Abstract
The apolipoprotein E (APOE) e4 allele is the most prevalent genetic risk factor for Alzheimer's disease (AD). Hippocampal volumes are generally smaller in AD patients carrying the e4 allele compared to e4 noncarriers. Here we examined the effect of APOE e4 on hippocampal morphometry in a large imaging database—the Alzheimer's Disease Neuroimaging Initiative (ADNI). We automatically segmented and constructed hippocampal surfaces from the baseline MR images of 725 subjects with known APOE genotype information including 167 with AD, 354 with mild cognitive impairment (MCI), and 204 normal controls. High‐order correspondences between hippocampal surfaces were enforced across subjects with a novel inverse consistent surface fluid registration method. Multivariate statistics consisting of multivariate tensor‐based morphometry (mTBM) and radial distance were computed for surface deformation analysis. Using Hotelling's T 2 test, we found significant morphological deformation in APOE e4 carriers relative to noncarriers in the entire cohort as well as in the nondemented (pooled MCI and control) subjects, affecting the left hippocampus more than the right, and this effect was more pronounced in e4 homozygotes than heterozygotes. Our findings are consistent with previous studies that showed e4 carriers exhibit accelerated hippocampal atrophy; we extend these findings to a novel measure of hippocampal morphometry. Hippocampal morphometry has significant potential as an imaging biomarker of early stage AD. Hum Brain Mapp 35:3903–3918, 2014. © 2014 Wiley Periodicals, Inc.
Keywords: Alzheimer's disease, hippocampus, APOE e4, MRI, multivariate tensor‐based morphometry
INTRODUCTION
The decline of cognitive skills to a functionally disabling degree is a sign of the clinical onset of Alzheimer's disease (AD), but optimizing disease modification strategies requires early intervention against appropriate therapeutic targets that may vary with disease stage. Current therapeutic failures in patients with symptomatic memory loss may reflect intervention that is too late, or else targets that represent secondary effects less relevant to disease initiation and early progression (Hyman, 2011). For therapy to be successful, timing may be critical.
In presymptomatic subjects, determining whether AD is present is challenging. The apolipoprotein E (APOE) e4 allele is the most prevalent risk factor for AD (Corder et al., 1993; Saunders et al., 1993), and is present in roughly 20–25% of North Americans and Europeans (Gerdes et al., 1992). This discovery has made it possible to study large numbers of genetically at‐risk individuals before the onset of symptomatic memory impairment and has led to the concept of preclinical stage AD (Sperling et al., 2011), a concept validated in autopsy studies of nondemented elderly subjects with neuropathological evidence of AD at autopsy (Bennett et al., 2009; Caselli et al., 2010; Dickson et al., 1992; Gouras et al., 1997; Kok et al., 2009), magnetic resonance imaging (MRI) studies of infants at differential genetic risk (Dean et al., 2013; Knickmeyer et al., 2013), fluorodeoxyglucose positron emission tomography (PET) studies of APOE e4 carriers that have revealed AD‐like patterns of reduced CMRglucose (Reiman et al., 1996, 2005), amyloid ligand binding studies using Pittsburgh Imaging Compound B that show evidence of cerebral amyloidosis in APOE e4 carriers (Reiman et al., 2009), cerebrospinal fluid levels of beta amyloid that begin to fall, suggesting the onset of AD, in the early 50s in e4 carriers (Morris et al., 2010), and neuropsychological studies showing the accelerated decline of memory scores in a gene‐dose pattern in APOE e4 carriers beginning between age 55 and 60 (Caselli et al., 2009) that is further accelerated in APOE e4 homozygotes by cerebrovascular risk factors (Caselli et al., 2011).
So far we lack a widely available, highly objective brain imaging biomarker that can identify abnormal degrees of cerebral atrophy and accelerated rate of atrophy in preclinical individuals at high risk of AD for whom early intervention is needed. A biologically grounded approach is vital to identify reliable biomarkers, consolidate all information, reduce the sheer number of statistical tests, and improve statistical power. In AD research, structural magnetic resonance imaging (MRI)‐based measures include whole‐brain (Chen et al., 2007; Fox et al., 1999; Stonnington et al., 2010), entorhinal cortex (Cardenas et al., 2011), hippocampus (den Heijer et al., 2010; Jack et al., 2003; Reiman et al., 1998; Thompson et al., [Link]; Wolz et al., 2010), and temporal lobe volumes (Hua et al., 2010), as well as ventricular enlargement (Jack et al., 2003; Thompson et al., [Link]; Wang et al., 2011). These correlate closely with differences and changes in cognitive performance, supporting their validity as markers of disease progression. Although many current studies examine cortical and substructural volumes (den Heijer et al., 2010; Dewey et al., 2010; Holland et al., 2009; Jack et al., 2004, 2003; Ridha et al., 2008; Vemuri et al., 2008a, 2008b; Wolz et al., 2010), recent research (Apostolova et al., 2010b, 2010c; Chou et al., 2009; Costafreda et al., 2011; Ferrarini et al., 2008; Madsen et al., 2010; Morra et al., 2009b; Qiu et al., 2010; Styner et al., 2005; Thompson et al., [Link]) has demonstrated that surface‐based subregional structure analysis offers advantages over volume measures, in some respects. For precise analysis of MRI patterns of hippocampal deformation for preclinical AD research, a subregional analysis would be beneficial.
Recently, we introduced surface multivariate tensor‐based morphometry (mTBM) system (Shi et al., 2013a, 2013b; Wang et al., 2012, 2011, 2010) and applied it to study AD effects on hippocampal morphometry (Shi et al., 2013a; Wang et al., 2011). Based on our experience assessing APOE e4 effects in preclinical populations (Caselli et al., 2011, 2009, 2010; Reiman et al., 2001, 1996, 2009; Stein et al., 2012) and the relatively large sample size in the Alzheimer's Disease Neuroimaging Initiative (ADNI; Jack et al., 2008; Miller, 2009; Mueller et al., 2005a, 2005b) dataset, we applied a novel hippocampal morphometry method to a large cohort of MR images of individuals with known genotype. Subregional variations in hippocampal surfaces in 725 subjects (167 AD, 354 MCI, and 204 controls) were examined for relationships with APOE e4 dose information, from APOE e4 noncarriers, people heterozygous and homozygous for APOE e4 allele. We hypothesized that the degree of hippocampal deformation would relate to genetic risk groups for AD, including adults who carry one or two copies of the APOE e4 allele, a major AD susceptibility gene.
SUBJECTS AND METHODS
Subjects
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non‐profit organizations, as a $60 million, 5‐year publicprivate partnership. The primary goal of ADNI has been to test whether serial magneticresonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California – San Francisco. ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 subjects but ADNI has been followed by ADNI‐GO and ADNI‐2. To date these three protocols have recruited over 1500 adults, ages 55 to 90, to participate in the research, consisting of cognitively normal older individuals, people with early or late MCI, and people with early AD. The follow up duration of each group is specified in the protocols for ADNI‐1, ADNI‐2 and ADNI‐GO. Subjects originally recruited for ADNI‐1 and ADNI‐GO had the option to be followed in ADNI‐2. For up‐to‐date information, see http://www.adni-info.org.
At the time of downloading (September 2010), among the 843 subjects in the baseline dataset, 738 subjects were genotyped and classified as APOE e4 carriers or noncarriers. All subjects underwent thorough clinical and cognitive assessment at the time of acquisition, including the Mini‐Mental State Examination (MMSE) score (Folstein et al., 1975), Clinical Dementia Rating (Berg, 1988), and Delayed Logical Memory Test (Wechsler, 1987).
In this study, all T1‐weighted images from ADNI baseline dataset were automatically segmented using FIRST software1 to segment the hippocampus substructure. We reconstructed hippocampal surfaces based on binary segmentations (Shi et al., 2013a). As a quality control, we manually checked all the constructed meshes and excluded five AD, five MCI, and three healthy control subjects with wrong surface topologies (Shi et al., 2013a). As a result, a total of 725 ADNI baseline subjects with APOE information, including 167 AD (age: 75.5 ± 7.6 years), 354 MCI (age: 75.1 ± 7.2 years), and 204 controls (age: 76.2 ± 4.9 years) were studied using the new system for this article. Table 1 gives detailed demographic data information on the subjects.
Table 1.
