Abstract
Because of their high-energy metabolism, neurons are highly dependent on mitochondria, which generate cellular ATP through oxidative phosphorylation. The mitochondrial genome encodes for critical components of the oxidative phosphorylation pathway machinery, and therefore mutations in mitochondrial DNA (mtDNA) cause energy production defects that frequently have severe neurological manifestations. Here, we review the principles of mitochondrial genetics and focus on prototypical mitochondrial diseases to illustrate how primary defects in mtDNA or secondary defects in mtDNA due to nuclear genome mutations can cause prominent neurological and multisystem features. In addition, we discuss the pathophysiological mechanisms underlying mitochondrial diseases, the cellular mechanisms that protect mitochondrial integrity, and the prospects for therapy.
INTRODUCTION
Although all cells require ATP to maintain homeostasis, neurons have special metabolic needs. To continually transmit electrical signals, neurons must generate ATP for a number of energy-consuming processes: control of membrane potential by the Na+/K+ ATPase pump, regulation of intracellular Ca++, and exocytosis/recycling of synaptic vesicles. The latter process has been shown to be a particularly high consumer of synaptic ATP (Rangaraju et al., 2014). Synaptic ATP generation is stimulated by electrical activity and is generated by both glycolysis and mitochondrial function (Rangaraju et al., 2014). Fly mutants with defects in transport of axonal mitochondria show synaptic defects (Guo et al., 2005; Stowers et al., 2002; Verstreken et al., 2005) due to depletion of ATP at the nerve terminal (Guo et al., 2005). In humans, mitochondrial dysfunction is often associated with pathology affecting the central and peripheral nervous systems (Schon and Przedborski, 2011).
Mitochondria are the source of oxidative phosphorylation (OXPHOS), a metabolic pathway that is critical for the efficient extraction of energy from food sources (Scheffler, 2009). Unique to all the biochemical processes within animal cells, the OXPHOS pathway is under dual genetic control. Its components are largely encoded by the nuclear genome, but a handful of subunits are encoded by the small mitochondrial genome (mtDNA), a semiautonomous, circular and multi-copy DNA present within mitochondria (Figure 1). Although mitochondria are best known for their role in OXPHOS, they also play additional metabolic roles through the citric acid cycle, the urea cycle, and β-oxidation of fatty acids. Beyond metabolism, mitochondria have important functions in iron-sulfur cluster assembly, intracellular calcium handling, reactive oxygen species signaling (ROS), apoptosis, and innate immunity (Scheffler, 2009).
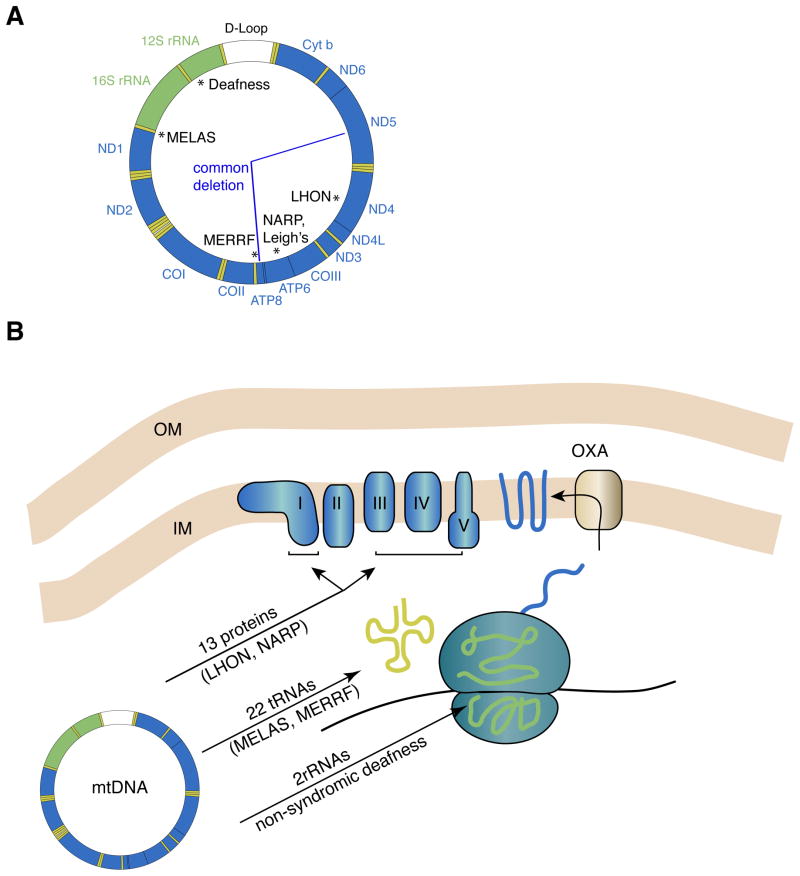
Figure 1. The human mtDNA genome and oxidative phosphorylation.
(A) Schematic of the circular mtDNA genome, showing the 13 protein coding genes (blue), the 2 rRNAs (green) and the 22 tRNAs (yellow). At the top is the non-coding D-loop (white), also known as the control region. This region is involved in mtDNA replication and transcriptional initiation. Classic examples of point mutations associated with prototypical mitochondrial encephalomyopathies are noted with asterisks. The “common deletion” removes 4977 bp of mtDNA and is one of many deletions that have been associated with sporadic KSS, PEO, and PS. (B) Oxidative phosphorylation and mtDNA gene products. The five enzyme complexes constituting the OXPHOS machinery reside in the mitochondrial inner membrane and consist of components encoded by both the nuclear and mitochondrial genomes. The 13 mtDNA proteins are transmembrane subunits of the enzyme complexes I, III, IV, and V. They are translated in the matrix of the mitochondrion and inserted into the inner membrane via the oxidase assembly (OXA) machinery. Mitochondrial ribosomes have polypeptides encoded by the nuclear genome. These polypeptides assemble into large and small ribosomal subunits that form complexes with rRNAs encoded by the mtDNA. The assembled ribosomes use mtDNA-encoded tRNAs to decode the messenger RNA. Examples of diseases caused by mutations in mtDNA-encoded proteins, tRNAs, and rRNAs are indicated.
Although mtDNA gene products are directly required only in OXPHOS, mitochondria with defective mtDNA have secondary defects beyond OXPHOS due to the diverse functions of mitochondria, which in turn can have wide-ranging effects in tissues and have been implicated in the pathogenesis of many diseases. This review focuses on the function of the mitochondrial genome and how defects in this genome can lead to neurological disease. We first review the general principles of mitochondrial genetics and discuss how mtDNA mutations affect mitochondrial function. This is followed by a description of the major classes of mtDNA disease, which can originate from either primary mtDNA mutations or mtDNA mutations secondary to nuclear DNA defects. Finally, we discuss the pathogenic mechanisms underlying mtDNA disease and the prospects for therapy. Throughout, we emphasize the impact of defective mtDNA on the central and peripheral nervous systems.
MITOCHONDRIAL GENETICS
Organization of the mtDNA genome
The mammalian mtDNA genome is 16.6 kilobases in length and encodes 13 polypeptides that are essential OXPHOS components (Figure 1A) (Anderson et al., 1981). The OXPHOS system is organized into five enzymatic complexes that reside in the mitochondrial inner membrane. These are the respiratory chain components NADH-ubiquinone oxidoreductase (Complex I), succinate-ubiquinone oxidoreductase (Complex II), ubiquinone-cytochrome c oxidoreductase (Complex III), cytochrome c reductase (Complex IV), and the ATP synthase (Complex V). The nuclear genome provides the majority of the OXPHOS components, and Complex II is entirely encoded by the nuclear genome. The other four complexes have one or more essential subunits encoded by the mtDNA (Figure 1B).
To generate these 13 mtDNA gene products, the mitochondrial genome has an additional 24 genes that support a dedicated translational system utilizing a slightly different genetic code. 22 transfer RNAs and 2 ribosomal RNAs encoded by mtDNA are necessary for the function of the mitochondrial ribosomes (Figure 1B). As a result, all 37 mtDNA genes are essential for normal levels of OXPHOS activity. The mtDNA genome in mammals is highly compact compared to the organization of the nuclear genome. The only significant noncoding segment is the control region that regulates transcription and replication. As would be expected, mutations in mtDNA frequently cause respiratory chain dysfunction.
Maternal inheritance of the mtDNA genome
Inherited diseases caused by mtDNA mutations are passed through the maternal lineage (Schon et al., 2012). This feature arises from the fact that almost all eukaryotes show uniparental inheritance of mtDNA, and in the case of mammals, it is the maternal mtDNA that is exclusively passed onto the offspring. Exclusive maternal inheritance of mtDNA is facilitated by the large size of the egg in comparison to the sperm. However, there also appears to be active mechanisms to ensure removal of paternal mtDNA. Mammalian mitochondria are ubiquitinated, and this mark has been suggested to target the organelles for degradation upon fertilization (Sutovsky et al., 1999). In nematodes, paternal mitochondria are removed by autophagy (Al Rawi et al., 2011; Sato and Sato, 2011), a process involving the trafficking of cellular material to that autophagosome and then the lysosome for degradation. In fertilized mammalian eggs, some autophagy markers are associated with the paternal mitochondria (Al Rawi et al., 2011). However, a recent study has argued that autophagy is not involved in degradation of paternal mitochondria in mice (Luo et al., 2013), and more work is needed to understand how fertilized mammalian embryos remove paternal mitochondria.
mtDNA segregation during maternal transmission
Maternal transmission of mtDNA has two unusual features. First, the mtDNA population undergoes a bottleneck phenomenon during oogenesis, such that only a small population of mtDNA molecules, estimated to be approximately 200, are amplified and transmitted (Jenuth et al., 1996). This feature can lead to rapid segregation of mtDNA variants within a few generations. For example, a rare mtDNA haplotype in the mother can become predominant in one of her offspring (Koehler et al., 1991). The molecular mechanism of the bottleneck is poorly understood and has been attributed to a numerical reduction in mtDNA molecules in the developing oocyte (Cree et al., 2008) or to selective amplification of a small pool of mtDNA molecules (Cao et al., 2009; Wai et al., 2008). The latter mechanism is plausible because mtDNA replication is not synchronized with the cell cycle, allowing variation in the number of times a particular mtDNA genome is replicated. Thus, a subpopulation of mtDNA molecules may be expanded at the expense of others due to selective replication or selective degradation.
The second unusual feature of maternal transmission is the presence of a quality control mechanism, termed purifying selection, that removes oocytes with a significant load of severe mtDNA mutations. Mice carrying a severe frameshift mutation in the ND6 mtDNA gene show progressive loss of the mutation in each subsequent generation (Fan et al., 2008). Analysis of oocyte DNA indicates that oocytes contain lower levels of the mutated mtDNA compared to the mother’s somatic tissues. In contrast, a milder mtDNA mutation that had a less severe effect on OXPHOS is retained through multiple generations. In another study, the presence of selection in the female germline was detected through statistical analysis of inherited mtDNA mutations (Stewart et al., 2008b). The mtDNA mutator mice, engineered to express a proofreading-defective mtDNA polymerase, contain high levels of mtDNA point mutations. Analysis of the maternal transmission of these mtDNA mutations revealed bias in the position of the mutations in the offspring. In the mtDNA protein-coding regions, synonymous mutations were more common than non-synonymous ones. In particular, there was greater suppression of mutations in the first and second codon positions, compared to the third position. Taken together, these two studies strongly imply purifying selection in the female germline as a mechanism to limit the inheritance of pathogenic mutations (Stewart et al., 2008a). The molecular basis of purifying selection is unclear but it appears that this mechanism can sense and eliminate oocytes with low levels of mtDNA mutations that normally do not result in respiratory chain defects, at least when such mtDNA mutations are studied in experimental cell culture systems.
This phenomenon may explain why very severe mtDNA mutations, such as those involving large deletions, are rarely inherited. Less severe mutations, such as point mutations affecting the tRNA genes, are commonly transmitted maternally. Such mutations may escape purifying selection because they must accumulate to very high levels before a respiratory chain defect is apparent, although more work is required to understand this effect.
Instability of the mtDNA genome
Sequence data from human populations indicate that the mitochondrial genome accumulates inherited mutations at a rate several orders of magnitude higher than that of nuclear DNA (Pakendorf and Stoneking, 2005). The mitochondrial genome also shows a several-hundred fold increase in the spontaneous mutation rate compared to the nuclear genome (Khrapko et al., 1997). Many explanations have been proposed for this high mutability: continuous mtDNA replication independent from the cell cycle, high ROS exposure generated as a by-product of the OXPHOS machinery, less efficient protection of mtDNA by DNA packaging proteins, and the rather limited mtDNA repair system.
Low levels of mtDNA heteroplasmy (the presence of more than one mtDNA haplotype within a cell) can be detected by deep sequencing in virtually any individual. A portion of these heteroplasmic variants can be found in the mother and therefore appear to be maternally inherited. The rest are presumed to be de novo mutations that accumulated in somatic tissues with age (He et al., 2010; Payne et al., 2013). It has been estimated that one in every 200 healthy newborn carries a common pathogenic mtDNA mutation at a level below clinical manifestation (Elliott et al., 2008). At a low frequency, such mutations, either inherited or de novo, will lead to disease if the mutation load rises to a sufficiently high level. Epidemiological studies estimate the prevalence of mtDNA disease as 1 in 5000–10,000 (Chinnery et al., 2000; Darin et al., 2001; Schaefer et al., 2008; Skladal et al., 2003).
