Abstract
Recent research has deepened our understanding of the ancient, conserved chemosensory array that detects small molecule attractants and repellents, and directs the chemotaxis of bacterial and archaeal cells towards an optimal chemical environment. Here we review advances towards a molecular description of the ultrastable lattice architecture and ultrasensitive signal transduction mechanism of the chemosensory array, as well as controversies and challenges requiring further research. Ultimately, a full molecular understanding of array structure and on-off switching will foster (i) the design of novel therapies that block pathogenic wound seeking and infection, (ii) the development of highly specific, sensitive, stable biosensors, and (iii) the elucidation of general functional principles shared by receptor patches in all branches of life.
Keywords: two-component signaling pathway, chemotaxis, chemosensory receptor, methyl-accepting chemotaxis protein, histidine kinase, Tsr, Tar, CheA, CheW, ultrastable
The Bacterial Chemosensory Pathway
The bacterial chemosensory pathway is an ancient, ubiquitious signaling system that controls cell migration up or down chemical attractant or repellent gradients, respectively, towards an optimal living environment (reviewed in (1-4)). The resulting chemotactic behavior is essential for cell survival under nutrient stress, and to pathogenic processes including wound-seeking (5-7). The pathway components are localized within a two-dimensional, hexagonal signaling lattice termed the chemosensory array, which is remarkable for its highly sensitive detection of one or a few ligand molecules, its positive cooperativity involving dozens of coupled receptors, its multi-week kinetic stability, and its millisecond signaling speed (3,8-13). In short, the chemosensory array functions as an ultrasensitive, ultrastable biological integrated circuit or sensory chip. The array is conserved throughout Bacteria and Archaea (14-16), and it belongs to the ubiquitous two-component family of prokaryotic signaling pathways. However, its unique specializations for array structure and function set it apart from other family members, and yield similarities to clustered receptor patches now recognized as important elements in eukaryotic signaling (17-20). The present review focuses on the best-characterized bacterial chemosensory arrays of Escherichia coli, Salmonella typhimurium, and Thermatoga maritima.
Chemosensory Array Components, Assembly, Architecture, and Ultrastability
Components
All components of the bacterial chemotaxis pathway localize to the chemosensory array, which typically contains five protein functional classes: (i) transmembrane chemoreceptors for attractants and repellents, (ii) a pair of His and Asp protein kinases that form a phosphotransfer-relay (CheA and CheY, respectively), (iii) an adaptor protein that stabilizes array architecture (CheW), (iv) adaptation enzymes that covalently modify the chemoreceptors (CheR, CheB), and (v) a phosphatase that speeds hydrolysis of phospho-proteins as required for rapid responses to changing stimuli (CheZ) (1-4). High-resolution structures are available for all functional classes, and for several multi-protein complexes (1,4,21-25).
Core Unit
The first three of the above components constitute the structural and functional core of the array (1,4,26-28). Specifically, the core unit contains two receptor oligomers, the homodimeric His-kinase CheA, and two copies of the adaptor protein CheW (Figures 1A-C). As the receptor oligomer is a trimer-of-homodimers, the resulting receptor:kinase:adaptor stoichiometry is 2:1:2 (oligomers), or 12:2:2 (polypeptide chains). In the current model of core unit architecture, the kinase and adaptor proteins bridge the cytoplasmic tips of the two receptor oligomers (Figs 1B,C) (1,21-28). Core units have been directly detected by cryo-EM imaging of reconstituted complexes on bacterial membranes (1). Furthermore, core units reconstituted in a water-soluble nanodisc system are stable for hours and possess near-native, receptor-regulated kinase activity (29).
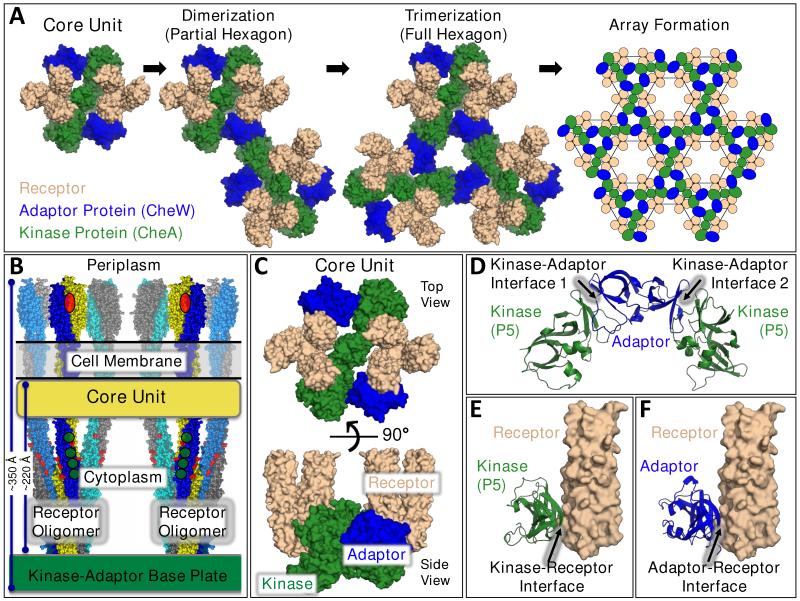
Figure 1. Chemosensory Array Components, Assembly, Architecture, and Stabilizing Contacts.