Demographic information of studied subjects in ADNI baseline dataset
| ApoE genotype | Number of subjects | Gender (M/F) | Education | Age | MMSE at baseline | |
|---|---|---|---|---|---|---|
| AD | e2/e2 | 0 | — | — | — | — |
| e2/e3 | 4 | 1/3 | 15.00 ± 2.24 | 74.25 ± 8.26 | 22.00 ± 1.58 | |
| e2/e4 | 4 | 0/4 | 15.75 ± 1.79 | 79.25 ± 5.12 | 24.75 ± 2.17 | |
| e3/e3 | 52 | 27/25 | 15.15 ± 2.05 | 76.96 ± 8.58 | 23.23 ± 2.05 | |
| e3/e4 | 73 | 44/29 | 14.62 ± 3.16 | 75.93 ± 6.43 | 23.42 ± 2.00 | |
| e4/e4 | 34 | 20/14 | 14.71 ± 2.67 | 71.92 ± 7.17 | 23.44 ± 1.83 | |
|
Control |
e2/e2 | 1 | 1/0 | 16 | 70 | 30 |
| e2/e3 | 24 | 12/12 | 15.83 ± 3.14 | 76.13 ± 5.68 | 28.83 ± 1.14 | |
| e2/e4 | 2 | 2/0 | 13.00 ± 1.00 | 76.50 ± 5.50 | 27.50 ± 2.50 | |
| e3/e3 | 125 | 69/56 | 16.20 ± 2.71 | 76.29 ± 4.83 | 29.18 ± 0.89 | |
| e3/e4 | 48 | 25/23 | 16.13 ± 2.73 | 76.50 ± 4.48 | 29.25 ± 0.83 | |
| e4/e4 | 4 | 2/2 | 16.75 ± 1.92 | 73.75 ± 3.34 | 29.00 ± 0.71 | |
|
MCI |
e2/e2 | 0 | — | — | — | — |
| e2/e3 | 15 | 7/8 | 15.93 ± 2.86 | 76.67 ± 7.44 | 27.60 ± 1.50 | |
| e2/e4 | 10 | 7/3 | 16.50 ± 2.33 | 74.20 ± 8.58 | 28.00 ± 1.26 | |
| e3/e3 | 145 | 95/50 | 15.81 ± 2.94 | 76.20 ± 7.71 | 27.23 ± 1.77 | |
| e3/e4 | 141 | 91/50 | 15.61 ± 3.06 | 74.82 ± 6.63 | 26.94 ± 1.76 | |
| e4/e4 | 43 | 25/18 | 15.81 ± 2.57 | 71.81 ± 5.91 | 26.84 ± 1.95 |
In our study, following prior work (Morra et al., 2009a, 2009b; Shi et al., 2013a), we pooled both the subjects who are heterozygous APOE e4 carriers (e3/e4) and homozygous APOE e4 carriers (e4/e4) together to form the APOE e4 carriers group and correlated presence of the APOE e4 allele with hippocampal morphometry, both (1) in the entire sample and (2) in nondemented (pooled MCI and controls) subjects. Throughout the article, we call these two populations as the full ADNI cohort and nondemented cohort, respectively.
Processing Pipeline
Figure 1 shows our overall sequence of processing. First, given the three‐dimensional (3D) MRI scans from the ADNI baseline dataset, hippocampal substructures were segmented with FIRST (Patenaude et al., 2011) and hippocampal surfaces were automatically reconstructed based on the segmentations (Han et al., 2003). Second, a conformal grid was generated for each surface with the holomorphic 1‐form based surface conformal parameterization (Wang et al., 2011). With this conformal grid, we computed the conformal representation of the surface (Gu and Vemuri, 2004), i.e., the conformal factor and mean curvature, which represent the intrinsic and extrinsic features of the surface, respectively. The “feature image” of a surface was computed by combining the conformal factor and mean curvature and linearly scaling the dynamic range into [0, 255]. Third, we registered the feature image of each surface in the dataset to a common template with an inverse consistent fluid registration algorithm (Shi et al., 2013a). With conformal parameterization, we essentially converted a 3D surface registration problem into a 2D image registration problem. The flow induced in the parameter domain establishes high‐order correspondences between 3D surfaces. Finally, we studied the differences between different diagnostic groups with the mTBM statistics (Leporé et al., 2008; Wang et al., 2010), which retain the full tensor information of the deformation Jacobian matrix, together with the radial distance, which retains information on the deformation along the surface normal direction.
Figure 1.
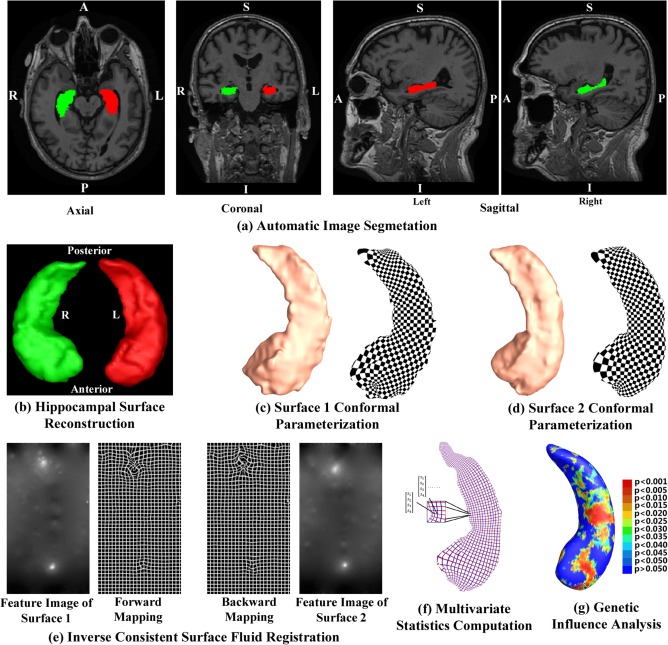
A chart showing the key steps in our system. MR images were automatically segmented by FIRST to extract the hippocampal substructure (a). After the hippocampal surfaces were constructed from FIRST segmentations (b), we computed their conformal parameterizations with holomorphic 1‐forms (c and d). Then feature images were generated by combining the local conformal factor and mean curvature that were computed from the conformal parameterizations. After the inverse consistent fluid registration was done in the feature image domain, we deformed the surfaces using the obtained displacements (e). The new statistics consisting of radial distance and multivariate TBM were computed at each point on the resultant matching surface (f). Then the Hotelling T 2 test was applied to study genetic influence of APOE e4 allele (g). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Hippocampus Segmentation and Surface Reconstruction
All T1‐weighted MR images were automatically segmented using FIRST (Patenaude et al., 2011). FIRST is a model‐based subcortical structure segmentation and registration tool developed as part of the FSL library, which is written mainly by members of the Analysis Group, FMRIB, Oxford, UK. We ran the run_first_all routine with default parameters tuned by FIRST as optimal for hippocampal segmentation. Among the results of the routine, we took the three‐phase image which contains the labels of the left and right hippocampi as shown in Figure 1a. Then the binary image for each side was obtained by a simple thresholding process. Hippocampal surfaces were constructed with a topology‐preserving level set method based on the binary segmentations (Han et al., 2003) and triangular surface meshes were obtained based on the marching cubes algorithm (Lorensen and Cline, 1987). After mesh refinement (Shi et al., 2013a), we obtained smooth surfaces that are suitable for generating conformal grids as shown in Figure 1b. Finally each of the smoothed meshes was aligned into the MNI standard space using a global affine transformation with a nine‐parameter (three parameters for translation, three parameters for rotation, and three parameters for scaling) matrix that was computed by FIRST (Patenaude et al., 2011).
Conformal Grid Generation
To facilitate hippocampal shape analysis, we generated a conformal grid on each surface and used it as a canonical space for the following surface registration and multivariate statistical analysis. To generate a planar surface conformal parameterization for a closed hippocampal surface, we applied an automatic algorithm, topological optimization, to introduce two cuts on a hippocampal surface to convert it into a genus zero surface with two open boundaries (Shi et al., 2013a). The locations of the two cuts are at the front and back of the hippocampal surface, representing its anterior junction with the amygdala, and its posterior limit as it turns into the white matter of the fornix. They are biologically valid and consistent landmarks across subjects. Given the hippocampal tube‐like shape, these landmark curves are automatically determined by checking the extreme points when searching along the first principle direction of geometric moments of surface (Elad et al., 2004; Wang et al., 2011; Zhang and Lu, 2004). For quality control purposes, we have manually checked the consistency of all landmark curves. Then the exact 1‐form basis was computed with the open boundary surface (Wang et al., 2010). Later we computed the basis for all closed but nonexact 1‐forms. The harmonic 1‐form basis is the union of the exact 1‐form basis and the closed but nonexact 1‐form basis. By solving a linear system with the harmonic 1‐form basis, we obtained the conjugate of the exact 1‐form basis. The exact 1‐form basis and its conjugate 1‐form form the holomorphic 1‐form basis, which induces a conformal grid on the hippocampal surface. Figure 1c,d shows two example hippocampal surfaces with their conformal grids. In both pictures, the overlaid checkerboard texture is used to demonstrate the angle preserving property, i.e., the right angles on the planar checkerboard texture are well preserved after they are overlaid on hippocampal surfaces.
