SUMMARY
Obesity develops when energy intake chronically exceeds energy expenditure. Because brown adipose tissue (BAT) dissipates energy in the form of heat, increasing energy expenditure by augmenting BAT-mediated thermogenesis may represent an approach to counter obesity and its complications. The ability of BAT to dissipate energy is dependent on expression of mitochondrial protein Uncoupling Protein 1 (UCP1). To facilitate the identification of pharmacological modulators of BAT UCP1 levels, which may have potential as anti-obesity medications, we have developed a transgenic model in which luciferase activity faithfully mimics endogenous UCP1 expression and its response to physiologic stimuli. Phenotypic screening of a library using cells derived from this model yielded a small-molecule that increases UCP1 expression in brown fat cells and mice. Upon adrenergic stimulation, compound-treated mice showed increased energy expenditure. These tools offer an opportunity to identify pharmacologic modulators of UCP1 expression and uncover new regulatory pathways that impact BAT-mediated thermogenesis.
Keywords: Obesity, brown adipose tissue, UCP1, in vivo imaging, small-molecule screening
INTRODUCTION
The epidemic of obesity poses a dire public health problem, for obesity is a major risk factor for development of insulin resistance, type 2 diabetes, cardiovascular disease, and cancer. Obesity is the result of a sustained energy imbalance in which intake exceeds expenditure. Current anti-obesity drugs work by limiting energy intake, either through suppression of appetite or inhibition of intestinal lipid absorption (Kim et al., 2014). These medications are effective, but side effects often associated with long-term use, such as depression or steatorrhea, limit patient compliance. The discovery of brown adipose tissue (BAT) in adult humans and its correlation with body mass index (Cypess et al., 2009; Saito et al., 2009; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009) suggests that active BAT may protect from obesity. BAT dissipates energy in the form of heat, thus increasing energy expenditure. It is thought that pharmacological activation of BAT thermogenesis may be an alternative approach to alter energy balance, one complementary to existing obesity medications (Nedergaard and Cannon, 2010; Kajimura and Saito, 2014).
The ability of BAT to produce heat is dependent on expression of the BAT-specific protein Uncoupling Protein 1 (UCP1). In response to exposure to cold or a high-fat diet, UCP1 reduces the mitochondrial membrane potential and uncouples cellular respiration from ATP synthesis, thereby generating heat. Because other UCP proteins, (e.g. UCP2, UCP3) do not contribute to adaptive thermogenesis (Golozoubova et al., 2001), UCP1 is thought to be solely responsible for adaptive non-shivering thermogenesis. UCP1 null mice are intolerant to cold (Enerback et al., 1997) and develop obesity at thermoneutral conditions (Feldmann et al., 2009). In contrast, transgenic expression of UCP1 in fat increases oxygen consumption in BAT and epididymal white adipose tissue (WAT) and reduces body weight gain (Kopecky et al., 1995). Contrary to the mechanism of action of small-molecule mitochondrial uncouplers such as 2,4-dinitrophenol that proved too toxic as weight loss agents (Grundlingh et al., 2011), UCP1-mediated uncoupling is a highly regulated process that requires direct binding of long-chain free fatty acids to UCP1 in response to physiologic cAMP signaling (Fedorenko et al., 2012). A pharmacological approach to increase UCP1 expression and activity in adipose tissue is thus likely to constitute a safer avenue to enhance whole-body thermogenic capacity and energy expenditure. To test this concept, it is of great interest to identify small molecules that can stimulate UCP1 expression in fat tissue.
Phenotypic screens using adipocytes have proven to be a powerful method to isolate small molecules that ameliorate the symptoms of metabolic syndrome through novel mechanisms of action (Waki et al., 2007; Dominguez et al., 2014). To facilitate the identification of pharmacologic agents to modulate UCP1 expression in adipocytes, we have generated a transgenic reporter mouse, ThermoMouse, in which luciferase activity recapitulates the pattern of expression of UCP1 in vivo and allows real-time visualization and quantification of UCP1 expression in live animals. A chemical screen using brown adipocytes derived from this model yielded a compound that can induce UCP1 expression in cells and enhance UCP1 expression in vivo. In response to adrenergic stimulation, mice treated with this compound show increased energy expenditure. These results demonstrate the utility of these models to identify pharmacological modulators of UCP1 levels. Discovery of compounds with this ability is an important stride towards the goal of enhancing BAT function in obese individuals with drug-like molecules.
RESULTS
Development of a transgenic model to image UCP1 expression in vivo
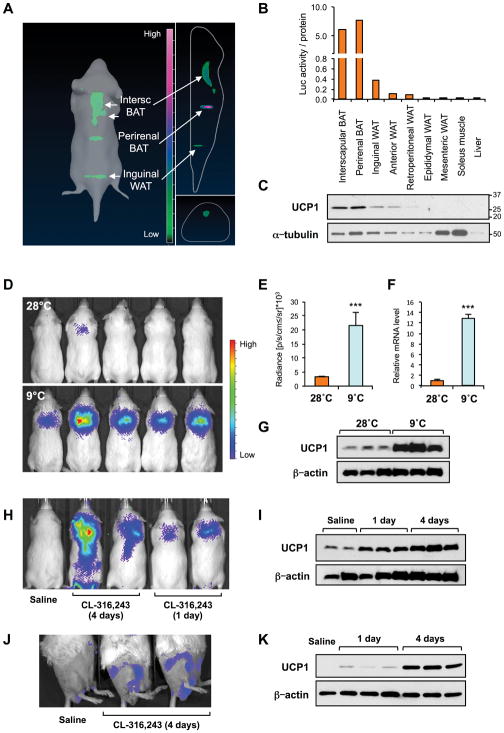
To develop an in vivo reporter system to monitor endogenous UCP1 expression in a non-invasive manner, we generated transgenic mice that express luciferase2 under the control of the Ucp1 genetic locus. A luciferase2-T2A-tdTomato cassette was inserted at the initiation codon of the Ucp1 gene in a 98.6 kb bacterial artificial chromosome (BAC) containing the entire Ucp1 gene locus (Fig. S1A) and proper targeting confirmed (Fig. S1B). Next, we used the IVIS Spectrum Imaging System to monitor luciferase activity in transgenic mice generated using this BAC construct. Three-dimensional imaging detected robust luciferase signals in interscapular BAT, perirenal BAT, and inguinal WAT (Fig. 1A). No signals were detected in adipose depots of wild type mice (Fig. S1C). To assess if luciferase activity recapitulated endogenous UCP1 protein levels, we measured luciferase activity and UCP1 protein in BAT, WAT, liver, and muscle. A tight correlation was found between luciferase activity and endogenous UCP1 protein expression (Fig. 1B and C), indicating that the reporter model mirrors the tissue distribution of UCP1.