Effect of mtDNA mutations
In inherited mtDNA diseases, affected offspring inherit a maternal load of mtDNA mutations. Individuals with mtDNA disease are typically heteroplasmic and harbor both wildtype and pathogenic mtDNA molecules. Cells can tolerate a high load of mtDNA mutations before encountering bioenergetic failure. The understanding of the pathological effects of mtDNA mutations has been greatly facilitated by the development of cybrid technology. In this approach, host cells lacking mtDNA serve as the recipient of mitochondria from mutant cells (King and Attardi, 1989), thereby allowing the pathological effects of mtDNA variants to be studied in the context of a uniform nuclear background. Cybrid cell models have been developed for numerous mtDNA mutations associated with mitochondrial encephalomyopathies (Chomyn et al., 1991; Hayashi et al., 1991). Systematic analyses of cybrid clones containing varying levels of mtDNA mutations indicates that OXPHOS failure does not manifest until mtDNA mutations accumulate to greater than 60–90% of total mtDNA (Chomyn et al., 1992; Rossignol et al., 2003). Studies in mice also support this idea. In a mouse model heteroplasmic for a 4 kilobase mtDNA deletion, which removed multiple protein-encoding and tRNA genes, tissues did not show COX deficiency until the pathogenic mtDNA level reached 85% (Nakada et al., 2001). Cells therefore have a high threshold for mtDNA mutations, with the threshold dependent on the exact nature of the mutation. As a result, patients with mtDNA disease have a mosaic distribution of respiratory chain deficiency. In skeletal muscle, for example, histochemical analysis may reveal a patchy distribution of OXPHOS-negative fibers intermingling with functionally normal fibers (Figure 2A and B). These features arise from the high-copy number of mtDNA and the need to accumulate mtDNA mutations to high levels before cellular dysfunction is evident.
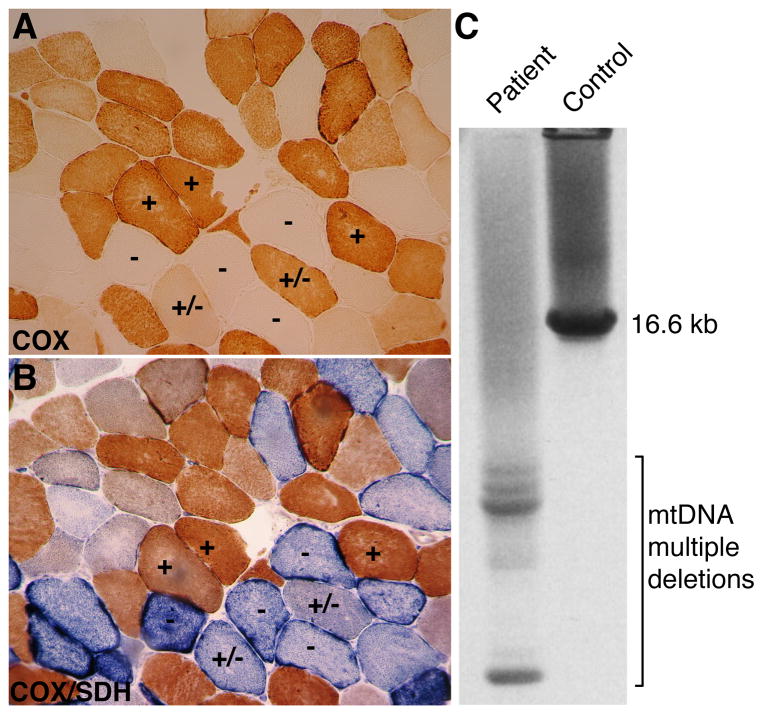
Figure 2. Muscle fiber defects and mtDNA deletions associated with a Polg mutation.
COX and COX/SDH stains are used to clinically evaluate mitochondrial dysfunction in muscle. (A) COX staining of a transverse muscle section reveals a mosaic pattern, with some fibers showing full enzymatic reaction (+), partial reaction (+/−) and no reaction (−). (B) The double COX/SDH staining of an adjacent section shows that the COX-positive fibers (+) display a brownish color, slightly darker compared to COX alone. In contrast, the COX-negative fibers (−) are intensely stained by SDH (blue) with frequent subsarcolemmal enhancement, and the COX-partial fibers (+/−) are intermediate, with preponderant SDH blue color. (C) Long-range PCR reveals a single band for wild-type mtDNA in the control subject, and multiple smaller bands denoting multiple mtDNA deletions in the patient. Both the histological sections and the mtDNA analysis are from a patient with compound heterozygous Polg mutations and SANDO phenotype. Images are courtesy of Dr. Maria Lucia Valentino.
mtDNA diseases show a progressive, age-related clinical course. In affected cells, the mutational load is generally quite high and often homoplasmic. These observations have led to the idea that inherited mtDNA mutations undergo random genetic drift during cell division (mitotic segregation), such that some cell lineages eventually acquire mutational loads that surpass the threshold for bioenergetic failure (DiMauro and Schon, 2003). Because of the variables of inherited mutational load, mosaicism, and genetic drift, even the same mtDNA mutation can lead to different clinical outcomes in affected individuals. Post-mitotic tissues such as skeletal muscle, cardiac muscle, brain and peripheral nerves are the most frequently affected by mtDNA pathogenic mutations, due to their high energy requirements (DiMauro et al., 2013).
Because mtDNA gene products are essential for the function of OXPHOS components, mutations in mtDNA reduce energy production, and this deficiency probably accounts for most of the clinical phenotypes. In addition to this bioenergetic effect, some mutations may also have secondary effects on apoptosis or ROS production. Mice with extensive mtDNA mutations (Kujoth et al., 2005) or mtDNA depletion (Wang et al., 2001) show widespread apoptosis, and cell death in the nervous system has been reported in various human mtDNA disorders (Leigh, 1951).
Mitochondrial dynamics
In considering the function of mtDNA and the effects of mtDNA mutations, it is important to note that the mitochondria within a cell are dynamic and continually engage in fusion and fission (division) (Chan, 2012). In mitochondrial fusion, two mitochondria merge into a single, larger organelle. Because mitochondria have double membranes, fusion involves the sequential fusion of the outer and inner membranes, and separate machinery have been identified for these processes (Figure 3A). Outer membrane fusion requires mitofusin GTPases (Mfn1 and Mfn2) located in the mitochondrial outer membrane. Inner membrane fusion requires the OPA1 GTPase associated with the mitochondria inner membrane. Mitochondrial fusion is balanced by the opposing process of mitochondrial fission, which requires yet another GTPase, dynamin-related protein 1 (Drp1). These dynamic processes are essential for proper mitochondrial function and serve to homogenize the mitochondrial population.
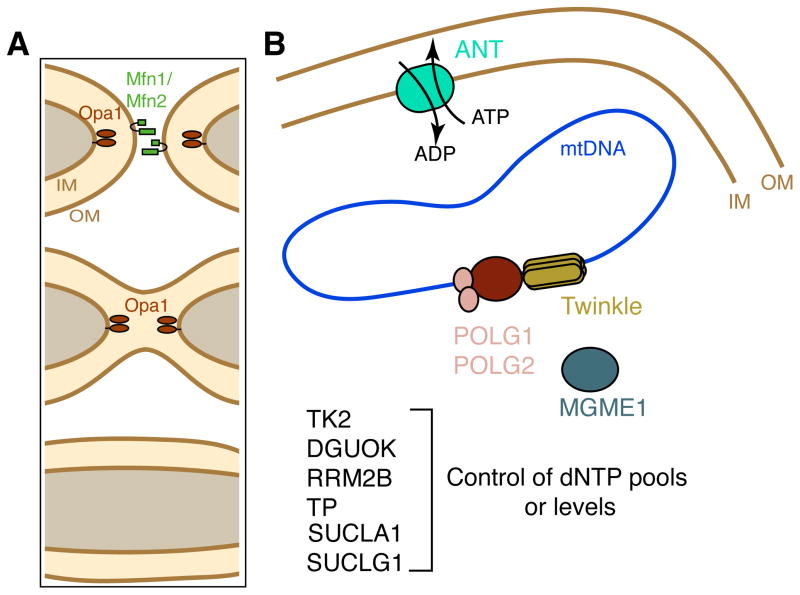
Figure 3. Nuclearly encoded proteins involved in Mendelian disorders of mtDNA maintenance.
(A) Molecules involved in mitochondrial fusion. Mitochondria are dynamic organelles that continually undergo fusion and fission. The balance of these opposing actions controls mitochondrial morphology and enables mixing of the mitochondrial population. Because mitochondria have 2 membranes, mitochondrial fusion is a multi-step process. Outer membrane (OM) fusion requires the mitofusins 1 and 2 (green), transmembrane GTP hydrolyzing enzymes. After outer membrane fusion, inner membrane (IM) fusion requires OPA1 (brown ovals), another large GTPase that is localized to the inner membrane. Mitofusins and OPA1 belong to the dynamin superfamily of GTPases. Mutations in OPA1 and Mfn2 can lead to mtDNA deletions. (B) Nuclearly encoded genes important for mtDNA replication and maintenance. The mitochondrial genome (represented by the closed loop) is replicated by the Polg DNA polymerase, a heterotrimeric enzyme complex composed of the catalytic subunit (POLG1) and 2 accessory subunits (POLG2). Twinkle (C10orf2/PEO1) is a mitochondrial DNA helicase that is thought to unwind mtDNA during at the replication fork (Milenkovic et al., 2013; Tyynismaa et al., 2004). Mgme1 (mitochondrial genome maintenance exonuclease 1) cleaves single-stranded DNA and is essential for mtDNA maintenance. The adenine nucleotide translocase (ANT) in the inner membrane (IM) exchanges ATP for ADP. Several additional proteins, listed at the bottom, are important for regulating dNTP pools or levels in the mitochondria and are important for mtDNA maintenance (see Table I). TK2, DGUOK, SUCLA1, SUCLG1 are mitochondrial proteins; RRM2B is nuclear and TP is cytosolic.
MITOCHONDRIAL DISEASES
Multiple genetic origins of mitochondrial disease: maternally inherited, sporadic, or Mendelian
Mitochondrial encephalomyopathies are a group of diseases due to a defect in mitochondrial function, and they often have central and peripheral nervous system involvement (Schon et al., 2012; Shapira et al., 1977). Many prototypical mitochondrial encephalomyopathies are due to mutations in mtDNA: Kearn-Saryre-Syndrome (KSS) (Kearns and Sayre, 1958); mitochondrial encephalomyopathy, lactic acidosis and stroke-like syndrome (MELAS) (Pavlakis et al., 1984); myoclonic epilepsy with ragged red fibers (MERRF) (Fukuhara et al., 1980); and Leber’s hereditary optic neuropathy (LHON) (Leber, 1871). KSS is sporadic, and the latter three diseases are maternally inherited. In 1988, there were breakthrough descriptions of the molecular defects responsible for some of these clinical entities. Single mtDNA macrodeletions were associated mitochondrial myopathies (Holt et al., 1988) and with KSS (Zeviani et al., 1988). In contrast, an mtDNA point mutation in the complex I subunit ND4 was associated with LHON (Wallace et al., 1988). These findings established that mtDNA mutations cause both maternally inherited and sporadic mitochondrial disease. In the last 25 years, the field of mitochondrial medicine (Luft, 1994) has grown exponentially, and a plethora of mtDNA mutations have been identified for a large range of clinical phenotypes (Ruiz-Pesini et al., 2007; Schon et al., 2012). A catalog of human mtDNA mutations can be found at http://mitomap.org/MITOMAP.
In addition, it soon became clear that other mtDNA diseases are transmitted as Mendelian traits, indicating that nuclear genetic defects can drive mtDNA mutagenesis and pathologic maintenance. These disorders are characterized by the accumulation of multiple mtDNA deletions (Zeviani et al., 1989) or the severe reduction of mtDNA copy number (Moraes et al., 1991).
We consider the major classes of mtDNA disorders, focusing on prototypic phenotypes that illustrate their effects on the central and peripheral nervous systems (Table 1).
Table I.
The major classes of mitochondrial disease, illustrated with prototypical examples.
| Disease | Gene Mutation | Neuromuscular features |
|---|---|---|
| Maternally inherited mtDNA mutations | ||
| LHON | ND4, ND1, ND6 | optic atrophy |
| MELAS | tRNALeu, and other tRNAs and ND subunits | muscle weakness including eye muscles and exercise intolerance, cardiomyopathy, hearing loss, headaches, epilepsy, stroke-like episodes, brain atrophy and cognitive deterioration |
| MERRF | tRNALys, and other tRNAs | muscle weakness including eye muscles and exercise intolerance, myoclonic jerks, epilepsy, cerebellar atrophy and ataxia, hearing loss |
| non-syndromic deafness | 12S rRNA, and tRNAs | hearing loss |
| NARP/MILS | ATPase 6 | retinitis pigmentosa, sensory neuropathy, ataxia, seizures, hearing loss, mental retardation and cognitive deterioration, bilateral necrotizing lesion of basal ganglia with spastic dystonia |
| Sporadic mtDNA deletions | ||
| CPEO | mtDNA deletion | weakness of eye muscles |
| KSS | mtDNA deletion | weakness of eye muscles, pigmentary retinopathy, cerebellar atrophy and ataxia, cardiac conduction abnormalities |
| PS | mtDNA deletion | pancitopenia |
| Mendelian disorders affecting mtDNA stability (multiple deletions) and maintenance (depletion) | ||
| Polg-associated phenotypes | POLG1 | See Table II |
| Autosomal dominant and recessive PEO | POLG1, POLG2, ANT1, Twinkle, MPV17, TK2, DGUOK, RRM2B, DNA2, MGME1 | weakness of eye muscles variably associated with hearing loss, peripheral neuropathy, Parkinsonism, psychiatric disturbances |
| MINGIE | thymidine phosphorylase | weakness of eye muscles, peripheral neuropathy, hearing loss, gastrointestinal dysmotility |
| DOA “plus” | Opa1, Mfn2 | optic atrophy, hearing loss, peripheral neuropathy, weakness of eye muscles |
MATERNAL INHERITED DISEASES
Maternally inherited mtDNA point mutations in respiratory chain subunits: LHON
As the first maternally inherited disease to be associated with an mtDNA point mutation, LHON was initially associated with mutation m.11778/G>A in the ND4 subunit of complex I (Wallace et al., 1988). Since then, 14 mutations have been confirmed as pathogenic for this disorder in the Mitomap website (Achilli et al., 2012; Ruiz-Pesini et al., 2007), and many others, all affecting ND subunits of complex I, have been associated with LHON complicated by phenotypes overlapping with MELAS and Leigh syndromes (discussed later) (Carelli et al., 2009).
