Summary
Throughout evolution, primate genomes have been modified by waves of retrotransposon insertions1,2,3. For each wave, the host eventually finds a way to repress retrotransposon transcription and prevent further insertions. In mouse embryonic stem cells (mESCs), transcriptional silencing of retrotransposons requires TRIM28 (KAP1) and it’s repressive complex, which can be recruited to target sites by KRAB zinc finger proteins such as murine-specific ZFP809 which binds to integrated murine leukemia virus DNA elements and recruits KAP1 to repress them4,5. KZNF genes are one of the fastest growing gene families in primates and this expansion is hypothesized to enable primates to respond to newly emerged retrotransposons6,7. However, the identity of KZNF genes battling retrotransposons currently active in the human genome, such as SINE-VNTR-Alu (SVA)8 and Long Interspersed Nuclear Element-1 (L1)9, is unknown. We find that two primate-specific KZNF genes rapidly evolved to repress these two distinct retrotransposon families shortly after they began to spread in our ancestral genome. ZNF91 underwent a series of structural changes 8-12 MYA that enabled it to repress SVA elements. ZNF93 evolved earlier to repress the primate L1 lineage until ~12.5 MYA when the L1PA3-subfamily escaped ZNF93’s restriction through purge of the ZNF93 binding site. Our data support a model where KZNF gene expansion limits the activity of newly emerged retrotransposon classes, and this is followed by mutations in these retrotransposons to evade repression, a cycle of events that could explain the rapid expansion of lineage-specific KZNF genes.
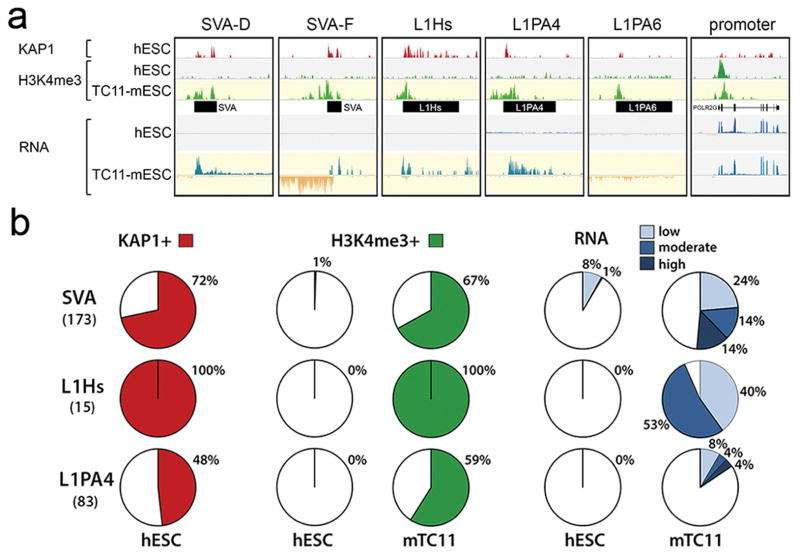
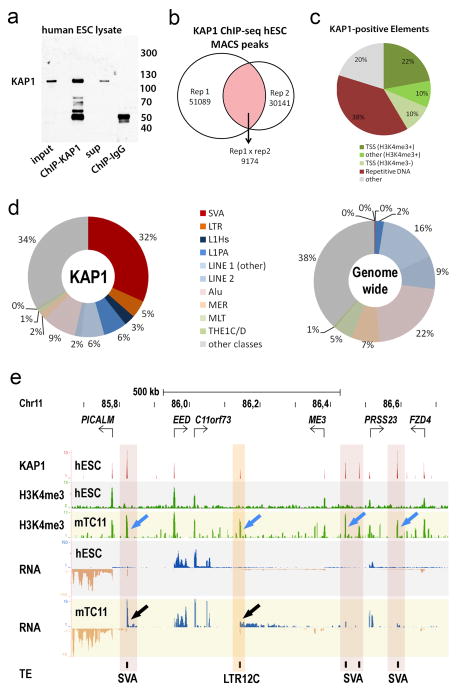
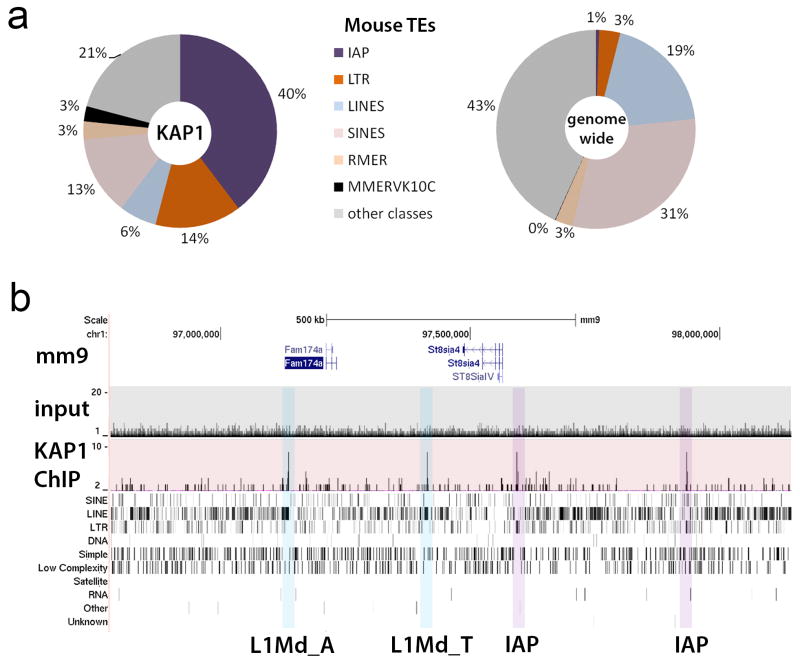
KAP1 mediates transcriptional silencing of retrotransposons and protects genome integrity through repression of retrotransposition activity10,11. ChIP-seq analysis revealed that in human ESCs, KAP1 predominantly associates with active primate-specific classes of retrotransposons such as SVA and L1Hs (Extended Data Fig. 1)11,12. Similarly, in mouse ESCs KAP1 primarily associates with mouse lineage-specific retrotransposon classes (Extended Data Fig. 2)12. These data support the hypothesis that species-specific KZNFs recruit KAP1 to species-specific retrotransposon classes that recently invaded the host’s genome7,13. To test this, we determined the fate of primate-specific retrotransposons in a non-primate background using mouse trans-chromosomic ESCs that contain a copy of human chromosome 11 (E14(hChr11) cells14, hereafter termed Trans-Chromosomic 11 (TC11)-mESCs)). In the TC11-mESC cellular environment, primate-specific retrotransposons, including SVA and L1PA elements, are de-repressed and gain activating H3K4me3 histone marks (Fig. 1a, b; Extended Data Fig. 1e). As a result of this de-repression, a majority of SVA (51%), L1Hs (93%) and some L1PA elements, such as L1PA4 (16%), become aberrantly transcribed. These findings suggest primate-specific retrotransposons harbor a transcriptional potential15,16 that is repressed by primate-specific factors.
Figure 1. SVAs and L1PAs are de-repressed in a non-primate cellular environment.
a, KAP1, H3K4me3 ChIP-seq and RNA-seq coverage tracks for a selection of KAP1-bound primate-specific retrotransposons de-repressed in TC11-mESCs (yellow) relative to human ESCs (grey). H3K4me3 signal on promoters is similar in hESCs and TC11-mESCs. b, Percentages of SVA, L1Hs and L1PA elements on human chromosome 11 positive for KAP1, H3K4me3 (MACS ChIP-seq analysis) and arbitrary levels of transcription (see Methods) in hESC and TC11-mESCs. Total elements of each type on human chromosome 11 in parentheses.