Figure 1. Luciferase imaging of UCP1 expression in vivo.
A: 3D reconstruction of luciferase signals in Ucp1-luciferase reporter mice. Adipose depots with specific luciferase signal are indicated.
B: Quantification of luciferase in adipose depots, skeletal muscle, and liver of mice kept at room temperature. Values normalized to protein content.
C: UCP1 protein expression in tissue lysates from the mouse in (B).
D: Luciferase activity in Ucp1-luciferase reporter mice kept at 28°C and subsequently kept at 9°C for 24 hr. Representative mice are shown.
E: Quantification of luciferase signal in interscapular BAT of Ucp1-luciferase reporter mice shown in (D) (n=9). *** P < 0.001. Data are expressed as mean ± SEM.
F: Ucp1 mRNA expression in interscapular BAT of Ucp1-luciferase reporter mice kept at 28°C and 9°C for 24 hr. *** P < 0.001. Data are expressed as mean ± SEM.
G: UCP1 protein expression in interscapular BAT of transgenic mice analyzed in (F).
H: Representative image of luciferase signal in Ucp1-luciferase reporter mice treated with saline or CL-316,243 for 1 day (acute) or 4 days (subchronic) (n=3).
I: UCP1 protein expression in BAT of transgenic mice analyzed in (H).
J: Representative images of luciferase signal in inguinal WAT depots of Ucp1-luciferase reporter mice treated with saline or CL-316,243 for 4 days.
K: UCP1 protein expression in inguinal WAT depots of Ucp1-luciferase mice treated with saline or CL-316,243 for 1 day or 4 days.
To examine if reporter mice responded to physiologic stimuli known to induce UCP1 expression and BAT activity, we monitored changes in luciferase activity during cold adaptation (Fig. 1D). Luciferase signal in interscapular BAT was low in mice maintained at 28 °C (Fig. S1D), but increased robustly in mice kept at 9 °C for 24 hours (Fig. 1D and E). Luciferase induction correlated with increases in UCP1 mRNA (Fig. 1F) and protein levels (Fig. 1G). Similarly, subchronic (4 days) or acute (1 day) treatment of reporter mice kept at 28 °C with a specific β3 adrenergic receptor agonist (CL-316,243; 1 mg/kg) dramatically enhanced the luciferase signal and endogenous UCP1 protein levels in BAT (Fig. 1H and I). To evaluate the response of beige cells in inguinal WAT, imaging was performed from a side angle. Strong luciferase activity was detected in inguinal WAT in response to chronic CL-316,243 treatment (Fig. 1J). The increase in signal was paralleled by robust induction of UCP1 protein in this depot (Fig. 1K). These results show that our Ucp1-luciferase reporter accurately indicates changes in UCP1 elicited by physiological responses and is an useful tool to quantify changes in UCP1 expression in vivo.
A cell-based screening platform to monitor UCP1 protein expression
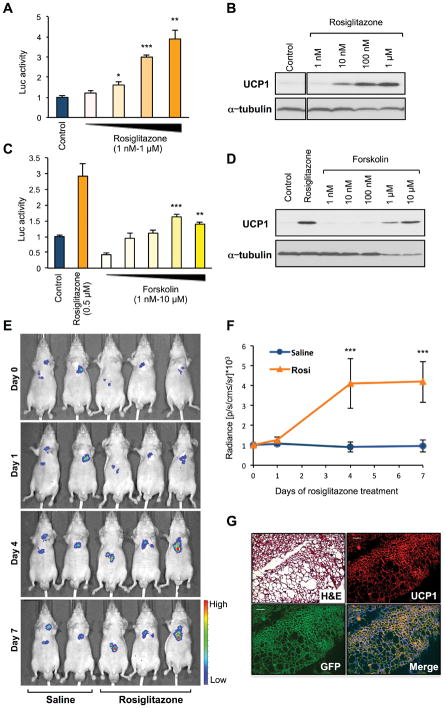
Next, to establish a cell-based system for quantifying UCP1 expression in fat cells suited for screening of small-molecule or genomic libraries, we generated immortalized preadipocyte lines from Ucp1-luciferase transgenics. From six cell lines derived from interscapular BAT, we selected one that showed high adipogenic capacity and significant levels of UCP1 and luciferase expression. Because PPARγ ligands can stimulate UCP1 expression (Sears et al., 1996), we tested the response of this line to rosiglitazone. Luciferase activity was induced in a dose-dependent manner and correlated tightly with endogenous UCP1 protein levels (Fig. 2A and B). Treatment with the cAMP signaling activator forskolin also increased luciferase activity and UCP1 protein expression (Fig. 2C and D).
Figure 2. Cell-based system to monitor UCP1 expression.
A: Luciferase activity in immortalized Ucp1-luciferase brown adipocytes. Differentiated adipocytes were treated with DMSO (control) or rosiglitazone for 5 days (n=3).
B: UCP1 protein expression in cells analyzed in (A).
C: Luciferase activity in differentiated Ucp1-luciferase brown adipocytes treated with DMSO (control), forskolin, or rosiglitazone (0.5 μM) for 5 days (n=3).
D: UCP1 protein expression in cells analyzed in (C).
E: Luciferase activity monitored at day 0, 1, 4, and 7 after the start of rosiglitazone treatment in mice implanted with Ucp1-luciferase immortalized preadipocytes. Saline (n=4) or rosiglitazone (n=6; 10 mg/kg) treatment started 6 days post-implantation. Representative mice are shown.
F: Sequential changes in luciferase activity measured in fat transplants from mice in (E).
G: Haematoxylin/Eosin and immunofluorescent stains for UCP1 or GFP (i.e., tdTomato) in transplants from mice treated with rosiglitazone. Merged UCP1 and GFP image counterstained with DAPI. Bar: 50 μm.
*P <0.05, ** P <0.01, *** P < 0.001. Data are expressed as mean ± SEM.
To confirm that the Ucp1-luciferase reporter cell line preserved the response to BAT activators, and that it could report sequential changes in UCP1 expression, nude mice were implanted with Ucp1-luciferase preadipocytes subcutaneously and treated 6 days post-transplantation with saline or rosiglitazone (10 mg/kg per day) for 7 days. Luciferase activity in transplants of rosiglitazone-treated mice was increased significantly at 4 days of treatment and thereafter (Fig. 2E and F). Transplanted preadipocytes in mice treated with rosiglitazone formed discrete adipose tissue containing multilocular adipocytes (Fig. 2G, upper left). These adipocytes were positive for UCP1 protein (Fig. 2G, upper right) and GFP (i.e., tdTomato; Fig. 2G, lower left), indicating that transplanted cells retained brown adipogenic capacity in vivo. These observations indicate that this cell-based system is a robust surrogate to assess endogenous UCP1 expression.