A) A core unit (far left) is comprised of the three core components: two receptor oligomers (each a trimer-of-homodimers, tan), a homodimeric His-kinase (CheA, green), and two copies of a monomeric adaptor protein (CheW, blue) (28-31,56,63). Core units are hypothesized to associate, forming dimers and then trimers during assembly of individual hexagons (1). Continued assembly forms a hexagonal array (far right) with receptor oligomers located at the vertices (21-23,26,27). In this array, the kinase and adaptor proteins are arranged in a system of interconnecting rings that stabilize individual hexagons and, more globally, the full lattice. In each hexameric ring, the structurally homologous kinase regulatory domain (P5) and the adaptor protein alternate, yielding pseudo-6-fold symmetry with contacts to each of the six surrounding receptor oligomers. Signals can be transmitted within individual core units, or between core units via the kinase-adaptor ring system. B) Schematic side view of the core unit, showing the two receptor trimers-of-dimers, the periplasmic attractant binding sites (red ovals), the cell membrane, the cytoplasmic adaptation sites (green circles), and the kinase-adaptor base plate region (4,23,26,27,46). C) Top view (from periplasm, upper) and side view (lower) of the molecular core unit model, focusing on the receptor protein interaction region and the kinase and adaptor proteins that stably bind to this region (21-23,26,27). D) Current models for the two types of interfaces, 1 and 2, between the kinase regulatory domain (P5) and the adaptor protein in the kinase-adaptor ring system (22,23,26,27). E), F) Current models for the kinase-receptor and adaptor-receptor interfaces, respectively (21,23,26,27).
Array Assembly
Figure 1A shows a hypothesized array assembly mechanism in which core units join together to form partial hexagons, then full hexagons, and ultimately the full array, which may contain hundreds of core units and hexagons. The proposed early intermediates have been detected by cryo-EM studies of in vitro reconstitutions that assemble the three core proteins on bacterial membranes (1).
Array Architecture
The structural framework of the array is provided by the cell membrane, by the long, three-pronged transmembrane receptor oligomers, and by the kinase-adaptor base plate located in a plane near the receptor cytoplasmic tips (Figs. 1B,C) (1,4,21-28). Diffusion of the receptors in the membrane is prevented by their tight coupling to the base plate via specific contacts with both the kinase and adaptor proteins. The base plate is an interconnected network of six-membered, kinase-adaptor rings, each of which possesses three kinase regulatory domains (P5) alternating with three adaptor proteins (Fig. 1A, right). Since the kinase regulatory domain is structurally homologous to the adaptor protein (both are dual-SH3-like-fold domains (30,31)), the six-membered ring possesses pseudo-6-fold symmetry, and each member binds to one of the surrounding six receptor oligomers.
The thickness of the E. coli array is ~350 Å from receptor tip-to-tip (Fig. 1B) (27). The cytoplasmic region resembles a proto-organelle wherein an aqueous, pseudo-compartment of height ~220 Å lies between the membrane and kinase-adaptor base plate (15); this height can vary between species possessing receptor cytoplasmic domains of different lengths (15,32). The edges of the array and the centers of the base plate hexagons remain open to the cytoplasm, but their narrowness can limit the rate of macromolecular diffusion between bulk cytoplasm and the internal compartment (26,27). Within this compartment the adaptation enzymes diffuse between receptors and modify their covalent adaptation sites. The resulting negative-feedback adaptation system enables the array to zero out a constant background concentration of attractant while responding to a superimposed attractant gradient (4).
Some species, such as Rhodobacter sphaeroides (33), possess soluble, double-layered cytoplasmic arrays in which two hexagonal lattices formed by soluble receptors, kinases and adaptor proteins are sandwiched back-to-back. Similar double-layered, soluble arrays can be formed by reconstituting the isolated cytoplasmic domains of transmembrane receptors with the kinase and adaptor proteins (1). In these soluble arrays the two-layered architecture appears to provide the structural integrity normally provided by the membrane in cell surface arrays.
Protein-Protein Contacts at the Array Vertices
Cryo-EM has provided a wealth of information about global array architecture (1,26,27). At higher resolution, crystallographic and NMR studies of complexes between core protein fragments have provided valuable insights into key array protein-protein contacts, while disulfide mapping has identified essential contacts in functional, full-length, membrane-bound core complexes (21-25). Four types of repeating contacts anchor the array at its vertices. Some or all of these contacts must also transmit signals between proteins in the same or different core units (see below). (i) The kinase-receptor contact is formed by the tight coupling between the kinase regulatory domain and the N-terminal helix of the receptor protein-interaction region (Figs. 1E, and 2B below). Structural studies of core protein fragments originally provided two alternative models of this contact (23,26). Disulfide mapping studies carried out in functional, full-length, membrane-bound, reconstituted core complexes revealed the correct model, illustrating the synergistic information provided by high resolution studies of soluble fragment complexes and disulfide mapping of intact, membrane-bound core complexes (21,23). (ii) The adaptor-receptor contact is believed to be structurally homologous to the kinase-receptor contact, based on structural studies of fragment complexes and the homologous folds of the kinase regulatory domain and the adaptor protein (Fig. 1F) (23,26). (iii) The structure of kinase-adaptor interface 1 has been fully defined by crystallographic and disulfide mapping studies (22,23). This core unit interface also provides half of the contacts within the kinase-adaptor ring (Fig. 1D). (iv) Kinase-adaptor interface 2 provides the other kinase-adaptor ring contacts (crystallographic model in Fig. 1D) and bridges adjacent core units in higher order assemblies (23,26,27).
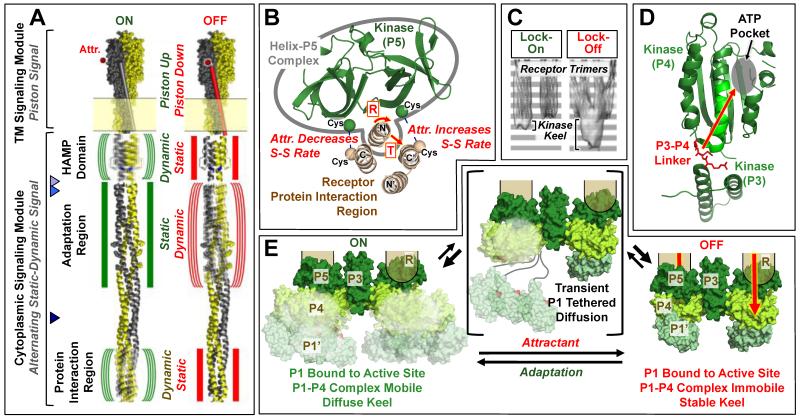
Figure 2. Models of signal transduction in the membrane-bound core unit.
