Abstract
In the post-genomic era, a pressing challenge to biological scientists is to understand the organization of gene functions, the interaction between gene and nutrient environment, and the genesis of phenotypes. Metabolomics, the quantitation of low molecular weight compounds, has been used to provide a phenotypic description of a cell or tissue by a set of metabolites. Gene function is hypothesized from its correlation with the corresponding set of macromolecules by transcriptomics or proteomics. Another approach to genotype–phenotype correlation is by the reconstruction of genome-scale metabolic maps. The utilization of specific pathways as predicted by reaction network analysis provides the phenotypic characterization of a cell, which can be plotted on a phenotypic phase plane. Tracer based metabolomics is the experimental approach to reaction network analysis using stable isotope tracers. The redistribution of the isotope tracer among metabolic intermediates is used to identify a finite number of pathways, the utilization of which is characteristic of the phenotypic behavior of cells. In this paper, we review tracer based metabolomic methods for the construction of phenotypic phase plane plots, and discuss the functional implications of phenotypic phase plane analysis. Examples of phenotypic changes in response to differentiation, inhibition of signaling pathways and perturbation in nutrient environment are provided.
KEY WORDS: tracer based metabolomics, reaction network, phenotypic phase plane analysis, constraint based modeling
Introduction
Since the completion of the Human Genome Project, it is increasingly recognized that the phenotypes of many inherited disorders are often not predictable from genome sequence-based analysis, and the pathogenesis of disease is often the result of complex interaction between gene and environment. A fundamental challenge to biologists is to develop an experimental approach to establish the phenotypic properties of a cell that allows the understanding of the organization of gene functions, the effect of nutrient environment, and the role of cell to cell interaction in a multicellular organism. In this effort, metabolomics (metabonomics), the quantitation of low molecular weight compounds, plays an important role in providing a set of metabolites in a cell or tissue (Harrigan and Goodacre, 2003; Goodacre et al., 2004) which is complementary to the set of macromolecules determined by transcriptomics or proteomics. It is anticipated from the theory of metabolic control analysis (MCA) that changes in levels of metabolic intermediates of a sequential series of reactions are often more pronounced than the changes in enzyme kinetics or individual fluxes (Kell and Westerholf, 1986; Fell, 1996). For this reason, metabolomics is considered to be a sensitive tool for detecting genetic mutations and genotype–phenotype correlation. Many successful applications of metabonomics have been reported in the area of toxicology research from the Consortium on Metabonomics in Toxicology (COMET) (Nicholson et al., 2002, Lindon et al., 2005). Metabolomics has also been applied to characterize phenotype of organisms with genetic mutations (Raamsdonk et al., 2001; Allen et al., 2003). However, despite such successes we are still far from fully understanding the inner workings (functional phenotype) of an organism (Voit and Almeida, 2004).
Limitation of analyses substrates and fluxes
Quantitation of metabolic intermediates and determination of reaction kinetics are the basic tools of classical biochemistry. The availability of high throughput analytical methodologies of NMR or mass spectrometry has greatly expanded the number of compounds that can be detected and quantitated by orders of magnitude. Metabolomics, the quantitation of low molecular weight compounds, and fluxomics (Sanford et al., 2002), the determination of fluxes, are natural extension of classical biochemistry except at much larger scales. To understand the limitation of classical biochemistry as a tool for the investigation of system properties of a cell, let us consider the example of a metabolic network shown in figure 1 (top).
Figure 1.
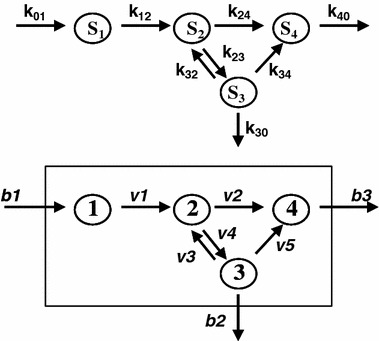
Metabolic (reaction) network from two different perspectives. A system of biochemical reactions can be analyzed using classical distribution analysis (top) with emphasis on the determination of individual flux (k ij’s) and pool sizes (S i’s)(concentrations). The metabolic network can also be studied from a bioengineering point of view (bottom), i.e., the response characteristics of the system of fluxes (v’s) enclosed within the boundary.
The example is a system of four variables and eight parameters indicated as four substrate pools (1 to 4) and the respective eight kinetic constants kij. The functional properties of the system is represented by a set of flux equations:
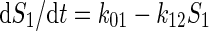 |
1 |
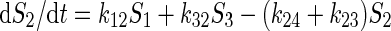 |
2 |
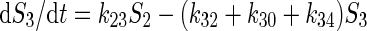 |
3 |
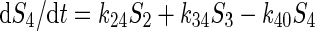 |
4 |
Solving the system requires four metabolite measurements and eight flux measurements, not all of which can be determined at any one time. Thus, even with a complete analysis of metabolic intermediates (metabolomics) or a complete analysis of individual fluxes (fluxomics), the set of equations still remain inherently underdetermined, and the behavior of the system in response to changes of substrate or kinetic parameter cannot be accurately predicted. Metabolomics and fluxomics have the same limitation of classical biochemistry in addition to the burden of the computational challenges of large data set (bioinformatics) such as principal component analysis (PCA), partial least squares analysis (PLS) and other clustering analyses (Brown et al., 2005; Harrigan et al., 2005).
