SUMMARY
Nutrient intake is often measured with substantial error both in commonly used surrogate instruments such as a food frequency questionnaire (FFQ) as well as in gold standard type instruments such as a diet record (DR). If there is correlated error between the FFQ and DR, then standard measurement error correction methods based on regression calibration can produce biased estimates of the regression coefficient (λ) of true intake on surrogate intake. However, if a biomarker exists and the error in the biomarker is independent of the error in the FFQ and DR, then the method of triads can be used to obtain unbiased estimates of λ, provided that there is replicate biomarker data on at least a subsample of validation study subjects. Since biomarker measurements are expensive, for a fixed budget one can either use a design where a large number of subjects have 1 biomarker measure and only a small subsample is replicated, or have a smaller number of subjects and have most or all subjects validated. The purpose of this paper is to optimize the proportion of subjects with replicated biomarker measures, where optimization is with respect to minimizing the variance of ln(λ̂). The methodology is illustrated using vitamin C intake data from the EPIC study where plasma vitamin C is the biomarker. In this example, the optimal validation study design is to have 21% of subjects with replicated biomarker measures.
Keywords: measurement error, biomarker, method of triads
1. INTRODUCTION
In nutritional epidemiology, the weighed diet record (DR) is considered the gold standard for assessing nutrient intake. However, it is expensive to obtain diet records and the food frequency questionnaire (FFQ) is usually used as an instrument to obtain dietary intake data from large numbers of people. It is well known that the FFQ and other dietary assessment methods have appreciable measurement error. To correct for measurement error, a validation study is often performed where both the FFQ (Z) and DR (X) are administered to the same subjects. The regression calibration factor estimated by the regression coefficient of DR on FFQ can then be used as an unbiased estimate of the regression coefficient (λ) of true dietary intake (T) on Z, which can then be used for measurement error correction. However, this is only valid if measurement error in the DR and FFQ are uncorrelated, an assumption which may be violated. To address this issue this design is often enhanced with additional biomarker measurements (W). If the error in W is uncorrelated with the error in Z and X, then correlated error methods [1] can be used to estimate the regression calibration factor λ. The only requirement is that there be available replicate biomarker measurements on at least a subset of participants.
However, since biomarker measurements are expensive, it would be desirable to estimate the optimal proportion of subjects (θ) with replicate values of W, given a fixed total number of biomarker measures (B). The goal of this paper is to obtain a closed-form expression for var(λ̂) and to use it to estimate the optimal value of θ.
2. METHODS
2.1 Balanced Design
We let Zij = surrogate measure for the jth replicate from the ith subject, j=1, …, mz, i=1, …, N; Xik = gold standard measure for the kth replicate from the ith subject, k=1, …, mx, i=1, …, N; Wil = biomarker for the lth replicate from the ith subject, l=1, …, mw ≥2, i=1, …, N.
Thus, each subject provides mz replicates for the surrogate, mx replicates for the gold standard and mw replicates for the biomarker.
From Spiegelman, Zhao and Kim [1] we consider the model
| (1) |
where xi = true intake for the ith subject
ri= person-specific bias in the surrogate measure
si= person-specific bias in the gold standard measure
ezij, exik, ewil are distributed and are mutually independent of each other. , and ri, si are mutually independent of ezij, exik, ewil. Our goal is to estimate the regression calibration factor (λx|Z) = regression coefficient of x on Z. It can be shown from (1) that the MLE of λx|Z is given by
| (2) |
We have found that, in simulation studies, that the distribution of λ̂x|Z is generally skewed, while the distribution of ln(λ̂x|Z) is approximately normal. Hence, two-sided 100% × (1-α) confidence limits for λx|Z are obtained from [exp(c1), exp(c2)], where and zp = pth percentile of a N(0,1) distribution. It remains to derive an analytic expression for var[ln(λ̂x|Z)]. For this purpose, we take the natural log of each side of equation 2 and obtain:
| (3) |
Thus,
| (4) |
We derive var(A). The other components can be derived in a similar manner. For notational purposes, it will be useful to introduce the notation:
μabc =E[(Zij − Z̄)a(Xik − X̄)b(Wil − W̄)c] which we estimate by
| (5) |
where a* = 1 if a ≥ 1,=0 else, b* =1 if b ≥ 1,=0 else and c* =1 if c ≥ 1,=0 else. Using the delta method, we have that
| (6) |
Furthermore,
| (7) |
where k1 ≠ k2 and j1 ≠ j2. We can write
| (8) |
Similarly, we can write
In general, we introduce the notation
where j1 ≠ j2 ≠ ··· ≠ jr, k1 ≠ k2 ≠ ··· ≠ ks, and l1 ≠ l2 ≠ ··· ≠ lt.
