Abstract
Modern molecular genetics studies necessitate the manipulation of genes in their endogenous locus, but most of the current methodologies require an inefficient donor-dependent homologous recombination step to locally modify the genome. Here we describe a methodology to efficiently generate Drosophila knock-in alleles by capitalizing on the availability of numerous genomic MiMIC transposon insertions carrying recombinogenic attP sites. Our methodology entails the efficient PhiC31-mediated integration of a recombination cassette flanked by unique I-SceI and/or I-CreI restriction enzyme sites into an attP-site. These restriction enzyme sites allow for double-strand break−mediated removal of unwanted flanking transposon sequences, while leaving the desired genomic modifications or recombination cassettes. As a proof-of-principle, we mutated LRRK, tau, and sky by using different MiMIC elements. We replaced 6 kb of genomic DNA encompassing the tau locus and 35 kb encompassing the sky locus with a recombination cassette that permits easy integration of DNA at these loci and we also generated a functional LRRKHA knock in allele. Given that ~92% of the Drosophila genes are located within the vicinity (<35 kb) of a MiMIC element, our methodology enables the efficient manipulation of nearly every locus in the fruit fly genome without the need for inefficient donor-dependent homologous recombination events.
Keywords: Drosophila, genome editing, homologous recombination, MiMIC
The continued development of novel genetic tools to manipulate gene function in Drosophila has boosted the use of this organism to study in vivo biological mechanisms and to model disease (Bier 2005; Yamamoto et al. 2014). Ideally genes are modified in their endogenous locus by the use of homologous recombination (Rong and Golic 2000; Chan et al. 2012), but this process is rather tedious and inefficient. Hence, methodologies that facilitate Drosophila genome editing and allow for efficient repetitive targeting of a locus without the need for homologous recombination are eagerly welcomed (Gao et al. 2008; Choi et al. 2009; Huang et al. 2009; Weng et al. 2009).
Improvements in technology to target loci by homologous recombination have recently been made. These are based on generating a double-strand break (DSB) in the target DNA near or in the locus of interest and then allow manipulations via repair mechanisms, including donor dependent repair by homologous recombination. These DSBs can be introduced by nucleases, including zinc finger nucleases (ZFN) (Bibikova et al. 2003), TAL effector nucleases (TALEN) (Liu et al. 2012), or CRISPR-associated protein9 (Cas9)(Jinek et al. 2012; Cong et al. 2013; Gratz et al. 2013) that are designed to recognize their target DNA in a sequence-dependent manner. The ability to introduce a DSB increases the efficiency of homologous recombination by donor dependent repair by 10- to 100-fold (Rong et al. 2002; Bibikova et al. 2003). However, a given endonuclease or Cas9/gRNA pair may find promiscuous cut sites in the genome (Hsu et al. 2013; Mali et al. 2013; Pattanayak et al. 2013) and in addition, every locus in the genome requires the design and the testing of new nucleases (TALEN, ZFN) or guide RNA (Cas9) molecules to target the locus of interest. Finally, the identification of a modified genomic locus in the Drosophila genome ideally requires the integration or the excision of selection markers and such strategies often result in leaving behind recombination recognition sites (FRT, LoxP, attR, …) (Gao et al. 2008; Choi et al. 2009; Huang et al. 2009; Weng et al. 2009; Wesolowska and Rong 2013). These sites would then still need to be removed from the genome in an independent step. Hence, a strategy that does not require the need to test tools locus-by-locus would allow one to efficiently modify the locus of interest without the need for homologous recombination and without leaving exogenous sequences could serve as a parallel alternative to the current genome editing methodologies.
Compared with classical homologous recombination using a donor construct, homologous recombination by single strand annealing is 100-fold more efficient (Rong and Golic 2000; Rong et al. 2002). Single-strand annealing entails homologous recombination between two regions of homology located on the same chromosome. Hence, if a homology cassette that harbors a homology arm is provided nearby the locus of interest, a DSB flanking this cassette induces homologous recombination by single-strand annealing between the regions of homology with high efficiency (up to 85%) (Rong and Golic 2000). Hence, an alternative methodology for genome editing would (1) facilitate the integration of such a homology cassette nearby the locus of interest, using a methodology different from donor-dependent homologous recombination; and (2) use single-strand annealing to resolve this cassette, leaving mutations or other functional sequences behind.
Here we describe a methodology that allows for efficient genome editing in nearly every Drosophila locus without the need for donor dependent homologous recombination. Our methodology capitalizes on the ongoing efforts of the Drosophila Gene Disruption Project that has generated numerous MiMIC transposon insertions nearby or in many genes in the Drosophila genome (Venken et al. 2011). MiMIC transposons that carry attP sites allow for effective PhiC31-mediated integration of a recombination cassette that we flanked by unique restriction enzyme sites. These sites allow one to efficiently generate DSBs followed by homologous recombination by single-strand annealing, thereby locally editing the genome. We inserted an HA tag in the LRRK gene by using a MiMIC in a neighboring gene, and we replaced 6 kb and 35 kb encompassing the tau and sky loci, respectively, with a cassette that allows for recombination-mediated DNA exchange in these loci. The high efficiency of our approach allowed us to screen for correct modification of the loci by simple genomic polymerase chain reaction (PCR) of a limited number of individual fly lines. Given that MiMIC insertions are widely present in the fly genome (Venken et al. 2011), our method enables to manipulate almost every Drosophila gene without the need for donor-dependent homologous recombination.