Reaction network analysis
In order to handle the problem of finding the solution to a large underdetermined set of differential equations, biologists have turned to the well developed engineering model of reaction network analysis (Schilling et al., 1999, 2000). The engineering approach to reaction network analysis is fundamentally different from that of classical biochemistry. It begins with a definition of the system’s boundary (figure 1, bottom). The system’s boundary separates two sets of kinetic parameters of substrate fluxes (bi’s and vj’s). The vj’s are internal fluxes subject to stoichiometric constraints and the bi’s are the individual system input and output of the various substrates (Palsson et al., 2003). The arrangement of these equations in terms of bi’s and vj’s permits the application of linear programming algorithm to solve for values satisfying the optimization (maximization or minimization) of an objective function. The solution of this underdetermined set of equations, subject to non-zero constraints of substrates and products, gives a convex conical solution space (Famili and Palsson, 2003) representing all potential phenotypes of the reaction network (cell). Essentially the problem of describing the phenotypic behavior of a cell (its metabolic network) is converted to an input–output analysis of the system. The use of reaction network analysis in the studies of cellular metabolism has been extensively reviewed (Papin et al., 2003; Price et al., 2003; Reed and Palsson, 2003). The phenotypic behavior of a whole cell system can be understood in terms of its response to substrate environment changes (b’s) or to transcriptional regulation of the reaction rates of metabolic pathways (v’s). This is illustrated by the example in figure 2. In this arbitrary example, there are three principal pathways (P1, P2 and P3) in the reaction network. The table accompanying figure 2 provides four different scenarios where some of the individual fluxes v’s are set to zero, and their respective effect on the system’s output in P’s. The first column contains values of P1, P2 and P3 and the total input at the basal condition. Input is maintained at 12 units for conditions in column 2 (v2=0) and 4 (v5=0). The input is changed 6 units in column 3 with v4 set to zero.
Figure 2.
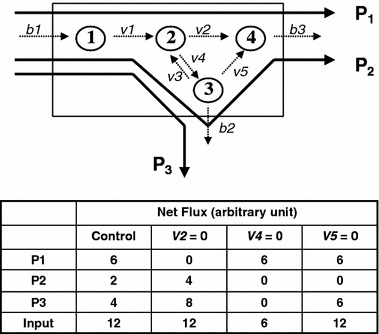
The identification of the paths (sequence of reactions (v’s)) within the reaction network by which inputs of precursors are converted to specific products (b’s). The input and output characteristics of four scenarios of specific changes in the kinetic parameters are provided in the inset.
Metabolic phenotypic phase plane analysis
The solution of reaction network analysis is a high dimension convex flux cone (Schilling and Palsson, 1998). The phenotype of a cell is represented by a point in the flux cone and is a highly abstract concept. In order to fully understand the significance of reaction network analysis, the conical solution space is resolved into a number of phenotypic phase planes where the line of optimality for the observed basal condition in the particular plane can be drawn. It should be noted that this plane is not infinite and is bounded by the maximum possible input and output of the network. The metabolic network properties of different areas on the plane can be predicted when the network is fully reconstructed from genomic database using a constrained-based model (Edwards et al., 2002; Schilling et al., 2000).
Phenotypic phase plane analysis is an important aspect of reaction network analysis. It generally falls into three categories: input–output analysis, output–output analysis and input–input analysis. Such analysis provides insightful information regarding the metabolic properties of the network and their relations to phenotypic changes. To illustrate, the examples given in figure 2 are plotted onto an input-output phase plane (figure 3) and an output–output phase plane (figure 4). The example in figure 2 has only one input and there is no input–input phase plane. In reality, the inputs into a cellular system are always multiple, and the input–input phase plane analysis is very useful to demonstrate the complementarities of substrates.
Figure 3.
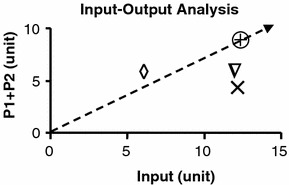
The plot of output (P1+P2) against input as phenotype in phase plane analysis. The basal phenotype is indicated by
 . The condition v
5=0 is indicated by Δ, v
2=0 indicated by ×, and v
4=0 indicated by
. The condition v
5=0 is indicated by Δ, v
2=0 indicated by ×, and v
4=0 indicated by
 . The slope of the line indicates the amount of input required for a certain amount of output.
. The slope of the line indicates the amount of input required for a certain amount of output.
Figure 4.
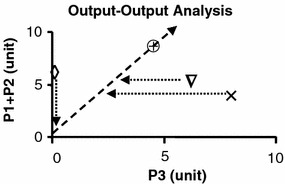
The plot of output (P1+P2) against P3 as phenotype in phase plane analysis. The basal phenotype is indicated by
 . The condition v
5=0 is indicated by Δ, v
2=0 indicated by × and v
4=0 indicated by
. The condition v
5=0 is indicated by Δ, v
2=0 indicated by × and v
4=0 indicated by
 . The slope of the line indicates the relative proportion of each product under optimal condition. Such a proportion is the result of optimal function of the network. The lines drawn in parallel to each of the axis intersect with the line of optimality reflecting restrictions on optimal phenotype.
. The slope of the line indicates the relative proportion of each product under optimal condition. Such a proportion is the result of optimal function of the network. The lines drawn in parallel to each of the axis intersect with the line of optimality reflecting restrictions on optimal phenotype.