A) Schematic chemoreceptor on- and off-states (green and red symbols, respectively), shown for simplicity in a single chemoreceptor homodimer by omitting the other two dimers of the receptor oligomer. The transmembrane and cytoplasmic signaling modules signal via an established piston mechanism (4,46) and a hypothesized alternating static-dynamic mechanism, respectively (42,43,48-52). The transmembrane signal begins in the periplasm where attractant binds to the apo-receptor and displaces the transmembrane signaling helix (TM2) towards the cytoplasm, thereby driving the piston-up, kinase-activating state into the piston-down, kinase-inhibiting state. The cytoplasmic HAMP domain converts this piston signal into a static-dynamic signal, such that the piston-up, kinase-on state possesses greater HAMP dynamics, while the piston-down, kinase-off state possesses less HAMP dynamics. The model further proposes that in the adaptation region the static-dynamic states are reversed in polarity relative to HAMP, and another polarity reversal is postulated in the protein interaction region. Triangles point to two proposed hinge regions: the major proteolysis site (53) and the nearby hotspot Gly (54) (light and medium blue, respectively), and the multi-Gly hinge (55) (dark blue). B) Shown is the tight complex formed between the kinase regulatory domain (P5) and the N-terminal helix of the receptor protein interaction region (see also Fig. 1E), as well as the attractant-triggered displacement of this complex (either a rotation or translation) detected by disulfide mapping in functional core complexes (21,23). This displacement is believed to transmit, at least in part, the on-off switching signal from the receptor to the kinase. C) Cryo-EM average density maps for signaling arrays containing a lock-on (I241E) or lock-off (A413T) mutant receptor, respectively (40). Within the average core unit, the two receptor trimers of dimers are easily observed, and an additional “keel” of protein density below the base plate region is less evident in locked-on arrays than in locked-off arrays. D) Shown is a hypothesized Class I (see text) allosteric on-off signal, based on NMR studies of CheA kinase, transmitted from the linker between the dimerization (P3) and catalytic (P4) domains, through the catalytic domain to the active site (39). E) Three-state picture of kinase on-off switching compatible with Class I and II models (see text). In Class I, the substrate domain (P1’) of one subunit is proposed to be bound much of the time to the active site on the catalytic domain of the other subunit (P4) awaiting trans-autophosphorylation, yielding a stable P1’-P4 complex regulated by an allosteric on-off signal traveling through the kinase to the P4 active site (28,67). In Class II, the substrate domain P1’ is bound most of the time either to the active site or to a separate inhibitory site on P4 in the on- and off-states, respectively, such that on-off signals regulate the ratio of binding to the two sites (64). In both Class I and II models, the keel domains including the P1’-P4 complex are proposed to be more mobile in the on-state than in the off-state, explaining the low keel density of the on-state in cryo-EM (see (C)) (40). Loss of mobility in the off-state could involve additional constraints on inter-domain linkers, or steric hindrance due to changes in the relative positions of kinase, adaptor and receptor regions. In both models, tethered diffusion of the substrate domain via the long P1-P2 and/or P2-P3 linkers could occur as shown, but would likely be transient to explain the known proteolytic protection of the linkers (10,11,34).
Ultrastability
Arrays formed in cells then isolated in bacterial membranes, as well as core complexes reconstituted on isolated bacterial membranes, are kinetically ultrastable and continue to exhibit receptor-regulated kinase activity for long periods up to weeks at room temperature (10,11). This remarkable long term stability is believed to arise from the extensive network of protein contacts provided by core unit and lattice organization (10,11). Eventually, the kinase activity decays as the bound kinase molecules are clipped by a protease endogenous to the membranes, leading to loss of the N-terminal substrate domain and kinase function (10). Reconstituted core complexes exhibit both quasi- and ultra-stable components with lifetimes of ~1.5 days and ~3 weeks, respectively. Incorporation of a tryptophan bump at the kinase-receptor interface or at kinase-adaptor interface 1 preferentially destabilizes the ultrastable component, such that the entire population becomes quasi-stable. In contrast, a disulfide bond covalently coupling the latter interface shifts the quasi-stable component into the ultra-stable state, such that the entire population becomes ultrastable with a longer exponential lifetime spanning at least 6 weeks (34). Overall, the evidence indicates that ultrastability protects the kinase from proteolysis, and requires well-formed contacts within the core unit.
Complementary Systems for Studying Core Units and Arrays
Rapid progress in the field has been facilitated by a diverse set of core unit and array preparations that each offer unique advantages as well as limitations. Native, functional hexagonal arrays formed in cells can be analyzed in vivo by cryo-EM or specialized chemical methods (1,26,27,40,46,68,69), or can be isolated in cell membranes for ex vivo studies (11). Membrane-bound core complexes can be reconstituted by combining isolated, receptor-containing cell membranes with purified CheA kinase and CheW adaptor protein (11,21,22,34). The resulting, functional complexes allow incorporation of chemically and chromophore-modified components, enabling unfettered disulfide chemistry and spectroscopic analysis of core unit structure, assembly and signaling mechanism. The reconstituted core complexes range in size from core units and small oligomers, including individual hexagons formed by three core units, up to small hexagonal arrays (1). Present also are nonfunctional receptor aggregates termed zippers, but stoichiometric analysis indicates that at least 2/3 of the receptors are in functional core complexes; moreover, the zippers are invisible in the standard kinase activity-based assays (11,21,22,34). Individual core units can be reconstituted in the nanodisc bilayer system that provides the simplest functional complex, eliminates interactions between core units, and also enables incorporation of modified components (4,29). Most recently, studies of soluble, membrane-bound, and crystallized complexes of core component fragments have utilized EPR, NMR, mass spec and crystallographic methods to probe core complex structure (1,23-26,33,39,52,61). Although these fragment systems lack attractant regulation, they provide the highest resolution structural information.
