Abstract
The incidences of chronic inflammatory disorders have increased significantly over the past three decades1. Recent shifts in dietary consumption are believed to have contributed importantly to this surge, but how dietary consumption modulates inflammatory disease is poorly defined. Pstpip2cmo mice that express a homozygous L98P missense mutation in the Pombe Cdc15 homology (PCH) family proline-serine-threonine phosphatase interacting protein 2 (PSTPIP2) phosphatase spontaneously develop osteomyelitis that resembles chronic recurrent multifocal osteomyelitis (CRMO) in humans2-4. Recent reports demonstrated osteomyelitis to critically rely on IL-1β, but deletion of the inflammasome components caspase-1 and NLRP3 failed to rescue Pstpip2cmo mice from inflammatory bone disease5,6. Thus, the upstream mechanisms controlling IL-1β production in Pstpip2cmo mice remain to be identified. In addition, the environmental factors driving IL-1β-dependent inflammatory bone erosion are unknown. Here, we show that the intestinal microbiota of diseased Pstpip2cmo mice was characterized by an outgrowth of Prevotella. Notably, Pstpip2cmo mice that were fed a diet rich in fat and cholesterol maintained a normal body weight, but were markedly protected against inflammatory bone disease and bone erosion. Diet-induced protection against osteomyelitis was accompanied by marked reductions in intestinal Prevotella levels and significantly reduced proIL-1β expression in distant neutrophils. Furthermore, proIL-1β expression was also decreased in antibiotics-treated Pstpip2cmo mice, and in wildtype mice that were kept under germfree conditions. We further demonstrated that combined deletion of caspases 1 and 8 was required for protection against IL-1β-dependent inflammatory bone disease, whereas deletion of each caspase alone, elastase or neutrophil proteinase-3 failed to prevent inflammatory disease. Collectively, this work reveals diet-associated changes in the intestinal microbiome as a critical factor regulating inflammasome- and caspase-8-mediated maturation of IL-1β and osteomyelitis in Pstpip2cmo mice.
Keywords: NLR, Pstpip2, inflammasome, IL-1β, caspase-1, caspase-8, RIP3, dysbiosis, autoinflammation, microbiome, neutrophils
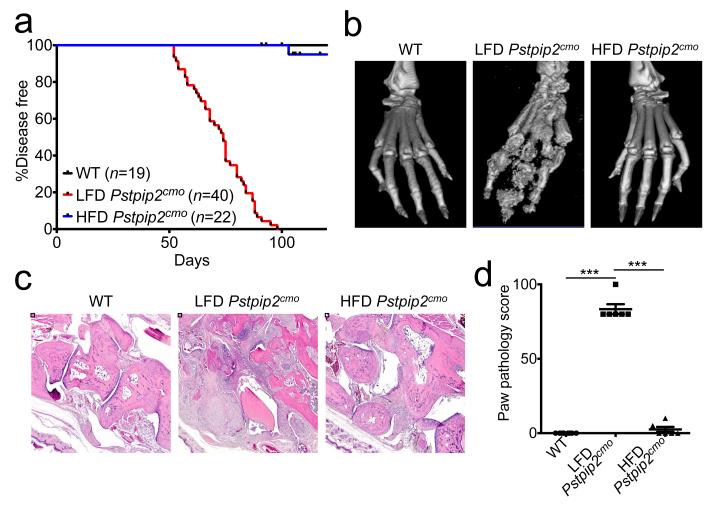
Changes in diet are known to determine susceptibility to common autoimmune diseases such as atherosclerosis, coronary heart disease and type II diabetes7. To address whether dietary intake affects osteomyelitis in Pstpip2cmo mice, a cohort of animals were fed ad libidum a diet rich in high saturated fats and cholesterol (HFD), and disease progression was compared to that of Pstpip2cmo mice placed on a regular lean fat diet (LFD). As expected, all animals on a LFD (n=40) had developed inflammatory bone disease by day 100 (Fig. 1a), as evidenced by the red and swollen appearance of their hind paws (Extended Data Fig. 1a), the significant bone erosion and deformity seen in representative isosurface micro-CT micrographs (Fig. 1b), and the increased size of draining popliteal lymph nodes (Extended Data Fig. 1b). In marked contrast, Pstpip2cmo mice that were fed a HFD (n=22) were largely protected from osteomyelitis, and these mice resembled healthy wild-type (WT) mice in terms of hind paw inflammation, bone erosion, and lymph node size (Fig. 1a,b and Extended Data Fig. 1b). In agreement, haematoxylin & eosin (H&E)-stained sections of the hind paws and tails of Pstpip2cmo mice that were fed a HFD were devoid of infiltrating inflammatory cells and lacked signs of osteolytic bone destruction (Fig. 1c,d and Extended Data Fig. 1c,d). In sharp contrast, Pstpip2cmo mice that were fed a regular LFD diet displayed significant bone destruction and inflammatory cell infiltration in H&E-stained paw (Fig. 1b-d) and tail sections (Extended Data Fig. 1c,d). In agreement, profound reductions in the numbers of infiltrating neutrophils and macrophages were evident in the footpads of HFD-fed Pstpip2cmo mice compared to LFD-fed Pstpip2cmo mice (Extended Data Fig. 1e). Consumption of a HFD was also found to rescue hyperinflammatory cytokine production in Pstpip2cmo mutant mice (Extended Data Fig. 2a,b). As expected for mice on a BALB/cJ genetic background8, Pstpip2cmo mice retained a normal body weight during these studies, regardless of whether they were fed a LFD or HFD (Extended Data Fig. 3a,b). Collectively, these observations demonstrate that the dietary composition determines to a large extent whether genetically susceptible Pstpip2cmo mice develop osteomyelitis independently of gross changes in body weight.
Figure 1. Changes in diet limit the development of inflammatory bone disease in Pstpip2cmo mutant mice.