Materials and Methods
Molecular genetics and Drosophila maintenance
The recombination cassette to be integrated Mi{MIC}GluRIIEMI01886 was created by chimeric PCR of LRRK from the BAC clone CH322-120O10 with the following primers: 5′-CAG GTA CCA GTT ACG CTA GGG ATA ACA GGG TAA TAT AGG CCC AAG ATG AAC ATG TTG TGC-3′, introducing an I-SceI restriction site and a KpnI restriction site 1095 bp upstream of the LRRK stop codon; 5′-CGC CAA GCA CTG GAC CTA CCC ATA CGA CGT ACC AGA TTA CGC TTA CCC ATA CGA CGT ACC AGA TTA CGC TTA CCC ATA CGA CGT ACC AGA TTA CGC T-3′ and 5′-TAC CCA TAC GAC GTA CCA GAT TAC GCT TAC CCA TAC GAC GTA CCA GAT TAC GCT TAC CCA TAC GAC GTA CCA GAT TAC GCT TAA ATT CGA TCT CAT TCA AAA TAT TTG-3′, introducing a 3xHA tag in front of the stop codon of LRRK; and 5′CAA TCG GCA TCC GAT AAG TGC AAA ACG TCG TGA GAC AGT TTG GTG GTA CC-3′, introducing an I-CreI restriction site and a KpnI restriction site 639 bp downstream of the LRRK stop codon. This PCR product was subsequently cloned in the pABC plasmid with the use of restriction-ligation with KpnI (Choi et al. 2009); pABC harbors two attB sites that allow for PhiC31-mediated integration in the attP sites of a MiMIC element. This plasmid was injected (GenetiVision, Houston, TX) in Drosophila embryos carrying both the Mi{MIC}GluRIIEMI01886 and an embryonic source of PhiC31 (Bischof et al. 2007).
Positive integration events were selected by scoring for the absence of y+ followed by PCR to assess the orientation of the insert using the following primers: 5′-GCG ATT GAT GAG CAT GTG AAC-3′ (forward primer the end of the LRRK gene) and 5′-GTT ACG CTA GGG ATA ACA GG-3′ (reverse primer in the I-SceI restriction site). The “targeting plasmid,” pSV001, was generated by ligating a synthetic DNA fragment carrying multiple restriction sites, an I-SceI recognition site, and an F3 recombination site (Integrated DNA Technologies) in pFL44S{w+}-attB linearized by KpnI and XbaI and treated with “Alkaline Phosphatase, Calf intestinal” (New England Biolabs). The sequence of the synthetic KpnI-ISceI-AgeI-SmaI-AvrII-F3-XbaI fragment was 5′-ATG CGG TAC CGG ATA GGG ATA ACA GGG TAA TAT AGA CCG GTC CCG GGC CTA GGG AAG TTC CTA TAC TAT TTG AAG AAT AGG AAC TTC GGA ATA GGA ACT TCT CTA GAA TGC-3′. Homology arms for tau were PCR amplified with following primers, tau (fw) 5′-CGC GAC CGG TCT AAG TGC AAC AAC GCC GAG ATT TGG-3′ (with an AgeI site), and tau (rev) 5′-GCG CCC TAG GGC CGA AAT GCA TGT CGA GCT GTA TC-3′ (with an AvrII site) respectively and cloned into pSV001. Homology arms for sky were PCR amplified with the following primers: sky (fw) 5′-GAC TGG ATC CTA GGG ATA ACA GGG TAA TAC CGG TTC TAG ACT CGA GCG GCA GTC TGG TCT TGT TTC-3′ (with an I-SceI site) and sky (rev) 5′-GTC ACT GCA GGA AGT TCC TAT ACT ATT TGA AGA ATA GGA ACT TCG GAA TAG GAA CTT CAC TAG TGG CGC GCC AAG CTT CTA TTT CAT TCT TCT AGG GGC-3′ (with an F3 site) and directly cloned into pFL44S{w+}-attB. These plasmids were injected into the following MiMICs harboring transgenic flies (BestGene Inc): Mi{MIC}skyMI04695 and Mi{MIC}tauMI03440.
