Summary
Intraspecific genetic incompatibilities prevent the assembly of specific alleles into single genotypes and influence genome- and species-wide patterns of sequence variation. A common incompatibility in plants is hybrid necrosis, characterized by autoimmune responses due to epistatic interactions between natural genetic variants. By systematically testing thousands of F1 hybrids of Arabidopsis thaliana strains, we identified a small number of incompatibility hotspots in the genome, often in regions densely populated by NLR immune receptor genes. In several cases, these immune receptor loci interact with each other, suggestive of conflict within the immune system. A particularly dangerous locus is a highly variable cluster of NLR genes, DANGEROUS MIX2 (DM2), which causes multiple, independent incompatibilities with genes that encode a range of biochemical functions, including NLRs. Our findings suggest that deleterious interactions of immune receptors at the front lines of host-pathogen co-evolution limit the combinations of favorable disease resistance alleles accessible to plant genomes.
Keywords: hybrid incompatibility, hybrid necrosis, epistasis, autoimmunity, NLR, Arabidopsis thaliana
Introduction
When independently diverging genomes meet in hybrids, the differences that have accumulated over evolutionary time can have detrimental consequences. The ensuing incompatibilities were formally described by Bateson, Dobzhansky and Muller, who proposed a scenario under which complementary changes occur in two different populations; the individual changes are innocuous in their native genomic contexts, and they reduce viability or fertility only when combined in hybrids (Coyne and Orr, 2004). This type of deleterious, or negative, epistasis has been most prominently studied in interspecific crosses, where the interacting alleles are fixed in the different populations (Maheshwari and Barbash, 2011; Presgraves, 2010; Rieseberg and Blackman, 2010). More recent work has begun to focus on deleterious epistasis within species, where the interacting alleles are polymorphic and segregate in a single intermating population (Corbett-Detig et al., 2013; Hou et al., 2014; Seidel et al., 2008). One can envision a series of evolutionary forces responsible for the emergence of interacting alleles. On the one hand, genetic drift could play a role, with segregating alleles that are neutral or merely mildly deleterious on their own giving rise to synthetic deleterious interactions (Bikard et al., 2009; Phillips and Johnson, 1998). At the other end of the spectrum, adaptation and intragenomic conflicts have been implicated as factors that may contribute to a reduction in hybrid viability or fertility (Crespi and Nosil, 2013; Cutter, 2012; Lachance and True, 2010). Regardless of the ultimate cause, high levels of sequence divergence at incompatibility loci appear to be positively correlated with deleterious interactions. Ultimately, as lineages diverge and become genetically more differentiated, segregating variants may rise to high frequency in specific populations and thereby reduce gene flow among them.
Genes of the immune system are particularly polymorphic in many organisms, because of an evolutionary arms race between hosts and pathogens (Quintana-Murci and Clark, 2013; Sackton et al., 2007; Vilches and Parham, 2002). This is also true for plants. Prominent, highly variable components of the plant immune system are the nucleotide-binding domain and leucine-rich repeat (NLR) proteins, with plant genomes often encoding hundreds of NLRs (Cao et al., 2011; Jacob et al., 2013). Plant NLRs typically function as immune receptors that confer disease resistance by monitoring the integrity of important plant proteins or the presence of pathogen effector proteins (Collier and Moffett, 2009).
Apart from having to keep up with the ongoing evolution of individual pathogens, hosts must also accumulate resistance against as many different pathogens as possible. This in turn entails its own dangers, in the form of genetic variants that lead to enhanced immunity, but at the same time reduce growth or health due to autoimmunity (Todesco et al., 2010; Trowsdale and Knight, 2013). In plants, especially severe autoimmune phenotypes have been observed in hybrids descended from phenotypically normal parents. This syndrome, hybrid necrosis, is found in intra- and interspecific crosses and it spans a range of severity, from cases where only a small subset of F2 progeny is weakly affected to ones in which all F1 hybrids die. The lesions and reduced growth of necrotic hybrids are often alleviated at higher temperature, greatly facilitating the genetic analysis of severe cases (Bomblies and Weigel, 2007).
To date, four hybrid necrosis cases due to two-locus epistasis have been studied at the molecular level in tomato, lettuce and rice. Of the six causal loci that have been positively identified, one encodes an NLR and another one a known NLR-interactor. In addition, the mapping interval for one of the remaining loci includes several NLRs (Chen et al., 2014; Jeuken et al., 2009; Krüger et al., 2002; Yamamoto et al., 2010). Similarly, the first hybrid necrosis gene positively identified in Arabidopsis thaliana, DANGEROUS MIX1 (DM1), encodes an NLR. It interacts with the DM2 locus, which was mapped to a polymorphic cluster of RPP1 NLR genes that is probably also responsible for an independent F2 incompatibility (Alcázar et al., 2009; Bomblies et al., 2007). In natural accessions of A. thaliana, the RPP-subfamily of NLR genes is particularly variable, both in sequence and copy number. The high diversity of RPP loci, which recognize different strains of the oomycete pathogen Hyaloperonospora arabidopsidis ex parasitica (Hpa) in an allele-specific manner, points to these genes as actors in an active co-evolutionary tug-of-war between host and pathogen (Allen et al., 2004; Bakker et al., 2006; Holub and Beynon, 1997).
While there is increasing evidence for natural variation in immunity potentially leading to genetic incompatibilities in plants, species-wide patterns of immune-related deleterious epistasis remain unknown. For example, are specific immune loci especially likely to be involved in deleterious epistasis? Do they interact more often with other immune loci than with non-immune genes? And is deleterious epistasis correlated with geographic and genetic distance? To systematically investigate which factors in the plant immune system contribute to intraspecific incompatibility, we have examined F1 progeny from thousands of A. thaliana crosses, including all combinations among 80 accessions that represent much of the common genetic diversity in the species (Cao et al., 2011). We have mapped several hybrid necrosis loci to regions of the genome that contain multiple NLR genes in tandem arrays, with different allelic variants at DM2/RPP1 being responsible for several independent incompatibilities. We also found cases where different alleles at a locus interact with each other, or where independent pairs of alleles at two loci are incompatible with each other. Because many interactions are between components of the plant immune system, we propose that epistatic fitness effects limit the extent to which favorable immune alleles can be combined.
Results
A systematic resource for the discovery of genetic incompatibilities
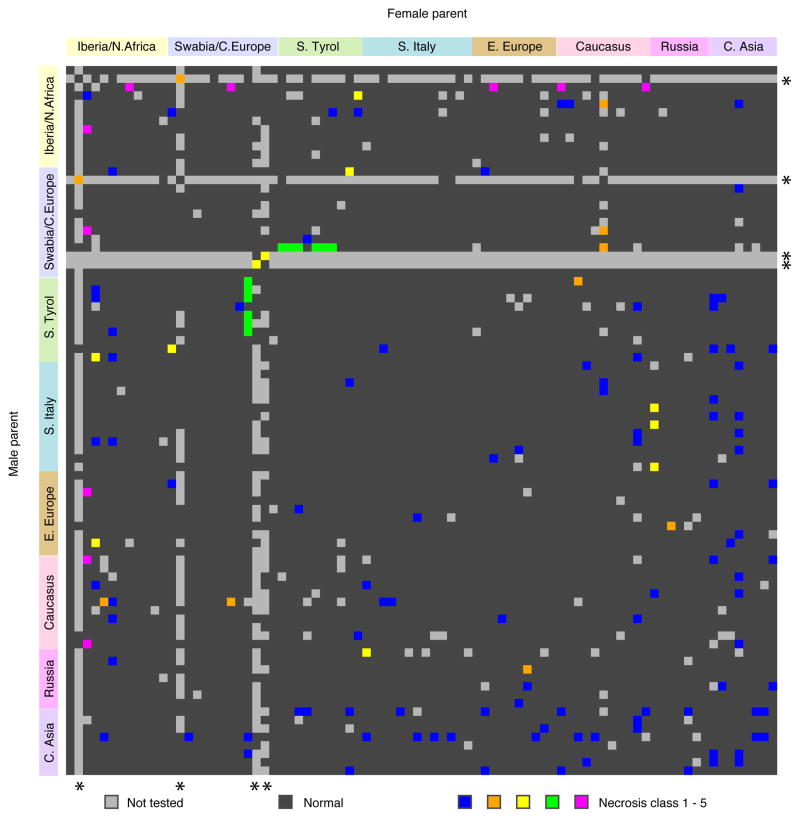
About two percent of crosses between randomly chosen accessions of A. thaliana suffer from F1 hybrid necrosis when grown at 16°C (Bomblies et al., 2007). To determine the incidence of hybrid necrosis and other F1 weakness syndromes more systematically, and to obtain insights into how genetic kinship affects the probability of hybrid necrosis, we turned to 80 accessions that had their genomes sequenced in the first phase of the 1001 Genomes project (Cao et al., 2011). These 80 accessions represent much of the common diversity across the species’ native range, and can thus provide insights into the distribution of hybrid necrosis alleles in both the global and in local populations. To facilitate the large number of crosses, male-sterile lines were derived by knocking down the floral homeotic gene AP3 (Wuest et al., 2012). Together with additional crosses that mostly involved accessions carrying known DM alleles, we assembled a total of 6,409 crosses. This collection comprised 3,330 unique parental combinations; 3,160 of these made up a complete half-diallel of the 80 accessions (Table S1).