Structural Controversies and Challenges
The robust combination of cryo-EM imaging of native arrays, crystallographic and NMR studies of soluble fragment complexes, and disulfide mapping studies of intact, functional, reconstituted core complexes on membranes has revealed many key elements of array architecture. Not yet tested, however, is the hypothesized array assembly mechanism. In addition, the proposed adaptor-receptor and kinase-adaptor 2 contacts have not yet been tested by disulfide mapping in intact core complexes. Perhaps the main outstanding architectural question is whether the adaptor protein fills the empty hexagons in the array with a symmetric 6-membered ring homologous to the pseudo-symmetric kinase-adaptor ring (Fig. 1A, right). Turning to ultrastability, the importance of contacts within core units is now well established, but the contributions of contacts between core units has not been tested. Moreover, the available evidence suggests that in live cells the array-bound kinase and adaptor components exchange with bulk cytoplasmic copies more rapidly than the bulk exchange observed for complexes on isolated cell membranes (10,11,35). This faster exchange is hypothesized to arise from an unidentified cytoplasmic array disassembly machinery, perhaps Hsp70 or Hsp90 (36,37), that is lost when membranes are isolated. In cells, such machinery would ensure that the ultrastable array does not exhibit unregulated growth when protein synthesis is operating.
Signal Transduction within Core Units, and Cooperative Signaling Between Core Units
Minimal Effects of Signaling on Global Array Structure
In the absence of attractant, apo-receptors stimulate the His-kinase activity of native arrays and of core complexes built from the three core components, while attractant binding to the chemoreceptors inhibits the kinase (4). Cryo-EM comparison of the on- and off-states of native arrays has shown that the receptor and base plate lattices are maintained and do not undergo major, detectable rearrangements during on-off switching (38), consistent with previous evidence that receptor on-off switching involves subtle structural and/or dynamical changes (39-43). Detectable changes are observed near the cytoplasmic surface of the base-plate, likely arising from receptor-regulated changes in the positions or dynamics of CheA kinase domains outside the base-plate (40). The lattice structure likely constrains the magnitudes of conformational changes that can occur during on-off switching.
Signaling within the Core Unit
Receptor-regulated, kinase on-off switching is an inherent feature of the core unit in isolated nanodiscs, in reconstituted core complexes, and in native arrays. In all three systems, in the absence of attractant the activated kinases exhibit similarly large specific activities, whereas the addition of attractant inhibits these specific kinase activities about 102-fold in each system (11,29). On-off switching in all three systems can be regulated by covalent receptor adaptation as well as attractant binding (4). In short, the signaling properties of the core unit and higher order assemblies, including arrays, are similar – except in attractant titrations wherein the arrayed assemblies exhibit significantly higher positive cooperativity (2,9,44,45).
Receptor on-off switching mechanism
For simplicity, the mechanism of on-off switching is discussed herein for a single chemoreceptor homodimer, noting its native context is the receptor trimer-of-dimers within a core unit. Broadly, the receptor can be divided into (i) a transmembrane signaling module containing the periplasmic ligand binding domain and the transmembrane helices, and (ii) a cytoplasmic signaling module containing the HAMP, adaptation, Gly hinge, and protein interaction regions (Figure 2A) (4).
Multiple, independent lines of evidence support a conserved, piston-type signaling mechanism in the transmembrane signaling module of chemoreceptors (4,46). In the apo state of the receptor, the attractant binding site is, on average, closed, and the transmembrane signaling helix (TM2) is in its “up” position. During a thermal fluctuation that opens the pocket, attractant binds and traps the transmembrane signaling helix in its “down” position translocated ~1.6 Å towards the cytoplasm (Fig. 2A). The signal can be asymmetric due to negative cooperativity between the two attractant binding sites of the homodimer, resulting in attractant binding and displacement of the TM2 helix in one subunit only. Alternatively, the signal can be symmetric and displace TM2 in both subunits. Other mechanisms, including scissors and rotational, are disfavored by extensive data (4,46). In contrast to the array-forming chemoreceptors, the other major class of two-component pathway receptors – sensor His-kinase receptors – are structurally distinct from chemoreceptors, generally do not form arrays, and are proposed to undergo a large-amplitude scissors rearrangement during transmembrane signaling (47). Thus, CheA-linked chemoreceptors and CheA-independent sensor His-kinase receptors appear to possess different signaling mechanisms evolved for their different contexts within an array, or in the absence of an array, respectively.
In the long, cytoplasmic signaling module an alternating static-dynamic mechanism (also termed yin-yang) has been proposed for signal transmission through adjacent structural regions (42,43,48-52). The model postulates that when one region switches from its more tightly packed, more static state to its less tightly packed, more dynamic state, the adjacent region makes the opposite transition from dynamic to static, or vice-versa. Up to three cytoplasmic regions – the HAMP domain, the adaptation region, and the protein interaction region – are hypothesized to be coupled in this way, such that the kinase-activating and -inhibiting states possess dynamic-static-dynamic and static-dynamic-static configurations in these three regions, respectively. Notably, the three regions are separated by flexible regions important for signal transmission: (i) the junction between HAMP and the adaptation region is susceptible to proteolysis (53) and contains a Gly hotspot where large sidechains lock the receptor in the kinase-activating on-state (54), and (ii) the junction between the adaptation and protein interaction regions contains a multi-Gly hinge where substitutions can trap either the on- or off-state (55).
The alternating static-dynamic model has been tested independently by multiple methods in intact, membrane-bound, core complexes and arrays. Mutational, functional and structural studies of HAMP support the model, as do local charge engineering, disulfide trapping and engineered socket studies of the adaptation region, and an engineered socket study of the protein interaction region (42,43,49-52). In addition, the latter socket study (50) indicates that the crystal structure of the protein interaction region (56) represents the kinase off-state stabilized by multiple socket (knob-in-holes) interactions between pairs of helices both within and between the two identical subunits. The loss of even one socket interaction, which would presumably increase local helix dynamics, typically prevents attractant-triggered kinase inhibition as predicted by the alternating static-dynamic model (50).