Clinically, LHON is a mono-symptomatic blinding disease with extreme neuronal selectivity—the only neuron undergoing degeneration is the retinal ganglion cell (RGC) (Carelli et al., 2004; Yu-Wai-Man et al., 2011). The affected individuals experience subacute/acute loss of central vision at a young/adult age (Figure 4A–G), which evolves in about one year to a stable chronic condition characterized by profound visual impairment (Carelli et al., 2004; Yu-Wai-Man et al., 2011). Further peculiarities include male prevalence and incomplete penetrance, notwithstanding that the mtDNA pathogenic mutation is homoplasmic along the maternal line in almost all LHON families. Unaffected carriers may remain asymptomatic lifelong, although many actually demonstrate subtle abnormalities at ophthalmological examination.
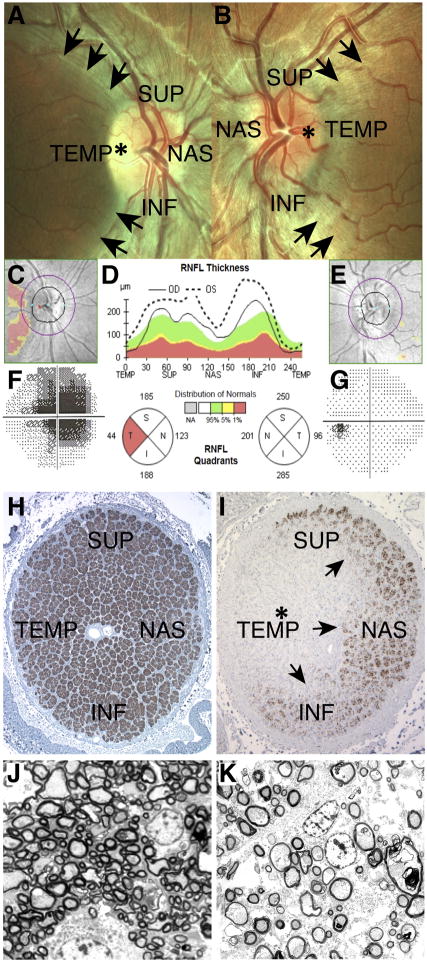
Figure 4. Neurological features in LHON.
Fundus images of the right eye (OD) and left eye (OS) of an LHON patient are shown in (A) and (B), respectively. In (A), temporal pallor (pale yellow region, asterisk) of the optic disc (bright circle near the center) indicates initial atrophy of the nerve. This defect is accompanied by the complete loss of fibers of the papillomacular bundle (loss of the translucent stripes in the dark area delimited by the arrows). The remaining quadrants--superior (SUP), inferior (INF) and nasal (NAS)--are characterized by pseudoedema of the retinal fibers, visible as translucent stripes converging to the optic disc, blurring its margins. In (B), the eye is at a preclinical stage, characterized by the still intact papillomacular bundle (presence of the translucent stripes in the area delimited by the arrows) and pseudoedematous appearance of the retinal fibers in all quadrants. The optic disc is hyperemic (congested due to engorgement with blood, asterisk), and retinal vessels are tortuous and frequently blurred by the pseudoedematous nerve fibers. Images courtesy of Dr. Piero Barboni. In (C), (D) and (E), optical coherence tomography (OCT) is used to measure the thickness of the retinal nerve fiber layer (RNFL) for OD (C) and OS (E) of the same patient. In (D), the green area denotes the normal range of retinal fiber layer thickness, whereas the red region indicates a pathological reduction of thickness (atrophy). The OD scan is indicated by the continuous line, which presents a pathological reduction only in the temporal sector (red sector on the circular graph). This is visualized by the pink area of atrophy in (C). OS is indicated by the dotted line and displays an overall increased thickness, being over the green range in all sectors, which denotes preclinical swelling of the retinal fibers due to pseudoedema. This is reflected in a still normal pattern in (E). Images courtesy of Dr. Piero Barboni. (F) and (G) Computerized Humphrey visual fields for OD and OS, respectively. In (F), a central scotoma (dark region) is evident. This defect correlates with the loss of retinal fibers of the papillomacular bundle at fundus observation (A) and detected by OCT measurements (C and D). In (G) the visual field is still unaffected. This is consistent with the preclinical stage of this eye, which has intact papillomacular bundle fibers visible at fundus observation (B) and detected by OCT measurements (D and E). Images courtesy of Dr. Piero Barboni. (H) and (I) Light microscopy appearance of optic nerve cross-sections from control (H) and LHON (I) individuals. In (H), the normal optic nerve is densely packed, with about 1.2 million axons organized in bundles. In (I), there is complete loss of axons (asterisk) in the temporal quadrant (TEMP) corresponding to the papillomacular bundle, and profound depletion of fibers also in the superior (SUP), inferior (INF) and nasal (NAS) sectors with a clear transition zone indicated by the arrows. Images courtesy of Dr. Alfredo A. Sadun and Fred Ross-Cisneros. (J) and (K) Electron microscopy of optic nerve cross-sections from control (J) and LHON (K) individuals. In (J), there is a normal density of axons, with prevalent small caliber ones. In (K), there is profound depletion of axons, in particular the smaller caliber population. The spared axons frequently show a thinner ring of myelin. Images courtesy of Dr. Alfredo A. Sadun and Fred Ross-Cisneros.
A large body of biochemical and cell biology investigations, often involving cybrid cell models, demonstrate that the LHON mutations in complex I subunits cause chronically increased ROS production, due to impaired ATP synthesis. As a result, cells harboring the mutation are prone to undergo apoptotic death (Carelli et al., 2004; Yu-Wai-Man et al., 2011). Recent breakthroughs provide evidence that estrogens may mitigate the cellular pathologies by up-regulating mitochondrial biogenesis, thus suggesting a possible explanation for the disease prevalence in males (Giordano et al., 2011). Furthermore, a variable level of compensatory mitochondrial biogenesis occurs in LHON carriers. This increased biogenesis may account for the incomplete penetrance in both genders and may be modulated by a combination of genetic and environmental modifying factors (Giordano et al., 2014). In fact, tobacco smoking appears to be a major environmental trigger favoring visual loss in LHON (Kirkman et al., 2009).
The pattern of RGC degeneration in LHON likely relates to their unique biology. The natural history of LHON, as defined by longitudinal studies with optical coherence tomography (OCT) (Barboni et al., 2010), as well as by post-mortem investigations on retinal and optic nerve specimens (Pan et al., 2012), is characterized by progression of a neurodegenerative front along a gradient determined by the axonal diameter (Figure 4H–K). The small axons on the temporal quadrant of the optic nerve, belonging to the papillomacular bundle deputed to central vision, are the earliest target. The disease progressively involves larger axons, but usually spares the largest axons on the nasal quadrant (Barboni et al., 2010; Pan et al., 2012). Mathematical modeling of the degenerative pattern suggests large axons have a more favorable surface/volume ratio that allows a higher capacity to increase mitochondrial mass that can alleviate reduced OXPHOS (Pan et al., 2012). The available evidence from human studies therefore suggests that axonal pathology precedes loss of axons and soma, although this conclusion is limited by the small number of documented post-mortem studies. In a recent mouse model of LHON, pathological changes in axons—including swelling, accumulation of abnormal mitochondria, and demyelination—are also observed well before axonal loss (Lin et al., 2012).
The special sensitivity of RGCs to OXPHOS defects also arises from the structural organization of myelin in this axonal system. The proximal portion of the RGC axon is devoid of myelin, and therefore this region of the neuron is particularly energy-dependent and sensitive to mitochondrial dysfunction. The axons become myelinated only after passing the anatomical structure known as the lamina cribrosa. Thereafter, axon potentials can propagate via the more energy efficient mode of saltatory conduction.
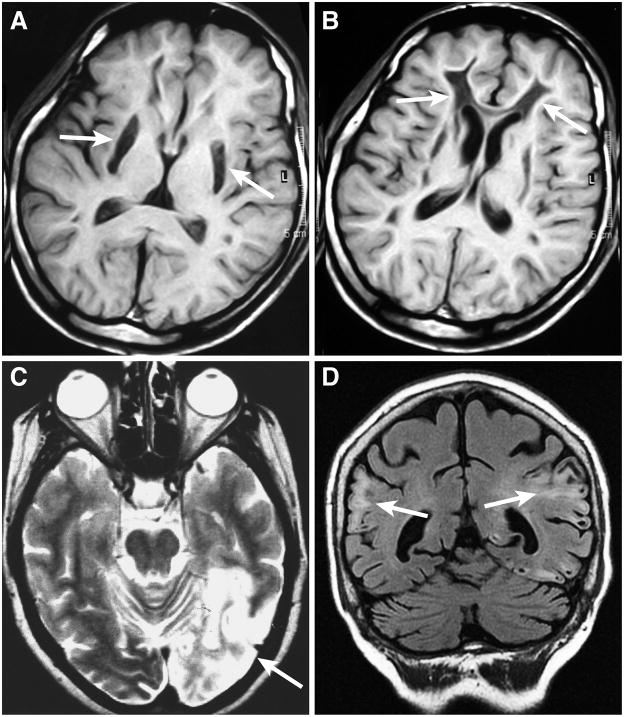
Another mutation, m.8993T>G or C in the ATPase 6 subunit gene, is associated with a maternally inherited syndrome characterized by peripheral neuropathy, ataxia, and pigmentary retinopathy (NARP), but when the mutant load exceeds 90–95% the patient’s clinical phenotype switches to maternally inherited Leigh’s syndrome (MILS), a severe subacute necrotizing encephalopathy that leads to bilateral lesions in basal ganglia and brainstem (Figure 5A, B) (Holt et al., 1990; Tatuch et al., 1992; de Vries et al., 1993). Leigh’s disease can be caused by either mtDNA mutations or nuclear mutations, and is a common clinical outcome of any severe OXPHOS dysfunction, particularly complex I or IV deficiency (DiMauro et al., 2013).
Figure 5. Brain magnetic resonance images (MRI) of mitochondrial encephalomyopathies.
Two axial brain T1-weighted images from the same patient with Leigh’s syndrome due to complex I deficiency are shown in (A) and (B). In (A), bilateral necrotizing striatal lesions (arrows) and widespread leukoencephalopathy are visible. In (B), the leukoencephalopathy presents with cavitations in the frontal white matter (arrows). Brain images from two MELAS patients are shown in (C) and (D). Axial T2-weighted image (C) shows a large hyperintense area extending in the occipital and parietal lobes of the left hemisphere (right side of image, arrow), the classical posterior location for stroke-like lesions in a patient with the most frequent MELAS m.3243A>G/tRNALeu(UUR) mutation. In another MELAS patient (D), with a complex I mtDNA mutation, multiple and bilaterally distributed cortical signal changes with cavitations (arrows) are evident on a coronal FLAIR (Fluid attenuated inversion recovery) image. These MELAS lesions do not obey the distribution of a major arterial vascular territory. In fact they are not due to a true ischemic infarct of cerebral tissue but to tissue edema and may be transient and partially reversible.
Maternally inherited mtDNA point mutations in tRNA and rRNA genes: MELAS, MERRF, and aminoglycoside-induced deafness
Disruption of the mitochondrial protein translation machinery encoded by mtDNA causes a diverse set of diseases that features neurological symptoms. The MELAS and MERRF multi-systemic syndromes are the most representative and studied examples of maternally inherited encephalomyopathies due to heteroplasmic point mutations of tRNA genes in the mtDNA. Most frequently, they are associated with the m.3243A>G/tRNALeu(UUR) mutation for MELAS (Goto et al., 1990) and the m.8344A>G/tRNALys mutation for MERRF (Shoffner et al., 1990). Additional point mutations have been associated with both MELAS and MERRF, affecting different positions in the tRNALeu(UUR) and tRNALys hotspot genes, as well as in other tRNAs (Ruiz-Pesini et al., 2007). It could be expected that all these mutations should produce similar pathogenic outcomes due to the impaired translation of mtDNA-encoded proteins. In practice, the clinical phenotypes of MELAS versus MERRF are strikingly distinct. Furthermore, the proband’s maternal lineage can show an extraordinary variety of other frequently overlapping phenotypes, ranging from milder and incomplete forms of the syndromes to the most severe Leigh’s syndrome (Chae et al., 2004; Howell et al., 1996; Mancuso et al., 2014; Mancuso et al., 2013; Moraes et al., 1993; Silvestri et al., 1993). A major reason for such clinical variability has been ascribed to variable heteroplasmic loads of the mutant mtDNA within and among tissues (Chinnery et al., 1997). However, the reason for the inherent difference in the clinical presentation between the m.8344A>G/tRNALys and m.3243A>G/tRNALeu mutations remains unclear, and therefore the heterogeneous presentation of mtDNA mutations remains a mysterious aspect of mitochondrial disease.
Both MELAS and MERRF are characterized by severe neurological symptoms. A peculiar clinical hallmark of MELAS is the repeated occurrence of stroke-like episodes (Betts et al., 2006; Iizuka et al., 2007) that result in brain damage (Figure 5C, D). The episodes appear to be due to pathology of small blood vessels. Due to respiratory chain dysfunction and low plasma levels of citrulline and l-arginine, there is decreased capacity for nitric oxide (NO) dependent vasodilation (El-Hattab et al., 2014; Koga et al., 2012). The inability to physiologically match cerebral blood supply to metabolic needs may lead to episodes of metabolic failure in large sections of the brain tissue. Such episodes may be triggered and worsened by stressful conditions such as neuronal hyperexcitability during epileptic discharges or the “spreading depression” of migrainous attacks. Thus, migraine is a frequent prodromal feature in MELAS for stroke-like episodes, which may affect brain regions corresponding to the migraine area (Betts et al., 2006; El-Hattab et al., 2014; Iizuka et al., 2007; Koga et al., 2012). In contrast, the MERRF mutation has a predilection for myoclonus and cerebellar dysfunction (Mancuso et al., 2013). In both disorders, muscle weakness and neurological defects are prevalent as the disease progresses with age.