Input–output analysis
In figure 3, values for P1+P2 are plotted against total inputs. The line of optimality is arbitrarily defined as the line drawn through the point for the basal state (
 ) corresponding to conditions satisfying the objective function. The slope of the line describes the optimal level of product formation of P1 and P2 for a given level of input. In bacteria and yeasts systems, the increase in cell mass is often used as the objective function. In mammalian cell systems such as in liver cells, the objective functions can be the production of substrates or elimination of toxins. The line of optimality bisects the phenotypic phase plane. The area above the line represents substrate deficient and the area below the line, substrate excess condition. It is clear that changes in internal fluxes (v’s) affect the cell’s phenotype as indicated by the position on the phenotypic phase plane. When v5 (indicated by Δ) and v2 (indicated by ×) are set to zero, the input is excessive for the amount of P1+P2 produced. Under such a condition, it is expected that the substrate input have to be dissipated through other metabolic processes, in this example the production of P3. The converse is true for phenotype located above the line of optimality when v4=0 (indicated by
). In the situation where v4 is set to zero, the survival of this phenotype depends on the external supply of P3. An interesting application of this phase plane analysis is in the prediction of cell-to-cell competition in co-culture systems. For example, cells with defects in v2, v4 or v5 are expected not to compete well with the wild-type (basal) when they are co-cultured because they are metabolically less efficient. However, cells with defect in v4 can thrive in co-culture with cells having defect in v2 or v5. This is due to the metabolic complementarities among cells with defect in v4 and those with defect in v2 or v5. The excess production of P3 by cells with defect in v2 or v5 can be used to support the growth of cells with defect in v4. Cooperation among cells with defect in v2 and those with defect in v5 is not possible since they both produce excess P3.
) corresponding to conditions satisfying the objective function. The slope of the line describes the optimal level of product formation of P1 and P2 for a given level of input. In bacteria and yeasts systems, the increase in cell mass is often used as the objective function. In mammalian cell systems such as in liver cells, the objective functions can be the production of substrates or elimination of toxins. The line of optimality bisects the phenotypic phase plane. The area above the line represents substrate deficient and the area below the line, substrate excess condition. It is clear that changes in internal fluxes (v’s) affect the cell’s phenotype as indicated by the position on the phenotypic phase plane. When v5 (indicated by Δ) and v2 (indicated by ×) are set to zero, the input is excessive for the amount of P1+P2 produced. Under such a condition, it is expected that the substrate input have to be dissipated through other metabolic processes, in this example the production of P3. The converse is true for phenotype located above the line of optimality when v4=0 (indicated by
). In the situation where v4 is set to zero, the survival of this phenotype depends on the external supply of P3. An interesting application of this phase plane analysis is in the prediction of cell-to-cell competition in co-culture systems. For example, cells with defects in v2, v4 or v5 are expected not to compete well with the wild-type (basal) when they are co-cultured because they are metabolically less efficient. However, cells with defect in v4 can thrive in co-culture with cells having defect in v2 or v5. This is due to the metabolic complementarities among cells with defect in v4 and those with defect in v2 or v5. The excess production of P3 by cells with defect in v2 or v5 can be used to support the growth of cells with defect in v4. Cooperation among cells with defect in v2 and those with defect in v5 is not possible since they both produce excess P3.
Output–output analysis
The values of P1+P2 are plotted against those of P3 in figure 4. The slope of the line of optimality represents the optimal ratio of the products (P1+P2) to P3. This phase plane analysis provides information on the relative importance of the two products to the phenotype of the cell. The information in figure 4 complements that presented in figure 3. The relative excess and deficiency of P3 is readily apparent. When a line is drawn from a phenotype in parallel to the major axis, the intersection between the line of optimality and the parallel line indicates the degree of optimality relative to the basal state. In this example, the condition where
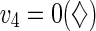 is shown as a non-viable condition.
is shown as a non-viable condition.
Tracer-based metabolomics 1
Reaction network analysis has its experimental counterpart in tracer-based metabolomics (Boros et al., 2002a, b). When a 13C labeled substrate is introduced into a biological system, 13C is incorporated into a wide range of metabolites of the metabolome either through exchange or by direct synthesis. The incorporation of a labeled carbon molecule into a metabolic product generates a “mass” signature (a difference in molecular weight from the naturally existing compound), which permits detection by mass spectrometry or by NMR. The intracellular reaction network that a labeled carbon traverses, determines the distribution of the isotope in its metabolic products. Therefore, the metabolic phenotype determines tracer distribution within individual compounds and distribution among compounds. Such a distribution represents the metabolic functions of the cell and defines its metabolic phenotype as would be predicted by reaction network analysis.
Figure 5 shows an example of a tracer-based metabolomic study of the tricarboxylic acid (TCA) cycle (Lee, 1993). In this TCA cycle subsystem, pyruvate is the input and glutamate is the output. Pyruvate is first converted to oxaloacetate by pyruvate carboxylase (PC) or acetyl-CoA pyruvate dehydrogenase (PDH). These two products ultimately contribute the necessary 3-carbon or 2-carbon units in the synthesis of glutamate. In addition,
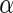 -ketoglutarate, an intermediate in the glutamate synthesis, may be recycled through the TCA cycle. In fact, there are three major paths that pyruvate can contribute to glutamate synthesis. These are indicated as P1 (blue line), P2 (red line) and P3 (dotted line) in figure 5. If the carbon atoms in position 2 and 3 of pyruvate are substituted with 13C, the three paths result in distinct isotopomer products (Boros et al., 2002b). [4, 5−13C2]glutamate is the product of P1, [2, 3−13C2]glutamate, the product of P2, and [1−13C]-, [2−13C] and [3−13C]-glutamate, the products of P3. The relative abundance of these products can be determined using gas chromatography/mass spectrometry. The mass isotopomer analysis of glutamate for products of P1, P2 and P3 is shown in figure 6. The trifluoroacetamide-butyl-ester derivative of glutamate gives two major fragments. The fragment at m/z 198 contains carbons C2–C5, and the fragment at m/z 152, the carbons of C2-C4 (Lee et al., 1996). By simple arithmetic manoeuvre, the relative contribution of P1, P2 and P3 are determined.
