Abstract
The daily life of photosynthetic plants revolves around sugar production, transport, storage and utilization, and the complex sugar metabolic and signaling networks integrate internal regulators and environmental cues to govern and sustain plant growth and survival. Although diverse sugar signals have emerged as pivotal regulators from embryogenesis to senescence, glucose is the most ancient and conserved regulatory signal that controls gene and protein expression, cell-cycle progression, central and secondary metabolism, as well as growth and developmental programs. Glucose signals are perceived and transduced by two principal mechanisms: direct sensing through glucose sensors and indirect sensing via a variety of energy and metabolite sensors. This review focuses on the comparative and functional analyses of three glucose-modulated master regulators in Arabidopsis thaliana, the hexokinase1 (HXK1) glucose sensor, the energy sensor kinases KIN10/KIN11 inactivated by glucose, and the glucose-activated target of rapamycin (TOR) kinase. These regulators are evolutionarily conserved, but have evolved universal and unique regulatory wiring and functions in plants and animals. They form protein complexes with multiple partners as regulators or effectors to serve distinct functions in different subcellular locales and organs, and play integrative and complementary roles from cellular signaling and metabolism to development in the plant glucose signaling networks.
Keywords: hexokinase, energy sensor kinase, target of rapamycin kinase, glucose signaling networks
Introduction
Glucose fuels life from bacteria, yeasts, plants to humans. Despite the fundamental and multifaceted regulatory roles of glucose in gene and protein expression, physiology, metabolism, proliferation, growth and development, and connections to human diseases, the molecular and cellular mechanisms underlying glucose signaling remain largely elusive in multicellular plants and animals. Research on plant regulatory networks over the past half-century has emphasized hormonal and peptide signals that are perceived at minute quantities by high affinity receptors (Santner et al., 2009; Katsir et al., 2011; Lee et al., 2013), whereas physiological nutrient and metabolite signals can act at several orders of magnitude higher levels via mostly unknown sensors (Rolland et al., 2002; Gibson, 2005; Rolland et al., 2006; Polge and Thomas, 2007; Ramon et al, 2008; Smeekens et al., 2010; Laplante and Sabatini, 2012; Robaglia et al., 2012; Urano et al., 2012; Dobrenel et al., 2013; Eveland and Jackson, 2013; Yuan et al., 2013; Xiong and Sheen, 2014).
The research on glucose signaling has been quite challenging due to the difficulties in separating glucose metabolism from glucose sensing and signaling events, elucidating and connecting primary and long-term responses, or creating powerful genetic tools to dissect the distinct roles of regulators whose essential functions are manifested in mutant lethality and functional redundancy. Over the past decade, integrated genetic, cellular, chemical, proteomic and genomic approaches in the reference plant Arabidopsis thaliana have begun to unravel the surprisingly broad range of functions and actions of three glucose-modulated master regulators, HXK1, KIN10/11 and TOR (Fig. 1). These regulators control the expression of thousands of plant genes involved in a wide spectrum of cellular functions from signaling, transcription, anabolism, catabolism, transport, to growth, development and stress adaptation in response to altered glucose signals (Rolland et al., 2002; Halford et al., 2003; Rolland et al., 2006; Polge and Thomas, 2007; Baena-González and Sheen 2008; Ramon et al., 2008; Sheen, 2010; Smeekens et al., 2010; Robaglia et al., 2012; Dobrenel et al., 2013; Xiong and Sheen, 2014).
Fig. 1.
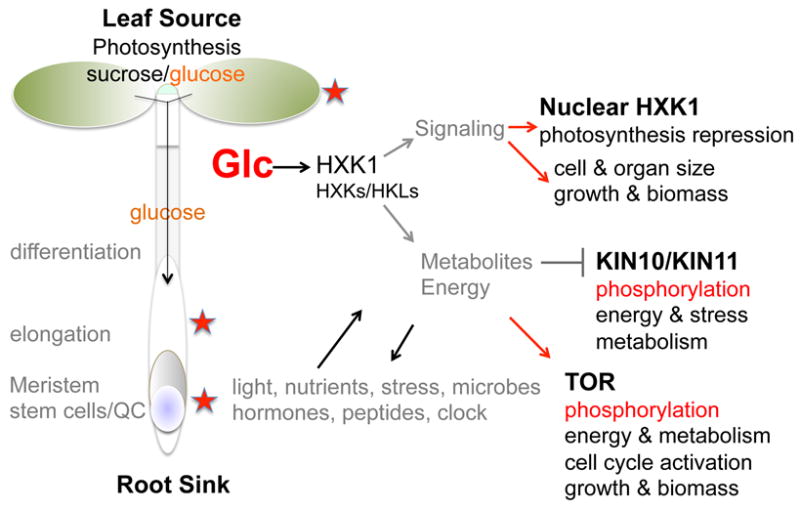
Arabidopsis glucose-signaling networks. Glucose is generated from the photosynthetic or storage source and transported as sucrose or glucose to the sink tissues and organs to promote cell proliferation, elongation, expansion, and to maintain energy and metabolic homeostasis. The regulatory mechanisms and functions of three master regulators, HXK1, KIN10/11 and TOR, modulated by glucose signals are shown. The glucose signaling networks are intertwined with the signaling pathways controlled by environmental light, nutrients, stresses and microbes, as well as internal hormones, peptides and clock. Red stars mark the action sites of glucose signaling. Glc, glucose; HXK, hexokinase; HKLs, hexokinase-like; KIN, Arabidopsis protein kinase; QC, quiescent center; TOR, target of rapamycin.
Arabidopsis HXK1 acts as the direct glucose sensor mediating multiple functions in the glucose repression and glucose promotion of transcription and growth (Xiao et al., 2000; Moore et al., 2003; Yanagisawa et al., 2003; Cho et al., 2006; Cho et al., 2009). The protein kinase activity of KIN10/11 is repressed by glucose (Baena-González et al., 2007), whereas TOR kinase is activated by glucose (Xiong and Sheen, 2012, Xiong et al., 2013). KIN10/11 and TOR sense opposite energy levels and govern the partially overlapping plant transcriptional networks, which are intimately connected to glucose-derived energy and metabolite signaling tightly associated with glycolysis and mitochondrial bioenergetics, but are mostly uncoupled from the HXK1 actions as a glucose sensor (Baena-González et al., 2007; Xiong et al., 2013). However, HXK1 and other metabolic enzymes also contribute to the generation of energy and metabolite signals derived from glucose (Moore et al., 2003; Kim et al., 2006; Granot, 2007; Paul et al., 2008; Cho et al., 2009; Karve et al., 2010; Nilsson et al., 2011; Kim et al., 2013). For example, the Arabidopsis transcription factor genes, bZIP1 and TZF1 (TANDEM ZINC FINGER1), are KIN10 and TOR target genes (Baena-González et al., 2007; Xiong et al., 2013), but their expression is partially insensitive to glucose repression in the HXK1 mutant gin2 (glucose insensitive2)(Kang et al., 2010; Lin et al., 2010). It is important to consider multiple regulatory criteria by applying specific and informative molecular, cellular, genetic, genomic and chemical tools to distinguish the direct and indirect mechanisms in diverse glucose responses (Fig. 1)(Xiao et al., 2000; Moore et al., 2003; Price et al., 2004; Blasing et all, 2005; Gonzali et al., 2006; Li et al., 2006; Baena-González et al., 2007; Sheen, 2010; Xiong et al., 2013; de Jong et al., 2014). This review focuses on the progresses in understanding the novel functions and the emerging molecular mechanisms of Arabidopsis HXK1, KIN10/11 and TOR actions over the past decade.
Direct Glucose Sensing and Signaling via HXK1
Distinct HXK1 Functions
The discovery of unique and global repression of photosynthesis genes by glucose in photoautotrophic plants led to the identification of Arabidopsis HXK1 as the first plant glucose sensor with uncoupled sensor and metabolic functions (Sheen, 1990; Jang et al., 1997; Xiao et al., 2000; Rolland et all, 2002; Moore et al., 2003; Cho et al., 2006; Rolland et al., 2006; Ramon et al., 2008; Li and Sheen, unpublished). The glucose repression of photosynthesis genes and photosynthetic organ development mediated by HXK1 and the functional orthologs from other plants is conserved, which serves as a physiological feedback loop in sugar production and is promoted by glucose availability but antagonized by nitrogen signals (Martin et al., 2002; Moore et al., 2003; Price et al., 2004; Granot, 2007; Zhang et al., 2010; Cho et al., 2009; Cho et al., 2010; Kelly et al., 2012; Kim et al., 2013). The reported variability in leaf glucose responses is likely due to different plant architecture, developmental stage, as well as carbon and nitrogen storage strategies or use efficiency under various natural or artificial growth conditions in different plant species. For instance, tobacco, tomato and maize are large plants and their leaves are more prone to nitrogen deficiency to conspicuously display glucose repression, whereas potato plants with strong tuber sink for sugar and starch storage may require different growth conditions to manifest glucose repression (Sheen, 1990; Xiao et al., 2000; Moore et al., 2003; Yanagisawa et al., 2003; Granot 2007; Kelly et al., 2012; Kim et al., 2013). Although the distantly related cyanobacterial glucokinase partially complement the leaf phenotypes in gin2 (Ryu et al, 2008), other HXK1 functions remain unfulfilled (Li and Sheen, unpublished). The closely related yeast HXK2 complements the catalytic function but not the sensor function of HXK1 in transgenic Arabidopsis (Jang et al., 1997; Yanagisawa 2003; Moore et al., 2003; Li and Sheen, unpublished). To further elucidate the conserved or distinct molecular and cellular mechanisms of glucose signaling, it is important to optimize diverse glucose response assays tailored to each plant species by recognizing and considering prominent and unique molecular, physiological and morphological features (Moore et al., 2003; Granot, 2007; Ryu et al, 2008; Cho et al., 2009; Cho et al, 2010; Zhang et al., 2010; Karve et al., 2010; Nilsson et al., 2011; Kelly et al., 2012; Kim et al., 2013).