The most common morphological defects seen in F1 hybrids at 16°C were dwarfism and tissue necrosis, which fell into five classes of increasing severity, including two new extreme classes (Figures 1 and S1). In the previously described cases, morphological defects largely disappeared at 23°C (Bomblies et al., 2007). Class 4 phenotypes were only suppressed at 28°C, while class 5 individuals died as seedlings at all temperatures tested. Most necrotic hybrids had only mild defects (103 cases in class 1), 29 cases were intermediate (classes 2 to 4), and 10 cases were not genetically tractable due to lethality (class 5). Our threshold for identifying necrosis was quite stringent, and there might well be many more weak cases.
Figure 1. Crosses for Detection of Hybrid Incompatibilities.
Accessions are color coded by region of origin. Accessions that are not part of the 80 accessions from (Cao et al., 2011) are marked with asterisks.
Mapping and identification of incompatibility loci
From the 142 F1 hybrid necrosis cases described here and the 27 identified previously (Bomblies et al., 2007), we chose seven for further genetic analysis. In addition to obvious phenotypes, we prioritized cases where at least one of the causal alleles was likely to be present in multiple genetic backgrounds, as judged by one parent producing similar F1 phenotypes with several other parents. Thus, the selected cases are likely to represent 31 of the 48 intermediate, genetically tractable hybrid necrosis examples in our collection.
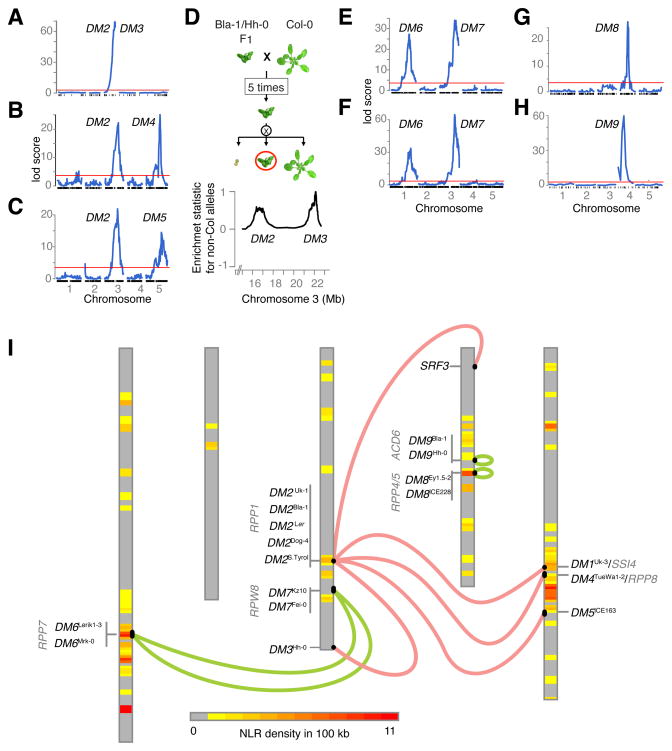
The fraction of affected individuals in F2 populations indicated that hybrid phenotypes in five cases were due to pairwise interactions between genetically separable loci (Table S2). Segregation ratios in Ey1.5-2 x ICE228 and for the Bla-1 x Hh-0 lesioning trait were consistent with effects arising from heterozygous disadvantage at single regions of the genome (Table S2). We mapped causal loci mostly using Genotyping-By-Sequencing (GBS) of individual F2 plants (Elshire et al., 2011; Poland et al., 2012) and quantitative trait locus (QTL) methods (Table S3; Figures 2A–H). For leaf-twisting in Bla-1 x Hh-0, we used whole-genome sequencing of pooled DNA from F1-like individuals segregating in selfed BC5 progeny (Figures 2A, 2D and S2B).
Figure 2. Linkage Mapping of Seven Hybrid Incompatibilities.
Hybrid necrosis QTL maps in (A) Bla-1/Hh-0 (leaf twisting), (B) TueWa1-2/ICE163, (C) Dog-4/ICE163, (E) Fei-0/Lerik1-3, (F) KZ10/Mrk-0, (G) Ey1.5-2/ICE228, and (H) Bla-1/Hh-0 (late-onset lesioning). Red lines mark significance threshold (p=0.05, 1,000 permutations); vertical marks along the X-axes indicate marker positions.
(D) Scheme for Illumina sequencing of bulk segregants to delineate DM2 and DM3.
(I) Genomic location of DM alleles compared to NLR gene density (1 Mb windows) on the five chromosomes of the reference strain Col-0. SRF3, DM2Ler and DM1/SSI4 have been reported before (Alcázar et al., 2010; Alcázar et al., 2009; Bomblies et al., 2007). DM2/RPP1 interactions in red, others in green.
See also Figure S2, Tables S2, S3 and S4.
Our analyses identified seven new hybrid necrosis loci, DM3 to DM9 (Figures 2 and S2A; Table 1), with final mapping intervals between 110 and 969 kb (Table S4). The DM2 region was represented in multiple crosses: DM3, DM4, and DM5 all interacted with DM2 alleles from different strains, as do the previously identified DM1 and SRF3 loci (Alcázar et al., 2010; Bomblies et al., 2007). Thus, at least five out of nine A. thaliana hybrid necrosis cases include DM2 as one of the interactors. Two cases mapped to different pairs of DM6 and DM7 alleles, and two involved heterozygous effects at single loci, DM8 and DM9.
Table 1.
Candidate Genes for Hybrid Necrosis
| Cross | Class* | Locus A | Evidence† | Locus B | Evidence† |
|---|---|---|---|---|---|
| Uk-1/Uk-3 | 3 | DM2 (RPP1) | Genomic/amiRNA | DM1 (SSI4) | Genomic/amiRNA†† |
| Bla-1/Hh-0 F3a | 2 | DM2 (RPP1) | Genomic/amiRNA | DM3 (At3g61540) | Genomic/MIGS |
| Bla-1/Hh-0 F2b | 2 | DM9 (ACD6) | Genomic/amiRNA§ | DM9 (ACD6) | Genomic/amiRNA§ |
| TueWa1-2/ICE163 | 4 | DM2 (RPP1) | Map only | DM4 (RPP8) | Map only |
| Dog-4/ICE163 | 2 | DM2 (RPP1) | Map only | DM5 | Map only |
| KZ10/Mrk-0 | 3 | DM6 (RPP7) | Map only | DM7 (RPW8) | Genomic |
| Fei-0/Lerik1-3 | 2 | DM6 (RPP7) | amiRNA | DM7 (RPW8) | Map only |
| Ey1.5-2/ICE228 | 3 | DM8 (RPP4/5) | amiRNA | DM8 (RPP4/5) | amiRNA |
Classes 1 to 3 as described (Bomblies et al., 2007). Class 4 hybrids arrest as seedlings with only cotyledons and severe necrosis at 16°C, which were recovered at 28°C to set seeds.
‘Genomic’ refers to genomic fragment reproducing hybrid necrosis in transgenic plants.
Described previously (Bomblies et al., 2007).
Described in detail elsewhere (Todesco et al., 2014).
We identified the DM3 prolyl aminopeptidase (At3g61540) from Hh-0 as an interactor of DM2h from Bla-1 using transformation with genomic fragments and artificial miRNA (amiRNA) knockdown (Table 1; Figures 2 and S2B–E). In two crosses, KZ10 x Mrk-0 and Fei-0 x Lerik1-3, one of the causal loci, DM7, mapped to the RESISTANCE TO POWDERY MILDEW8 (RPW8) region, which contains a variable tandem array of genes encoding coiled-coil proteins (Xiao et al., 2001). Transgenic experiments revealed that RPW8.1KZ10 was sufficient to induce necrosis in the Mrk-0 background (Table 1; Figures 2 and S2F–G). Despite similar genomic locations of the incompatibility genes, KZ10 is compatible with Lerik1-3, and Mrk-0 with Fei-0 (Table S1), indicating that these incompatibilities likely include different pairs of alleles at DM6 and DM7.
We mapped the DM9 locus in two crosses, Bla-1 x Hh-0 (this work) and Mir-0 x Se-0. A detailed analysis of the causal locus, ACCELERATED CELL DEATH6 (ACD6, is reported elsewhere (Todesco et al., 2014).