a-d, Wild-type (WT) and Pstpip2cmo mutant mice were fed a lean fat diet (LFD) or a high fat and cholesterol diet (HFD). a, Incidence of inflammatory bone disease. b-d, Representative isosurface micro-CT paw scans (b), H&E sections (c) and pathology scores (d) for hind paw samples from 12-14 week old WT, LFD Pstpip2cmo and HFD Pstpip2cmo mice. Each point represents an individual mouse, and the line represents the mean ± s.e.m. (***P < 0.001; Student’s t-test)
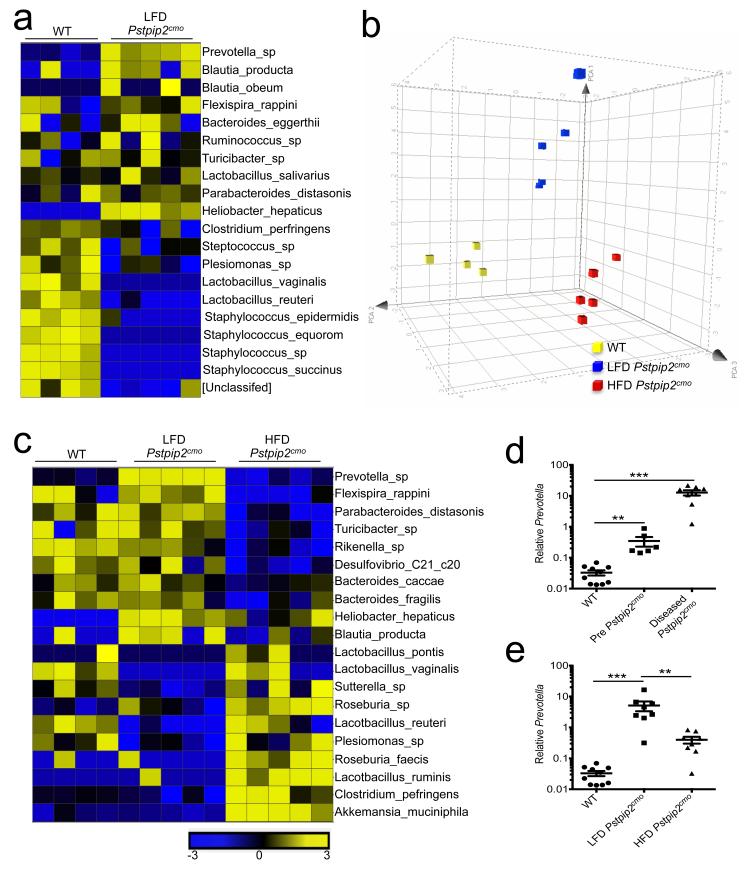
Diets high in fat and cholesterol induce large-scale changes in the host microbiota composition9,10. We made use of 16S ribosomal RNA coding gene (rDNA) metagenomic sequencing to address whether inflammatory bone disease in Pstpip2cmo mice was associated with intestinal dysbiosis that was rescued by a HFD regimen. The commensal intestinal ecology of Pstpip2cmo mice that were fed a regular LFD was markedly different from the microbiota of healthy age- and sex-matched WT mice (Fig. 2a). Notable alterations included the outgrowth of Prevotella and concomitant reductions in Lactobacillus genera in LFD-fed Pstpip2cmo mice (Fig. 2a). A HFD regimen induced remarkable changes in the colonic microbiota that was characterized by a suppression of disease-associated commensals (Fig. 2b,c). Most strikingly, LFD-fed Pstpip2cmo mice displayed a time-dependent increase in Prevotella levels (Fig. 2d), which was significantly reduced in Pstpip2cmo mice that were kept on a HFD (Fig. 2e). The latter group was further characterized by an expansion of Lactobacillus species in their intestinal tract (Fig. 2c). Diet-induced changes in the microbiota composition were not accompanied by readily detectable intestinal inflammation (Extended Data Fig. 3c-e). Moreover, we failed to detect bacteria in the peripheral organs of LFD-fed Pstpip2cmo mice (Extended Data Fig. 3f). Together, these results show that inflammatory bone disease in Pstpip2cmo mice is specifically characterized by an outgrowth of inflammation-associated intestinal commensals, which is suppressed by a HFD regimen.
Figure 2. Alterations in commensal microbiota landscape that are associated with Pstpip2cmo-mediated osteomyelitic disease can be modified by changes in diet.
a-c, Fecal samples were collected from WT, LFD Pstpip2cmo and HFD Pstpip2cmo mice at 10-12 weeks of age and 16S rRNA metagenomic sequencing was conducted. a, Heat map of fold differences in relative abundance of commensal bacteria. b, Principal coordinated analysis (PCoA) plot of fecal microbiota. c, Heat map of the top 20 commensal genera and species that differ between LFD Pstpip2cmo and HFD Pstpip2cmo mice are presented. d, Prevotella 16S rDNA copy numbers in WT and Pstpip2cmo mice before (pre-disease: 3-6 weeks of age) and after (diseased: 10-16 weeks of age) the development of osteomyelitis. Each point represents an individual mouse, and the line represents the mean ± s.e.m. Data are representative of four independent experiments. e, 16S rDNA analysis of Prevotella abundance. Data are representative of four independent experiments. (**P < 0.01, ***P < 0.001; Student’s t-test)
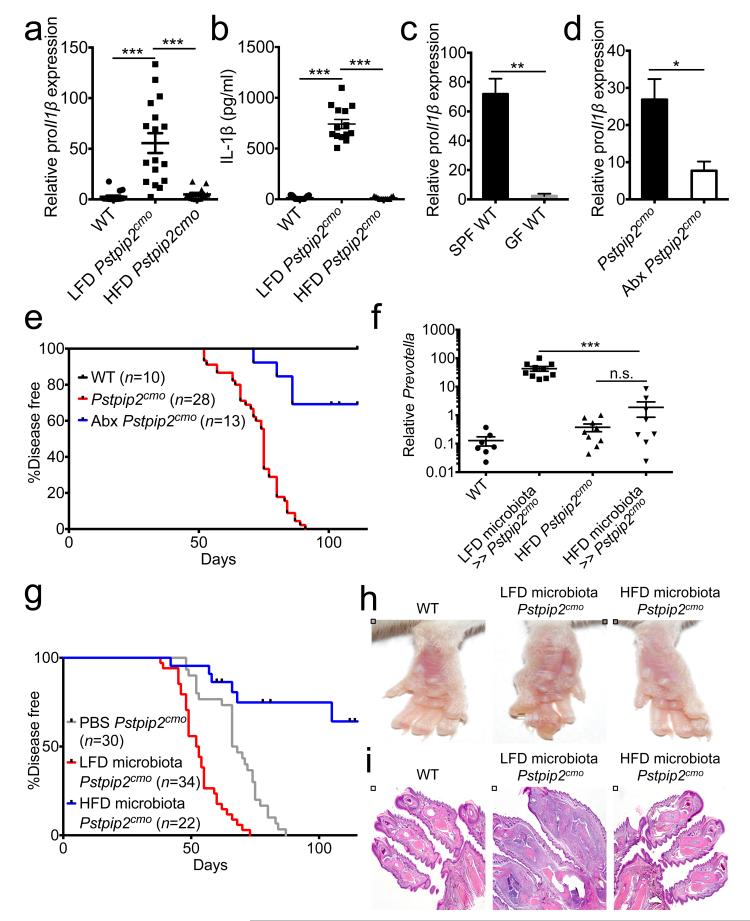
We and others have previously shown that inflammatory bone disease in Pstpip2cmo mice critically relies on IL-1β5,6. Given that Pstpip2cmo mice on a HFD were markedly resistant to disease progression, we addressed whether HFD dampened IL-1β levels. Pstpip2cmo mice that were fed a LFD had proIl1β mRNA levels that were on average 60-fold higher than in footpads of healthy WT mice (Fig. 3a). In sharp contrast, HFD-fed Pstpip2cmo mice had markedly suppressed local proIl1β transcript levels that were comparable to those of healthy WT mice (Fig. 3a). In agreement with these observations, IL-1β protein concentrations were significantly increased in the footpads of LFD-fed Pstpip2cmo mice, whereas those of HFD-fed Pstpip2cmo mice were comparable to healthy controls (Fig. 3b and Extended Data Fig. 4a). Together, this suggests that HFD suppressed osteomyelitis in Pstpip2cmo mice by dampening proIL-1β expression.