In the presence of sufficient nitrate, increased light, photosynthesis or glucose has revealed the second distinct function of HXK1 glucose sensor in promoting the growth of vegetative and reproductive organs, also independent of glucose metabolism in Arabidopsis plants. This growth promotion function of HXK1 appears to be more complex, as it is integrated in each organ to maximize cell and organ size and whole plant biomass, which is proportional to available glucose signals (Moore et al., 2003; Cho et al., 2009; Hall and Sheen, unpublished). Furthermore, diverse isoforms of HXK and HXK-like (HKL) genes have been found in the genome of all land plants from the nonflowering moss and lycophyte to seed plants, including rice, maize, sorghum, poplar, tobacco, tomato, grape and Arabidopsis (Moore et al., 2003; Granot, 2007; Karve et al., 2008, 2010; Nilsson et al., 2011; Karve et al., 2012; Kim et al., 2013). The amplification and diversification of the HXK superfamily suggests more complex modes of plant glucose signaling and metabolism in different subcellular compartments and organs evolved to support a wide spectrum of plant growth strategies and architectures dictated by sugar availability and other essential nutrients (Moore et al., 2003; Claeyssen and Rivoal, 2007; Granot, 2007; Karve et al., 2008; Cho et al., 2009; Zhang et al., 2010; Karve et al., 2010; Nilsson et al., 2011; Karve et al., 2012; Kim et al., 2013).
HXK1 and Hormone Interactions
Extensive genetic, molecular, cellular, and biochemical analyses have uncovered the intimate links between HXK1-mediated glucose signaling and multiple hormones, including but not limited to ethylene, abscisic acid (ABA), auxin and cytokinin (Rolland et al., 2002; Leon and Sheen, 2003; Moore et al., 2003; Yanagisawa et al., 2003; Rolland et al., 2006; Huang et al., 2008; Ramon et al., 2008; Cho et al., 2010; Karve et al., 2012; Hsu et al., 2014). In maize and Arabidopsis leaf cells, Arabidopsis HXK1 but not the yeast HXK promotes the proteasome-dependent degradation of nuclear EIN3 (ETHYLENE INSENSITIVE3) and EIL1 (EIN3-LIKE1) through the C-terminus, which is stabilized by ethylene. Consistently, ein3 as well as etr1, ein2 and mkk9 are hypersensitive to glucose but insensitive to ethylene (Yanagisawa et al., 2003; Yoo et al., 2008). HXK1 also modulates the glucose repression of ERF1 (ETHYLENE RESPONSE FACTOR1) in ethylene signaling, and the glucose activation of ABA2 in ABA synthesis and ABI4/ABI5 (ABA INSENSITIVE) transcription factors in ABA signaling (Rolland et al., 2002; Leon and Sheen, 2003; Rolland et al., 2006; Ramon et al., 2008; Cho et al., 2010; Hsu et al., 2014). More detailed molecular mechanisms underlying sugar regulation of hormonal signaling and biosynthesis remained to be further resolved using defined HXK1 mutants, as distinct signaling and metabolic effects of sugars may be involved (Moore et al., 2003; Ohto et al., 2006; Loreti et al., 2008; Mishra et al., 2009; Mudgil et al., 2009; LeClere et al., 2010; Lin et al., 2010). Recent findings in sucrose-specific stabilization of the DELLA repressors support an antagonistic relationship between sucrose and gibberellin signaling and the opposite roles in anthocyanin biosynthesis uncoupled from HXK1 signaling (Loreti et al., 2008; Li et al., 2013). Comprehensive studies in the rice and barley aleurones indicate that gibberellin and SNRK1A (SNF1-RELATED PROTEIN KINASE1A) antagonize sugar repression and promoting the nuclear translocation and activity of MYBS1 and MYBGA, which is also enhanced by nitrogen and phosphate deficiency but mostly uncoupled from the HXK1 glucose sensor functions (Lu et al., 2007; Hong et al., 2012).
The finding that the gin2 mutant is glucose insensitive but fructose sensitive has led to the identification of the fructose-specific signaling pathway (Cho and Yoo, 2011; Li et al., 2011). Cell-based screens and genetic analyses lead to the identification of FRUCTOSE-1,6-BIPHOSTASE as a nuclear fructose sensor independent of its catalytic activity (Cho and Yoo, 2011). Interestingly, a gain-of-function transcription factor ANAC089 lacking the membrane-bound domain specifically and dominantly suppresses fructose but not glucose sensitivity in Arabidopsis (Li et al., 2011). It will be exciting to further elucidate the fructose-specific signaling pathway by linking the nuclear sensor to the membrane-bound transcription factor. Although different nuclear sensors mediate glucose and fructose responses, their actions converge downstream on the activation of ABA signaling but the repression of ethylene signaling (Cho and Yoo, 2011; Li et al., 2011). In future research aiming at understanding and predicting the signaling crosstalk and distinct sugar signaling pathways, it is informative to elucidate the precise molecular node, mechanism and degree of connections between glucose, fructose, sucrose and each hormonal signaling pathway to assess and discover the complex and dynamic regulators and their influences on plant growth and developmental programs. As different levels of glucose and hormones could lead to distinct or even opposite consequences in plant responses, application of physiologically relevant regulatory signals with defined and specific mutant manipulations not limited to overexpression is critical for logical interpretation of diverse sugar responses.
Molecular Actions of HXK1
To elucidate the molecular and cellular mechanisms of novel HXK1 actions in distinct glucose responses, several complementary approaches have been developed, including targeted mutagenesis and functional analyses of Arabidopsis HXK1 in gin2, biochemical and structural analyses of glucose binding and phosphorylation activities, genetic manipulation of subcellular localization, purification and characterization of the nuclear HXK1 protein complexes, identification of HXK1 interacting proteins, and establishing HXK1-dependent transcriptome reprogramming and chromatin association (Xiao et al., 2000; Moore et al., 2003; Yanagisawa et al., 2003; Cho et al., 2006; Baena-González et al., 2007; Balasubramanian et al., 2007; Cho et al., 2009; Cho et al., 2010; Karve et al., 2012; Xiong et al., 2013). Comprehensive analyses of the most informative HXK1 mutation, S177A without detectable catalytic activity but with full glucose binding, have provided compelling evidence for the glucose sensing and signaling activities critical for diverse glucose responses uncoupled from glucose phosphorylation and metabolism (Moore et al., 2003; de Jong et al., 2014; Li and Sheen, unpublished). Importantly, the catalytically inactive mutations of the rice orthologs, OsHXK5 and OsHXK6, provide supporting evidence for the conserved glucose signaling functions similar to Arabidopsis HXK1S177A in transgenic gin2 plants (Cho et al., 2009).
As observed in Arabidopsis, maize, tomato, tobacco and moss, the majority of HXK1 and closely related HXK proteins are attached to the outer membrane of mitochondria serving the conventional function in glycolysis (Kandel-Kfir et al., 2006; Kim et al., 2006; Balasubramanian et al., 2007; Granot, 2007; Karve et al., 2008; Cho et al., 2009; Nilsson et al., 2011; Kim et al., 2013). Virus-induced gene-silencing studies in Nicotiana benthamiana suggest a role of tobacco HXKs in the control of programmed cell death (Kim et al., 2006), which is reminiscent of the glucokinase function in mice (Danial et al., 2003). However, several studies have identified Arabidopsis and rice HXK glucose sensor proteins in the nucleus, especially when the N-terminal region responsible for the association with the mitochondria outer membrane was deleted (Yanagisawa et al., 2003; Cho et al., 2006; Cho et al., 2009). Studies by genetic complementation analyses in gin2 indicate that the association of HXK1 with mitochondria is dispensable for the nuclear glucose sensor functions in transgenic plants (Li and Sheen, unpublished). Furthermore, extensive efforts have led to the biochemical isolation of the nuclear HXK1 protein complexes in Arabidopsis leaves and the identification of multiple HXK1 interacting protein partners. Genetic analyses of two of these HXK1 interacting proteins, a scaffold vacuolar H+-ATPase B1 subunit and an AAA-ATPase subunit of the 19S regulatory particle of proteasome, support their roles in glucose repression of photosynthesis gene repression in leaves and in glucose promotion of plant growth in low nutrient conditions (Cho et al., 2006). Transcriptome analyses by RNA-sequencing have supported a central and unique role of Arabidopsis HXK1 in the global transcriptional program encompassing over 2000 genes in rapid leaf responses to physiological glucose signals (Li and Sheen, unpublished). Future genetic and biochemical characterization as well as genomic analyses of the transcription factors and RNA regulators in the nuclear HXK1 complexes will potentially illuminate novel molecular mechanisms of the HXK1 actions on the chromatin in glucose signaling (Cho et al., 2006; Hsu et al., 2014). The application of super-resolution fluorescence microscopy (Doksani et al., 2013) will provide dynamic view of regulatory proteins in HXK1-mediated glucose signaling at single-molecule details in different subcellular compartments. It will be interesting to learn whether the E3 ligase, protein kinase and histone acetyltransferase identified through Arabidopsis sugar-response mutants may act downstream or independent of HXK1-mediated glucose signaling (Huang et al., 2010; Heisel et al., 2013: Huang et al., 2013).