A small molecule screen identifies a regulator of UCP1 expression
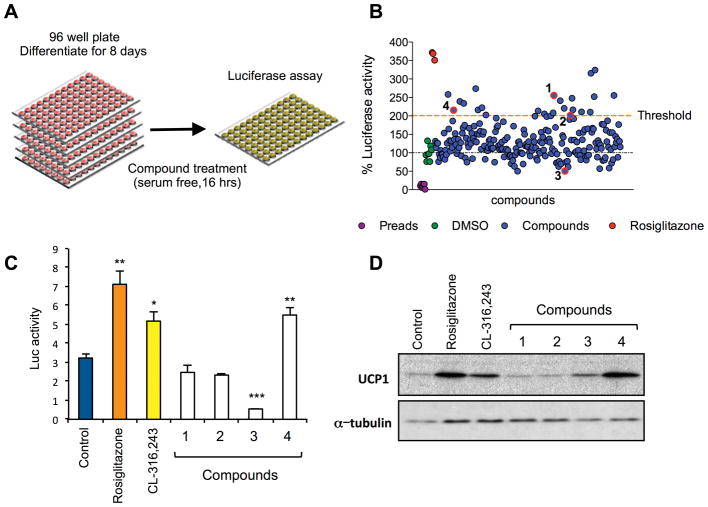
To test the ability of our monitoring systems to identify new regulators of UCP1 expression, we performed a phenotypic screen using a modestly-sized library of small molecules, primarily carbamates and triazole ureas (Adibekian et al., 2011; Bachovchin et al., 2010). Ucp1-luciferase brown preadipocytes were seeded and differentiated in 96 well plates. Mature adipocytes (day 8) were incubated with compounds and luciferase activity quantified 16 hours later (Fig. 3A). To evaluate the power of the assay to identify compounds that modulate UCP1 levels in either direction, four hits with differing properties (activators and inhibitors) were selected for study (Fig. 3B). As it is often the case with reporter-based screens, two hits could not be validated and may represent, for example, non-specific stabilizers of luciferase signal. Of those that confirmed, compound 4 robustly increased luciferase activity (Fig. 3C). In contrast, compound 3 significantly reduced luciferase signal. Importantly, compound 4 increased endogenous UCP1 protein expression to a similar level to that induced by rosiglitazone (Fig. 3D). None of the compounds had any effect on brown preadipocytes (Fig. S2).
Figure 3. Small-molecule screen to identify regulators of UCP1 expression.
A: Screen scheme. Ucp1-luciferase brown preadipocytes were differentiated in 96 well plates and treated at day 8 with compounds for 16 hr (n=3).
B: Screen performance. Mean luciferase activity of compounds (blue circles) plotted relative to the value of DMSO-treated Ucp1-luciferase adipocytes (100%; green circles). Rosiglitazone served as positive control (red circles). Ucp1-luciferase preadipocytes (purple circles) were used to determine background and induction of signal upon differentiation. Hits selected for evaluation (blue circles, red outline) are numbered according to the scheme used in validation experiments.
C: Luciferase activity in differentiated Ucp1-luciferase brown adipocytes treated with compounds for 5 days. Rosiglitazone (0.5 μM) and CL-316,243 (10 nM) served as positive controls (n=3). * P <0.05, ** P <0.01, *** P < 0.001 vs. control. Data expressed as means ± SEM.
D: UCP1 protein expression in cells from (C).
Compound 4 (WWL113 in original nomenclature) was also a hit in a different, image-based screen we recently described in which white preadipocytes were treated chronically (8 days) during differentiation (Dominguez et al., 2014). WWL113 inhibits two serine hydrolases expressed in adipocytes, carboxylesterase 3 (Ces3 or Ces1d) and Ces1f (CesML1). However, the ability of WWL113 to induce UCP1 expression in brown adipocytes does not appear to be mediated by Ces3/1f inhibition, for a structurally distinct Ces3 inhibitor (WWL229) failed to have the same effect on UCP1 expression, while the urea version of WWL113 (WWL113U), which does not inhibit Ces3, retained the ability to induce UCP1 (Fig. S3A). Nonetheless, we concluded that WWL113 could be a valuable chemical probe to further validate our reporter systems.
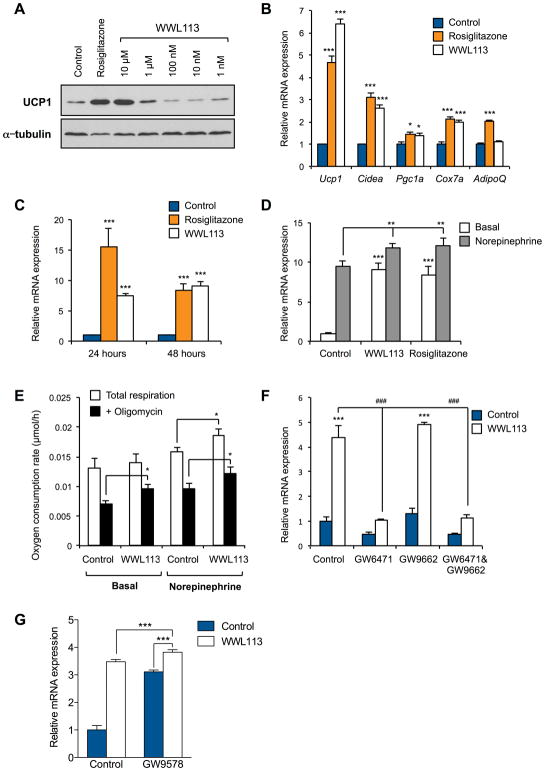
Induction of UCP1 expression by WWL113 relies on PPARα signaling
To characterize the effect of WWL113 on endogenous UCP1 expression, we treated cultured brown adipocytes with several doses of WWL113. At a dose of 1 μM and higher, WWL113 significantly increased UCP1 protein expression (Fig. 4A). The effect of WWL113 on UCP1 protein levels was largely due to activation of Ucp1 transcription, because WWL113 powerfully increased expression of Ucp1 mRNA (Fig. 4B). WWL113 treatment also stimulated expression of mRNAs for other thermogenic genes, such as Cidea, Pgc1a, and Cox7a1 (Fig. 4B). WWL113 treatment had no effect on expression of the adipogenic marker Adiponectin. WWL113 treatment for 24 hours was sufficient to activate endogenous Ucp1 mRNA expression without affecting expression of multiple adipogenic markers (Fig. 4C and Fig. S3B), indicating that WWL113 enhanced the BAT-selective thermogenic gene program in a cell-autonomous manner without affecting adipogenesis per se. To test the functional consequences of WWL113 treatment, we examined the extent to which WWL113 could sensitize brown adipocytes to physiologic activators of BAT such as norepinephrine. WWL113 pre-treatment enhanced the increase in Ucp1 mRNA expression normally induced by norepinephrine (Fig. 4D). More importantly, WWL113-treated cells showed greater total and uncoupled respiration in response to norepinephrine, and greater uncoupled basal respiration (Fig. 4E and Fig. S3C).