We confirmed genes from the RPP7 and RPP4/5 NLR clusters as causal for DM6 and DM8 using amiRNAs (Table 1). DM4 also overlapped the location of an RPP cluster, RPP8 (Table S4). Four out of nine DM loci, accounting for ten of the causal alleles, thus map to highly variable RPP clusters (Figure 2I), which are the major sources for resistance to Hpa in A. thaliana (Nemri et al., 2010). In one DM2 case, discussed in detail below, we have direct evidence for interactions between two different NLR genes. Three other interactions, DM2/DM4, DM2/DM5, and the DM8 heterozygous incompatibility, also likely involve interactions between NLR genes, while the DM6/DM7 interactions involve an NLR candidate and a complex non-NLR immune locus, RPW8 (Xiao et al., 2001). Finally, the heterozygous interaction at DM9 is caused by distinct alleles of another complex non-NLR immune locus ACD6 (Lu et al., 2003). Our systematic mapping efforts therefore indicate that NLR alleles along with other polymorphic immune genes located in tandem arrays are responsible for most intraspecific F1 incompatibilities in A. thaliana.
Multiple incompatibilities due to the complex RPP1/DM2 NLR locus
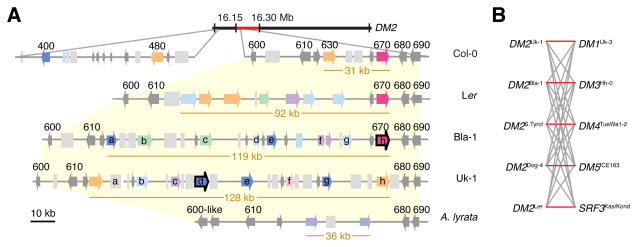
To understand how incompatibilities at RPP clusters evolve, we studied the RPP1/DM2 locus from the accessions Uk-1 and Bla-1 in detail. We first assembled genomic sequences of the Uk-1 and Bla-1 DM2 clusters from overlapping fosmid clones, and compared these with sequences from the Ler and Col-0 accessions and the related species A. lyrata. In the A. thaliana reference strain Col-0, the DM2 locus contains two RPP1 paralogs that span 31 kb and that are part of a 180-kb RPP1 supercluster. The DM2 regions are much larger in Bla-1 and Uk-1, 119 kb and 128 kb (Figures 3A and S3). Both accessions contain eight RPP1 paralogs, similar to the 92-kb DM2Ler cluster, which includes seven complete and at least one truncated RPP1 homolog (Alcázar et al., 2009). Not a single RPP1-like gene is identical between accessions, consistent with the pattern of accession-specific incompatibilities (Figure 3B).
Figure 3. The DM2 Cluster in Arabidopsis.
(A) DM2 clusters in four A. thaliana accessions and in A. lyrata MN47. The mapping interval for DM2 in Bla-1 is indicated in red. Genes are indicated with colored arrows, pseudogenes with colored boxes and transposons with light grey boxes. Non-NLR genes are in dark grey. NLR genes are colored according to their phylogenetic history (see Figure S4), with unresolved relationships indicated in light blue. Numbers above arrows indicate the last three digits of At3g44XXX and are given only when there are homologs in the reference genome. The two incompatibility genes are outlined in black. The DM2Ler cluster was re-annotated based on GenBank FJ446580.1. The sizes in kb refer to the core DM2 clusters, defined as the coding regions of all NLRs (colored arrows) in a cluster.
(B) Test crosses between DM2 carriers and interacting allele carriers. Red and grey lines indicate incompatible and compatible interactions, respectively.
See also Figure S3.
To test which of the RPP1-like genes in Uk-1 and Bla-1 are responsible for hybrid necrosis, we first knocked down individual DM2 genes with allele-specific amiRNAs (Figure S4A and Supplemental Experimental Procedures). We also introduced DM2 genomic clones into the incompatible parents Uk-3, which carries a DM2 interactor at DM1, and Hh-0, which carries a DM2 interactor at DM3 (Table 1). These experiments identified single genes in each accession, DM2dUk-1 and DM2hBla-1, as necessary and sufficient for hybrid necrosis (Figures 4A and S4B–G).
Figure 4. Identification of DM2dUk-1 and DM2hBla-1 as Hybrid Necrosis Genes.
(A) Arabidopsis thaliana F1 hybrids and rescued siblings expressing amiRNAs at 16 °C.
(B) Reconstitution of incompatibilities in Col-0, with transgenic F1 hybrids at 16°C. Transgenic effects were often stronger than F1 hybrid phenotypes.
(C) HR-like cell death in N. benthamiana induced by co-expression of A. thaliana DM proteins at 23°C. mDM2 indicates P-loop mutant versions (GIGKTT to GIAATT), which should not be able to bind and hydrolyze ATP (Chung et al., 2011).
Scale bars in (A) and (B) represent 1 cm.
See also Figure S4.
We asked whether autoimmunity depended on additional factors specific to the incompatible accessions. We first reconstituted the DM2Uk-1/DM1Uk-3 and DM2Bla-1/DM3Hh-0 interactions in the Col-0 background by crossing lines with the respective transgenes; in both cases doubly transgenic lines were severely necrotic (Figure 4B). Next, we transiently expressed each pair in Nicotiana benthamiana leaves. We observed necrosis symptoms that mimicked the hypersensitive response (HR) seen upon recognition of a pathogen by a plant host when incompatible alleles of DM2 and DM1, or DM2 and DM3 were combined (Figure 4C). Importantly, DM2 genes closely related to either DM2dUk-1 or DM2hBla-1 did not trigger HR-like necrosis in N. benthamiana when combined with DM1Uk-3 or DM3Hh-0 (Figure 4C), confirming that HR is not simply induced by any combination of foreign NLR genes. Furthermore, enzymatic activity of DM2 was required for HR in this system (Figure 4C), indicating that DM2 directly couples to downstream signaling events (Chung et al., 2011). These results demonstrate that incompatible pairs of DM proteins are sufficient to initiate cell death signaling conserved between species.
Distinct evolutionary histories of two causal DM2 alleles
Not all clades in a DM2 phylogeny (Figures S4H–J) include DM2 genes from all accessions. In addition, relationships among accessions within one clade often differ from those in another clade. Thus, likely as a result of independent cycles of local gene duplication and loss along with illegitimate recombination and gene conversion (Table S5), DM2 clusters from different lineages show little conserved synteny, vary in size, and are poorly conserved outside NLR gene fragments (Figure 3A). This is consistent with patterns reported for major immune receptor gene clusters throughout flowering plants (Jacob et al., 2013).
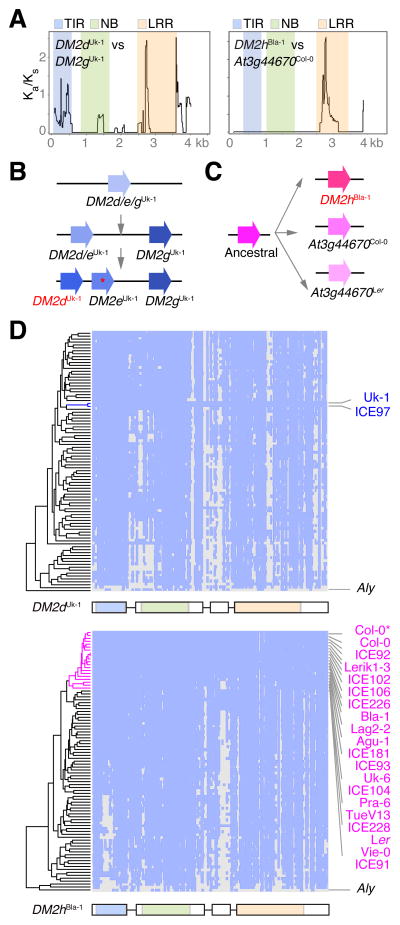
Despite the overall similarity of DM2h/At3g44670 alleles among several accessions (Figure S4H–J), the LRR region, which is likely responsible for pathogen recognition, showed signs of diversifying selection (Figures 5A and S5A–B; average Ka/Ks = 4.2 for codons encoding the putative solvent-exposed residues). We thus hypothesized that rare allelic differences in a rapidly evolving portion of this gene gave rise to the incompatible behavior of DM2hBla-1. We localized residues responsible for incompatibility with DM3 using domain swaps. We first mapped the incompatible sequences to the C-terminus of the DM2hBla-1 protein, which includes the LRRs (Figure S5C). By engineering polymorphisms from DM2hBla-1 that are rare in other accessions into the At3g44670Col-0 reference allele (Figures S5A and S5B), we identified two adjacent residues in the putative solvent exposed site of LRR4 that can confer partial necrosis-inducing activity when combined with C-terminal sequences (Figure S5D). This result highlights the potential of natural NLR variants for the identification of residues that increase protein activity, which would inform efforts to engineer semi-synthetic NLRs (Harris et al., 2013; Segretin et al., 2014).
Figure 5. Origin and Variation of DM2 Hybrid Necrosis Genes.
(A) Ka/Ks ratios of closely related DM2 genes (window length 150 bp, step size 9 bp).
(B) Most parsimonious path for evolution of DM2dUk-1 paralogs.
(C) Most parsimonious path for evolution of DM2hBla-1 orthologs.