Figure 3. Microbiota-mediated regulation of IL-1β expression shapes inflammatory bone disease.
a, qRT-PCR analysis of relative proIl1β expression in the footpads of 12-16 week old WT, LFD Pstpip2cmo and HFD Pstpip2cmo mice. Each point represents an individual mouse, and the line represents the mean ± s.e.m. Combined data from three independent experiments. b, Protein levels of IL-1β in the hind paws. Combined data from two independent experiments. c, Relative proIl1β mRNA expression levels in CD45+ cells isolated from the colons of specific pathogen free (SPF) and germ-free (GF) WT mice. Two biological replicates, with two technical replicates each. d-e, Pstpip2cmo mice were treated with a cocktail of broad-spectrum antibiotics in their drinking water (Abx). d, qRT-PCR analysis of colonic Il1β expression levels from 12-14 week old Pstpip2cmo mice that received either regular drinking water (n=15) or Abx water (n=9). e, Incidence of inflammatory bone disease. f-i, Young Pstpip2cmo mice (3 wks old) received PBS or fecal microbiota from diseased LFD Pstpip2cmo or disease-free HFD Pstpip2cmo mice by oral transplantation. f, 16S rDNA analysis of Prevotella copy numbers. g, Incidence of inflammatory bone disease. h-i, Representative footpad images (h) and H&E micrographs (i). (*P < 0.05, **P < 0.01, ***P < 0.001; Student’s t-test)
Given that HFD skewed the intestinal microbiota composition of Pstpip2cmo mice (Fig. 2), we next asked whether the microbiota controlled proIl1β expression. We found that proIl1β levels in CD45+ cells that were isolated from the colons of germ-free (GF) WT mice were significantly reduced when compared to those of WT that were kept under specific pathogen-free (SPF) conditions (Fig. 3c). Moreover, the levels of proIL-1β protein were appreciably diminished in the hind paws of GF WT mice (Extended Data Fig. 4b). However, the expression of Il1β mRNA by CD45.2+ cells isolated from GF mice was greatly enhanced following in vitro stimulation with LPS suggesting that these GF mice do not have any intrinsic defects in Il1β mRNA expression (Extended Data Fig. 4c). Notably, broad-spectrum antibiotics that significantly reduced Prevotella and Flexispira levels in LFD-fed Pstpip2cmo mice (Extended Data Fig. 5a), also substantially decreased the levels of colonic proIl1β in these mice (Fig. 3d). In addition, broad-spectrum antibiotics significantly protected LFD-fed Pstpip2cmo mice from developing osteomyelitis (Fig. 3e). To further address the role of the intestinal microbiota, we performed fecal microbiota transplantation studies. Transplantation of the microbiota of diseased Pstpip2cmo mice into WT mice failed to cause disease (Extended Data Fig. 5b). Similarly, LFD-fed Pstpip2cmo mice also failed to transfer disease to co-housed WT and Il1β-deficient Pstpip2cmo mice (Extended Data Fig. 5c,d). However, transplantation of the fecal microbiota of diseased (LFD-fed) Pstpip2cmo mice to young LFD-fed Pstpip2cmo mice by oral gavage promoted the expansion of Prevotella (Fig. 3f), and significantly accelerated the development of osteomyelitis relative to PBS-operated controls (Fig. 3g-i). Conversely, transplanting the microbiota of HFD-fed Pstpip2cmo mice into young LFD-fed Pstpip2cmo mice greatly limited Prevotella outgrowth (Fig. 3f), and significantly protected mice from developing osteomyelitis (Fig. 3g-i). Although re-derivation of Pstpip2cmo mice under germ-free conditions is needed to provide conclusive proof that commensal-derived factors are required to promote inflammatory bone disease, our findings clearly support the notion that diet-induced modulation of the microbiota composition regulates proIL-1β expression and osteomyelitis development in disease-susceptible Pstpip2cmo mice.
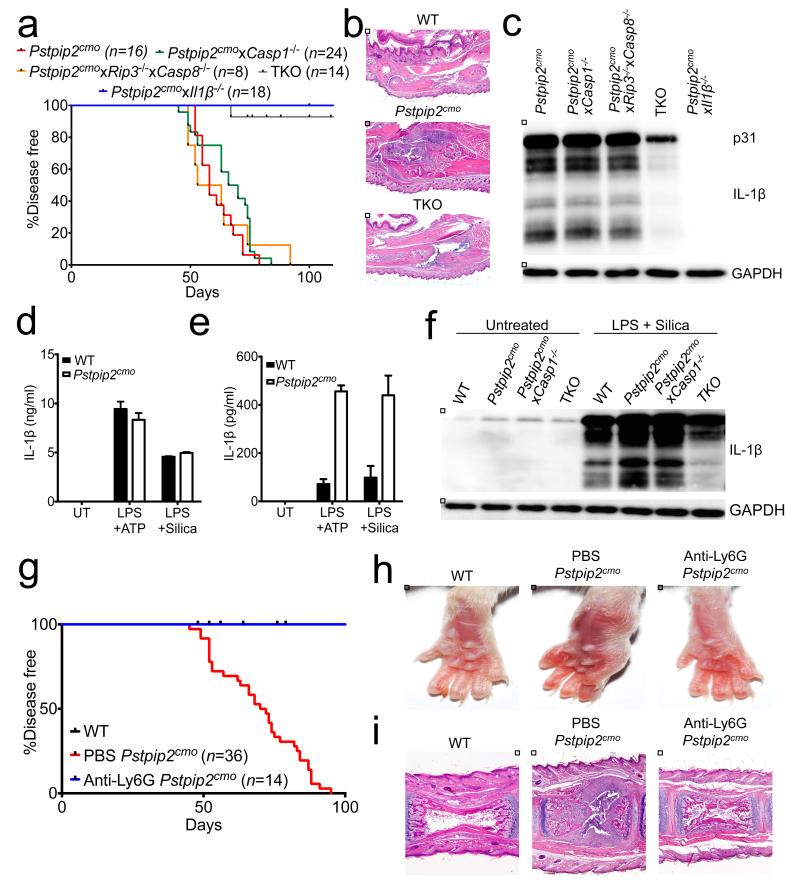
ProIL-1β is produced as a biologically inactive molecule that resides in the cytosol and needs to be proteolytically converted into mature IL-1β to gain biological activity. Caspase-1, a protease that is activated by inflammasome complexes, is the principal protease responsible for IL-1β maturation11. Neutrophil proteinase 3, elastase and caspase-8 were recently shown to also convert proIL-1β into its bioactive form12-18. Genetic deletion of caspase-1 and the related protease caspase-11 failed to rescue Pstpip2cmo mice from inflammatory bone disease5,6. We therefore addressed the role of additional proteases in IL-1β-dependent osteomyelitis. To this end, Pstpip2cmo mice were bred onto mice with gene-targeted deletions in neutrophil proteinase 3 and elastase. However, deletion of neither neutrophil proteinase 3 nor elastase rescued or delayed inflammatory bone disease in Pstpip2cmo mice (Extended Data Fig. 6a,b). We next sought to examine the role of caspase-8 in Pstpip2cmo-associated osteomyelitis. Mice deficient in caspase-8 are embryonic lethal19-21, and this lethality is rescued by further deleting the necroptosis-regulating kinase RIP322,23. We thus bred Casp8−/−Rip3−/− mice onto Pstpip2cmo mice. Caspase-8 may act redundantly with caspase-1 in proIL-1β conversion under particular conditions12-14,16, which we addressed by further deleting caspase-1 in Casp8/Rip3-deficient Pstpip2cmo mice. As expected, Pstpip2cmo mice gradually developed inflammatory bone disease, with all mice being afflicted by 80 days (Fig. 4a). As reported5,6, Pstpip2cmo mice lacking IL-1β were fully resistant to osteomyelitis development (Fig. 4a). Rip3-deficient, Casp1-deficient and Casp8/Rip3-deficient Pstpip2cmo mice developed osteomyelitis with similar kinetics as Pstpip2cmo mice (Fig. 4a and Extended Data Fig. 7a), which was also reflected in the extent of bone erosion and histopathology seen in these mice (Extended Data Fig. 7b,c). Remarkably, combined deletion of caspases 1 and 8 provided significant protection against osteomyelitic disease (Fig. 4a,b). In agreement, proIL-1β expression levels were reduced and IL-1β maturation was virtually blunted in footpads of Pstpip2cmo mice lacking both caspases (Fig. 4c and Extended Data Fig. 7d). In marked contrast, we observed spontaneous IL-1β maturation in footpads of Pstpip2cmo mice, as well as in mice lacking either caspases 1 or 8 (Fig. 4c).