Positive integration events were selected by scoring for w+ progeny (BestGene inc) followed by PCR to assess which attR were generated with following primers, I-SceI primer: 5′-CGG-TAT TAC CCT GTT ATC CC-3′and sky and tau primers, 5′-CTG CGG CTG CAA TTT ATT TC-3′ (sky), 5′-GCA AGT AGG TCG CAT CGG CC-3′ (tau), respectively. DSBs were generated by 1 hr 37° heat shock in second instar larvae. Primers used for PCR screening after single-strand annealing are as follows: for loss of the I-SceI site: 5′-GTT ACG CTA GGG ATA ACA GG-3′ (reverse primer I-SceI restriction site) and 5′-CAC ATT CAT TGC CTG CTG TGG-3′ (forward primer LRRK 3′), and for loss of the I-CreI site: 5′-CGT CGT GAG ACA GTT TGG-3′ (reverse primer I-CreI restriction site) and 5′-GCG ATT GAT GAG CAT GTG AAC-3′ (forward primer end of LRRK gene), and finally for the integration of the HA-tag: 5′-CCA TTA GTG TTT TCC GAC C-3′ (forward primer LRRKHA) and 5′-ACT CCT CAG CGA ATA TAC C-3′ (reverse primer LRRKHA). To screen for flies that carry sky and tau deletions, PCR was performed from the novel RMCE over the duplication to the endogenous DNA with following primers: sky (fw) 5′-CAG AAA ACG GCG TGC GTA AG-3′ sky (rev) 5′-GAA TAG GAA CTT CGG AAT AGG-3′ tau (fw) 5′-AGG TGG CTC TGT TGG AGT TC-3′ tau (rev) 5′-GTT CCT ATT CCG AAG TTC CTA TTC-3′ and sequence verified. All crosses and stocks were maintained on standard cornmeal and molasses media at 25° and fly genetics and crossing schemes are shown in Supporting Information, Figure S1 and Figure S4.
Western blot
Adult flies were decapitated with a razor blade. Heads were homogenized on ice using a motorized pellet pestle in lysis buffer containing 50 mM Tris-HCl pH 6.8, 130 mM NaCl, 1% Triton, 1 mM MgCl, and protease inhibitor complete (Roche). After a 30-min extraction on ice, the homogenate was cleared by centrifugation for 20 min at 3000 × g. Supernatant was resuspended in LDS sample buffer (Invitrogen) and denatured for 10 min at 70°. A volume of supernatant that corresponds to 15 heads was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis in 3–8% NuPAGE Tris-Acetate gels (Invitrogen) and blotted onto polyvinylidene fluoride transfer membrane (Millipore). Protein bands were visualized with a Ponceau S stain (0.1% Ponceau S and 0.5% acetic acid). Blots were blocked in TBST+5% milk and the membrane was incubated overnight with mouse anti-HA (Clone 16B12, Covance) diluted 1:500 in TBST+1% BSA or with antisynapsin [3C11 (anti-SYNORF1), Developmental Studies Hybridoma Bank, Iowa, City, IA] diluted 1:500 in TBST+1% BSA. Peroxidase-conjugated secondary antibodies and ECL plus system (Pierce) were used for detection.
Bioinformatics
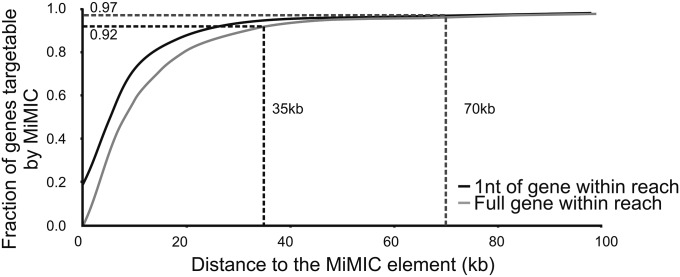
The bioinformatics analysis is based on MiMIC insertion site list (release version 27-02-2013) (Venken et al. 2011) and the Flybase (http://flybase.org) Drosophila melanogaster genome annotation version 5.51 (FlyBase Genome 2013). The analysis determined for all region sizes between 0 and 100 (with steps of 10) how many genes are either fully within the specified distance (full gene) of a MiMIC insert site, or have an overlap of at least one nucleotide (1nt). The results are combined into a total number of unique genes encompassed by or overlapping with the MiMIC region and plotted in Figure 4. The calculations were performed in an iPython (http://ipython.org/notebook.html) by use of the Pandas data analysis library (http://pandas.pydata.org/). The notebook containing the full analysis is available as a source ipynb file or a PDF (File S2).
Figure 4.
MiMICs allow the targeting of the majority of the Drosophila genes. Graph of distance to the MiMIC elements (kb) in function of fraction of genes targetable by a given MiMIC. Black line represents targetable genes where one nucleotide of the gene is within reach or gray line where the full gene is within reach. The dashed line at 35 kb distance to a MiMIC indicates that 92% of the Drosophila genes are within this distance to a MiMIC, whereas the dashed line at 70 kb indicates that 97% of the Drosophila genes are within this distance and within reach to perform single strand annealing (or one-ended invasion crossover) experiments (Wesolowska and Rong 2013).