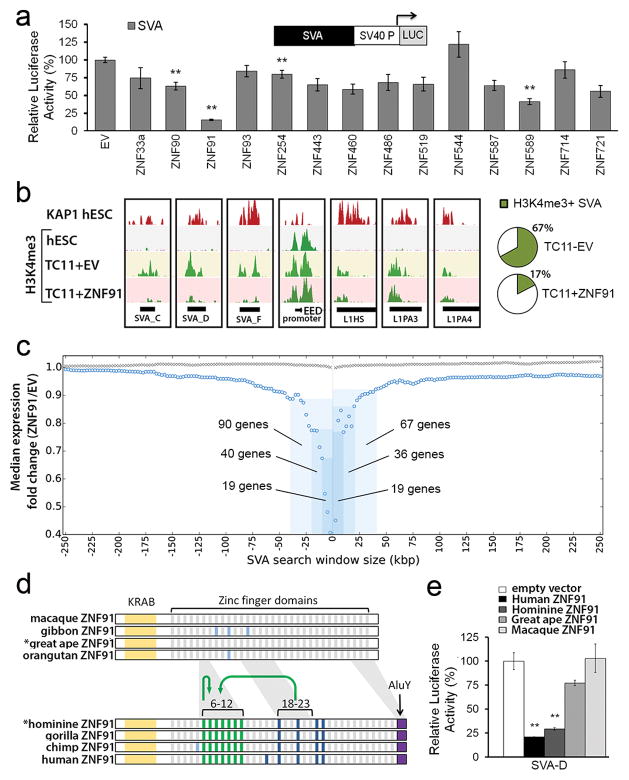
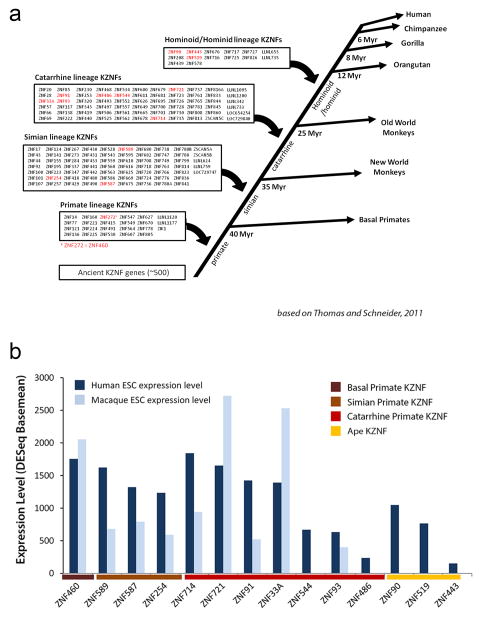
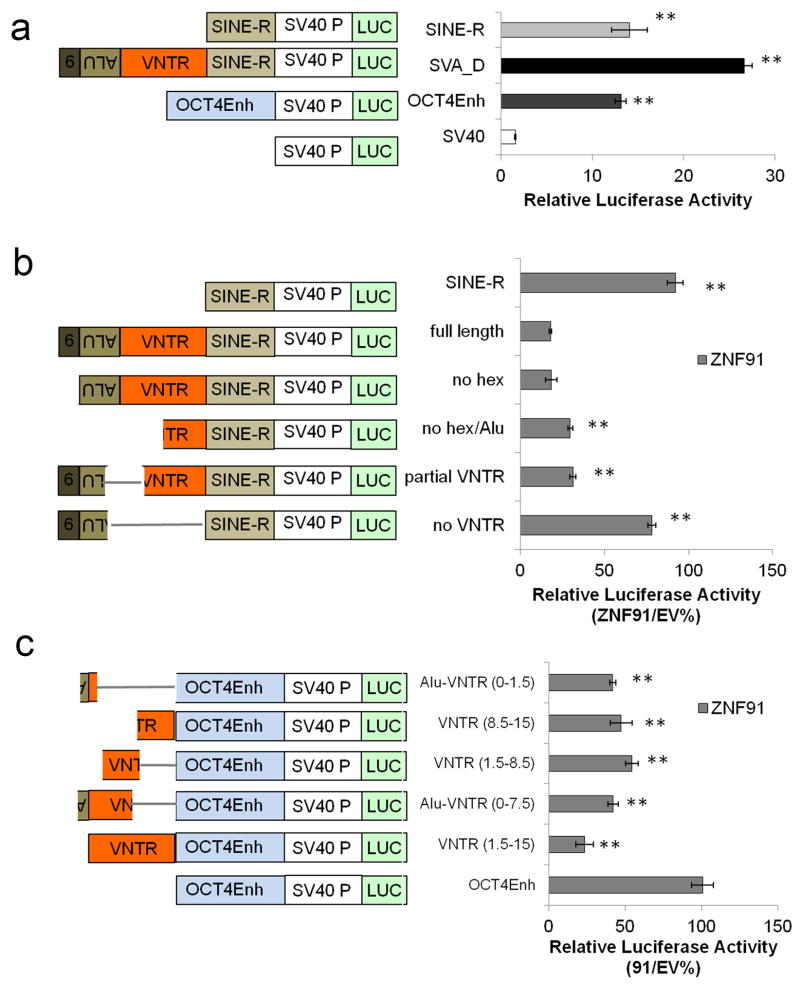
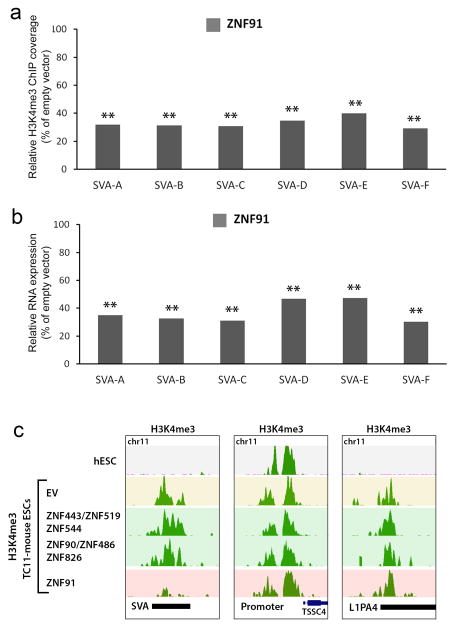
Good candidates for these factors are ~170 KZNF genes that emerged during primate evolution7 (Extended Data Fig. 3a). We reasoned that a KZNF gene responsible for protecting genome integrity, most critical in the germ line, must be highly expressed in human ESCs. So we focused on 14 highly expressed, primate-specific KZNF genes (Extended Data Fig. 3b) and tested each candidate for a role in repressing SVA retrotransposons, which first appeared in great apes 18–25 million years ago (MYA)8, and are still active17. We set up a luciferase assay-based screen in mESCs in which an SVA element cloned upstream of a minimal SV40 promoter strongly enhances luciferase activity (Extended Data Fig. 4a). Each candidate KZNF was co-expressed with the SVA-luciferase construct to determine its effect on reporter activity. Of all KZNFs tested, ZNF91 most dramatically decreased SVA-driven luciferase activity, reducing activity to 16 +/− 4% relative to empty vector (EV)-transfected control (Fig. 2a). Some other KZNFs had modest effects on this reporter, but were not further analyzed as those with the strongest effect also inhibited the OCT4 enhancer (Extended Data Fig. 7a). Structure-function analysis of SVA revealed that the variable number tandem repeat (VNTR) domain is necessary and sufficient for ZNF91-mediated repression of luciferase activity (Extended Data Fig. 4b, 4c). Furthermore, transfection of TC11-mESCs with human ZNF91 restored the repression of deregulated SVAs on human chromosome 11, causing a strong decrease of aberrant H3K4me3 ChIP-seq signal at SVAs, while leaving other de-repressed elements such as L1Hs or L1PAs unaffected (Fig. 2b, Extended Data Fig. 5a). Transfection of ZNF91 also significantly repressed aberrant transcription of SVA repeats, indicating that ZNF91 is sufficient to restore transcriptional silencing of SVAs. (Extended Data Fig. 5b). No such effects were observed for other primate KZNFs (ZNF90, ZNF93, ZNF486, ZNF826, ZNF443, ZNF544, ZNF519) transfected in TC11-mESCs, validating the specificity of the ZNF91-SVA interaction (Extended Data Fig. 5c). Cellular genes near SVAs on human chromosome 11 in TC11-mESCs were also repressed by ZFN91, with distance of a gene to an SVA as the major factor in governing the amount of bystander repression (Fig. 2c), supporting the hypothesis that the host response to retrotransposon insertion has significantly impacted human gene expression patterns11,15,16.
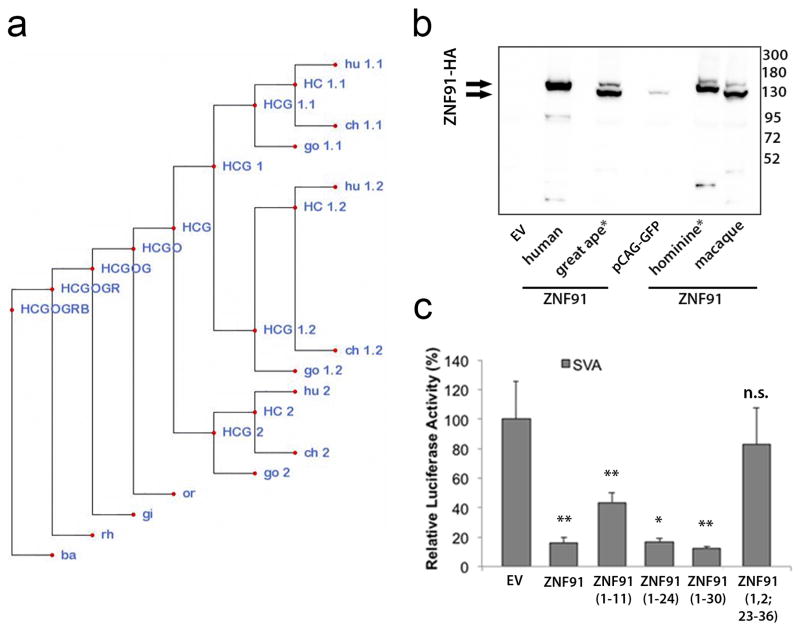
Figure 2. SVA elements are repressed by primate-specific ZNF91.
a, Relative luciferase activity of a SVA_D-SV40-luciferase reporter after co-transfection of KZNFs in mESCs. b, KAP1 and H3K4me3 ChIP-seq coverage tracks for a selection of loci in hESCs and TC11-mESCs transfected with EV or ZNF91. Pie charts: percentages of H3K4me3-positive SVAs on human chromosome 11. c, Median fold expression change (ZNF91/EV), for genes with (blue circles) or without (gray crosses) an SVA within the indicated genomic distance among the 994 expressed human chromosome 11 genes. d, ZNF91 structural evolution. Green stripes: duplicated zinc fingers; Blue stripes: zinc fingers that changed contact residues in the lineage to humans (dark blue) or in other lineages (light blue). Green arrows: segmental duplications. *= reconstructed ancestral protein. e, Relative SVA_D-SV40-luciferase activity in the presence of various ZNF91 proteins. Error bars: SEM; ** p<0.01
ZNF91 emerged in the last common ancestor (LCA) of human and old world monkeys and has undergone dramatic structural changes, including the addition of 7 zinc fingers, in the LCA of human and gorilla18 (Fig. 2c). We reconstructed ancestral ZNF91 versions by parsimony analysis (Extended Data Fig. 6a, b) and found that ZNF91 as it likely existed in the LCA of human and gorilla (ZNF91hominine) was able to repress the SVA-Luciferase reporter similar to human ZNF91 (Fig. 2d). However, ZNF91 as it existed in the LCA of human and orangutan (ZNF91great ape) only reduced luciferase activity to ~80% and macaque ZNF91 completely lacked the ability to repress SVA-driven luciferase activity. The importance of the 7 newly added hominine zinc fingers was further supported by deletion analysis of ZNF91 (Extended Data Fig. 6c). These findings suggest that the changes in ZNF91 between 8-12 MYA have significantly optimized the protein’s ability to bind and repress SVA.