Figure 4. Effect of WWL113 on UCP1 expression and cellular respiration.
A: UCP1 protein expression in differentiated Ucp1-luciferase brown adipocytes treated with WWL113 for 5 days. Rosiglitazone (0.5 μM) served as positive control.
B: Expression of thermogenic genes in differentiated Ucp1-luciferase brown adipocytes treated with WWL113 (10 μM) for 5 days. Rosiglitazone (0.5 μM) was served as positive control (n=3). * P <0.05, *** P < 0.001 vs. control.
C: Ucp1 mRNA expression in primary differentiated brown adipocytes treated with WWL113 (10 μM) or rosiglitazone (5 μM) for 24 or 48 hours (n=3). *** P < 0.001 vs. control.
D: Ucp1 mRNA expression in primary differentiated brown adipocytes treated with WWL113 (10 μM) or rosiglitazone (5 μM) for 48 hr. Norepinephrine (0.1 μM) was added 8 hr prior to harvest (n=3). ** P <0.01, *** P <0.001 vs. control.
E: Total and uncoupled (oligomycin-insensitive) respiration of differentiated brown adipocytes (5 × 105 cells/sample) treated with WWL113 (10 μM) in the presence or absence of norepinephrine (0.1 μM) (n=3–4). * P <0.05 vs. control.
F: Ucp1 mRNA expression in differentiated brown adipocytes treated with vehicle or WWL113 (10 μM) for 24 hr with or without 30 min pre-treatment with a PPARα-selective antagonist (GW6471; 3 μM) and/or a PPARγ-selective antagonist (GW9662; 10 μM) (n=3). *** P < 0.001 vs. control. ### P< 0.001 vs. WWL113-treated cells.
G. Ucp1 mRNA expression in differentiated brown adipocytes treated with vehicle, WWL113 (10 μM), a PPARα-selective agonist (GW9578), or the combination for 24 hr (n=4). * P < 0.05, *** P < 0.001.
Next, we explored the mechanism by which WWL113 increased Ucp1 transcription. Because PPARα is a critical regulator of thermogenic gene expression in BAT (Barbera et al., 2001), we hypothesized that the action of WWL113 in BAT could be mediated via PPARα. To test this notion, primary brown adipocytes were treated with a selective PPARα antagonist (GW6471) and/or a PPARγ antagonist (GW9662) in the presence or absence of WWL113. The capacity of WWL113 to increase Ucp1 mRNA expression was largely blunted by treatment with GW6471, but not with GW9662 (Fig. 4F). The inhibitory effect of GW6471 on WWL113-induced Ucp1 mRNA expression was not altered when cells were co-treated with GW9662. These results indicate that the ability of WWL113 to enhance Ucp1 expression is principally dependent on PPARα, but not PPARγ, activity. Because WWL113 is not a direct PPARα activator (Dominguez et al., 2014), we tested the extent to which co-treatment of differentiated brown adipocytes with WWL113 and a PPARα agonist (GW9578) would further boost Ucp1 mRNA levels. Co-treatment with WWL113 and GW9578 modestly but significantly increased expression of Ucp1 relative to treatment with either compound alone (Fig. 4G), suggesting that WWL113 cooperates with the PPARα pathway to enhance UCP1 expression.
WWL113 increases UCP1 expression and the thermogenic response in vivo
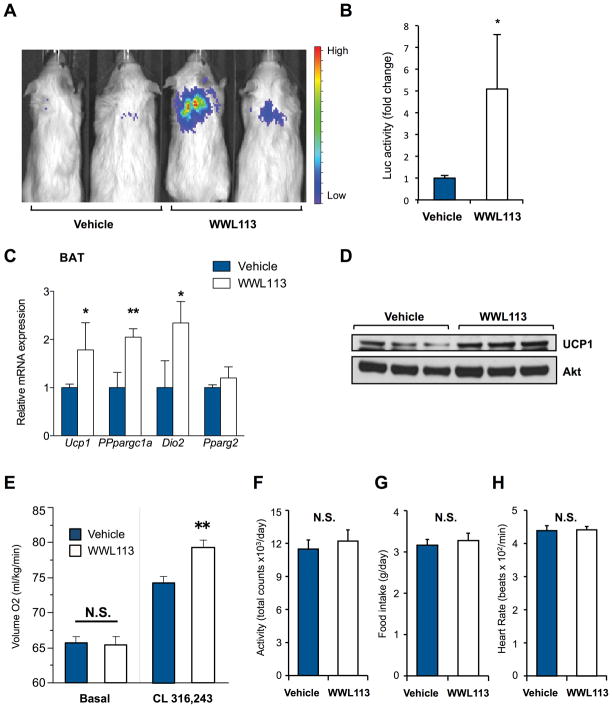
We next tested if WWL113 could enhance UCP1 expression in vivo. Ucp1-luciferase mice were treated with vehicle or WWL113 (50 mg/kg once daily) for 5 days. This dose has been shown to be effective in mice (Dominguez et al., 2014). A robust and significant increase (5 fold) in Ucp1-driven luciferase expression was detected in the interscapular BAT depots of transgenic mice treated with WWL113 (Fig. 5A and B). To confirm these findings, we examined the ability of WWL113 to enhance thermogenic gene expression in C57BL/6J mice treated with the compound for 5 days. WWL113 treatment induced significant increases in mRNA expression of Ucp1 and thermogenic genes, such as Pgc1a and Dio2, in the BAT of wild-type mice (Fig. 5C). No change in the general marker Pparγ was seen. Importantly, UCP1 protein expression was highly induced in vivo by WWL113 (Fig. 5D). In contrast, no difference in mRNA expression of Ucp1, Pgc1a, Dio2, and Pparγ was observed in inguinal WAT of WWL113-treated mice (Fig. S5A) indicating that the effects of WWL113 on the thermogenic gene program may be specific to brown fat.
Figure 5. WWL113 increases UCP1 expression in mice.
A: Luciferase activity in Ucp1-luciferase reporter mice treated daily with WWL113 (50 mg/kg) or vehicle for 5 days (n=5). Representative mice are shown.
B: Quantification of luciferase signal in interscapular BAT of mice treated as in (A). Values normalized to protein content and shown as fold change relative to vehicle.