Glucose Repression of Energy Sensor Kinase
Central Integrators in Energy and Stress Signaling
The evolutionarily conserved genes encoding the catalytic subunit of energy sensor kinases in eukaryotes have been identified for more than two decades, including yeast SNF1 (SUCROSE-NON-FERMENTATION1), mammalian AMPK (AMP-ACTIVATED PROTEIN KINASE) and plant SNRK1 (Celenza and Carlson, 1986; Halford and Hardie, 1998; Bhalerao et al., 1999; Halford et al., 2003; Hardie et al., 2012). The most prevailing studies of SNRK1 have been focused on the regulation of cytoplasmic enzymes, such as nitrate reductase and sucrose phosphate synthase, involved in nitrogen and sugar metabolism (Halford and Hardie, 1998; Sugden et al., 1999; Halford et al., 2003). By establishing a combination of cellular, biochemical, functional genomic and genetic tools in Arabidopsis thaliana, ample evidence now supports novel functions of the redundant Arabidopsis KIN10 (SNRK1.1) and KIN11 (SNRK1.2) as central integrators of transcriptional networks in stress and energy signaling (Baena-González et al., 2007; Baena-González and Sheen, 2008; Smeekens et al., 2010). The glucose-repressed SNRK1 is likely conserved in all plants and the orthologous genes encoding the catalytic subunit complement the yeast snf1 mutant, supporting the conserved roles in glucose signaling (Bhalerao et al., 1999; Halford et al., 2003; Lovas et al., 2003; Polge and Thomas, 2007; Baena-González and Sheen, 2008; Ramon et al., 2008).
Studies on the regulation of universal DARK-INDUCIBLE (DIN) genes in Arabidopsis have provided crucial evidence to molecularly connect glucose-repressible transcription to plant energy sensor kinases and diverse stress conditions in a HXK1 independent manner (Fujiki et al., 2001; Baena-González et al., 2007). Transient elevation of KIN10 activity mediates the activation and repression of over 1000 genes involved in transcription, signaling, catabolism and anabolism. Remarkably, 600 of these genes are either positively correlated with stress and starvation genes or negatively correlated with genes activated by sucrose, glucose, and elevated CO2 in cultured cells, seedlings and adult leaves in genome-wide analyses (Price et al., 2004; Contento et al, 2004; Lin and Wu, 2004; Thimm et al., 2004; Blasing et al., 2005; Buchanan-Wollaston et al., 2005; Baena-González et al., 2007; Baena-González and Sheen, 2008). The high correlations between KIN10 target genes and the transcriptome reprogramming in multiple physiological responses provide compelling evidence to support a critical and convergent role of KIN10 in glucose repression triggered by darkness, starvation or diverse stress conditions (Baena-González et al., 2007; Baena-González and Sheen, 2008). It has been shown that potato SNRK1 is responsible for the sucrose activation of a potato gene encoding sucrose synthase (SUS4) in specific antisense transgenic lines (Purcell et al., 1998). However, different from KIN10/11 and another potato gene (StubSNF1), this PKIN1 gene does not complement the yeast snf1 mutant (Lovas et al., 2003). While StubSNF1 and KIN10/11 are closely related to the SNRK1a subgroup genes that are conserved in plants, PKIN1 bears more resemblance with the SNRK1b subgroup that is more typical for cereal species, and may have evolved to acquire specific functions, distinct from the universal ones proposed for Arabidopsis KIN10/11 (Halford and Hardie, 1998; Bhalerao et al., 1999; Halford et al., 2003; Lovas et al., 2003; Baena-González et al., 2007). It would be informative to directly compare the functions of PKIN1 and StubSNF1 in potato. Interesting studies have demonstrated the essential role of rice SNRK1A but not SNRK1B in the transcription control and phosphorylation of MYBS1 transcription factor and the regulation of αAMY3 (α–AMYLASE) promoter repressed by glucose in transient assays and in transgenic and mutant rice plants. Importantly, the snrk1a but not the snrk1b rice mutant also affects normal seed germination and seedling development (Lu et al., 2007). It will be fascinating to further explore and compare the various SNRK1 signaling pathways in diverse plant species.
Diverse Physiological Functions of KIN10/11
Overexpression of KIN10 in transgenic Arabidopsis plants significantly alters seedling growth, starvation response, anthocyanin accumulation and vegetative-to-reproductive phase transition. The flowering time is significantly delayed especially in reduced photoperiod with striking promotion of plant longevity (Baena-González et al., 2007). Leaf longevity in transgenic Arabidopsis plants is promoted by the wild-type rice OsSNRK1 but not the inactive OsSnRK1K43M, which also prevents the induction of hypoxia marker genes, ALCOHOL DEHYDROGENASE1 and PYRUVATE DECARBOXYLASE1, in flooding stress response and leads to plant death (Cho et al., 2012). In germinating rice seedlings, molecular and genetic studies have identified a specific calcineurinB-like-interacting protein kinase CIPK15 acting upstream of OsSNRK1A to coordinate sugar and oxygen deficiency enabling flood tolerance (Lee et al., 2009). The identification of an Arabidopsis seed transcription factor FUSCA3 (FUS3) as a potential KIN10 substrate provides a novel link to the regulatory program in seed development and germination. As the fus3-3 mutant partially rescues the phase transition and organ development defects in KIN10 overexpression transgenic plants, FUS3 may possess regulatory roles beyond embryo development (Tsai and Gazzarrini, 2012). It will be important to complement the fus3-3 mutant in KIN10 overexpression plants with a wild-type FUS3 gene to support its molecular function beyond seeds and determine FUS3 expression patterns in meristems responsible for organ development and phase transitions. KIN10 and KIN11 also interact with another transcription factor ATAF1 in the NAC gene family. Unexpectedly, T2 silencing transgenic plants with an ATAF1 overexpression construct exhibit severe developmental defects, supporting a positive regulatory function in plant development (Kleinow et al., 2009). The identification of more KIN10/KIN11 substrates and their functional characterization through integrated genetic, cellular, biochemical and genomic approaches will bring exciting new insight into how the energy and stress signaling networks are connected to plant developmental programs.
Although the single kin10 and kin11 mutants mostly resemble wild-type plants, the virus-induced kin10 kin11 double mutants display small deformed leaves, inflorescences and flowers, short petioles, reduced stress and starvation gene activation and starch degradation in the dark, and are eventually lethal even under constant light (Baena-González et al., 2007). Many of the complex phenotypes are consistent with the analyses of the snf1a snf1b double mutants in the moss Physcomitrella patens, including developmental abnormalities in caulonemal and chloronemal filaments and leafy shoots, premature senescence, hypersensitivity to auxin, and reduced response to cytokinin. As the double snf1a snf1b mutants in moss are unable to grow in a normal day-night light cycle, SNRK1 is required for metabolic changes to cope with darkness (Thelander et al., 2004). These studies reveal essential roles of the conserved energy sensor kinase not only in stress and energy signaling, but also in previously unknown functions governing normal plant growth and development (Thelander et al., 2004; Baena-González et al., 2007; Kleinow et al., 2009; Tsai and Gazzarrini, 2012).
Changes observed in SNRK1-repressed pea seeds indicate the regulation of cytokinin levels for cotyledon and leaf formation, and ABA levels critical for seed maturation. It will be important to investigate the regulatory links between altered hormone levels and the perturbation in organic and amino acids (Radchuk et al., 2010). Unexpectedly, overexpression of KIN10 also confers ABA hypersensitivity (Radchuk et al., 2010; Cho et al., 2012), which is supported by recent findings that KIN10 interacts with and is inactivated by several protein phosphatase 2Cs (PP2Cs) that are negative regulators in ABA signaling. As KIN10 activation and ABA signaling share some overlapping target genes, specific PP2Cs appear to serve as the converged hub for the coordinated activation of ABA and energy/stress signaling pathways (Rodrigues et al., 2013).
Interestingly, the glucose activation of pathogenesis-related genes, PR1, PR2 and PR5, which are regulated by HXK catalytic activity through downstream metabolites (Xiao et al., 2000), is strongly diminished in KIN10 overexpression plants, suggesting a link between energy and stress signaling and plant defense (Jossier et al., 2009). Other findings suggesting a role of SNRK1 in plant defense include viral inactivation of tobacco SNRK1 important for resistance against tomato golden mosaic virus and beet curly top virus (Hao et al., 2003), sugar reallocation for herbivory tolerance (Schwachtje et al., 2006), Xanthomonas AvrBsT effector targeting SNRK1 to suppress AvrBs1-induced immunity in pepper (Szczesny et al., 2010), and tomato protein kinase Adi3 interacting with a specific SNRK1 β-subunit to suppress cell death in resistance response (Avila et al., 2012). As SNRK1 regulates many enzymes and large sets of plant genes, it will be important to uncover the specific downstream molecular responses underlying SNRK1-mediated plant immunity.