The MELAS mutation has been thoroughly studied for over two decades, mainly in vitro by exploiting cybrid cell models. Multiple pathogenic mechanisms have been proposed for the tRNA mutation, including impairment of mitochondrial transcription termination (Hess et al., 1991), increased steady-state levels of the aberrant transcript RNA (Kaufmann et al., 1996), defective aminoacylation of the tRNA (Chomyn et al., 2000), and defective modification of the wobble base (Yasukawa et al., 2000). Some of these mechanisms have also been proposed for the MERRF mutation, such as defective aminoacylation (Enriquez et al., 1995) and wobble modification (Yasukawa et al., 2001). Overall, both MELAS and MERRF mutations lead to a global defect of the respiratory chain in patient-derived tissues as well as in cultured cells (King et al., 1992; Masucci et al., 1995). Neurons differentiated from MELAS patient-derived induced pluripotent stem cells (iPSCs) have a prevalent complex I defect (Hamalainen et al., 2013). This last result fits with the observation that many mtDNA point mutations affecting complex I cause phenotypes overlapping those of MELAS (Carelli et al., 2009; Ruiz-Pesini et al., 2007).
Point mutations in the rRNA subunits can also cause depression of mitochondrial protein translation. This is the case for the point mutation m.1555/A>G, which affects the 12S rRNA gene. This mutation induces non-syndromic sensorineural deafness and causes sensitivity to aminoglycoside-related ototoxicity (Prezant et al., 1993). This latter feature is related to the ancestral bacterial origin of “endosymbiotic” mitochondria, which can be rendered more susceptible to anti-bacterial drugs by mtDNA polymorphisms or mutations (Pacheu-Grau et al., 2010). The m.1555/A>G mutation is usually homoplasmic along the maternal line, and its penetrance is very variable among the mutation carriers (Prezant et al., 1993). Cybrid studies demonstrate the important modifying effect of the nuclear background (Guan et al., 2001). Nuclear genetic modifiers seem to regulate the clinical severity of the rRNA mutation (Guan et al., 2006; Raimundo et al., 2012) and the susceptibility to aminoglycosides.
SPORADIC DISEASES
Sporadic single large-scale deletions: Kearn-Sayre syndrome (KSS), Chronic Progressive External Ophthalmoplegia (CPEO), and Pearson syndrome (PS)
Single mtDNA macrodeletions, removing one or more mtDNA genes, underlie a number of mitochondrial diseases (Holt et al., 1988; Zeviani et al., 1988). In most cases they are sporadic and not transmitted, probably because the mutations arise de novo in the somatic lineage during early embryogenesis. Chronic progressive external ophthalmoplegia (CPEO) is characterized by inability to move the eyes and eyebrows, a sign of muscle weakness. KSS is a complex multisystem disorder characterized by the invariant triad of CPEO, pigmentary retinopathy, and onset before 20 years of age (Kearns and Sayre, 1958). Frequent additional symptoms include poor growth, progressive cerebellar syndrome, heart block, and increased protein content (above 100 mg/dl) in the cerebrospinal fluid (CSF). RRF and COX-negative fibers are the morphological hallmarks of muscle in both isolated CPEO with mitochondrial myopathy and in KSS. PS is a condition of early infancy characterized mainly by sideroblastic anemia or pancytopenia. In some cases, individuals who survive into childhood later develop KSS or even Leigh syndrome (Larsson et al., 1990; Santorelli et al., 1996).
Single mtDNA deletions in most cases are flanked by direct repeats, whose molecular rearrangement causes the formation of deletions (Samuels et al., 2004; Schon et al., 1989; Shoffner et al., 1989). Clonal expansion of the deleted species may be favored by their shorter replication time (Fukui and Moraes, 2009; Krishnan et al., 2008). However, deletions without repeats at the deletion boundaries also exist (Damas et al., 2014a; Damas et al., 2014b). Single mtDNA deletions may coexist with mtDNA duplications (Poulton et al., 1989) and which rearranged mtDNA is pathogenic has been questioned (Manfredi et al., 1997). Furthermore, mtDNA single deletions can occasionally be maternally inherited (Ballinger et al., 1992; Shanske et al., 2002). It has been suggested that the duplicated mtDNA is the molecular mtDNA species passing through the germline, and that the duplicated form regenerates single deletions in somatic tissues of the newborn (Ballinger et al., 1994). More recently, it has been shown that double strand breaks favor the occurrence of mtDNA deletions through a recombinogenic mechanism (Srivastava and Moraes, 2005). These results suggest that DNA repair and not replication generates the mtDNA deletions. The age of onset and progression of disease burden are correlated with the size of the deletion, the deletion heteroplasmy level in skeletal muscle, and the location of the deletion within the genome (Grady et al., 2014). These correlations may provide some predictive tools for prognosis.
MENDELIAN DISEASES
Multiple deletions and depletion of mtDNA in Mendelian disorders of mtDNA maintenance
Shortly after the first mtDNA mutations were identified in 1988, an autosomal dominant disorder characterized by CPEO and myopathy was associated with multiple mtDNA deletions (Figure 2C) (Zeviani et al., 1989). A second group of recessive syndromes, characterized by infantile mitochondrial myopathy or hepatopathy or kidney failure, was associated with mtDNA depletion in the affected tissues (Moraes et al., 1991). This combination of Mendelian inheritance and mtDNA defects suggested that nuclear mutations can cause mtDNA instability syndromes. Furthermore, mtDNA point mutations, multiple deletions and depletion were found in post-mitotic tissues in a complex, multisystem syndrome combining muscle, brain, and gastrointestinal symptoms (mitochondrial neuro-gastro-intestinal encephalomyopathy or MNGIE) (Nishigaki et al., 2003; Nishino et al., 2000). In this latter disorder, a nuclear defect caused both mtDNA mutations and defective maintenance.
Over the last decade, many different nuclear mutations have been shown to cause mtDNA multiple deletions and/or depletion, along with a wide range of neuromuscular symptoms (see Table I). These findings indicate that a large set of nuclear genes are involved in maintaining mtDNA integrity. Most of the diseases involve proteins directly implicated in mtDNA replication (Figure 3B) (POLG1 and 2 (components of Polg), Twinkle, DNA2, MGME1) or in maintaining the balanced supply of nucleotides (dNTP) necessary for mtDNA synthesis (TP, TK2, DGUOK, RRM2B, SUCLA2, SUCLG1) (Copeland, 2012; DiMauro et al., 2013; Spinazzola and Zeviani, 2005). In addition, mutations in OPA1 and MFN2, core components of the mitochondrial fusion machinery (Figure 3A), can also cause accumulation of mtDNA deletions in post-mitotic tissues (Chan, 2012; Renaldo et al., 2012; Rouzier et al., 2012; Yu-Wai-Man et al., 2010).
The copy number and stability of mtDNA play crucial roles for neuronal survival and brain metabolism. Mice with mutations in TFAM (Larsson et al., 1998; Wang et al., 1999; Wredenberg et al., 2002) or Polg (Larsson et al., 1998; Trifunovic et al., 2004; Wang et al., 1999; Wredenberg et al., 2002) have mtDNA depletion or accumulation of mtDNA mutations. These mouse models show features not only of mitochondrial diseases, but also of ageing (Trifunovic et al., 2004) and age-related neurodegenerative disorders such as Parkinson disease (Ekstrand et al., 2007).
A complete description of the phenotype associated with this still growing list of genetic defects is beyond the scope of the present review. Instead, we focus on the most striking examples-- the mitodynamics pathologies associated with OPA1 and MFN2 mutations and the many syndromes associated with Polg mutations.
Disorders of mitochondrial dynamics affect mtDNA maintenance: OPA1 and MFN2
The population of mitochondria within a cell undergoes cycles of fusion and fission events that promote mixing of mitochondria and control their shape and function (Chan, 2012). The biomedical relevance of these dynamic processes was highlighted by the observation that mutations in OPA1 and MFN2 can be deleterious for mtDNA maintenance, leading to mtDNA instability and depletion syndromes (Amati-Bonneau et al., 2008; Hudson et al., 2008; Rouzier et al., 2012). Similar to what is observed for the Polg mutations (see next section), there has been an expanding spectrum of phenotypes associated with different mutations in the OPA1 gene. These phenotypes may range from classical dominant optic atrophy (DOA), to the association of optic atrophy and sensorineural deafness, to a more complex and multi-systemic phenotype recognized as DOA plus (Yu-Wai-Man et al., 2010). The latter is frequently due to missense mutations affecting the GTPase domain of OPA1 and often manifests with CPEO. Its hallmark is the accumulation of mtDNA multiple deletions in post-mitotic tissues, particularly in skeletal muscle. Central and peripheral nervous systems are also affected.
Mutations in the MFN2 gene were identified in Charcot-Marie-Tooth type 2A, a peripheral sensorimotor neuropathy (Zuchner et al., 2004). As with OPA1, some mutations are now being associated with unusual and more severe phenotypes, including association with optic atrophy, or mtDNA instability or depletion (Boaretto et al., 2010; Renaldo et al., 2012; Rouzier et al., 2012). Studies in mice indicate that mitochondrial fusion is important for organization of mtDNA into nucleoids (Chen et al., 2007) and for maintaining mtDNA levels (Chen et al., 2010).
Polg syndromes: from mtDNA multiple deletions to depletion
Mutations in DNA polymerase gamma (Polg), the master enzyme for mtDNA replication, cause a remarkably wide range of mitochondrial diseases with defective mtDNA maintenance. Polg mutations were found in dominant and subsequently recessive syndromes characterized by late-onset CPEO and mitochondrial myopathy with mtDNA multiple deletions (Figure 2) (Lamantea et al., 2002; Van Goethem et al., 2001). A more severe syndrome termed SANDO (sensory-ataxia neuropathy with dysarthria and ophthalmoplegia) is almost invariably associated with compound heterozygote Polg mutations (Van Goethem et al., 2003). In addition to these adult-onset mitochondrial diseases, Polg mutations can also lead to severe, childhood neurologic disorders, such as Alpers-Huttenlocher hepatopathic poliodystrophy (Naviaux and Nguyen, 2004).
Polg mutations cause an extraordinary spectrum of clinical phenotypes (Table II), in part because they cause a wide range of molecular lesions in mtDNA. These defects include mtDNA base substitutions, deletions (Figure 2C) and/or depletion, ultimately resulting in dysfunctional OXPHOS complexes and/or their depletion. Recently, there has been an effort to map pathogenic mutations in Polg to functional clusters, to establish genotype-phenotype relationships (Farnum et al., 2014).
Table II.
Diversity of clinical syndromes caused by Polγ mutations.
| Syndrome | Neuromuscular and systemic features |
|---|---|
| Alpers-Huttenlocher syndrome (AHS) | the most severe phenotype characterized by childhood-onset, progressive and severe encephalopathy with intractable epilepsy and hepatic failure |
| Childhood myocerebrohepatopathy spectrum (MCHS) | presents in early infancy (up to three years) with developmental delay, lactic acidosis, myopathy with failure to thrive; variably associated with liver failure, renal tubular acidosis, pancreatitis, cyclic vomiting, and hearing loss |
| Myoclonic epilepsy myopathy sensory ataxia (MEMSA) | spectrum of disorders with epilepsy, myopathy, and ataxia without ophthalmoplegia |
| Ataxia neuropathy spectrum (ANS) | recessive ataxia, neuropathy, dysphagia, seizures, ophthalmoplegia |
| Late-onset autosomal dominant CPEO and myopathy | sensorineural hearing loss, axonal neuropathy, ataxia, depression, Parkinsonism, hypogonadism, and cataracts |
The functional separation of the proofreading from the polymerase activity in Polg has been exploited to generate an mtDNA “mutator” mouse model, where proofreading is defective, but replicative capacity is intact (Trifunovic et al., 2004). This mutator mouse model displayed reduced lifespan and premature onset of ageing-related phenotypes, including weight loss, reduced fat, alopecia, kyphosis, osteoporosis, anemia, reduced fertility and heart failure. This model has been interpreted as a strong indication of a causative link between mtDNA mutations, which are well documented to accumulate somatically with age in humans (Cortopassi et al., 1992; Soong et al., 1992), and ageing. More recently, this mouse model has been revisited as a “progeroid” phenotype with precocious somatic stem cell dysfunction (Ahlqvist et al., 2012). The significance of the somatic accumulation of mtDNA mutations in human ageing remains a hot topic of investigation and discussion (Bratic and Larsson, 2013).
Ageing-related somatic accumulation of mtDNA mutations: the case of Parkinson’s disease
mtDNA is continuously replicated independently from the cell cycle (Birky, 1994). Given the high mutability of mtDNA, a lifetime of mtDNA replication can result in the somatic accumulation of age-related mutations. These mutations include both point mutations and deletions, similar to those observed in mtDNA instability syndromes (Bratic and Larsson, 2013). Certain networks of neuronal cells in the brain, such as the dopaminergic system, seem prone to suffer an enhanced accumulation of these somatic mtDNA mutations. The accumulation in single cells presumably results from clonal expansion of pathogenic mtDNA mutations. In neurons with a high mutational load, one can imagine a cascade of additional pathogenic processes. These may include impaired clearance of dysfunctional mitochondria by autophagy/mitophagy and inefficient transport of mitochondria to dendrites and axons (Li et al., 2004; Sheng, 2014).
The pathologic accumulation of mtDNA multiple deletions and their clonal expansion in single dopaminergic neurons has been elegantly demonstrated by laser capture analysis of dopaminergic neurons in the substantia nigra in the elderly and, as an enhanced process, in patients with sporadic Parkinson’s disease (Bender et al., 2006; Kraytsberg et al., 2006). The frequent feature of Parkinsonism complicating the mtDNA instability syndromes further highlights the pathogenic link between mtDNA deletions and Parkinson disease. Thus, the direct role of mtDNA in ageing-related neurodegenerative disorders is an important topic that is increasingly investigated (Schon and Przedborski, 2011).