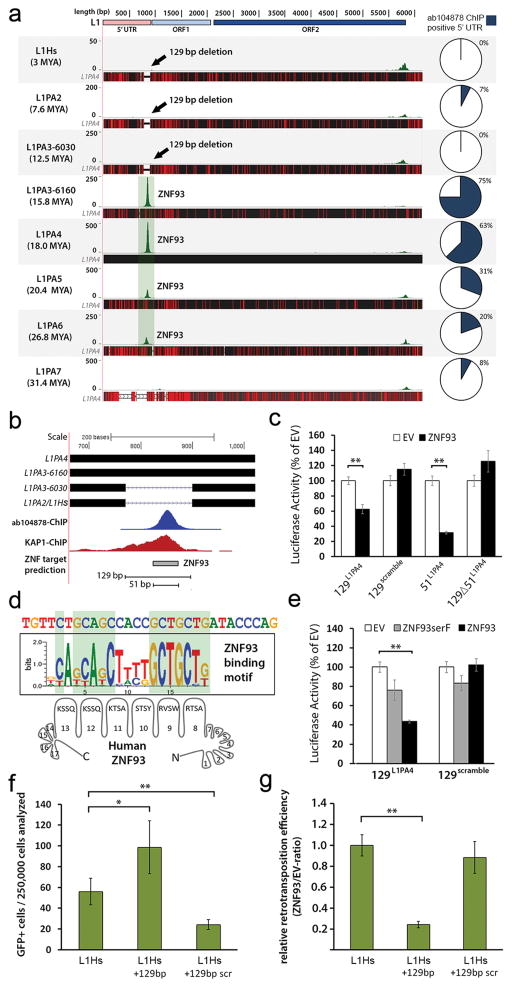
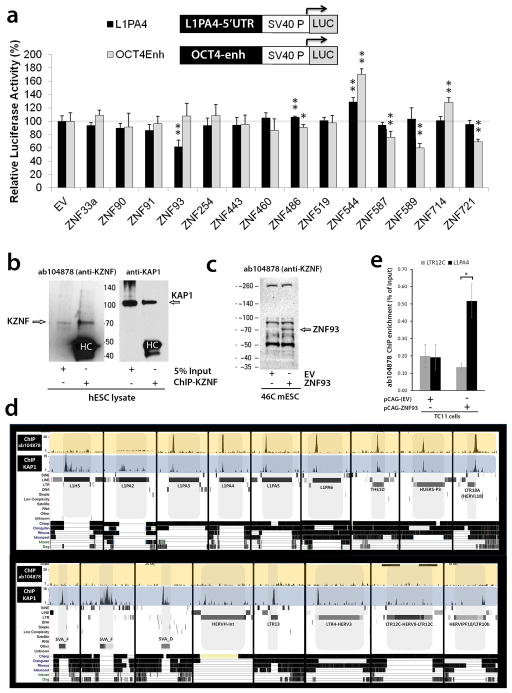
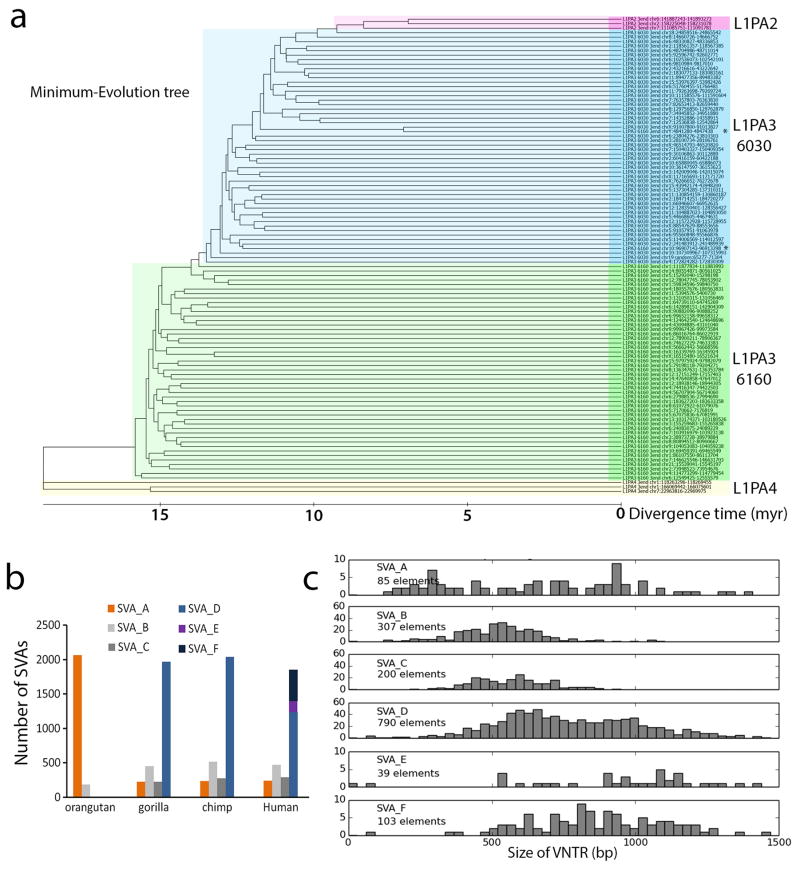
In our KAP1 ChIP experiments, KAP1 also showed a strong association with the 5′UTR of L1PA elements. None of the 14 KZNFs had a significant effect on the 5′UTR of the current active human L1 subtype L1Hs9,19 cloned upstream of the luciferase reporter when tested in mESCs. However, ZNF93 significantly reduced luciferase activity of a reporter with the 5′UTR of a KAP1-positive L1PA4 element (62 +/− 10%, Extended Data Fig. 7a). To verify the recruitment of ZNF93 to L1PA4 elements on the human genome, we performed ChIP-seq analysis on hESCs using antibody ab104878, which recognizes ZNF93 and co-immunoprecipitates KAP1 (Extended Data Fig 7b, c). We found that ZNF93 binds to the 5′end of L1PA4, its predecessors L1PA6 and L1PA5 and its successor L1PA3 (Fig. 3a; Extended Data Fig 7d). To validate that the ab104878-ChIP-seq signal on L1PAs is derived from ZNF93, we performed ab104878-ChIP analysis followed by quantitative PCR on TC11-mESC transfected with ZNF93 or EV and found significant enrichment of the L1PA4 5′UTR compared to a LTR12C control element (Extended Data Fig 7e). No consistent ZNF93 binding was detected at L1PA7 or older subtypes nor at the most recently evolved L1PA2 and L1Hs (Fig 3a). Comparative sequence analysis revealed that the absence of ZNF93 binding in L1Hs and L1PA2 can be explained by a 129 bp deletion in the 5′UTR that spans the ChIP-determined ZNF93 and KAP1 binding site (Fig. 3b). The deletion is also present in ~50% of L1PA3 elements, resulting in distinct subgroups of shorter (L1PA3-6030) and longer (L1PA3-6160) L1PA3 elements, but is not present in L1PA4-6 families.
Figure 3. L1PA elements are repressed by primate-specific ZNF93.
a, Green peaks represent genome-wide ab104878-ChIP-seq peak-summits mapped to L1PA consensus sequences. Black horizontal bars: alignment to L1PA4, red lines: divergent positions. b, The 129 bp deletion and predicted 51 bp ZNF93 binding motif (grey bar) relative to L1PA4. c, Relative activity of OCT4-enhancer-luciferase reporters after co-transfection of EV or ZNF93. d, Consensus central sequence of ab104878-ChIP-seq summits for L1PA4, aligned with the predicted recognition motif of ZNF93 zinc fingers 8-13. e, Relative activity for OCT4-enhancer-luciferase reporters after co-transfection of EV, ZNF93SerF or ZNF93. f, Number of GFP-positive cells derived from retrotransposition events of L1Hs, L1Hs+129 and L1Hs+129-scrambled constructs in HEK cells (n=7). g, Same as (f) but showing the ratio of retrotransposition events after co-transfection with ZNF93 compared to EV. Error bars: SEM. *= p<0.05; **= p<0.01
To investigate the interaction of ZNF93 with the 129 bp L1PA element, we tested a series of L1PA4 segments cloned upstream of an OCT4-Enhancer-SV40 promoter luciferase reporter in mESCs (Fig. 3c). Both the 129 bp element and a 51 bp sub-fragment were sufficient to confer ZNF93-mediated repression of the luciferase reporter, and this repression was abolished by elimination of the 51 bp portion in the 129 bp fragment (129Δ51L1PA4). The 51 bp element encompasses a computationally predicted DNA binding motif for ZNF93’s 17 fingers20 and the central 18bp of this region displays strong similarity to the predicted recognition motif of zinc fingers 8-13 of human ZNF93 (Fig. 3d). A ZNF93 variant that has all contact residues in zinc fingers 8-13 replaced by serine residues (ZNF93SerF), a modification that abolishes DNA binding selectivity21, was unable to repress luciferase activity of the L1PA4 elements (Fig. 3e), suggesting that fingers 8-13 of ZNF93 are important for recognition of the 129bp element in L1PA6-3 retrotransposons.
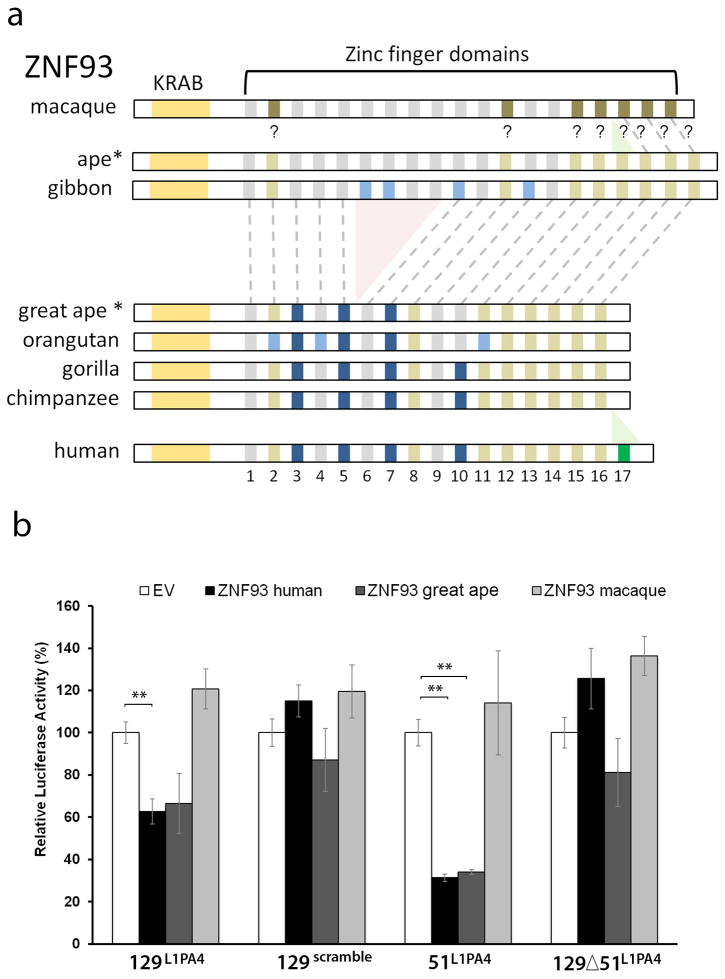
ZNF93 emerged in the LCA of apes and old world monkeys and reconstruction of the evolutionary history of the ZNF93 protein by parsimony suggests that dramatic changes took place in the LCA of orangutan and human between 12-18 MYA (ZNF93great ape, Extended Data Fig. 8a). Indeed macaque ZNF93 does not have the ability to repress the 129bp or 51bp element of L1PA4 in the luciferase assay, but ZNF93great ape represses at levels similar to ZNF93human (Extended Data Fig. 8b), suggesting changes in the ape lineage likely enabled ZNF93 to regulate L1 activity.