Results
Direct targeting of LRRK using a MiMIC insertion in GluRIIE
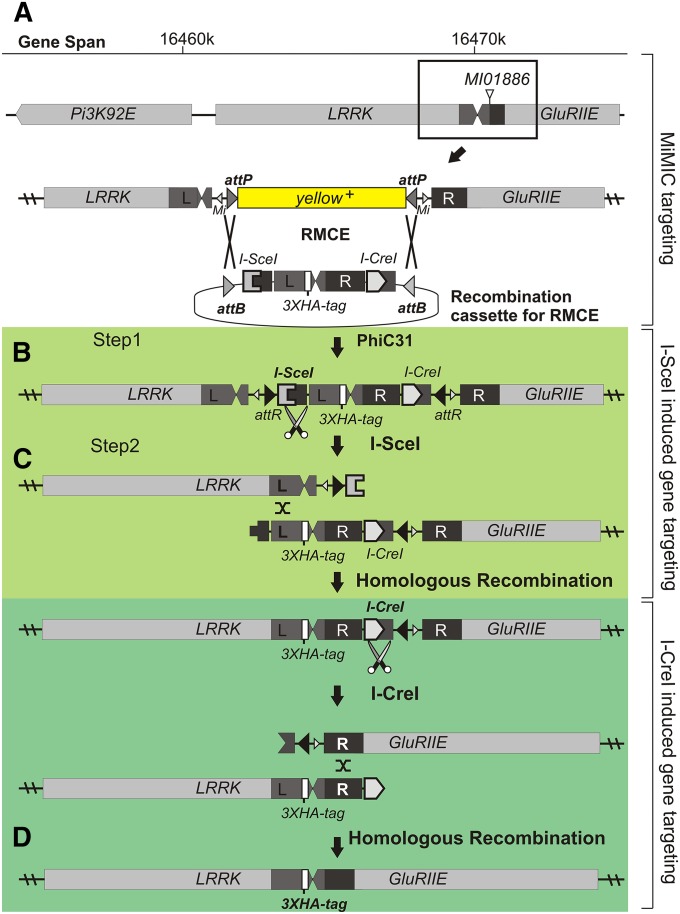
Our methodology to manipulate the Drosophila genome entails a multistep process in which we first target an attP site in a MiMIC transposon inserted close to or in our gene of interest with a “recombination cassette” and in a second phase we resolve the cassette by single-strand annealing, thereby bridging and modifying the nearby genome while removing unwanted transposon sequences. To provide a proof of principle for our targeting methodology, we used it to knock in an HA tag in the LRRK gene (Figure 1). First we selected a MiMIC insertion (Mi{MIC}GluRIIEMI01886) in the GluRIIE gene located 3′ of the LRRK gene (Figure 1A). Second, we generated a recombination cassette that consists of two 500-bp stretches of sequence; one that encompasses the 3′ end of the LRRK gene (dark gray in Figure 1A, marked by ‘L’) and one identical to the sequence immediately 3′ of the MiMIC insertion site (black in Figure 1A, marked by ‘R’). In this cassette we inserted a sequence coding for an HA tag in front of the LRRK stop codon and flanked the construct with an I-SceI and an I-CreI endonuclease site as well as with attB sites on either side (Figure 1A, methods). This recombination cassette was injected into embryos that harbor Mi{MIC}GluRIIEMI01886 and that express PhiC31 recombinase. PhiC31-mediated recombination between the attB sites in the recombination cassette and attP sites in the MiMIC replaced the yellow+(y+) marker in the MiMIC with the I-SceI and I-CreI flanked recombination cassette. Consistent with previous reports on recombination mediated cassette exchange in different contexts (Bateman et al. 2006; Venken et al. 2011), this first step of our methodology was very efficient and about 20% of the injected animals integrated the recombination cassette. Using genomic PCR we verified the orientation of the cassette and 50% of them are inserted with the LRRK-homology arm in the recombination cassette oriented toward the LRRK gene (i.e., 1/10 of the injected animals; Figure 1B). Hence, using a single set of germ line injections (100 embryos) we correctly integrated the recombination cassette in Mi{MIC}GluRIIEMI01886.
Figure 1.
Genome editing using MiMICs through two consecutive double-strand breaks. Schematic representation of (A) gene span around the MiMIC [Mi{MIC}GluRIIEMI01886 harboring a yellow+ marker flanked by attP sites and Minos element arms (Mi)] downstream of LRRK, which is being targeted with a targeting construct consisting of a duplication of part of LRRK, an HA-tag and a duplication of part of GluRIIE and flanked by an I-SceI and I-CreI endonuclease sites. (B) Phase 1: PhiC31-mediated integration using attP sites in the MiMIC and the attB sites of the targeting plasmid replacing the yellow+ marker by recombinase-mediated cassette exchange (RMCE) sequence. (C) Phase 2: two consecutive double-strand breaks, by I-SceI (light green) and I-CreI (dark green) followed by repair through single-strand annealing to remove the unwanted flanking sequences whereas (D) leaving a triple HA-tag in the endogenous LRRK locus. Bold text marks which sites/enzymes/mechanisms are being used. Green colors indicating I-SceI/I-CreI−induced gene targeting, and this color scheme matches that used in the crossing scheme in Figure S1.