Thus, we have:
| (9) |
Similarly,
and
| (10) |
Upon combining equations 6–10, we obtain
| (11) |
The other components in equation 4 are obtained similarly and are provided in Web Appendix A.
Upon combining equations A1–A10, we obtain var[ln(λ̂x|Z)] in equation 4.
To obtain confidence limits for λx|Z we assume asymptotic normality of ln(λ̂x|Z) whereby a two-sided 100% × (1-α) CI for λx|Z is given by [exp(c1), exp (c2)], where
| (12) |
and z1−α/2= upper α/2 percentile of a N(0,1) distribution.
2.2 Unbalanced Design
We now consider the unbalanced design situation. In this case, we assume all subjects have the same number of replicates for the surrogate dietary instrument (e.g., FFQ) and the gold standard dietary instrument (e.g., DR) denoted by mz and mx, respectively. However, since biomarker measurements are the most expensive, we assume that ng of the subjects have g biomarker measurements, where g = 1,2 and n1 + n2 = N. Also, let bi = the number of replicate biomarker measurements for the ith subject and let . Finally, let θ = proportion of biomarker measurements that are replicated =2n2/M where 0 ≤ θ ≤ 1. We assume that M is fixed due to budgetary constraints and we wish to determine the value of θ that minimizes var[ln(λ̂x|Z)] in equation 4.
We will derive var(A) in the unbalanced case and present the results for the other components of equation 4 in Appendix B. In the unbalanced case, we estimate μabc by
| (13) |
where a* and b* are defined in equation 5.
We have
| (14) |
where μ101 is estimated using equation 13.
We have:
| (15) |
Furthermore,
| (16) |
If we denote by M(2) and combine equations 14, 15 and 16, we obtain
| (17) |
Note, if there are a total of N subjects of whom n1 have one replicate and n2 have two replicates, then M = 2n2 + n1, M(2) = 4n2 + n1, and M(2) − M =2n2. In this case, equation 17 reduces to:
| (18) |
Derivation of the other components of equation 4 under an unbalanced design are obtained similarly and are provided in Web Appendix B.
Finally, a large sample 100% × (1-α) CI for λx|Z is given by [exp(c1), exp(c2)] where
2.3 Optimization
We wish to minimize var[ln(λ̂x|Z)] in equation 4 in the setting where bi= 1 or 2. We can re-express equation B.1 in Web Appendix B as a function of θ as follows:
| (19) |
where
Similarly,
| (20) |
| (21) |
| (22) |
| (23) |
| (24) |
| (25) |
| (26) |
| (27) |
| (28) |
The expressions for f1A, …, fCD are given in Appendix C.
Note that in general if there is positive correlation among replicate Z, X and W values, then it can be shown that f2A > 0, f2B > 0, fC > 0, fD > 0, f2,AB > 0, fAD > 0, fBD > 0, and fCD > 0. Hence, var (A), var (B), var (D), cov(A, B), cov(A, D), cov(B, D) and cov(C, D) are minimized if θ = 0, i.e., all subjects have only one biomarker measurement, since this will maximize the number of subjects. Conversely, var(C) is minimized if θ = 1; where all subjects have two biomarker measurements.
Assume all subjects have either one or two biomarker measurements. If we combine equations 4 and 19–28, we obtain:
| (29) |
where
If we differentiate V(θ) with respect to θ in equation 29 and collect terms, we obtain the 4th degree polynomial equation as follows:
| (30) |
where
Although it is possible to obtain an exact solution to this equation, it is simpler to use a polynomial equation solver (e.g., the POLYROOT function of SAS) to determine the solution that satisfies 0 < θ < 1.