C: Expression of thermogenic genes in interscapular BAT of C57BL/6 mice treated daily with WWL113 (50 mg/kg) or vehicle for 5 days (n=5). * P <0.05, ** P <0.01
D: UCP1 protein expression in interscapular BAT of C57BL/6 mice analyzed in (C).
E: VO2 of wild-type mice treated daily with WWL113 (50 mg/kg) or vehicle for 7 days (n=6). CL316,243 (1 mg/kg) was injected to examine the response to adrenergic stimulation. ** P <0.01. Data expressed as means ± SEM.
F: Locomotor activity of mice in (E).
G: Food intake of mice in (E).
H: Heart rate of mice in (E).
Finally, we examined the effects of WWL113 on whole-body energy expenditure. Mice treated with WWL113 for 7 days showed no differences in basal energy expenditure, but upon adrenergic stimulation (CL-316,243 injection), they responded with a more robust increase in energy expenditure than controls (Fig. 5E). WWL113 did not affect locomotor activity (Fig. 5F), food intake (Fig. 5G), or heart rate (Fig. 5H). These data indicate that WWL113 increased UCP1 levels in vivo to a functionally meaningful degree, increasing the adaptive thermogenic capacity of treated mice.
DISCUSSION
We have developed an in vivo monitoring system that allows to quantitatively track sequential changes in UCP1 expression within the same individual. Because UCP1 expression shows a high level of inter-individual variation (Boeuf et al., 2002), and changes dynamically during circadian oscillation (Gerhart-Hines et al., 2013), seasonal changes (Au-Yong et al., 2009), and aging (Rogers et al., 2012), this model may prove of wide utility. 18Fluoro-labeled 2-deoxy-glucose positron emission tomography (18FDG-PET) scanning has been applied to assess BAT activity in rodents and humans (Cypess et al., 2009; Saito et al., 2009; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009), but detection of 18FDG-PET signals in BAT depends entirely on glucose uptake. In contrast, the level of UCP1 expression in BAT is a more direct measure of the thermogenic capacity of this tissue (Nedergaard and Cannon, 2010). Hence, the transgenic UCP1 reporter we describe provides an opportunity to identify novel signaling pathways and transcriptional events that control thermogenic capacity in brown adipocytes in vivo. Unlike prior UCP1 models (Cassard-Doulcier et al., 1993), it also enables characterization of modulators of BAT function in real time in live animals.
Ucp1-luciferase brown adipocyte lines derived from this transgenic retain the characteristics of bona-fide brown fat cells. A phenotypic screen using these cells identified a compound, WWL113, that can increase UCP1 expression in vitro and in vivo. It is important to note that UCP1’s uncoupling activity is dependent on sympathetic nerve activation and increased intracellular cAMP levels. As BAT activity is stimulated by cold exposure, long-chain free fatty acids supplied from cAMP-induced lipolysis and from the circulation directly bind UCP1 and serve as a substrate to transport protons into the mitochondrial matrix (Fedorenko et al., 2012). Thus, small-molecules that solely stimulate UCP1 expression are unlikely to induce substantial thermogenesis. In agreement with this notion, we found that the effect of WWL113 treatment on cellular respiration was greater when cells were stimulated with norepinephrine. In that setting, this chemical probe significantly increased total and uncoupled cellular respiration. More importantly, mice treated with WWL113 showed considerably enhanced energy expenditure, but this increase required adrenergic stimulation. WWL113-treated mice showed no differences in locomotor activity, food intake, or heartbeat, indicating that the compound does not enhance basal sympathetic drive. Together with the finding that WWL113 increases UCP1 expression in brown fat cells in a cell-autonomous manner, these results suggest that WWL113 boosts energy expenditure primarily by increasing the content of UCP1 in BAT that can be activated by physiologic stimuli such as cold.
We previously reported that chronic administration (2 months) of WWL113 to obese-diabetic mice reduced body weight gain, improved systemic glucose and lipid homeostasis, and cleared hepatic steatosis (Dominguez et al., 2014). We ascribed the effects of WWL113 to inhibition of Ces3 in WAT and liver. Using a distinct phenotypic screen as a starting point, in this study we have found that WWL113 can have Ces3-independent effects in BAT. WWL113 is not a direct activator of PPARα (Dominguez et al., 2014), but its effects on UCP1 expression in brown fat cells depend to a large extent, though not completely, on PPARα signaling. Although the molecular target(s) for WWL113’s action in BAT remains to be clarified, this tool compound has nonetheless demonstrated the utility of our UCP1 monitoring systems to identify pharmacologic activators of UCP1 expression.
BAT is the major adipose depot that contains UCP1 positive adipocytes (classical brown adipocytes), but rodents and humans also possess an inducible type of thermogenic fat cells, termed beige or brite adipocytes (Sharp et al., 2012; Wu et al., 2012; Cypess et al., 2013; Lidell et al., 2013). UCP1-positive beige adipocytes emerge within WAT in response to external cues, such as sustained cold exposure or exercise. Beige adipocytes are considered promising reservoirs for enhancing energy expenditure, but current technologies (e.g., 18FDG-PET, MRI scans) do not possess enough resolution to detect beige cells in vivo. Our data indicates that our Ucp1-luciferase mouse may serve as a tool to monitor the effects of compounds and biological factors on beige cells.
Although they share the thermogenic ability of brown adipocytes, beige adipocytes have a distinct, heterogenous developmental origin that is not fully understood. Beige adipocytes in multiple WAT depots can arise from Myf5-positive and negative cells (Sanchez-Gurmaches and Guertin, 2014), and a subset of inguinal beige adipocytes originates from a smooth-muscle lineage (Long et al., 2014). Prior work has shown that regulation of Ucp1 is distinct in the two types of thermogenic adipocytes (Guerra et al., 1998; Koza et al., 2000; Xue et al., 2007). We found that WWL113 could activate UCP1 expression in BAT but not in inguinal WAT, implying that WWL113-initiated signaling events that regulate UCP1 expression may be unique to brown adipocytes.
BAT and liver are the major organs responsible for triglyceride uptake (Bartelt et al., 2011) and emerging evidence points to a close connection between BAT activity, liver function, and systemic lipid homeostasis. For example, defects in thermogenesis caused by BAT-specific knockout of the enzyme, euchromatic histone-lysine N-methyltransferase 1 (EHMT1) resulted in hepatic steatosis and insulin resistance even when weight-matched mice were compared (Ohno et al., 2013). Conversely, activation of BAT thermogenesis, for example by overpression of PTEN, reduced hepatic lipid accumulation and enhanced systemic insulin sensitivity (Ortega-Molina et al., 2012). Thus, the therapeutic benefits of increased BAT thermogenesis may not be limited to effects on weight, but could include improvements in lipid homeostasis and whole-body insulin sensitivity. The set of tools we have developed should aid the development of pharmacologically tractable approaches to activate BAT function as a therapy against obesity and its complications.