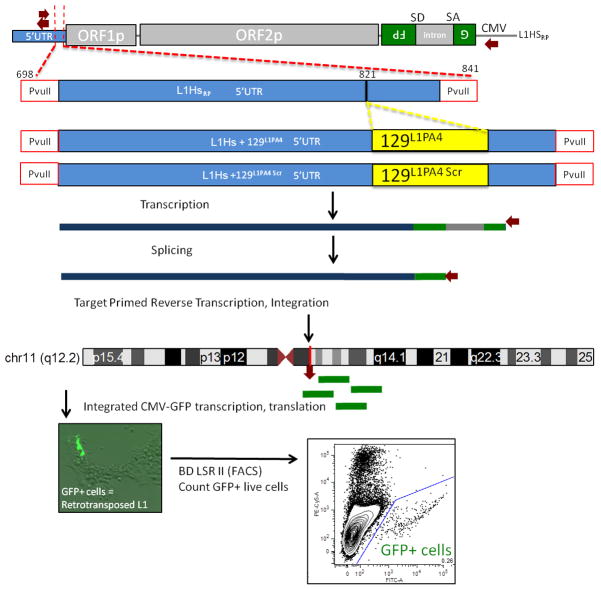
To explore the function of the lost 129 bp element, we created a version of L1Hs with this sequence restored in its 5′UTR (L1Hs+129), or a scrambled version of this 129 bp sequence (L1Hs+129scramble) as a control, and compared retrotransposition efficiencies in HEK293FT cells in an in vitro retrotransposition assay22 using L1RP23 as the ‘wild type’ L1Hs. In this assay, a retrotransposition event results in green fluorescent protein (GFP) expression (Extended Data Fig. 9). L1Hs+129 shows a 1.76 (+/−0.45 SEM) fold higher retrotransposition activity compared to wild type L1Hs, an effect not seen with L1Hs+129scramble (Fig. 3f), suggesting this 129 bp sequence promotes retrotransposition. Importantly, co-expression of ZNF93 significantly reduced retrotransposition of L1Hs+129 to just 24% (+/−3% SEM) relative to L1Hs, but had no significant effect on L1Hs+129scramble (Fig. 3g).
These data suggest the 129bp sequence, as it once existed in the 5′UTR of L1PA subfamilies, may have been beneficial to L1 mobilization, but since ZNF93 evolved to bind this element, losing it allowed the L1 lineage to escape ZNF93 mediated repression, providing net selective advantage. Indeed, phylogenetic analysis of L1PA3 elements and calculation of the average distance of L1PA3-6030 and L1PA3-6160 elements from the respective consensus sequences, suggests that L1PA3-6030 elements lacking the 129 bp element have expanded more recently in our genome than L1PA3-6160 elements, showing an estimated age of 12.5 and 15.8 MYA respectively (Extended Data Fig. 10a). This strongly suggests that loss of the ZNF93 binding site, and thereby the evasion of the host repression, propagated a new wave of L1 insertions in great ape genomes.
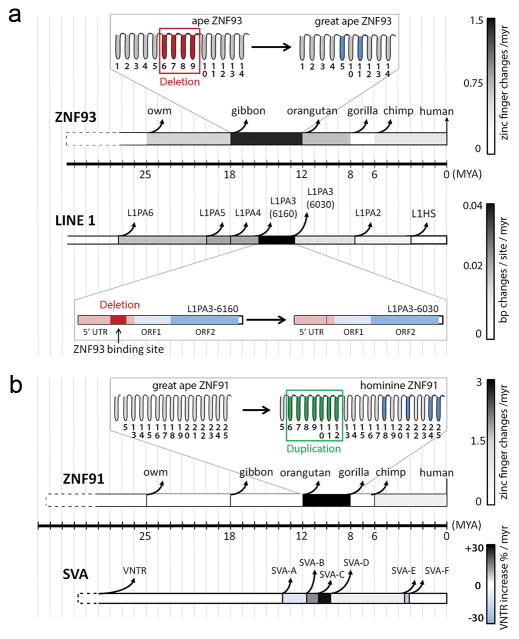
Repeated turnover of the 5′UTR occurred in early L1PA evolution9 and was previously thought to be associated with competition for host factors24. Our results suggest turnover was instead driven by avoidance of host factors. The surgical removal of the ZNF93 binding site likely took place soon after ZNF93 had a series of structural changes, suggesting the deletion may have been driven by improved host repression of L1PA activity (Fig. 4a). In a similar fashion, the structural changes in ZNF91 allowing it to repress SVA elements may have driven the further evolution of new and different SVA-subtypes in gorilla, chimpanzee and human, a pattern that is not observed in orangutan, which diverged before ZNF91 had undergone its structural changes (Extended Data Fig. 10b). Interestingly, the size of the VNTR region of SVA, the prime interaction site of ZNF91, has increased during the timeframe of structural changes to ZNF91 (Fig. 4b, Extended Data Fig. 10c).
Figure 4. Dynamic patterns of co-evolution between ZNFs and target retrotransposons.
Schematic showing the evolution of L1PA9 and SVA8 retrotransposons parallel to the structural evolution of ZNF93 and ZNF91 along an evolutionary timescale. Red zinc fingers: deletion; blue zinc fingers: change in contact residues; green zinc fingers: duplication. Coloring of ZNF91/ZNF93 horizontal bars: Zinc finger changes (changes in DNA-contacting residues, zinc finger deletions and duplications)/myr during the time interval indicated. Coloring of TE horizontal bars: basepair substitutions/deletions/insertions per site/myr (L1PA), or percentage increase in VNTR size/myr (SVA). myr = million years; owm = old world monkey
Our data support a model where modifications to lineage-specific KZNF genes are utilized by the host to repress new families of retrotransposons as they emerge, which in turn drives the evolution of newer families of retrotransposons, in a continuing arms race. Because repression affects nearby genes, KZNFs have likely been co-opted for other functions that persisted long after the original transposon expansion they first evolved to repress had subsided25, fueling the relentless evolution of more complex gene regulatory networks. Unlike an arms race with an external pathogen, retrotransposons are host DNA, suggesting that a mammalian genome is itself in an internal arms race with its own DNA, and thereby inexorably driven toward greater complexity.
Methods
Embryonic Stem Cell culture and ZNF overexpression analysis
Human (H9) ESC colonies were maintained as described (http://www.wicell.org). Colonies were manually passaged at a 1:3 ratio onto plates containing mitomycin-C-treated mouse embryonic fibroblasts that were seeded at a density of 35 k cells/cm2 on 0.25% gelatin coated plates (porcine; Sigma) the day before. Mouse transchromosomic E14(hChr11) (TC11) embryonic stem cells were cultured on mouse embryonic fibroblast feeder layers as described14. For transfections, cells were cultured on gelatin for 2 passages and transfected with 24 ug of ZNF and 1 ug of GFP expression vectors per 10 cm plate of cells, using lipofectamine 2000 (Invitrogen). Cells were cultured for an additional 40 hours, harvested with trypleE reagent (Life technologies) and washed three times and collected in FACS buffer (1 x PBS, 2% fetal bovine serum (FBS), 5 mM EDTA). GFP-positive cells were sorted using a FACSAria III (BD Biosciences) and samples were used for RNA isolation and ChIP analysis.
RNA-seq library preparation
RNA was treated with RQ1 DNAseI (Promega) for 1 hour at 37 C and total RNA was cleaned up using the RNAeasy Mini kit (Qiagen). For each sample, the non-ribosomal fraction of 5 ug of total RNA was isolated using a Ribo-Zero rRNA removal Kit (Epicentre) following the manufacturer’s protocol (Lit. #309-6/2011). For the non-ribosomal fraction of RNA, double stranded (ds) cDNA was synthesized as described previously28 using dUTP in the second strand synthesis and USER digest before amplification to retain strand specificity. Clean-up steps were performed using RNA Clean & Concentrator or DNA Clean & Concentrator kits (Zymo research). Double stranded cDNA was used for library preparation following the Low Throughput Guidelines of the TruSeq DNA Sample Preparation kit (Illumina), with the following additions. Size selections were performed before and after cDNA amplification on an E-gel Safe Imager (Invitrogen) using 2% E-gel SizeSelect gels (Invitrogen). The cDNA fraction of 300–400 bp in size (including adapters) was isolated and purified. For adapter ligations, 1 ul instead of 2.5 ul of DNA Adapter Index was used. Indexed libraries were pooled and sequenced on the Illumina HiSEQ platform. 2 biological replicate samples were analyzed for EV transfected cells and ZNF91 transfected cells, 3 biological replicate samples were analyzed for human ESCs and 2 for rhesus LYON-ES1 ESCs. Data can be viewed on the UCSC browser: http://goo.gl/5RItX24
Mapping and analysis of RNA-seq data
All samples were mapped using Tophat229 with Bowtie230 as the underlying alignment tool. The input Illumina fastq files consisted of paired end reads with each end containing 100bp. The target genome assembly for the human samples was GRCh37/UCSC-hg19 for hESCs, or a hybrid target genome of mm9-hChr11 for TC11-mESCs, and Tophat was additionally supplied with a gene model (using its “-GTF” parameter) with data from the hg19 UCSC KnownGenes track31. For multiply-mapped fragments, only the highest scoring mapping determined by Bowtie2 was kept. Only mappings with both read ends aligned were kept. Potential PCR duplicates (mappings of more than one fragment with identical positions for both read ends) were removed with the samtools “rmdup”32 function, keeping only one of any potential duplicates. The final set of mapped paired-end reads for a sample were converted to position-by-position coverage of the relevant genome assembly using the bedtools “genomeCoverageBed”33 function. To determine the count of fragments mapping to a gene, the position-by-position coverage was summed over the exonic positions of the gene. This gene total coverage was divided by the factor 200, to account for the 200 bp of coverage induced by each mapped paired-end fragment (100bp from each end), and rounded to an integer. For the human samples, this was calculated for each gene in the UCSC Known Gene set. For input to DESeq34 all genes with nonzero counts in any sample were considered. Two replicates of each sample were combined per the DESeq methodology.