Figure 4. Compensatory processing of IL-1β by caspase-1 and caspase-8 in neutrophils drives inflammatory bone disease.
a, Incidence of osteomyelitic disease in Pstpip2cmo, Pstpip2cmoxCasp1−/−, Pstpip2cmoxRip3−/−xCasp8−/−, Pstpip2cmoxCasp1−/−xRip3−/−xCasp8−/− (TKO) and Pstpip2cmoxIl1β−/− mice over time. b, Representative H&E staining of hind paw sections. c, Western blot analysis of IL-1β regulation in the footpads of 10-12 week old Pstpip2cmo, Pstpip2cmoxCasp1−/−, Pstpip2cmoxRip3−/−xCasp8−/−, Pstpip2cmoxCasp1−/−xRip3−/−xCasp8−/− (TKO) and Pstpip2cmoxIl1β−/− mice. d-f, WT and Pstpip2cmo bone marrow-derived macrophages (d) or neutrophils (e-f) were left untreated (UT) or were first primed with LPS for 3 hrs followed by stimulation with ATP (30 mins) or silica (12 hrs). d-e, Secretion of IL-1β was measured by ELISA. Bar graphs depict mean ± s.e.m. One out of three biological replicates, with two-three technical replicates each. f, Western blot analysis of IL-1β in WT, Pstpip2cmo, Pstpip2cmoxCasp1−/− and Pstpip2cmoxCasp1−/−xRip3−/−xCasp8−/− (TKO) neutrophils. g-i, WT and Pstpip2cmo mice received either PBS or anti-Ly6G antibody every 4-5 days starting at 6 weeks of age. g, Incidence of inflammatory bone disease. h-i, Representative footpad images (h) and tail H&E micrographs (i).
Pstpip2cmo hematopoietic cells were recently shown to be sufficient to induce osteomyelitis in WT donor mice5, suggesting that bone marrow-derived cell populations are likely responsible for aberrant IL-1β production in Pstpip2cmo mice. We first evaluated the production of IL-1β by macrophages and neutrophils because these are the predominant immune cell types found in active osteoinflammatory lesions (Extended Data Fig. 1e). As reported6, stimulation of LPS-primed Pstpip2cmo macrophages with NLRP3 inflammasome triggers such as ATP and silica triggered normal levels of secreted IL-1β (Fig. 4d and Extended Data Fig. 8a). In contrast, levels of IL-1β secreted by Pstpip2cmo neutrophils that were stimulated with these agents were at least four-fold higher than those of WT cells (Fig. 4e,f). Importantly, neutrophils of HFD-fed Pstpip2cmo mice expressed less proIL-1β (Extended Data Fig. 8b), and IL-1β maturation was markedly affected when compared to neutrophils of LFD-fed Pstpip2cmo mice (Extended Data Fig. 8c). In contrast, proIL-1β production and IL-1β maturation were not significantly different in macrophages of LFD- and HFD-fed Pstpip2cmo mice (Extended Data Fig. 8d). To further ascertain the role of neutrophils in IL-1β-dependent osteomyelitis, Pstpip2cmo mice were treated with anti-Ly6G antibodies to deplete neutrophils. Anti-Ly6G treatment led to marked reductions in circulating neutrophil counts (Extended Data Fig. 9a-c). Importantly, neutrophil ablation conferred significant protection from clinical disease progression (Fig. 4g,h) and histopathological tissue damage (Fig. 4i).
Collectively, our findings presented here show that dietary intake determines the composition of the intestinal microbiota, and greatly influences disease outcome in osteomyelitis-susceptible Pstpip2cmo mice by upregulating proIL-1β levels. We further show that activation of caspases 1 and 8 in these mice result in spontaneous induction of IL-1β-driven neutrophilic osteomyelitis in Pstpip2cmo mice (Extended Data Fig. 10). These results suggest that diet-induced changes in the intestinal microbiota composition may promote autoinflammatory disease in susceptible individuals by increasing proIL-1β levels available for conversion by caspases 1 and 8.
Methods
Mice
Pstpip2cmo3, Il1β−/−24, Casp1−/−25, Casp8−/−26, Rip3−/−27, Elastase−/−28 and NePr3−/−29 mice were previously described. Pstpip2cmo were purchased from The Jackson Laboratory and are on the BALB/c background. All other mutant mice are on the C57BL/6 background. To generate the necessary controls and experimental mice for these experiments, mice that were heterozygous for both the Pstpip2 and KO allele(s) were used as breeders. Littermate controls were utilized to evaluate whether genetic deletions influence immune responses, IL-1β regulation and osteomyelitic disease development. Germ-free mice were obtained from Taconic. All mice were kept in specific pathogen-free conditions within the Animal Resource Center at St. Jude Children’s Research Hospital. Animal studies were conducted under protocols approved by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital.
Diet
Feed that was high in fat and cholesterol was purchased from Research Diets Incorporated (Stock number: D12107) and consisted of 40% fat and 0.5% cholesterol. Standard low fat diet was obtained from LabDiet (Stock number: 5013) and consisted of 5% fat and 0% cholesterol.
Histopathology
Formalin-preserved paws and tails were processed and embedded in paraffin according to standard procedures. Haematoxylin and eosin (H&E) sections (5 μm) were examined by a pathologist blinded to the experimental groups. Tail and paw H&E sections were scored based on the extent and severity of inflammation, pyogranulomatous, osteolysis, and osteogenesis in a blinded fashion by a veterinary pathologist.
Micro-CT
Micro-CT micrographs of paws and tails fixed in formalin were made using an ex vivo micro-CT scanner (LocusSP Specimen CT, GE Healthcare) at 28-μm isotropic voxel size, with 720 projections, an integration time of 1,700 msec, photon energy of 80 keV, and a current of 70 μA.
16S rRNA microbiome analysis
Fifty nanograms of purified DNA was prepared using Nextlex 16s v4 Amplicon-seq kit according to the manufacturer's instructions (Bioo Scientific Austin, TX.). Briefly, PCR primers targeted the fourth hypervariable domain of microbial 16s ribosomal RNA genes and simultaneously introduced sequences required for sequencing demultiplexing. Ampure XP PCR purification was used to clean up the PCR reactions (Beckman Coulter Brea, CA.) PCR products were quantified using the Quant-iT PicoGreen assay (Invitrogen Carlsbad, CA), normalized, and pooled. Pooled samples were sequenced on a MiSeq sequencer (Illumina San Diego, CA) according to manufacturer's instructions with modifications specified in the Nextflex 16s v4 kit. The 16S primers targeting the V4 region were aligned to the full set of sequences from the Greengenes database v13.5 using exonerate. Each sequence was truncated to include only the V4 region, the primer-matching regions, and an additional 40 bases on either side. Duplicate V4 regions were removed from the dataset. All taxa labels from the removed duplicates were associated with the remaining representative V4 region sequence. Reads from each sample were aligned exhaustively to the non-redundant V4 sequences using USEARCH allowing a minimum sequence identity of 90%. All taxon labels associated with the top-scoring V4 region(s) were used to determine the taxon assignment of each read. The highest resolution non-conflicting taxon from all taxa associated with the top-scoring V4 region was assigned as the taxa for a read.