EXPERIMENTAL PROCEDURES
Animals
Experiments were approved by the Institutional Animal Care and Use Committees of the University of California, San Francisco and The Scripps Research Institute. Unless stated, mice were kept at room temperature. WWL113 was administered either intraperitoneally once daily (50 mg/kg in a 4:1 PEG300:Tween 80 vehicle solution) or orally (50 mg/kg in 0.5% hydroxypropylmethylcellulose). At the conclusion of treatment, BAT, WAT, skeletal muscle, and liver were snap frozen for luciferase assays and RNA and protein analysis. Energy balance studies were performed over the course of 7 days using a Comprehensive Lab Animal Monitoring System (Columbus Instruments) as described (Ohno et al., 2013). Data was normalized to body weight, as there were no differences between groups. Heart rate in the conscious state was measured by the indirect tail cuff method with a SC1000 MSP (Hatteras Instruments, Inc.).
Generation of Ucp1-luciferase reporter mice
A 98.6 kb BAC (bMQ353d13; Source BioScience) containing the entire Ucp1 gene locus was obtained and a luciferase2-T2A-tdTomato reporter cassette (Addgene 32904) inserted at the initiation codon of the Ucp1 coding sequence located in exon 1 using BAC recombineering techniques (Warming et al., 2005). Ucp1-luciferase BAC DNA was microinjected into single-cell FVB embryos and transgenic founders and their offspring identified by PCR (primers provided in Supplementary Table). Segregation patterns indicate that the transgene inserted into the Y-chromosome. Transgenics display no decrease in fertility or any other abnormalities.
In vivo luciferase imaging
Luciferase activity was monitored using an IVIS Spectrum Instrument (Caliper Life Sciences). For 3D reconstruction, six images (Exposure time: 180 sec., Binning: L, F/Stop:1, Emission filters: 560 to 660 nm, Field of view: C) were collected starting 8 minutes after injection of 150 mg/kg luciferin (Goldbio). In other experiments, one image per scan (Exposure time: 300 sec., Binning: M, F/Stop:1, Emission filters: Open, Field of view: D) was acquired 15 minutes after luciferin injection. 3D reconstructions and luciferase activity were calculated using Living Image Software (Caliper Life Sciences).
Immortalized Ucp1-luciferase adipocyte lines
The stromal vascular fraction from interscapular BAT of 3-week-old male UCP1-luciferase transgenic mice was isolated (Ohno et al., 2012) and cells infected with a retrovirus expressing large T antigen (pBabe SV40 Large T antigen; Addgene) and selected in puromycin (2 μg/ml). Immortalized preadipocytes were cultured in DMEM, 10% FBS, penicillin, and streptomycin. Upon 100% confluence (day 0), differentiation was induced with medium containing 10% FBS, 5 μg/ml insulin, 1 nM T3, 0.125 mM indomethacin, 2 μg/ml dexamethasone, and 0.125 mM IBMX for 2 days. From day 2 on, cells were cultured only in the presence of insulin and T3.
Phenotypic screen
A library of approximately 300 compounds (Adibekian et al., 2011; Bachovchin et al., 2010) was screened. Immortalized Ucp1-luciferase brown preadipocytes were seeded in 96 well plates and differentiated as described above. At day 8, cells were exposed to compounds (5 μM for carbamates, 2.5 μM for triazole ureas) in serum-free DMEM. Rosiglitazone (0.5 μM) served as positive control. After 16 hours, media was replaced with Glo Lysis Buffer (Promega) and luciferase activity quantified in a PHERAstar reader (BMG Labtech). Activity was normalized relative to the signal in DMSO-treated cells in each plate. Assays were performed in triplicate. Compounds inducing >50% increase or decrease in luciferase activity were selected for follow-up.
Luciferase assays
Cells or tissues were lysed in Cell Culture Lysis Reagent (Promega) and luciferase activity quantified using the Luciferase Assay System (Promega). Activity was measured in an Optocomp I reader (MGM Instruments) and normalized to total protein content.
Gene expression
Total RNA was isolated using RiboZol reagents (AMRESCO), reversed transcribed using an iScript cDNA synthesis kit (Bio-Rad) and quantitative real-time PCR performed using SYBR green and an ABI ViiA™ 7 machine. Relative mRNA expression was determined by the ΔΔ-Ct method using TATA-binding protein as a normalization control. Primer sequences provided in Supplementary Table.
Metabolic studies
Whole body energy expenditure was measured at ambient temperature using a Comprehensive Lab Animal Monitoring System (CLAMS, Columbus Instruments) at the UCSF animal metabolic core facility. Wild-type mice were treated daily with WWL113 (50 mg/kg) or vehicle for 7 days (n=6). The mice were injected intraperitoneally with a β3-adrenergic receptor-specific agonist CL316,243 at a dose of 1 mg/kg to examine the response to adrenergic stimulation. VO2 was normalized by body weight.
Western analysis
Proteomes were separated by SDS-PAGE. Antibodies used: UCP1 (Abcam, ab10983), Akt (Cell signaling, 9272), α-tubulin (Sigma, T8203), and β-actin (Sigma, AC-15).
Cellular respiration
Oxygen consumption rate was measured in a MT200A Cell Respirometer (Strathkelvin), as previously described (Kajimura et al., 2009). Briefly, differentiated brown adipocytes treated with DMSO or WWL113 (10 μM) for 48 hours were trypsinized and incubated in serum free medium in the presence or absence of norepinephrine. Uncoupled and non-mitochondrial cellular respiration were measured using oligomycin (1 μM) and antimycin A (1 μM).
Fat transplantation
Preadipocytes were implanted as described (Kajimura et al., 2009). Briefly, cultured immortalized Ucp1-luciferase preadipocytes were trypsinized, washed, and resuspended in PBS. Preadipocytes in a volume of 300 μl (~4 × 107 cells) were injected subcutaneously into NUCR mice (Taconic). Six days after transplantation, mice were injected with either saline (n=4) or rosiglitazone (n=6; 10 mg/kg) twice daily for 7 days. Luciferase activity was monitored on day 0, 1, 4 and 7.
Histology
Tissues were fixed in 4 % PFA and embedded in paraffin. Sections (7 μm) were analyzed with Haematoxylin/Eosin staining and immunofluorescence to detect UCP1 (ab10983 1:1000 in 5% goat serum) or GFP (GFP-1020 1:500 in 5% goat serum) as described (Sharp et al., 2012).
