Abstract
Background
Identification of primary melanoma patients at the highest risk of recurrence remains a critical challenge, and monitoring for recurrent disease is limited to costly imaging studies. We recently reported our array-based discovery of prognostic serum miRNAs in melanoma. In the current study, we examine the clinical utility of these serum-based miRNAs for prognosis as well as detection of melanoma recurrence.
Methods
Serum levels of 12 miRNAs were tested using qRT-PCR at diagnosis in 283 melanoma patients (training cohort n=201, independent validation n=82; median FU 68.8 months). A refined miRNA signature was chosen and evaluated. We also tested the potential clinical utility of the miRNAs in early detection and monitoring of recurrence using multiple longitudinal samples (pre- and post-recurrence) in a subset of 82 patients (n=225). In addition, we integrated our miRNA signature with publicly available TCGA data to examine the relevance of these miRNAs to melanoma biology.
Results
Four miRNAs (miR-150, miR-30d, miR-15b, and miR-425) in combination with stage separated patients by recurrence-free and overall survival and improved prediction of recurrence over stage alone in both training and validation cohorts (training RFS and OS p<0.001, validation RFS p<0.001, OS p=0.005). Serum miR-15b levels significantly increased over time in recurrent patients (p<0.001), adjusting for endogenous controls as well as age, gender, and initial stage. In non-recurrent patients, miR-15b levels were not significantly changed with time (p=0.17).
Conclusion
Data demonstrate that serum miRNAs can improve melanoma patient stratification over stage and support further testing of miR-15b to guide patient surveillance.
Keywords: Melanoma, MicroRNAs, Biomarkers, Serum Markers, Recurrence
BACKGROUND
Melanoma, the deadliest of skin cancers, is increasing in incidence and remains virtually incurable in the metastatic setting, despite recent advances in targeted therapies1. While clinical stage is used to stratify patients at time of primary diagnosis2, stage alone cannot account for heterogeneity in patients’ outcomes, highlighting the need for novel molecular biomarkers that are easily measurable at the time of diagnosis3. In addition, there is a need for biomarkers to monitor primary melanoma patients for disease recurrence, as regular imaging studies are not cost-efficient with estimated costs of over $20,000 per recurrence in patients diagnosed with stage 1 and II disease4.
Blood-based biomarkers are attractive due to the ease and minimal invasiveness of collecting multiple blood samples compared to repeat tissue sampling. In this regard, we recently reported our array-based screening of 355 microRNAs (miRNAs) to identify miRNAs with potential as predictors of recurrence5. While the signature was particularly reliable in stage II patients, the study was limited by an imbalanced patient cohort by stage, making the applicability of the signature to the general patient population uncertain. In addition, as the signature was focused on discovery rather than on clinical applicability, it did not include endogenous controls for normalization of miRNA levels.
In the current study, we examined the levels of 12 pre-selected miRNAs in the serum of melanoma patients collected at the time of primary diagnosis, using expanded training and validation cohorts with extended prospective follow up. In addition, we examined the potential utility of the chosen miRNAs as markers of recurrence and melanoma progression and performed bioinformatics analysis to predict the functional relevance of the identified serum-based miRNAs.
PATIENTS AND METHODS
Patient Accrual and Sample Collection
A cohort of 283 primary (AJCC stage I-III2) melanoma patients prospectively enrolled in the New York University Interdisciplinary Melanoma Cooperative Group (IMCG) from September 2002 to September 2011 was identified. All recurred patients in the database with primary serum samples available were included. To ensure a reasonable balance of each clinical stage in the non-recurred group, patients were identified according to clinical stage (I, II, and III), with target accrual for stage II and stage III patients proportionally higher than in the general melanoma population. Within each stage, patients with available primary serum samples were identified based on length of follow-up. A subset of 82 patients was reserved for an independent validation cohort. Patient demographics and primary tumor pathological characteristics were obtained for each patient at the time of enrollment. Recurrence and survival information are prospectively obtained via active follow-up every six months for all patients. The study was approved by the Institutional Research Board at NYU in accord with a Federalwide Assurance approved by the Department of Health and Human Services, and all patients provided informed written consent at the time of enrollment.
Blood was collected in BD Vacutainer serum tubes from each patient at the time of diagnosis and immediately stored at 4°C. Additionally, in a subset of 82 patients from the training and validation cohorts, multiple longitudinal blood samples drawn during regular clinical follow-up visits were collected. Within two hours of collection, samples were centrifuged for 10 minutes at 750 x g. The supernatant serum was divided into 1 mL aliquots and stored in cryovials at −80°C. Levels of 18-pre selected miRNAs (12 as biomarker candidates, 5 as normalizer candidates, and 1 to assess sample hemolysis) were measured using qPCR (see Supporting Information section for detailed methods). The 12 candidate biomarker miRNAs were all identified in our previously published data as predictive of recurrence in multivariable discovery models and/or as statistically associated with disease recurrence longitudinally5. The 5 potential normalizers were identified as stably expressed or unaffected by hemolysis in our previous array data and validated in preliminary testing in control and melanoma patients (Supporting Information Table S1). Following the miRNA qPCR, two miRNAs, miR-30c-5p and miR-181a-5p, were selected as the most stable miRNA using NormFinder6. Test miRNA Cp values were normalized to the average Cp of the two most stable miRNAs.