3. SIMULATION STUDY
We simulated data from a hypothetical dataset with a similar correlation structure as in our example with (Z1, Z2, X1, X2, W1, W2)~N(μ, Σ) where μ = (100, 100, 100, 100, 50, 50) and
We then estimated ln(λx|Z) in equation 3, its variance in equation 4 and a 95% CI for λx|Z in equation 12 from 4,000 simulated samples. The results are given in Table 1. We see that there is good agreement between the mean theoretical variances and covariances considered in equation 4 and derived in Appendix B and the corresponding empirical variances and covariances obtained from the 4,000 simulated samples. Also, the overall estimate of λx|Z has little bias and the estimated 95% confidence intervals have approximately (94.1%) coverage.
4. EXAMPLE
We analyzed data from the EPIC-Norfolk study [2]. Individuals were seen at a baseline visit and at a 4-year follow-up visit as part of the study. At both baseline and follow-up, a food frequency questionnaire (FFQ) and a 1-week diet record (DR) were obtained. In addition, a blood sample was obtained at both the baseline and 4-year follow-up visit. In this example, we focus on dietary vitamin C and assess the regression coefficient of true dietary vitamin C intake (xi in equation 1) on FFQ vitamin C intake (Zij in equation 1) which is given by λ̂x|Z in equation 2 using plasma vitamin C as a biomarker. We refer to λ̂x|Z as the estimated regression calibration factor. For this example, we assume that true dietary intake has not changed over four years, but allow for the possibility of correlated error between FFQ and DR intake (ρrs in equation 1). We also assume that there is no systematic error in the biomarker and that the random error in FFQ intake, DR intake and plasma vitamin C are uncorrelated. The marginal and joint distribution of FFQ intake (Zij), DR intake (Xij) and plasma vitamin C (Wij) are given in Table II. There is moderate correlation between dietary vitamin C (Z, X) and plasma vitamin C (W) which are similar for the FFQ and DR when the intake assessments at year 4 are compared with the biomarker values at baseline (which provides the most appropriate assessment of their relative measurement of long-term intake). For the purpose of better approximating a normal distribution, the log transform was used for each of dietary vitamin C from FFQ (Zij) and DR (Xik) in subsequent analyses.
Table II.
Marginal and Joint Distribution of FFQ vitamin C, DR vitamin C and plasma vitamin C in the EPIC-Norfolk study
| variable
|
mean
|
sd
|
correlation matrix | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
Zi1
|
Zi2
|
Xi1
|
Xi2
|
Wi1
|
Wi2
|
||||
|
|
134.4 | 54.5 | 1.0 | 0.60 | 0.47 | 0.42 | 0.25 | 0.18 | |
|
|
135.6 | 58.7 | 1.0 | 0.45 | 0.57 | 0.25 | 0.27 | ||
|
|
90.5 | 50.1 | 1.0 | 0.59 | 0.40 | 0.23 | |||
|
|
94.6 | 52.0 | 1.0 | 0.28 | 0.34 | ||||
|
|
57.7 | 21.2 | 1.0 | 0.43 | |||||
|
|
64.8 | 23.2 | 1.0 | ||||||
Zi1, Zi2 = baseline and 4-year calorie-adjusted FFQ vitamin C intake (mg/day)
Xi1, Xi2 = baseline and 4-year calorie-adjusted DR vitamin C intake (mg/day)
Wi1, Wi2 = baseline and 4-year plasma vitamin C intake (μmol/L)
Computer program: :/proj/stross/stros0a/example_usevitc.sas 09/19/13
In Table III we provide the point estimate and 95% CI for λx|Z as well as the individual components used in equation 12. We see that the estimated regression calibration factor (λx|Z) is 0.308 with 95% confidence limits from 0.201 to 0.471. The point estimate implies that there is substantial measurement error in the assessment of dietary vitamin C. For example if the estimated hazard ratio based on observed vitamin C is 1.2 then the deattenuated estimate would be 1.21/0.308 = 1.8, indicating substantial deattenuation. The degree of measurement error in the FFQ will vary depending on the nutrients/foods being considered. In general, beverage intake has less measurement error, while food intake can have considerable measurement error. Dietary vitamin C is derived mainly from fruits and vegetables which have moderate measurement error.