(D) DM2dUk-1- and DM2hBla-1-type profiling using Illumina reads from accessions with one mismatch. Grey indicates uncovered regions. Accessions carrying a DM2h-type are labeled in magenta.
See also Figure S5, Tables S5 and S6.
RPP1-type proteins recognize and associate with proteins encoded by the Hpa ATR effector locus in an allele-specific manner (Krasileva et al., 2010). The hybrid necrosis-inducing residues in DM2hBla-1 mapped near a modeled docking site of ATR1 onto RPP1-WsB (Steinbrenner et al., 2012), suggesting that these variants have arisen from an arms-race between an immune receptor and a pathogen ligand.
The topology of genes in the DM2Uk-1 cluster, as well as the phylogenetic relationships between DM2 genes, suggested that DM2dUk-1 arose by two within-cluster duplications and involved at least three gene conversion events (Figure 5B; Table S5). The two closest paralogs within the Uk-1 cluster are DM2e and DM2g, with DM2e having suffered mutations that truncate the encoded protein (Figures 5B, S3 and S4H–J). Despite being within-cluster duplicates, DM2dUk-1 and DM2gUk-1 differ at many non-synonymous sites, partly due to sequence exchanges with different paralogs (Figure 5A; Table S5). We visualized broader patterns of variation by using DM2dUk-1 as a target for mapping of Illumina reads from 87 accessions. Accessions with very similar sequences across the entire gene were rare (Figure 5D). Moreover, similarity did not predict incompatibility: although ICE97 from Southern Italy has a DM2d copy that is very similar to that of Uk-1, ICE97 was not incompatible with Uk-3 (Table S1). The rarity of DM2dUk-1 is consistent with the hypothesis that DM2d is a rapidly evolving type I plant NLR gene, characterized by frequent sequence exchanges with other members of the same cluster (Kuang et al., 2004).
Unlike DM2dUk-1, DM2hBla-1 shows a clear orthologous relationship with DM2 genes in other accessions, At3g44670Col-0 and At3g44670Ler (Figures 5C and S4H–J), a pattern typical for type II plant NLR genes (Kuang et al., 2004). Alleles at type II genes, which can be rare or common, diversify mostly by point mutations in the LRR region, rather than by sequence exchanges between paralogs. All three DM2h-type orthologs, DM2hBla-1, At3g44670Col-0 and At3g44670Ler, are located at the 3′ end of the DM2 cluster and mark the beginning of a syntenic region of at least 22 kb that is well conserved between Ler, Col-0 and Bla-1, but that is not found in Uk-1 or A. lyrata (Figures 3A and 6A). DM2 hybrid necrosis alleles thus have arisen both as diversified orthologs and as paralogs within the tandemly arrayed gene cluster, accompanied by diversifying selection.
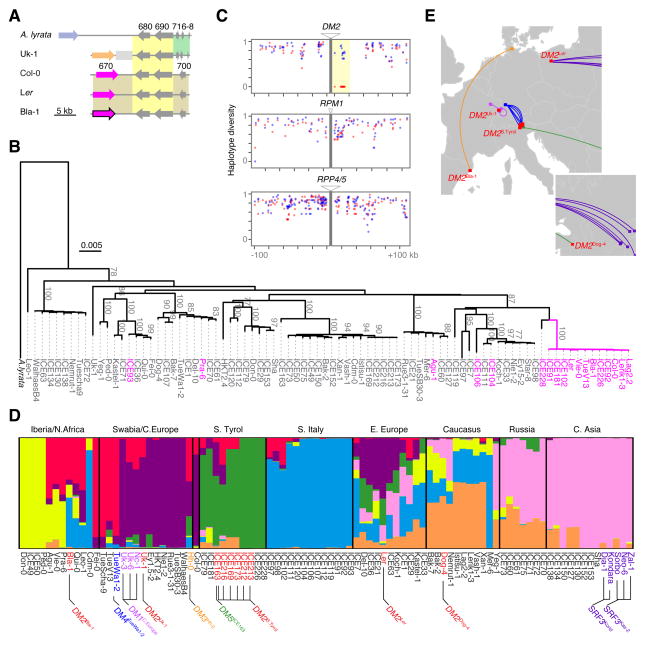
Figure 6. Haplotype Sharing Around DM2h and Geographic Distribution of DM2 alleles.
(A) Syntenic overview of the region beyond the DM2 clusters in A. thaliana accessions and in A. lyrata MN47. See Fig. 3A legend for color and number code.
(B) Phylogeny of 8-kb conserved sequences spanning At3g44680 and At3g44690 (yellow shade in panel A). DM2h-type carriers are magenta, non-carriers grey. Bootstrap values over 70% are indicated.
(C) Haplotype diversity based on groups of 10 adjacent SNPs in the regions flanking NLR loci DM2, RPM1 and RPP4/5. Twelve DM2h-type carriers in red and 12 non-carriers in blue.
(D) STRUCTURE analysis (k = 7) of hybrid necrosis risk allele carriers together with a selection of global accessions. At the bottom, accessions carrying different DM2 hybrid necrosis alleles in red, and those carrying DM2 interacting alleles in other colors. These colors are unrelated to the ones used to identify membership in STRUCTURE clusters on top.
(E) Carriers of DM2 hybrid necrosis alleles in red, carriers of interacting alleles in other colors. DM2/SRF3 interactions are from (Alcázar et al., 2010).
See also Figure S6.
To assess the prevalence of haplotype sharing in this region, we asked whether the 3′ syntenic region is present in 16 additional accessions with a DM2h-type gene (Figure 5D; Table S6). Reconstruction of phylogenetic relationships across an 8 kb region demonstrated that at least 12 of the DM2h carriers shared very similar sequences in this region (Figure 6B). Close relationships were not evident on the other side of the DM2 cluster (Figure S6A), arguing against reduced haplotype diversity being simply a consequence of suppressed recombination, as reported for some NLR clusters (Chin et al., 2001). Such haplotype sharing among the 12 carriers, which extended over a region of 16 kb downstream of DM2h/At3g44670 (Welch two sample t-test, P<0.0001), was not observed next to two other NLR loci, the RPM1 single-gene locus and the RPP4/5 cluster (Figure 6C). We further confirmed haplotype sharing at DM2 among the DM2h carriers using the FST statistic as a proxy for genetic differentiation (Figure S6B). Together with the observation that the 12 accessions are otherwise not particularly related either (Figure 6D), this suggests that the pattern of reduced haplotype diversity is not due to a recent population bottleneck.
Geographic distribution of hybrid necrosis risk alleles
Two proteins that can produce hybrid necrosis in combination with DM2 alleles from Uk-3 and Ler have been previously identified, the NLR protein DM1 from Uk-1 (Bomblies et al., 2007) and the kinase SRF3 from Central Asian accessions (Alcázar et al., 2010). In this study, we identified the prolyl aminopeptidase DM3 from Hh-0 as an interactor of DM2h from Bla-1 (Figures 6E and S2B–E). In addition, the DM2 cluster from Dog-4 interacts with an unknown gene at the DM5 locus from ICE163, while the DM2 clusters from several South Tyrolean accessions including ICE163 interact with DM4 from TueWa1-2 (Figure 6E). We analyzed the genome-wide differentiation of DM2 risk allele carriers among the 80 accessions that served as parents of many of our hybrid crosses plus other known carriers. As expected, accessions with different DM2 alleles did not cluster with each other, but rather with other accessions from the same geographical regions (Figure 6D), supporting independent origins of the different risk alleles. One of the DM2 risk alleles was present in multiple strains from South Tyrol (Figure 6D; Table S1). This is similar to SRF3, for which incompatibility alleles are found throughout Central Asia (Alcázar et al., 2010), although overall population differentiation appears to be lower in Central Asia than in South Tyrol (Figures 6D, E) (Cao et al., 2011).
Discussion
The extent to which non-additive interactions between segregating alleles affect fitness related traits in both outcrossing and selfing organisms is a central question in genetics (Corbett-Detig et al., 2013; Mackay, 2014; Phillips, 2008). We have used a new, systematically structured population of F1 hybrids to determine the prevalence and causes of a common form of deleterious epistasis in plants, hybrid necrosis. A main finding is that interactions among immune genes are the most common cause of hybrid necrosis; this observation indicates that there are limits to the assembly of potentially favorable immune gene alleles in the same genetic background.