-ketoglutarate, an intermediate in the glutamate synthesis, may be recycled through the TCA cycle. In fact, there are three major paths that pyruvate can contribute to glutamate synthesis. These are indicated as P1 (blue line), P2 (red line) and P3 (dotted line) in figure 5. If the carbon atoms in position 2 and 3 of pyruvate are substituted with 13C, the three paths result in distinct isotopomer products (Boros et al., 2002b). [4, 5−13C2]glutamate is the product of P1, [2, 3−13C2]glutamate, the product of P2, and [1−13C]-, [2−13C] and [3−13C]-glutamate, the products of P3. The relative abundance of these products can be determined using gas chromatography/mass spectrometry. The mass isotopomer analysis of glutamate for products of P1, P2 and P3 is shown in figure 6. The trifluoroacetamide-butyl-ester derivative of glutamate gives two major fragments. The fragment at m/z 198 contains carbons C2–C5, and the fragment at m/z 152, the carbons of C2-C4 (Lee et al., 1996). By simple arithmetic manoeuvre, the relative contribution of P1, P2 and P3 are determined.
Figure 5.
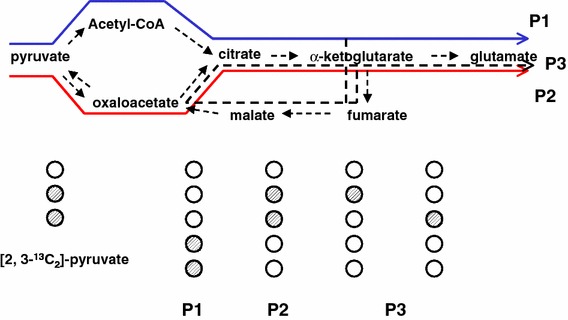
The tricarboxylic acid cycle (TCA) subsystem for the production of glutamate from pyruvate. Three major paths relating output to input are shown. P1 (in blue) is series of reaction that convert pyruvate to glutamate through the pyruvate dehydrogenase (PDH) pathway. The product from each path has a specific “mass” signature when specific labeled precursor is used.
Figure 6.
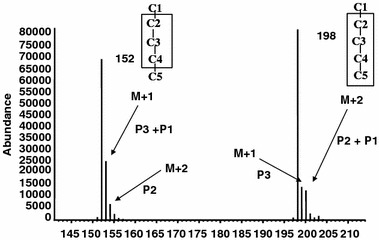
Mass spectrum of trifluoroacetamide butyl-ester of glutamate showing the two fragments corresponding to C2–C4 and C2–C5 of glutamate with specific mass shift corresponding to P1, P2 and P3 due to the presence of 13C carbons.
In the past decade, many labeling approaches have been used in whole cell systems including 13C labeled glucose (Marin et al., 2004), lactate (Xu et al., 2002, 2003), acetate (Lee et al., 1996; Garg et al., 2005), butyrate (Boren et al., 2003), propionate (Jones et al., 1997) and fatty acids (Lee et al., 1995; Lee et al., 1998a; Wong et al., 2004). Mass isotopomer analyses of products from these labeled substrates have been reviewed (Boros et al., 2002b). In addition to providing information regarding specific pathways, the results from each labeled precursor can be used in metabolic phenotypic phase plan analysis, and inference on the metabolic efficiency can be made of the cellular system. The methodology of mass isotopomer analysis is the experimental tool for phenotypic characterization with tracer based metabolomics and network analysis is the theoretical foundation for the interpretation of metabolic phenotypes (figure 7). Tracer-based metabolomics has been applied to characterize phenotypic changes in response to differentiation (Boros et al., 2002c), activation (Boros et al., 2000) and inhibition (Boren et al., 2001) of signaling pathways and perturbation in nutrient environment. An example of each is presented below.
Figure 7.
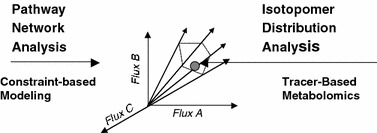
The relationship between pathway network analysis and isotopomer distribution analysis. Pathway network is fully reconstructed from genomic database using a constrained-based model. Linear programming is then used to solve for all possible solutions, the result of which is a metabolic phenotype space in the form of a convex cone. Tracer based metabolomics is the experimental approach by which a specific metabolic phenotype can be defined.
Tracer-based metabolomics has been applied to characterize phenotypic changes in cell differentiation of immature lung fibroblasts (Boros et al., 2002c). Immature rat lung fibroblasts are characterized by the presence of an adipogenic biomarker (adipose differentiation related protein ADRP) and the capacity for lipogenesis. When these cells are exposed to high oxygen tension, they lose the adipogenic biomarker and trans-differentiate into a myofibroblast like phenotype. This trans-differentiation is illustrated by the change in location in the ribose synthesis phase plane in figure 8. There are two major branches of the pentose phosphate pathways: the oxidative by glucose-6-phosphate dehydrogenase pathway and the non-oxidative by the transketolase/transaldolase pathways. The oxidation of [1, 2−13C2]-glucose results in M+1 species of ribose while the non-oxidative synthesis of ribose results in mostly M+2 species of ribose (Lee et al., 1998b).2 The relative contribution of oxidative and non-oxidative branch of the pentose cycle to ribose synthesis can be estimated from the ratio of these molecular species. When immature lung fibroblasts were incubated with [1, 2−13C2]-glucose, the transdifferentiated phenotype was shown to utilize the non-oxidative pathway of pentose synthesis more than the oxidative pathway (figure 8). Consequently, for the same glucose uptake, less reducing equivalents are generated from the oxidative pathway resulting in less de novo lipogenesis. The reduced lipogenesis from glucose also makes available glucose for the non-oxidative pathway of pentose synthesis suggesting a proliferative phenotype.
Figure 8.