For Figure 2c, the median fold change in expression (ZNF91/EV, vertical axis) for genes with an SVA element within some distance (blue circles) and genes without an SVA element within the same distance (gray crosses) were plotted against the up or downstream distance from each gene. A total of 994 expressed genes were considered. Points were computed every 2.5 kbp, For every window size starting at 2.5kbp and progressing cumulatively up to 250kbp in 2.5kbp intervals upstream and downstream of genes on chromosome 11, we identified the set of genes with and without at least one SVA element within the window. For the two sets (genes with SVA and genes without SVA), at every window size we calculated the median fold change in gene expression (ZNF91/EV) using the DESeq results from TC11-mESCs transfected with either ZNF91 or empty vector (EV). The python script to generate the figure and the associated data are available at http://hgwdev.sdsc.edu/~ewingad/Tc11SVAFig2e.tar.gz
Chromatin immunoprecipitation (ChIP), ChIP-qPCR and ChIP-seq library preparation
Human (H9) and mouse ESCs (46C and transchromosomic TC11) were crosslinked in 1% formaldehyde for 10 minutes on ice by adding 1/10 volume of freshly prepared 11X cross-linking solution (50 mM Hepes (pH 8.0); 0.1 M NaCl; 1 mM EDTA; 0.5 mM EGTA; 11% formaldehyde). The crosslinking reaction was quenched by adding glycine to a final concentration of 0.125 M and incubating for 5 minutes on ice. For KAP1-ChIP and ChIP with the KZNF antibody ab104878, cells were washed 3x in PBS + 0.1% BSA and dissolved in 10 packed cell volumes 0.3 % SDS-lysis buffer (10 mM Tris (pH 8.0); 1 mM EDTA (pH 8.0); 0.3% (w/v) SDS + Complete proteinase inhibitor cocktail (Roche)). Cells were incubated on ice for 20 minutes and cells were lysed in a pre-chilled dounce homogenizer by ten strokes with pestle B. Cell lysate was transferred to a 15 ml conical (human ESC) or 1.5 ml tube (mouse ESC) and chromatin was sheared to an average size of ~500 bp in a Bioruptor Sonicator (Diagenode) (settings: HIGH; 30″ on; 60″ off; 10–12 cycles). Sonicated lysate was transferred to 2 ml tubes and 3 volumes of immunoprecipitation buffer (50 mM Tris-HCl (pH 8.0); 150 mM NaCl; 5 mM MgCl; 0.5 mM EDTA; 0.2% NP-40; 5% glycerol; 0.5 mM DTT); Complete Protease Inhibitor Cocktail was added. Debris was pelleted by centrifugation for 15 minutes at 12,000 g at 4C and supernatant was transferred to a new 2 ml vial. Supernatant was precleared with 50 ul of Sheep-anti-Rabbit (M-280) Dynabeads (Invitrogen) for 4 hours at 4C. Dynabeads (Invitrogen) were blocked with BSA according to the Dynabeads manual. Precleared lysate was incubated with 10 ul of dynabeads suspension pre-bound for 4 hours with an excess of anti-KAP1 antibody (ab10484), or anti-KRAB ZNF-antibody (ab104878). Immunoprecipitation was performed overnight at 4C on a rotator. Immune-complexes were washed 6 times in freshly prepared RIPA buffer (50 mM Hepes (ph8.0); 1 mM EDTA (ph8.0); 1% (v/v) NP40; 0.7% (w/v) deoxycholate; 0.5 M LiCl; Complete Proteinase Inhibitor Cocktail) and 1 time in TE buffer (10 mM Tris-HCl (pH 8.0); 1 mM EDTA (pH 8.0)). H3K4me3-ChIP (H3K4me3 antibody: Milipore; catalog# 07-473; lot# JBC1888194) was performed as described previously by the Ren Lab (http://commonfund.nih.gov/sites/default/files/ChIP_Broad_ChIP_REMC_Protocol_v01B.pdf). Immune-complexes were eluted from the beads by incubation at 67 C for 20 minutes in ChIP elution buffer (TE + 1% SDS) and vortexing every 2 minutes and cross-linking was reversed by incubation at 67 C overnight. ChIP DNA was treated with RNAse A/T for 2 hours at 37 C and Proteinase K for 2 hours at 55 C. NaCl was added to a final concentration of 200 mM and ChIP DNA was extracted twice with Phenol/Chloroform/Iso-amyl-alcohol (25:24:1) and twice with Chloroform/Iso-amyl-alcohol (24:1). ChIP DNA was ethanol precipitated and dissolved in nuclease free water. ChIP DNA was cleaned up one extra time using ZYMO PCR purification columns.
To determine the genome-wide binding of ZNF93, we performed chromatin immunoprecipitation (ChIP) analysis, using a KRAB ZNF antibody (ab104878) which was originally raised against a peptide in ZNF486 that displays 88% identity to ZNF93 and we show is capable of recognizing ZNF93 (Extended Data Fig S7a, b). Notably, the size of the protein immunoprecipitated by ChIP from human ESC lysates corresponds to the size of ZNF93 and not ZNF486, arguing that this antibody predominantly immunoprecipitates the highly expressed ZNF93. To establish that ZNF93 can direct ab104878 to the L1PA4 5′UTR, ChIP-quantitative PCR was performed on ab104878-ChIP-DNA derived from 3 biological replicates of TC11-mESCs transfected with either pCAG-EV or pCAG-ZNF93. qPCR was performed on a Roche LightCycler 480 II, using primers to amplify an amplicon in the 5′UTR of L1PA4 (f: CATTTGCGGTTCACCAATATC; r: GCTAGAGGTCCACTCCAGAC) and LTR12C (f: GCACTTGAGGAGCCCTTCAG; r: ACACCTCCCTGCAAGCTGAG).
For ChIP-seq analysis, ChIP DNA was used for library preparation following the Low Throughput Guidelines of the TruSeq DNA Sample Preparation kit (Illumina), with the following minor additions. Size selections were performed before and after amplification on an E-gel Safe Imager (Invitrogen) using 2% E-gel SizeSelect gels (Invitrogen). The ChIP-DNA fraction of 300–400 bp in size (including adapters) was isolated and purified. For adapter ligations, 1 ul instead of 2.5 ul of DNA Adapter Index was used. Indexed libraries were pooled and sequenced on the Illumina HiSEQ platform. For ChIP-seq analysis in hESCs, 3 biological replicates of KAP-ChIP, 2 biological replicates of H3K4me3 ChIP and 2 biological replicates of ab104878 ChIP were analyzed, and for H3K4me3 ChIP-seq analysis in TC11-mESCs, 2 biological replicate samples were analyzed for EV transfected cells and ZNF91 transfected cells, and one sample was analyzed for other KZNF genes reported in Extended Data Figure 5c. Data can be viewed on the UCSC browser: http://goo.gl/5RItX2
MACS ChiP-seq peak calling
All samples were mapped using Bowtie30 using input Illumina fastq files consisting of paired end reads. The human samples were mapped to the GRCh37/UCSC hg19 genome assembly. Only fully paired end, uniquely mapping reads were kept. Potential PCR duplicates (mappings of more than one fragment with identical positions for both read ends) were removed with the samtools “rmdup”32 function, keeping only one of any potential duplicates. Based on the paired end mappings, the median length of the fragments was determined for each sample. For input to MACS 1.435 only the read1 mappings were used and the median fragment length was used to determine the “-shiftsize” parameter. For each ChIP sample mappings, the corresponding input DNA sample mappings were used as a control. The UCSC table browser36 was used to select MACS peaks that were called in both biological replicates. The overlap between KAP1 ChIP-seq replicates is ~30%, which is lower than expected and can probably be best explained by numerous retrotransposon and promoter regions on the genome displaying a low level of (transient?) KAP1 binding, that may be below threshold in the one, and above threshold in the other replicate.
Quantification of ChIP-seq and RNA-seq data for Figure 1b and 2b: For specific retrotransposon classes, the percentage of elements on human chromosome 11 (a total of 173 SVA elements; 15 full length L1Hs elements; 84 full length L1PA4 elements) that overlapped with KAP1 ChIP-seq peaks and H3K4me3 ChIP-seq peaks in hESCs and mouse TC11-mESCs was determined using the UCSC table browser. Only L1PAs >5700 bp were considered to select (near) full length L1 elements for the analysis. Transcription derived from individual SVA, full length L1Hs and full length L1PA4 human chromosome 11 elements in human ESCs and mouse TC11-mESCs was scored manually based on the RNA-seq coverage track uploaded in the UCSC browser, using a fixed scale that was normalized for relative sequencing depth. Level of transcription was divided in four categories: no (~0–10 reads), low (~10–30 reads), moderate (~30–50 reads) and high transcription (>50 reads). Isolated reads were not counted as transcription, nor were elements scored as transcribed when the transcription covering the retrotransposon was clearly part of exonic or intronic expression of genes. For Figure 2b, only H3K4me3 ChIP-seq peaks that had a minimal ‘score’ of 100 for both EV-transfected and ZNF91 transfected mouse TC11-mESCs were considered. The ‘score’ is a value defined by MACS analysis representing the ‘height’ of each ChIP-seq signal, and the score of ‘100’ is an arbitrary cut off that we choose. This provides a quantitative measure of the percentage of SVAs on chromosome 11 that display a reduction of H3K4me3 signal. For the pie charts in Figure 3a, we used the UCSC table browser to determine the percentage of full-length L1PA elements on chromosome 11 that overlapped with an ab104878-ChIP-seq peak in the 5′UTR (5′-most 1000 bp of each individual L1PA element). This analysis was based on 15 L1Hs, 54 L1PA2, 29 L1PA3-6030, 36 L1PA3-6160, 83 L1PA4, 39 L1PA5, 41 L1PA6, 50 L1PA7 and 14 L1PA8 full-length elements. The following should be noted about the small fraction of L1PA2 (7%) and L1PA7 (8%) that overlap with ab104878-ChIP-seq peaks in the 5′UTR, whereas based on the repeat browser no ab104878 ChIP-summit is observed. The annotation of L1PAs on the Repeat Masker track is based on ~500 bp in the 3′ UTR only, whereas the L1PA reference sequences in the repeat browser we used to generate the ChIP-seq summit tracks in Fig3a are based on the consensus of full length L1PA sequences. In the repeat masker track that was used to make the pie-charts, we have noticed incidental misannotations for these highly similar L1PA subfamilies. In particular, some L1PAs appear to be one subtype on the 3′ end (based on which they were categorized) yet are annotated as a different subfamily on the 5′ end. In fact, manual analysis of the 7% of Repeatmasker-annotated L1PA2 fragments positive for KZNF-ChIP, revealed that all are misannotations and based on the consensus of the full length L1PA sequence should have been categorized as L1PA4 or L1PA3.
Immunoblotting
Human ESC (H9) and ZNF-transfected mouse transchromosomic ES and HEK cells were lysed in 50 mM Tris-HCl (pH 8.0); 150 mM NaCl; 5 mM MgCl; 0.5 mM EDTA; 0.2% NP-40; 5% glycerol; 0.5 mM DTT and Complete protease inhibitor cocktail (Roche) and centrifuged at max speed for 10 minutes at 4C to remove debris. Cleared lysates were subjected to SDS-PAGE on Nupage (Invitrogen) 4–12% protein gels for SDS-PAGE and transferred to nitrocellulose as described in the Nupage manual. Blots were incubated overnight in 5% non-fat dried milk in PBS-T and incubated with 1:1000 anti-KAP1 antibody (ab10484), 1:1000 anti-KZNF antibody (ab104878) or 1:1000 anti-HA (ab9110) antibody in PBS for 3 hours and Goat-anti-rabbit-HRP secondary antibody for 30 minutes at room temperature (RT). Blots were incubated with SuperSignal West Dura Extended Duration Substrate (Thermo Scientific) and visualized on a Biorad Chemidoc MP system.
Plasmids
KZNF cDNAs were amplified from hESC cDNA, isolated from IMAGE clones or synthesized (Genscript) and cloned into pCAG EN (Addgene 11160) for transient transfections. For generation of the luciferase constructs, SVA_D (Hg19: chr11:65,529,663-65,531,199) was synthesized (Genscript); the OCT4 enhancer region (OCT4Enh; Hg19: chr6:31,139,549-31,141,393) was amplified by PCR from hESC gDNA, and L1PA4-5′UTR (chr11:74,005,653-74,006,113) was synthesized (IDT, gBlock) and were cloned upstream of a pGL4CP-SV4036 luciferase reporter construct. Retrotransposition assay constructs were modified from pCPE4-L1RP-GFP22. Detailed plasmid descriptions and sequences of inserts can be found at (http://compbio.soe.ucsc.edu/arms-race-znfs-retrotransposons/methods-plasmids.docx).