The conformal parameterization of a surface contains a number of geometric features about the surface. In our system, we computed the local conformal factor and mean curvature, which uniquely determine a closed surface in 3D, up to a rigid motion (Gu et al., 2004). Conformal factor is the area ratio of an infinitesimal region around a point on the surface and an infinitesimal region around the same point on the planar parameter domain. It represents the intrinsic features of a surface. By contrast, the mean curvature represents the extrinsic features of a surface. Both measurements, the conformal factor and mean curvature, are local features which are defined on each surface vertex. Since the conformal factor and mean curvature encode both intrinsic structure and 3D embedding information, we call them the surface conformal representation. In our framework, conformal representation is adopted as surface features for automated surface registration. As shown in Figure 1e, we summed up the conformal factor and mean curvature and linearly scaled the dynamic range of the summation into [0, 255] to form the feature image of the surface.
Hippocampal Surface Registrations
Similar to other tensor‐based morphometry (TBM) work (e.g., Chung, 2012; Davatzikos et al., 1996; Hua et al., 2011), we need to register each individual hippocampal surface to a common template surface for morphometric analysis. With the conformal parameterization and conformal representation, we convert the 3D surface registration problem into a 2D image registration problem. The well‐studied image fluid registration algorithm (Bro‐Nielsen and Gramkow, 1996; D'Agostino et al., 2003) can be easily applied to induce a deformation flow in the parameter domain, which in turn enforces a high‐order correspondence in 3D. We introduced a correction term in the traditional Navier–Stokes equation to compensate for the parameterization area distortion. With conformal parameterization, the correction term was simply the conformal factor and the surface fluid registration can be easily developed by extending the Navier–Stokes equation to drive flows on general surfaces, regardless of the underlying parameterizations. We call this method surface fluid registration (Shi et al., 2013a).
Furthermore, most image registration algorithms in the literature are not symmetric, i.e., the correspondences established between the two images depend on which image is assigned as the deforming image and which is the nondeforming target image. An asymmetric algorithm can be problematic as they tend to penalize the expansion of image regions more than shrinkage (Rey et al., 2002). Thus, in our system, we further extended the surface fluid registration method into an inverse consistent framework (Leow et al., 2005). The obtained surface registration is diffeomorphic. An example is as shown in Figure 1e. For details of our inverse consistent surface fluid registration method, we refer to (Shi et al., 2013a).
Surface Multivariate Morphometry Statistics
Our multivariate morphometry statistical analysis consists of mTBM (Leporé et al., 2008; Wang et al., 2009) and radial distance analysis (Pizer et al., 1999; Thompson et al., [Link]). This combines complementary information from mTBM, which measures deformation within surfaces, and radial distance, which measures hippocampal size in terms of the surface normal direction.
The mTBM statistics have been carefully studied in brain structure morphology analyses and they can demonstrate improved signal detection power relative to more standard Jacobian matrix statistics (Shi et al., 2013a, 2013b; Wang et al., 2012, 2011, 2013). As mTBM retains the full information in the deformation tensor fields, it is very sensitive to deformations such as rotation, dilation, and shear along the surface tangent direction, which is perpendicular to the surface normal. Given the hippocampal tube‐like shape, its atrophy and enlargement directly affect the distance from each surface point to its medial core (analogous to the center line in a tube). We call this distance the radial distance of a hippocampal surface. Radial distance mainly describes morphometric changes along the surface normal direction and has been applied in many subcortical studies (Bansal et al., 2000; Gerig et al., 2001; Morra et al., 2009b; Pizer et al., 1999; Thompson et al., [Link]). Thus, these two statistics are complementary to each other. In this article, we adopted the multivariate statistics proposed in (Wang et al., 2011) to study shape differences between groups with different diagnosis, APOE e4 dose, and healthy controls.
As in our prior work (Wang et al., 2011), the mTBM was computed as a 3 × 1 vector consisting of the “Log‐Euclidean metric” (Arsigny et al., 2006), computed as the matrix logarithm of the deformation tensor. Given a hippocampal surface with the conformal parameterization as described in Conformal Grid Generation section, the radial distance was computed as the distance from each parametric surface point to the center of 3D positions of the iso‐u curves in the parameter domain (Wang et al., 2011), as shown by the red curves in Figure 1f. We formed the new multivariate surface morphometry statistic as a 4 × 1 vector consisting of the mTBM and radial distance.
Statistical Group Difference
To assess group differences with multivariate statistics, we applied Hotelling's T 2 test (Cao and Worsley, 1999; Hotelling, 1931; Kim et al., 2012; Thirion et al., 2000) on sets of values in the log‐Euclidean space of the deformation tensors. For each surface vertex, given two groups of n × 4‐dimensional vectors, , we used the Mahalanobis distance M to measure the group mean difference,
where N S and N T are the numbers of subjects in the two groups, and are the means of the two groups and Σ is the combined covariance matrix of the two groups (Leporé et al., 2008; Wang et al., 2011, 2010).
Specifically, for each hippocampal surface point, we ran a permutation test with 10,000 random assignments of subjects to different groups to estimate the statistical significance of the areas with group difference in surface morphometry. We also used a predefined statistical threshold of P = 0.05 at each surface point to estimate the overall significance of the group difference maps by nonparametric permutation testing (Holmes et al., 1996; Nichols and Holmes, 2002). In each case, the covariate (group membership) was permuted 10,000 times and a null distribution was developed for the area of the average surface with group difference statistics above the predefined threshold in the significance map. The overall significance of the map is defined as the probability of finding, by chance alone, a statistical map with at least as large a surface area beating the predefined statistical threshold of P = 0.05. This omnibus P value is commonly referred to as the overall significance of the map (or the features in the map), corrected for multiple comparisons. It basically quantifies the level of surprise in seeing a map with this amount of the surface exceeding a predefined threshold, under the null hypothesis of no systematic group differences. The permutation test on the overall rejection areas is used to evaluate the significance of overall experimental results and correct the overall significant P values for multiple comparisons. Figure 1g shows an example of the significance p‐map with uncorrected P values, which is used to visualize the surface regions with significant differences between groups.
RESULTS
Effects of APOE e4 Genotype
To explore whether the presence of the APOE e4 allele was associated with greater hippocampal atrophy, we conducted two experiments to study the effects of APOE e4 genotype on hippocampal morphometry in two populations:
APOE e4 carriers versus noncarriers in the full ADNI cohort;
APOE e4 carriers versus noncarriers in the nondemented cohort.
The experiments aimed to determine if the APOE e4 allele was associated with hippocampal atrophy in all subjects or in subjects who have not yet developed AD. By contrast with (Shi et al., 2013a), the study in this article is more rigorous as the APOE e4 noncarriers are those subjects who are homozygous noncarriers (e3/e3). Subjects with one e2 allele, i.e., e2/e3 and e2/e4 were excluded due to the possible protective effect of e2 allele for AD (Morra et al., 2009b).
In the 725 subjects of known APOE e4 genotype, there were 322 noncarriers (all homozygous for APOE e3) and 343 APOE e4 carriers. The nondemented cohort consisted of 506 subjects who were either MCI or control subjects, including 270 e4 noncarriers and 236 e4 carriers. Figure 2 shows the statistical p‐map for the full ADNI cohort (N = 665; 322 noncarriers and 343 carriers). Nonblue colors show vertices with statistical differences at the nominal 0.05 level, uncorrected for multiple comparisons. As shown in Figure 2, the APOE e4 carriers differed significantly from the noncarriers (P < 0.0002). Figure 3 shows the p‐map for the nondemented cohort (N = 506; 270 noncarriers and 236 carriers). After correcting for multiple comparisons, the difference remained highly significant (P < 0.0027).
Figure 2.
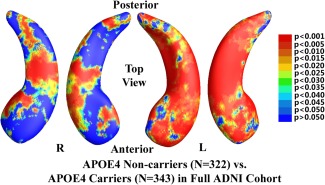
Illustration of local shape differences (P values) between the APOE e4 noncarriers (e3/e3, N = 322) and carriers (e3/e4 and e4/e4, N = 343) in the full ADNI cohort. Nonblue colors show vertices with statistical differences, at the nomial 0.05 level, uncorrected. The overall significance after multiple comparisons with permutation test is P < 0.0002. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Figure 3.
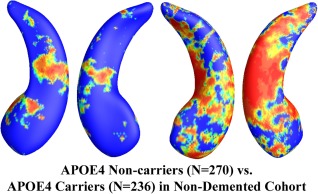
Illustration of local shape differences (P values; a) between the APOE e4 noncarriers (e3/e3, N = 270) and carriers (e3/e4 and e4/e4, N = 236) in the nondemented cohort (MCI and controls). Nonblue colors show vertices with statistical differences, at the nomial 0.05 level, uncorrected. The overall significance after multiple comparisons with permutation test is P < 0.0027. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
APOE e4 Dose Effects: Difference Comparison Between Heterozygous and Homozygous APOE e4 Carriers
To explore whether APOE e4 allele dose affects hippocampal surface morphometry and how this atrophy is related to normal aging, we studied hippocampal morphometry between persons homozygous for the APOE e4 allele and those heterozygous for this allele. We studied group differences between heterozygous and homozygous APOE e4 subjects in the full ADNI cohort, and in the nondemented APOE e4 carrier cohort in ADNI baseline dataset.