The crosses we investigated included parental pairs that were from the same location and sometimes closely related throughout the genome, as well as geographically and genetically distant parents. We found that genome-wide genetic differentiation, which is correlated with geographic distance in A. thaliana (Cao et al., 2011), is not a good predictor for hybrid incompatibility. We interpret the occurrence of incompatibilities between accessions from the same geographic region as a sign that the incompatibilities on their own do not greatly affect the frequency of the individual causal alleles in the population. The genetic architectures we uncovered include interactions between one locus and several other distinct loci (DM2 with DM1, DM3, DM4, DM5 and SRF3), between different pairs of alleles at the same two loci (DM6 with DM7), and between different alleles at the same locus (at DM8 and at DM9). This highlights that particular loci are disproportionately dangerous, and can repeatedly cause independent deleterious epistatic interactions. Another important finding is that a large fraction of the hybrid necrosis alleles map to plant NLR immune receptor loci. While we do not yet have proof that any of the specific alleles we have identified are beneficial in nature, the extreme variability of a subset of immune genes is in itself thought to be advantageous, particularly where resistance genes co-evolve with extant pathogens (Holub, 2001; Michelmore and Meyers, 1998; Yang et al., 2013). Moreover, crop breeders have actively selected hybrid necrosis genes because they confer agronomically relevant resistances (Bomblies and Weigel, 2007; Krüger et al., 2002). This specific connection to the immune system sets our study apart from intraspecific studies in other systems where the evolutionary forces that give rise to deleterious epistasis remain largely unknown (Corbett-Detig et al., 2013).
It may not appear surprising that many hybrid necrosis genes encode NLR proteins, but two findings were unexpected: that RPP genes, which correspond to the major regions of the A. thaliana genome that encode resistance to the pathogen Hpa (Holub and Beynon, 1997; Nemri et al., 2010) are over-represented, and that a single locus, DM2/RPP1, is involved in over half of all hybrid necrosis cases mapped to date. Among RPP genes, RPP1 appears to be the most versatile locus, with alleles conferring resistance against many different Hpa genotypes and mediating different necrosis phenotypes (Holub and Beynon, 1997). That DM2/RPP1 is at the same time a frequent trigger of autoimmunity underscores the potential dangers of a rapidly evolving immune system, both with respect to new mutations at this locus, and upon outcrossing between accessions.
Because we found several interactions between different alleles at the same locus, our observations have implications not only for what has been called statistical epistasis, which is concerned with interactions between segregating polymorphisms, but also for functional epistasis, which refers to the allowed mutational paths of individual loci (Weinreich et al., 2005). Similar to DM2/RPP1, NLR genes are often arranged in tandem arrays. In a single tandem array, mutations could arise that cause deleterious interactions between proteins encoded by different members of such an array, but these would presumably be selected against, thereby limiting diversification within the array. In this context, it is of interest that the hybrid necrosis activity of the DM2hBla-1 allele was apparently acquired stepwise, as deduced from our experiments with domain swaps.
Perhaps the most important conclusion from our findings is that autoimmunity in hybrids might limit the assembly of superior immune alleles into a single genotype, because of the interactions between NLRs and other loci involved in immunity. We note that the self-fertilizing mating system of A. thaliana is not a barrier to the rapid reassortment of genetic diversity. In the native range of the species, identical multi-locus haplotypes are generally confined to individual small stands, and outcrossing rates in nature are sufficient to frequently generate new genetic combinations (Bomblies et al., 2010). It is noteworthy that we identified several accessions that carry multiple hybrid necrosis risk alleles. For example, the ICE163 accession carries both a DM2 risk allele that is incompatible with DM4 from TueWa1-2, and a risk allele at DM5 that is incompatible with DM2 from Dog-4. Similarly, hybrid necrosis alleles at both DM2 and DM9 are found in Bla-1, at both DM4 and DM7 in TueWa1-2, and at both DM7 and an unmapped class 5 locus in TueScha-9 (Table S1). Multiple incompatibility risk alleles in the same genome would increase the chances of deleterious epistasis between immune genes upon crosses with other genotypes.
While most hybrid necrosis alleles appear to be rare, we emphasize that we have limited our analyses to cases that are associated with strong morphological defects. These cases are almost certainly only the extremes of a wider range of interactions that lead to autonomous activation of the immune system. This argument follows from several observations: First, the F1 hybrid necrosis cases display a range of severity, with some dying without flowering and others being able to set seeds as dwarves, and one of the DM2 cases described in the literature is expressed only in the F2 generation (Alcázar et al., 2009). Similarly, in our diallel among the 80 fully sequenced accessions, we have observed dozens of weakly necrotic F1 cases, several of which showed stronger symptoms in the F2 generation. Thus, it is likely that in addition to the interactions we have reported here there are others that cause milder immune phenotypes, but still limit growth and development in a manner that is disadvantageous in the wild. Second, expression of hybrid necrosis symptoms can be modulated by genetic background (Alcázar et al., 2009), suggesting that more complex crossing strategies than the biparental design used here may reveal additional cases of hybrid necrosis.
Epistatic interactions between components of the immune system are likely to be relevant in other kingdoms as well. Such interactions have, for example, been observed in mammals, where there is evidence for positive selection acting on specific combinations of KIR-type receptors and MHC class I ligands (Single et al., 2007). Allelic variation at these loci is also responsible for autoimmune syndromes (Trowsdale and Knight, 2013). In A. thaliana, it seems perhaps unlikely that the interactions between the specific hybrid necrosis risk alleles we have described are beneficial, but it is conceivable that interaction between other alleles at these loci have positive effects on immune function. This can in principle be addressed using population genomic data, but because of the extreme variability of many of these loci, current whole-genome resequencing datasets are insufficient to ask directly whether specific alleles co-occur more often than expected by chance.
An important question for the future will be the biochemical nature of the interactions between hybrid necrosis risk alleles, and how they differ from interactions between non-risk alleles. DM2 risk alleles trigger hybrid necrosis when combined with alleles at loci that encode a broad range of biochemical functions, including at least one NLR, DM1, consistent with other cases of NLR proteins that act in pairs (Eitas and Dangl, 2010). Two other natural DM2 interactors encode a kinase (SRF3, (Alcázar et al., 2010)) and a prolyl aminopeptidase (DM3, this work). In addition, a chemically induced allele at a gene encoding a cysteine synthase can combine with a natural DM2 allele to cause necrosis (Tahir et al., 2012). Outside of A. thaliana, hybrid necrosis alleles encode a cysteine protease (Krüger et al., 2002), a kinase (Yamamoto et al., 2010) and a subtilisin-like protease (Chen et al., 2014). Enzyme-encoding genes are clearly enriched, but in most cases we do not know yet how they modulate the activity of NLRs.
In conclusion, we propose that the study of hybrid necrosis can provide important insights into the role of epistatic interactions, particularly between immune genes, in the evolution of genotypes with multiple resistances to diverse pathogens. That hybrid necrosis alleles can increase functional disease resistance in crop breeding programs suggests that greater immune effectiveness may be tied to autoimmune risk. Understanding the relationship between effectiveness in pathogen recognition and autoimmunity will have applications in crop breeding, where it can help to guide strategies for maximizing disease resistance while minimizing yield penalties. Finally, it will be important to investigate whether the immune system of obligatory outbreeding species is more or less constrained than that of self-compatible species, and whether it therefore produces adverse effects in progeny less or more often.
Experimental Procedures
Plant material
F1 hybrids were grown at 16°C, under long days (16 hours of light). They were monitored for signs of autoimmunity-associated morphological defects for the first 12 weeks of growth. Afterwards, plants were transferred to 23°C, and F2 seeds were harvested from individual plants that did not carry the sterility inducing AP3 amiRNA transgene. Class 4 and 5 hybrids were additionally grown at 28 °C.
Genotyping and QTL analyses
A GBS approach was used for genotyping F2 mapping populations, with PstI/MseI double digested tags. Sequence tags and segregating SNPs for bulk segregants were generated either on Illumina GAIIx or Illumina HiSeq 2000 (Illumina, San Diego, CA). Filtered markers were used for further QTL analyses (Table S3).
DM sequence analyses
A total of six fosmid clones each were Sanger shotgun sequenced to assemble the DM2 locus in Bla-1 and Uk-1. Illumina reads of 87 A. thaliana accessions (from (Cao et al., 2011) plus Col-0, Ler, Uk-1, Uk-3, Uk-6, Nc-1, Bla-1) and A. lyrata MN47 were trimmed to 36 bp in length and aligned to DM2dUk-1 and DM2hBla-1 open reading frames as reference using GenomeMapper (Schneeberger et al., 2009). One mismatch and zero gaps were allowed. Matrices were generated by assigning a value of one to each position covered by at least one read, and a value of zero to the remaining positions. Resulting profiles were clustered using complete linkage clustering with Euclidean distance.
Population genetic analyses
One-hundred-kb regions upstream or downstream of three NLR loci were extracted from a genome variant matrix (Cao et al., 2011). Only positions with allele frequency above 0.1 were retained, and ten consecutive SNPs were binned for further calculations. The ‘compute’ program, based on the ‘libsequence’ C++ library (Thornton, 2003), was used to calculate haplotype diversity.
Accession numbers
Short reads of Uk-3, Uk-6, KZ10 and Mrk-0 have been deposited in the European Nucleotide Archive under the accession number ERP005469. Sequences of the DM2 regions from Bla-1 and Uk-1, of DM3 from Hh-0, and of the RPW8 region from KZ10 have been deposited in GenBank under accession numbers KJ454428, KJ45449, KJ634210 and KJ634211.