A recent MD simulation of the protein interaction region has revealed a Phe ring flip at the buried homodimer interface, involving the most highly conserved residue in chemoreceptors (F396 Tsr, F394 Tar) (57). The ring flip is correlated with distance changes between adjacent helices, and with the receptor signaling state. The conserved Phe is located near the contact surface thought to transmit information from receptor to kinase, and this same position is the location of the only engineered, intersubunit disulfide bond in the region that locks the kinase-on state (50). These findings suggest that the conserved Phe may play an important role in receptor signal transmission, and could be directly involved in static-dynamic or conformational on-off switching.
Many chemoreceptors detect small molecule attractants (such as Ser, Asp for the two commonly studied amino acid receptors) with small binding energies, while sending a signal over 300 Å to the kinase (4,46). Both the transmembrane piston and cytoplasmic alternating static-dynamic signals are small amplitude, low energy transitions well-suited for such signal transmission (22,50,58,59). In the piston model the incompressibility of an α-helix (which is much easier to bend than to compress), together with the isoenergetic nature of the 1.5 Å piston displacement, ensures that the small displacement is carried the length of the signaling helix with little damping. Such a small helix sliding movement costs little energy since specific side chain contacts with adjacent helices are maintained by side chain flexibility (60). Similarly, the alternating static-dynamic model proposes a nearly isoenergetic transition between adjacent receptor regions, since each state possesses a set of tight and loose helix-helix interactions that are simply swapped during on-off switching.
The signal exiting the receptor protein interaction region can, in principle, be transmitted to other receptor dimers, and to a bound kinase or adaptor protein (4,46). The apo receptor homodimer possesses two-fold rotational symmetry about its long axis, but when incorporated into the trimer-of-dimers this symmetry is lost. One face binds the two other receptors in the same oligomer, leaving the opposite face open to bind a kinase or adaptor protein. Each of these faces is a potential site of signal transmission from a receptor dimer to other core proteins.
Receptor-Kinase Signal Transfer
In principle, receptor signals could be transmitted to the kinase by either a direct receptor-to-kinase path, or an indirect receptor-to-adaptor-to-kinase path, or both (61). Disulfide mapping studies of the receptor-kinase interface in functional core complexes have revealed an attractant triggered displacement of the tight complex formed between the receptor protein interaction region and the kinase regulatory domain (Fig. 2C) (34). It follows that the direct receptor-to-kinase path is utilized, but the contribution of the indirect path remains unknown.
Kinase on-off switching mechanism
Each subunit of the homodimeric kinase possesses five structural and functional domains: the substrate domain containing the His autophosphorylation site (P1), the response regulator binding domain (P2), the dimerization domain (P3), the ATP-binding catalytic domain (P4), and the kinase regulatory domain (P5) (4,30,62,63). When the kinase is active, it catalyzes a trans-autophosphorylation reaction in which the catalytic domain transfers the γ-phosphate of ATP to the target His on the substrate domain of the other subunit. Subsequently this same phosphate is transferred to a response regulator protein, either the motor regulator CheY or the adaptation enzyme CheB.
A recent cryo-EM study compared native arrays in kinase-on and -off states trapped by signal locking receptor mutations, revealing detectable protein density, termed a “keel”, projecting below the cytoplasmic surface of the base plate (40). The keel is presumably comprised by large, mobile CheA kinase domains including the P1 substrate and P4 catalytic domains. Notably, the on-state keel appears significantly smaller than the off-state keel, indicating the on-state keel domains are more difficult to detect, most likely because they are more mobile, or exhibit static, heterogeneous positions (Fig. 2C).
Currently, two classes of models (which are not mutually exclusive) exist for the mechanism of kinase on-off switching. In Class I models, the signal is allosteric and travels through protein domains to the kinase active site, as proposed by a recent NMR study (Fig. 2D,E) (39). In Class II models, the signal regulates the mobility or location of the P1 substrate domain, as for example in a model in which P1 is bound to the active site of the P4 catalytic domain in the on-state, but is bound to a separate inhibitory site on P4 in the off-state (Fig. 2E) (64). Both Class I and Class II models would explain changes in apparent keel density (Fig. 2C) by proposing that the keel domains are more mobile or heterogeneous in the kinase activating on-state than in the kinase inhibiting off-state (see legend Fig. 2E). Moreover, in both Class I and II models, it appears likely that tethered diffusion of the P1 substrate domain via the exposed, extended conformations of the P1-P2 and P2-P3 linkers is limited to transient events (Fig. 2E) given the striking inaccessibility of these linkers to proteases in ultrastability (10,34).
Cooperative Signaling between Core Units
The ultrasensitivity of the chemosensory array arises in part from extensive positive cooperativity between receptors, which yields an extremely steep dependence of kinase activity on attractant concentration (Hill coefficients up to ~ 15) (9,44,45,65). Circuit modeling studies suggest this cooperativity is generated by signal spread through the array, enabling attractant binding at a single receptor homodimer to downregulate 20-30 receptors and their associated kinases (2,3,8). On a molecular level, the receptor trimers-of-dimers are not believed to contact each other directly in the array, thus positive cooperativity is hypothesized to spread through the array via the network of interconnected kinase-adaptor rings. Presumably such signals must be transmitted through kinase-adaptor interface 2, which bridges different core units, but little is currently known about this type of signaling.
Mechanistic Controversies and Challenges
Significant progress has been made in elucidating the mechanism of receptor transmembrane signaling, and the piston model is widely accepted (4,46). Strong evidence supporting the alternating static-dynamic hypothesis of signal transduction in the receptor cytoplasmic domain has been generated (42,43,50-52), but additional direct structural and dynamical studies in functional core complexes are needed to fully test this hypothesis. One laboratory continues to champion an alternative, rotational model (66). Analysis of receptor switching mechanisms face technical challenges: studies of receptors in intact core complexes are less amenable to high resolution analysis but ensure retention of native membrane interactions, packing constraints and switching mechanism, while studies of receptor fragments enable high resolution analysis but may lack native constraints and mechanism.
Direct signal transmission from receptor to kinase has been detected (21), but it is not yet clear whether or not an indirect transmission route from receptor to adaptor to kinase also exists, and if so whether both routes are essential for receptor regulation of kinase activity. Further research is also needed to elucidate the mechanism of kinase on-off switching. Even less well understood is the mechanism of signal spread between core units underlying positive cooperativity and ultrasensitivity.