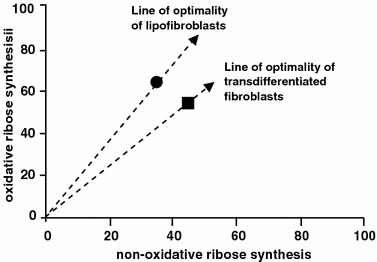
Non-oxidative and oxidative ribose synthesis phase plane (output–output analysis). Under high oxygen exposure, lipofibroblasts (•) from immature lung differentiate into myofibroblasts (
 ). The metabolic phenotype change is indicated by the increased use of the non-oxidative pathways for ribose synthesis and its restriction on the production of reducing equivalents from the oxidative pathway. Data are adapted from article of Boros et al. (2002c).
). The metabolic phenotype change is indicated by the increased use of the non-oxidative pathways for ribose synthesis and its restriction on the production of reducing equivalents from the oxidative pathway. Data are adapted from article of Boros et al. (2002c).
Metabolic changes during the activation or inhibition of signaling pathways can also be studied using tracer-based metabolomics. An example of such application is provided by the study of a tyrosine kinase inhibitor (Gleevac) on myeloid leukemic cells (Boren et al., 2001). A significant effect of Gleevac on the metabolism of myeloid leukemic cells is the reduction in glucose utilization and new palmitate synthesis per cell (figure 9). When these parameters are analyzed by phenotypic phase plane analysis, it is apparent that MIA cells and the myeloid leukemic cells have distinct metabolic phenotype characterized by the differences in glucose utilization and palmitate synthesis.3 Under Gleevac treatment, the utilization of glucose and palmitate synthesis progressively diminishes. The new phenotypes are located below the line of optimality meaning that there is relatively more reduction in palmitate synthesis than glucose utilization. The reduction in glucose utilization was supported by the reduction in hexose kinase activity and the diminished fatty acid synthesis by the impressive decrease in G6PDH activity (Boren et al., 2001). The phenotypic phase plane analysis demonstrates that changes in the activity of these enzymes are probably the primary effects of the inhibition of tyrosine kinase signaling pathway by Gleevac.
Figure 9.
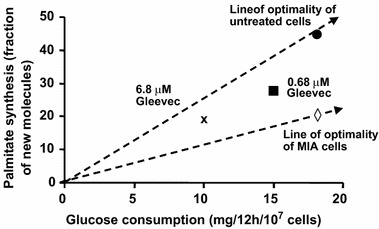
Glucose consumption (input) and palmitate synthesis (output) phase plane. Phenotype of treated and untreated MIA cells is shown as (
 ). Phenotype of untreated myeloid leukemic cells are shown as •. Phenotype of myeloid leukemic cells treated with low dose and high dose Gleevac are shown as
). Phenotype of untreated myeloid leukemic cells are shown as •. Phenotype of myeloid leukemic cells treated with low dose and high dose Gleevac are shown as
 and x. MIA cells and myeloid leukemic cells are cancer cells but have different metabolic requirement for their growth as indicated by different lines of optimality. Under Gleevac treatment, the phenotype of leukemics cells deviate from the line of optimality and exhibit reduced growth due to metabolic inefficiency. Data are adapted from article of Boren et al. (2001).
and x. MIA cells and myeloid leukemic cells are cancer cells but have different metabolic requirement for their growth as indicated by different lines of optimality. Under Gleevac treatment, the phenotype of leukemics cells deviate from the line of optimality and exhibit reduced growth due to metabolic inefficiency. Data are adapted from article of Boren et al. (2001).
The influence of substrate environment on the phenotypic behavior of cells can be seen from the example of colon cancer cells (HT29 cells) in response to butyrate treatment (Boren et al., 2003). Colonic epithelial cells are highly adapted to and dependent on butyrate of the colonic environment as a nutrient substrate. Colonic epithelium is often noted to be atrophic in the colon stumps of colostomy subjects when butyrate is absent. In malignant transformation, HT29 cells acquired the capability of using glucose for cell proliferation. Therefore, the utilization of glucose and butyrate by HT29 cells is different from that of normal colon cells. This is shown in the phenotypic phase plane analysis of butyrate/glucose utilization for acetyl-CoA (figure 10). HT29 cells possess the metabolic machinery for the utilization of glucose as a substrate. Thus at low butyrate environment, these cells (marked by x) convert glucose to acetyl-CoA. Under high butyrate environment, despite the same availability of glucose, HT29 cells (marked by ) preferentially utilize butyrate as a nutrient source for acetyl-CoA production. On the other hand, pancreatic cancer (MIA) cells have the same metabolic characteristics as HT29 cells when cultured with low (
) preferentially utilize butyrate as a nutrient source for acetyl-CoA production. On the other hand, pancreatic cancer (MIA) cells have the same metabolic characteristics as HT29 cells when cultured with low (
 ) or high (circled
) or high (circled
 ) butyrate. In a high butyrate environment, HT29 and MIA cells are capable of using butyrate, and less glucose is used. Since HT29 cells treated with high dose of butyrate expressed the biomarker for differentiation (Boren et al., 2003), one can imagine a line in this phenotypic phase plane below which HT29 cells would undergo differentiation to assume a less proliferative phenotype of normal colon cells.
) butyrate. In a high butyrate environment, HT29 and MIA cells are capable of using butyrate, and less glucose is used. Since HT29 cells treated with high dose of butyrate expressed the biomarker for differentiation (Boren et al., 2003), one can imagine a line in this phenotypic phase plane below which HT29 cells would undergo differentiation to assume a less proliferative phenotype of normal colon cells.
Figure 10.
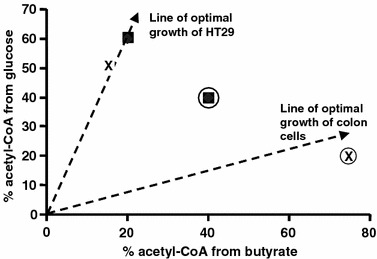
Glucose and butyrate utilization phase plane (output-output analysis). Utilization of glucose and butyrate under low and high butyrate (circled) conditions by HT29 colonic cancer cells (x) and MIA cells (□) are plotted. Under high butyrate condition, the growth of HT29 cells was reduced and cells expressed alkaline phosphatase as a differentiation marker suggesting different line of optimality for HT29 cells and normal colonic cells.