Statistical Methods
Using the training cohort (n=201), the 12 candidate miRNAs were ranked by univariate association of serum levels with recurrence via logistic regression analysis with adjustment for tumor stage (I, II, or III) as a categorical variable. All miRNAs were standardized into mean 0 and unit variance variables for scale consistency across cohorts. The 4 miRNA-signature was selected by minimizing Akaike’s information criterion (AIC) of the logistic regression model through backward stepwise selection. The linear combination of model predictors weighted by regression coefficients was defined as the risk score. The selected miRNA signature was evaluated by identifying the area under the Receiver Operating Characteristic (ROC) curve (AUC). An optimal risk score cutoff, maximizing specificity subject to a minimum value for sensitivity at 80% was chosen to classify patients into high and low risk groups7–9. Kaplan-Meier survival curves and log-rank tests were used to compare the recurrence free survival and overall survival distributions of the two groups. Regression coefficients of the logistic regression model were then applied to an independent validation cohort (n=82) to classify patients. The same cutoff was used to classify patients into high- and low-risk groups. All statistical analyses were performed in R 2.14.0.
To test the ability of the signature miRNAs to monitor for melanoma recurrence, we measured miRNA levels, including the signature miRNAs and the normalizers (miR-30c and miR-181a), in 82 patients (34 recurred, 48 non-recurred) with at least two serum samples available, drawn at different time points (n=225 samples total). All available samples from the 82 patients were analyzed; of note, 26 recurred patients had serum samples tested both pre- and post-recurrence. For each normalized miRNA in the identified signature, we used a linear mixed effect model to analyze the normalized serum levels of the miRNA as a function of time since recurrence (negative values representing sampling time pre-recurrence), adjusting for age, gender, and initial tumor stage at diagnosis in the recurred patients. In addition, we fit a separate linear mixed effect model in the non-recurred patients to analyze miRNA levels as a function of time since diagnosis.
To identify messenger RNAs (mRNAs) most closely associated with the signature miRNAs, we obtained the 20,531 mRNA (level 3 normalized RNAseq results on gene levels) and 1,046 miRNAs (level 3 normalized results) on 325 Skin Cutaneous Melanoma (SKCM) cases from the Cancer Genome Atlas (TCGA) Data Portal (http://tcga-data.nci.nih.gov/tcga/findarchives.htm). We first filtered mRNAs with no between-sample variations (n=405 mRNAs). All miRNAs and mRNAs were first log2 transformed, then standardized into mean 0 and unit variance variables for scale consistency in the multivariate regression. An L1 penalized regression approach (Lasso) was used to identify the mRNAs associated with each miRNA with the miRNA as dependent variable. A linear coefficient and p-value were estimated for each mRNA predictor in the Lasso regression, reflecting the mRNA’s association strength and significance to the miRNA. A threshold of association correlation = 0.1 and p=0.05 were used to define the set of significantly associated mRNAs of each miRNA. The Lasso regression was implemented by glmnet() function in R. As additional analysis to identify potential direct mRNA targets, we also filtered the results to only those mRNAs (a) negatively associated with each serum miRNA and (b) predicted to be targeted by the serum miRNA using TargetScan software10.
For each miRNA, we analyzed the selected potential target mRNAs using gene set enrichment analysis software package (GSEA; http://www.broadinstitute.org/gsea/index.jsp) to find the enriched molecular pathways. Pathways were selected with significant p-values (<0.01) and FDR (<0.01). Key genes in each selected pathway were identified.
RESULTS
Table 1 illustrates the demographic and clinicopathological characteristics for the training (n=201) and independent validation (n=82) cohorts, demonstrating that the two cohorts are well-matched in all categories, including recurrence and survival status. The median time of follow-up for survivors was 90.9 months (range, 17.5–128.9 months) in the training cohort, and 39.0 months (range, 0.8–95.0 months) in the validation cohort.
Table 1.
