in light of genome editing technology. Development. 2014;141:3103–3104. doi: 10.1242/dev.112003. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Hibi M. Formation and patterning of the forebrain and olfactory system by zinc-finger genes Fezf1 and Fezf2. Dev. Growth Differ. 2009;51:221–231. doi: 10.1111/j.1440-169X.2009.01088.x. [DOI] [PubMed] [Google Scholar]
- Sinn R, Wittbrodt J. An eye on eye development. Mech. Dev. 2013;130:347–358. doi: 10.1016/j.mod.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger R, Harland R. Early Development of Xenopus Laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 2000. [Google Scholar]
- Staudt N, Houart C. The prethalamus is established during gastrulation and influences diencephalic regionalization. PLoS Biol. 2007;5:e69. doi: 10.1371/journal.pbio.0050069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigloher C, Ninkovic J, Laplante M, Geling A, Tannhäuser B, Topp S, Kikuta H, Becker TS, Houart C, Bally-Cuif L. Segregation of telencephalic and eye-field identities inside the zebrafish forebrain territory is controlled by Rx3. Development. 2006;133:2925–2935. doi: 10.1242/dev.02450. [DOI] [PubMed] [Google Scholar]
- Suzuki K-IT, Isoyama Y, Kashiwagi K, Sakuma T, Ochiai H, Sakamoto N, Furuno N, Kashiwagi A, Yamamoto T. High efficiency TALENs enable F0 functional analysis by targeted gene disruption in Xenopus laevis embryos. Biol. Open. 2013;2:448–452. doi: 10.1242/bio.20133855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain RK, Katoh M, Medina A, Steinbeisser H. Xenopus frizzled-4 S, a splicing variant of Xfz4 is a context-dependent activator and inhibitor of Wnt/β-catenin signaling. Cell Commun. Signal. 2005;3:12. doi: 10.1186/1478-811X-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada K, Kitayama A, Kanamoto T, Ueno N, Furukawa T. Nucleosome regulator Xhmgb3 is required for cell proliferation of the eye and brain as a downstream target of Xenopus rax/Rx1. Dev. Biol. 2006;291:398–412. doi: 10.1016/j.ydbio.2005.12.029. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhusha VV, Kuznetsova IM, Stepanenko OV, Zaraisky AG, Shavlovsky MM, Turoverov KK, Uversky VN. High Stability of Discosoma DsRed As Compared to Aequorea EGFP†. Biochemistry (Mosc.) 2003;42:7879–7884. doi: 10.1021/bi034555t. [DOI] [PubMed] [Google Scholar]
- Voronina VA, Kozhemyakina EA, O’Kernick CM, Kahn ND, Wenger SL, Linberg JV, Schneider AS, Mathers PH. Mutations in the Human RAX Homeobox Gene in a Patient with Anophthalmia and Sclerocornea. Hum. Mol. Genet. 2004;13:315–322. doi: 10.1093/hmg/ddh025. [DOI] [PubMed] [Google Scholar]
- Voronina VA, Kozlov S, Mathers PH, Lewandoski M. Conditional alleles for activation and inactivation of the mouse Rx homeobox gene. Genesis. 2005;41:160–164. doi: 10.1002/gene.20109. [DOI] [PubMed] [Google Scholar]
- Wang Q, Chen S, Esumi N, Swain PK, Haines HS, Peng G, Melia BM, McIntosh I, Heckenlively JR, Jacobson SG, et al. QRX, a novel homeobox gene, modulates photoreceptor gene expression. Hum. Mol. Genet. 2004;13:1025–1040. doi: 10.1093/hmg/ddh117. [DOI] [PubMed] [Google Scholar]
- Wang Z-B, Boisvert E, Zhang X, Guo M, Fashoyin A, Du Z-W, Zhang S-C, Li X-J. Fezf2 regulates telencephalic precursor differentiation from mouse embryonic stem cells. Cereb. Cortex. 2011;21:2177–2186. doi: 10.1093/cercor/bhr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JS, Salinger A, Justice MJ. Optimal N-ethyl-N-nitrosourea (ENU) doses for inbred mouse strains. Genesis. 2000;26:230–233. [PubMed] [Google Scholar]
- Wilson SW, Houart C. Early steps in the development of the forebrain. Dev. Cell. 2004;6:167–181. doi: 10.1016/s1534-5807(04)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R, Mizutani-Koseki Y, Koseki H, Takahashi N. Requirement for Mab21l2 during development of murine retina and ventral body wall. Dev. Biol. 2004;274:295–307. doi: 10.1016/j.ydbio.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Zhang L, Mathers PH, Jamrich M. Function of Rx, but not Pax6, is essential for the formation of retinal progenitor cells in mice. Genesis. 2000;28:135–142. [PubMed] [Google Scholar]
- Zuber ME. Chapter two - Eye Field Specification in Xenopus laevis. In: Cagan Ross L., Reh Thomas A., editors. Current Topics in Developmental Biology. Academic Press; 2010. pp. 29–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.