Supplementary Material
Necrotic F1 Hybrids in A. thaliana, Related to Figure 1
(A) DM2-related hybrids with different severity of necrosis, class 2 to 4.
(B) Class 1 example.
(C) Class 3 example. Can be rescued to set seeds at 23 °C.
(D) Class 4 example. Can be rescued to set seeds at 28 °C.
(E) Class 5 example. Seedling lethal at all temperatures tested.
Plants shown were grown at 16°C for four (A–D) or two weeks (E). Blue arrowheads in (D) and (E) point to growth arrested seedlings. Control plants on the right side in each panel (B–E) are half-siblings sharing one parent with the necrotic hybrids. Scale bars represent 1 cm.
Two-dimensional Genome Scans for QTL Analyses and Identification of DM3 and DM7, Related to Figure 2
(A) LOD scores from a two-dimensional, two-QTL genome scan of each F2 population. Values in the upper left triangle indicate LOD for the test of epistasis (LODi); values in the lower right triangle indicate joint two-locus LOD scores (LODf). In the color scale on the right, numbers to the left and right correspond to LODi and LODf, respectively. Plots show that there is little evidence for genomic regions other than DM loci contributing to the phenotypic variance.
(B) Flowchart of DM3 identification. We narrowed down the final list of candidates by searching for non-synonymous SNPs from Hh-0 that were rare in other accessions, since the causal allele should occur in low frequency based on our crosses (Table S1).
(C) Dot plot of alignment of DM3 regions in Col-0 versus Hh-0, indicating absence of large structural variants in the interval.
(D) Rescue of hybrid phenotype by MiRNA-Induced Gene Silencing (MIGS) (de Felippes et al., 2011). The silencing constructs were introduced into BC5 of Bla-1/Hh-0 hybrids. B, Bla-1; C, Col-0; H, Hh-0. First pair of letters indicates genotype at DM2, second pair of letters at DM3. “+” or “−” indicates presence or absence of the MIGS transgene.
(E) Recapitulation of Bla-1/Hh-0 hybrid phenotype with a 3.7 kb genomic fragment of At3g61540 from Hh-0 introduced into Bla-1 (I). Genomic fragment II did not induce hybrid necrosis.
(F) Diagram of the RPW8 region in Col-0, Ms-0, in which the RPW8 powdery mildew resistance locus was initially identified, and KZ10. HR genes and RPW8 genes are similar to each other (Xiao et al., 2001). The incompatibility gene RPW8.1 in KZ10 is outlined in black.
(G) Recapitulation of KZ10/Mrk-0 hybrid phenotypes with a 1.9-kb genomic fragment of RPW8.1KZ10 introduced into Col-0 and T1 plant crossed to Mrk-0. Scale bars in (D), (E) and (G) represent 1 cm.
Domains of DM2 proteins, Related to Figure 3
Protein sequences were deduced based on intron positions in Col-0 homologs. When exon-intron junctions were uncertain, the Softberry algorithm (http://linux1.softberry.com) was used to predict protein sequences. Domains were predicted using NCBI’s conserved domain search module (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). LRRs were refined with HHrepIP (http://toolkit.tuebingen.mpg.de/hhrepid). Only LRRs with a P-value <1.0e−10 were indicated as orange ovals. Only LRRs carrying the consensus LXXLXLXXNCSSLVXLPXSIX3-6 (Figure S5A) were numbered. Differences in N-terminal sequences divide the DM2 proteins into two groups; a group including a 5′ intron (*) and the other group without the intron, generating a predicted membrane localization signal (MLS). The two hybrid necrosis DM2 proteins are labeled in red.
Identification and Analysis of DM2 Hybrid Necrosis Genes, Related to Figure 4
(A) Target predictions for the two rescuing amiRNAs.
(B) qRT-PCR analysis of DM2 expression in the Bla-1/Hh-0 F1 hybrid and rescued sibling shown in Figure 4A. Only DM2hBla-1 was efficiently downregulated in the rescued sibling. qRT-PCR analysis in the rescued Uk-1/Uk-3 hybrids could not be performed due to the close paralogous relationship between DM2UK-1d/e/g and DM2 genes in Uk-3. Relative value of each technical replicate is shown as individual bar.
(C) Uk-3 transformed with DM2dUk-1 (left); Uk-3 control plant (right). Plants were grown at 16°C for four weeks.
(D) DM2dUk-1 Uk-3 transgenic plants, grown at 23°C. Photos were taken at indicated days after sowing.
(E) Phenotypic classes of DM2hBla-1 transgenic plants at 16°C.
(F) Phenotypic distribution of T1 plants in different genetic backgrounds. Hh-0 shows the highest sensitivity to the transgene.
(G) Correlation between transgene expression and phenotypic classes, measured by qRT-PCR in the Uk-1 background that does not carry DM2hBla-1 type. Relative value of each technical replicate is shown as individual bar.
(H) Neighbor-joining tree inferred from nucleotide sequences of TIR and NBS encoding regions.
(I) Neighbor-joining tree of LRR-encoding region.
(J) Neighbor-joining tree of the whole genic region.
Numbers in (H), (I) and (J) indicate bootstrap support values above 50%.
Colors define well-supported clades in (H), and the same color code was used to identify members of each clade in (I). At3g25510Col-0, which encodes a TIR-NLR protein, was used as outgroup in (J). DM2 genes in Col-0 were labeled with the last three digits of At3g44XXX in (H) and (I).
Scale bars in (C), (D) and (E) represent 1 cm.
Mapping of Causal Polymorphisms in DM2hBla-1, Related to Figure 5
(A) Consensus sequences of LRRs from all DM2 homologs. Putative solvent-exposed regions are indicated as light purple bars. Residues conserved across LRRs are in black. Indel positions are shown with asterisks for single events or with numbers for multiple events.
(B) Alignment of select LRRs (light purple region in A) of DM2hBla-1 type genes. Only LRRs with nonsynonymous changes between the hybrid necrosis gene from Bla-1 (boxed) and other genes are shown. Residues found only in one allele in red, in two in blue, and variable positions without a clear minority allele in grey. Bla-1 has four rare changes, more than any of the other genes.
(C) Identification of causal residues in DM2hBla-1 using chimeras with At3g44670Col-0, with amino acid changes and indels indicated in red.
(D) Hybrid necrosis induced by At3g44670, DM2 and chimeras expressed under the promoter of At3g44670Col-0. The LRR mutation changes the LQQL motif in LRR4 of At3g44670Col to LRKL. A small fraction of T1 Col-0 plants carrying the At3g44670Col transgene had a dark-green or pointy-leaf phenotype but without necrosis (purple class).
Genetic Differentiation Around DM2 Cluster, Related to Figure 6
(A) Neighbor-joining tree inferred from nucleotide sequences of At3g44610 and At3g44620, which are 5′ adjacent to the first NLR gene in the DM2 cluster. Accessions carrying the DM2h-type orthologs are labeled in magenta. Bootstrap values over 70% are indicated.
(B) Extended haplotype sharing at DM2 among 12 DM2hBla-1 carriers, but not at the NLR loci RPM1 and RPP4/5, as revealed by FST statistics in comparison with 12 non-carriers. Values showing significant genetic differentiation (P<0.01) between the two subgroups after 10,000 resampling are highlighted in bright green.
Table S1. F1 Hybrid Phenotypes of Different Crosses, Related to Figure 1
Table S2. F2 Segregation Ratios at 16°C, Related to Figure 2
Table S3. F2 Populations for Genetic Mapping, Related to Figure 2
Table S4. Mapping Intervals for DM Loci, Related to Figure 2
Table S5. GENECONV Analyses, Related to Figure 5
Table S6. PCR Survey of LRR Polymorphisms in DM2hBla-1 Type Genes, Related to Figure 5
Acknowledgments
We thank Rubén Alcázar, Jane Parker and Maarten Koornneef for information regarding RPP1, Jesse Poland for GBS advice, Frank Wellmer for the AP3 amiRNA, Eui-Hwan Chung and Jeffery Dangl for pointers on cell death assays, William Ho, Paula Sancha-Vilchez and Josip Perkovic for technical support, and Jeffery Dangl, Ya-Long Guo, Daniel Koenig, George Wang and Jun Cao for discussion. We especially thank the anonymous reviewers, who greatly helped us with the evolutionary framing of our work. This work was supported by an NIH Ruth Kirschstein NRSA (K.B.), an HFSP Long-Term Fellowship (R.A.L.), an Alexander von Humboldt Foundation Fellowship (B.A.R.), an HFSPO Grant (RGP 57/2007), a Gottfried Wilhelm Leibniz Award of the DFG and the Max Planck Society (D.W.).
