Abstract
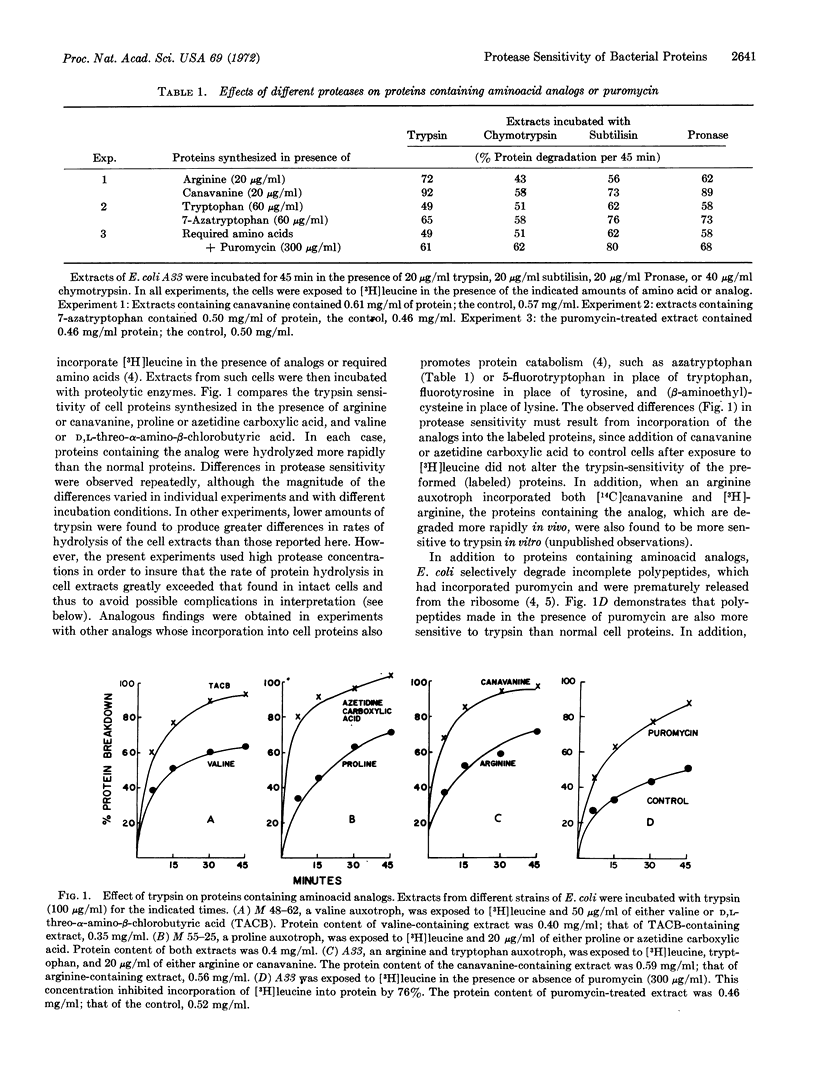
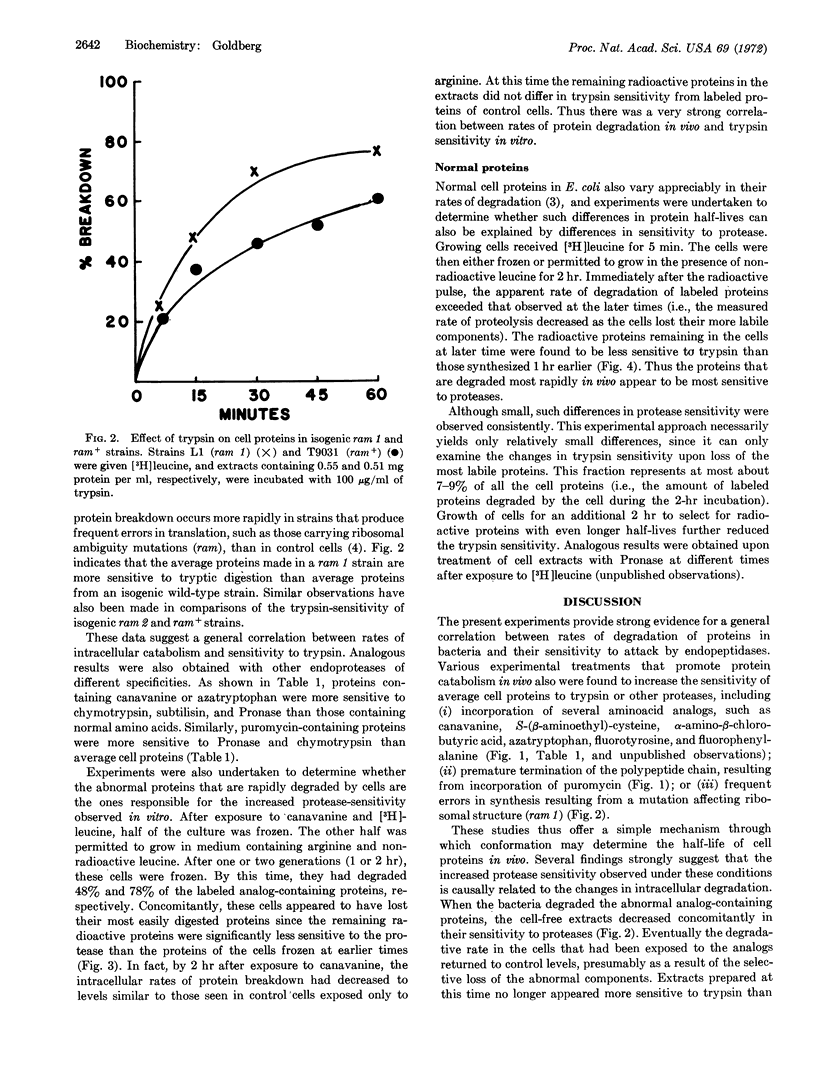
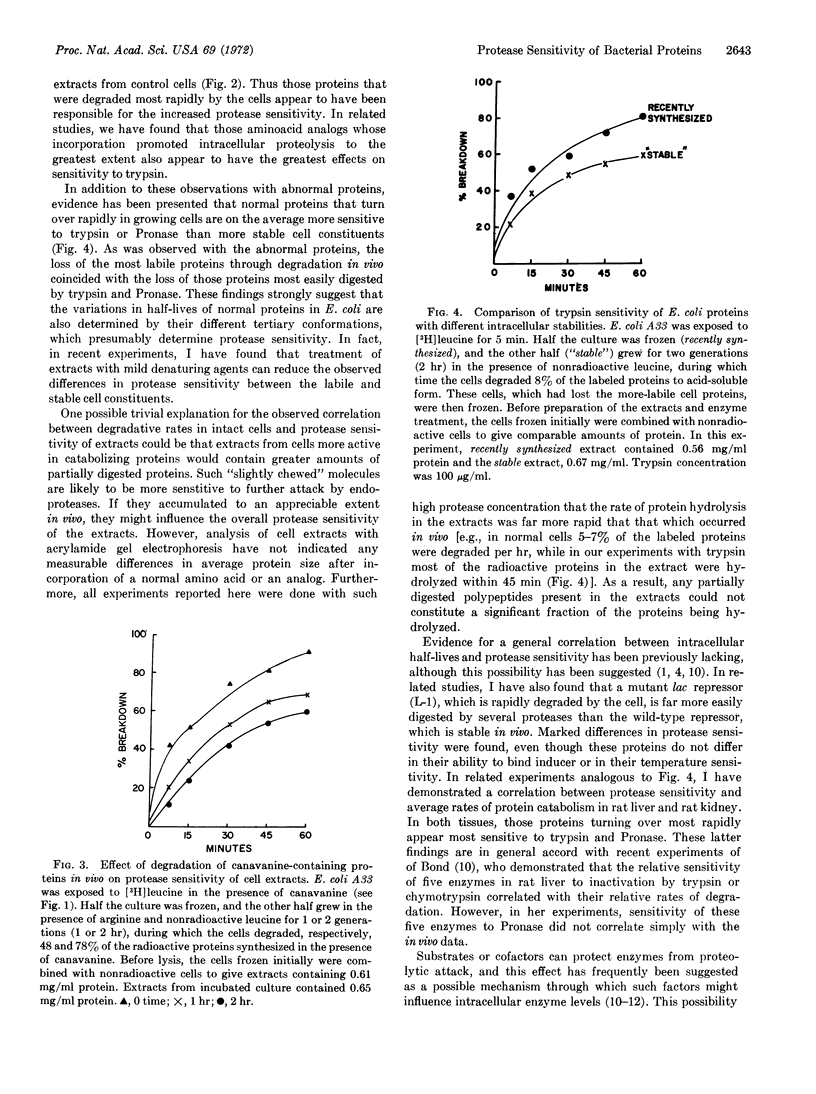
Experiments were undertaken to test whether the relative rates of degradation of proteins in vivo might correlate with their sensitivity to proteases. Various experimental conditions that promote degradation of proteins in Escherichia coli increased sensitivity of average cell proteins to trypsin or other endoproteases, including incorporation of several aminoacid analogs into the proteins, incorporation of puromycin into the polypeptide chain, or frequent errors in translation. Those abnormal proteins that were degraded most rapidly within the cell appear responsible for the increased protease sensitivity of the cell extract. Normal E. coli proteins that rapidly turn over in growing cells are, on the average, more sensitive to trypsin or Pronase than cell proteins that turn over more slowly. The inherent sensitivity of a protein to proteolytic digestion is thus a major determinant of protein half-lives in vivo.