Statistics
Statistical significance was defined as P < 0.05 and determined by 2-tailed Student t tests, Wilcoxon or ANOVA, with Dunne’s Multiple Comparison post hoc analysis.
Supplementary Material
Figure S1 related to Figure 1. Generation of Ucp1-luciferase reporter mice
A: Genomic structure of the Ucp1-luciferase BAC transgene. The coding sequence of luciferase2-T2A-tdTomato followed by a bovine growth hormone (bGH)-derived polyadenylation signal (polyA) was inserted into the initiation codon of Ucp1 located in exon 1.
B: BAC transgenic mice (Tg) were identified by PCR analysis using the primers shown as red arrows in (A). The BAC construct (BAC) was used as a positive control. Mouse wild-type (wt) DNA and water (H20) were used as negative controls.
C: Luciferase signal in wild-type (wt) and Ucp1-luciferase transgenic mice (Tg) imaged using the IVIS Spectrum Imaging system at 9°C.
D: Long-exposure image of luciferase signal in Ucp1-luciferase reporter mice maintained at 28°C.
Figure S2 related to Figure 3. Effect of screening hits on Ucp1-luciferase preadipocytes
Subconfluent Ucp1-luciferase preadipocytes were treated with selected screen hits (10 μM) or rosiglitazone (0.5 μM) for 24 hr prior to luciferase and gene expression analysis (n=4). ND, not detectable.
Figure S3 related to Figure 3. Induction of Ucp1 expression and cellular respiration by WWL113 is not mediated by Ces3 inhibition
A. Ucp1 mRNA in differentiated primary brown adipocytes treated with WWL113, WWL113U or WWL229 (all at 10 μM) for 24 hr. WWL113 and WWL229 inhibit Ces3, while WWL113U does not (n=4). Structures are shown on the right. *** P < 0.001 vs. control.
B: Gene expression in differentiated primary brown adipocytes treated with vehicle, WWL113 (10 μM) or rosiglitazone (0.5 μM) for 48 hr (n=3). * P < 0.05, ** P < 0.01, *** P < 0.001 vs. control.
C: Total and uncoupled (oligomycin-insensitive) respiration measured in differentiated brown adipocytes (5 × 105 cells/sample) treated with WWL113 (10 μM) in the presence or absence of norepinephrine (0.1 μM) (n=3–4). Data is expressed as relative to basal for each condition. * P <0.05 vs. control.
Figure S4 related to Figure 5. WWL113 does not enhance Ucp1 expression in inguinal WAT
A: Expression of thermogenic genes in inguinal WAT of C57BL/6 mice treated daily with WWL113 (50 mg/kg) or vehicle for 5 days (n=5).
B: Core body temperature in wild-type mice treated with vehicle or WWL113 (50 mg/kg) for 7 days (n=6).
Supplementary Table related to Experimental Procedures: Primer sequences
Acknowledgments
We thank Haemin Hong, Kathleen Jay, Christophe Paillart, and Denis Glenn for assistance. Work was supported by NIH grants DK087853 and DK97441 to S.K. and DK99810 and CA197489 to E.S. S.K. acknowledges support from the DERC center grant (DK63720), UCSF PBBR program, the Pew Charitable Trust, and PRESTO from Japan Science and Technology Agency. S.B.S. was supported by a fellowship from the Alfred Benzon Foundation and A.G. by fellowship 14POST18200019 from the American Heart Association.
Footnotes
AUTHOR CONTRIBUTIONS
A.G., S.B.S, E.S., and S.K. designed experiments. J.W.C. and B.F.C. provided libraries and synthesized compounds. S.K. and S.B.S. generated transgenic model and cell lines. A.G. performed screen and follow-up. A.G., S.B.S., S.K., Y.H., K.S., and S.K. performed in vivo and vitro experiments. L.Z.S., and I.L. provided technical help. A.G., S.B.S., E.S, and S.K. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adibekian A, Martin BR, Wang C, Hsu KL, Bachovchin DA, Niessen S, Hoover H, Cravatt BF. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nature chemical biology. 2011;7:469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au-Yong IT, Thorn N, Ganatra R, Perkins AC, Symonds ME. Brown adipose tissue and seasonal variation in humans. Diabetes. 2009;58:2583–2587. doi: 10.2337/db09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera MJ, Schluter A, Pedraza N, Iglesias R, Villarroya F, Giralt M. Peroxisome proliferator-activated receptor alpha activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. The Journal of biological chemistry. 2001;276:1486–1493. doi: 10.1074/jbc.M006246200. [DOI] [PubMed] [Google Scholar]
- Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, et al. Brown adipose tissue activity controls triglyceride clearance. Nature medicine. 2011;17:200–205. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- Boeuf S, Keijer J, Franssen-Van Hal NL, Klaus S. Individual variation of adipose gene expression and identification of covariated genes by cDNA microarrays. Physiological genomics. 2002;11:31–36. doi: 10.1152/physiolgenomics.00051.2002. [DOI] [PubMed] [Google Scholar]
- Cassard-Doulcier AM, Gelly C, Fox N, Schrementi J, Raimbault S, Klaus S, Forest C, Bouillaud F, Ricquier D. Tissue-specific and β-adrenergic regulation of the mitochondrial uncoupling protein gene: control by cis-acting elements in the 5′-flanking region. Molecular Endocrinology. 1993;7:497–506. doi: 10.1210/mend.7.4.8388995. [DOI] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. The New England journal of medicine. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, Huang TL, Roberts-Toler C, Weiner LS, Sze C, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nature medicine. 2013;19:635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez E, Galmozzi A, Chang JW, Hsu KL, Pawlak J, Li W, Godio C, Thomas J, Partida D, Niessen S, et al. Integrated phenotypic and activity-based profiling links Ces3 to obesity and diabetes. Nature chemical biology. 2014;10:113–121. doi: 10.1038/nchembio.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerback S, Jacobsson A, Simpson EM, Guerra C, Yamashita H, Harper ME, Kozak LP. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. 1997;387:90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- Fedorenko A, Lishko PV, Kirichok Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell. 2012;151:400–413. doi: 10.1016/j.cell.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell metabolism. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Feng D, Emmett MJ, Everett LJ, Loro E, Briggs ER, Bugge A, Hou C, Ferrara C, Seale P, et al. The nuclear receptor Rev-erbalpha controls circadian thermogenic plasticity. Nature. 2013;503:410–413. doi: 10.1038/nature12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golozoubova V, Hohtola E, Matthias A, Jacobsson A, Cannon B, Nedergaard J. Only UCP1 can mediate adaptive nonshivering thermogenesis in the cold. FASEB J. 2001;15:2048–2050. doi: 10.1096/fj.00-0536fje. [DOI] [PubMed] [Google Scholar]