Table III.
Estimation of Regression Calibration factor in EPIC data example, n=323
| A* | 2.998 |
| B* | 5.003 |
| C* | 263.319 |
| D* | 0.1852 |
| var(A) | 0.034 |
| var(B) | 0.019 |
| var(C) | 0.019 |
| var(D) | 0.007 |
| cov(A,B) | 0.018 |
| cov(A,C) | 0.013 |
| cov(A,D) | 0.009 |
| cov(B,C) | 0.011 |
| cov(B,D) | 0.004 |
| cov(C,D) | 0.003 |
| λ̂x|Z | 0.308 |
| log(λ̂x|Z) | −1.179 |
| var[log(λ̂x|Z)] | 0.0473 |
| 95% CI for λx|Z | (0.201,0.471) |
A = cov(Zij, Wik); B = cov(Xij, Wik); C = cov(Wil1, Wil2); D = var(Zij)
Computer run: :/proj/stross/stros0c/measurmentErrBio/example1/example_usevitc.sas 6/4/12
5. OPTIMIZATION
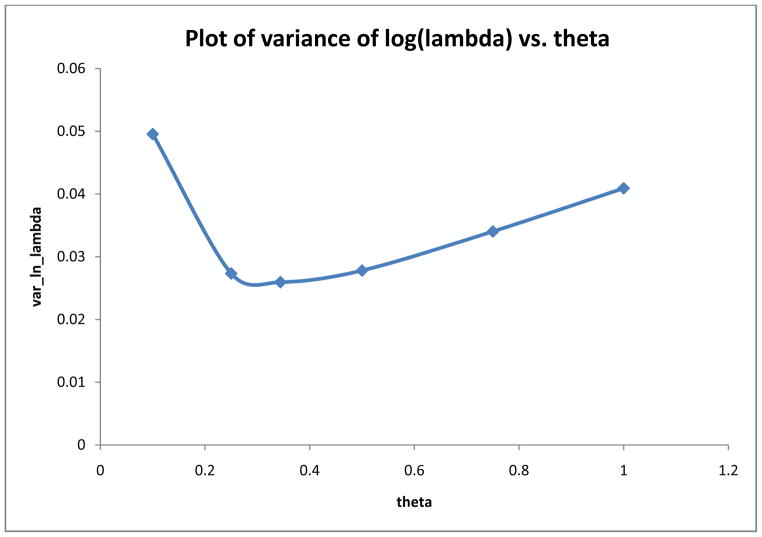
We also used the EPIC data to estimate the optimal proportion of replicated biomarker measurements based on equations 29 and 30. The results are presented in Table IV. The estimated parameters (C1, C2, C3), (d1, d2) in equations 29 and 30 are given in the left side of the table. The solution using the POLYROOT function of SAS was θ̂ = 0.349 = the optimal proportion of replicated biomarker measurements (i.e., 2n2/(n1 + 2n2)). It follows directly that the optimal estimate of n2/n1 = 0.349/[2(0.651)] = 0.268 or equivalently n2/(n1 + n2) = 0.268/1.268 = 0.211. Thus, the optimal design (i.e., min var[ln(λ̂x|Z)]) is for approximately 21% of the sample to have replicated biomarker measurements given a fixed total of M biomarker measurements. To assess the sensitivity of var[ln(λ̂x|Z)] to variation in θ we computed var[ln(λ̂x|Z)] for different values of θ. The results are given in the right hand side of Table IV and are plotted in Figure 1. We see that the variance function is fairly flat between θ̂ = 0.2 – 0.5 corresponding to a proportion of subjects with replicated biomarkers of 0.14 to 0.33. However, the variance increases moderately outside these limits.
Table IV.