Relative proportions (p) of microbial taxa for each sample were assembled from the highest resolution sequence counts into a matrix with samples as columns and taxa as rows with proportions in cells. Columns were also assigned to a WT/KO group according to the design. All relative proportions are transformed to near normality with a shifted logit-p transformation.
Unpaired T-test with unequal variance for the normalized proportions (Ptransformed) and a two-factor ANOVA model was used to investigate significant taxa. The transformed values are then normalized for each taxa to produce a signal to noise ratio (SNR).
Signal to noise ratios are depicted in heatmap plots and PCA analyses generated with spotfire.
ELISA
Paw samples were snap frozen in liquid nitrogen and protein lysates were generated in RIPA lysis buffer supplemented with complete protease inhibitor cocktail (Roche) and PhosSTOP (Roche) using a tissue homogenizer. Debris was pelleted and the supernatants were assessed via ELISA according to manufacturers’ instructions (Milliplex and eBioscience).
Real-time PCR
Hind paw samples were snap frozen in liquid nitrogen and stored at −80°C for later use. Tissue was homogenized in Trizol using a tissue homogenizer. Total RNA was isolated from the hind paws with Trizol (Invitrogen) according to the manufacturer’s instructions. Briefly, 200μl of chloroform was added to the 1 ml of Trizol tissue lysate and the samples were incubated at room temperate for 5 mins following vortexing. After centrifugation, the aqueous phase was transferred to a new tube and equal volumes of isopropanol were added. After incubation at room temperature for 10 mins, the RNA was pelleted by centrifugation and then the RNA was washed two times in 70% ethanol before resuspension in ultrapure water. 1 μg of RNA was reverse-transcribed to cDNA with random RNA-specific primers using the high-capacity cDNA reverse transcription kit (Applied Biosystems). Transcript levels of Il1β, KC, Il6, 16S Prevotella, 16S universal bacteria, βactin and Gapdh were analyzed using SYBR-Green (Applied Biosystems) on an ABI7500 real-time PCR machine according to the manufacturers’ recommendations. Relative expression was calculated using the delta-delta Ct standardization method.
Commensal bacteria depletion
Mice were treated with a broad-spectrum antibiotics regimen that contained 125 mg/L ciprofloxacin, 1 g/L bacitracin, 2 g/L streptomycin, 1.5 g/L metronidazole and 172 mg/L gentamycin in their drinking water.
In vitro macrophage stimulation
Bone marrow derived macrophages (BMDMs) were generated by culturing bone marrow cells in L-cell-conditioned IMDM medium supplemented with 10% FBS, 1% nonessential amino acid, and 1% penicillin-streptomycin for 5 days. BMDMs were seeded in 12-well cell culture plates and cultured overnight. To evaluate IL-1β production, BMDMs were primed with 2 μg/ml ultrapure Escherichia coli-derived LPS (Invivogen) for 3 hours followed by 5 mM ATP (Sigma-Aldrich) for an additional 30 min. To measure IL-1β processing and production in response to stimulation with LPS+silica, cells were first primed with 2 μg/ml ultrapure Escherichia coli-derived LPS (Invivogen) for 3 hours and then were further activated with 500 μg/ml Min-U-Sil-5 silica (US Silica) for an additional 5-12 hours.
Neutrophil isolation and stimulation
Bone marrow cells were flushed from the femurs and tibias. Total bone marrow cells were passed through a 70μm cell strainer and purified neutrophils were isolated from the interface of a 62.5% Percoll (GE Healthcare) gradient.
Western blotting
Hind paw protein lysates were collected in RIPA lysis buffer supplemented with complete protease inhibitor cocktail (Roche) and PhosSTOP (Roche) using a tissue homogenizer. Samples were clarified with at least two centrifugation steps to remove cellular debris. Lysates were resolved by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes via electroblotting. Membranes were blocked in 5% non-fat milk and incubated overnight at 4°C with primary antibodies. The following primary antibodies were used in our studies: anti-IL-1β clone D3H1Z (Cell Signaling) and anti-GAPDH (Cell Signaling). The membranes were then probed with horseradish peroxidase (HRP)-tagged secondary antibodies at room temperature for 1 hr. Immunoreactive proteins were visualized using the ECL method (Pierce).
Fecal transplantation
Fresh fecal samples were obtained from LFD-fed or HFD-fed Pstpip2cmo mice and pellets were homogenized in PBS. Debris was pelleted by microcentrifugation and commensal bacteria was transplanted into young Pstpip2cmo mice by oral transplantation every 2-4 days. Fecal reconstitution was confirmed by evaluating the intestinal abundance of Prevotella by 16S rDNA analysis in fecal microbiota transplantation mice 4-8 weeks later.
Neutrophil depletion
WT and Pstpip2cmo mice received either PBS or 500 μg/mouse anti-Ly6G antibody (clone IA8) by intraperitoneal injection every 4-5 days starting at 6 weeks of age and the incidence of inflammatory bone disease was evaluated over time. Depletion of neutrophils was confirmed by FACs staining for CD45.2+CD11b+Gr-1+ cells in the peripheral blood.
Statistical analysis
All results are presented as means ± standard errors. We performed statistical analysis using the two-tailed Student’s t-test. P values are denoted by *P<0.05, **P<0.01, ***P<0.001, ****P < 0.0001.
Extended Data
Extended Data Figure 1. Placing Pstpip2cmo mice on a high fat and cholesterol diet limits the development of inflammatory bone disease.
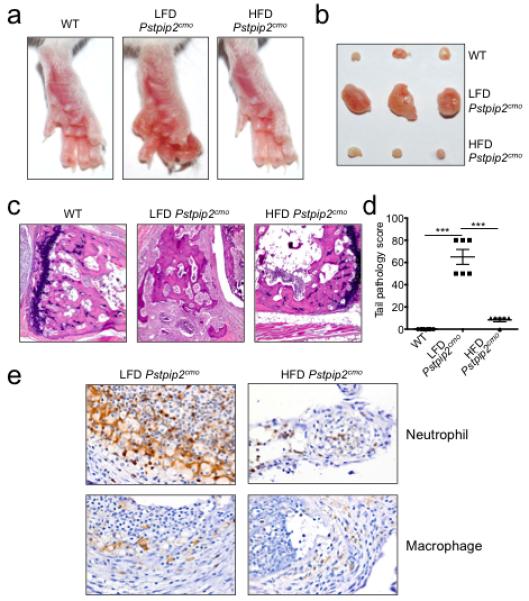
a-e, Wild-type (WT) and Pstpip2cmo mutant mice were fed a lean fat diet (LFD) or a high fat and cholesterol diet (HFD). Representative hind paw images (a) and representative pictures of popliteal lymph nodes (b) from WT, LFD Pstpip2cmo and HFD Pstpip2cmo mice at 12-14 weeks of age. c-d, Hematoxylin and eosin (H&E) staining (c) and pathology scores (d) of tail samples from 12-14 week old WT, LFD Pstpip2cmo and HFD Pstpip2cmo mice. Pathology scores were assigned in a blinded fashion by a veterinary pathologist based on the extent and severity of inflammation, osteolysis and osteogenesis. e, Representative immunostaining of neutrophils and macrophages in hind paw sections from 14-18 weeks old Pstpip2cmo mice that were fed either a LFD or a HFD. (***P < 0.001; Student’s t-test)
Extended Data Figure 2. Consumption of a HFD limits hyperinflammatory cytokine production in Pstpip2cmo mutant mice.