Molecular Mechanisms of KIN10/11 Actions and Regulations
Genome-wide expression profiling of KIN10 target genes supports a key role of the energy sensor kinase in orchestrating the transcriptional network promoting catabolism but repressing anabolism (Baena-González et al., 2007; Baena-González and Sheen, 2008). Detailed molecular studies on the promoter of a primary KIN10 target gene, DIN6 encoding asparagine synthase, have defined a specific G-box DNA motif to be responsible for its convergent transcriptional activation by darkness, hypoxia stress, herbicide, and elevated KIN10 activity in Arabidopsis leaf cells. By establishing a physiologically relevant and cell-based functional genomic screen for predicted transcription factors, four closely related Arabidopsis bZIP transcription factors (bZIP1, bZIP2/GBF5, bZIP11/ATB2 and bZIP53) in the S subgroup have been shown to function synergistically with KIN10 to activate the DIN6 promoter and many endogenous KIN10 target genes (Jakoby et al., 2002; Baena-González et al., 2007; Baena-González and Sheen, 2008). Gain- and loss-of-function approaches indicate a critical role of bZIP53 in the ternary complex with bZIP10, bZIP25 and ABI3 involved in the transcription of seed maturation genes (Alonso et al., 2009). The findings support the possibility that SNRK1 is involved in the seed maturation process, which is linked to ABA signaling and stress tolerance (Alonso et al., 2009; Radchuk et al., 2010; Tsai and Gazzarrini, 2012; Rodrigues et al., 2013). Innovative systems approach has identified bZIP1 as a key regulator in nitrogen and light regulation (Gutierrez et al., 2008; Obertello et al., 2010), and transgenic plants overexpressing bZIP1, bZIP11 and bZIP53 share similar growth defects, activation of stress and energy starvation genes, and dark-induced amino acid accumulation (Hanson et al., 2008; Alonso et al., 2009; Kang et al., 2010; Dietrich et al., 2011). However, these related bZIP transcription factors are regulated differentially at transcriptional and posttranscriptional levels through multiple sugar signaling pathways and show overlapping but distinct interactions with different C-group partners (bZIP9, bZIP10, bZIP44 and bZIP63) (Jakoby et al., 2002; Baena-González et al., 2007; Hanson et al., 2008; Alonso et al., 2009; Rahmani et al., 2009; Kang et al., 2010; Dietrich et al., 2011). Besides transcriptional controls, the studies of energy deprivation responses in the dcl1-9 (dicer-like1) mutant with miRNA biogenesis defects have demonstrated a role of miR319 in the stress regulation of TCP mRNA abundance (Confraria et al., 2013). Although SNRK1 phosphorylates and regulates enzymes in the cytoplasm (Sugden et al., 1999), direct association of the Arabidopsis KIN10 and rice SNRK1 with the target genes promoter chromatin in the nucleus has been shown by chromatin-immunoprecipitation PCR analyses (Cho et al., 2011). Future investigation will require the identification of diverse SNRK1 substrates for mediating transcriptional and post-transcriptional regulations in the stress and energy signaling networks in multiple subcellular compartments (Baena-González et al., 2007; Bitrian et al., 2011; Cho et al., 2012).
Extensive research has identified diverse KIN10-interacitng proteins that act as positive or negative regulators, as well as substrates/effectors in the energy signaling networks (Farras et al., 2001; Halford et al., 2003; Polge and Thomas, 2007; Ananieva et al., 2008; Kleinow et al., 2009; Tsai and Gazzarrini, 2012; Ramon et al., 2013). The SNRK1 heterotrimeric protein complexes, consisting of the catalytic kinase α subunit and the β and γ regulatory subunits, are conserved from yeasts to plants, animals and humans (Halford et al., 2003; Polge and Thomas, 2007; Hardie et al., 2012; Ramon et al., 2013). Arabidopsis KIN10 directly interacts with KINβ1 KINβ2 or KINβ3 but the hybrid four-CBS-Domain KINβγ instead of the previously predicted KINγ interacts with KIN10 and the three KINβ subunits and functionally complements the yeast snf4 mutant (Ramon et al., 2013). Comprehensive and high-resolution phylogenetic reconstruction supports the evolutionarily significant findings on the divergence of the predicted KINγ protein from the monophyletic clade including the canonical γ regulatory subunits from yeast SNF1, animal and human AMPK, and KINβγ from all plant species. Consistently, the Arabidopsis kinβγ and kin10 kin11 mutant are lethal, but the null kinγ mutant does not exhibit detectable phenotypes. Moreover, partial silencing of KINβγ significantly compromises the expression of several KIN10 target genes (Baena-González et al., 2007; Ramon et al., 2013; Ramon and Sheen, unpublished). Future studies will illuminate the specific functions of the conserved KINβ subunits and KINβγ, as well plant-specific regulators in modulating the SNRK1 activity in different biological contexts.
Interestingly, plants appear to have evolved novel regulators of SNRK1, including myoinositol polyphosphate 5-phosphatase and cyclin-dependent kinase E1 as positive regulators (Ananieva et al., 2008; Ng et al., 2013), and a WD protein PRL1 (pleiotropic regulatory locus1), as well as ribose-5-phosphate, fructose-1,6-bisphosphate, 3-phosphoglycerate, glucose-6-phosphate and trehalose-6-phosphate as potential negative regulators (Bhalerao et al., 1999; Toroser et al., 2000; Zhang et al., 2009; Piattoni et al., 2011). Extensive genetic and biochemical analyses have supported a central role of trehalose-6-phosphate or trehalose metabolism in embryogenic, vegetative and inflorescence growth and branching, as well as the regulation of flowering (van Dijken et al., 2004; Schluepmann et al., 2004; Satoh-Nagasawa et al., 2006; Paul et al., 2007; Ramon et al., 2007; Chary et al., 2008; Zhang et al., 2009; Gomez et al., 2010; Delatte et al., 2011; Wahl et al., 2013). As putative trehalose metabolism enzymes are encoded by 22 genes and many are differentially regulated by sugar levels in Arabidopsis thaliana, it will be informative to dissect the metabolic and regulatory functions of each annotated trehalose-6-phosphate syntheses and trehalose-6-phosphate phosphatases (Avonce et al., 2004; Thimm et al., 2004; Baena-González et al., 2007; Ramon and Fillip, 2007; Ramon et al., 2009) to establish the genetic and molecular link between sugar metabolism and plant development in the energy signaling networks.
Glucose Activation of TOR kinase
Discovering Plant TOR Functions and Regulations
TOR is an exceptionally large protein kinase (2481 aa, TAIR10) with multiple repeats and regulatory domains in the N-terminus and an evolutionarily conserved Ser/Thr protein kinase domain at the C-terminus. In yeast and mammals, TOR forms at least two structurally and functionally distinct complexes, TORC1 (TOR complex1) and TORC2. Each complex contains shared and distinct TOR interacting partners, and recruits and differentially regulates diverse TOR kinase substrates to control a variety of biological processes (Wullschleger et al., 2006; Laplante and Sabatini, 2012; Robaglia et al., 2012; Cornu et al., 2013; Kang et al., 2013; Yuan et al., 2013; Xiong and Sheen, 2014). The precise compositions of the TOR kinase complexes have not been systematically characterized in plants. However, some of the mTORC1 components and downstream effectors have been identified in photosynthetic eukaryotes by sequence similarity search, including Arabidopsis RAPTOR1/2 (REGULATORY ASSOCIATE PROTEIN OF TOR), LST8-1/2 (LETHAL WITH SEC-13 PROTEIN8), S6K1/2 (RIBOSOMAL PROTEIN S6 KINASE), RPS6a/b (RIBOSOME PROTEIN SMALL SUBUNIT6) and TAP46 (TYPE 2A-PHOSPHATASE-ASSOCIATED PROTEIN 46 KD)(Anderson et al., 2005; Deprost et al., 2005; Mahfouz et al., 2006; Ahn et al., 2011; Moreau et al., 2012; Ren et al., 2012; Xiong and Sheen, 2014).
The function of TOR in coupling nutrient and energy availability with other environmental signals to coordinate growth, development and survival is likely conserved in yeasts, plants, animals and humans (Wullschleger et al., 2006; Laplante and Sabatini, 2012; Robaglia et al., 2012; Cornu et al., 2013; Yuan et al., 2013; Xiong and Sheen, 2014). However, due to the lack of facile molecular and biochemical assays for endogenous TOR kinase activity, the embryo lethality of null Arabidopsis tor mutants, and the prevailingly perceived rapamycin resistance in land plants (Menand et al., 2002; Ren et al., 2011; Robaglia et al., 2012), the molecular functions and the dynamic regulatory mechanisms of TOR kinase remained largely unclear in photosynthetic plants. Recent breakthroughs enabled by integrating sensitive cellular, chemical, genetic, genomic, and metabolomic analyses are beginning to reveal that glucose and auxin activate TOR kinase, which regulates plant growth, development, flowering, senescence and life span by modulating translation, transcription, autophagy, and primary and secondary metabolism (Deprost et al., 2007, Liu and Bassham, 2010; Ahn et al., 2011; Ren et al., 2011; Moreau et al., 2012; Ren et al., 2012; Robaglia et al., 2012; Xiong and Sheen, 2012; Caldana et al., 2013; Dobrenel et al., 2013; Xiong et al., 2013; Xiong and Sheen, 2014). Glucose-activated TOR is essential for transcriptome reprogramming, meristem activation and plant growth (Xiong and Sheen, 2012; Xiong et al., 2013), hereas auxin enhances TOR activity to promote the translation reinitiation of uORF ntaining mRNAs via S6K1 phosphorylation of elF3h (Schepetilnikov et al., 2013). nterestingly, TOR also enters the nucleus through a nuclear localization sequence in the kinase domain to directly bind to the 45S rRNA promoter and 5’-external transcribed spacer elements to regulate rRNA expression (Ren et al., 2011).