Baseline demographic and primary tumor characteristics of Training and Independent Validation Cohorts
| Characteristic n (%) | Training Cohort (n=201) | Independent Validation Cohort (n=82) | Total (n=283) |
|---|---|---|---|
| Gender | |||
| Male | 112 (56) | 52 (63) | 164 (58) |
| Female | 89 (44) | 30 (37) | 119 (42) |
| Age, median (range) | 61 (21–96) | 59.5 (25–88) | |
| Thickness, median (range) | 1.5 (0.16–30) | 1.3 (0.27–28) | |
| Ulceration | |||
| Present | 65 (32) | 24 (29) | 89 (31) |
| Absent | 136 (68) | 58 (71) | 194 (69) |
| Mitoses | |||
| Present | 141 (70) | 60 (73) | 201 (71) |
| Absent | 58 (29) | 22 (27) | 80 (28) |
| Unclassified | 2 (1) | 0 (0) | 2 (1) |
| Initial Stage & Recurrence Status | |||
| I | 89 (44) | 41 (50) | 130 (46) |
| Recurrent | 14 (16) | 6 (15) | 20 (15) |
| Non-Recurrent | 75 (84) | 35 (85) | 110 (85) |
| II | 52 (26) | 20 (24) | 72 (25) |
| Recurrent | 15 (29) | 7 (35) | 22 (31) |
| Non-Recurrent | 37 (71) | 13 (65) | 50 (69) |
| III | 60 (30) | 21 (26) | 81 (29) |
| Recurrent | 33 (55) | 13 (62) | 46 (57) |
| Non-Recurrent | 27 (45) | 8 (38) | 35 (43) |
| Follow-up time in months, median (range) | 90.9 (17.5–128.9) | 39.0 (0.8–95.0) | 68.8 (0.8, 128.9) |
| Status as last follow-up | |||
| Alive | 157 (78) | 62 (76) | 219 (77) |
| Died, of Melanoma | 39 (19) | 18 (22) | 57 (20) |
| Died, Other Causes | 5 (2) | 2 (2) | 7 (2) |
A serum-based miRNA signature predicts recurrence
Using serum miRNA data from a training cohort of 201 melanoma patients, we developed a prognostic model of melanoma recurrence, which we subsequently validated in an independent patient cohort. The 4-miRNA signature (miR-150, miR-15b, miR-425, miR-30d) normalized by two endogenous controls (miR-30c and miR-181a) can effectively distinguish recurrent cases from non-recurrent cases. In combination with stage, this miRNA classifier displayed an area under the receiver operating characteristic (ROC) curve AUC=0.760 (95% CI= (0.687, 0.833)) in the training cohort. Applying this classifier to the independent validation (n=82) cohort yielded AUC of 0.790 (95% CI = (0.684, 0.897). In contrast, using the same patient cohorts, a model using clinical stage alone achieved AUCs of 0.704 and 0.759, respectively. AUC values of the miRNA-based predictive model within each clinical stage can be found in Supporting Information, Table S2.
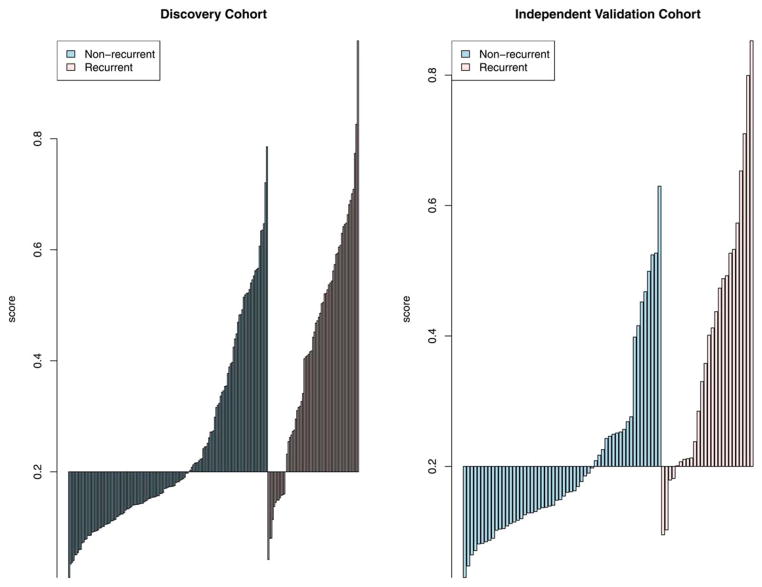
We defined risk scores for individual patients as the linear combination of model predictors weighted by regression coefficients. Based on the cut-off value selected to classify patients into high- and low-risk groups, the sensitivity was 80.9% and 84.6% and the specificity was 60.1% and 66.1%, respectively, in the two cohorts (Figure 1). For patients predicted to be negative for recurrence by the miRNA signature in the two cohorts (negative predictive value), 87.4% and 90.2% patients have not recurred, which supports the clinical sensitivity of the signature.
Figure 1. Sensitivity and specificity for prediction of recurrence using a risk score derived from miRNA-based predictive model.
An optimal risk score cutoff (horizontal line) maximizing specificity subject to a minimum value for sensitivity at 80% was chosen to classify patients into high and low risk groups. (A) Training cohort, (B) Independent validation cohort.