Luciferase Assay
Luciferase assay was carried out according to Promega dual-luciferase kit instructions and as previously published36. 46C37 mESCs were plated in the afternoon on gelatin coated 24 well plates at 35k/cm2. The next morning, media was changed and 200ng of pCAG ZNF was co-transfected with 20ng of SV40-Luciferase reporter and 2ng of pRL-TK-Renilla (a 10:1 firefly to Renilla ratio) per 24-well using Lipofectamine2000 in duplicate wells. 24 hours after transfection, wells were washed 1x with PBS, harvested with 100ul of Passive Lysis Buffer for 15 minutes on a room temperature rocker. Each well is then read in duplicate as 40ul of lysate was transferred twice to a 96 well white opti-plate and combined with 50ul of LARII substrate and read on a Perkin-Elmer luminometer and Wallace Victor Light software counting 1s/well. Next, lysate and substrate was combined with 50ul of Stop & Glo reagent to quench and measure Renilla activity to control for transfection efficiency. Data were normalized in Microsoft Excel by dividing firefly/renilla and the average of 4 technical replicate measurements was taken as a raw value of activity. This activity was further normalized against SV40-Luciferase control for each KZNF pCAG construct. Final values are displayed where for each biological replicate, pCAG EV (empty vector) is set to 100%. Statistical testing was done by Student’s T-Test, two tailed; equal variance and statistical differences of P<0.01 were indicated in the figures as **. The following number of biological replicates were used: Figure 2a: EV (n=42); ZNF90 (n=6); ZNF91 (n=17); ZNF93 (n=9); ZNF254 (n=10); ZNF443/ZNF460/ZNF486/ZNF519/ZNF 544/ZNF 587/ZNF589/ZNF714/ZNF721/ZNF33a (n=3). Figure 2e: EV (n=6); Human ZNF91 (n=3); Hominine ZNF91 (n=3); Great ape ZNF91 (n=3); Macaque ZNF91 (n=3). Figure 3c: EV (n=6); ZNF93 (n=3). Figure 3e: EV (n=6); ZNF93 (n=4); ZNF93SerF (n=6). Extended data figure S4a: n=6. Extended data figure S4b: ‘no VNTR’ (n=9); ‘partial VNTR’ (n=3); ‘ no hex/Alu’ (n=2); ‘no hex’ (n=2); ‘full length SVA’ (n=15); ‘SINE-R’ (n=3). Extended data figure S4c: n=3. Extended Data Figure 6c : EV (n = 42): ZNF91 (1-11) (n = 4); ZNF91 (1-24) (n = 7); ZNF91 (1-30) (n =4 ); ZNF91 (1,2; 23-36) (n = 3). Extended Data Figure 7a: n=3 Extended Data Figure 8b: (n=4)
Retrotransposition assay
The full length L1Hs retrotransposition reporter construct (Ostertag et al 2000 NAR), was modified to have the 129 bp element of L1PA4 (L1Hs-129L1PA4 ) or a scrambled 129 bp sequence (L1Hs+129 scramble) inserted at the corresponding position where the 129 bp element is present in L1PA4 and lost in L1PA3-6030. See http://compbio.soe.ucsc.edu/arms-race-znfs-retrotransposons/methods-plasmids.docx for more details on the cloning of these constructs. Retrotranspositon assay of L1Hs and related 129L1PA4 containing constructs was carried out based on established protocols 22, 38. HEK293FT cells were plated at 35k/cm2 on 6 well plates and incubated overnight in DMEM+FBS (no pen/strep). The next day cells were transfected with 300 ng of L1HS reporter and 1ug of pCAG EV or pCAG ZNF93 using lipofectamine 2000/Optimem (Invitrogen), and media was changed after 6 hours per manufacturer recommendations. Cells were maintained and on day 4 cells were harvested with TrypLE, washed 2x with PBS, placed on ice and incubated with propidium iodide. 250,000 cells per transfection were analyzed for GFP positive and dead cells on a BD LSR II. Data were gated and analyzed in FlowJo software to determine the number of live, GFP positive cells. Biological replicates: n=7; Statistical testing: Student’s T-Test, two tailed; equal variance.
Repeat Browser
We constructed a consensus sequence of SVA_D and L1PA elements. To remove extremely short and long copies, we first eliminated the longest 2% of the copies in the genome, then took the 50 longest sequences annotated by repeatmasker (www.repeatmasker.org) in the UCSC genome39, aligned them with muscle and constructed a consensus sequence from the multiple alignment. We created a version of the UCSC genome browser using this consensus as a reference sequence. MACS summits of KZNF(ab104878)-ChIP-seq and KAP1-ChIP-seq were mapped to the Repeat browser for Figure 3a and 3b. (Link to Repeat browser: http://goo.gl/4Xxnny)
Multi-species ZNF91 and ZNF93 nucleotide sequence identification
We focused on finding homologs in other species for the fourth exon of human ZNF91 and ZNF93, which contains all the important functional domains of the genes, including the KRAB domains and all the zinc finger domains. Using BLAT from the UCSC genome browser toolset to align the human ZNF91 (ENST00000300619) genomic nucleotide sequence (UCSC hg19 chr19:23,539,498-23,579,269, from 1 kb upstream to 1 kb downstream), we identified best reciprocal hit ZNF91 sequences in the chimpanzee (panTro4), gorilla (gorGor3), orangutan (ponAbe2), gibbon (nomLeu3), rhesus (rheMac2) and baboon (papAnu2) genomes. Of note, for rhesus, we used the rheMac2 assembly because we have identified a potential assembly error in the ZNF91 fourth exon region of the latest assembly, rheMac3, which resulted in an early stop codon. The ZNF91 sequence obtained from rheMac2 was validated by RNA-seq data.
For ZNF93, the human fourth exon is located at: UCSC hg19, chr19:20,043,993-20,045,627. We extracted the homologous regions in other species using the UCSC 100 vertebrate species multiple sequence alignment (UCSC browser (genome.ucsc.edu), Multiz Alignments of 100 Vertebrates track). To refine the alignments, we independently aligned the human ZNF93 fourth exon nucleotide sequence to these homologous regions together with their immediate upstream and downstream regions (using BLAT) and manually analyzed and ensured the quality of the alignments. We obtained homologs for chimpanzee (panTro4 chr19:20,255,111-20,256,670), gorilla (partial homolog due to missing information, gorGor3 chr19:20,328,848-20,330,482), orangutan (partial homolog due to missing information, ponAbe2 chr19_random:3,818,660-3,820,506), green monkey (chlSab1 chr6:18,428,342-18,430,231), rhesus (rheMac3 chr3:73,136,331-73,137,882), crab-eating macaque (macFas5 chr19:20,589,892-20,591,781) and baboon (papHam1 scaffold15384:40,473-42,362). We aligned these sequences back to the human genome and validated that ZNF93 was their best match. We used RAxML to construct a phylogenetic tree for these sequences and sequences of human ZNF93 and its close relatives ZNF90, ZNF737 and ZNF626. The results confirmed that these sequences were closest to human ZNF93. To check for reciprocal best matches, we aligned the human ZNF93 fourth exon sequence to the species genome assemblies. Due to high repetitiveness of the zinc-finger domains and high diversity of the sequences across species, the alignments resulted in a large number of matches, many of which spanned large regions (i.e. false positive matches with large “introns”). We manually analyzed these alignments and confirmed that the regions listed above were the best matches.
The ZNF93 match in gibbon (nomLeu3 chr10:54,583,066-54,586,723) contains long insertions, indicating that there are potential errors in the gibbon reference assembly, and/or that the exon is broken into multiple exons in gibbon, and/or that the gibbon exon contains extra bases). In the next section, we explain how we used PCR to correct assembly errors in the gibbon reference to obtain a valid gibbon homolog.
Closing gaps in the Orangutan and Gorilla assemblies overlapping ZNF91 and ZNF93 exon 4 and correcting sequencing errors in the Orangutan assembly overlapping ZNF91 exon 4 and in the Gibbon assembly overlapping ZNF93 exon 4
Alignments of both translated amino acid and nucleotide sequences revealed that the identified Orangutan and Gorilla sequences had scaffold gaps within the fourth exon of the gene ZNF91, which includes crucial zinc fingers. To fill in the gaps and correct assemblies we used gDNA from orangutan and gorilla fibroblasts (Coriell, San Diego Zoo), and performed PCR using a selection of primers that are provided at http://compbio.soe.ucsc.edu/arms-race-znfs-retrotransposons/Gorilla-Orangutan-Gibbon-Assembly-Gap-Primers.docx. Cloned PCR-products were Sanger sequenced and sequences were aligned to the corresponding assemblies as well as to the human genome using BLAT. Only clones that mapped uniquely with at least 90% coverage to the corresponding regions were kept. Similarly, Orangutan and Gorilla sequences had scaffold gaps within the fourth exon of the gene ZNF93. We used gDNA from Sumatran Orangutan and Gorilla fibroblasts (San Diego Zoo) to fill in these gaps.
We identified potential assembly errors in the Gibbon reference assembly (nomLeu3). To obtain a confident homolog of ZNF93 exon 4 in gibbon, we used gDNA of Gibbon species Hylobates pileatus, Hyloplates gabriellae, Nomascus leucogenys, which were a kind gift from Lucia Carbone (Oregon Health Sciences Univeristy Primate Center) and purchased from Coriell Cell Repositories. Purified PCR products were ligated into PCR4-TOPO (Invitrogen) and sequenced. The resulting sequences were aligned to the gibbon reference assembly (nomLeu3) and were manually analyzed and assembled into the consensus gibbon ZNF93 fourth exon sequence. The reference gibbon assembly nomLeu3 contains one tandem duplication (of the corresponding human domains 6-12) and one long insertion (~1kb), both were refuted by sequence evidence obtained from this experiment.