PATHOGENIC MECHANISMS
Clonal expansion of mtDNA
To understand the pathogenesis of mtDNA diseases, it is critical to consider how a pathogenic mtDNA variant can over time accumulate to high enough levels to cause disease. One aspect of this issue involves random genetic drift, which can result in a daughter cell inheriting a higher load of the pathogenic variant. In some cases, however, a single mtDNA variant can be clonally expanded to become homoplasmic. Clonal expansion of mtDNA mutations, along with declining mitochondrial function, has long been associated with aging (Chomyn and Attardi, 2003). In aged skeletal muscle, analysis of transverse sections shows an increased incidence of muscle fibers showing loss of OXPHOS, as indicated by lack of cytochrome c oxidase activity and increased succinate dehydrogenase activity (analogous to Figure 2A and B). Serial histological sectioning and reconstruction indicate that the defective muscle fibers have segmental loss of OXPHOS activity (Wanagat et al., 2001). In other words, when viewed longitudinally, a discrete segment of the muscle fiber has defective OXPHOS activity. Analysis of mtDNA from these affected segments reveals segmental homoplasmy of a defective mtDNA genome, typically containing an internal deletion. The accumulation of an mtDNA genome with a deletion results in the loss of OXPHOS activity. SDH activity is paradoxically increased because of compensatory mitochondrial biogenesis. Because SDH activity is entirely encoded by the nuclear genome, its function is not disrupted by mutation of mtDNA. As described above, clonal expansion is also well documented in dopaminergic neurons in the substantia nigra from aged individuals (Bender et al., 2006; Kraytsberg et al., 2006).
It is unclear how an mtDNA deletion that arises de novo in somatic cells is subsequently expanded at the expense of wild-type genomes. One model postulates that clonal expansion is driven by the faster replication time for a smaller mtDNA genome (Wallace, 1989). However, in contrast to a prediction of this model, the sizes of clonally expanded segments in the muscle fibers do not correlate with the extent of the mtDNA deletion (Campbell et al., 2014). Other possible explanations are reduced turnover of mitochondria containing deleted genomes (de Grey, 1997) and random genetic drift coupled with relaxed replication of mtDNA (Elson et al., 2001). In muscle fibers, clonal expanded mtDNA genomes are found in segments. These segments presumably represent snapshots of the expansion process. Muscle fibers are long, multinucleated cells, and the segmental nature of clonal expansion suggests that mitochondrial mixing is relatively restricted. It seems likely that the dynamic processes of fusion and fission would affect the dimensions of such segments.
Role of dynamics in tolerance of mtDNA mutations
In mouse models heteroplasmic for mtDNA containing an internal deletion, respiratory chain defects do not manifest in cells until the level of the pathogenic mtDNA approaches 85% (Nakada et al., 2001). Therefore, low levels of the wildtype mtDNA genome are sufficient to complement the pathogenic molecules. This threshold effect has led to the proposal that content exchange between mitochondria can complement recessive mtDNA mutations (Nakada et al., 2009). In a mouse model containing increased mtDNA mutations, mitochondrial fusion was found to be a protective factor. Removal of Mfn1, a GTPase required for mitochondrial fusion, greatly exacerbated the phenotype of the mice and promoted respiratory chain deficiency (Chen et al., 2010). In addition, mitochondrial fusion is required for maintenance of mtDNA levels (Chen et al., 2007; Chen et al., 2010).
Role of mitophagy
Mitophagy is the degradation of mitochondria through autophagy (Youle and Narendra, 2011). Although mitochondria can be degraded as part of a nonspecific autophagy response, mitophagy can also be selective for dysfunctional mitochondria. As a result, mitophagy, by culling out aged and damaged mitochondria, may be an important quality control mechanism for maintaining the function of the mitochondrial population. Mitochondria with degenerative morphologies have been reported to accumulate in cells when core components of the autophagy machinery are removed (Takamura et al., 2011). However, it is unclear whether this is a direct result of the failure to turnover mitochondria, and therefore the physiological functions of mitophagy remain to be clarified. In some pathological states, dysfunctional mitochondria are persistent and are not removed by mitophagy.
How does the autophagy machinery recognize mitochondria? Perhaps the clearest example exists in yeast, where a mitochondrial outer membrane protein links mitochondria to the autophagy machinery. Yeast cells grown with a nonfermentable carbon source show enhanced mitophagy in post-log phase. In a screen of yeast mutants, the mitochondrial outer membrane protein ATG32 was identified as a receptor for the mitophagy machinery (Kanki et al., 2009; Okamoto et al., 2009). ATG32 expression is induced during post-log phase growth and physically interacts with ATG11, an adaptor for the autophagy machinery. ATG32-deficient yeast has a selective defect in mitophagy, whereas the degradation of other cellular components by autophagy is unaffected.
Mitochondrial fission has been linked to mitophagy, because inhibition of mitochondrial fission reduces the efficiency of mitophagy (Frank et al., 2012; Tanaka et al., 2010). In yeast, the autophagy adaptor ATG11 physically associates with Dnm1 (Mao et al., 2013), a large GTPase that is the central player in mitochondrial fission. These observations suggest that the onset of mitophagy is coordinated with mitochondrial fission. The role of fission may be to help segregate mitochondria into smaller physical units that can be readily engulfed by autophagosomes.
Studies in the last several years have linked Parkinson’s disease (PD) to mitophagy. Pink1 and Parkin are genes mutated in some inherited forms of PD. As first revealed in fly studies, both genes are critical for maintenance of mitochondrial function (Clark et al., 2006; Park et al., 2006; Yang et al., 2006). Studies in mammalian cell culture suggest that they operate in a linear pathway to remove dysfunctional mitochondria. When Parkin is over-expressed, it localizes to the surface of depolarized mitochondria and promotes their degradation by mitophagy (Narendra et al., 2008). Pink1 is a serine/threonine kinase that is required to localize Parkin onto the surface of dysfunctional mitochondria (Narendra et al., 2010b). Pink1 is normally kept at low levels on mitochondria due to degradation by the PARL protease, but accumulates on the mitochondrial surface upon depolarization of the inner membrane (Jin et al., 2010). This accumulation allows recruitment of Parkin onto dysfunctional mitochondria. Once Parkin is recruited, it causes widespread ubiquitination of mitochondrial outer membrane proteins (Chan et al., 2011). Some autophagy adaptors, such as NBR1 and p62, bind to ubiquitin, but there are conflicting reports about their involvement in Parkin-mediated mitophagy (Geisler et al., 2010; Narendra et al., 2010a; Okatsu et al., 2010). Ubiquitination causes mitochondrial outer membrane protein degradation, an event that is required for the subsequent degradation of mitochondria by autophagosomes (Chan et al., 2011; Tanaka et al., 2010).
The involvement of Pink1 and Parkin in mitophagy suggests the intriguing hypothesis that some forms of PD may result from a loss of mitochondrial quality control, leading to the accumulation of dysfunctional mitochondria. PD has long been linked to mitochondrial dysfunction (Abou-Sleiman et al., 2006). Cell culture experiments suggest that Parkin can influence the segregation of mtDNA mutations in a heteroplasmic cell, biasing the population towards functional mtDNA (Suen et al., 2010). However, our knowledge of the Pink1/Parkin system in mitophagy is still preliminary. In some neuronal cultures, Parkin recruitment to depolarized mitochondria is not robust (Van Laar et al., 2011) or occurs with slower kinetics compared to commonly used cell lines, such as HeLa cells (Cai et al., 2012). In an experimental mouse model of mitochondrial dysfunction leading to neurodegeneration, Parkin is not recruited to damaged mitochondria and does not appear to play a significant protective role (Sterky et al., 2011). The latter result may also reflect differences between the role of Parkin in mice versus humans, because mice lacking Parkin or Pink1 do not show neurodegenerative changes. Therefore, it will be important to clarify the physiological functions of Parkin and mitophagy in mitochondrial disease.
Adaptive selection of mtDNA variants: haplogroups as multifaceted modulators of healthiness and disease
Due to its very high mutational rate, the small mtDNA molecule is extraordinarily variable among human populations. Many variants have been extensively studied in the last two decades to understand how human populations evolved, migrated, and colonized the continents (Torroni et al., 2006). A large fraction of this variation may have been selected by environmental adaptation. In particular, two driving forces for adaptation, climate and diet, have been postulated to be major contributors in shaping regional mtDNA genetic variation in human populations (Wallace, 2013). This mtDNA genetic variation, as exemplified by classifying mtDNA genomes into different haplogroups, generates intra-species variability in terms of adaptation to environment and protection or predisposition to pathological conditions, thus impinging on the ageing process (Wallace, 2013).
The exponential increase in studies showing specific mtDNA haplogroups associated with human pathologies provides mounting evidence that “normal” variation of the mtDNA background sequence may predispose to diseases. Furthermore, mtDNA variation may act as modifying factor of clinical severity or penetrance in the case of mtDNA-related genetic disorders. The most replicated case is the association of specific branches of the Caucasian haplogroup J with penetrance in LHON (Carelli et al., 2006; Hudson et al., 2007). Similarly, mtDNA haplogroup K has been consistently associated across different studies with protection from developing Parkinson disease (Ghezzi et al., 2005; Hudson et al., 2013; van der Walt et al., 2003).
THERAPEUTIC STRATEGIES FOR mtDNA DISEASE
Historically, mitochondrial diseases related to defective mtDNA have been treated empirically with variable combinations of co-factors and vitamins, a “mito-cocktail” frequently including antioxidants such as quinones (CoQ and idebenone), lipoic acid, vitamins E and C, and molecules boosting bioenergetics such as creatine and carnitine (Pfeffer et al., 2012) The efficacy of these treatments has been unclear due to the intrinsic difficulties in running properly designed controlled trials with rare diseases, with mitochondrial disorders posing additional problems due to their clinical heterogeneity and loosely defined natural history (Pfeffer et al., 2013).
At the genetic level, the lack of tools to manipulate the multi-copy mtDNA genome, delimited by a double membrane, has been a major obstacle. However, major breakthroughs have been achieved recently, opening a new era for the therapy of mitochondrial disorders. A general strategy, supported by translational evidence from both patients (Giordano et al., 2014) and animal models (Wredenberg et al., 2002) is the compensatory activation of mitochondrial biogenesis. Multiple approaches have converged on activating the transcriptional co-activator PGC1α, the master regulator of mitochondrial biogenesis (Cerutti et al., 2014; Khan et al., 2014) These results provide hope for rapid translation into clinical trials in human patients.
Another major achievement is based on the simple idea of shifting heteroplasmy towards wild-type mtDNA to restore under-threshold heteroplasmy in the key tissues. Using either mitochondria-targeted TALEN (mitoTALEN) nucleases (Bacman et al., 2013) or mitochondria-targeted obligate heterodimeric zinc finger nucleases (mtZFNs) (Gammage et al., 2014) for site-specific elimination of mutant mtDNA, it has been possible to provide proof of principle that these strategies are feasible. Another proposed approach to counteract mtDNA mutations is the allotopic nuclear re-expression of the wild type mtDNA subunit gene, engineered for mitochondrial import from the cytosol (Guy et al., 2002; Manfredi et al., 2002) This strategy will be soon tested in human clinical trials for LHON (Guy et al., 2002). On a similar line, it has been shown that the carboxy-terminal domain of human mitochondrial leucyl-tRNA synthetase can be used to correct mitochondrial dysfunctions caused by mt-tRNA mutations (Perli et al., 2014) The nuclear expression of such small peptides, engineered for mitochondrial import, may become a universal therapeutic approach for encephalomyopathies such as MELAS and MERRF.
Finally, to minimize germline transmission of mutant mtDNA, there have been advances in nuclear DNA transfer techniques designed to reduce mutant mtDNA from patient cells. The spindle-chromosomal complex (Tachibana et al., 2009) or the polar body (Wang et al., 2014) of an affected oocyte, or the pronuclei (Craven et al., 2010) of an affected zygote, can be used as the source of nuclear genome to be transferred into an enucleated recipient with wild-type mtDNA. The resulting embryo will generate a so-called three-parent offspring, carrying the correct complement of nuclear genes from the natural parents, and normal mtDNA from a third-parent. These transfer techniques differ in their efficiency at reducing or eliminating mutant mtDNA from the offspring. In vitro and animal experiments in primates and mice support the feasibility of the nuclear DNA transfer approach, and important steps towards the first application in humans have been taken, with a large ongoing discussion on the ethical implications (Amato et al., 2014)
CONCLUSION
Belying its small size, the mitochondrial genome plays a central role in cellular metabolism, and defects in mtDNA result in an extraordinary range of human diseases. Because of their high metabolic requirements, neurons in both the central and peripheral nervous systems are among the most commonly affected cell types in mitochondrial disease. A full understanding of these diseases will require more insight into the basic biology of mitochondria, including the mechanisms that maintain mitochondrial dynamics, cull defective organelles, and protect mtDNA integrity during maternal inheritance and cell division. A deeper understanding of the basic biology of mitochondria holds promise for developing effective therapies, which for most mitochondrial diseases currently remain at the level of palliative and symptomatic approaches.