To circumvent the embryo lethality of tor mutants in Arabidopsis (Menand et al., 2002; Ren et al., 2011), ethanol or estradiol inducible TOR silencing based on RNA interference or artificial miRNA have allowed conditional control of the TOR gene expression to different degrees in transgenic Arabidopsis plants. All these transgenic Arabidopsis lines together with rapamycin and the new generation of ATP-competitive TOR kinase inhibitors have provided invaluable genetic and chemical tools to start deciphering the TOR signaling networks in plants (Deprost et al., 2007; Liu et al., 2012; Ren et al., 2012; Xiong and Sheen, 2012; Montane and Menand, 2013; Caldana et al., 2013; Xiong et al., 2013). Long-term (3-6 days) and partial TOR inhibition cause limited gene expression changes, which corroborate with some of the observed metabolite accumulation revealed by large-scale profiling. For example, reduction of TOR expression or kinase activity, or a mutation of LST8-1 encoding a conserved WD40 protein in TOR complexes, leads to retarded growth and accumulation of starch, triacylglycerides, amino acids, TCA (tricarboxylic acid cycle) intermediates, and secondary metabolites. Partial TOR deficiency reduces gene expression in the anabolic and biosynthetic pathways, but activates genes involved in the catabolic, stress and defense processes (Deprost et al., 2007; Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013). The growth defects and broad changes in transcripts and metabolites support the central roles of TOR signaling in plant growth and development (Menand et al., 2002; Anderson et al., 2005; Deprost et al., 2005, 2007; Ren et al., 2011; Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013; Xiong and Sheen 2014). As the consequences of long-term TOR silencing are more complex, the opposite senescence phenotypes and different raffinose and galactinol accumulation in various conditional tor mutants and lst8 plants remain to be resolved (Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013).
Dynamic Glucose-TOR Signaling
To better understand the molecular landscape of the TOR signaling networks and the dynamic links between TOR signaling and transcriptional regulation, the rapid global transcriptome changes stimulated by 2-h glucose (15 mM) treatment was investigated in WT and the estradiol-inducible null tor-es seedlings at the photoautotrophic transition checkpoint. The minimal endogenous glucose level maximizes the detection sensitivity upon glucose induction, and leads to the identification of 1318 up- and 1050 down-regulated genes to be differentially affected by a physiological level of glucose (Xiong et al., 2013). This swift global transcriptional reprogramming induced by glucose is completely blocked in the tor-es mutant, which has not been reported previously in TOR signaling from extensive mammalian and plant studies (Laplante and Sabatini, 2012; Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013; Cornu et al., 2013; Xiong et al., 2013; Yuan et al., 2013). Glucose-TOR signaling activates large sets of genes involved in glycolysis, the TCA cycle, mitochondria functions, the mitochondrial electron transport chain, ribosomal proteins, and protein synthesis machineries, suggesting a universal and conserved TOR function in controlling translation, and central carbon and energy metabolism (Urban et al., 2007; Laplante and Sabatini, 2012; Moreau et al., 2012; Cornu et al., 2013; Caldana et al., 2013; Xiong et al., 2013; Yuan et al., 2013). Glucose-TOR signaling also controls plant specific genes involved in the seed germination program, the synthesis of cell-wall polymers/proteins, S-assimilation, and lignin and flavonoid synthesis that are uniquely required for plant growth, defense or communication to promote adaptation, fitness and survival (Keurentjes et al., 2006; Moreau et al., 2012; Caldana et al., 2013; Xiong et al., 2013). The transcriptome reprogramming mediated by glucose-TOR signaling at high nitrogen levels shows striking correlations with the gene expression profiles regulated by glucose, sucrose and CO2 in seedlings and adult leaves (Blasing et al., 2005; Gonzali et al., 2006; Li et al., 2006), which partially overlap with KIN10 target genes (Baena-González et al., 2007; Xiong et al., 2013) but are mostly distinct from HXK1 target genes (Li and Sheen, unpublished), suggesting distinct glucose signaling mechanisms. Importantly, TOR senses and transduces shoot photosynthesis-derived glucose signals through the glycolysis and mitochondria energy relay, to specifically control root meristem cell proliferation. Other sugars (fructose, xylose and galactose), amino acids (mix or glutamine) or plant growth hormones (auxin, cytokinin, gibberellin and brassinosteroid) are unable to substitute for glucose in TOR signaling, reinforcing the role of glucose as the main nutrient mediator derived from source leaf photosynthesis in systematic gene regulation and root growth (Xiong et al., 2013).
TOR kinase is activated by photosynthesis-derived glucose signals to promote the exit of quiescent root meristems, and many primary glucose-TOR activated genes match the typical G1- and S-phase genes as the putative targets of Arabidopsis E2Fa transcription factor (Vandepoele et al., 2005; de Jager et al., 2009; Naouar et al., 2009; Xiong et al., 2013). Significantly, direct phosphorylation of E2Fa by TOR kinase is essential for activation of S-phase genes. This finding uncovers a previously unrecognized TOR function in direct transcriptional regulation of cell cycle and provides a compelling novel mechanism for how glucose-TOR signaling controls transcription of S-phase genes and cell cycle to promote root meristem activation and growth (Fig. 2). This molecular mechanism of TOR-E2Fa signaling is distinct from the conventional functions of TOR in stimulating the translation of proteins involved in cell cycle progression through S6K1 and 4E-BP in mammals (Dowling et al., 2010; Laplante and Sabatini, 2012; Xiong et al., 2013; Xiong and Sheen, 2013; Yuan et al., 2013; Xiong and Sheen, 2014). The studies on glucose-TOR signaling illustrate how localized stem/progenitor cell proliferation is activated through inter-organ nutrient coordination to control developmental transition and growth (Xiong et al., 2013; Xiong and Sheen, 2013; Xiong and Sheen, 2014).
Fig. 2.
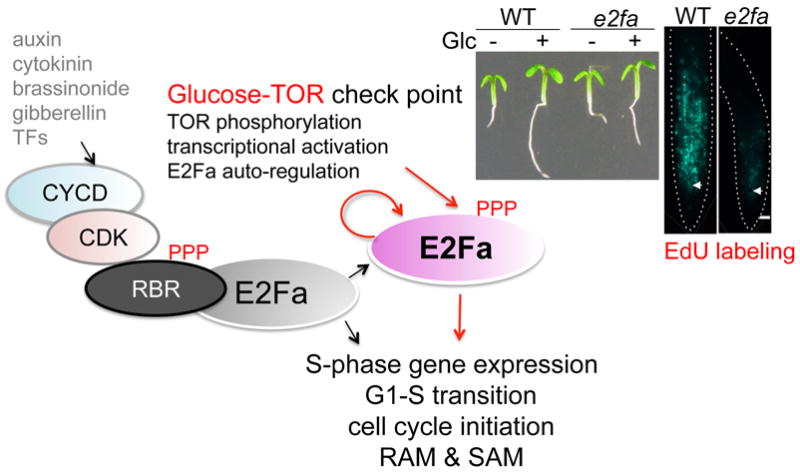
Glucose-TOR-E2Fa signaling regulates the G1 to S transition and promotes cell cycle initiation in meristems. TOR kinase activated by photosynthesis-derived glucose directly phosphorylates E2Fa transcription factor, which leads to the transcriptional activation of S1-phase genes involved in DNA replication in root meristems. The e2fa mutant shows defects in glucose-activated G1-S phase transition and rapid root growth. The glucose-TOR signaling pathway occurs in most cells in the root meristem and is distinct from the conventional pathway mediated by plant growth hormones or cell-specific transcription factors, which activate the expression of CYCD. Elevated CYCD activates CDK, which phosphorylates the RBR suppressor to release E2Fa. CYCD, cyclinD; CDK, cyclin-activated protein kinase; EdU, 5-ethyle-2’-deoxyuridine; RAM, root apical meristem; RBR: retinoblastoma-related; SAM, shoot apical meristem.
Future Perspectives
Understanding how Arabidopsis HXK1 glucose sensor, KIN10/11 energy sensor kinase, and TOR kinase act as master regulators to modulate a myriad of cellular activities and orchestrate the expression of thousands of target genes via multiple partners and effectors in complex signaling networks will continue to be fascinating challenges (Fig. 1). The tools, strategies and knowledge developed in the reference plant system will facilitate future research on glucose signaling networks in other plant species, as well as in animals and humans. Further advances will be enabled by the integration of powerful functional genomic and chemical screens, sophisticated bioinformatic search and prediction, sensitive proteomic/phosphoproteomic analyses, super-resolution microscopy, as well as creative genetic design and research. The identification, characterization and visualization of HXK1 signaling partners and the phosphorylation substrates of KIN10/11 and TOR kinases will unravel the dynamic actions of glucose and energy signaling complexes in various subcellular compartments and in diverse biological contexts. It will be interesting and important to dissect the molecular signaling pathways stimulated by glucose, sucrose, fructose, trehalose/trehalose-6-phosphate, metabolites and other nutrients perceived by different sensors to explore the diverse sugar responses discovered in plants (Schluepmann and Paul, 2009; Wind et al., 2010; Cho and Yoo, 2011; Li et al., 2011; Urano et al, 2012; Eveland and Jackson, 2013; Wahl et al., 2013; de Jong et al., 2014). Future studies will lead to more surprising regulatory mechanisms underlying sugar, energy and hormone connections in different cell types and plant organs intertwined with developmental programs and environmental stimulation.
Acknowledgments
I thank Q Hall, Li Li, Lei Li, K-H Liu, M Ramon, F Rolland, Y Xiong, P. Giavalisco and C. Caldana for unpublished information and discussions. I am grateful to the NSF (IOS-0843244), NIH (R01 GM060493 and R01 GM070567), and WJC Special Project (PJ009106) RDA-Korea for supporting the research in the Sheen lab. Former and current lab members are greatly appreciated for sharing their passion, knowledge and discoveries.