As summarized herein, recent findings in the field have emphasized the importance of both conformational changes and dynamics changes in receptor and kinase on-off switching. In reality, switching mechanisms are likely to generally involve changes in both average conformation and dynamics. A major challenge will be to distinguish whether one of these two components is more important to native regulation, or whether both are essential. In short, the pursuit of a molecular understanding of signal transduction will continue to provide controversies and challenges for years to come.
HIGHLIGHTS.
-
–
A conserved, ultrasensitive, ultrastable chemosensory array guides bacterial motility
-
–
Three core proteins form the array framework: receptor, His-kinase, and adaptor
-
–
Complementary approaches are developing a molecular model of array architecture
-
–
Recent progress has furthered the mechanistic understanding of receptor signaling
-
–
Early studies are investigating the mechanism of kinase on-off switching
ACKNOWLEDGEMENTS
The authors gratefully acknowledge funding by NIH / NIGMS grant R01 GM-040731 (to JJF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
Special (*) or Outstanding
(**) Interest
- 1**.Briegel A, Wong ML, Hodges HL, Oikonomou CM, Piasta KN, Harris MJ, Fowler DJ, Thompson LK, Falke JJ, Kiessling LL, Jensen GJ. New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography. Biochemistry. 2014;53:1575–1585. doi: 10.1021/bi5000614. First cryo-EM analysis of array assembly, providing images of multiple assembly intermediates including intact, membrane-bound core units and assemblies of core units ranging up to small hexagonal arrays. Newly described soluble arrays constructed from receptor fragments lack the native constraints provided by the membrane and other receptor domains, but yield higher resolution images than the membrane-associated core units.
- 2.Tu Y. Quantitative modeling of bacterial chemotaxis: signal amplification and accurate adaptation. Annu Rev Biophys. 2013;42:337–359. doi: 10.1146/annurev-biophys-083012-130358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sourjik V, Wingreen NS. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol. 2012;24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaparro AP, Ali SK, Klose KE. The ToxT-dependent methyl-accepting chemoreceptors AcfB and TcpI contribute to Vibrio cholerae intestinal colonization. FEMS Microbiol Lett. 2010;302:99–105. doi: 10.1111/j.1574-6968.2009.01835.x. [DOI] [PubMed] [Google Scholar]
- 6.Raterman EL, Welch RA. Chemoreceptors of Escherichia coli CFT073 play redundant roles in chemotaxis toward urine. PLoS One. 2013;8:e54133. doi: 10.1371/journal.pone.0054133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grim CJ, Kozlova EV, Sha J, Fitts EC, van Lier CJ, Kirtley ML, Joseph SJ, Read TD, Burd EM, Tall BD, Joseph SW, Horneman AJ, Chopra AK, Shak JR. Characterization of Aeromonas hydrophila wound pathotypes by comparative genomic and functional analyses of virulence genes. MBio. 2013;4:e00064–00013. doi: 10.1128/mBio.00064-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray D, Levin MD, Morton-Firth CJ. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 9.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 10*.Slivka PF, Falke JJ. Isolated Bacterial Chemosensory Array Possesses Quasi- and Ultrastable Components: Functional Links between Array Stability, Cooperativity, and Order. Biochemistry. 2012;51:10218–10228. doi: 10.1021/bi301287h. Quantitative analysis of core complex ultrastability, revealing nearly equal populations of quasi- and ultrastable with bound state lifetimes of days and weeks, respectively.
- 11.Erbse AH, Falke JJ. The core signaling proteins of bacterial chemotaxis assemble to form an ultrastable complex. Biochemistry. 2009;48:6975–6987. doi: 10.1021/bi900641c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segall JE, Manson MD, Berg HC. Signal processing times in bacterial chemotaxis. Nature. 1982;296:855–857. doi: 10.1038/296855a0. [DOI] [PubMed] [Google Scholar]
- 13.Khan S, Jain S, Reid GP, Trentham DR. The fast tumble signal in bacterial chemotaxis. Biophys J. 2004;86:4049–4058. doi: 10.1529/biophysj.103.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 15.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010;3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balagopalan L, Coussens NP, Sherman E, Samelson LE, Sommers CL. The LAT story: a tale of cooperativity, coordination, and choreography. Cold Spring Harb Perspect Biol. 2010;2:a005512. doi: 10.1101/cshperspect.a005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scarselli M, Annibale P, Radenovic A. Cell type-specific beta2-adrenergic receptor clusters identified using photoactivated localization microscopy are not lipid raft related, but depend on actin cytoskeleton integrity. J Biol Chem. 2012;287:16768–16780. doi: 10.1074/jbc.M111.329912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palczewski K. Oligomeric forms of G protein-coupled receptors (GPCRs) Trends Biochem Sci. 2010;35:595–600. doi: 10.1016/j.tibs.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y, Smoligovets AA, Groves JT. Modulation of T cell signaling by the actin cytoskeleton. J Cell Sci. 2013;126:1049–1058. doi: 10.1242/jcs.098210. [DOI] [PubMed] [Google Scholar]
- 21**.Piasta KN, Ulliman CJ, Slivka PF, Crane BR, Falke JJ. Defining a Key Receptor-CheA Kinase Contact and Elucidating Its Function in the Membrane-Bound Bacterial Chemosensory Array: A Disulfide Mapping and TAM-IDS Study. Biochemistry. 2013;52:3866–3880. doi: 10.1021/bi400385c. Disulfide mapping and live cell study that unambiguously determines the receptor-kinase interface, shows that the kinase regulatory domain binds to the N-terminal helix of the cytoplasmic receptor four-helix bundle, and that the resulting tight helix-kinase complex moves as a unit during on-off switching.