Acknowledgments
Work in the authors’ laboratories is supported by HHMI (D.C.C.), NIH grants GM062967 (D.C.C.) and GM110039 (D.C.C.), Telethon grants GGP11182 (V.C.) and GPP10005 (V.C), the Emilia-Romagna region program ER-MITO (V.C.), support of Fondazione Galletti (V.C.) and support from the patient’s associations MITOCON, UMDF, IFOND, Struggling Within Leber’s and The Poincenot Family (V.C.). We are grateful to Maria Lucia Valentino (University of Bologna), Piero Barboni (Università Vita-Salute San Raffaele), Alfredo A. Sadun (Doheny Eye Institute, UCLA), and Fred Ross-Cisneros (Doheny Eye Institute, UCLA) for providing clinical and histological images used in the figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Abou-Sleiman PM, Muqit MM, Wood NW. Expanding insights of mitochondrial dysfunction in Parkinson’s disease. Nature reviews Neuroscience. 2006;7:207–219. doi: 10.1038/nrn1868. [DOI] [PubMed] [Google Scholar]
- Achilli A, Iommarini L, Olivieri A, Pala M, Hooshiar Kashani B, Reynier P, La Morgia C, Valentino ML, Liguori R, Pizza F, Barboni P, Sadun F, De Negri AM, Zeviani M, Dollfus H, Moulignier A, Ducos G, Orssaud C, Bonneau D, Procaccio V, Leo-Kottler B, Fauser S, Wissinger B, Amati-Bonneau P, Torroni A, Carelli V. Rare primary mitochondrial DNA mutations and probable synergistic variants in Leber’s hereditary optic neuropathy. PLoS One. 2012:7. doi: 10.1371/journal.pone.0042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlqvist KJ, Hamalainen RH, Yatsuga S, Uutela M, Terzioglu M, Gotz A, Forsstrom S, Salven P, Angers-Loustau A, Kopra OH, Tyynismaa H, Larsson NG, Wartiovaara K, Prolla T, Trifunovic A, Suomalainen A. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell metabolism. 2012;15:100–109. doi: 10.1016/j.cmet.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- Amati-Bonneau P, Valentino ML, Reynier P, Gallardo ME, Bornstein B, Boissiere A, Campos Y, Rivera H, de la Aleja JG, Carroccia R, Iommarini L, Labauge P, Figarella-Branger D, Marcorelles P, Furby A, Beauvais K, Letournel F, Liguori R, La Morgia C, Montagna P, Liguori M, Zanna C, Rugolo M, Cossarizza A, Wissinger B, Verny C, Schwarzenbacher R, Martin MA, Arenas J, Ayuso C, Garesse R, Lenaers G, Bonneau D, Carelli V. OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus’ phenotypes. Brain. 2008;131:338–351. doi: 10.1093/brain/awm298. [DOI] [PubMed] [Google Scholar]
- Amato P, Tachibana M, Sparman M, Mitalipov S. Three-parent in vitro fertilization: gene replacement for the prevention of inherited mitochondrial diseases. Fertility and sterility. 2014;101:31–35. doi: 10.1016/j.fertnstert.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Pinto M, Peralta S, Moraes CT. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nature medicine. 2013;19:1111–1113. doi: 10.1038/nm.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger SW, Shoffner JM, Gebhart S, Koontz DA, Wallace DC. Mitochondrial diabetes revisited. Nat Genet. 1994;7:458–459. doi: 10.1038/ng0894-458. [DOI] [PubMed] [Google Scholar]
- Ballinger SW, Shoffner JM, Hedaya EV, Trounce I, Polak MA, Koontz DA, Wallace DC. Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat Genet. 1992;1:11–15. doi: 10.1038/ng0492-11. [DOI] [PubMed] [Google Scholar]
- Barboni P, Carbonelli M, Savini G, Ramos CdVF, Carta A, Berezovsky A, Salomao SR, Carelli V, Sadun AA. Natural history of Leber’s hereditary optic neuropathy: longitudinal analysis of the retinal nerve fiber layer by optical coherence tomography. Ophthalmology. 2010;117:623–627. doi: 10.1016/j.ophtha.2009.07.026. [DOI] [PubMed] [Google Scholar]
- Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, Taylor RW, Turnbull DM. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- Betts J, Jaros E, Perry RH, Schaefer AM, Taylor RW, Abdel-All Z, Lightowlers RN, Turnbull DM. Molecular neuropathology of MELAS: level of heteroplasmy in individual neurones and evidence of extensive vascular involvement. Neuropathology and applied neurobiology. 2006;32:359–373. doi: 10.1111/j.1365-2990.2006.00731.x. [DOI] [PubMed] [Google Scholar]
- Birky CW. Relaxed and Stringent Genomes: Why Cytoplasmic Genes Don’t Obey Mendel’s Laws. J Hered. 1994;85:355–365. [Google Scholar]
- Boaretto F, Vettori A, Casarin A, Vazza G, Muglia M, Rossetto MG, Cavallaro T, Rizzuto N, Carelli V, Salviati L, Mostacciuolo ML, Martinuzzi A. Severe CMT type 2 with fatal encephalopathy associated with a novel MFN2 splicing mutation. Neurology. 2010;74:1919–1921. doi: 10.1212/WNL.0b013e3181e240f9. [DOI] [PubMed] [Google Scholar]
- Bratic A, Larsson NG. The role of mitochondria in aging. J Clin Invest. 2013;123:951–957. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Zakaria HM, Simone A, Sheng ZH. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Current biology : CB. 2012;22:545–552. doi: 10.1016/j.cub.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G, Krishnan KJ, Deschauer M, Taylor RW, Turnbull DM. Dissecting the mechanisms underlying the accumulation of mitochondrial DNA deletions in human skeletal muscle. Human molecular genetics. 2014 doi: 10.1093/hmg/ddu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Shitara H, Sugimoto M, Hayashi J, Abe K, Yonekawa H. New evidence confirms that the mitochondrial bottleneck is generated without reduction of mitochondrial DNA content in early primordial germ cells of mice. PLoS genetics. 2009;5:e1000756. doi: 10.1371/journal.pgen.1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli V, Achilli A, Valentino ML, Rengo C, Semino O, Pala M, Olivieri A, Mattiazzi M, Pallotti F, Carrara F, Zeviani M, Leuzzi V, Carducci C, Valle G, Simionati B, Mendieta L, Salomao S, Belfort R, Jr, Sadun AA, Torroni A. Haplogroup effects and recombination of mitochondrial DNA: novel clues from the analysis of Leber hereditary optic neuropathy pedigrees. Am J Hum Genet. 2006;78:564–574. doi: 10.1086/501236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli V, La Morgia C, Valentino ML, Barboni P, Ross-Cisneros FN, Sadun AA. Retinal ganglion cell neurodegeneration in mitochondrial inherited disorders. Biochim Biophys Acta. 2009;1787:518–528. doi: 10.1016/j.bbabio.2009.02.024. [DOI] [PubMed] [Google Scholar]
- Carelli V, Ross-Cisneros FN, Sadun AA. Mitochondrial dysfunction as a cause of optic neuropathies. Progress in retinal and eye research. 2004;23:53–89. doi: 10.1016/j.preteyeres.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Cerutti R, Pirinen E, Lamperti C, Marchet S, Sauve AA, Li W, Leoni V, Schon EA, Dantzer F, Auwerx J, Viscomi C, Zeviani M. NAD(+)-dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell metabolism. 2014;19:1042–1049. doi: 10.1016/j.cmet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae JH, Hwang H, Lim BC, Cheong HI, Hwang YS, Kim KJ. Clinical features of A3243G mitochondrial tRNA mutation. Brain & development. 2004;26:459–462. doi: 10.1016/j.braindev.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Human molecular genetics. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery PF, Howell N, Lightowlers RN, Turnbull DM. Molecular pathology of MELAS and MERRF. The relationship between mutation load and clinical phenotypes. Brain. 1997;120 (Pt 10):1713–1721. doi: 10.1093/brain/120.10.1713. [DOI] [PubMed] [Google Scholar]
- Chinnery PF, Johnson MA, Wardell TM, Singh-Kler R, Hayes C, Brown DT, Taylor RW, Bindoff LA, Turnbull DM. The epidemiology of pathogenic mitochondrial DNA mutations. Ann Neurol. 2000;48:188–193. [PubMed] [Google Scholar]
- Chomyn A, Attardi G. MtDNA mutations in aging and apoptosis. Biochemical and biophysical research communications. 2003;304:519–529. doi: 10.1016/s0006-291x(03)00625-9. [DOI] [PubMed] [Google Scholar]
- Chomyn A, Enriquez JA, Micol V, Fernandez-Silva P, Attardi G. The mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episode syndrome-associated human mitochondrial tRNALeu(UUR) mutation causes aminoacylation deficiency and concomitant reduced association of mRNA with ribosomes. J Biol Chem. 2000;275:19198–19209. doi: 10.1074/jbc.M908734199. [DOI] [PubMed] [Google Scholar]
- Chomyn A, Martinuzzi A, Yoneda M, Daga A, Hurko O, Johns D, Lai ST, Nonaka I, Angelini C, Attardi G. MELAS mutation in mtDNA binding site for transcription termination factor causes defects in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:4221–4225. doi: 10.1073/pnas.89.10.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn A, Meola G, Bresolin N, Lai ST, Scarlato G, Attardi G. In vitro genetic transfer of protein synthesis and respiration defects to mitochondrial DNA-less cells with myopathy-patient mitochondria. Molecular and cellular biology. 1991;11:2236–2244. doi: 10.1128/mcb.11.4.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Copeland WC. Defects in mitochondrial DNA replication and human disease. Crit Rev Biochem Mol Biol. 2012;47:64–74. doi: 10.3109/10409238.2011.632763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortopassi GA, Shibata D, Soong NW, Arnheim N. A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:7370–7374. doi: 10.1073/pnas.89.16.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven L, Tuppen HA, Greggains GD, Harbottle SJ, Murphy JL, Cree LM, Murdoch AP, Chinnery PF, Taylor RW, Lightowlers RN, Herbert M, Turnbull DM. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature. 2010;465:82–85. doi: 10.1038/nature08958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cree LM, Samuels DC, de Sousa Lopes SC, Rajasimha HK, Wonnapinij P, Mann JR, Dahl HH, Chinnery PF. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet. 2008;40:249–254. doi: 10.1038/ng.2007.63. [DOI] [PubMed] [Google Scholar]
- Damas J, Carneiro J, Amorim A, Pereira F. MitoBreak: the mitochondrial DNA breakpoints database. Nucleic Acids Res. 2014a;42:1261–1268. doi: 10.1093/nar/gkt982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J, Samuels DC, Carneiro J, Amorim A, Pereira F. Mitochondrial DNA rearrangements in health and disease--a comprehensive study. Human mutation. 2014b;35:1–14. doi: 10.1002/humu.22452. [DOI] [PubMed] [Google Scholar]
- Darin N, Oldfors A, Moslemi AR, Holme E, Tulinius M. The incidence of mitochondrial encephalomyopathies in childhood: clinical features and morphological, biochemical, and DNA anbormalities. Ann Neurol. 2001;49:377–383. [PubMed] [Google Scholar]
- de Grey AD. A proposed refinement of the mitochondrial free radical theory of aging. BioEssays : news and reviews in molecular, cellular and developmental biology. 1997;19:161–166. doi: 10.1002/bies.950190211. [DOI] [PubMed] [Google Scholar]
- de Vries DD, van Engelen BG, Gabreels FJ, Ruitenbeek W, van Oost BA. A second missense mutation in the mitochondrial ATPase 6 gene in Leigh’s syndrome. Ann Neurol. 1993;34:410–412. doi: 10.1002/ana.410340319. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. The New England journal of medicine. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA, Carelli V, Hirano M. The clinical maze of mitochondrial neurology. Nat Rev Neurol. 2013;9:429–444. doi: 10.1038/nrneurol.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand MI, Terzioglu M, Galter D, Zhu S, Hofstetter C, Lindqvist E, Thams S, Bergstrand A, Hansson FS, Trifunovic A, Hoffer B, Cullheim S, Mohammed AH, Olson L, Larsson NG. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hattab AW, Emrick LT, Chanprasert S, Craigen WJ, Scaglia F. Mitochondria: role of citrulline and arginine supplementation in MELAS syndrome. Int J Biochem Cell Biol. 2014;48:85–91. doi: 10.1016/j.biocel.2013.12.009. [DOI] [PubMed] [Google Scholar]
- Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet. 2008;83:254–260. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson JL, Samuels DC, Turnbull DM, Chinnery PF. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am J Hum Genet. 2001;68:802–806. doi: 10.1086/318801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez JA, Chomyn A, Attardi G. MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNA(Lys) and premature translation termination. Nat Genet. 1995;10:47–55. doi: 10.1038/ng0595-47. [DOI] [PubMed] [Google Scholar]
- Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, Macgregor GR, Wallace DC. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnum GA, Nurminen A, Kaguni LS. Mapping 136 pathogenic mutations into functional modules in human DNA polymerase gamma establishes predictive genotype-phenotype correlations for the complete spectrum of POLG syndromes. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbabio.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Duvezin-Caubet S, Koob S, Occhipinti A, Jagasia R, Petcherski A, Ruonala MO, Priault M, Salin B, Reichert AS. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim Biophys Acta. 2012;1823:2297–2310. doi: 10.1016/j.bbamcr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Tokiguchi S, Shirakawa K, Tsubaki T. Myoclonus epilepsy associated with ragged-red fibres (mitochondrial abnormalities ): disease entity or a syndrome? Light-and electron-microscopic studies of two cases and review of literature. J Neurol Sci. 1980;47:117–133. doi: 10.1016/0022-510x(80)90031-3. [DOI] [PubMed] [Google Scholar]
- Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Human molecular genetics. 2009;18:1028–1036. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammage PA, Rorbach J, Vincent AI, Rebar EJ, Minczuk M. Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO molecular medicine. 2014;6:458–466. doi: 10.1002/emmm.201303672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature cell biology. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Ghezzi D, Marelli C, Achilli A, Goldwurm S, Pezzoli G, Barone P, Pellecchia MT, Stanzione P, Brusa L, Bentivoglio AR, Bonuccelli U, Petrozzi L, Abbruzzese G, Marchese R, Cortelli P, Grimaldi D, Martinelli P, Ferrarese C, Garavaglia B, Sangiorgi S, Carelli V, Torroni A, Albanese A, Zeviani M. Mitochondrial DNA haplogroup K is associated with a lower risk of Parkinson’s disease in Italians. European journal of human genetics : EJHG. 2005;13:748–752. doi: 10.1038/sj.ejhg.5201425. [DOI] [PubMed] [Google Scholar]