Footnotes
Author’s Contributions
This review has a narrow focus and is based on the 2013 lectures on plant glucose signaling presented by the author, who takes the full responsibility for the selected contents and discussions.
References
- Ahn CS, Han JA, Lee HS, Lee S, Pai HS. The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell. 2011;23:185–209. doi: 10.1105/tpc.110.074005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso R, Oñate-Sánchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W. A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell. 2009;21:1747–1761. doi: 10.1105/tpc.108.062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananieva EA, Gillaspy GE, Ely A, Burnette RN, Erickson FL. Interaction of the WD40 domain of a myoinositol polyphosphate 5-phosphatase with SnRK1 links inositol, sugar, and stress signaling. Plant Physiol. 2008;148:1868–1882. doi: 10.1104/pp.108.130575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GH, Veit B, Hanson MR. The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 2005;3:12. doi: 10.1186/1741-7007-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila J, Gregory OG, Su D, Deeter TA, Chen S, Silva-Sanchez C, Xu S, Martin GB, Devarenne TP. The β-subunit of the SnRK1 complex is phosphorylated by the plant cell death suppressor Adi3. Plant Physiol. 2012;159:1277–1290. doi: 10.1104/pp.112.198432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avonce N, Leyman B, Mascorro-Gallardo JO, van Dijck P, Thevelein JM, Iturriaga G. The Arabidopsis trehalose 6-phosphtae synthase AtTPS1 gene is a regulator of glucose, abscisic acid and stress signalling. Plant Physiol. 2004;136:3649–59. doi: 10.1104/pp.104.052084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein J, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J. Convergent energy and stress signaling. Trends in Plant Science. 2008;13:474–481. doi: 10.1016/j.tplants.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R, Karve A, Kandasamy M, Meagher RB, Moore B. A role of F-actin in hexokinase-mediated glucose signaling. Plant Physiol. 2007;145:1423–1434. doi: 10.1104/pp.107.108704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bako L, Okresz L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C. Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci USA. 1999;96:5322–5327. doi: 10.1073/pnas.96.9.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitrian M, Roodbarkelari F, Horvath M, Koncz C. BAC-recombineering for studying plant gene regulation: developmental control and cellular localization of SnRK1 kinase subunits. Plant J. 2011;65:829–842. doi: 10.1111/j.1365-313X.2010.04462.x. [DOI] [PubMed] [Google Scholar]
- Blasing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidainski J, Ishizaki K, Leaver CJ. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- Caldana C, Li Y, Leisse A, Zhang Y, Bartholomaeus L, Fernie AR, Willmitzer L, Giavalisco P. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 2013;73:897–909. doi: 10.1111/tpj.12080. [DOI] [PubMed] [Google Scholar]
- Celenza JL, Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- Chary SN, Hicks GR, Choi YG, Carter D, Raikhel NV. Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiology. 2008;146:97–107. doi: 10.1104/pp.107.107441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell. 2006;127:579–589. doi: 10.1016/j.cell.2006.09.028. [DOI] [PubMed] [Google Scholar]
- Cho JI, Ryoo N, Eom JS, Lee DW, Kim HB, Jeong SW, Lee YH, Kwon YK, Cho MH, Bhoo SH, Hahn TR, Park YI, Hwang I, Sheen J, Jeon JS. Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 2009;149:745–759. doi: 10.1104/pp.108.131227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Sheen J, Yoo SD. Low glucose uncouples HXK1-dependent sugar signaling from stress and defense hormone ABA and C2H4 responses in Arabidopsis. Plant Physiol. 2010;152:1180–1182. doi: 10.1104/pp.109.148957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD. Signaling role of fructose mediated by FINS1/FBP in Arabidopsis thaliana. PLOS Genet. 2011;7:e1001263. doi: 10.1371/journal.pgen.1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Hong JW, Kim EC, Yoo SD. Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 2012;158:1955–1964. doi: 10.1104/pp.111.189829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeyssen E, Rivoal J. Isozymes of plant hexokinase: occurrence, properties and functions. Phytochem. 2007;68:709–731. doi: 10.1016/j.phytochem.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Confraria A, Martinho C, Elias A, Rubio-Somoza I, Baena-González E. miRNAs mediate SnRK1-dependent energy signaling in Arabidopsis. Front Plant Sci. 2013;4:197. doi: 10.3389/fpls.2013.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento AL, Kim SJ, Bassham DC. Transcriptome profiling of the response of Arabidopsis suspension culture cells to suc starvation. Plant Physiol. 2004;135:2330–2347. doi: 10.1104/pp.104.044362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu M, Albert V, Hall MN. mTOR in aging, metabolism, and cancer. Curr Opin Genet Dev. 2013;23:53–62. doi: 10.1016/j.gde.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, Data SR, Greenberg ME, Licklider LJ, Lowell BB, Gigi SP, Korsmeyer SJ. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424:952–956. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- De Jager SM, Scofield S, Huntley RP, Robinson AS, den Boer BG, Murray JA. Dissecting regulatory pathways of G1/S control in Arabidopsis: common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol Biol. 2009;71:345–365. doi: 10.1007/s11103-009-9527-5. [DOI] [PubMed] [Google Scholar]
- De Jong F, Thodey K, Lejay LV, Bevan MW. Glucose elevates NITRATE TRANSPORTER2.1 protein levels and nitrate transport activity independently of its HEXOKINASE1-mediated stimulation of NITRATE TRANSPORTER2.1 expression. Plant Phyiol. 2014;164:308–320. doi: 10.1104/pp.113.230599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte TL, Sedijani P, Kondou Y, Matsui M, de Jong GJ, Somsen GW, Wiese-Klinkenberg A, Primavesi LF, Paul MJ, Schluepmann H. Growth arrest by trehalose-6-phosphate: an astonishing case of primary metabolite control over growth by way of the SnRK1 signaling pathway. Plant Physiology. 2011;157:160–174. doi: 10.1104/pp.111.180422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Truong HN, Robaglia C, Meyer C. An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem Biophys Res Commun. 2005;326:844–850. doi: 10.1016/j.bbrc.2004.11.117. [DOI] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolai M, Bedu M, Robaglia C, Meyer C. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8:864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich K, Weltmeier F, Ehlert A, Weiste C, Stahl M, Harter K, Dröge-Laser W. Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell. 2011;23:381–395. doi: 10.1105/tpc.110.075390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Marchive C, Azzopardi M, Clement G, Moreau M, Sormani R, Robaglia C, Meyer C. Sugar metabolism and the plant target of rapamycin kinase: a sweet operaTOR? Front Plant Sci. 2013;4:93. doi: 10.3389/fpls.2013.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveal TRF2-dependent T-loop formation. Cell. 2013;155:345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, Kozma SC, Thomas G, Sonenberg N. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveland AL, Jackson DP. Sugars, signaling, and plant development. J Exp Bot. 2013;63:3367–3377. doi: 10.1093/jxb/err379. [DOI] [PubMed] [Google Scholar]
- Farras R, Ferrando A, Jasik J, Kleinow T, Okresz L, Tiburcio A, Salchert K, del Pozo C, Schell J, Koncz C. SKP1-SnRK proten kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 2001;20:2742–2756. doi: 10.1093/emboj/20.11.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Yoshikawa Y, Sato T, Inada N, Ito M, Nishida I, Watanabe A. Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol Plant. 2001;111:345–352. doi: 10.1034/j.1399-3054.2001.1110312.x. [DOI] [PubMed] [Google Scholar]
- Gibson SI. Control of plant development and gene expression by sugar signaling. Curr Opin Plant Biol. 2005;8:93–102. doi: 10.1016/j.pbi.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Gonzali S, Loreti E, Solfanelli C, Novi G, Alpi A, Perata P. Identification of sugar-modulated genes and evidence for in vivo sugar sensing in Arabidopsis. J Plant Res. 2006;119:115–123. doi: 10.1007/s10265-005-0251-1. [DOI] [PubMed] [Google Scholar]
- Gomez LD, Gilday A, Feil R, Lunn JE, Graham IA. AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J. 2010;64:1–13. doi: 10.1111/j.1365-313X.2010.04312.x. [DOI] [PubMed] [Google Scholar]
- Granot D. Role of tomato hexose kinases. Functional Plant Biol. 2007;34:564–570. doi: 10.1071/FP06207. [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari M, Tanurdzic M, Dean A, Nero DC, McClung CR, Coruzzi GM. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA. 2008;105:4939–4944. doi: 10.1073/pnas.0800211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hardie DG. SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol Biol. 1998;37:735–748. doi: 10.1023/a:1006024231305. [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y. Metabolic signaling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J Exp Bot. 2003;54:467–475. doi: 10.1093/jxb/erg038. [DOI] [PubMed] [Google Scholar]