Results of Optimization Procedure based on EPIC dataset
| Parameter | Value | θ | var[ln(λ̂x|Z)] | n2/(n1 + n2) |
|---|---|---|---|---|
|
|
|
|||
| C1 | 0.04120 | 0.10 | 0.0496 | 0.053 |
| C2 | 0.00472 | 0.25 | 0.0273 | 0.143 |
| C3 | −0.00664 | 0.349 | 0.0259 | 0.208 |
| d1 | 3.72420 | 0.500 | 0.0278 | 0.333 |
| d2 | 0.45839 | 0.75 | 0.0340 | 0.600 |
| θ̂ | 0.349 | 0.90 | 0.0382 | 0.818 |
Computer program:
:/proj/stross/stros0c/measurmentErrBio/example1/example2_usevitc.sas 9/30/13
:/proj/stross/stros0c/measurmentErrBio/example1/test_getLambda.sas 9/30/13
Figure 1.
6. DISCUSSION
Correlated error between gold standard dietary measures such as a diet record and surrogate measures such as a food frequency questionnaire can bias standard techniques for correcting for measurement error such as regression calibration. The method of triads using a biomarker in addition to the above dietary instruments is an effective method for eliminating this bias. However, it requires replicate measurements on the biomarker for at least a subset of study participants [1]. In the current paper, we derive a closed form expression for the variance estimate of the Spiegelman, Zhao and Kim estimator of the regression calibration factor (λx|Z) and associated 95% confidence limits for both balanced (same number of biomarker replicates per subject) and unbalanced (different number of biomarker replicates per subject) designs.
Ideally, all subjects in a validation study would have replicated biomarker measurements; however, these measures are usually expensive. Thus, in this paper, we derive an expression for the optimal proportion of validation study subjects with replicated biomarker measures given a fixed total number of biomarker measures (M), where optimality is defined as minimizing var[ln(λ̂x|Z)]. In the EPIC example, this was about 21%, but would be expected to vary for other biomarkers or in other studies.
The algorithms used to derive var[ln(λ̂x|Z)] and associated confidence limits and the optimal design formulas in equations 29 and 30 are available in the form of SAS macros from the authors upon request.
Supplementary Material
Table I.
Simulation Study Results, 4000 replications
| Component | Theoretical value* | Empirical estimate | Coverage probability |
|---|---|---|---|
| var(A) | 0.0351 | 0.0389 | |
| var(B) | 0.0204 | 0.0233 | |
| var(C) | 0.0234 | 0.0234 | |
| var(D) | 0.0041 | 0.0041 | |
| cov(A,B) | 0.0169 | 0.0181 | |
| cov(A,C) | 0.0109 | 0.0116 | |
| cov(A,D) | 0.0048 | 0.0049 | |
| cov(B,C) | 0.0108 | 0.0116 | |
| cov(B,D) | 0.0022 | 0.0023 | |
| cov(C,D) | 0.0009 | 0.0010 | |
| cov(Zij, Wik) | 48.0 | 48.0 | |
| cov(Xij, Wik) | 64.0 | 63.9 | |
| cov(Wil1, Wil2) | 40.0 | 39.8 | |
| var(Zij) | 400.0 | 399.6 | |
| var[ln(λ̂x|Z)] | 0.0609 | 0.0672 | |
| λ̂x|Z | 0.192 | 0.194** | 0.941 |
Based on Web Appendix B
median
Computer program :/proj/stross/stros0c/measurmentErrBio/Undesignx4000a.sas 09/27/13
:/proj/stross/stros0c/measurmentErrBio/all_new2.txt 09/27/13
Acknowledgments
We acknowledge the support of R01 CA50597, R01 CA077398 and U54 CA155626 from the National Institutes of Health in performing this work. We also acknowledge programming support of Rong Chen and Marion McPhee. Sara Hendrickson was supported in part by training grants a 5 T32 CA09001 and R25 CA098566.
References
- 1.Spiegelman D, Zhao B, Kim J. Correlated errors in biased surrogates: study designs and methods for measurement error correction. Statistics in Medicine. 2005;24(11):1657–82. doi: 10.1002/sim.2055. [DOI] [PubMed] [Google Scholar]
- 2.Rosner B, Michels KB, Chen YH, Day NE. Measurement error correction for nutritional exposures with correlated measurement error: use of the method of triads in a longitudinal setting. Statistics in Medicine. 2008;27(18):3466–89. doi: 10.1002/sim.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.