We repeated the training and validation processes in the subsets of recurrent patients and non-recurrent patients with at least 3 years of follow-up of recurrence free survival (n=200 and 59 in the training and validation cohorts, respectively). The resulting miRNA signature identified was the same as those selected when all patients were used. Using this follow-up restriction, the AUCs obtained with the same miRNA + stage classifier applied to the training and validation cohorts were 76 and 80%, respectively. Collectively, these results demonstrate that serum levels of a small set of miRNAs can improve prediction of recurrence when used in combination with clinical stage.
Four-miRNA classifier accurately stratifies patients according to recurrence-free survival and overall survival
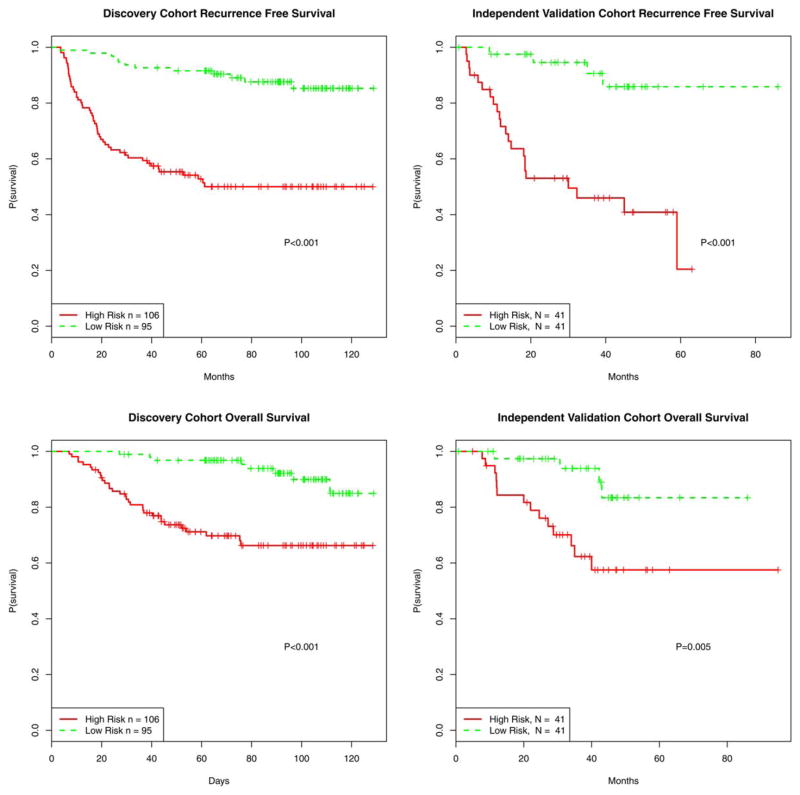
To assess the ability of the miRNA signature to stratify patients by survival, high- and low-risk groups were plotted in Kaplan-Meier survival curves and statistical analysis performed by log-rank tests. High- and low-risk groups displayed clear differences in recurrence free survival (p<0.001) and overall survival (p<0.001) (Figure 2). When applying the same cutoff risk scores to the independent validation cohort (n=82), a significant segregation of patients based on recurrence free survival (p<0.001) and overall survival (p=0.005) was observed (Figure 2). These analyses underscore the robustness of our miRNA-based classifier in predicting melanoma patient outcome.
Figure 2. Recurrence-free and overall survival of patients stratified by the miRNA-based predictive model.
Using the risk score previously defined by the miRNA-based predictive model, high and low risk groups were plotted in Kaplan-Meier survival curves and statistical analysis performed by log-rank tests.
miR-15b levels are significantly increased over time from pre-recurrence to post-recurrence
To test the ability of the signature miRNAs to monitor for melanoma recurrence, we measured miRNA serum levels in 82 patients with at least two serum samples available (n=225 samples total), drawn at different time points pre- and post-recurrence. Starting pre-recurrence and moving to post-recurrence, serum levels of miR-15b were significantly increased over time in recurrent patients (p<0.001, Figure 3; see also Supporting Information, Figure S1), adjusting for levels of endogenous controls as well as patients’ age, gender, and initial tumor stage at diagnosis in the linear mixed effect model. In non-recurrent patients, miR-15b levels were not significantly changed with time since diagnosis (p=0.17, Supporting Information Figure S2), suggesting that miR-15b has potential to act as a specific early marker for melanoma recurrence.
Figure 3. Normalized levels of miR-15b measured in multiple serum samples from recurrent patients over time.