Phenotypic phase plane analysis is an important application of tracer-based metabolomics and reaction network analysis. The metabolic phenotype of a cell is resolved into a finite number of phase planes whose axes are pair-wise inputs and outputs4 of metabolites from a cell through particular series of reaction pathways. The phenotype of cells under a given nutrient environment is represented by a point in a phenotypic phase plane. Under basal conditions, the line through the point of the phenotype from the origin represents the line of optimality. The line of optimality has important implications on the function of the metabolic reaction network. Under the same culture conditions, cells from different tissue types often have different lines of optimality suggesting different efficiencies in specific substrate utilization (figure 10). As demonstrated in figures 9 and 10, the phenotype of cells deviates from the line of optimality in response to changes in nutrient environment, or to the activation or inhibition of signaling pathways.
Concluding remarks
Metabolomics has its origin in the profiling of small molecules and has been shown to be useful for phenotypic characterization by the metabolite composition of samples obtained from subjects. The main purpose of this approach is to distinguish one population from another by such characteristics, and it has been recognized as an important field in systems biology complementary to those of transcriptomics and proteomics. Tracer based metabolomics is the confluence of reaction network analysis and metabolite profiling. It takes advantage of the analytical methods of metabolite profiling and the dynamic information of flux analysis. A major distinction between tracer-based metabolomics, and NMR analysis in metabonomics, is the use of a conceptual or cellular boundary in tracer based metabolomics; and the results of tracer based metabolomics are analyzed as inputs and outputs from the cell according to metabolic network analysis from a systems biology perspective. Instead of characterizing phenotype as a collection of reactions and metabolites, tracer-based metabolomics characterizes phenotype as the input–output characteristics of the whole metabolic network (cell). Phenotypic behavior of a cell is the consequence of the function of the reaction network within the cell. The phenotypic behavior of a cell is the response of the cell to substrate inputs and environmental changes, and is constrained by the genetic makeup of the cell including the transcriptional regulation by cell signals and environmental cues. The combined use of multiple tracers in separate experiments provides a more comprehensive view of the metabolic phenotype in the form of an array (SIDMAParray®) (Boros et al., 2004; Harrigan et al., 2005) and the impact of gene and nutrient changes on the performance of the metabolic network can be gleaned from such a data set.
A prevailing assumption in biology is that there is a one-to-one correspondence between genotype and phenotype. Phenotypic variations such as susceptibility to diseases are often attributed to genetic polymorphism. Genetic manipulations by over-expression or knockout of genes are considered to be the primary tools for the investigation of phenotypes. However, by inspection of the set of differential equations for the reaction network (figure 1), it is clear that without taking into account of its intracellular substrate concentrations, the dynamic behavior of a whole cell system cannot be predicted by the kinetic parameters as determined by its genotype alone. One-to-one correspondence between genotype and phenotype is often the exception rather than the rule. The functional behavior of a cell is a function of both its genetic and nutrient/environmental constraints. Thus, for a given genotype, there can be potentially many phenotypes. The observed phenotype of a cell is dependent on its nutrient/environment context. Just as existing species of living things are the products of natural selection, the observed phenotypes of cells are the products of metabolic selection (Ibarra et al., 2002; Lee and Go, 2005) 5. There are potentially four important phenotypes that a cell can assume, namely, proliferation, differentiation, cell cycle arrest and apoptosis. Each of these phenotypes is associated with a typical metabolic profile. We have previously shown that metabolic constraints are major determinant of phenotypes (Boros et al., 2002a). The interaction of cells and their nutrient environment according to the rules of metabolic selection results in the observed histological changes of hyperplasia, dysplasia and hypoplasia in human diseases. In conclusion, tracer based metabolomics is a unique experimental tool for the studies of phenotype of cells as the collective function of genes and their interaction with nutrient environment in a multicellular organism.
Acknowledgments
The author would like to thank Dr. Bill Go for many of his comments and suggestions during the preparation of the manuscript. This work was supported in part by PHS M01-RR00425 of the General Clinical Research Unit and P01-CA42710 of the UCLA Clinical Nutrition Research Unit Stable Isotope Core.
Footnotes
The concept of tracer has its origin from the early application of radioactive isotope where the amount of labeled material is small relative to the unlabeled trace. Since the application of stable isotopes, the term tracer is used to designate a labeled compound. When the amount of the labeled compound used is in the “physiological” range, the concept of tracer is preserved. However, in experiments where the concentration of the labeled substrate is non-physiological, such qualification should be indicated.
Ribose molecules having one or two 13C substitutions are designated as M+1 and M+2 ribose.
Fractional synthesis of palmitate was determined by dividing the total enrichment of palmitate by the enrichment of the acetyl-CoA using mass isotopomer distribution analysis (MIDA) (Lee et al., 1994).
Output from a cell is not limited to metabolites secreted by the cell. Included with the output is the increase in biomass from cell proliferation.
Metabolic selection of cells in response to changes in nutrient environment has been demonstrated in bacterial culture. Such adaptive evolution can be predicted by constraint based modeling of pathways (Ibarra et al., 2002).