Among the APOE e4 carriers, 81 subjects were homozygous (e4/e4) and 262 were heterozygous (e3/e4) for APOE e4 allele. Figure 4 shows the statistical p‐map for all APOE e4 subjects. The e4 heterozygotes differed significantly from the e4 homozygotes (P < 0.0129 after multiple comparisons correction with the permutation test). Excluding those APOE e4 carriers in the AD group, the nondemented APOE e4 carrier group consisted of 189 e4 heterozygotes and 47 homozygotes. Figure 5 shows the statistical p‐map for nondemented APOE e4 carriers. However, after correcting for multiple comparisons, the effect was not significant (P = 0.142). (There may be some subthreshold difference for the right hippocampus, but a larger sample size would be needed to detect it, if present.)
Figure 4.
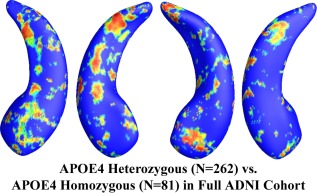
Illustration of local shape differences (P values) between the heterozygous APOE e4 carriers (e3/e4, N = 262) and the homozygous APOE e4 carriers (e4/e4, N = 81) in the full ADNI cohort. Nonblue colors show vertices with statistical differences, at the nomial 0.05 level, uncorrected. The overall significance after multiple comparisons with permutation test is P < 0.0129. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Figure 5.
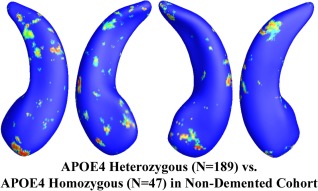
Illustration of local shape differences (P values) between the heterozygous APOE e4 carriers (e3/e4, N = 189) and the homozygous APOE e4 carriers (e4/e4, N = 47) in the nondemented cohort. Nonblue colors show vertices with statistical differences, at the nomial 0.05 level, uncorrected. The overall significance after multiple comparisons with permutation test is P < 0.142. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
APOE e4 Dose Effects: Difference Comparison Between APOE e4 Noncarriers and Carriers With Different APOE e4 Dose
To further study the APOE e4 dose effects, we divided the subjects into three groups, APOE e4 homozygotes, heterozygotes, and noncarriers. We performed group difference analysis between two groups and compared the statistical power. We hypothesized that morphometric differences would be greater in APOE e4 homozygotes than heterozygotes, who would in turn show greater deformities compared to e4 noncarriers.
Figures 6 and 7 show how APOE e4 noncarriers differ in hippocampal shape from APOE e4 heterozygotes and homozygotes in the full ADNI cohort and the nondemented cohort, respectively. Figure 6 shows the statistical p‐map for the full ADNI cohort. Nonblue colors show vertices with statistical differences, at the nominal 0.05 level, uncorrected. As shown in Figure 6a, the APOE e4 heterozygotes differed from e4 noncarriers (P < 0.0031). Figure 6b shows the statistical p‐map for the full ADNI cohort and demonstrates that the APOE e4 homozygotes differed from e4 noncarriers (P < 0.0001). Figure 6b also shows more extensive statistically significant areas of difference than those in Figure 6a, for both the left and right hippocampal surfaces.
Figure 6.
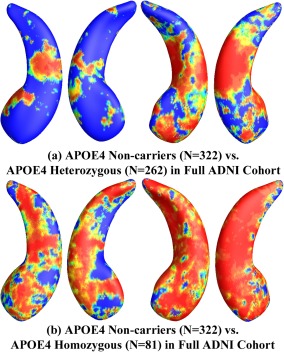
Illustration of local shape differences (P values) between the APOE e4 noncarriers (e3/e3, N =322) and heterozygous carriers (e3/e4, N = 262; a), between the APOE e4 noncarriers (e3/e3, N = 322) and homozygous carriers (e4/e4, N = 81; b), in the full ADNI cohort. Nonblue colors show vertices with statistical differences, at the nomial 0.05 level, uncorrected. The overall significances after multiple comparisons with permutation test are P < 0.0031 for (a) and P < 0.0001 for (b). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Figure 7.
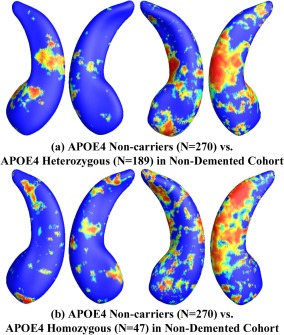
Illustration of local shape differences (P values) between the APOE e4 noncarriers (e3/e3, N = 270) and heterozygous carriers (e3/e4, N = 189; a), between the APOE e4 noncarriers (e3/e3, N = 270) and homozygous carriers (e4/e4, N = 47; b), in the nondemented cohort. Nonblue colors show vertices with statistical differences, at the nomial 0.05 level, uncorrected. The overall significances after multiple comparisons with permutation test are P < 0.017 for (a) and P < 0.006 for (b). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
After excluding AD subjects from these three groups, we repeated the group difference analysis among APOE e4 noncarriers (e3/e3, N = 270), e4 heterozygotes (e3/e4, N = 189), and e4 homozygotes (e4/e4, N = 47). Figure 7a shows the statistical p‐map for the nondemented cohort [N = 459; 270 noncarriers (e3/e3) and 189 APOE e4 heterozygous carriers (e3/e4)]. The APOE e4 heterozygotes differed from the e4 noncarriers (P < 0.017). Figure 7b shows the p‐map for the nondemented cohort [N = 317; 270 noncarriers (e3/e3) and 47 APOE e4 homozygous carriers (e4/e4)] and showed that the APOE e4 homozygotes differed from the e4 noncarriers (P < 0.006). Similar to Figure 6, the homozygous vs. noncarrier comparison showed more extensive areas of difference in the uncorrected p‐maps.
In Figure 8, the cumulative distribution functions of the P values observed for the contrast of APOE e4 carriers versus noncarriers are plotted against the corresponding P value that would be expected, under the null hypothesis of no group difference, for the four experiments shown in Figures 6 and 7. For null distributions, the cumulative distribution of P values is expected to fall approximately along the dotted line. Large deviations from that curve are associated with significant signal, and greater effect sizes represented by larger deviations. The theory of false discovery rates (Benjamini and Hochberg, 1995) gives formulae for thresholds that tend to control false positives at a known rate. This protocol was adopted in several of our prior articles (Shi et al., 2013a, 2013b; Wang et al., 2011, 2013, 2010) as an empirical standard to compare effects in group difference analysis. We note that the deviation of the statistics from the null distribution generally increases from heterozygotes vs. noncarriers to homozygotes vs. noncarriers in both the full ADNI cohort and nondemented cohort studies.
Figure 8.
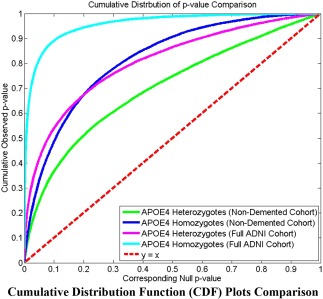
Illustration of cumulative distribution functions of the P values observed for the contrast of APOE e4 carriers versus noncarriers, plotted against the corresponding P value that would be expected under null hypothesis of no group difference, for the four experiments shown in Figures 6 and 7. We note that the deviation of the statistics from the null distribution generally increases from heterozygotes vs. noncarriers to homozygotes vs. noncarriers in both the full ADNI cohort and nondemented cohort studies, suggesting that the APOE e4 allele dose may be associated with more accelerated atrophy of hippocampus. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
As such, although more rigorous statistical tests are certainly necessary, from the p‐maps and CDF plots, we can observe the trend that in all groups, APOE e4 homozygotes appear to differ more from noncarriers than do e4 heterozygotes, suggesting a clear APOE e4 dose effect.
DISCUSSION
Prior studies of APOE e4 carriers (Caselli et al., 2011, 2009, 2010; Reiman, 2007) have helped to define and characterize preclinical AD with possible implications for primary AD prevention research. Essential to this effort are sensitive biomarkers that can track disease progression in the absence of symptoms. Imaging endophenotypes are promising, but further refinement of their relevance in early stage disease is needed (Frisoni et al., 2010). MRI hippocampal morphometry may help move disease detection earlier and evaluate the effectiveness of promising disease‐slowing and prevention therapies in a shorter time and a more cost‐effective way. Much recent research has used brain imaging to study how APOE e4 allele affects hippocampal morphometry in patients and cognitively normal people (Farrer et al., 1997; Lehtovirta et al., 1995; Lemaitre et al., 2005; Morra et al., 2009a, 2009b; Mueller and Weiner, 2009; Pievani et al., 2011; Qiu et al., 2009; Reiman et al., 1996; Shi et al., 2013a).
Our study has two main findings. First, as one of the largest hippocampal morphometry studies to date, involving 725 baseline ADNI subjects, we found that, for the nondemented subjects, the APOE e4 genotype is associated with greater hippocampal deformation. Second, our novel hippocampal surface morphometry method (Shi et al., 2013a, 2013b), which involves conformal mapping, inverse consistent surface fluid registration and multivariate statistical analysis, automatically processed all ADNI baseline imaging data and was as or more sensitive to APOE e4 effects than some previously reported methods (e.g., Morra et al., 2009a, 2009b), that have used the ADNI dataset.