- Hanson J, Hanssen M, Wiese A, Hendrik MMWB, Smeekens S. The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J. 2008;53:935–949. doi: 10.1111/j.1365-313X.2007.03385.x. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature Rev Mol Cell Biol. 2012;13:251–261. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Wang H, Sunter G, Bisaro DM. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell. 2003;15:1034–1048. doi: 10.1105/tpc.009530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisel TJ, Li CY, Grey KM, Gibson SI. Mutations in HISTONE ACETYLETRANSFERASE1 affect sugar response and gene expression in Arabidopsis. Front Plant Sci. 2013;4:245. doi: 10.3389/fpls.2013.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YF, Ho THD, Wu CF, Ho SL, Yeh RH, Lu CA, Chen PW, Yu LC, Chao A, Yu SM. Convergent starvation signals and hormone crosstalk in regulating nutrient mobilization upon germination in cereals. Plant Cell. 2012;24:2857–2873. doi: 10.1105/tpc.112.097741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li CY, Biddle KD, Gibson SI. Identification, cloning and characterization of sis7 and sis10 sugar-insensitive mutants of Arabidopsis. BMC Plant Biol. 2008;8:104. doi: 10.1186/1471-2229-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li CY, Pattison DL, Gray WM, Park S, Gibson SA. SUGAR-INSENSITIVE3, a ring E3 ligase, is a new player in plant sugar response. Plant Physiol. 2010;152:1889–1900. doi: 10.1104/pp.109.150573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li CY, Park S, Gibson SI. SIS8, as putative mitogen-activated protein kinase kinase kinase, regulates sugar resistant seedling development in Arabidopsis. Plant J. 2013 doi: 10.1111/tpj.12404. [DOI] [PubMed] [Google Scholar]
- Hsu YF, Chen YC, Hsiao YC, Wang BJ, Lin SY, Cheng WH, Jauh GY, Harada JJ, Wang CS. AtRH57, a DEAD-box RNA helicase, is involved in feedback inhibition of glucose-mediated abscisic acid accumulation during seedling development and additively affects pre-ribosomal RNA processing with high glucose. Plant J. 2014;77:119–135. doi: 10.1111/tpj.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Jang JC, Leon P, Zhou L, Sheen J. Hexokinase as a sugar sensor in higher plants. Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossier M, Bouly J-P, Meimoun P, Arjmand A, Lessard P, Hawley S, Hardie DG, Martine Thomas M. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signaling in Arabidopsis thaliana. Plant J. 2009;59:316–328. doi: 10.1111/j.1365-313X.2009.03871.x. [DOI] [PubMed] [Google Scholar]
- Kandel-Kfir M, Damari-Weissler H, German MA, Gidoni D, Mett A, Belausov E, Petreikov M, Adir N, Granot D. Two newly identified membrane-associated and plastidic tomato HXKs: characteristics, predicted structure and intracellular localization. Planta. 2006;224:1341–1352. doi: 10.1007/s00425-006-0318-9. [DOI] [PubMed] [Google Scholar]
- Kang SG, Price J, Lin PC, Hong JC, Jang JC. The Arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol Plant. 2010;3:361–373. doi: 10.1093/mp/ssp115. [DOI] [PubMed] [Google Scholar]
- Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB, Sabatini DM. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013;341 doi: 10.1126/science.1236566. 1236566-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karve A, Rauh BL, Xia X, Kandasamy M, Meagher RB, Sheen J, Moore BD. Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta. 2008;228:411–425. doi: 10.1007/s00425-008-0746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karve R, Lauria M, Virnig A, Xia X, Rauh BL, Moore BD. Evolutionary lineages and functional diversification of plant hexokinases. Mol Plant. 2010;3:334–346. doi: 10.1093/mp/ssq003. [DOI] [PubMed] [Google Scholar]
- Karve A, Xia X, Moore BD. Arabidopsis hexokinase-like1 and hexokinase1 form a critical node in mediating plant glucose and ethylene responses. Plant Physiol. 2012;158:1965–1975. doi: 10.1104/pp.112.195636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Davies KA, Bergmann DC, Laux T. Peptide signaling in plant development. Curr Biol. 2011;21:R356–R364. doi: 10.1016/j.cub.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly G, David-Schartz R, Sade N, Moshelion M, Levi A, Alchanatis V, Granot D. The pitfalls of transgenic selection and new roles of AtHXK1: a high level of AtHXK1 expression uncouples hexokinas1-dependent sugar signaling from exogenous sugar. Plant Physiol. 2012;159:47–51. doi: 10.1104/pp.112.196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, de Vos CH, Lommen A, Hall RD, Bino RJ, van der Plas LH, Jansen RC, Vreugdenhil D, Koornneef M. The genetics of plant metabolism. Nat Genet. 2006;38:842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- Kim M, Lim JH, Shn CS, Park K, Kim GT, Kim WT, Pai HS. Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell. 2006;18:2341–2355. doi: 10.1105/tpc.106.041509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Heinzel N, Giese JO, Koeber J, Melzer M, Rutten T, von Wiren N, Sonnewald U, Hajirezaei MR. A dual role of tobacco hexokinase1 in primary metabolism and sugar sensing. Plant cell Env. 2013;36:1311–1327. doi: 10.1111/pce.12060. [DOI] [PubMed] [Google Scholar]
- Kleinow T, Himbert S, Krenz B, Jeske H, Koncz C. NAC domain transcription factor ATAF1 interacts with SNF1-related kinases and silencing of its subfamily causes severe developmental defects in Arabidopsis. Plant Sci. 2009;177:360–370. [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere S, Schmelz EA, Chourey PS. Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol. 2010;153:306–318. doi: 10.1104/pp.110.155226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Chen PW, Lu CA, Chen S, Ho TH, Yu SM. Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci Signal. 2009;2:ra61. doi: 10.1126/scisignal.2000333. [DOI] [PubMed] [Google Scholar]
- Lee H, Chah OK, Plotnikov J, Sheen J. Stem Cell Signaling in Immunity and Development. CSHL Symposium on Quantitative Biology; 2013. Online. [DOI] [PubMed] [Google Scholar]
- Leon P, Sheen J. Sugar and hormone connections. Trends in Plant Science. 2003;8:110–116. doi: 10.1016/S1360-1385(03)00011-6. [DOI] [PubMed] [Google Scholar]
- Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res. 2006;16:414–427. doi: 10.1101/gr.4237406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Wind JJ, Shi X, Zhang H, Hanson J, Smeekens SC, Teng S. Fructose sensitivity is suppressed in Arabidopsis by the transcription factor ANAC089 lacking the membrane-bound domain. Proc Natl Acad Sci USA. 2011;108:3436–3441. doi: 10.1073/pnas.1018665108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JF, Wu SH. molecular events in senescing Arabidopsis leaves. Plant J. 2004;39:612–628. doi: 10.1111/j.1365-313X.2004.02160.x. [DOI] [PubMed] [Google Scholar]
- Lin PC, Pomeranz MC, Jikumaru Y, Kang SG, Hah C, Fujioka S, Kamiya Y, Jang JC. The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J. 2010;65:253–268. doi: 10.1111/j.1365-313X.2010.04419.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Bassham DC. TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS One. 2010;5:e11883. doi: 10.1371/journal.pone.0011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kirubakaran S, Hur W, Niepel M, Westover K, Thoreen CC, Wang J, Ni J, Patricelli MP, Vogel K, Riddle S, Waller DL, Traynor R, Sanda T, Zhao Z, Kang SA, Zhao J, Look AT, Sorger PK, Sabatini DM, Gray NS. Kinome-wide selectivity profiling of ATP-competitive mammalian target of rapamycin (mTOR) inhibitors and characterization of their binding kinetics. J Biol Chem. 2012;287:9742–9752. doi: 10.1074/jbc.M111.304485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Povero G, Novi G, Solfanelli C, Alpi A, Perat P. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 2008;179:1004–1016. doi: 10.1111/j.1469-8137.2008.02511.x. [DOI] [PubMed] [Google Scholar]
- Lova A, Sos-Hegedus A, Bimbo A, Banfalvi Z. Functional diversity of potato SNF1-related kinases tested in Saccharomyces cerevisiae. Gene. 2003;321:123–129. doi: 10.1016/j.gene.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Lu CA, Lin CC, Lee KW, Chen JL, Huang LF, Ho SL, Liu HJ, Hsing YI, Yu SM. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell. 2007;19:2484–2499. doi: 10.1105/tpc.105.037887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM, Kim S, Delauney AJ, Verma DP. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell. 2006;18:477–490. doi: 10.1105/tpc.105.035931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T, Oswald O, Graham IA. Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol. 2002;128:472–481. doi: 10.1104/pp.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci U S A. 2002;99:6422–6427. doi: 10.1073/pnas.092141899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra BS, Singh M, Aggrawal P, Laxmi A. Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PLOS one. 2009;4:e4502. doi: 10.1371/journal.pone.0004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montane MH, Menand B. ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J Exp Bot. 2013;64:4361–4374. doi: 10.1093/jxb/ert242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- Moreau M, Azzopardi M, Clement G, Dobrenel T, Marchive C, Renne C, Martin-Magniette ML, Taconnat L, Renou JP, Robaglia C, Meyer C. Mutations in the Arabidopsis homolog of LST8/GbetaL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell. 2012;24:463–481. doi: 10.1105/tpc.111.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil Y, Uhrig J, Zhou J, Temple B, Jiang K, Jones AM. Arabidopsis N-MYC DOWNREGULATED-LIKE1, a positive regulator of auxin transport in a G protein–mediated pathway. Plant Cell. 2009;21:3591–3609. doi: 10.1105/tpc.109.065557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naouar N, Vandepoele K, Lammens T, Casneuf T, Zeller G, van Hummelen P, Weigel D, Ratsch G, Inze D, Kuiper M, De Veylder L, Vuylsteke M. Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J. 2009;57:184–194. doi: 10.1111/j.1365-313X.2008.03662.x. [DOI] [PubMed] [Google Scholar]