References
- Allen J., Davey H.M., Broadhurst D., Heald J.K., et al. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 2003;21:692–696. doi: 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- Boren J., Cascante M., Marin S., Comin-Anduix B., Centelles J.J., Lim S., Bassilian S., Ahmed S., Lee W.N., Boros L.G. Gleevec (STI571) influences metabolic enzyme activities and glucose carbon flow toward nucleic acid and fatty acid synthesis in myeloid tumor cells. J. Biol. Chem. 2001;276:37747–37753. doi: 10.1074/jbc.M105796200. [DOI] [PubMed] [Google Scholar]
- Boren J., Lee W.N., Bassilian S., Centelles J.J., Lim S., Ahmed S., Boros L.G., Cascante M. The stable isotope-based dynamic metabolic profile of butyrate-induced HT29 cell differentiation. J. Biol. Chem. 2003;278:28395–28402. doi: 10.1074/jbc.M302932200. [DOI] [PubMed] [Google Scholar]
- Boros L.G., Lee W.N., Go V.L. A metabolic hypothesis of cell growth and death in pancreatic cancer. Pancreas. 2002a;24:26–33. doi: 10.1097/00006676-200201000-00004. [DOI] [PubMed] [Google Scholar]
- Boros L.G., Cascante M., Lee W.N. Metabolic profiling of cell growth and death in cancer: applications in drug discovery. Drug Discov. Today. 2002b;7:364–372. doi: 10.1016/S1359-6446(02)02179-7. [DOI] [PubMed] [Google Scholar]
- Boros L.G., Serkova N.J, Cascante M., Lee W.N. Use of metabolic pathway flux information in targeted cancer drug design. Drug Discov. Today Therap. Strat. 2004;1:435–443. doi: 10.1016/j.ddstr.2004.11.016. [DOI] [Google Scholar]
- Boros L.G., Torday J.S., Lim S., Bassilian S., Cascante M., Lee W.N. Transforming growth factor beta2 promotes glucose carbon incorporation into nucleic acid ribose through the nonoxidative pentose cycle in lung epithelial carcinoma cells. Cancer Res. 2000;60:1183–1185. [PubMed] [Google Scholar]
- Boros L.G., Torday J.S., Lee W.N., Rehan V.K. Oxygen-induced metabolic changes and transdifferentiation in immature fetal rat lung lipofibroblasts. Mol. Genet. Metab. 2002c;77:230–236. doi: 10.1016/S1096-7192(02)00140-3. [DOI] [PubMed] [Google Scholar]
- Brown M., Dunn W.B., Ellis D.I., et al. A metabolome pipeline: from concept to data to knowledge. Metabolomics. 2005;1:39–52. doi: 10.1007/s11306-005-1106-4. [DOI] [Google Scholar]
- Edwards J.S., Ramakrishna R., Palsson B.O. Characterizing the metabolic phenotype: a phenotype phase plane analysis. Biotechnol. Bioeng. 2002;77:27–36. doi: 10.1002/bit.10047. [DOI] [PubMed] [Google Scholar]
- Famili I., Palsson B.O. The convex basis of the left null space of the stoichiometric matrix leads to the definition of metabolically meaningful pools. Biophys. J. 2003;85:16–26. doi: 10.1016/S0006-3495(03)74450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell D.A. Understanding the Control of Metabolism. London: Portland Press; 1996. [Google Scholar]
- Garg M., Bassilian S., Bell C., Lee S., Lee W.N. Hepatic de novo lipogenesis in stable low-birth-weight infants during exclusive breast milk feedings and during parenteral nutrition. JPEN J. Parenter Enteral Nutr. 2005;29:81–6. doi: 10.1177/014860710502900281. [DOI] [PubMed] [Google Scholar]
- Goodacre R., Vaidyanathan S., Dunn W.B., Harrigan G.G., Kell D.B. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Harrigan G.G., Goodacre R., editors. Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis. Boston: Kluwer Academic Publishers; 2003. [Google Scholar]
- Harrigan G.G., Brackett D.J., Boros L.G. Medicinal chemistry, metabolic profiling and drug target discovery: a role for metabolic profiling in reverse pharmacology and chemical genetics. Mini. Rev. Med. Chem. 2005;5:13–20. doi: 10.2174/1389557053402800. [DOI] [PubMed] [Google Scholar]
- Ibarra R.U., Edwards J.S., Palsson B.O. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- Jones J.G., Naidoo R., Sherry A.D., Jeffrey F.M., Cottam G.L., Malloy C.R. Measurement of gluconeogenesis and pyruvate recycling in the rat liver: a simple analysis of glucose and glutamate isotopomers during metabolism of [1,2,3-(13)C3]propionate. FEBS Lett. 1997;412:131–137. doi: 10.1016/S0014-5793(97)00764-3. [DOI] [PubMed] [Google Scholar]
- Kell D.B., Westerhoff H.V. Metabolic control Theory: its rolein microbiology and biotechnology. FEMS Microbiol. Rev. 1986;39:305–320. [Google Scholar]
- Lee W.N. Appendix. Analysis of tricarboxylic acid cycle using mass isotopomer ratios. J. Biol. Chem. 1993;268:25522–25526. [PubMed] [Google Scholar]
- Lee W.N., Bassilian S., Guo Z., Schoeller D., Edmond J., Bergner E.A., Byerley L.O. Measurement of fractional lipid synthesis using deuterated water (2H2O) and mass isotopomer analysis. Am. J. Physiol. 1994;266:E372–E383. doi: 10.1152/ajpendo.1994.266.3.E372. [DOI] [PubMed] [Google Scholar]
- Lee W.N., Byerley L.O., Bassilian S., Ajie H.O., Clark I., Edmond J., Bergner E.A. Isotopomer study of lipogenesis in human hepatoma cells in culture: contribution of carbon and hydrogen atoms from glucose. Anal. Biochem. 1995;226:100–112. doi: 10.1006/abio.1995.1197. [DOI] [PubMed] [Google Scholar]