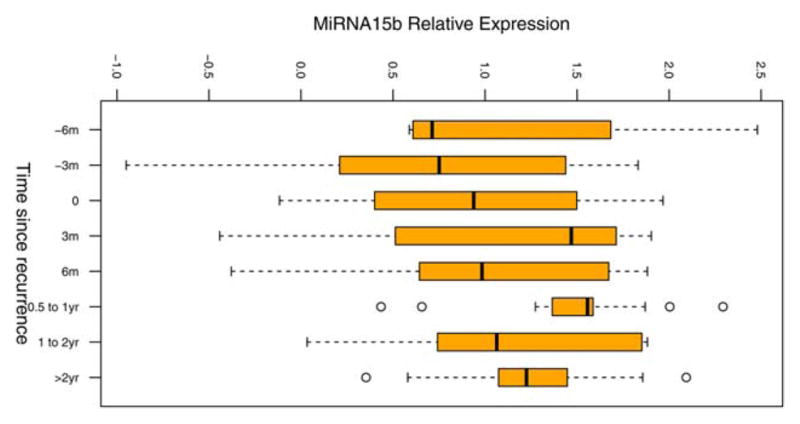
Serum miR-15b levels were measured in a subset of patients. Recurrent patients (n=34) are shown. Each patient had at least two serum samples, drawn at different time points, tested. t=0 at date of first recurrence. Serum level of miR-15b increases leading up to recurrence.
Signature miRNAs are associated with immune signaling, melanogenesis, and the cell cycle
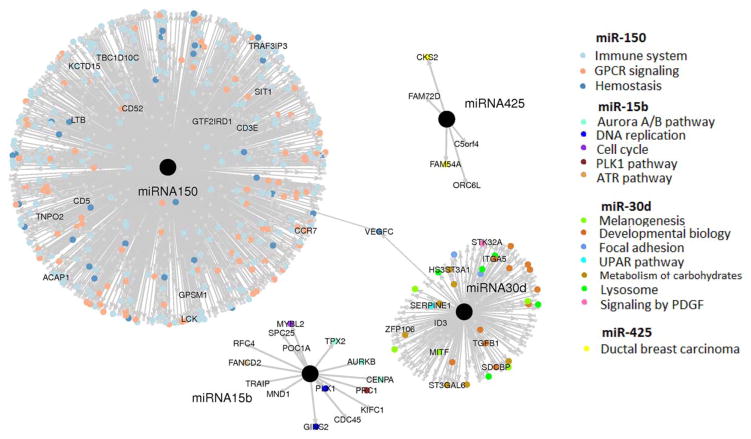
To understand the biological relevance of the signature miRNAs, we performed data mining of the TCGA Skin Cutaneous Melanoma dataset to define the set of significantly associated mRNAs with each miRNA (Figure 4). In order to identify all downstream effects, positively and negatively associated mRNAs were initially included11. We further identified the key functional roles of the miRNA-associated mRNAs (Table 2). Genes most strongly associated with miR-150 include immune system related genes, cytokine signaling genes, and numerous G protein-coupled receptor signaling genes. miR-30d was associated with MITF, tyrosinase and other genes involved in melanogenesis and developmental biology. miR-15b was associated with aurora B kinase, centromere protein A, and several other factors involved in the cell cycle and DNA replication. Finally, miR-425 had a small number of associated mRNAs (n=2) that included CKS2 and FAM54A, genes which have been implicated in breast cancer12.
Figure 4. Key mRNAs and functional roles associated with signature miRNAs as identified in TCGA skin cutaneous melanoma dataset.
An L1 penalized regression approach (Lasso) was used to identify the mRNAs associated with each miRNA. A linear coefficient and p-value were estimated for each mRNA predictor in the Lasso regression, reflecting the mRNA’s association strength and significance to the miRNA. mRNA targets with the strongest association with signature miRNAs are identified by colored dots representing their predicted functional significance according to GSEA analysis.
Table 2.
Summary of major functional pathways associated with signature miRNAs
| microRNA | Gene Set Description | # of Genes | Key Genes | P-value | FDR Q-Value |
|---|---|---|---|---|---|
| miR-150 | Immune System | 107 | VAV1, RASGRP1/2 | <0.001 | <0.001 |
| Cytokine Signaling in Immune System | 27 | PTAFR | <0.001 | <0.001 | |
| Signaling by GPCR | 53 | CCXR3, CCL21 | <0.001 | <0.001 | |
| miR-30d | Melanogenesis | 5 | MITF, GNAI2, TYR | <0.001 | 0.005 |
| Developmental Biology | 11 | TGFB1, ITGA5 | <0.001 | <0.001 | |
| miR-15b | Cell Cycle | 6 | AURKB, CENPA | <0.001 | <0.001 |
| Cell Cycle (mitotic) | 6 | AURKB, CENPA | <0.001 | <0.001 | |
| DNA Replication | 5 | AURKB, CENPA | <0.001 | <0.001 | |
| miR-425 | Up-regulated in ductal carcinoma tumor cells | 2 | CKS2, FAM54A | <0.001 | 0.005 |
We further narrowed the analysis to identify potential direct targets of the key miRNAs. As miRNAs typically repress the expression of specific target mRNAs, we focused on those mRNAs that were (a) negatively associated with each serum miRNA and (b) predicted to be targeted by the serum miRNA using TargetScan software10. This analysis yielded a number of interesting putative target mRNAs for miR-30d in particular, including ITGA5, SERPINE1, and ADAM19.