Our work is related to the shape modeling of hippocampal surfaces. The Large Deformation Diffeomorphic Metric Mapping (LDDMM; Joshi and Miller, 2000) has been used to deform labeled anatomical templates of the hippocampus onto new images, using a combination of manual landmarking of points on the hippocampus and 3D fluid image registration (Csernansky et al., 2000; Haller et al., 1996; Wang et al., 2007). In the LDDMM method, the surface of the hippocampus is parcellated a priori using a neuroanatomical template into three zones to approximate the locations of underlying subfields, and LDDMM is used to generate the hippocampal surfaces of all subjects and to register the surface zones across subjects. Another important shape modeling approach models the hippocampal surface using spherical harmonic functions (Gutman et al., 2009; Shen et al., 2009; Styner et al., 2004), and uses the coefficients of the harmonic expansion to infer shape differences between patient groups and controls. Other methods (Van Leemput et al., 2009; Wang et al., 2006, 2003; Yassa et al., 2010; Yushkevich et al., 2010) segment hippocampus into different regions and analyze the volume and shape changes in these subfields. These methods compute volumetric image registration between template and individual subject and translate and visualize the deformation on surfaces. In hippocampal subfield shape analysis work (Apostolova et al., 2010a; Cho et al., 2011; Morra et al., 2009a; Qiu et al., 2009; Shi et al., 2013a; Thompson et al., [Link]), the morphometry comparison was performed by registering hippocampal surfaces with geometric feature analysis. This type of methods affords the benefits of high resolution information from the hippocampal surface representation and efficient numerical solutions to register and analyze surface deformation across subjects.
In our APOE e4 carrier vs. noncarriers experiments, comparisons with both the nondemented and the full ADNI cohorts yielded significant differences that were apparently more pronounced on the left hippocampal surface. A prior study (Morra et al., 2009b), which conducted similar experiments with a smaller number of images in ADNI baseline dataset (N = 490) was only able to achieve significance for the left hippocampal surfaces on the full ADNI cohort but did not detect significant differences in the nondemented cohort. That aside, our finding of more significant areas on the left than on the right side, agree with (Morra et al., 2009b), despite differences in our image segmentation methods, surface parameterization and registration algorithms, and statistics. Our results also agree with another APOE e4 study with manually segmented hippocampal surfaces (Pievani et al., 2011).
To our knowledge, this is the first study to use a surface‐based approach to study APOE e4 gene dose effects on the hippocampal morphometry differences among subjects with two copies, one copy and no copies of the APOE e4 allele. Although most current approaches use cortical and substructural volume measures (den Heijer et al., 2010; Dewey et al., 2010; Holland et al., 2009; Jack et al., 2004, 2003; Ridha et al., 2008; Vemuri et al., 2008a, 2008b; Wolz et al., 2010), recent research (Apostolova et al., 2010b, 2010c; Chou et al., 2009; Costafreda et al., 2011; Ferrarini et al., 2008; Luders et al., 2012; Madsen et al., 2010; Morra et al., 2009b; Qiu et al., 2010; Styner et al., 2005; Thompson et al., [Link]; Yang et al., 2012) has demonstrated that surface‐based subregional structure analysis can offer some advantages over volume measures. Our work identified significant differences in hippocampal shape between subjects heterozygous and homozygous for APOE e4 allele in the full ADNI study but did not detect such a difference on the nondemented cohort. When comparing these two groups with homozygous noncarriers (e3/e3), we detected significant differences in all four experiments. Furthermore, our empirical examination of the effect size clearly suggested that the homozygous APOE e4 carriers showed greater atrophy than heterozygous APOE e4 carriers in both full ADNI and nondemented cohorts.
Consistent with prior studies of APOE e4 effects on hippocampal surfaces (e.g., Morra et al., 2009b; Pievani et al., 2011), both heterozygous and homozygous APOE e4 patients show greater deformities for the left than the right hippocampus. By contrast with comparisons of APOE e4 carriers vs. noncarriers, differences between the heterozygous and homozygous APOE e4 groups were greater on the right side than on the left. Some prior research on hippocampal volume (Farrer et al., 1997; Lemaitre et al., 2005) also found that when comparing the heterozygous and homozygous APOE e4 groups, the right hippocampus may have more pronounced atrophy than the left side. However, contradictorily, when comparing these two groups with noncarriers, some other works (Farrer et al., 1997; Lemaitre et al., 2005) found greater atrophy on the right side. The inconsistency may be due to the algorithms used, as with surface‐based method and the new statistics we introduced, more local subtle changes may be captured, which may be missed by the global volume‐based method. More systematic validation and comparison studies are warranted.
Another important goal is to study differences between APOE e4 noncarriers and carriers within the cognitively normal group, i.e., healthy control subjects. As there were only two control subjects homozygous for the APOE e4 allele, tests of dose effects were underpowered in healthy control group. Thus we only compared APOE e4 carriers vs. noncarriers within the control group. Our experiments [N = 177, 125 noncarriers (e3/e3) vs. 52 APOE e4 carriers (e3/e4 or e4/e4)] identified greater deformities on the left hippocampus than the right side but no statistically significant differences were detected after multiple comparisons correction (P = 0.34). This is perhaps due to the low number of healthy control individuals with the APOE e4 genotype (Table 1). The association between APOE e4 and hippocampal atrophy may be detectable in a much larger sample.
In our work, we applied a nonparametric, multivariate permutation testing on Hotelling's T 2 statistics. Due to the Hotelling's T 2 test, comparisons of different genotype groups were only conducted pairwisely. However, the multivariate analysis of variance (MANOVA; Smith et al., 1962) may be also applicable to analyze our multivariate statistics. MANOVA is a generalized form of univariate analysis of variance. It is used when there are two or more dependent variables. MANOVA has been successfully applied to human brain mapping research (e.g., Bartley et al., 1997; Poline et al., 1996). It may be particularly useful for us to check the hippocampal morphometry differences among subjects with two copies, one copy and no copies of the APOE e4 allele. We plan to study MANOVA in our ongoing AD prevention research.
The current work focuses on describing structural differences at the group level and establishing the correlation between morphometry changes and genetic variance. The current work mainly explores the difference among multivariate statistics without considering the trends of the metric changes. Investigating the trend for multi‐variables as a whole (in the context of multivariate type analysis) might be difficult, but it would be useful for us to explore this “trend analysis” on individual statistics. For example, a medically interesting question could be that, among these three groups (noncarriers, heterozygotes and homozygotes of e4), which individual statistics is more monotonic. In our prior work (Shi et al., 2013b), we used TBM to visualize the metric trend for prematurity study but it may not be able to directly explain the outcome achieved with mTBM features. We plan to explore more along this direction in our future work. The answers to these questions will make our results more intuitive to be understood and eventually help our methods to be adopted quickly by the medical community.
This study has two limitations. First, as the participants are elderly, the ADNI subjects may not be the best representative of patient populations in clinical trials. Our ongoing work that studies the Arizona APOE cohort (Caselli et al., 2009) which consists of cognitively normal subjects with a wider age span may validate or extend our current ADNI findings. We have obtained another cohort dataset from Australian Imaging, Biomarkers and Lifestyle Study of ageing (AIBL, 2013; Ellis et al., 2009). In addition, our ongoing research on Enhancing Neuro Imaging Genetics through Meta‐Analysis (Thompson et al., in press) may potentially provide new and rich datasets for cross‐validation. Replication attempts will be made to corroborate our current biomedical discoveries. Second, because of the extremely low number of APOE e2 carriers in ADNI dataset (Table 1), we excluded them from our APOE e4 studies but did not perform any additional studies to show that APOE e2 might be protective.
CONCLUSIONS
We recently developed our MRI‐based computer‐assisted hippocampal surface morphometry system that uses conformal maps to induce well‐organized grids on surfaces. This simplifies a number of downstream computations of derivatives and metrics. In addition, the surface metric tensor, computable from the conformal grid, has a multivariate structure that contains a great deal of information on local surface geometry. Its components follow a log‐Euclidean law that affects their possible range of values and their statistical distributions. The resulting set of surface tensor methods encodes a great deal of information that would otherwise be inaccessible, or overlooked.
We applied our system to study hippocampal shape differences between subjects in the ADNI dataset with two copies, one copy, and no copies of the APOE e4 allele, a common susceptibility gene for late‐onset AD. We found significant differences between APOE e4 carriers and noncarriers in both full ADNI and nondemented cohorts, with more deformation of the left hippocampus than the right. Within the full ADNI cohort, the e4 homozygotes demonstrated more deformities than the e4 heterozygotes. Our work supports prior reports that the APOE e4 genotype is associated with accelerated brain atrophy along with disease progression, and that these differences can be mapped to morphological changes in subsections of the hippocampal surface. Future studies will test this framework in cognitively normal subjects for the detection of preclinical AD.
Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH‐12‐2‐0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol‐Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann‐La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
REFERENCES
- AIBL (2013): The Australian Imaging, Biomarkers & Lifestyle Flagship Study of Ageing, http://www.aibl.csiro.au/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Morra JH, Green AE, Hwang KS, Avedissian C, Woo E, Cummings JL, Toga AW, Jack CR Jr, Weiner MW, Thompson PM (2010a): Automated 3D mapping of baseline and 12‐month associations between three verbal memory measures and hippocampal atrophy in 490 ADNI subjects. Neuroimage 51:488–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Mosconi L, Thompson PM, Green AE, Hwang KS, Ramirez A, Mistur R, Tsui WH, de Leon MJ (2010b): Subregional hippocampal atrophy predicts Alzheimer's dementia in the cognitively normal. Neurobiol Aging 31:1077–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Thompson PM, Green AE, Hwang KS, Zoumalan C, Jack CR Jr, Harvey DJ, Petersen RC, Thal LJ, Aisen PS, Toga AW, Cummings JL, Decarli CS. (2010c): 3D comparison of low, intermediate, and advanced hippocampal atrophy in MCI. Hum Brain Mapp 31:786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsigny V, Fillard P, Pennec X, Ayache N (2006): Log‐Euclidean metrics for fast and simple calculus on diffusion tensors. Magn Reson Med 56:411–421. [DOI] [PubMed] [Google Scholar]
- Bansal R, Geiger B, Banihashemi A, Krishnan A (2000): Integrated segmentation, registration and visualization of multimodal medical image datasets In: IEEE Visualization. [Google Scholar]
- Bartley AJ, Jones DW, Weinberger DR (1997): Genetic variability of human brain size and cortical gyral patterns. Brain 120:257–269. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995): Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. [Google Scholar]
- Bennett DA, De Jager PL, Leurgans SE, Schneider JA (2009): Neuropathologic intermediate phenotypes enhance association to Alzheimer susceptibility alleles. Neurology 72:1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg L (1988): Clinical dementia rating (CDR). Psychopharmacol Bull 24:637–639. [PubMed] [Google Scholar]
- Bro‐Nielsen M, Gramkow C (1996): Fast fluid registration of medical images In: Visualization in Biomedical Computing (VBC'96). Germany: Springer; pp 267–276. [Google Scholar]
- Cao J, Worsley KJ (1999): The detection of local shape changes via the geometry of Hotelling's T 2 fields. Ann Statist 27:925–942. [Google Scholar]
- Cardenas VA, Chao LL, Studholme C, Yaffe K, Miller BL, Madison C, Buckley ST, Mungas D, Schuff N, Weiner MW (2011): Brain atrophy associated with baseline and longitudinal measures of cognition. Neurobiol Aging 32:572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli RJ, Dueck AC, Locke DE, Sabbagh MN, Ahern GL, Rapcsak SZ, Baxter LC, Yaari R, Woodruff BK, Hoffman‐Snyder C, Rademakers R, Findley S, Reiman EM. (2011): Cerebrovascular risk factors and preclinical memory decline in healthy APOE epsilon4 homozygotes. Neurology 76:1078–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli RJ, Dueck AC, Osborne D, Sabbagh MN, Connor DJ, Ahern GL, Baxter LC, Rapcsak SZ, Shi J, Woodruff BK, Locke DE, Snyder CH, Alexander GE, Rademakers R, Reiman EM. (2009): Longitudinal modeling of age‐related memory decline and the APOE epsilon4 effect. N Engl J Med 361:255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli RJ, Walker D, Sue L, Sabbagh M, Beach T (2010): Amyloid load in nondemented brains correlates with APOE e4. Neurosci Lett 473:168–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Reiman EM, Alexander GE, Caselli RJ, Gerkin R, Bandy D, Domb A, Osborne D, Fox N, Crum WR, Saunders AM, Hardy J. (2007): Correlations between apolipoprotein E epsilon4 gene dose and whole brain atrophy rates. Am J Psychiatry 164:916–921. [DOI] [PubMed] [Google Scholar]
- Cho Y, Seong JK, Shin SY, Jeong Y, Kim JH, Qiu A, Im K, Lee JM, Na DL (2011): A multi‐resolution scheme for distortion‐minimizing mapping between human subcortical structures based on geodesic construction on Riemannian manifolds. Neuroimage 57:1376–1392. [DOI] [PubMed] [Google Scholar]
- Chou Y, Lepore N, Avedissian C, Madsen SK, Parikshak N, Hua X, Shaw LM, Trojanowski JQ, Weiner MW, Toga AW, Thompson PM. (2009): Mapping correlations between ventricular expansion and CSF amyloid and tau biomarkers in 240 subjects with Alzheimer's disease, mild cognitive impairment and elderly controls. Neuroimage 46:394–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MK (2012): Computational Neuroanatomy: The Methods. World Scientific Publishing Company, Singapore. [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak‐Vance MA (1993): Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261:921–923. [DOI] [PubMed] [Google Scholar]
- Costafreda SG, Dinov ID, Tu Z, Shi Y, Liu CY, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, Wahlund LO, Spenger C, Toga AW, Lovestone S. (2011): Automated hippocampal shape analysis predicts the onset of dementia in mild cognitive impairment. Neuroimage 56: 212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Joshi S, Miller JP, Gado M, Kido D, McKeel D, Morris JC, Miller MI (2000): Early DAT is distinguished from aging by high‐dimensional mapping of the hippocampus. Dementia of the Alzheimer type. Neurology 55:1636–1643. [DOI] [PubMed] [Google Scholar]
- D'Agostino E, Maes F, Vandermeulen D, Suetens P (2003): A viscous fluid model for multimodal non‐rigid image registration using mutual information. Med Image Anal 7:565–575. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Vaillant M, Resnick SM, Prince JL, Letovsky S, Bryan RN (1996): A computerized approach for morphological analysis of the corpus callosum. J Comput Assist Tomogr 20:88–97. [DOI] [PubMed] [Google Scholar]
- Dean DC III, Jerskey BA, Chen K, Protas H, Thiyyagura P, Roontiva A, O'Muricheartaigh J, Dirks H, Waskiewicz N, Lehman K, Siniard AL, Turk MN, Hua X, Madsen SK, Thompson PM, Fleisher AS, Huentelman MJ, Deoni SCL, Reiman EM. (2013): Brain imaging differences in infants at differential genetic risk for late‐onset Alzheimer's disease. JAMA Neurol. doi: 10.1001/jamaneurol.2013.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Heijer T, van der Lijn F, Koudstaal PJ, Hofman A, van der Lugt A, Krestin GP, Niessen WJ, Breteler MMB (2010): A 10‐year follow‐up of hippocampal volume on magnetic resonance imaging in early dementia and cognitive decline. Brain 133:1163–1172. [DOI] [PubMed] [Google Scholar]
- Dewey J, Hana G, Russell T, Price J, McCaffrey D, Harezlak J, Sem E, Anyanwu JC, Guttmann CR, Navia B, Cohen R, Tate DF. (2010): Reliability and validity of MRI‐based automated volumetry software relative to auto‐assisted manual measurement of subcortical structures in HIV‐infected patients from a multisite study. Neuroimage 51:1334–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson DW, Crystal HA, Mattiace LA, Masur DM, Blau AD, Davies P, Yen SH, Aronson MK (1992): Identification of normal and pathological aging in prospectively studied nondemented elderly humans. Neurobiol Aging 13:179–189. [DOI] [PubMed] [Google Scholar]
- Elad M, Milanfar P, Golub GH (2004): Shape from moments—An estimation theory perspective. Trans Sig Proc 52:1814–1829. [Google Scholar]
- Ellis KA, Bush AI, Darby D, De Fazio D, Foster J, Hudson P, Lautenschlager NT, Lenzo N, Martins RN, Maruff P, Masters C, Milner A, Pike K, Rowe C, Savage G, Szoeke C, Taddei K, Villemagne V, Woodward M, Ames D. (2009): The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: Methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer's disease. Int Psychogeriatr 21:672–687. [DOI] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak‐Vance MA, Risch N, van Duijn CM (1997): Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta‐analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 278:1349–1356. [PubMed] [Google Scholar]
- Ferrarini L, Palm WM, Olofsen H, van der Landen R, van Buchem MA, Reiber JHC, Admiraal‐Behloul F (2008): Ventricular shape biomarkers for Alzheimer's disease in clinical MR images. Magn Reson Med 59:260–267. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR (1975): “Mini‐mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 12:189–198. [DOI] [PubMed] [Google Scholar]
- Fox NC, Scahill RI, Crum WR, Rossor MN (1999): Correlation between rates of brain atrophy and cognitive decline in AD. Neurology 52:1687–1689. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Fox NC, Jack CR, Scheltens P, Thompson PM (2010): The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol 6:67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes LU, Klausen IC, Sihm I, Faergeman O (1992): Apolipoprotein E polymorphism in a Danish population compared to findings in 45 other study populations around the world. Genet Epidemiol 9:155–167. [DOI] [PubMed] [Google Scholar]