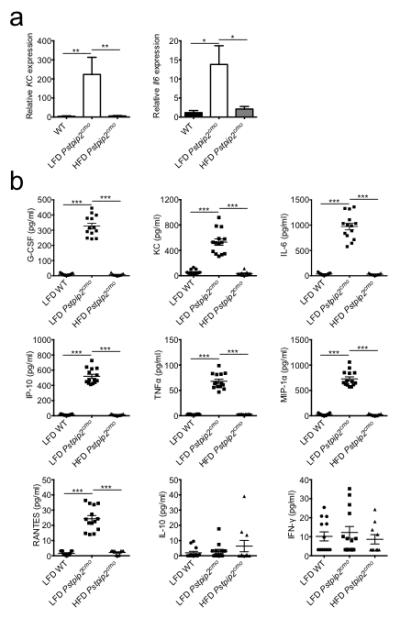
a, WT and Pstpip2cmo mutant mice were fed a lean fat diet (LFD) or a high fat diet (HFD) for 12 weeks. Relative expression of KC (WT n=8; LFD Pstpip2cmo n=4; HFD Pstpip2cmo n=9) and Il6 (WT n=11; LFD Pstpip2cmo n=10; HFD Pstpip2cmo n=8) in the hind paws was determined by qRT-PCR. The bar graphs depict combined data from two independent experiments. Data is shown as mean ± s.e.m. b, WT and Pstpip2cmo mutant mice were fed a LFD or a HFD for 12 weeks and cytokines levels in the hind paws were determined by ELISA. Combined data from two independent experiments. Each point represents an individual mouse, and the line represents the mean ± s.e.m. (*P < 0.05, **P < 0.01, ***P < 0.001; Student’s t-test)
Extended Data Figure 3. Placing Pstpip2cmo mice on a HFD does not cause abnormal weight gain, intestinal inflammation or extraintestinal translocation of commensal bacteria.
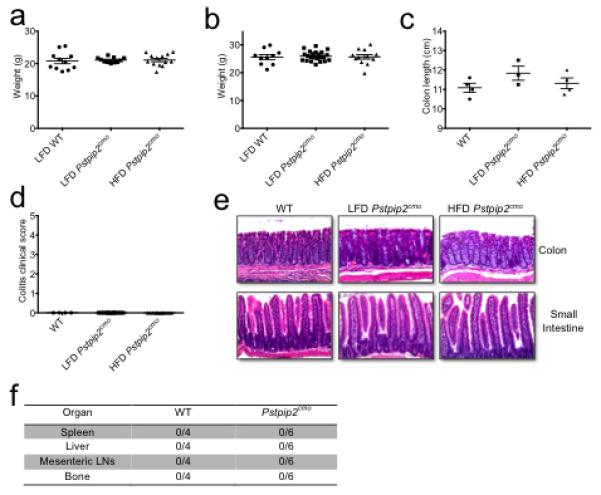
a-b, WT BALB/cJ and Pstpip2cmo mice were fed ad libidum a LFD or a HFD. Body weight was measured in age-matched female (a) and male (b) mice at 12-16 weeks of age. Each point represents an individual mouse and the line represents the mean ± s.e.m. Data were combined from three independent experiments. c-e, Colon length (c), colitis score based on rectal bleeding and stool consistency (d) and representative haematoxylin & eosin (H&E)-stained sections (e) of the intestinal tract of LFD- and HFD-fed Pstpip2cmo mice aged 14-18 weeks. f, Presence of commensal bacteria in the spleen, liver, mesenteric lymph nodes and bone of WT and diseased LFD-fed Pstpip2cmo mice was evaluated by Gram staining and 16S rDNA qPCR analysis of eubacteria.
Extended Data Figure 4. Dietary- and microbiota-associated factors influence the production of proIL-1β.
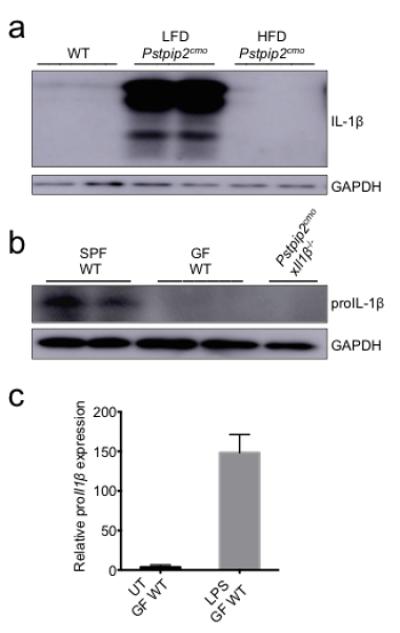
a, Footpad homogenates of 12-16 weeks old WT, LFD-fed Pstpip2cmo and HFD-fed Pstpip2cmo mice were immunoblotted for IL-1β. Data are representative of three independent experiments. b, Footpads samples were harvested from 10-14 week old specific pathogen free (SPF) WT, germ-free (GF) WT and Pstpip2cmoxIl1β−/− mice and proIL-1β protein levels were determined by Western blotting. c, CD45+ cells were isolated from the colons of GF WT mice and cells were left untreated (UT) or stimulated with LPS for 1 hr. Relative proIl1β mRNA expression levels were determined by qRT-PCR. Two biological replicates, with two technical replicates each.
Extended Data Figure 5. Co-housing does not alter disease progression of LFD-fed Pstpip2cmo mice.
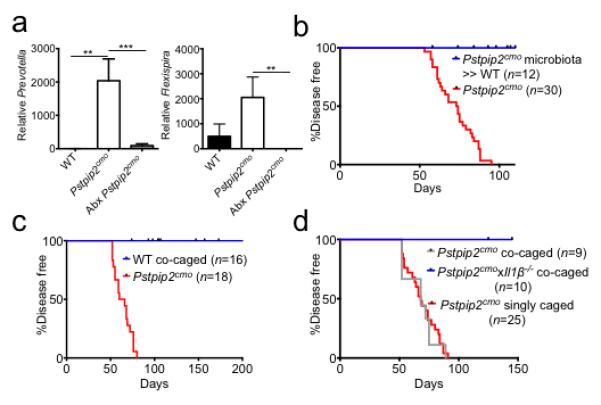
a, Pstpip2cmo mice were treated with a cocktail of broad-spectrum antibiotics in their drinking water (Abx). Fecal samples were collected from WT (n=5) and Pstpip2cmo mice that received either regular drinking water (n=5) or Abx water (n=11) 5-7 weeks later. Prevotella and Flexispira 16S rDNA copy numbers were quantified and normalized to total bacteria. The bar graphs depict the mean ± s.e.m. b, Fecal microbiota from diseased Pstpip2cmo mice was orally transplanted into WT mice (Pstpip2cmo microbiota » WT) and the incidence of inflammatory bone disease in control Pstpip2cmo and fecal transplantation mice was evaluated. c-d, Pstpip2cmo mice were singly housed or co-housed with WT (c) or Il1β-deficient Pstpip2cmo (d) mice. Clinical development of bone deformity and arthritic inflammation in hind paws and tails was monitored over time. (**P < 0.01, ***P < 0.001; Student’s t-test)
Extended Data Figure 6. The neutrophil associated proteases elastase and proteinase-3 are not required for Pstpip2cmo-mediated bone disease.