- Giordano C, Iommarini L, Giordano L, Maresca A, Pisano A, Valentino ML, Caporali L, Liguori R, Deceglie S, Roberti M, Fanelli F, Fracasso F, Ross-Cisneros FN, D’Adamo P, Hudson G, Pyle A, Yu-Wai-Man P, Chinnery PF, Zeviani M, Salomao SR, Berezovsky A, Belfort R, Ventura DF, Moraes M, Moraes Filho M, Barboni P, Sadun F, De Negri A, Sadun AA, Tancredi A, Mancini M, d’Amati G, Loguercio Polosa P, Cantatore P, Carelli V. Efficient mitochondrial biogenesis drives incomplete penetrance in Leber’s hereditary optic neuropathy. Brain. 2014;137:335–353. doi: 10.1093/brain/awt343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano C, Montopoli M, Perli E, Orlandi M, Fantin M, Ross-Cisneros FN, Caparrotta L, Martinuzzi A, Ragazzi E, Ghelli A, Sadun AA, d’Amati G, Carelli V. Oestrogens ameliorate mitochondrial dysfunction in Leber’s hereditary optic neuropathy. Brain. 2011;134:220–234. doi: 10.1093/brain/awq276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Nonaka I, Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- Grady JP, Campbell G, Ratnaike T, Blakely EL, Falkous G, Nesbitt V, Schaefer AM, McNally RJ, Gorman GS, Taylor RW, Turnbull DM, McFarland R. Disease progression in patients with single, large-scale mitochondrial DNA deletions. Brain. 2014;137:323–334. doi: 10.1093/brain/awt321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan MX, Yan Q, Li X, Bykhovskaya Y, Gallo-Teran J, Hajek P, Umeda N, Zhao H, Garrido G, Mengesha E, Suzuki T, del Castillo I, Peters JL, Li R, Qian Y, Wang X, Ballana E, Shohat M, Lu J, Estivill X, Watanabe K, Fischel-Ghodsian N. Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am J Hum Genet. 2006;79:291–302. doi: 10.1086/506389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan MX, Fischel-Ghodsian N, Attardi G. Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rRNA mutation. Human molecular genetics. 2001;10:573–580. doi: 10.1093/hmg/10.6.573. [DOI] [PubMed] [Google Scholar]
- Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47:379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Guy J, Qi X, Pallotti F, Schon EA, Manfredi G, Carelli V, Martinuzzi A, Hauswirth WW, Lewin AS. Rescue of a mitochondrial deficiency causing Leber Hereditary Optic Neuropathy. Ann Neurol. 2002;52:534–542. doi: 10.1002/ana.10354. [DOI] [PubMed] [Google Scholar]
- Hamalainen RH, Manninen T, Koivumaki H, Kislin M, Otonkoski T, Suomalainen A. Tissue- and cell-type-specific manifestations of heteroplasmic mtDNA 3243A>G mutation in human induced pluripotent stem cell-derived disease model. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3622–3630. doi: 10.1073/pnas.1311660110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi J, Ohta S, Kikuchi A, Takemitsu M, Goto Y, Nonaka I. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:10614–10618. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JF, Parisi MA, Bennett JL, Clayton DA. Impairment of mitochondrial transcription termination by a point mutation associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1991;351:236–239. doi: 10.1038/351236a0. [DOI] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- Howell N, Kubacka I, Smith R, Frerman F, Parks JK, Parker WD. Association of the mitochondrial 8344 MERRF mutation with maternally inherited spinocerebellar degeneration and Leigh disease. Neurology. 1996;46:219–222. doi: 10.1212/wnl.46.1.219. [DOI] [PubMed] [Google Scholar]
- Hudson G, Amati-Bonneau P, Blakely EL, Stewart JD, He L, Schaefer AM, Griffiths PG, Ahlqvist K, Suomalainen A, Reynier P, McFarland R, Turnbull DM, Chinnery PF, Taylor RW. Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: a novel disorder of mtDNA maintenance. Brain. 2008;131:329–337. doi: 10.1093/brain/awm272. [DOI] [PubMed] [Google Scholar]
- Hudson G, Carelli V, Spruijt L, Gerards M, Mowbray C, Achilli A, Pyle A, Elson J, Howell N, La Morgia C, Valentino ML, Huoponen K, Savontaus ML, Nikoskelainen E, Sadun AA, Salomao SR, Belfort R, Jr, Griffiths P, Man PY, de Coo RF, Horvath R, Zeviani M, Smeets HJ, Torroni A, Chinnery PF. Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background. Am J Hum Genet. 2007;81:228–233. doi: 10.1086/519394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G, Nalls M, Evans JR, Breen DP, Winder-Rhodes S, Morrison KE, Morris HR, Williams-Gray CH, Barker RA, Singleton AB, Hardy J, Wood NE, Burn DJ, Chinnery PF. Two-stage association study and meta-analysis of mitochondrial DNA variants in Parkinson disease. Neurology. 2013;80:2042–2048. doi: 10.1212/WNL.0b013e318294b434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka T, Sakai F, Ide T, Miyakawa S, Sato M, Yoshii S. Regional cerebral blood flow and cerebrovascular reactivity during chronic stage of stroke-like episodes in MELAS -- implication of neurovascular cellular mechanism. J Neurol Sci. 2007;257:126–138. doi: 10.1016/j.jns.2007.01.040. [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Developmental cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann P, Koga Y, Shanske S, Hirano M, DiMauro S, King MP, Schon EA. Mitochondrial DNA and RNA processing in MELAS. Ann Neurol. 1996;40:172–180. doi: 10.1002/ana.410400208. [DOI] [PubMed] [Google Scholar]
- Kearns TP, Sayre GP. Retinitis pigmentosa, external ophthalmophegia, and complete heart block: unusual syndrome with histologic study in one of two cases. AMA Arch Ophthalmol. 1958;60:280–289. [PubMed] [Google Scholar]
- Khan NA, Auranen M, Paetau I, Pirinen E, Euro L, Forsstrom S, Pasila L, Velagapudi V, Carroll CJ, Auwerx J, Suomalainen A. Effective treatment of mitochondrial myopathy by nicotinamide riboside, a vitamin B3. EMBO molecular medicine. 2014;6:721–731. doi: 10.1002/emmm.201403943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrapko K, Coller HA, Andre PC, Li XC, Hanekamp JS, Thilly WG. Mitochondrial mutational spectra in human cells and tissues. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13798–13803. doi: 10.1073/pnas.94.25.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- King MP, Koga Y, Davidson M, Schon EA. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNA(Leu(UUR)) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Molecular and cellular biology. 1992;12:480–490. doi: 10.1128/mcb.12.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman MA, Yu-Wai-Man P, Korsten A, Leonhardt M, Dimitriadis K, De Coo IF, Klopstock T, Chinnery PF. Gene-environment interactions in Leber hereditary optic neuropathy. Brain. 2009;132:2317–2326. doi: 10.1093/brain/awp158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler CM, Lindberg GL, Brown DR, Beitz DC, Freeman AE, Mayfield JE, Myers AM. Replacement of bovine mitochondrial DNA by a sequence variant within one generation. Genetics. 1991;129:247–255. doi: 10.1093/genetics/129.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga Y, Povalko N, Nishioka J, Katayama K, Yatsuga S, Matsuishi T. Molecular pathology of MELAS and L-arginine effects. Biochim Biophys Acta. 2012;1820:608–614. doi: 10.1016/j.bbagen.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- Krishnan KJ, Reeve AK, Samuels DC, Chinnery PF, Blackwood JK, Taylor RW, Wanrooij S, Spelbrink JN, Lightowlers RN, Turnbull DM. What causes mitochondrial DNA deletions in human cells? Nat Genet. 2008;40:275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Lamantea E, Tiranti V, Bordoni A, Toscano A, Bono F, Servidei S, Papadimitriou A, Spelbrink H, Silvestri L, Casari G, Comi GP, Zeviani M. Mutations of mitochondrial DNA polymerase gammaA are a frequent cause of autosomal dominant or recessive progressive external ophthalmoplegia. Ann Neurol. 2002;52:211–219. doi: 10.1002/ana.10278. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Holme E, Kristiansson B, Oldfors A, Tulinius M. Progressive increase of the mutated mitochondrial DNA fraction in Kearns-Sayre syndrome. Pediatr Res. 1990;28:131–136. doi: 10.1203/00006450-199008000-00011. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Leber T. Über hereditäre und congenital angelegte Sehnervenleiden. Arch Ophthalmol. 1871;17:249–291. [Google Scholar]
- Leigh D. Subacute necrotizing encephalomyelopathy in an infant. Journal of neurology, neurosurgery, and psychiatry. 1951;14:216–221. doi: 10.1136/jnnp.14.3.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Okamoto K, Hayashi Y, Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 2004;119:873–887. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Lin CS, Sharpley MS, Fan W, Waymire KG, Sadun AA, Carelli V, Ross-Cisneros FN, Baciu P, Sung E, McManus MJ, Pan BX, Gil DW, Macgregor GR, Wallace DC. Mouse mtDNA mutant model of Leber hereditary optic neuropathy. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20065–20070. doi: 10.1073/pnas.1217113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luft R. The development of mitochondrial medicine. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:8731–8738. doi: 10.1073/pnas.91.19.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo SM, Ge ZJ, Wang ZW, Jiang ZZ, Wang ZB, Ouyang YC, Hou Y, Schatten H, Sun QY. Unique insights into maternal mitochondrial inheritance in mice. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13038–13043. doi: 10.1073/pnas.1303231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso M, Orsucci D, Angelini C, Bertini E, Carelli V, Comi GP, Donati A, Minetti C, Moggio M, Mongini T, Servidei S, Tonin P, Toscano A, Uziel G, Bruno C, Ienco EC, Filosto M, Lamperti C, Catteruccia M, Moroni I, Musumeci O, Pegoraro E, Ronchi D, Santorelli FM, Sauchelli D, Scarpelli M, Sciacco M, Valentino ML, Vercelli L, Zeviani M, Siciliano G. The m.3243A>G mitochondrial DNA mutation and related phenotypes. A matter of gender? J Neurol. 2014;261:504–510. doi: 10.1007/s00415-013-7225-3. [DOI] [PubMed] [Google Scholar]
- Mancuso M, Orsucci D, Angelini C, Bertini E, Carelli V, Comi GP, Minetti C, Moggio M, Mongini T, Servidei S, Tonin P, Toscano A, Uziel G, Bruno C, Caldarazzo Ienco E, Filosto M, Lamperti C, Martinelli D, Moroni I, Musumeci O, Pegoraro E, Ronchi D, Santorelli FM, Sauchelli D, Scarpelli M, Sciacco M, Spinazzi M, Valentino ML, Vercelli L, Zeviani M, Siciliano G. Phenotypic heterogeneity of the 8344A>G mtDNA “MERRF” mutation. Neurology. 2013;80:2049–2054. doi: 10.1212/WNL.0b013e318294b44c. [DOI] [PubMed] [Google Scholar]
- Manfredi G, Fu J, Ojaimi J, Sadlock JE, Kwong JQ, Guy J, Schon EA. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nat Genet. 2002;30:394–399. doi: 10.1038/ng851. [DOI] [PubMed] [Google Scholar]
- Manfredi G, Vu T, Bonilla E, Schon EA, DiMauro S, Arnaudo E, Zhang L, Rowland LP, Hirano M. Association of myopathy with large-scale mitochondrial DNA duplications and deletions: which is pathogenic? Ann Neurol. 1997;42:180–188. doi: 10.1002/ana.410420208. [DOI] [PubMed] [Google Scholar]
- Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Developmental cell. 2013;26:9–18. doi: 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JP, Davidson M, Koga Y, Schon EA, King MP. In vitro analysis of mutations causing myoclonus epilepsy with ragged-red fibers in the mitochondrial tRNA(Lys)gene: two genotypes produce similar phenotypes. Molecular and cellular biology. 1995;15:2872–2881. doi: 10.1128/mcb.15.5.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic D, Matic S, Kuhl I, Ruzzenente B, Freyer C, Jemt E, Park CB, Falkenberg M, Larsson NG. TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Human molecular genetics. 2013;22:1983–1993. doi: 10.1093/hmg/ddt051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes CT, Ciacci F, Silvestri G, Shanske S, Sciacco M, Hirano M, Schon EA, Bonilla E, DiMauro S. Atypical clinical presentations associated with the MELAS mutation at position 3243 of human mitochondrial DNA. Neuromuscular disorders : NMD. 1993;3:43–50. doi: 10.1016/0960-8966(93)90040-q. [DOI] [PubMed] [Google Scholar]
- Moraes CT, Shanske S, Tritschler HJ, Aprille JR, Andreetta F, Bonilla E, Schon EA, DiMauro S. mtDNA depletion with variable tissue expression: a novel genetic abnormality in mitochondrial diseases. Am J Hum Genet. 1991;48:492–501. [PMC free article] [PubMed] [Google Scholar]
- Nakada K, Inoue K, Ono T, Isobe K, Ogura A, Goto YI, Nonaka I, Hayashi JI. Inter-mitochondrial complementation: Mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nature medicine. 2001;7:934–940. doi: 10.1038/90976. [DOI] [PubMed] [Google Scholar]
- Nakada K, Sato A, Hayashi J. Mitochondrial functional complementation in mitochondrial DNA-based diseases. Int J Biochem Cell Biol. 2009;41:1907–1913. doi: 10.1016/j.biocel.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010a;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS biology. 2010b;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux RK, Nguyen KV. POLG mutations associated with Alpers’ syndrome and mitochondrial DNA depletion. Ann Neurol. 2004;55:706–712. doi: 10.1002/ana.20079. [DOI] [PubMed] [Google Scholar]
- Nishigaki Y, Marti R, Copeland WC, Hirano M. Site-specific somatic mitochondrial DNA point mutations in patients with thymidine phosphorylase deficiency. J Clin Invest. 2003;111:1913–1921. doi: 10.1172/JCI17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino I, Spinazzola A, Papadimitriou A, Hammans S, Steiner I, Hahn CD, Connolly AM, Verloes A, Guimaraes J, Maillard I, Hamano H, Donati MA, Semrad CE, Russell JA, Andreu AL, Hadjigeorgiou GM, Vu TH, Tadesse S, Nygaard TG, Nonaka I, Hirano I, Bonilla E, Rowland LP, DiMauro S, Hirano M. Mitochondrial neurogastrointestinal encephalomyopathy: an autosomal recessive disorder due to thymidine phosphorylase mutations. Ann Neurol. 2000;47:792–800. [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Developmental cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS, Kimura M, Sato S, Hattori N, Komatsu M, Tanaka K, Matsuda N. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes to cells : devoted to molecular & cellular mechanisms. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheu-Grau D, Gomez-Duran A, Lopez-Perez MJ, Montoya J, Ruiz-Pesini E. Mitochondrial pharmacogenomics: barcode for antibiotic therapy. Drug discovery today. 2010;15:33–39. doi: 10.1016/j.drudis.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Pakendorf B, Stoneking M. Mitochondrial DNA and human evolution. Annu Rev Genomics Hum Genet. 2005;6:165–183. doi: 10.1146/annurev.genom.6.080604.162249. [DOI] [PubMed] [Google Scholar]