Reconstructing the evolutionary history of ZNF91
Multiple sequence alignments revealed a 588 base pair subsequence containing 7 extra zinc fingers in the human, chimpanzee and gorilla genomes that is not present in the orangutan, gibbon, rhesus and baboon genomes. This additional sequence corresponds to zinc fingers 6-12 of the human protein. Using BLAT to align the human copy of this sequence to the human genome, human zinc fingers 7-12 (2-7 of the subsequence) have best-reciprocal homology to zinc fingers 18-23 of human ZNF91, indicating that the subsequence was initially created by a local segmental duplication. Further analysis revealed human zinc finger 6 (the first zinc finger of the additional subsequence) is a near exact, best-reciprocal match of human zinc finger 7 (the 2nd zinc finger of the additional sequence), indicating that after the initial intra-gene segmental duplication there was a secondary tandem duplication of the first zinc finger. Blat analysis revealed the additional subsequence is not present anywhere in the orangutan and other outgroup genomes. To reconstruct a parsimonious nucleotide level evolutionary history of ZNF91, we constructed a global multiple sequence alignment using PRANK40 (http://tinyurl.com/prank-msa), which simultaneously aligns the sequences and infers the ancestral sequences using a realistic model of insertion, deletion and substitution evolution. To include the two inferred duplication events in this history we created edited versions of the human, chimpanzee and gorilla sequences with the additional duplicated sequence removed and included, for each species, as two extra input nucleotide sequences, one of the first additional zinc finger (zinc finger 6 in the human protein), and the second of the subsequent 6 additional zinc fingers (zinc fingers 7-12 in the human protein). As PRANK requires a phylogenetic tree, we gave it a tree that reflects the accepted species phylogeny, but which included the additional duplications branching off after the speciation from orangutan (Supplementary Figure S6a). There were 4 amino acid changes in DNA-contacting residues in the relatively short critical time 12-8 MYA after orangutan branched off and before the human-chimpanzee-gorilla split. This together with the duplications mentioned above gives an indication of positive selection. The full multiple species alignment is available at http://compbio.soe.ucsc.edu/arms-race-znfs-retrotransposons/znf91/znf91msa.html
Reconstructing the evolutionary history of ZNF93
Multiple sequence alignment and sequence analyses (Extended Data Figure S8a) revealed a deletion of four zinc finger domains (located between human domains 5 and 6) in the common ancestral great ape lineage after the split with gibbon (deleted in orangutan, gorilla, chimpanzee and human, but present in gibbon and old world monkeys (crab-eating monkey, rhesus, baboon and green monkey). Domains 5 and 6 (w.r.t. human) are identical to each other in the great ape species. Domain 13 (w.r.t human) is missing in old world monkeys and is identical to domain 12 in all apes, suggesting that this domain is likely the result of a tandem duplication event that occurred in the ape last common ancestor, after the split with non ape old world monkeys. Domain 17 (w.r.t. human) is present in human, crab-eating monkey and baboon (unknown in rhesus due to missing data) while missing in green monkey, gibbon, orangutan, gorilla and chimp. Analyzing the nucleotide sequences shows that one nucleotide insertion in the ape common ancestor (w.r.t old world monkeys) results in an early stop codon and the loss of this domain, and a compensatory deletion of four nucleotides in human (w.r.t. apes) nullifies the effect of the previous ape mutation and results in restoration of domain 17 in human. So human ZNF93 is not like the protein of other apes. The multiple sequence alignments were obtained and validated using MUSCLE41, MAFFT42 and PRANK40 and the ancestral reconstruction was constructed using PRANK. The full MSA is at http://compbio.soe.ucsc.edu/arms-race-znfs-retrotransposons/znf93/znf93msa.html.
Phylogenetic analysis and calculation of evolutionary divergence of L1PA3-6030 and L1PA3-6160 subclasses
50 sequences of L1PA3-6030, 50 sequences of L1PA3-6160, 3 sequences of L1PA2 and 3 sequences of L1PA4 were aligned by ClustalW in MEGA6 sofware package43. Only full length L1PAs were selected. For phylogenetic analysis, the sequence downstream of the 129 bp element (L1PA4 and L1PA3-6160), or the corresponding position (L1PA2 and L1PA3-6030) was used to generate phylogenetic trees. Multiple methods were used (Maximum Parsimony, Minimum Likelihood and Minimum Evolution) to generate trees with comparable outcome. The phylogenetic tree generated by the Minimum Evolution method27 was used to calculate the divergence times for all branching points with the RelTime method44.
To calculate the average divergence from consensus, first consensus sequences were calculated for L1PA3-6030 and L1PA3-6160, from 150 full length elements of each subclass using EMBOSS software (emboss.sourceforge.net). Each consensus sequence was aligned in MEGA6 with the respective 150 full-length elements by ClustalW. In order to be able to compare values for L1PA3-6030 and L1PA3-6160 to divergence values for other L1PA subfamilies, determined previously9 we used the 500 bp of the 3′ end of the L1PA3 subclasses, and excluded the polyA-stretch at the 3′-end of L1PAs. The pairwise distances for each of the 151 (500bp) sequences (150 individual L1PAs and 1 consensus) were calculated in MEGA6 and plotted in a distance-matrix. The average distance (divergence) from consensus was determined by calculating the mean distance (+/− SEM) from the consensus sequence to each individual L1PA3 element. The age of each L1PA3 subclass was estimated using a bp substitution rate of 0.17%/myr9.
VNTR size analysis for SVA-subfamilies
We extracted RepeatMasker SVA elements in the human genome as annotated in the UCSC Genome Browser RepeatMasker track (hg19.rmsk). Each element was annotated with Tandem Repeat Finder45 to identify all base pairs covered by a tandem repeat. While VNTR and HEX domains are both tandem repeats, we assumed that the length of the HEX region is a lot shorter and relatively fixed compared to the VNTR, so in the following we use the length of all base pairs masked by Tandem Repeat Finder as a proxy for the length of the VNTR. SVAs annotated by RepeatMasker as multiple adjacent SVA fragments can correspond to a single full length SVA element. Therefore, to restrict our analysis to unbroken full-length elements, we concentrated on elements that displayed an‘intact’ SVA structure, with at least 800 bp of sequence outside of the VNTR region, a size that corresponds to the sizes of Alu and SineR combined. For this enriched set of SVAs the histogram of VNTR lengths was plotted in Extended Data Figure S10c.
Determination of changes per million years for Figure 4
For ZNF91 and ZNF93, we counted the numbers of zinc fingers that have undergone structural changes that could affect DNA binding specificity for each of the evolutionary branchpoints, based on the multiple sequence analysis and ancestral reconstruction (see Methods sections “Reconstructing the evolutionary history of ZNF91“ and “Reconstructing the evolutionary history of ZNF93”). Changes in DNA binding residues, zinc finger deletions or zinc finger duplicaions/gains were all weighed equally and counted as ‘1’ because it is unpredictable how each of these changes may change target DNA recognition. The number of changes from one branchpoint to another was divided by the number of million years of that timeframe to determine the number of zinc fingers that changed per million years. For zinc fingers in ZNF93 that were different between macaque and gibbon, but conserved between Gibbon and great apes, we lacked an outgroup species necessary to determine when the changes have occurred. Therefore, to get a rough estimate, we divided the total number of changes between macaque and gibbon, by the amount of time on each of these lineages: From the point of divergence of old world monkeys to present day macaque is 25 myr, from the point of divergence of old world monkeys to the LCA of gibbon and great apes is 7 myr (25 myr – 18 myr). Therefore we estimated that about ¾ of the observed changes happened on the macaque lineage and ¼ of the changes on the lineage to the LCA of gibbon and great apes. Similarly, for L1PA elements the consensus sequences of each L1PA element was compared to its direct predecessor and successor, and bp substitutions, bp deletions or bp insertions were all counted as ‘1’. The number of bp changes per site within the 5′UTR (1000 bp) from one L1PA element and its successor was divided by the number of years within the time-frame each L1PA-subfamily was dominant9. (See methods section: Phylogenetic analysis and calculation of evolutionary divergence of L1PA3-6030 and L1PA3-6160 subclasses) to get the bp changes/site/myr values. For SVA, the percentage of VNTR increase/myr between SVA-subfamilies is indicated for the timeframe from the emergence of one SVA-subfamily to the successor. The average VNTR size for SVA-subtypes as determined in this study (Extended Data Fig. 10c) and the estimated timepoints of emergence previously reported for SVA-subfamilies12 were used to calculate the percentage increase of VNTR size/myr.
Extended Data
Extended Data Figure S1. KAP1 associates with recently emerged transposable elements.
a, Immunoblot incubated with anti-KAP1 antibody loaded with 1% input and eluates of KAP1-ChIP or IgG-ChIP derived from human ESC lysates. b, Diagram showing numbers of KAP1 peaks identified in two independent biological replicates and common peaks. c, Distribution of 9174 KAP1-ChIP-seq peaks over various DNA elements. d, Distribution of retrotransposon classes among KAP1-ChIP peaks from hESCs (left) or genome wide (right) e, KAP1 and H3K4me3 ChIP-seq and RNA-seq coverage tracks for a representative region on human chromosome 11 in hESCs (white-, grey-shaded) and TC11-mESCs (yellow-shaded). Blue arrows: de-repressed retrotransposons; black arrows: re-activated transcription; Red vertical shading: reactivated SVAs; orange shading: reactivated LTR12C. Blue and tan in RNA-seq tracks indicate positive and negative strand transcripts, respectively. Note that while the majority of SVAs display aberrant H3K4me3 signal, for unclear reasons not all SVAs display aberrant transcription in TC11-mESCs. sup: supernatant; Rep: biological replicate; TSS: transcription start site.
Extended Data Figure S2. Mouse KAP1 associates with mouse-specific retrotransposons in mouse ESCs.
a, Distribution of KAP1-ChIP-Seq reads from mouse ESCs (left) and the mouse genome (right) for retrotransposon families as defined by repeatmasker (RepeatMasker Open-3.0. http://www.repeatmasker.org. 1996–2010; Smit AFA, Hubley R, Green P.). b, UCSC Browser image displaying ChIP-seq tracks for input (grey shading) and KAP1 (red shading) as well as gene annotation and repeat element tracks for a region on mouse chromosome 1. Blue shading: KAP1-positive active mouse L1-subtypes26 Purple shading: KAP1-positive active IAP retrotransposons. TEs: transposable elements; IAP: Intracisternal A-particle.
Extended Data Figure S3. Selection of primate-specific KRAB ZNFs genes with high expression in hESCs.
a, Schematic of primate-specific KRAB zinc finger genes subdivided in different clades based on previous analysis7. KZNFs in (b) are in red. b, DESeq-calculated basemean expression levels for the 17 highest expressed KRAB zinc finger genes in hESCs (dark blue) and macaque ESCs (light blue), subdivided by clades.
Extended Data Figure S4. The SVA VNTR domain is necessary and sufficient for ZNF91-mediated repression of luciferase activity.
a–c, Schematic of SV40-Luciferase constructs used (left) and relative luciferase activity after transfection of the indicated constructs in mESCs (right). a, SVA and SINE-R are strong enhancers. (n = 6 biological replicates) b, Deletion analysis reveals the VNTR of SVA is required for ZNF91-mediated reporter regulation. Luciferase activity in the presence of ZNF91 expressed as a ratio of that observed for EV with the same reporter. Biological replicates: ‘no VNTR’ (n=9); ‘partial VNTR’ (n=3); ‘ no hex/Alu’ (n=2); ‘no hex’ (n=2); ‘full length SVA’ (n=15); ‘SINE-R’ (n=3). EV is set to 100% for comparison. c, 1.5 VNTR repeats is sufficient to confer ZNF91-mediated regulation on an OCT4Enh-SV40-luciferase reporter. Biological replicates: n=3. Student’s T-Test, two tailed; equal variance; Error bars: SEM. **p <0.01
Extended Data Figure S5. SVA is specifically repressed in vivo by ZNF91.
a, b Normalized DESEQ basemean values for H3K4me3 ChIP-seq (a) and RNA-seq (b) for retrotransposon classes that showed a significant change in ZNF91-transfected TC11-mESCs relative to EV. SVAs were the only transposable elements that showed a significant decrease (**= p-adj<0.01) in H3K4me3 and RNA-seq values. c, UCSC browser images for a representative SVA element, promoter and L1PA4 element, showing H3K4me3 ChIP-seq signal for hESC (grey) TC11-mESCs transfected with EV (yellow), pools of primate specific KRAB zinc fingers (green), and ZNF91 (red).