DISCUSSION
This study tested serum levels of a subset of pre-selected miRNAs in the blood of melanoma patients at the time of primary diagnosis and longitudinally over time. Our data demonstrate that a signature composed of four miRNAs combined with stage had superior prognostic capabilities over stage alone and accurately stratified patients by recurrence-free and overall survival. Second, our results demonstrated that serum levels of miR-15b can potentially be used for early detection and monitoring of disease recurrence in melanoma patients. Finally, our bioinformatics analysis of TCGA data revealed that the signature miRNAs are predicted to have involvement in key processes relevant to melanoma biology, including immunogenic signaling, melanogenesis, and DNA replication.
Our finding that serum-based miRNAs provide improved prognostic value in primary melanoma patients has important implications for the management of these individuals. The use of levels of circulating miRNAs in the clinical setting at the time of diagnosis could determine surveillance strategies by informing clinicians of the risk of disease recurrence. With the high sensitivity of our predictive model, patients with a low risk score can reasonably be spared aggressive adjuvant treatment regimens and costly imaging studies, even those with more advanced clinical stage. Meanwhile, for stage I patients, regular surveillance is currently highly cost-inefficient on a population level due to their relatively low recurrence rate4. Nevertheless, stage I cases account for a substantial proportion of total recurrences due to their high volume of between 56–71% of melanoma diagnoses13, 14, and identification of those specific patients at a higher risk could impart extraordinary cost savings without compromising patient care.
Our results also demonstrate that serum levels of miR-15b have potential utility for the early detection of melanoma recurrence when measured regularly following primary treatment. In our cohort, serum levels of miR-15b increased steadily in the six months prior to recurrence and then decreased shortly after recurrence, possibly reflecting early progression of disease followed by response to treatment. No significant change was observed in non-recurrent patients over time. While these results represent a small number of patients and require prospective validation in an independent cohort, this use of blood-based miR-15b has the potential to significantly improve the efficiency of follow-up care for primary melanoma patients. Evidence suggests that regular follow-up imaging studies alone are not efficient at detecting recurrent disease, due to high false-negative and false-positive rates as well as prohibitive cost when employed regularly4, 15. As such, a regularly administered blood-based miRNA assay can serve as an inexpensive and non-invasive method to identify patients at need for targeted imaging.
The increase of miR-15b levels just prior to recurrence is consistent with multiple lines of evidence suggesting a role of miR-15b in melanoma progression and metastasis. Genes associated with miR-15b revealed in our TCGA analysis include cell cycle and mitotic regulation genes, most notably aurora B kinase and centromere protein A. In addition, miR-15b presence in melanoma tissues has been associated with poorer recurrence-free and overall survival in a retrospective analysis16. Interestingly, despite our finding that miR-15b levels increased at the time of recurrence, our data show that lower levels of miR-15b are a negative prognostic factor at the time of diagnosis. These results suggest that miR-15b levels are involved in different functional processes at various stages of melanoma disease. Specifically, the increase in miR-15b at the time of recurrence is consistent with its putative functions in progression and metastasis, while the low levels at the time of diagnosis could reflect a different process that has yet to be identified.
The other miRNAs found in our signature predictive of recurrence are also predicted to play roles in key melanoma processes. Our group recently reported that miR-30d plays a role in melanoma invasion and metastasis17, a hypothesis which is further supported by our TCGA analysis revealing that miR-30d is associated with mRNA levels of microphthalmia-associated transcription factor (MITF), involved in melanogenesis and a potential melanoma oncogene18, 19. Furthermore, we identified a number of mRNAs predicted to be directly targeted by miR-30d, including ITGA5, SERPINE1 and ADAM19, which have been implicated in melanoma and/or other cancer progression20–22. For miR-150, our analysis showed that putative downstream mRNAs associated include numerous chemokine receptors and ligands that are heavily involved in immune signaling pathways and the development and activation of immune cells. Interestingly, it was recently shown in vivo that miR-150 is released in the external milieu upon T cell activation, resulting in a decrease of its intracellular levels and an increase of its levels in the serum, as a consequence of lymphocyte activation23. Accordingly, it is possible that miR-150 found in melanoma serum is reflective of tumor-infiltrating lymphocytes (TILs), accounting for the positive association between miR-150 upregulation and lower risk of recurrence, possibly via melanoma-related chemokines such as CXCR3 and CCL2124–26. Finally, although miR-425 has been mentioned in the context of non-melanoma cancers27, 28, its precise role in melanoma remains unclear and TCGA analysis revealed only a small number of connections to putative downstream genes (CKS2, FAM54A) that may play a role in breast cancer12. Our data suggest that further investigation of miR-425 is warranted as a novel potential biomarker in melanoma.