- Lee W.N., Edmond J., Bassilian S., Morrow J.W. Mass isotopomer study of glutamine oxidation and synthesis in primary culture of astrocytes. Dev. Neurosci. 1996;18:469–477. doi: 10.1159/000111442. [DOI] [PubMed] [Google Scholar]
- Lee W.N., Lim S., Bassilian S., Bergner E.A., Edmond J. Fatty acid cycling in human hepatoma cells and the effects of troglitazone. J. Biol. Chem. 1998a;273:20929–20934. doi: 10.1074/jbc.273.33.20929. [DOI] [PubMed] [Google Scholar]
- Lee W.N., Boros L.G., Puigjaner J., Bassilian S., Lim S., Cascante M. Mass isotopomer study of the nonoxidative pathways of the pentose cycle with [1,2–13C2]glucose. Am. J. Physiol. 1998b;274:E843–E851. doi: 10.1152/ajpendo.1998.274.5.E843. [DOI] [PubMed] [Google Scholar]
- Lee W.N., Go V.L. Nutrient-gene interaction: tracer based metabolomics. (2005) J. Nutr. 2005;135(12):3027S–3032S. doi: 10.1093/jn/135.12.3027S. [DOI] [PubMed] [Google Scholar]
- Lindon J.C., Keun H.C., Ebbels T.M., Pearce J.M., Holmes E., Nicholson J.K. The Consortium for Metabonomic Toxicology (COMET): aims, activities and achievements. Pharmacogenomics. 2005;6:691–699. doi: 10.2217/14622416.6.7.691. [DOI] [PubMed] [Google Scholar]
- Marin S., Lee W.N., Bassilian S., Lim S., Boros L.G., Centelles J.J., FernAndez-Novell J.M., Guinovart J.J., Cascante M. Dynamic profiling of the glucose metabolic network in fasted rat hepatocytes using [1,2–13C2]glucose. Biochem. J. 2004;381:287–294. doi: 10.1042/BJ20031737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J.K., Connelly J., Lindon J.C., Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- Palsson B.O., Price N.D., Papin J.A. Development of network-based pathway definitions: the need to analyze real metabolic networks. Trends Biotechnol. 2003;21:195–198. doi: 10.1016/S0167-7799(03)00080-5. [DOI] [PubMed] [Google Scholar]
- Papin J.A., Price N.D., Wiback S.J., Fell D.A., Palsson B.O. Metabolic pathways in the post-genome era. Trends Biochem. Sci. 2003;28:250–258. doi: 10.1016/S0968-0004(03)00064-1. [DOI] [PubMed] [Google Scholar]
- Price N.D., Papin J.A., Schilling C.H., Palsson B.O. Genome-scale microbial in silico models: the constraints-based approach. Trends Biotechnol. 2003;21:162–169. doi: 10.1016/S0167-7799(03)00030-1. [DOI] [PubMed] [Google Scholar]
- Raamsdonk L.M., Teusink B., Broadhurst D., Zhang N., Hayes A., Walsh M.C., Berden J.A., Brindle K.M., Kell D.B., Rowland J.J., Westerhoff H.V, van Dam K., Oliver S.G. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat. Biotechnol. 2001;19:45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- Reed J.L., Palsson B.O. Thirteen years of building constraint-based in silico models of Escherichia coli. J. Bacteriol. 2003;185:2692–2699. doi: 10.1128/JB.185.9.2692-2699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford K., Soucaille P., Whited G., Chotani G. Genomics to fluxomics and physiomics – pathway engineering. Curr. Opin. Microbiol. 2002;5:318–322. doi: 10.1016/S1369-5274(02)00318-1. [DOI] [PubMed] [Google Scholar]
- Schilling C.H., Palsson B.O. The underlying pathway structure of biochemical reaction networks. Proc. Natl. Acad. Sci. USA. 1998;95:4193–4198. doi: 10.1073/pnas.95.8.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling C.H., Schuster S., Palsson B.O., Heinrich R. Metabolic pathway analysis: basic concepts and scientific applications in the post-genomic era. Biotechnol. Prog. 1999;15:296–303. doi: 10.1021/bp990048k. [DOI] [PubMed] [Google Scholar]
- Schilling C.H., Letscher D., Palsson B.O. Theory for the systemic definition of metabolic pathways and their use in interpreting metabolic function from a pathway-oriented perspective. J. Theor. Biol. 2000;203:229–248. doi: 10.1006/jtbi.2000.1073. [DOI] [PubMed] [Google Scholar]
- Voit E.O., Almeida J. Decoupling dynamical systems for pathway identification from metabolic profiles. Bioinformatics. 2004;20:1670–1681. doi: 10.1093/bioinformatics/bth140. [DOI] [PubMed] [Google Scholar]
- Wong D.A., Bassilian S., Lim S., Lee W.N. Coordination of peroxisomal beta-oxidation and fatty acid elongation in HepG2 cells. J. Biol. Chem. 2004;279:41302–41309. doi: 10.1074/jbc.M406766200. [DOI] [PubMed] [Google Scholar]
- Xu J., Xiao G., Trujillo C., Chang V., Blanco L., Joseph S.B., Bassilian S., Saad M.F., Tontonoz P., Lee W.N., Kurland I.J. Peroxisome proliferator-activated receptor alpha (PPARalpha) influences substrate utilization for hepatic glucose production. J. Biol. Chem. 2002;277:50237–50244. doi: 10.1074/jbc.M201208200. [DOI] [PubMed] [Google Scholar]
- Xu J., Lee W.N., Xiao G., Trujillo C., Chang V., Blanco L., Hernandez F., Chung B., Makabi S., Ahmed S., Bassilian S., Saad M., Kurland I.J. Determination of a glucose-dependent futile recycling rate constant from an intraperitoneal glucose tolerance test. Anal. Biochem. 2003;315:238–246. doi: 10.1016/S0003-2697(02)00709-1. [DOI] [PubMed] [Google Scholar]