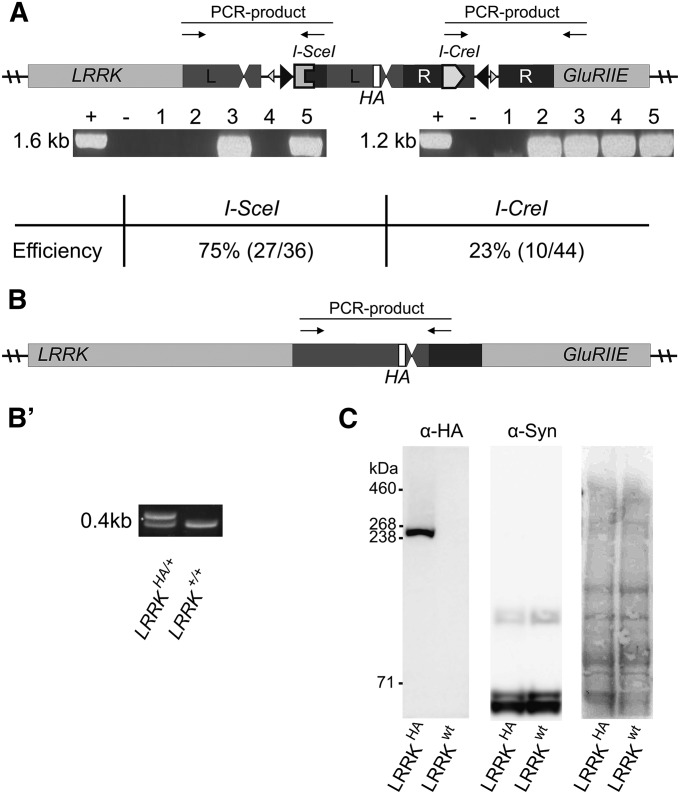
In a second phase we resolve the transposon sequence while leaving an HA tag in the LRRK gene (Figure 1C). In second instar larvae we generated a DSB adjacent to the recombination cassette using I-SceI that is expressed under control of a heat inducible promotor (Figure 1C). Repair of the DSB by recombination between the regions of homology removes the 5′ transposon sequence, while leaving the HA tag in LRRK. To assess the efficiency of this event we screened 36 individual lines by PCR. In 27 of these lines, the transposon sequence was lost and an HA tag was inserted in the LRRK locus (Figure 2A). We also confirmed the presence of the HA tag by sequencing. Next, we used single-strand annealing to remove the remainder of the 3′ MiMIC sequence in one of the HA-tagged LRRK lines and expressed I-CreI under control of a heat inducible promotor (Figure 1C). PCR screening of 44 individual lines revealed that the 5′ MiMIC sequence was removed in 10 lines (Figure 2A), suggesting that in this experiment, I-SceI−mediated single strand annealing was somewhat more efficient than I-CreI−mediated single strand annealing (Rong and Golic 2000; Rong et al. 2002). PCR and sequencing of the locus revealed no aberrations except for the insertion of an HA-tag at the 3′ end of the LRRK open reading frame (Figure 1D and Figure 2B and B′). Hence, these single-strand annealing steps allowed us to efficiently create a knock in allele without leaving any exogenous sequence (e.g., FRT, LoxP, attR…) in the locus (Figure 2B). The detailed crossing scheme we employed is also presented in Figure S1. All LRRKHA lines that we generated were viable, suggesting that the procedure did not induce second-site lethal aberrations.
Figure 2.
LRRKHA is expressed and functional. (A) Efficiency of the two heat-shock-induced endonuclease events (I-SceI and I-CreI) used to induce double-strand breaks, assessed by PCR using primers over the I-SceI site and in LRRK as well as using primers over I-CreI and in GluRIIE. When the I-SceI or I-CreI site is absent, no PCR product can be formed. (B) Schematic representation showing where primers anneal to generate PCR product over the introduced HA-tag and (B′) PCR products, the higher product indicates the presence of the triple HA tag, the lowest band indicates the product without the tag. (C) Western blot using HA antibody showing a 250-kDa large band of LRRKHA and using anti-synapsin antibody as a loading control. Right: Ponceau Red staining of the same blot.
To determine whether LRRK-HA is expressed, we used Western blotting of whole head extract of homozygous LRRKHA animals and probed the blots with anti-HA antibodies. Although we detected a clear band at 250 kDa that corresponded to LRRK-HA, this band was undetectable in control animals (Figure 2C). To further determine whether LRRKHA acts as a wild-type LRRK allele and recapitulates wild-type LRRK function, we also assessed synaptic vesicle endocytosis efficiency and measured the activity of the flies by using an automated monitoring system. Whereas LRRKP1 mutant flies show reduced synaptic vesicle endocytosis and activity defects, LRRKHA knock in animals are indistinguishable from controls (Figure S2, A−C, File S1). Hence, at this level of analysis, synaptic function is not disrupted in LRRKHA animals, and the LRRKHA knock-in allele we generated is functional.