- 22*.Natale AM, Duplantis JL, Piasta KN, Falke JJ. Structure, function, and on-off switching of a core unit contact between CheA kinase and CheW adaptor protein in the bacterial chemosensory array: A disulfide mapping and mutagenesis study. Biochemistry. 2013;52:7753–7765. doi: 10.1021/bi401159k. Disulfide mapping and live cell study that confirms one of the two types of kinase-adaptor contacts in the kinase-adaptor ring system proposed to stabilize the array and transmit signals.
- 23*.Li X, Fleetwood AD, Bayas C, Bilwes AM, Ortega DR, Falke JJ, Zhulin IB, Crane BR. The 3.2 A Resolution Structure of a Receptor:CheA:CheW Signaling Complex Defines Overlapping Binding Sites and Key Residue Interactions within Bacterial Chemosensory Arrays. Biochemistry. 2013;52:3852–3865. doi: 10.1021/bi400383e. Provides a high resolution view of the receptor-kinase interface in a soluble fragment complex.
- 24*.Wang X, Vu A, Lee K, Dahlquist FW. CheA-receptor interaction sites in bacterial chemotaxis. J Mol Biol. 2012;422:282–290. doi: 10.1016/j.jmb.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Vu A, Wang X, Zhou H, Dahlquist FW. The receptor-CheW binding interface in bacterial chemotaxis. J Mol Biol. 2012;415:759–767. doi: 10.1016/j.jmb.2011.11.043. NMR model of the receptor-adaptor interface in a soluble fragment complex.
- 26**.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1115719109. Cryo_EM and X-ray structural analysis of native, membrane-bound chemosensory arrays from E. coli, revealing the striking, hexagonal lattice architecture.
- 27**.Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci U S A. 2012;109:E1481–1488. doi: 10.1073/pnas.1200781109. Cryo_EM structural analysis of native, membrane-bound chemosensory arrays from E. coli, also revealing the hexagonal lattice architecture.
- 28*.Miller AS, Kohout SC, Gilman KA, Falke JJ. CheA Kinase of bacterial chemotaxis: chemical mapping of four essential docking sites. Biochemistry. 2006;45:8699–8711. doi: 10.1021/bi060580y. Engineered, fluorescein-modified Cys residues map out four docking surfaces on CheA kinase, providing the first glimpse of core unit architecture.
- 29.Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci U S A. 2011;108:9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 31.Griswold IJ, Zhou H, Matison M, Swanson RV, McIntosh LP, Simon MI, Dahlquist FW. The solution structure and interactions of CheW from Thermotoga maritima. Nat Struct Biol. 2002;9:121–125. doi: 10.1038/nsb753. [DOI] [PubMed] [Google Scholar]
- 32.Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci U S A. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Briegel A, Ladinsky MS, Oikonomou C, Jones CW, Harris MJ, Fowler DJ, Chang YW, Thompson LK, Armitage JP, Jensen GJ. Structure of bacterial cytoplasmic chemoreceptor arrays and implications for chemotactic signaling. Elife. 2014;3:e02151. doi: 10.7554/eLife.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34**.Piasta KN, Falke JJ. Increasing and Decreasing the Ultrastability of Bacterial Chemotaxis Core Signaling Complexes by Modifying Protein-Protein Contacts. Biochemistry. 2014 doi: 10.1021/bi500849p. In Press, Accelerated Publication, PMID: 25119814. Study reporting the discovery of an interfacial, engineered disulfide bond that enhances the scope and magnitude of ultrastability in reconstituted core complexes, largely by protecting the long linkers extending between the substrate domain and the catalytic core from proteases.
- 35.Schulmeister S, Ruttorf M, Thiem S, Kentner D, Lebiedz D, Sourjik V. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:6403–6408. doi: 10.1073/pnas.0710611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Press MO, Li H, Creanza N, Kramer G, Queitsch C, Sourjik V, Borenstein E. Genome-scale co-evolutionary inference identifies functions and clients of bacterial Hsp90. PLoS Genet. 2013;9:e1003631. doi: 10.1371/journal.pgen.1003631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seyffer F, Kummer E, Oguchi Y, Winkler J, Kumar M, Zahn R, Sourjik V, Bukau B, Mogk A. Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces. Nat Struct Mol Biol. 2012;19:1347–1355. doi: 10.1038/nsmb.2442. [DOI] [PubMed] [Google Scholar]
- 38.Briegel A, Beeby M, Thanbichler M, Jensen GJ. Activated chemoreceptor arrays remain intact and hexagonally packed. Mol Microbiol. 2011;82:748–757. doi: 10.1111/j.1365-2958.2011.07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Wang X, Vallurupalli P, Vu A, Lee K, Sun S, Bai WJ, Wu C, Zhou H, Shea JE, Kay LE, Dahlquist FW. The linker between the dimerization and catalytic domains of the CheA histidine kinase propagates changes in structure and dynamics that are important for enzymatic activity. Biochemistry. 2014;53:855–861. doi: 10.1021/bi4012379. NMR evidence for a Class I, allosteric structural change within CheA kinase that is proposed to be involved in receptor-regulated, kinase on-off switching.
- 40**.Briegel A, Ames P, Gumbart JC, Oikonomou CM, Parkinson JS, Jensen GJ. The mobility of two kinase domains in the Escherichia coli chemoreceptor array varies with signaling state. Mol Microbiol. 2013;89:831–41. doi: 10.1111/mmi.12309. Cryo-EM study of mutant arrays designed to mimic the on- and off-states, revealing a more pronounced protein “keel” below the base plate of the latter arrays, suggesting that the keel-associated kinase domains possess greater dynamic or static heterogeneity in the on-state.
- 41.Sferdean FC, Weis RM, Thompson LK. Ligand affinity and kinase activity are independent of bacterial chemotaxis receptor concentration: insight into signaling mechanisms. Biochemistry. 2012;51:6920–6931. doi: 10.1021/bi3007466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Q, Ames P, Parkinson JS. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Molecular microbiology. 2011;80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Ames P, Zhou Q, Parkinson JS. HAMP domain structural determinants for signalling and sensory adaptation in Tsr, the Escherichia coli serine chemoreceptor. Mol Microbiol. 2014;91:875–886. doi: 10.1111/mmi.12443. Most recent analysis of HAMP structure and dynamics, and their roles in alternating static-dynamic transitions within the receptor cytoplasmic domain.