Extended Data Figure S6. Evolutionary history of ZNF91.
a, The phylogenetic tree used in multiple sequence alignment and ancestral reconstruction of ZNF91 (http://compbio.soe.ucsc.edu/arms-race-znfs-retrotransposons/znf91/znf91msa.html). “hu 1.1”, “ch 1.1” and “go 1.1” represent human, chimpanzee and gorilla domain 6, respectively, “hu 1.2”, “ch 1.2”, “go 1.2” represent human, chimpanzee and gorilla domains 7-12, respectively, and “hu 2”, “ch 2” and “go 2” represent ZNF91 sequence from start to domain 5, a breakpoint, and from domain 13 to the end (see Methods). Ancestors are labeled with first letters of leaf species below them, e.g. HCG is human-chimp-gorilla ancestor. b, Immunoblot incubated with anti-HA antibody on lysates of HEK293FT cells transfected with HA-tagged human, great ape, hominine and macaque ZNF91 proteins or lysates transfected with EV and pCAG-GFP. *= reconstructed ancestral protein. c, ZNF91 domain deletion analysis showing relative luciferase activities on the SVA_D-SV40 luciferase reporter after transfection of EV or ZNF91 deletion constructs in mESCs. Error bars: stdev. Numbers in parenthesis indicate zinc fingers present in the ZNF91 deletion construct. Student’s T-Test, two tailed; equal variance; *P< 0.05; **P<0.01. Biological replicates: EV (n = 42): ZNF91 (1-11) (n = 4); ZNF91 (1-24) (n = 7); ZNF91 (1-30) (n =4 ); ZNF91 (1,2; 23-36) (n = 3). EV= empy vector.
Extended Data Figure S7. L1PA4 elements are repressed by primate-specific ZNF93.
a, Relative luciferase activity on a L1PA4- and a OCT4enhancer-SV40-luciferase reporter after transfection of 14 KZNFs in mESCs. Significance measured relative to EV. Student’s T-Test, two tailed; equal variance; Error bars: SEM; *=p<0.05; **=p<0.01; (biological replicates: n=3 ). b, Immunoblot showing ChIP with antibody ab104878 predominantly reacts with a protein of ~70 kDA (left panel) and co-immunoprecipitates KAP1 (right panel). HC: heavy chain of IgG. c, Immunoblot demonstrating that ChIP with ab104878 detects overexpressed ZNF93 in mouse 46c ESCs as a ~70 kDA protein. d, Repeat Browser (see Methods) displaying ChIP-seq coverage tracks for ab104878 (ZNF93) (yellow shading) and KAP1 (blue shading) for a selection of KAP1-bound retrotransposons. e, ChIP-qPCR for amplicons in L1PA4 and LTR12C elements on chromosome 11 in mouse TC11 mESCs after transfection with EV or ZNF93 and ChIP with ab104878. ChIP enrichment is plotted as percentage of input. n=3 biological replicates; error bars, SEM; Student’s T-Test, two tailed; equal variance, *= p<0.05
Extended Data Figure S8. Reconstruction of the evolutionary history of ZNF93.
a, Schematic based on the multiple sequence alignment of ZNF93 orthologs (http://compbio.soe.ucsc.edu/arms-race-znfs-retrotransposons/znf93/znf93msa.html). Red shaded area: deletion of zinc fingers. Green shaded area: gain of zinc fingers. Green stripes: gained zinc fingers; Dark blue stripes: zinc fingers that changed contact residues in the lineage to humans; light blue stripes: changes in other lineages. Brown stripes: zinc fingers with different binding residues between macaque and gibbon, with gibbon sharing the great ape conformation: for these zinc fingers, it is unknown (‘?’) whether the change happened in monkey or in the LCA of gibbon and great apes after divergence of old world monkey (see methods). *= reconstructed ancestral protein. b, Relative OCT4enhancer-SV40p-luciferase activity for reporters with the indicated L1PA4 derived sequences after co-transfection of EV or various ZNF93 constructs. Error bars: SEM; **=p<0.01.
Extended Data Figure S9. Schematic of L1Hs retrotranspostion assay.
a, Schematic of constructs tested indicating the site of 129L1PA4 transplant into L1Hs and concept of L1-GFP assay24 in which GFP expression marks cells where a transfected L1-episome has retrotransposed into a HEK293 cell’s chromosomes.
Extended Data Figure S10. Evolutionary history of L1PA3-6030, L1PA3-6160 and the VNTR size in SVA.
a, Phylogenetic tree, rooted on L1PA4, generated using the Minimum Evolution method27 for fifty 3′-end sequences of L1PA3-6030 and L1PA3-6160, and three 3′-end sequences for L1PA2 and L1PA4. b, Bargraphs showing the number of SVA-A through SVA-F insertions in each great ape genome. c, Distribution of VNTR size for untruncated SVA elements in the human genome plotted for each SVA-subfamily. Number of untruncated elements identified for each subtype is indicated.
Acknowledgments
This work was supported by California Institute of Regenerative Medicine (CIRM) facility awards (FA1-00617, CL1-00506-1.2) and scholar awards (TG2-01157) to FMJJ and DG and a Human Frontier Science Program Postdoctoral fellowship (LT000689) to FMJJ. DH is an Investigator of the Howard Hughes Medical Institute. SK is supported by the California Institute for Quantitative Biosciences, ADE by TCGA U24 24010-443720 and MH by EMBO ALTF 292-2011. We thank Florence Wianny and Colette Dehay (Lyon University) for the LYON-ES1 macaque embryonic stem cells; Mitsuo Oshimura and Toshiaki Inoue (Tottori University) for the E14(hChr11) transchromosomic ES cells, Nader Pourmand and the UCSC genome sequencing center; Bari Nazario (UCSC Institute for the Biology of Stem Cells) for flow cytometry assistance; Mark Batzer (LSU) and Kyudong Han (Dankook University) for L1CER sequences, Lucia Carbone (OHSU) for gibbon gDNA, Arian Smit (ISB, Seattle) for discussions on L1PA evolution, Dave Segal (UC Davis) for advice on ZNF-mutations, Haig Kazazian, Dustin Hancks, and John Goodier (JHMI) for retrotransposition plasmids and advice, Kristof Tygi, Casey Vizenor, Jimi Rosenkrantz, Will Novey, Sandrine Kyane, and Brad Mylenek for technical assistance and the entire Haussler lab for discussions and support.
Footnotes
Author contributions
FMJJ, DG, DH, SRS designed and analyzed the experiments, FMJJ performed RNA-seq, ChIP-seq and reintroduction of primate ZNFs in transchromosomic mouse ESCs; DG performed ZNF cloning, luciferase reporter and retrotransposition assays; NN, DG, ADE and BP performed resequencing and analysis to complete the ZNF91 and ZNF93 loci in various primates; NN and BP reconstructed the evolutionary history of ZNF91 and ZNF93 ZNF domains, MH generated a Repeatmasker UCSC-Browser and hub, ZNF-binding site predictions and VNTR length analysis, SK processed and analyzed RNA-seq and ChIP-seq data; ADE analyzed SVA numbers in great apes and SVA-gene expression correlations. FMJJ, DG, SRS and DH wrote the manuscript.
RNA-seq and ChIP-seq data is deposited in the GEO database, accession no XXX.
Reprints and permissions information is available at www.nature.com/reprints
The authors declare no competing financial interests.
References
- 1.Kazazian HH. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–32. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 2.Cordaux R, Batzer Ma. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 5.Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–4. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birtle Z, Ponting CP. Meisetz and the birth of the KRAB motif. Bioinformatics. 2006;22:2841–5. doi: 10.1093/bioinformatics/btl498. [DOI] [PubMed] [Google Scholar]
- 7.Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 2011;21:1800–12. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, et al. SVA elements: a hominid-specific retroposon family. J Mol Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 9.Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. 2006:78–87. doi: 10.1101/gr.4001406.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowe HM, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–40. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 11.Turelli P, et al. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 2014 doi: 10.1101/gr.172833.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castro-Diaz N, et al. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev. 2014;28:1397–409. doi: 10.1101/gad.241661.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huntley S, et al. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–77. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kai Y, et al. Enhanced apoptosis during early neuronal differentiation in mouse ES cells with autosomal imbalance. Cell Res. 2009;19:247–58. doi: 10.1038/cr.2008.305. [DOI] [PubMed] [Google Scholar]
- 15.Gifford WD, Pfaff SL, Macfarlan TS. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol. 2013;23:218–26. doi: 10.1016/j.tcb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward MC, et al. Latent regulatory potential of human-specific repetitive elements. Mol Cell. 2013;49:262–72. doi: 10.1016/j.molcel.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancks DC, Kazazian HH. Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellefroid EJ, et al. Emergence of the ZNF91 Krüppel-associated box-containing zinc finger gene family in the last common ancestor of anthropoidea. Proc Natl Acad Sci U S A. 1995;92:10757–10761. doi: 10.1073/pnas.92.23.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–27. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Persikov AV, Osada R, Singh M. Predicting DNA recognition by Cys2His2 zinc finger proteins. Bioinformatics. 2009;25:22–9. doi: 10.1093/bioinformatics/btn580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore M, Choo Y, Klug A. Design of polyzinc finger peptides with structured linkers. Proc Natl Acad Sci U S A. 2001;98:1432–6. doi: 10.1073/pnas.98.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–23. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimberland ML, et al. Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum Mol Genet. 1999;8:1557–60. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 24.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–29. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci U S A. 2007;104:8005–10. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naas TP, et al. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J. 1998;17:590–7. doi: 10.1093/emboj/17.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rzhetsky A, Nei M. A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol. 1992;9:945–967. [Google Scholar]
- 28.Parkhomchuk D, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. 2009;37 doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu F, et al. The UCSC Known Genes. Bioinformatics. 2006;22:1036–46. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 32.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Onodera CS, et al. Gene isoform specificity through enhancer-associated antisense transcription. PLoS One. 2012;7:e43511. doi: 10.1371/journal.pone.0043511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–6. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 38.Hancks DC, Mandal PK, Cheung LE, Kazazian HH. The minimal active human SVA retrotransposon requires only the 5′-hexamer and Alu-like domains. Mol Cell Biol. 2012;32:4718–26. doi: 10.1128/MCB.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kent WJ, et al. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Löytynoja A, Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008;320:1632–5. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- 41.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K, et al. Estimating divergence times in large molecular phylogenies. Proc Natl Acad Sci U S A. 2012;109:19333–8. doi: 10.1073/pnas.1213199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–80. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]