Footnotes
Author Contributions
Conceived and designed the experiments: E.C., K.B. and D.W. Performed the experiments: E.C., K.B., S.-T. K., M.Z., C.M.P., H.T., S.L., A.H.-M., M.D., C.L. Analyzed the data: E.C., K.B., S.-T. K., D.K., M.Z., S.O., R.A.L., B.A.R., G.R. Wrote the paper: E.C., K.B. and D.W. with contributions from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcázar R, García AV, Kronholm I, de Meaux J, Koornneef M, Parker JE, Reymond M. Natural variation at Strubbelig Receptor Kinase 3 drives immune-triggered incompatibilities between Arabidopsis thaliana accessions. Nat Genet. 2010;42:1135–1139. doi: 10.1038/ng.704. [DOI] [PubMed] [Google Scholar]
- Alcázar R, Garcia AV, Parker JE, Reymond M. Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc Natl Acad Sci USA. 2009;106:334–339. doi: 10.1073/pnas.0811734106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RL, Bittner-Eddy PD, Grenville-Briggs LJ, Meitz JC, Rehmany AP, Rose LE, Beynon JL. Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science. 2004;306:1957–1960. doi: 10.1126/science.1104022. [DOI] [PubMed] [Google Scholar]
- Bakker EG, Toomajian C, Kreitman M, Bergelson J. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell. 2006;18:1803–1818. doi: 10.1105/tpc.106.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson V. Heredity and variation in modern lights. In: Seward AC, editor. Darwin and Modern Science. Cambridge: Cambridge University Press; 1909. pp. 85–101. [Google Scholar]
- Bikard D, Patel D, Le Mette C, Giorgi V, Camilleri C, Bennett MJ, Loudet O. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science. 2009;323:623–626. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, Weigel D. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 2007;5:e236. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Weigel D. Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet. 2007;8:382–393. doi: 10.1038/nrg2082. [DOI] [PubMed] [Google Scholar]
- Bomblies K, Yant L, Laitinen RA, Kim ST, Hollister JD, Warthmann N, Fitz J, Weigel D. Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet. 2010;6:e1000890. doi: 10.1371/journal.pgen.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43:956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- Chen C, Chen H, Lin YS, Shen JB, Shan JX, Qi P, Shi M, Zhu MZ, Huang XH, Feng Q, et al. A two-locus interaction causes interspecific hybrid weakness in rice. Nat Commun. 2014;5:3357. doi: 10.1038/ncomms4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin DB, Arroyo-Garcia R, Ochoa OE, Kesseli RV, Lavelle DO, Michelmore RW. Recombination and spontaneous mutation at the major cluster of resistance genes in lettuce (Lactuca sativa) Genetics. 2001;157:831–849. doi: 10.1093/genetics/157.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EH, da Cunha L, Wu AJ, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL. Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe. 2011;9:125–136. doi: 10.1016/j.chom.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SM, Moffett P. NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 2009;14:521–529. doi: 10.1016/j.tplants.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Corbett-Detig RB, Zhou J, Clark AG, Hartl DL, Ayroles JF. Genetic incompatibilities are widespread within species. Nature. 2013;504:135–137. doi: 10.1038/nature12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- Crespi B, Nosil P. Conflictual speciation: species formation via genomic conflict. Trends Ecol Evol. 2013;28:48–57. doi: 10.1016/j.tree.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Cutter AD. The polymorphic prelude to Bateson-Dobzhansky-Muller incompatibilities. Trends Ecol Evol. 2012;27:209–218. doi: 10.1016/j.tree.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the Origin of Species. New York: Columbia University Press; 1937. [Google Scholar]
- Eitas TK, Dangl JL. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol. 2010;13:472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6:e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CJ, Slootweg EJ, Goverse A, Baulcombe DC. Stepwise artificial evolution of a plant disease resistance gene. Proc Natl Acad Sci USA. 2013;110:21189–21194. doi: 10.1073/pnas.1311134110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holub EB. The arms race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet. 2001;2:516–527. doi: 10.1038/35080508. [DOI] [PubMed] [Google Scholar]
- Holub EB, Beynon JL. Symbiology of mouse-ear cress (Arabidopsis thaliana) and oomycetes. Adv Bot Res. 1997;24:227–273. [Google Scholar]
- Hou J, Friedrich A, de Montigny J, Schacherer J. Chromosomal rearrangements as a major mechanism in the onset of reproductive isolation in Saccharomyces cerevisiae. Curr Biol. 2014;24:1153–1159. doi: 10.1016/j.cub.2014.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Vernaldi S, Maekawa T. Evolution and Conservation of Plant NLR Functions. Frontiers in immunology. 2013;4:297. doi: 10.3389/fimmu.2013.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeuken MJ, Zhang NW, McHale LK, Pelgrom K, den Boer E, Lindhout P, Michelmore RW, Visser RG, Niks RE. RIN4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell. 2009;21:3368–3378. doi: 10.1105/tpc.109.070334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva KV, Dahlbeck D, Staskawicz BJ. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell. 2010;22:2444–2458. doi: 10.1105/tpc.110.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang S, Mulder L, Jones JD. A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science. 2002;296:744–747. doi: 10.1126/science.1069288. [DOI] [PubMed] [Google Scholar]
- Kuang H, Woo SS, Meyers BC, Nevo E, Michelmore RW. Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell. 2004;16:2870–2894. doi: 10.1105/tpc.104.025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance J, True JR. X-autosome incompatibilities in Drosophila melanogaster: tests of Haldane’s rule and geographic patterns within species. Evolution. 2010;64:3035–3046. doi: 10.1111/j.1558-5646.2010.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Rate DN, Song JT, Greenberg JT. ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell. 2003;15:2408–2420. doi: 10.1105/tpc.015412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat Rev Genet. 2014;15:22–33. doi: 10.1038/nrg3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, Barbash DA. The genetics of hybrid incompatibilities. Annu Rev Genet. 2011;45:331–355. doi: 10.1146/annurev-genet-110410-132514. [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Isolating mechanisms, evolution and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- Nemri A, Atwell S, Tarone AM, Huang YS, Zhao K, Studholme DJ, Nordborg M, Jones JD. Genome-wide survey of Arabidopsis natural variation in downy mildew resistance using combined association and linkage mapping. Proc Natl Acad Sci USA. 2010;107:10302–10307. doi: 10.1073/pnas.0913160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC. Epistasis–the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC, Johnson NA. The population genetics of synthetic lethals. Genetics. 1998;150:449–458. doi: 10.1093/genetics/150.1.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland JA, Brown PJ, Sorrells ME, Jannink JL. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE. 2012;7:e32253. doi: 10.1371/journal.pone.0032253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. The molecular evolutionary basis of species formation. Nat Rev Genet. 2010;11:175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- Quintana-Murci L, Clark AG. Population genetic tools for dissecting innate immunity in humans. Nat Rev Immunol. 2013;13:280–293. doi: 10.1038/nri3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Blackman BK. Speciation genes in plants. Ann Bot. 2010;106:439–455. doi: 10.1093/aob/mcq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 2007;39:1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen JE, Weigel D, Andersen SU. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods. 2009;6:550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- Segretin ME, Pais M, Franceschetti M, Chaparro-Garcia A, Bos JI, Banfield MJ, Kamoun S. Single amino acid mutations in the potato immune receptor R3a expand response to Phytophthora effectors. Molecular plant-microbe interactions: MPMI. 2014 doi: 10.1094/MPMI-02-14-0040-R. [DOI] [PubMed] [Google Scholar]
- Seidel HS, Rockman MV, Kruglyak L. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 2008;319:589–594. doi: 10.1126/science.1151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, Kidd KK, Carrington M. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39:1114–1119. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- Steinbrenner AD, Goritschnig S, Krasileva KV, Schreiber KJ, Staskawicz BJ. Effector recognition and activation of the Arabidopsis thaliana NLR innate immune receptors. Cold Spring Harb Symp Quant Biol. 2012;77:249–257. doi: 10.1101/sqb.2012.77.014860. [DOI] [PubMed] [Google Scholar]
- Tahir J, Watanabe M, Jing HC, Hunter DA, Tohge T, Nunes-Nesi A, Brotman Y, Fernie AR, Hoefgen R, Dijkwel PP. Activation of R-mediated innate immunity and disease susceptibility is affected by mutations in a cytosolic O-acetylserine (thiol) lyase in Arabidopsis. Plant J. 2012 doi: 10.1111/tpj.12021. [DOI] [PubMed] [Google Scholar]
- Thornton K. Libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics. 2003;19:2325–2327. doi: 10.1093/bioinformatics/btg316. [DOI] [PubMed] [Google Scholar]
- Todesco M, Balasubramanian S, Hu TT, Traw MB, Horton M, Epple P, Kuhns C, Sureshkumar S, Schwartz C, Lanz C, et al. Natural allelic variation underlying a major fitness tradeoff in Arabidopsis thaliana. Nature. 2010;465:632–636. doi: 10.1038/nature09083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, Kim ST, Chae E, Bomblies K, Zaidem M, Smith LM, Weigel D, Laitinen RA. Activation of the Arabidopsis thaliana immune system by combinations of common ACD6 alleles. PLoS Genet. 2014;10:e1004459. doi: 10.1371/journal.pgen.1004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Watson RA, Chao L. Perspective: Sign epistasis and genetic constraint on evolutionary trajectories. Evolution. 2005;59:1165–1174. [PubMed] [Google Scholar]
- Wuest SE, O’Maoileidigh DS, Rae L, Kwasniewska K, Raganelli A, Hanczaryk K, Lohan AJ, Loftus B, Graciet E, Wellmer F. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc Natl Acad Sci USA. 2012;109:13452–13457. doi: 10.1073/pnas.1207075109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science. 2001;291:118–120. doi: 10.1126/science.291.5501.118. [DOI] [PubMed] [Google Scholar]
- Yamamoto E, Takashi T, Morinaka Y, Lin S, Wu J, Matsumoto T, Kitano H, Matsuoka M, Ashikari M. Gain of deleterious function causes an autoimmune response and Bateson–Dobzhansky–Muller incompatibility in rice. Mol Genet Genomics. 2010;283:305–315. doi: 10.1007/s00438-010-0514-y. [DOI] [PubMed] [Google Scholar]
- Yang S, Li J, Zhang X, Zhang Q, Huang J, Chen JQ, Hartl DL, Tian D. Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc Natl Acad Sci USA. 2013;110:18572–18577. doi: 10.1073/pnas.1318211110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Necrotic F1 Hybrids in A. thaliana, Related to Figure 1
(A) DM2-related hybrids with different severity of necrosis, class 2 to 4.