- Pan BX, Ross-Cisneros FN, Carelli V, Rue KS, Salomao SR, Moraes-Filho MN, Moraes MN, Berezovsky A, Belfort R, Jr, Sadun AA. Mathematically modeling the involvement of axons in Leber’s hereditary optic neuropathy. Investigative ophthalmology & visual science. 2012;53:7608–7617. doi: 10.1167/iovs.12-10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Pavlakis SG, Phillips PC, DiMauro S, De Vivo DC, Rowland LP. Mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes: a distinctive clinical syndrome. Ann Neurol. 1984;16:481–488. doi: 10.1002/ana.410160409. [DOI] [PubMed] [Google Scholar]
- Payne BAI, Wilson IJ, Yu-Wai-Man P, Coxhead J, Deehan D, Horvath R, Taylor RW, Samuels DC, Santibanez-Koref M, Chinnery PF. Universal heteroplasmy of human mitochondrial DNA. Human molecular genetics. 2013;22:384–390. doi: 10.1093/hmg/dds435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perli E, Giordano C, Pisano A, Montanari A, Campese AF, Reyes A, Ghezzi D, Nasca A, Tuppen HA, Orlandi M, Di Micco P, Poser E, Taylor RW, Colotti G, Francisci S, Morea V, Frontali L, Zeviani M, d’Amati G. The isolated carboxy-terminal domain of human mitochondrial leucyl-tRNA synthetase rescues the pathological phenotype of mitochondrial tRNA mutations in human cells. EMBO molecular medicine. 2014;6:169–182. doi: 10.1002/emmm.201303198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer G, Horvath R, Klopstock T, Mootha VK, Suomalainen A, Koene S, Hirano M, Zeviani M, Bindoff LA, Yu-Wai-Man P, Hanna M, Carelli V, McFarland R, Majamaa K, Turnbull DM, Smeitink J, Chinnery PF. New treatments for mitochondrial disease-no time to drop our standards. Nat Rev Neurol. 2013;9:474–481. doi: 10.1038/nrneurol.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer G, Majamaa K, Turnbull DM, Thorburn D, Chinnery PF. Treatment for mitochondrial disorders. The Cochrane database of systematic reviews. 2012;4:Cd004426. doi: 10.1002/14651858.CD004426.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton J, Deadman ME, Gardiner RM. Duplications of mitochondrial DNA in mitochondrial myopathy. Lancet. 1989;1:236–240. doi: 10.1016/s0140-6736(89)91256-7. [DOI] [PubMed] [Google Scholar]
- Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- Raimundo N, Song L, Shutt TE, McKay SE, Cotney J, Guan MX, Gilliland TC, Hohuan D, Santos-Sacchi J, Shadel GS. Mitochondrial stress engages E2F1 apoptotic signaling to cause deafness. Cell. 2012;148:716–726. doi: 10.1016/j.cell.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju V, Calloway N, Ryan TA. Activity-driven local ATP synthesis is required for synaptic function. Cell. 2014;156:825–835. doi: 10.1016/j.cell.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaldo F, Amati-Bonneau P, Slama A, Romana C, Forin V, Doummar D, Barnerias C, Bursztyn J, Mayer M, Khouri N, Billette de Villemeur T, Burglen L, Reynier P, Bernabe Gelot A, Rodriguez D. MFN2, a new gene responsible for mitochondrial DNA depletion. Brain. 2012;135:1–4. doi: 10.1093/brain/aws111. [DOI] [PubMed] [Google Scholar]
- Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T. Mitochondrial threshold effects. The Biochemical journal. 2003;370:751–762. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzier C, Bannwarth S, Chaussenot A, Chevrollier A, Verschueren A, Bonello-Palot N, Fragaki K, Cano A, Pouget J, Pellissier JF, Procaccio V, Chabrol B, Paquis-Flucklinger V. The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy ‘plus’ phenotype. Brain. 2012;135:23–34. doi: 10.1093/brain/awr323. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:823–828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DC, Schon EA, Chinnery PF. Two direct repeats cause most human mtDNA deletions. Trends Genet. 2004;20:393–398. doi: 10.1016/j.tig.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Santorelli FM, Barmada MA, Pons R, Zhang LL, DiMauro S. Leigh-type neuropathology in Pearson syndrome associated with impaired ATP production and a novel mtDNA deletion. Neurology. 1996;47:1320–1323. doi: 10.1212/wnl.47.5.1320. [DOI] [PubMed] [Google Scholar]
- Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- Schaefer AM, McFarland R, Blakely EL, He L, Whittaker RG, Taylor RW, Chinnery PF, Turnbull DM. Prevalence of mitochondrial DNA disease in adults. Ann Neurol. 2008;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- Scheffler IE. Mitochondria. 2. Hoboken, NJ: John Wiley & Sons, Inc; 2009. [Google Scholar]
- Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nature reviews Genetics. 2012;13:878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon EA, Przedborski S. Mitochondria: the next (neurode)generation. Neuron. 2011;70:1033–1053. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon EA, Rizzuto R, Moraes CT, Nakase H, Zeviani M, DiMauro S. A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science. 1989;244:346–349. doi: 10.1126/science.2711184. [DOI] [PubMed] [Google Scholar]
- Shanske S, Tang Y, Hirano M, Nishigaki Y, Tanji K, Bonilla E, Sue C, Krishna S, Carlo JR, Willner J, Schon EA, DiMauro S. Identical mitochondrial DNA deletion in a woman with ocular myopathy and in her son with pearson syndrome. Am J Hum Genet. 2002;71:679–683. doi: 10.1086/342482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira Y, Harel S, Russell A. Mitochondrial encephalomyopathies: a group of neuromuscular disorders with defects in oxidative metabolism. Isr J Med Sci. 1977;13:161–164. [PubMed] [Google Scholar]
- Sheng ZH. Mitochondrial trafficking and anchoring in neurons: New insight and implications. J Cell Biol. 2014;204:1087–1098. doi: 10.1083/jcb.201312123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell. 1990;61:931–937. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- Shoffner JM, Lott MT, Voljavec AS, Soueidan SA, Costigan DA, Wallace DC. Spontaneous Kearns-Sayre/chronic external ophthalmoplegia plus syndrome associated with a mitochondrial DNA deletion: a slip-replication model and metabolic therapy. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:7952–7956. doi: 10.1073/pnas.86.20.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri G, Ciafaloni E, Santorelli FM, Shanske S, Servidei S, Graf WD, Sumi M, DiMauro S. Clinical features associated with the A-->G transition at nucleotide 8344 of mtDNA (“MERRF mutation”) Neurology. 1993;43:1200–1206. doi: 10.1212/wnl.43.6.1200. [DOI] [PubMed] [Google Scholar]
- Skladal D, Halliday J, Thorburn DR. Minimum birth prevalence of mitochondrial respiratory chain disorders in children. Brain. 2003;126:1905–1912. doi: 10.1093/brain/awg170. [DOI] [PubMed] [Google Scholar]
- Soong NW, Hinton DR, Cortopassi G, Arnheim N. Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nat Genet. 1992;2:318–323. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- Spinazzola A, Zeviani M. Disorders of nuclear-mitochondrial intergenomic signaling. Gene. 2005;354:162–168. doi: 10.1016/j.gene.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Moraes CT. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Human molecular genetics. 2005;14:893–902. doi: 10.1093/hmg/ddi082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky FH, Lee S, Wibom R, Olson L, Larsson NG. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12937–12942. doi: 10.1073/pnas.1103295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JB, Freyer C, Elson JL, Larsson NG. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nature reviews Genetics. 2008a;9:657–662. doi: 10.1038/nrg2396. [DOI] [PubMed] [Google Scholar]
- Stewart JB, Freyer C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A, Larsson NG. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS biology. 2008b;6:e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Gorska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitin tag for sperm mitochondria. Nature. 1999;402:371–372. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sparman M, Sritanaudomchai H, Ma H, Clepper L, Woodward J, Li Y, Ramsey C, Kolotushkina O, Mitalipov S. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature. 2009;461:367–372. doi: 10.1038/nature08368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes & development. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatuch Y, Christodoulou J, Feigenbaum A, Clarke JT, Wherret J, Smith C, Rudd N, Petrova-Benedict R, Robinson BH. Heteroplasmic mtDNA mutation (T----G) at 8993 can cause Leigh disease when the percentage of abnormal mtDNA is high. Am J Hum Genet. 1992;50:852–858. [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Achilli A, Macaulay V, Richards M, Bandelt HJ. Harvesting the fruit of the human mtDNA tree. Trends Genet. 2006;22:339–345. doi: 10.1016/j.tig.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Human molecular genetics. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Haines JL, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH, Jr, Goetz CG, Small GW, Mastaglia F, Stajich JM, McLaurin AC, Middleton LT, Scott BL, Schmechel DE, Pericak-Vance MA, Vance JM. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Martin JJ, Dermaut B, Lofgren A, Wibail A, Ververken D, Tack P, Dehaene I, Van Zandijcke M, Moonen M, Ceuterick C, De Jonghe P, Van Broeckhoven C. Recessive POLG mutations presenting with sensory and ataxic neuropathy in compound heterozygote patients with progressive external ophthalmoplegia. Neuromuscular disorders : NMD. 2003;13:133–142. doi: 10.1016/s0960-8966(02)00216-x. [DOI] [PubMed] [Google Scholar]
- Van Laar VS, Arnold B, Cassady SJ, Chu CT, Burton EA, Berman SB. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Human molecular genetics. 2011;20:927–940. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47:365–378. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet. 2008;40:1484–1488. doi: 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial DNA mutations and neuromuscular disease. Trends Genet. 1989;5:9–13. doi: 10.1016/0168-9525(89)90005-x. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Bioenergetics in human evolution and disease: implications for the origins of biological complexity and the missing genetic variation of common diseases. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2013;368:20120267. doi: 10.1098/rstb.2012.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ, Nikoskelainen EK. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- Wanagat J, Cao Z, Pathare P, Aiken JM. Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2001;15:322–332. doi: 10.1096/fj.00-0320com. [DOI] [PubMed] [Google Scholar]
- Wang J, Silva JP, Gustafsson CM, Rustin P, Larsson NG. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4038–4043. doi: 10.1073/pnas.061038798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Bruning JC, Kahn CR, Clayton DA, Barsh GS, Thoren P, Larsson NG. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- Wang T, Sha H, Ji D, Zhang HL, Chen D, Cao Y, Zhu J. Polar body genome transfer for preventing the transmission of inherited mitochondrial diseases. Cell. 2014;157:1591–1604. doi: 10.1016/j.cell.2014.04.042. [DOI] [PubMed] [Google Scholar]
- Wredenberg A, Wibom R, Wilhelmsson H, Graff C, Wiener HH, Burden SJ, Oldfors A, Westerblad H, Larsson NG. Increased mitochondrial mass in mitochondrial myopathy mice. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15066–15071. doi: 10.1073/pnas.232591499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T, Suzuki T, Ishii N, Ohta S, Watanabe K. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 2001;20:4794–4802. doi: 10.1093/emboj/20.17.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T, Suzuki T, Ueda T, Ohta S, Watanabe K. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J Biol Chem. 2000;275:4251–4257. doi: 10.1074/jbc.275.6.4251. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nature reviews Molecular cell biology. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Wai-Man P, Griffiths PG, Chinnery PF. Mitochondrial optic neuropathies - disease mechanisms and therapeutic strategies. Progress in retinal and eye research. 2011;30:81–114. doi: 10.1016/j.preteyeres.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Wai-Man P, Griffiths PG, Gorman GS, Lourenco CM, Wright AF, Auer-Grumbach M, Toscano A, Musumeci O, Valentino ML, Caporali L, Lamperti C, Tallaksen CM, Duffey P, Miller J, Whittaker RG, Baker MR, Jackson MJ, Clarke MP, Dhillon B, Czermin B, Stewart JD, Hudson G, Reynier P, Bonneau D, Marques W, Lenaers G, McFarland R, Taylor RW, Turnbull DM, Votruba M, Zeviani M, Carelli V, Bindoff LA, Horvath R, Amati-Bonneau P, Chinnery PF. Multi-system neurological disease is common in patients with OPA1 mutations. Brain. 2010;133:771–786. doi: 10.1093/brain/awq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeviani M, Moraes CT, DiMauro S, Nakase H, Bonilla E, Schon EA, Rowland LP. Deletions of mitochondrial DNA in Kearns-Sayre syndrome. Neurology. 1988;38:1339–1346. doi: 10.1212/wnl.38.9.1339. [DOI] [PubMed] [Google Scholar]
- Zeviani M, Servidei S, Gellera C, Bertini E, DiMauro S, DiDonato S. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature. 1989;339:309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schroder JM, Vance JM. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]