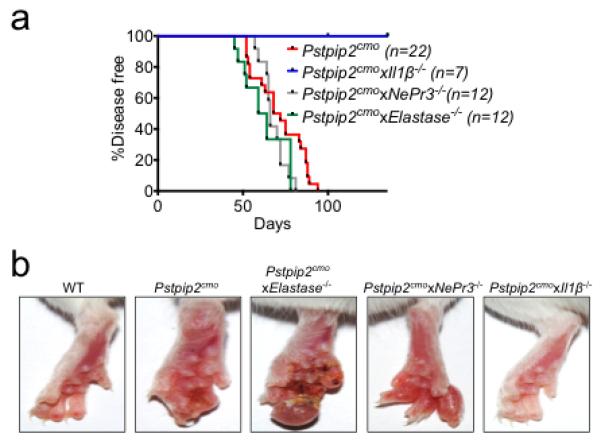
a, Incidence of inflammatory bone disease in Pstpip2cmo, Pstpip2cmoxElastase−/−, Pstpip2cmoxNePr3−/− and Pstpip2cmoxIl1β−/− mice. b, Representative footpad images from WT, Pstpip2cmo, Pstpip2cmoxElastase−/−, Pstpip2cmoxNePr3−/− and Pstpip2cmoxIl1β−/− mice.
Extended Data Figure 7. Combined deletion of RIP3 and Caspase-8 does not provide protection against Pstpip2cmo-mediated osteomyelitis.
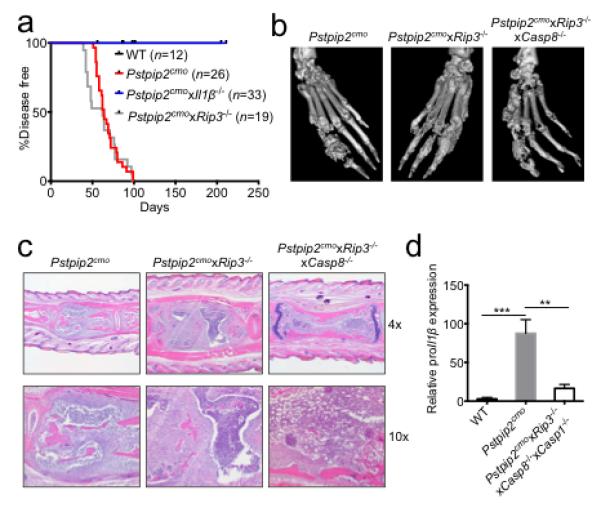
a, Incidence of osteomyelitic bone disease in WT, Pstpip2cmo, Pstpip2cmoxIl1β−/− and Pstpip2cmoxRip3−/− mice. b, Representative isosurface micro-computed tomography (micro-CT) images of hind paw samples from 12-18 week old Pstpip2cmo, Pstpip2cmoxRip3−/− and Pstpip2cmoxRip3−/−xCasp8−/− mice. c, Representative 4x and 10x H&E sections of inflammatory caudal vertebrae bone lesions in diseased Pstpip2cmo, Pstpip2cmoxRip3−/− and Pstpip2cmoxRip3−/−xCasp8−/− mice. d, qRT-PCR analysis of proIl1β expression in footpads of WT (n=7), Pstpip2cmo (n=7) and Pstpip2cmoxRip3−/−xCasp8−/−xCasp1−/− (n=7) mice aged 12-16 weeks. Data are expressed as mean ± s.e.m. of combined data from two independent experiments. (**P < 0.01, ***P < 0.001; Student’s t-test)
Extended Data Figure 8. Reduced proIL-1β expression and IL-1β maturation in neutrophils isolated from HFD-fed Pstpip2cmo mice.
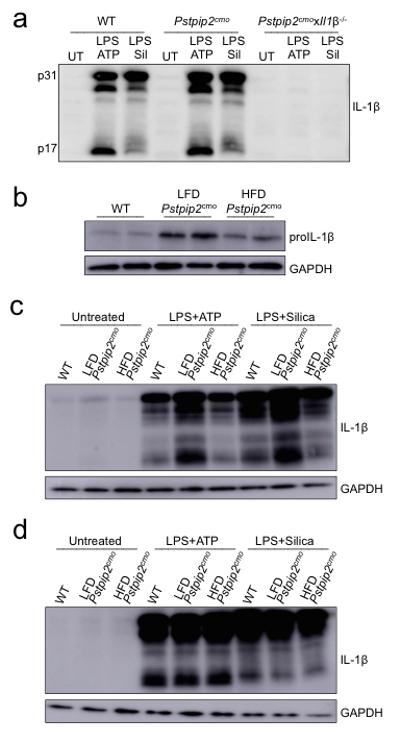
a, WT, Pstpip2cmo and Pstpip2cmoxIl1β−/− bone marrow-derived macrophages were left untreated (UT) or were first primed with LPS for 3 hrs followed by additional stimulation with ATP (30 mins) or silica (12 hours) and IL-1β processing was evaluated by Western blot. Data are representative of three independent experiments. b, Western blotting for proIL-1β in untreated neutrophils that were purified from WT, LFD-fed Pstpip2cmo and HFD-fed Pstpip2cmo mice. Data are representative of two independent experiments. b-c, Neutrophils (b) or macrophages (c) from WT, LFD-fed Pstpip2cmo and HFD-fed Pstpip2cmo mice were left untreated (UT), or primed with LPS for 3 hrs and then stimulated with ATP (30 min) or silica (12 hours) and IL-1β processing was evaluated by Western blotting. Data are representative of two independent experiments.
Extended Data Figure 9. Depletion of neutrophils in anti-Ly6G treated Pstpip2cmo mutant mice.
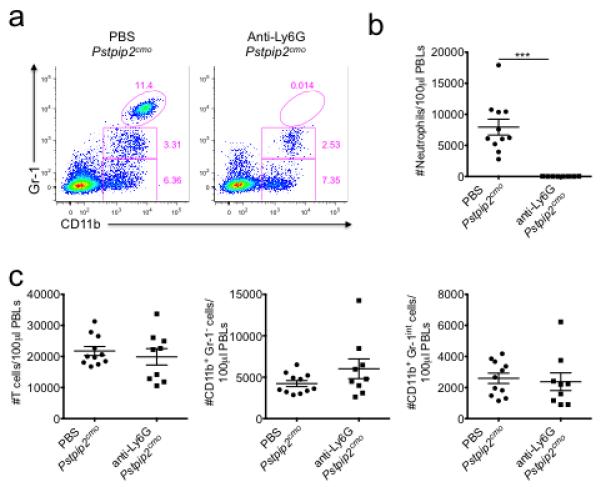
WT and Pstpip2cmo mice received either PBS or 500 μg/mouse anti-Ly6G antibody (clone IA8) by intraperitoneal injection every 4-5 days starting at 6 weeks of age. a-c, Two weeks following the first anti-Ly6G treatment, FACS analysis was performed on peripheral blood leukocytes. a, Representative FACs plots of Gr-1 and CD11b expression on CD45.2+ gated cells. b, Enumeration of CD45.2+Gr-1hiCD11b+ neutrophils in equal volumes of peripheral blood. c, Numbers of T cells (CD45.2+TCRβ+), CD45.2+Gr-1-CD11b+ monocytes/macrophages and CD45.2+Gr-1loCD11b+ cells in equal volumes of peripheral blood. Each point represents an individual mouse and the line represents the mean ± s.e.m. (***P < 0.001; Student’s t-test)
Extended Data Figure 10. Dietary modulation of the intestinal microbiota composition drives autoinflammatory osteomyelitis by setting proIL-1β levels available for maturation by caspases 1 and 8.