- 44.Bornhorst JA, Falke JJ. Attractant regulation of the aspartate receptor-kinase complex: limited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry. 2000;39:9486–9493. doi: 10.1021/bi0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 46.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47**.Molnar KS, Bonomi M, Pellarin R, Clinthorne GD, Gonzalez G, Goldberg SD, Goulian M, Sali A, DeGrado WF. Cys-Scanning Disulfide Crosslinking and Bayesian Modeling Probe the Transmembrane Signaling Mechanism of the Histidine Kinase. PhoQ, Structure. 2014 doi: 10.1016/j.str.2014.04.019. Disulfide trapping study utilizing novel Bayesian analysis providing the first view of scissors-type on-off switching in an intact, membrane-bound sensor His-kinase receptor, and noting that this class of receptors appears to possess a transmembrane signaling mechanism distinct from the piston mechanism of chemoreceptors.
- 48.Kim SH. “Frozen” dynamic dimer model for transmembrane signaling in bacterial chemotaxis receptors. Protein Sci. 3:159–165. doi: 10.1002/pro.5560030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Starrett DJ, Falke JJ. Adaptation mechanism of the aspartate receptor: electrostatics of the adaptation subdomain play a key role in modulating kinase activity. Biochemistry. 2005;44:1550–1560. doi: 10.1021/bi048089z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50**.Swain KE, Gonzalez MA, Falke JJ. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry. 2009;48:9266–9277. doi: 10.1021/bi901020d. Engineered socket study providing the first evidence for an alternating static-dynamic transition involving the receptor adaptation and protein interaction regions.
- 51**.Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol Microbiol. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. Mutational study providing the first evidence for an alternating static-dynamic transition involving the receptor HAMP and adaptation regions.
- 52**.Airola MV, Sukomon N, Samanta D, Borbat PP, Freed JH, Watts KJ, Crane BR. HAMP domain conformers that propagate opposite signals in bacterial chemoreceptors. PLoS Biol. 2013;11:e1001479. doi: 10.1371/journal.pbio.1001479. HAMP domain conformers proposed to mimic the native on- and off-states.
- 53.Swain KE, Falke JJ. Structure of the Conserved HAMP Domain in an Intact, Membrane-Bound Chemoreceptor: A Disulfide Mapping Study. Biochemistry. 2007;46:13684–13695. doi: 10.1021/bi701832b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trammell MA, Falke JJ. Identification of a site critical for kinase regulation on the central processing unit (CPU) helix of the aspartate receptor. Biochemistry. 1999;38:329–336. doi: 10.1021/bi981964u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coleman MD, Bass RB, Mehan RS, Falke JJ. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching. Biochemistry. 2005;44:7687–7695. doi: 10.1021/bi0501479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 57**.Ortega DR, Yang C, Ames P, Baudry J, Parkinson JS, Zhulin IB. A phenylalanine rotameric switch for signal-state control in bacterial chemoreceptors. Nature Communications. 2013;2013:2881. doi: 10.1038/ncomms3881. A study of the most conserved residue in chemoreceptor sequences – a Phe at the homodimer interface near the receptor surfaces involved in core unit formation – this Phe is observed to undergo ring-flip transitions in molecular dynamics simulations that are proposed to be involved in on-off switching.
- 58.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Falke JJ, Erbse AH. The piston rises again. Structure. 2009;17:1149–1151. doi: 10.1016/j.str.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gerstein M, Lesk AM, Chothia C. Structural mechanisms for domain movements in proteins. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- 61.Wang X, Wu C, Vu A, Shea JE, Dahlquist FW. Computational and experimental analyses reveal the essential roles of interdomain linkers in the biological function of chemotaxis histidine kinase CheA. J Am Chem Soc. 2012;134:16107–16110. doi: 10.1021/ja3056694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McEvoy MM, Muhandiram DR, Kay LE, Dahlquist FW. Structure and dynamics of a CheY-binding domain of the chemotaxis kinase CheA determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1996;35:5633–5640. doi: 10.1021/bi952707h. [DOI] [PubMed] [Google Scholar]
- 63.Mourey L, Da Re S, Pedelacq JD, Tolstykh T, Faurie C, Guillet V, Stock JB, Samama JP. Crystal structure of the CheA histidine phosphotransfer domain that mediates response regulator phosphorylation in bacterial chemotaxis. J Biol Chem. 2001;276:31074–31082. doi: 10.1074/jbc.M101943200. [DOI] [PubMed] [Google Scholar]
- 64.Hamel DJ, Zhou H, Starich MR, Byrd RA, Dahlquist FW. Chemical-shift-perturbation mapping of the phosphotransfer and catalytic domain interaction in the histidine autokinase CheA from Thermotoga maritima. Biochemistry. 2006;45:9509–9517. doi: 10.1021/bi060798k. [DOI] [PubMed] [Google Scholar]
- 65.Han XS, Parkinson JS. An unorthodox sensory adaptation site in the Escherichia coli serine chemoreceptor. J Bacteriol. 2014;196:641–649. doi: 10.1128/JB.01164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferris HU, Zeth K, Hulko M, Dunin-Horkawicz S, Lupas AN. Axial helix rotation as a mechanism for signal regulation inferred from the crystallographic analysis of the E. coli serine chemoreceptor. J Struct Biol. 2014;186:349–356. doi: 10.1016/j.jsb.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 67.Nishiyama S, Garzon A, Parkinson JS. Mutational analysis of the P1 phosphorylation domain in Escherichia coli CheA, the signaling kinase for chemotaxis. J Bacteriol. 2014;196:257–264. doi: 10.1128/JB.01167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khursigara CM, Wu X, Subramaniam S. Chemoreceptors in Caulobacter crescentus: trimers of receptor dimers in a partially ordered hexagonally packed array. J Bacteriol. 2008;190:6805–6810. doi: 10.1128/JB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci U S A. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]