(B) Class 1 example.
(C) Class 3 example. Can be rescued to set seeds at 23 °C.
(D) Class 4 example. Can be rescued to set seeds at 28 °C.
(E) Class 5 example. Seedling lethal at all temperatures tested.
Plants shown were grown at 16°C for four (A–D) or two weeks (E). Blue arrowheads in (D) and (E) point to growth arrested seedlings. Control plants on the right side in each panel (B–E) are half-siblings sharing one parent with the necrotic hybrids. Scale bars represent 1 cm.
Two-dimensional Genome Scans for QTL Analyses and Identification of DM3 and DM7, Related to Figure 2
(A) LOD scores from a two-dimensional, two-QTL genome scan of each F2 population. Values in the upper left triangle indicate LOD for the test of epistasis (LODi); values in the lower right triangle indicate joint two-locus LOD scores (LODf). In the color scale on the right, numbers to the left and right correspond to LODi and LODf, respectively. Plots show that there is little evidence for genomic regions other than DM loci contributing to the phenotypic variance.
(B) Flowchart of DM3 identification. We narrowed down the final list of candidates by searching for non-synonymous SNPs from Hh-0 that were rare in other accessions, since the causal allele should occur in low frequency based on our crosses (Table S1).
(C) Dot plot of alignment of DM3 regions in Col-0 versus Hh-0, indicating absence of large structural variants in the interval.
(D) Rescue of hybrid phenotype by MiRNA-Induced Gene Silencing (MIGS) (de Felippes et al., 2011). The silencing constructs were introduced into BC5 of Bla-1/Hh-0 hybrids. B, Bla-1; C, Col-0; H, Hh-0. First pair of letters indicates genotype at DM2, second pair of letters at DM3. “+” or “−” indicates presence or absence of the MIGS transgene.
(E) Recapitulation of Bla-1/Hh-0 hybrid phenotype with a 3.7 kb genomic fragment of At3g61540 from Hh-0 introduced into Bla-1 (I). Genomic fragment II did not induce hybrid necrosis.
(F) Diagram of the RPW8 region in Col-0, Ms-0, in which the RPW8 powdery mildew resistance locus was initially identified, and KZ10. HR genes and RPW8 genes are similar to each other (Xiao et al., 2001). The incompatibility gene RPW8.1 in KZ10 is outlined in black.
(G) Recapitulation of KZ10/Mrk-0 hybrid phenotypes with a 1.9-kb genomic fragment of RPW8.1KZ10 introduced into Col-0 and T1 plant crossed to Mrk-0. Scale bars in (D), (E) and (G) represent 1 cm.
Domains of DM2 proteins, Related to Figure 3
Protein sequences were deduced based on intron positions in Col-0 homologs. When exon-intron junctions were uncertain, the Softberry algorithm (http://linux1.softberry.com) was used to predict protein sequences. Domains were predicted using NCBI’s conserved domain search module (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). LRRs were refined with HHrepIP (http://toolkit.tuebingen.mpg.de/hhrepid). Only LRRs with a P-value <1.0e−10 were indicated as orange ovals. Only LRRs carrying the consensus LXXLXLXXNCSSLVXLPXSIX3-6 (Figure S5A) were numbered. Differences in N-terminal sequences divide the DM2 proteins into two groups; a group including a 5′ intron (*) and the other group without the intron, generating a predicted membrane localization signal (MLS). The two hybrid necrosis DM2 proteins are labeled in red.
Identification and Analysis of DM2 Hybrid Necrosis Genes, Related to Figure 4
(A) Target predictions for the two rescuing amiRNAs.
(B) qRT-PCR analysis of DM2 expression in the Bla-1/Hh-0 F1 hybrid and rescued sibling shown in Figure 4A. Only DM2hBla-1 was efficiently downregulated in the rescued sibling. qRT-PCR analysis in the rescued Uk-1/Uk-3 hybrids could not be performed due to the close paralogous relationship between DM2UK-1d/e/g and DM2 genes in Uk-3. Relative value of each technical replicate is shown as individual bar.
(C) Uk-3 transformed with DM2dUk-1 (left); Uk-3 control plant (right). Plants were grown at 16°C for four weeks.
(D) DM2dUk-1 Uk-3 transgenic plants, grown at 23°C. Photos were taken at indicated days after sowing.
(E) Phenotypic classes of DM2hBla-1 transgenic plants at 16°C.
(F) Phenotypic distribution of T1 plants in different genetic backgrounds. Hh-0 shows the highest sensitivity to the transgene.
(G) Correlation between transgene expression and phenotypic classes, measured by qRT-PCR in the Uk-1 background that does not carry DM2hBla-1 type. Relative value of each technical replicate is shown as individual bar.
(H) Neighbor-joining tree inferred from nucleotide sequences of TIR and NBS encoding regions.
(I) Neighbor-joining tree of LRR-encoding region.
(J) Neighbor-joining tree of the whole genic region.
Numbers in (H), (I) and (J) indicate bootstrap support values above 50%.
Colors define well-supported clades in (H), and the same color code was used to identify members of each clade in (I). At3g25510Col-0, which encodes a TIR-NLR protein, was used as outgroup in (J). DM2 genes in Col-0 were labeled with the last three digits of At3g44XXX in (H) and (I).
Scale bars in (C), (D) and (E) represent 1 cm.
Mapping of Causal Polymorphisms in DM2hBla-1, Related to Figure 5
(A) Consensus sequences of LRRs from all DM2 homologs. Putative solvent-exposed regions are indicated as light purple bars. Residues conserved across LRRs are in black. Indel positions are shown with asterisks for single events or with numbers for multiple events.
(B) Alignment of select LRRs (light purple region in A) of DM2hBla-1 type genes. Only LRRs with nonsynonymous changes between the hybrid necrosis gene from Bla-1 (boxed) and other genes are shown. Residues found only in one allele in red, in two in blue, and variable positions without a clear minority allele in grey. Bla-1 has four rare changes, more than any of the other genes.
(C) Identification of causal residues in DM2hBla-1 using chimeras with At3g44670Col-0, with amino acid changes and indels indicated in red.
(D) Hybrid necrosis induced by At3g44670, DM2 and chimeras expressed under the promoter of At3g44670Col-0. The LRR mutation changes the LQQL motif in LRR4 of At3g44670Col to LRKL. A small fraction of T1 Col-0 plants carrying the At3g44670Col transgene had a dark-green or pointy-leaf phenotype but without necrosis (purple class).
Genetic Differentiation Around DM2 Cluster, Related to Figure 6
(A) Neighbor-joining tree inferred from nucleotide sequences of At3g44610 and At3g44620, which are 5′ adjacent to the first NLR gene in the DM2 cluster. Accessions carrying the DM2h-type orthologs are labeled in magenta. Bootstrap values over 70% are indicated.
(B) Extended haplotype sharing at DM2 among 12 DM2hBla-1 carriers, but not at the NLR loci RPM1 and RPP4/5, as revealed by FST statistics in comparison with 12 non-carriers. Values showing significant genetic differentiation (P<0.01) between the two subgroups after 10,000 resampling are highlighted in bright green.
Table S1. F1 Hybrid Phenotypes of Different Crosses, Related to Figure 1
Table S2. F2 Segregation Ratios at 16°C, Related to Figure 2
Table S3. F2 Populations for Genetic Mapping, Related to Figure 2
Table S4. Mapping Intervals for DM Loci, Related to Figure 2
Table S5. GENECONV Analyses, Related to Figure 5
Table S6. PCR Survey of LRR Polymorphisms in DM2hBla-1 Type Genes, Related to Figure 5