One question that remains unresolved is the source of circulating miRNAs. For example, in our current signature, lower levels of miR-30d were associated with a higher risk of melanoma recurrence, despite earlier work from our group suggesting a role for miR-30d in tumor progression17. This discrepancy suggests that circulating miRNAs may not always be shed directly from tumor tissue. Indeed, other studies have also reported differences between miRNA levels in tumor tissue and circulating levels of miRNAs as measured in serum29, 30, and in our own group’s work in human melanoma tissues, we have observed only partial concordance between tissue-based and serum-based miRNAs31, 32. As such, the functional roles of miR-30d and the other signature miRNAs need to be further explored with regard to their levels in circulating fluids.
Similarly, our results have identified different miRNAs from other groups that have reported results from serum-based miRNAs in melanoma33–36. However, unlike the other reports in the literature, our study is unique in identifying miRNAs that are relevant to recurrence risk when measured at the time of primary diagnosis. Other studies have measured miRNAs in metastatic melanoma compared to disease-free patients33–35 or compared primary melanoma patients with healthy controls36; while these methodologies have important biological relevance, they do not speak to the potential utility of miRNAs to predict recurrence at the time of diagnosis.
CONCLUSION
Our data demonstrated the prognostic utility of a set of serum-based miRNAs in stratifying patients according to their risk of melanoma recurrence. miR-15b showed potential for early monitoring for recurrence with good specificity. Bioinformatics analysis revealed that the miRNAs predictive of recurrence may regulate several key aspects of melanoma biology, including immune signaling pathways, melanogenesis, and the cell cycle.
Supplementary Material
Acknowledgments
Research Support: This study was supported by the NYU Applied Research Support Fund, the NYU Clinical and Translational Science Institute Grant (IO), the NCI Cancer Center Support Grant (5 P30 CA 016087-27) (IO and JZ), the NIH grant 1R21 GM110450-01 (JZ), and the Marc Jacobs Campaign to support melanoma research.
Footnotes
The authors have no financial disclosures.
Informed consent was obtained from all subjects who participated in this investigation.
References
- 1.Diepgen TL, Mahler V. The epidemiology of skin cancer. Br J Dermatol. 2002;146 (Suppl 61):1–6. doi: 10.1046/j.1365-2133.146.s61.2.x. [DOI] [PubMed] [Google Scholar]
- 2.Balch CM, Gershenwald JE, Soong SJ, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gogas H, Eggermont AM, Hauschild A, et al. Biomarkers in melanoma. Ann Oncol. 2009;20(Suppl 6):vi8–13. doi: 10.1093/annonc/mdp251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leiter U, Marghoob AA, Lasithiotakis K, et al. Costs of the detection of metastases and follow-up examinations in cutaneous melanoma. Melanoma Res. 2009;19:50–57. doi: 10.1097/CMR.0b013e32831bc41c. [DOI] [PubMed] [Google Scholar]
- 5.Friedman EB, Shang S, de Miera EV, et al. Serum microRNAs as biomarkers for recurrence in melanoma. J Transl Med. 2012;10:155. doi: 10.1186/1479-5876-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 7.Schaefer A, Jung M, Mollenkopf HJ, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 8.Vermont J, Bosson JL, Francois P, Robert C, Rueff A, Demongeot J. Strategies for graphical threshold determination. Comput Methods Programs Biomed. 1991;35:141–150. doi: 10.1016/0169-2607(91)90072-2. [DOI] [PubMed] [Google Scholar]
- 9.Gallop RJ, Crits-Christoph P, Muenz LR, Tu XM. Determination and Interpretation of the Optimal Operating Point for ROC Curves Derived Through Generalized Linear Models. Understanding Statistics. 2003;2:219–242. [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 12.Liberal V, Martinsson-Ahlzen HS, Liberal J, et al. Cyclin-dependent kinase subunit (Cks) 1 or Cks2 overexpression overrides the DNA damage response barrier triggered by activated oncoproteins. Proc Natl Acad Sci U S A. 2012;109:2754–2759. doi: 10.1073/pnas.1102434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang AE, Karnell LH, Menck HR. The National Cancer Data Base report on cutaneous and noncutaneous melanoma: a summary of 84,836 cases from the past decade. The American College of Surgeons Commission on Cancer and the American Cancer Society. Cancer. 1998;83:1664–1678. doi: 10.1002/(sici)1097-0142(19981015)83:8<1664::aid-cncr23>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 14.Malignant Melanoma (C43), Proportion of Cases Diagnosed at Each Stage, Adults (Aged 15–99), Former Anglia Cancer Network, 2006–2010. Skin cancer incidence statistics: Cancer Research UK. 2013 [Google Scholar]