- Grundlingh J, Dargan PI, El-Zanfaly M, Wood DM. 2,4-dinitrophenol (DNP): a weight loss agent with significant acute toxicity and risk of death. Journal of medical toxicology: official journal of the American College of Medical Toxicology. 2011;7:205–212. doi: 10.1007/s13181-011-0162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Koza RA, Yamashita H, Walsh K, Kozak LP. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. The Journal of clinical investigation. 1998;102:412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Saito M. A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annual review of physiology. 2014;76:225–249. doi: 10.1146/annurev-physiol-021113-170252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GW, Lin JE, Blomain ES, Waldman SA. Antiobesity pharmacotherapy: new drugs and emerging targets. Clinical pharmacology and therapeutics. 2014;95:53–66. doi: 10.1038/clpt.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky J, Clarke G, Enerback S, Spiegelman B, Kozak LP. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. The Journal of clinical investigation. 1995;96:2914–2923. doi: 10.1172/JCI118363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koza RA, Hohmann SM, Guerra C, Rossmeisl M, Kozak LP. Synergistic gene interactions control the induction of the mitochondrial uncoupling protein (Ucp1) gene in white fat tissue. The Journal of biological chemistry. 2000;275:34486–34492. doi: 10.1074/jbc.M002136200. [DOI] [PubMed] [Google Scholar]
- Lidell ME, Betz MJ, Leinhard OD, Heglind M, Elander L, Slawik M, Mussack T, Nilsson D, Romu T, Nuutila P, et al. Evidence for two types of brown adipose tissue in humans. Nature medicine. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, et al. A Smooth Muscle-Like Origin for Beige Adipocytes. Cell metabolism. 2014;19:1–11. doi: 10.1016/j.cmet.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard J, Cannon B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell metabolism. 2010;11:268–272. doi: 10.1016/j.cmet.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013;504:163–167. doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARgamma agonists Induce a White-to-Brown Fat Conversion through Stabilization of PRDM16 Protein. Cell metabolism. 2012;15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Molina A, Efeyan A, Lopez-Guadamillas E, Munoz-Martin M, Gomez-Lopez G, Canamero M, Mulero F, Pastor J, Martinez S, Romanos E, et al. Pten positively regulates brown adipose function, energy expenditure, and longevity. Cell metabolism. 2012;15:382–394. doi: 10.1016/j.cmet.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Rogers NH, Landa A, Park S, Smith RG. Aging leads to a programmed loss of brown adipocytes in murine subcutaneous white adipose tissue. Aging cell. 2012;11:1074–1083. doi: 10.1111/acel.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, Iwanaga T, Miyagawa M, Kameya T, Nakada K, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J, Guertin DA. Adipocyte lineages: tracing back the origins of fat. Biochimica et biophysica acta. 2014;1842:340–351. doi: 10.1016/j.bbadis.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears IB, MacGinnitie MA, Kovacs LG, Graves RA. Differentiation-dependent expression of the brown adipocyte uncoupling protein gene: regulation by peroxisome proliferator-activated receptor gamma. Molecular and cellular biology. 1996;16:3410–3419. doi: 10.1128/mcb.16.7.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp LZ, Shinoda K, Ohno H, Scheel DW, Tomoda E, Ruiz L, Hu H, Wang L, Pavlova Z, Gilsanz V, et al. Human BAT possesses molecular signatures that resemble beige/brite cells. PloS one. 2012;7:e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. The New England journal of medicine. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, et al. Functional brown adipose tissue in healthy adults. The New England journal of medicine. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Waki H, Park KW, Mitro N, Pei L, Damoiseaux R, Wilpitz DC, Reue K, Saez E, Tontonoz P. The small molecule harmine is an antidiabetic cell-type-specific regulator of PPARgamma expression. Cell metabolism. 2007;5:357–370. doi: 10.1016/j.cmet.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic acids research. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Rim JS, Hogan JC, Coulter AA, Koza RA, Kozak LP. Genetic variability affects the development of brown adipocytes in white fat but not in interscapular brown fat. Journal of lipid research. 2007;48:41–51. doi: 10.1194/jlr.M600287-JLR200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 related to Figure 1. Generation of Ucp1-luciferase reporter mice
A: Genomic structure of the Ucp1-luciferase BAC transgene. The coding sequence of luciferase2-T2A-tdTomato followed by a bovine growth hormone (bGH)-derived polyadenylation signal (polyA) was inserted into the initiation codon of Ucp1 located in exon 1.
B: BAC transgenic mice (Tg) were identified by PCR analysis using the primers shown as red arrows in (A). The BAC construct (BAC) was used as a positive control. Mouse wild-type (wt) DNA and water (H20) were used as negative controls.
C: Luciferase signal in wild-type (wt) and Ucp1-luciferase transgenic mice (Tg) imaged using the IVIS Spectrum Imaging system at 9°C.
D: Long-exposure image of luciferase signal in Ucp1-luciferase reporter mice maintained at 28°C.
Figure S2 related to Figure 3. Effect of screening hits on Ucp1-luciferase preadipocytes
Subconfluent Ucp1-luciferase preadipocytes were treated with selected screen hits (10 μM) or rosiglitazone (0.5 μM) for 24 hr prior to luciferase and gene expression analysis (n=4). ND, not detectable.
Figure S3 related to Figure 3. Induction of Ucp1 expression and cellular respiration by WWL113 is not mediated by Ces3 inhibition
A. Ucp1 mRNA in differentiated primary brown adipocytes treated with WWL113, WWL113U or WWL229 (all at 10 μM) for 24 hr. WWL113 and WWL229 inhibit Ces3, while WWL113U does not (n=4). Structures are shown on the right. *** P < 0.001 vs. control.
B: Gene expression in differentiated primary brown adipocytes treated with vehicle, WWL113 (10 μM) or rosiglitazone (0.5 μM) for 48 hr (n=3). * P < 0.05, ** P < 0.01, *** P < 0.001 vs. control.
C: Total and uncoupled (oligomycin-insensitive) respiration measured in differentiated brown adipocytes (5 × 105 cells/sample) treated with WWL113 (10 μM) in the presence or absence of norepinephrine (0.1 μM) (n=3–4). Data is expressed as relative to basal for each condition. * P <0.05 vs. control.
Figure S4 related to Figure 5. WWL113 does not enhance Ucp1 expression in inguinal WAT
A: Expression of thermogenic genes in inguinal WAT of C57BL/6 mice treated daily with WWL113 (50 mg/kg) or vehicle for 5 days (n=5).
B: Core body temperature in wild-type mice treated with vehicle or WWL113 (50 mg/kg) for 7 days (n=6).
Supplementary Table related to Experimental Procedures: Primer sequences