- Ng S, Giraud E, Duncan O, Law SR, Wang Y, Xu L, Narsai R, Carrie C, Walker H, Day DA, Blanco NE, Strand A, Whelan J, Ivanova A. Cyclin-dependnet kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J Biol Chem. 2013;288:3449–3459. doi: 10.1074/jbc.M112.416727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson A, Olsson T, Ulfstedt M, Thelander M, Ronne H. Two novel types of hexokinases in the moss Physcomitrella patens. BMC Plant Biol. 2011;11:32. doi: 10.1186/1471-2229-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obertello M, Krouk G, Katari MS, Runko SJ, Coruzzi GM. Modeling the global effect of the basic-leucine zipper transcription factor 1 (bZIP1) on nitrogen and light regulation in Arabidopsis. BMC Sys Biol. 2010;4:111. doi: 10.1186/1752-0509-4-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M, Hayashi S, Sawa S, Hashimoto-Ohta A, Nakamura K. Involvement of HLS1 in sugar and auxin signaling in Arabidopsis leaves. Plant cell Physiol. 2006;47:1603–1611. doi: 10.1093/pcp/pcl027. [DOI] [PubMed] [Google Scholar]
- Paul MJ, Primavesi LF, Jhurreea D, Zhang Y. Trehalose metabolism and signaling. Annu Rev Plant Biol. 2008;59:417–441. doi: 10.1146/annurev.arplant.59.032607.092945. [DOI] [PubMed] [Google Scholar]
- Piattoni CV, Bustos DM, Guerrero SA, Iglesias AA. Nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase is phosphorylated in wheat endosperm at serine-404 by an SNF1-related protein kinase allosterically inhibited by ribose-5-phosphate. 2011;156:1337–1350. doi: 10.1104/pp.111.177261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C, Thomas M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007;12:20–28. doi: 10.1016/j.tplants.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC. Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell. 2004;16:2128–2150. doi: 10.1105/tpc.104.022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell PC, Smith AM, Halford NG. Antisense expression of a sucrose non-fermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. The Plant Journal. 1998;14:195–202. [Google Scholar]
- Radchuk R, Emery RJN, Weier D, Vigeolas H, Geigenberger P, Lunn JE, Feil R, Weschk W, Weber H. Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. Plant J. 2009;61:324–338. doi: 10.1111/j.1365-313X.2009.04057.x. [DOI] [PubMed] [Google Scholar]
- Rahmani F, Hummel M, Schuurmans J, Wiese-Klinkenberg A, Smekens S, Hanson J. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol. 2009;50:1356–1367. doi: 10.1104/pp.109.136036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M, Rolland F. Plant development: introducing trehalose metabolism. Trend Plant Sci. 2007;12:185–188. doi: 10.1016/j.tplants.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Ramon M, Rolland F, Sheen J. Sugar sensing and signaling in Arabidopsis. The Arabidopsis Book. 2008 doi: 10.1199/tab.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M, De Smet I, Vandesteene L, Naudts M, Leyman B, Van Dijck P, Rolland F, Beeckman T, Thevelein JM. Extensive expression regulation and lack of heterologous enzymatic activity of the Class II trehalose metabolism proteins from Arabidopsis thaliana. Plant, Cell and Environment. 2009;32:1015–1032. doi: 10.1111/j.1365-3040.2009.01985.x. [DOI] [PubMed] [Google Scholar]
- Ramon M, Ruelens P, Li Y, Sheen J, Geuten K, Rolland F. The hybrid Four-CBS-Domain KINβγ-subunit functions as the canonical γ-subunit of the plant energy sensor SnRK1. Plant J. 2013;75:11–25. doi: 10.1111/tpj.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Qiu S, Venglat P, Xiang D, Feng L, Selvaraj G, Datla R. Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 2011;155:1367–1382. doi: 10.1104/pp.110.169045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Venglat P, Qiu S, Feng L, Cao Y, Wang E, Xiang D, Wang J, Alexander D, Chalivendra S, Logan D, Mattoo A, Selvaraj G, Datla R. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell. 2012;24:4850–4874. doi: 10.1105/tpc.112.107144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia C, Thomas M, Meyer C. Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr Opin Plant Biol. 2012;15:301–307. doi: 10.1016/j.pbi.2012.01.012. [DOI] [PubMed] [Google Scholar]
- Rodrigues A, Adamo M, Crozet P, Margalha L, Confrairia A, Martinho C, Elias A, Rabissi A, Lumbreras V, González-Guzmán M, Antoni R, Rodriguez PL, Baena-González E. ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell. 2013;25:3871–3884. doi: 10.1105/tpc.113.114066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J. Sugar sensing and signaling in plants. Plant Cell. 2002;14(Suppl):S185–205. doi: 10.1105/tpc.010455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- Ryu JY, Jeong SW, Kim SY, Ko Y, Yoon S, Choi SB, Park YI. Cyanobacterial glucokinase complements the glucose sensing role of Arabidopsis thaliana hexkinase1. Bioch Bioph Res Comm. 2008;374:454–459. doi: 10.1016/j.bbrc.2008.07.041. [DOI] [PubMed] [Google Scholar]
- Santner A, Calderon-Villalobos LI, Estelle M. Plant hormones are versatile chemical regulators of plant growth. Nat Chem Biol. 2009;5:301–307. doi: 10.1038/nchembio.165. [DOI] [PubMed] [Google Scholar]
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature. 2006;441:227–30. doi: 10.1038/nature04725. [DOI] [PubMed] [Google Scholar]
- Sheen J. Metabolic repression of transcription in higher plants. Plant Cell. 1990;2:1027–1038. doi: 10.1105/tpc.2.10.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Discover and connect cellular signaling. Plant Physiol. 2010;154:562–566. doi: 10.1104/pp.110.161364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martinez E, Geldreich A, Keller M, Ryabova LA. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013;32:1087–1102. doi: 10.1038/emboj.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H, van Dijken A, Aghdasi M, Wobbes B, Paul M, Smeekens S. Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiology. 2004;135:879–890. doi: 10.1104/pp.104.039503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H, Paul M. Trehalose metabolites in Arabidopsis-elusive, active and central. Arabidopsis Book. 2009;7:e0122. doi: 10.1199/tab.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtje J, Minchin PEH, Jahnke S, van Dongen JT, Schittko U, Baldwin IT. SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc Natl Acad Sci USA. 2006;103:12935–12940. doi: 10.1073/pnas.0602316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S, Ma J, Hanson J, Rolland F. Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol. 2010;13:274–279. doi: 10.1016/j.pbi.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Szczesny R, Büttner D, Escolar L, Schulze S, Seiferth A, Bonas U. Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytol. 2010;187:1058–1074. doi: 10.1111/j.1469-8137.2010.03346.x. [DOI] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 1999;120:257–274. doi: 10.1104/pp.120.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander M, Olsson T, Ronne H. Snf1-related protein kinase 1 is needed for growth in a normal day–night light cycle. EMBO J. 2004;23:1900–1910. doi: 10.1038/sj.emboj.7600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibson Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Toroser D, Plaut Z, Huber SC. Regulation of a plant SNF1-related protein kinase by glucose-6-phosphate. Plant Physiol. 2000;123:403–412. doi: 10.1104/pp.123.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AYL, Gazzarrini S. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 2012;69:809–821. doi: 10.1111/j.1365-313X.2011.04832.x. [DOI] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Urano D, Phan N, Jones JC, Yang J, Huang J, Grigston J, Taylor JP, Jones AM. Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nature Cell Biol. 2012;14:1079–1088. doi: 10.1038/ncb2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijken AJH, Schluepmann H, Smeekens SCM. Arabidopsis trehalose 6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol. 2004;135:969–77. doi: 10.1104/pp.104.039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GT, Gruissem W, Van de Peer Y, Inze D, De Veylder L. Genome-wide identification of potential plant E2F target genes. Plant Physiol. 2005;139:316–328. doi: 10.1104/pp.105.066290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Frank A, Feil R, Lunn JE, Stitt M, Schmid M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science. 2013;339:704–707. doi: 10.1126/science.1230406. [DOI] [PubMed] [Google Scholar]
- Wind J, Smeekens S, Hanson J. Sucrose: metabolite and signaling molecule. Phytochem. 2010;71:1610–1614. doi: 10.1016/j.phytochem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Xiao W, Sheen J, Jang JC. Multiple sugar signal transduction pathways in plants. Plant Mol Biol. 2000;44:451–461. doi: 10.1023/a:1026501430422. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem. 2012;287:2836–2842. doi: 10.1074/jbc.M111.300749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J. Glucose-TOR signaling reprograms the transcriptome and activates meristems. Nature. 2013;496:181–186. doi: 10.1038/nature12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. Moving beyond translation: glucose-TOR signaling in the transcriptional control of cell cycle. Cell Cycle. 2013;12:1989–1990. doi: 10.4161/cc.25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 2014;164:499–512. doi: 10.1104/pp.113.229948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J. Differential regulation of EIN3 stability by glucose and ethylene signaling in plants. Nature. 2003;425:521–525. doi: 10.1038/nature01984. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature. 2008;451:789–795. doi: 10.1038/nature06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan HX, Xiong Y, Guan KL. Nutrient sensing, metabolism, and cell growth control. Mol Cell. 2013;49:379–387. doi: 10.1016/j.molcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andraloj PJ, Mitchell RAC, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 2009;149:1860–1871. doi: 10.1104/pp.108.133934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZW, Yaun S, Xu F, Yang H, Zhang NH, Cheng J, Lin HH. The plastid hexokinase pHXK: a node of convergence for sugar and plastid signals in Arabidopsis FEBS Lett. 2010;584:3573–35790. doi: 10.1016/j.febslet.2010.07.024. [DOI] [PubMed] [Google Scholar]