Keywords: protein conformation, abnormal proteins, aminoacid analogs, mistranslation
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bond J. S. A comparison of the proteolytic susceptibility of several rat liver enzymes. Biochem Biophys Res Commun. 1971 Apr 16;43(2):333–339. doi: 10.1016/0006-291x(71)90757-1. [DOI] [PubMed] [Google Scholar]
- Fritz P. J., Vesell E. S., White E. L., Pruitt K. M. The roles of synthesis and degradation in determining tissue concentrations of lactate dehydrogenase-5. Proc Natl Acad Sci U S A. 1969 Feb;62(2):558–565. doi: 10.1073/pnas.62.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRISOLIA S. THE CATALYTIC ENVIRONMENT AND ITS BIOLOGICAL IMPLICATIONS. Physiol Rev. 1964 Oct;44:657–712. doi: 10.1152/physrev.1964.44.4.657. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L. Degradation of abnormal proteins in Escherichia coli (protein breakdown-protein structure-mistranslation-amino acid analogs-puromycin). Proc Natl Acad Sci U S A. 1972 Feb;69(2):422–426. doi: 10.1073/pnas.69.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. L. Effects of protease inhibitors on protein breakdown and enzyme induction in starving Escherichia coli. Nat New Biol. 1971 Nov 10;234(45):51–52. doi: 10.1038/newbio234051a0. [DOI] [PubMed] [Google Scholar]
- Goldschmidt R. In vivo degradation of nonsense fragments in E. coli. Nature. 1970 Dec 19;228(5277):1151–1154. doi: 10.1038/2281151a0. [DOI] [PubMed] [Google Scholar]
- Hershko A., Tomkins G. M. Studies on the degradation of tyrosine aminotransferase in hepatoma cells in culture. Influence of the composition of the medium and adenosine triphosphate dependence. J Biol Chem. 1971 Feb 10;246(3):710–714. [PubMed] [Google Scholar]
- LINDERSTRØM-LANG K. Structure and enzymatic break-down of proteins. Cold Spring Harb Symp Quant Biol. 1950;14:117–126. doi: 10.1101/sqb.1950.014.01.016. [DOI] [PubMed] [Google Scholar]
- Markus G. Protein substrate conformation and proteolysis. Proc Natl Acad Sci U S A. 1965 Jul;54(1):253–258. doi: 10.1073/pnas.54.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimore G. E., Mondon C. E. Inhibition by insulin of valine turnover in liver. Evidence for a general control of proteolysis. J Biol Chem. 1970 May 10;245(9):2375–2383. [PubMed] [Google Scholar]
- Nath K., Koch A. L. Protein degradation in Escherichia coli. I. Measurement of rapidly and slowly decaying components. J Biol Chem. 1970 Jun 10;245(11):2889–2900. [PubMed] [Google Scholar]
- Nath K., Koch A. L. Protein degradation in Escherichia coli. II. Strain differences in the degradation of protein and nucleic acid resulting from starvation. J Biol Chem. 1971 Nov 25;246(22):6956–6967. [PubMed] [Google Scholar]
- Pine M. J. Heterogeneity of protein turnover in Escherichia coli. Biochim Biophys Acta. 1965 Jul 8;104(2):439–456. doi: 10.1016/0304-4165(65)90349-1. [DOI] [PubMed] [Google Scholar]
- Pine M. J. Response of intracellular proteolysis to alteration of bacterial protein and the implications in metabolic regulation. J Bacteriol. 1967 May;93(5):1527–1533. doi: 10.1128/jb.93.5.1527-1533.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T., Miller J. H., Weber K. In vivo degradation of mutant lac repressor. Nature. 1970 Dec 19;228(5277):1154–1156. doi: 10.1038/2281154a0. [DOI] [PubMed] [Google Scholar]
- Prouty W. F., Goldberg A. L. Effects of protease inhibitors on protein breakdown in Escherichia coli. J Biol Chem. 1972 May 25;247(10):3341–3352. [PubMed] [Google Scholar]
- RABINOVITZ M., FISHER J. M. CHARACTERISTICS OF THE INHIBITION OF HEMOGLOBIN SYNTHESIS IN RABBIT RETICULOCYTES BY THREO-ALPHA-AMINO-BETA-CHLOROBUTYRIC ACID. Biochim Biophys Acta. 1964 Oct 16;91:313–322. doi: 10.1016/0926-6550(64)90255-5. [DOI] [PubMed] [Google Scholar]
- Schimke R. T., Sweeney E. W., Berlin C. M. Studies of the stability in vivo and in vitro of rat liver tryptophan pyrrolase. J Biol Chem. 1965 Dec;240(12):4609–4620. [PubMed] [Google Scholar]
- Segal H. L., Matsuzawa T., Haider M., Abraham G. J. What determines the half-life of proteins in vivo? Some experiences with alanine aminotransferase of rat tissues. Biochem Biophys Res Commun. 1969 Aug 22;36(5):764–770. doi: 10.1016/0006-291x(69)90675-5. [DOI] [PubMed] [Google Scholar]
- Sussman A. J., Gilvarg C. Protein turnover in amino acid-starved strains of Escherichia coli K-12 differing in their ribonucleic acid control. J Biol Chem. 1969 Nov 25;244(22):6304–6306. [PubMed] [Google Scholar]