Targeting sky and tau with an RMCE cassette using a MiMIC
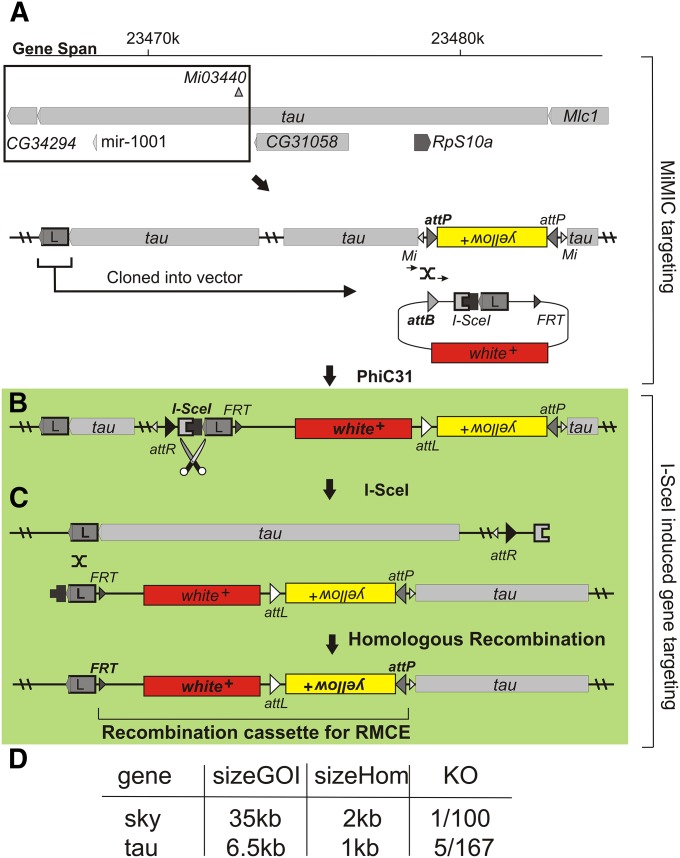
Next we expanded on our methodology and targeted two additional genes, tau and sky. We also adapted our method and replaced these loci with a recombination-mediated-cassette-exchange (RMCE) module. The knock-in of such a module would, in a later phase, facilitate the very efficient integration of multiple modified DNA sequences in these loci. This adapted strategy builds on the methodology we used to create LRRKHA but rather than targeting both attP sites in the MiMIC element to replace the y+ marker, we are now targeting only one attP site to integrate a “targeting plasmid.” This plasmid harbors an I-SceI−flanked 1 or 2 kb homology arm identical to the sequence adjacent to the region we wish to delete, an FRT site and a white+ (w+) eye marker (tau: Figure 3, A and B and sky: Figure S3A). The integration of this plasmid into the MiMIC elements using a genomic source of PhiC31 is very efficient. PCR verification indicates that for the MiMIC in tau, seven integrations were in the correct attP (the one proximal to the region duplicated in the 500 bp homology arm; Figure 3B); seven integrations occurred in both attP sites, and four were in the wrong attP site. Although for the MiMIC in sky, PCR verification indicates seven integrations were in the correct attP; 10 integrations occurred in both attP sites, and two were in the wrong attP site.
Figure 3.
Targeting tau with an recombinase-mediated cassette exchange (RMCE) cassette using a MiMIC element. (A) Targeting one attP site in the MiMIC (Mi{MIC}tauMI03440 harboring a yellow+ marker flanked by attP sites and Minos element arms (Mi)) using PhiC31-mediated integration with the pSV001 plasmid containing 5′- an attB site, an I-SceI restriction site, a homology arm with a stretch of DNA downstream of tau, an FRT site and a w+ marker -3′. (B) I-SceI expression introduces double-strand breaks and repair (green). (C) Repair removes part of tau locus leaving an RMCE cassette that in a later step can be used to replace the locus with different DNA sequences. (D) Efficiencies of our approach when targeting sky and tau genes, size GOI (gene of interest) indicates the size of the deletion generated, size Hom (size homology arm) indicates the length of the used homology arm and KO (knock-out) indicates the amount of deletions per number of flies screened. Bold text marks which sites/enzymes are being used. Green color indicating I-SceI−induced gene targeting and the color scheme matches that used in Figure S4.
In a next step, we expressed I-SceI under a heat-inducible promotor in flies where the “targeting plasmid” is inserted in the correct attP site, thereby inducing a DSB (Figure 3C and Figure S3). The subsequent recombination event deletes the region of interest but leaves an FRT-attP−flanked RMCE cassette marked by the w+ and y+ visible markers. As indicated in Figure 3D and Figure S3B, our methodology to create these alleles of tau and sky was efficient, deleting 35 kb of sky and 6.5 kb of tau (Figure 3D) while replacing the deleted region with an RMCE module that will allow, in a later step, to insert modified genes at these loci. Hence, the combination of the efficient PhiC31-enabled integration of a homology arm in a MiMIC element close to a site of interest, together with restriction endonuclease-induced single strand annealing makes for a very powerful combination that allows us to manipulate and edit the genome at high efficiency over relatively long distances. A detailed crossing scheme is give in supplemental Figure S4.