- 15.Morton RL, Craig JC, Thompson JF. The role of surveillance chest X-rays in the follow-up of high-risk melanoma patients. Ann Surg Oncol. 2009;16:571–577. doi: 10.1245/s10434-008-0207-5. [DOI] [PubMed] [Google Scholar]
- 16.Satzger I, Mattern A, Kuettler U, et al. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer. 2010;126:2553–2562. doi: 10.1002/ijc.24960. [DOI] [PubMed] [Google Scholar]
- 17.Gaziel-Sovran A, Segura MF, Di Micco R, et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell. 2011;20:104–118. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 19.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Feng J, Huang C, Diao X, et al. Screening biomarkers of prostate cancer by integrating microRNA and mRNA microarrays. Genet Test Mol Biomarkers. 2013;17:807–813. doi: 10.1089/gtmb.2013.0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein RM, Bernstein D, Higgins SP, Higgins CE, Higgins PJ. SERPINE1 expression discriminates site-specific metastasis in human melanoma. Exp Dermatol. 2012;21:551–554. doi: 10.1111/j.1600-0625.2012.01523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007;98:621–628. doi: 10.1111/j.1349-7006.2007.00434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Candia P, Torri A, Gorletta T, et al. Intracellular Modulation, Extracellular Disposal and Serum Increase of MiR-150 Mark Lymphocyte Activation. PLoS One. 2013;8:e75348. doi: 10.1371/journal.pone.0075348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monteagudo C, Martin JM, Jorda E, Llombart-Bosch A. CXCR3 chemokine receptor immunoreactivity in primary cutaneous malignant melanoma: correlation with clinicopathological prognostic factors. J Clin Pathol. 2007;60:596–599. doi: 10.1136/jcp.2005.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thanarajasingam U, Sanz L, Diaz R, et al. Delivery of CCL21 to metastatic disease improves the efficacy of adoptive T-cell therapy. Cancer Res. 2007;67:300–308. doi: 10.1158/0008-5472.CAN-06-1017. [DOI] [PubMed] [Google Scholar]
- 26.Mullins IM, Slingluff CL, Lee JK, et al. CXC chemokine receptor 3 expression by activated CD8+ T cells is associated with survival in melanoma patients with stage III disease. Cancer Res. 2004;64:7697–7701. doi: 10.1158/0008-5472.CAN-04-2059. [DOI] [PubMed] [Google Scholar]
- 27.Rio-Machin A, Ferreira BI, Henry T, et al. Downregulation of specific miRNAs in hyperdiploid multiple myeloma mimics the oncogenic effect of IgH translocations occurring in the non-hyperdiploid subtype. Leukemia. 2013;27:925–931. doi: 10.1038/leu.2012.302. [DOI] [PubMed] [Google Scholar]
- 28.Di Leva G, Piovan C, Gasparini P, et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 2013;9:e1003311. doi: 10.1371/journal.pgen.1003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selth LA, Townley SL, Bert AG, et al. Circulating microRNAs predict biochemical recurrence in prostate cancer patients. Br J Cancer. 2013;109:641–650. doi: 10.1038/bjc.2013.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wulfken LM, Moritz R, Ohlmann C, et al. MicroRNAs in renal cell carcinoma: diagnostic implications of serum miR-1233 levels. PLoS One. 2011;6:e25787. doi: 10.1371/journal.pone.0025787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segura MF, Belitskaya-Levy I, Rose AE, et al. Melanoma MicroRNA signature predicts post-recurrence survival. Clin Cancer Res. 2010;16:1577–1586. doi: 10.1158/1078-0432.CCR-09-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanniford D, Gaziel A, Zhong J, et al. A microRNA-based signature predicts melanoma brain metastasis at the time of diagnosis. Pigment Cell Melanoma Res; Society for Melanoma Research 2013 Congress; Philadelphia, PA. 2013. p. 959. [Google Scholar]
- 33.Shiiyama R, Fukushima S, Jinnin M, et al. Sensitive detection of melanoma metastasis using circulating microRNA expression profiles. Melanoma Res. 2013 doi: 10.1097/CMR.0b013e328363e485. [DOI] [PubMed] [Google Scholar]
- 34.Alegre E, Sanmamed MF, Rodriguez C, Carranza O, Martin-Algarra S, Gonzalez A. Study of Circulating MicroRNA-125b Levels in Serum Exosomes in Advanced Melanoma. Arch Pathol Lab Med. 2014;138:828–832. doi: 10.5858/arpa.2013-0134-OA. [DOI] [PubMed] [Google Scholar]
- 35.Greenberg E, Besser MJ, Ben-Ami E, et al. A comparative analysis of total serum miRNA profiles identifies novel signature that is highly indicative of metastatic melanoma: a pilot study. Biomarkers. 2013;18:502–508. doi: 10.3109/1354750X.2013.816777. [DOI] [PubMed] [Google Scholar]
- 36.Kanemaru H, Fukushima S, Yamashita J, et al. The circulating microRNA-221 level in patients with malignant melanoma as a new tumor marker. J Dermatol Sci. 2011;61:187–193. doi: 10.1016/j.jdermsci.2010.12.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.