- Gerig G, Styner M, Jones D, Weinberger D, Lieberman M (2001). Shape analysis of brain ventricles using SPHARM In: IEEE Workshop on Mathematical Methods in Biomedical Image Analysis (MMBIA'01). [Google Scholar]
- Gouras GK, Relkin NR, Sweeney D, Munoz DG, Mackenzie IR, Gandy S (1997): Increased apolipoprotein E epsilon 4 in epilepsy with senile plaques. Ann Neurol 41:402–404. [DOI] [PubMed] [Google Scholar]
- Gu X, Vemuri B (2004): Matching 3D Shapes Using 2D Conformal Representations. Med Image Comp Comput Assist Interv 3216:771–780. [Google Scholar]
- Gu X, Wang Y, Yau S‐T (2004): Geometric compression using riemann surface structure. Commun Inf Syst 3:171–182. [Google Scholar]
- Gutman B, Wang Y, Morra J, Toga AW, Thompson PM (2009): Disease classification with hippocampal shape invariants. Hippocampus 19:572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller JW, Christensen GE, Joshi SC, Newcomer JW, Miller MI, Csernansky JG, Vannier MW (1996): Hippocampal MR imaging morphometry by means of general pattern matching. Radiology 199:787–791. [DOI] [PubMed] [Google Scholar]
- Han X, Xu C, Prince JL (2003): A topology preserving level set method for geometric deformable models. IEEE Trans Pattern Anal Mach Intell 25:755–768. [Google Scholar]
- Holland D, Brewer JB, Hagler DJ, Fenema‐Notestine C, Dale AM (2009): Subregional neuroanatomical change as a biomarker for Alzheimer's disease. Proc Natl Acad Sci USA 106:20954–20959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AP, Blair RC, Watson JD, Ford I (1996): Nonparametric analysis of statistic images from functional mapping experiments. J Cereb Blood Flow Metab 16:7–22. [DOI] [PubMed] [Google Scholar]
- Hotelling H (1931): The generalization of Student's ratio. Ann Math Statist 2:360–378. [Google Scholar]
- Hua X, Gutman B, Boyle C, Rajagopalan P, Leow AD, Yanovsky I, Kumar AR, Toga AW, Jack CR Jr, Schuff N, Alexander GE, Chen K, Reiman EM, Weiner MW, Thompson PM. (2011): Accurate measurement of brain changes in longitudinal MRI scans using tensor‐based morphometry. Neuroimage 57:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Lee S, Hibar DP, Yanovsky I, Leow AD, Toga AW, Jack CR Jr, Bernstein MA, Reiman EM, Harvey DJ, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM. (2010): Mapping Alzheimer's disease progression in 1309 MRI scans: Power estimates for different inter‐scan intervals. Neuroimage 51:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman BT (2011): Amyloid‐dependent and amyloid‐independent stages of Alzheimer disease. Arch Neurol 68:1062–1064. [DOI] [PubMed] [Google Scholar]
- Jack CR Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, Whitwell JL, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox‐Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. (2008): The Alzheimer's disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging 27:685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr, Shiung MM, Gunter JL, O'Brien PC, Weigand SD, Knopman DS, Boeve BF, Ivnik RJ, Smith GE, Cha RH, Tangalos EG, Petersen RC. (2004): Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology 62:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr, Slomkowski M, Gracon S, Hoover TM, Felmlee JP, Stewart K, Xu Y, Shiung M, O'Brien PC, Cha R, Knopman D, Petersen RC. (2003): MRI as a biomarker of disease progression in a therapeutic trial of milameline for AD. Neurology 60:253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SC, Miller MI (2000): Landmark matching via large deformation diffeomorphisms. IEEE Trans Image Process 9:1357–1370. [DOI] [PubMed] [Google Scholar]
- Kim WH, Pachauri D, Hatt C, Chung MK, Johnson SC, Singh V (2012): Wavelet based multi‐scale shape features on arbitrary surfaces for cortical thickness discrimination In: NIPS. pp 1250–1258. [PMC free article] [PubMed] [Google Scholar]
- Knickmeyer RC, Wang J, Zhu H, Geng X, Woolson S, Hamer RM, Konneker T, Lin W, Styner M, Gilmore JH (2013): Common variants in psychiatric risk genes predict brain structure at birth. Cereb Cortex. DOI: 10.1093/cercor/bhs401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok E, Haikonen S, Luoto T, Huhtala H, Goebeler S, Haapasalo H, Karhunen PJ (2009): Apolipoprotein E‐dependent accumulation of Alzheimer disease‐related lesions begins in middle age. Ann Neurol 65:650–657. [DOI] [PubMed] [Google Scholar]
- Lehtovirta M, Laakso MP, Soininen H, Helisalmi S, Mannermaa A, Helkala EL, Partanen K, Ryynanen M, Vainio P, Hartikainen P, Riekkinen PJ Sr. (1995): Volumes of hippocampus, amygdala and frontal lobe in Alzheimer patients with different apolipoprotein E genotypes. Neuroscience 67:65–72. [DOI] [PubMed] [Google Scholar]
- Lemaitre H, Crivello F, Dufouil C, Grassiot B, Tzourio C, Alperovitch A, Mazoyer B (2005): No epsilon4 gene dose effect on hippocampal atrophy in a large MRI database of healthy elderly subjects. Neuroimage 24:1205–1213. [DOI] [PubMed] [Google Scholar]
- Leow A, Huang SC, Geng A, Becker J, Davis S, Toga A, Thompson P (2005): Inverse consistent mapping in 3D deformable image registration: Its construction and statistical properties. Inf Process Med Imaging 19:493–503. [DOI] [PubMed] [Google Scholar]
- Lepore N, Brun C, Chou Y‐Y, Chiang M‐C, Dutton RA, Hayashi KM, Luders E, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM. (2008): Generalized tensor‐based morphometry of HIV/AIDS using multivariate statistics on deformation tensors. IEEE Trans Med Imag 27:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorensen WE, Cline HE (1987): Marching cubes: A high resolution 3D surface construction algorithm. SIGGRAPH Comput Graph 21:163–169. [Google Scholar]
- Luders E, Thompson PM, Kurth F, Hong JY, Phillips OR, Wang Y, Gutman BA, Chou YY, Narr KL, Toga AW (2012): Global and regional alterations of hippocampal anatomy in long‐term meditation practitioners. Hum Brain Mapp 34:3369–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen SK, Ho AJ, Hua X, Saharan PS, Toga AW, Jack CR Jr, Weiner MW, Thompson PM (2010): 3D maps localize caudate nucleus atrophy in 400 AD, MCI, and healthy elderly subjects. Neurobiol Aging 31:1312–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G (2009): Alzheimer's biomarker initiative hits its stride. Science 326:386–389. [DOI] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR Jr, Schuff N, Weiner MW, Thompson PM. (2009a): Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Hum Brain Mapp 30:2766–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Toga AW, Jack CR Jr, Schuff N, Weiner MW, Thompson PM. (2009b): Automated mapping of hippocampal atrophy in 1‐year repeat MRI data from 490 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Neuroimage 45:S3–S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, Mintun MA (2010): APOE predicts amyloid‐beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol 67:122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW (2009): Selective effect of age, Apo e4, and Alzheimer's disease on hippocampal subfields. Hippocampus 19:558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, Trojanowski JQ, Toga AW, Beckett L (2005a): The Alzheimer's disease neuroimaging initiative. Neuroimag Clin N Am 15:869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L (2005b): Ways toward an early diagnosis in Alzheimer's disease: The Alzheimer's disease neuroimaging initiative (ADNI). Alzheimers Dement 1:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP (2002): Nonparametric permutation tests for functional neuroimaging: A primer with examples. Hum Brain Mapp 15:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude B, Smith SM, Kennedy DN, Jenkinson M (2011): A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage 56:907–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pievani M, Galluzzi S, Thompson PM, Rasser PE, Bonetti M, Frisoni GB (2011): APOE4 is associated with greater atrophy of the hippocampal formation in Alzheimer's disease. Neuroimage 55:909–919. [DOI] [PubMed] [Google Scholar]
- Pizer S, Fritsch D, Yushkevich P, Johnson V, Chaney E (1999): Segmentation, registration, and measurement of shape variation via image object shape. IEEE Trans Med Imag 18:851–865. [DOI] [PubMed] [Google Scholar]
- Poline JB, Vandenberghe R, Holmes AP, Friston KJ, Frackowiak RS (1996): Reproducibility of PET activation studies: Lessons from a multi‐center European experiment. EU concerted action on functional imaging. Neuroimage 4:34–54. [DOI] [PubMed] [Google Scholar]