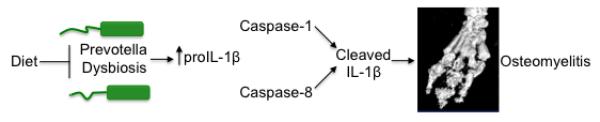
Proposed model highlighting how dysbiosis and processing of IL-1β by caspase-1/8 contribute to the development of inflammatory bone disease.
Acknowledgements
We thank Drs. D. Chaplin, V. Dixit, R. Flavell, D. Green and C. Pham for the generous supply of mutant mice. We thank J. Kim in the St. Jude Small Animal Imaging Center for helping to acquire and analyze the μCT data. We thank S. Olsen, D. Roeber and the Genome Sequencing Facility in the Hartwell Center at St. Jude Children’s Research Hospital for performing metagenomics sequencing of 16S rRNA. ML is supported by grants from Ghent University (BOF 01N02313 and 01J11113) and the European Research Council (Grant 281600). LVW is a postdoctoral fellow of the Fund for Scientific Research-Flanders. This work was supported by: the National Institute of Arthritis and Musculoskeletal and Skin Diseases, part of the National Institutes of Health, under Award Number AR056296 (T.-D.K); the National Cancer Institute, part of the National Institutes of Health, under Award Number CA163507 (T.-D.K); the National Institute of Allergy and Infectious Diseases, part of the National Institutes of Health, under Award Number AI101935 (T.-D.K); and ALSAC.
Abbreviations
- WT
wild-type
- IL
interleukin
- PCH
Pombe Cdc15 homology
- CRMO
chronic recurrent multifocal osteomyelitis
- PSTPIP2
PCH family phosphatase proline-serine-threonine phosphatase interacting protein 2
- LFD
lean fat diet
- HFD
high fat diet
- rRNA
ribosomal RNA
- GF
germ-free
- SPF
specific pathogen free
Footnotes
Conflict of interest
The authors declare no competing financial interests.
References
- 1.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. doi:10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferguson PJ, et al. A missense mutation in pstpip2 is associated with the murine autoinflammatory disorder chronic multifocal osteomyelitis. Bone. 2006;38:41–47. doi: 10.1016/j.bone.2005.07.009. doi:10.1016/j.bone.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chitu V, et al. Primed innate immunity leads to autoinflammatory disease in PSTPIP2-deficient cmo mice. Blood. 2009;114:2497–2505. doi: 10.1182/blood-2009-02-204925. doi:10.1182/blood-2009-02-204925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grosse J, et al. Mutation of mouse Mayp/Pstpip2 causes a macrophage autoinflammatory disease. Blood. 2006;107:3350–3358. doi: 10.1182/blood-2005-09-3556. doi:2005-09-3556 [pii]10.1182/blood-2005-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lukens JR, et al. Critical role for inflammasome-independent IL-1beta production in osteomyelitis. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1066–1071. doi: 10.1073/pnas.1318688111. doi:10.1073/pnas.1318688111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassel SL, et al. Inflammasome-independent IL-1beta mediates autoinflammatory disease in Pstpip2-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1072–1077. doi: 10.1073/pnas.1318685111. doi:10.1073/pnas.1318685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nature immunology. 2011;12:5–9. doi: 10.1038/ni0111-5. doi:10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 8.Montgomery MK, et al. Mouse strain-dependent variation in obesity and glucose homeostasis in response to high-fat feeding. Diabetologia. 2013;56:1129–1139. doi: 10.1007/s00125-013-2846-8. doi:10.1007/s00125-013-2846-8. [DOI] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. doi:10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 10.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. doi:10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamkanfi M, Vande Walle L, Kanneganti TD. Deregulated inflammasome signaling in disease. Immunological reviews. 2011;243:163–173. doi: 10.1111/j.1600-065X.2011.01042.x. doi:10.1111/j.1600-065X.2011.01042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maelfait J, et al. Stimulation of Toll-like receptor 3 and 4 induces interleukin-1beta maturation by caspase-8. The Journal of experimental medicine. 2008;205:1967–1973. doi: 10.1084/jem.20071632. doi:10.1084/jem.20071632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gringhuis SI, et al. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1beta via a noncanonical caspase-8 inflammasome. Nat Immunol. 2012;13:246–254. doi: 10.1038/ni.2222. doi:ni.2222 [pii]10.1038/ni.2222. [DOI] [PubMed] [Google Scholar]
- 14.Antonopoulos C, El Sanadi C, Kaiser WJ, Mocarski ES, Dubyak GR. Proapoptotic chemotherapeutic drugs induce noncanonical processing and release of IL-1beta via caspase-8 in dendritic cells. J Immunol. 2013;191:4789–4803. doi: 10.4049/jimmunol.1300645. doi:10.4049/jimmunol.1300645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurung P, et al. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J Immunol. 2014;192:1835–1846. doi: 10.4049/jimmunol.1302839. doi:10.4049/jimmunol.1302839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Man SM, et al. Salmonella infection induces recruitment of Caspase-8 to the inflammasome to modulate IL-1beta production. J Immunol. 2013;191:5239–5246. doi: 10.4049/jimmunol.1301581. doi:10.4049/jimmunol.1301581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joosten LA, et al. Inflammatory arthritis in caspase 1 gene-deficient mice: contribution of proteinase 3 to caspase 1-independent production of bioactive interleukin-1beta. Arthritis Rheum. 2009;60:3651–3662. doi: 10.1002/art.25006. doi:10.1002/art.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karmakar M, Sun Y, Hise AG, Rietsch A, Pearlman E. Cutting edge: IL-1beta processing during Pseudomonas aeruginosa infection is mediated by neutrophil serine proteases and is independent of NLRC4 and caspase-1. J Immunol. 2012;189:4231–4235. doi: 10.4049/jimmunol.1201447. doi:10.4049/jimmunol.1201447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varfolomeev EE, et al. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity. 1998;9:267–276. doi: 10.1016/s1074-7613(00)80609-3. [DOI] [PubMed] [Google Scholar]
- 20.Yeh WC, et al. FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science. 1998;279:1954–1958. doi: 10.1126/science.279.5358.1954. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Cado D, Chen A, Kabra NH, Winoto A. Fas-mediated apoptosis and activation-induced T-cell proliferation are defective in mice lacking FADD/Mort1. Nature. 1998;392:296–300. doi: 10.1038/32681. doi:10.1038/32681. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser WJ, et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471:368–372. doi: 10.1038/nature09857. doi:10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oberst A, et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature. 2011;471:363–367. doi: 10.1038/nature09852. doi:10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shornick LP, et al. Mice deficient in IL-1beta manifest impaired contact hypersensitivity to trinitrochlorobenzone. The Journal of experimental medicine. 1996;183:1427–1436. doi: 10.1084/jem.183.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanneganti TD, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. doi:10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 26.Salmena L, et al. Essential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunity. Genes & development. 2003;17:883–895. doi: 10.1101/gad.1063703. doi:10.1101/gad.1063703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton K, Sun X, Dixit VM. Kinase RIP3 is dispensable for normal NF-kappa Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Molecular and cellular biology. 2004;24:1464–1469. doi: 10.1128/MCB.24.4.1464-1469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belaaouaj A, et al. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nature medicine. 1998;4:615–618. doi: 10.1038/nm0598-615. [DOI] [PubMed] [Google Scholar]
- 29.Kessenbrock K, et al. Proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating antiinflammatory progranulin. The Journal of clinical investigation. 2008;118:2438–2447. doi: 10.1172/JCI34694. doi:10.1172/JCI34694. [DOI] [PMC free article] [PubMed] [Google Scholar]