The MiMIC insertion collection permits to target most genes in the Drosophila genome
To determine how widely applicable this methodology would be, we calculated the number of genes located in regions of increasing size around the MiMIC insertions currently present in the Drosophila genome (Venken et al. 2011) (6507 insertions in release version 2014-06-13). A gene is determined to be in the vicinity of a MiMIC insertion site either by the most proximal nucleotide of the gene to the MiMIC insertion site (1nt in Figure 4) or by the most distal nucleotide of the gene (full gene in Figure 4). Assuming our methodology allows to efficiently bridge 35 kb, we find that 92% of the Drosophila genes are within this vicinity of a MiMIC insertion site (Figure 4). Hence, the methodology we describe can be used as an alternative to existing genome editing methodologies to modulate almost all loci in the fly genome.
Discussion
In this work, we provide proof of concept for a genome editing methodology that does not require homologous recombination by donor dependent repair and attains efficacies that are high enough to screen editing events by PCR using genomic DNA of a reasonable number of individuals. The high efficiency of genome targeting that we achieve stems from (1) the ability to insert a recombination cassette into a MiMIC transposon close to a gene of interest using PhiC31-mediated integration, and (2) resolving genomic sequence flanking the MiMIC insertion site using DSBs induced by I-SceI and/or I-CreI endonucleases. The power of the methodology further rests on the ever expanding collection of MiMIC transposon insertions (Venken et al. 2011) that harbor attP sites that can be used to insert a recombination cassette close to almost any Drosophila gene.
We replaced a region of up to 35 kb by an RMCE cassette or directly targeted a gene and introduced an HA-tag sequence using MiMICs in combination with single strand annealing or one-ended invasion crossover. Using the bridging of 35 kb as a cut off, we find that 92% of the genes are close enough to a MiMIC to be targeted using our strategy. However, in theory, single strand annealing or one-ended invasion crossover is still efficient over a 70-kb region (Wesolowska and Rong 2013) and using 70 kb as a cut off, 97% of all genes should be targetable using our strategy (Figure 4). These calculations indicate that our genome manipulation methodology is able to target most genes in the Drosophila genome. If the starting MiMIC insertion line does not harbor deleterious lesions, our methodology also allows us to edit the genome with very low risk of off-site genomic alterations.
Genome editing requires the insertion of exogenous DNA in the genome. Here, we used the very efficient PhiC31 integrase to insert a recombination cassette close to the locus of interest. Likewise, donor dependent homologous recombination-based methodologies also enable the insertion of exogenous DNA at the target locus, but the classical methodology is inefficient (Rong and Golic 2000; Rong et al. 2002). The recent development of sequence specific nucleases for the creation of DSBs at defined locations in the genome (CRISPR, TALEN, ZFN) (Bibikova et al. 2002; Liu et al. 2012; Gratz et al. 2013) significantly improve on the efficiency of donor dependent repair. Indeed direct editing by CRISPR/Cas9-mediated homologous recombination should permit genome modification in two generations (~1 month) (Figure S5A, dark pink). However there are two drawbacks to consider when using this direct approach: (1) the efficiency of direct targeting may not be high enough such that screening of successful events by PCR is not trivial. Integration of a visible marker would make this screening step easier, but removing this marker in a later step will add several generations of crosses (Figure S5, A and B). (2) When aiming to generate an allelic series, the direct strategy necessitates individual modifications of the genome per allele and will lead to a series of mutants that are not in an isogenic background. This is not an issue when first integrating an attP and creating an isogenic line that is then used to convert the attP site with the different alleles (Figure S5B) or when using the MiMIC methodology we describe. Hence, CRISPR/Cas9-mediated targeting usually starts by integrating attP sites with selection markers and then integrates DNA at these locations using the PhiC31 integrase (Gratz et al. 2013; Gratz et al. 2014). When including this extra “attP step,” CRISPR/Cas9 and the MiMIC-based targeting are rather similar in the time needed to create knock in alleles (compare Figure S4 and Figure S5B).
After initial targeting, both the MiMIC-based method and attP-CRISPR/Cas9 leave exogenous vector and/or marker sequence behind, and we provide a methodology to remove the unwanted vector and marker sequences that are present following the initial targeting event. Similar to the method described in Wesolowska and Rong (2013), we made use of single-strand annealing (or one-ended invasion crossover) to target genes close to an attP site (in our case the attP is provided in the MiMIC), but we postulate that this strategy can also be used when resorting to the attP-mediated CRISPR/Cas9 methodology. There are, however, also some differences. First, the method we describe here to generate an RMCE knock-in allele requires only the cloning of relatively small stretches of DNA (the homology arms); in (Wesolowska and Rong 2013) the entire locus was cloned. Of course, once the gene is targeted with an RMCE cassette and wild type or mutant genes need to be introduced, the entire locus needs to be cloned and manipulated in the methodology we describe here as well. However, these manipulations could be done in parallel with the gene targeting steps, thus saving significant amounts of time.