- Qiu A, Brown T, Fischl B, Ma J, Miller MI (2010): Atlas generation for subcortical and ventricular structures with its applications in shape analysis. IEEE Trans Image Process 19:1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Taylor WD, Zhao Z, MacFall JR, Miller MI, Key CR, Payne ME, Steffens DC, Krishnan KR (2009): APOE related hippocampal shape alteration in geriatric depression. Neuroimage 44:620–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM (2007): Linking brain imaging and genomics in the study of Alzheimer's disease and aging. Ann N Y Acad Sci 1097:94–113. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Caselli RJ, Chen K, Alexander GE, Bandy D, Frost J (2001): Declining brain activity in cognitively normal apolipoprotein E epsilon 4 heterozygotes: A foundation for using positron emission tomography to efficiently test treatments to prevent Alzheimer's disease. Proc Natl Acad Sci USA 98:3334–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Caselli RJ, Yun LS, Chen K, Bandy D, Minoshima S, Thibodeau SN, Osborne D (1996): Preclinical evidence of Alzheimer's disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. N Engl J Med 334:752–758. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J (2005): Correlations between apolipoprotein E epsilon4 gene dose and brain‐imaging measurements of regional hypometabolism. Proc Natl Acad Sci USA 102:8299–8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Liu X, Bandy D, Yu M, Lee W, Ayutyanont N, Keppler J, Reeder SA, Langbaum JB, Alexander GE, Klunk WE, Mathis CA, Price JC, Aizenstein HJ, DeKosky ST, Caselli RJ. (2009): Fibrillaramyloid‐beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer's disease. Proc Natl Acad Sci USA 106:6820–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Uecker A, Caselli RJ, Lewis S, Bandy D, de Leon MJ, De Santi S, Convit A, Osborne D, Weaver A, Thibodeau SN. (1998): Hippocampal volumes in cognitively normal persons at genetic risk for Alzheimer's disease. Ann Neurol 44:288–291. [DOI] [PubMed] [Google Scholar]
- Rey D, Subsol G, Delingette H, Ayache N (2002): Automatic detection and segmentation of evolving processes in 3D medical images: Application to multiple sclerosis. Med Image Anal 6:163–179. [DOI] [PubMed] [Google Scholar]
- Ridha BH, Anderson VM, Barnes J, Boyes RG, Price SL, Rossor MN, Whitwell JL, Jenkins L, Black RS, Grundman M, Fox NC. (2008): Volumetric MRI and cognitive measures in Alzheimer disease: Comparison of markers of progression. J Neurol 255:567–574. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George‐Hyslop PH, Pericak‐Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper‐MacLachlan DR, Alberts MJ, Hulette C, Crain B, Goldgaber D, Roses AD. (1993): Association of apolipoprotein E allele epsilon 4 with late‐onset familial and sporadic Alzheimer's disease. Neurology 43:1467–1472. [DOI] [PubMed] [Google Scholar]
- Shen L, Firpi HA, Saykin AJ, West JD (2009): Parametric surface modeling and registration for comparison of manual and automated segmentation of the hippocampus. Hippocampus 19:588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Thompson PM, Gutman B, Wang Y (2013a): Surface fluid registration of conformal representation: Application to detect disease burden and genetic influence on hippocampus. Neuroimage 78C:111–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang Y, Ceschin R, An X, Lao Y, Vanderbilt D, Nelson MD, Thompson PM, Panigrahy A, Lepore N (2013b): A multivariate surface‐based analysis of the putamen in premature newborns: Regional differences within the ventral striatum. PLoS One 8:e66736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H, Gnanadesikan R, Hughes JB (1962): Multivariate analysis of variance (MANOVA). Biometrics 18:22–41. [Google Scholar]
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR Jr, Kaye J, Montine TJ, Park DC, Reiman EM, Rowe CC, Siemers E, Stern Y, Yaffe K, Carrillo MC, Thies B, Morrison‐Bogorad M, Wagster MV, Phelps CH. (2011): Toward defining the preclinical stages of Alzheimer's disease: Recommendations from the National Institute on Aging‐Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 7:280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Toro R, Appel K, Bartecek R, Bergmann O, Bernard M, et al. (2012): Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet 44:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonnington CM, Chu C, Kloppel S, Jack CR Jr, Ashburner J, Frackowiak RS (2010): Predicting clinical scores from magnetic resonance scans in Alzheimer's disease. Neuroimage 51:1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styner M, Lieberman JA, McClure RK, Weinberger DR, Jones DW, Gerig G (2005): Morphometric analysis of lateral ventricles in schizophrenia and healthy controls regarding genetic and disease‐specific factors. Proc Natl Acad Sci USA 102:4872–4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styner M, Lieberman JA, Pantazis D, Gerig G (2004): Boundary and medial shape analysis of the hippocampus in schizophrenia. Med Image Anal 8:197–203. [DOI] [PubMed] [Google Scholar]
- Thirion JP, Prima S, Subsol G, Roberts N (2000): Statistical analysis of normal and abnormal dissymmetry in volumetric medical images. Med Image Anal 4:111–121. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer's disease. Neuroimage 22:1754–1766. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria M, Toro R, Jahanshad N, Schumann G, Franke B, et al. The ENIGMA Consortium: Large‐scale Collaborative Analyses of Neuroimaging and Genetic Data. Brain Imaging and Behavior. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinack J, Dickerson BC, Golland P, Fischl B (2009): Automated segmentation of hippocampal subfields from ultra‐high resolution in vivo MRI. Hippocampus 19:549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P, Gunter JL, Senjem ML, Whitwell JL, Kantarci K, Knopman DS, Boeve BF, Petersen RC, Jack CR Jr (2008a): Alzheimer's disease diagnosis in individual subjects using structural MR images: Validation studies. Neuroimage 39:1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P, Whitwell JL, Kantarci K, Josephs KA, Parisi JE, Shiung MS, Knopman DS, Boeve BF, Petersen RC, Dickson DW, Jack CR Jr. (2008b): Antemortem MRI based STructural Abnormality iNDex (STAND)‐scores correlate with postmortem Braak neurofibrillary tangle stage. Neuroimage 42:559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Beg F, Ratnanather T, Ceritoglu C, Younes L, Morris JC, Csernansky JG, Miller MI (2007): Large deformation diffeomorphism and momentum based hippocampal shape discrimination in dementia of the Alzheimer type. IEEE Trans Med Imaging 26:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Miller JP, Gado MH, McKeel DW, Rothermich M, Miller MI, Morris JC, Csernansky JG (2006): Abnormalities of hippocampal surface structure in very mild dementia of the Alzheimer type. Neuroimage 30:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Swank JS, Glick IE, Gado MH, Miller MI, Morris JC, Csernansky JG (2003): Changes in hippocampal volume and shape across time distinguish dementia of the Alzheimer type from healthy aging. Neuroimage 20:667–682. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chan TF, Toga AW, Thompson PM (2009): Multivariate tensor‐based brain anatomical surface morphometry via holomorphic one‐forms. Med Image Comp Comput Assist Interv 12:337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shi J, Yin X, Gu X, Chan TF, Yau S‐T, Toga AW, Thompson PM (2012): Brain surface conformal parameterization with the ricci flow. IEEE Trans Med Imag 31:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Song Y, Rajagopalan P, An T, Liu K, Chou YY, Gutman B, Toga AW, Thompson PM (2011): Surface‐based TBM boosts power to detect disease effects on the brain: An N = 804 ADNI study. Neuroimage 56:1993–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yuan L, Shi J, Greve A, Ye J, Toga AW, Reiss AL, Thompson PM (2013): Applying tensor‐based morphometry to parametric surfaces can improve MRI‐based disease diagnosis. Neuroimage 74:209–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang J, Gutman B, Chan TF, Becker JT, Aizenstein HJ, Lopez OL, Tamburo RJ, Toga AW, Thompson PM (2010): Multivariate tensor‐based morphometry on surfaces: Application to mapping ventricular abnormalities in HIV/AIDS. Neuroimage 49:2141–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D (1987): Wechsler Memory Scale‐Revised Manual. San Antonio, TX: Psychological Corporation. [Google Scholar]
- Wolz R, Heckemann RA, Aljabar P, Hajnal JV, Hammers A, Lötjönen J, Rueckert D (2010): Measurement of hippocampal atrophy using 4D graph‐cut segmentation: Application to ADNI. Neuroimage 52:109–118. [DOI] [PubMed] [Google Scholar]
- Yang X, Goh A, Chen SH, Qiu A (2012): Evolution of hippocampal shapes across the human lifespan. Hum Brain Mapp 34:3075–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa MA, Stark SM, Bakker A, Albert MS, Gallagher M, Stark CE (2010): High‐resolution structural and functional MRI of hippocampal CA3 and dentate gyrus in patients with amnestic mild cognitive impairment. Neuroimage 51:1242–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Wang H, Pluta J, Das SR, Craige C, Avants BB, Weiner MW, Mueller S (2010): Nearly automatic segmentation of hippocampal subfields in in vivo focal T2‐weighted MRI. Neuroimage 53:1208–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Lu G (2004): Review of shape representation and description techniques. Pattern Recognit 37:1–19. [Google Scholar]