Second, in the method we describe to generate an RMCE knock-in, the homology arm is the only DNA stretch identical to the endogenous locus and homologous recombination after I-SceI (or I-CreI)−induced DSB can only occur between the homology arm and the endogenous locus, thereby always deleting the DNA between the MiMIC insertion site and the region identical to the homology arm. In Wesolowska and Rong (2013), homologous recombination is possible throughout the length of the construct, and it is thus possible that these recombination events fail to target the gene of interest, lowering efficiency. Hence, our method builds on but is also different from pre-existing single strand annealing methods. In addition, we have prepared a plasmid harboring FRT(F3) and attB sites that allows for RMCE with our RMCE knock-in and this plasmid is available upon request. Recent work (Chan et al. 2013) suggests that these single-strand annealing (or one-ended invasion crossover) steps can be shortened in time as well and the I-SceI and I-CreI steps could potentially be combined into a single step.
Supplementary Material
Acknowledgments
We thank members of the Drosophila gene disruption project for generating MiMIC insertion lines and making these reagents available to the community as well as B. Hassan and members of the Verstreken lab for comments and Bruno André (Université Libre de Bruxelles) for the generous gift of the pFL44S plasmid. This work was supported by a Fonds Wetenschappelijk Onderzoek (FWO) fellowship [81207] to S.V.; a Flanders innovation agency (IWT) fellowship to R.V. [121324] and to I.M. [111352]; a Fundação para a Ciência e a Tecnologia (FCT) fellowship [SFRH/BD/70027/2010] to R.d.C.; Research fund KU Leuven (KUL) [GOA/13/017], CREA [ZKC6335]; European Research Council Starting Grant (ERC StG) [260678]; Fonds Wetenschappelijk Onderzoek (FWO) [G094011N, G095511N, G053913N, G079013N]; the Hercules Foundation [AKUL/09/37, AKUL/11/30]; Interuniversitaire Attractiepolen– Belgian Science Policy Office (IUAP-BELSPO) [P7/16]; and Vlaams Instituut voor Biotechnologie (VIB).
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.014803/-/DC1
Communicating editor: B. J. Andrews
Literature Cited
- Bateman J. R., Lee A. M., Wu C. T., 2006. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173: 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Golic M., Golic K. G., Carroll D., 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161: 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Beumer K., Trautman J. K., Carroll D., 2003. Enhancing gene targeting with designed zinc finger nucleases. Science 300: 764. [DOI] [PubMed] [Google Scholar]
- Bier E., 2005. Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Rev. Genet. 6: 9–23. [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. C., Scoggin S., Hiesinger P. R., Buszczak M., 2012. Combining recombineering and ends-out homologous recombination to systematically characterize Drosophila gene families: Rab GTPases as a case study. Commun. Integr. Biol. 5: 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. S., Huen D. S., Glauert R., Whiteway E., Russell S., 2013. Optimising homing endonuclease gene drive performance in a semi-refractory species: the Drosophila melanogaster experience. PLoS ONE 8: e54130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C. M., Vilain S., Langen M., Van Kelst S., De Geest N., et al. , 2009. Conditional mutagenesis in Drosophila. Science 324: 54. [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase Genome, 2013 Changes affecting gene model number or type in release 5.51 of the annotated D. melanogaster genome. http://flybase.org/reports/FBrf0220804.html.
- Gao G., McMahon C., Chen J., Rong Y. S., 2008. A powerful method combining homologous recombination and site-specific recombination for targeted mutagenesis in Drosophila. Proc. Natl. Acad. Sci. USA 105: 13999–14004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., et al. , 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196: 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., et al. , 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhou W., Dong W., Watson A. M., Hong Y., 2009. From the cover: directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. USA 106: 8284–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li C., Yu Z., Huang P., Wu H., et al. , 2012. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J. Genet. Genomics 39: 209–215. [DOI] [PubMed] [Google Scholar]
- Mali P., Aach J., Stranges P. B., Esvelt K. M., Moosburner M., et al. , 2013. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V., Lin S., Guilinger J. P., Ma E., Doudna J. A., et al. , 2013. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rong Y. S., Titen S. W., Xie H. B., Golic M. M., Bastiani M., et al. , 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16: 1568–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J., Schulze K. L., Haelterman N. A., Pan H., He Y., et al. , 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8: 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng R., Chen Y. W., Bushati N., Cliffe A., Cohen S. M., 2009. Recombinase-mediated cassette exchange provides a versatile platform for gene targeting: knockout of miR-31b. Genetics 183: 399–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesolowska N., Rong Y. S., 2013. Long-range targeted manipulation of the Drosophila genome by site-specific integration and recombinational resolution. Genetics 193: 411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Jaiswal M., Charng W. L., Gambin T., Karaca E., et al. , 2014. A Drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 159: 200–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.