Abstract
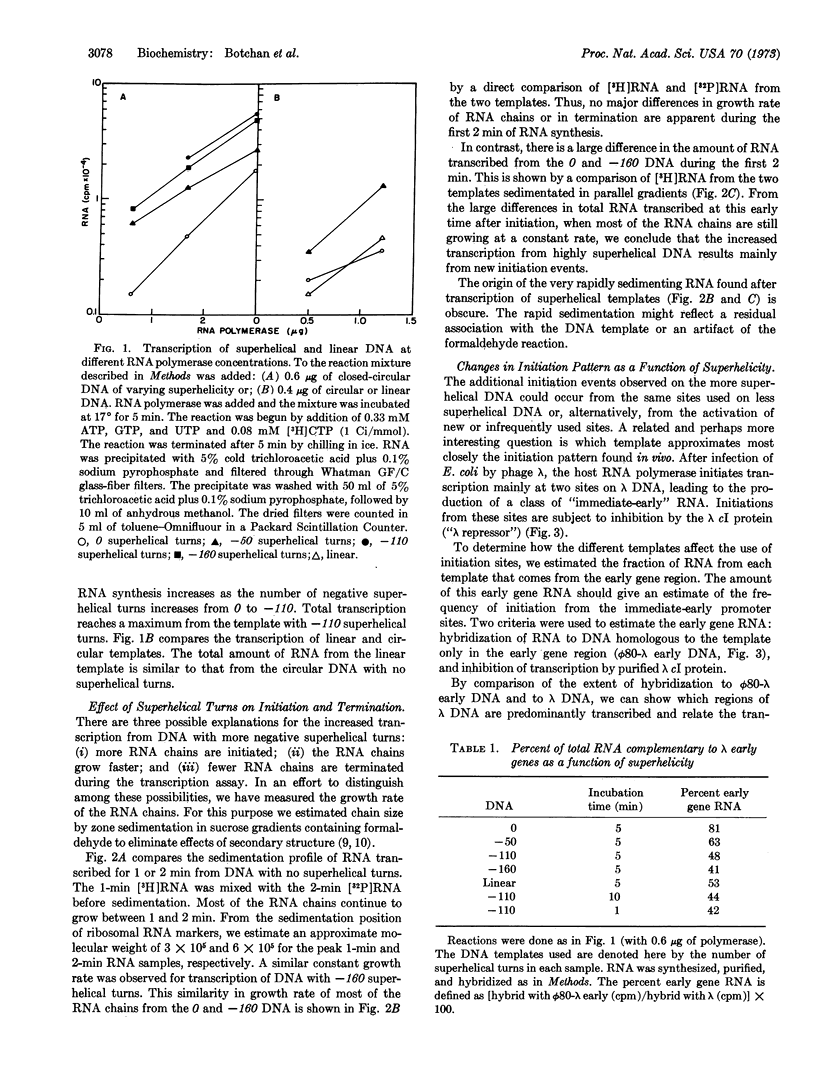
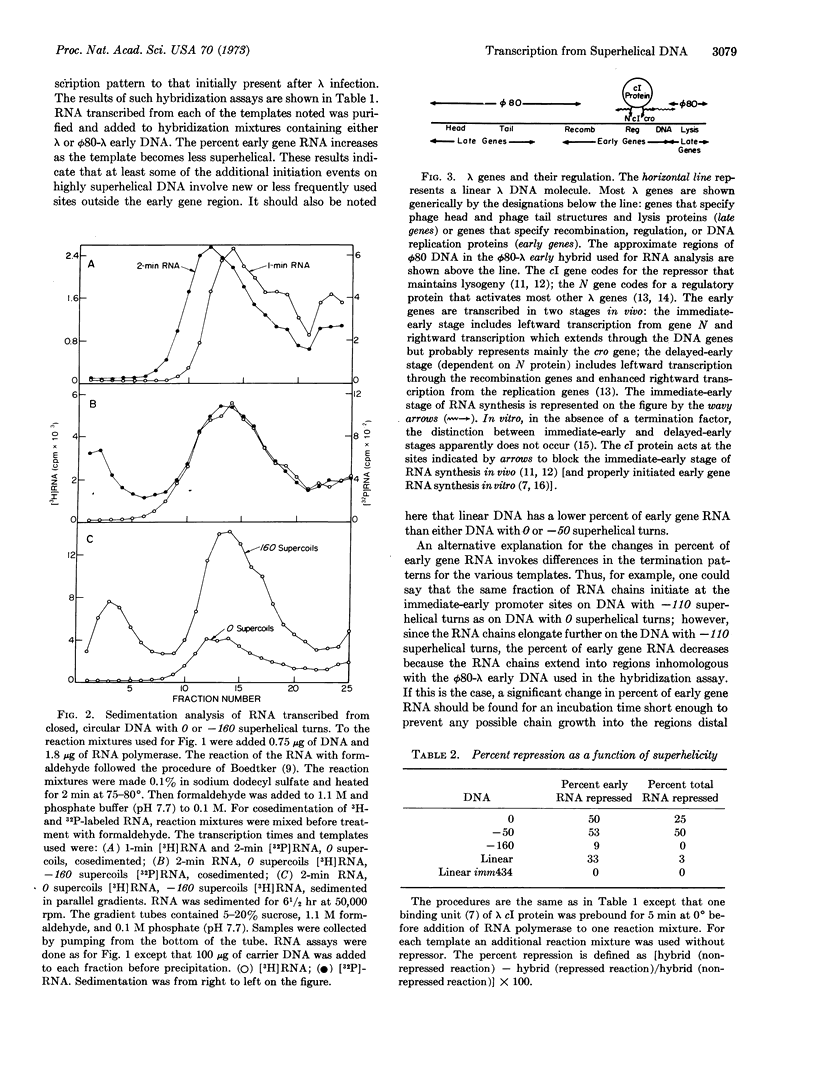
We studied RNA synthesis in vitro from closed-circular λ DNA molecules with varying degrees of superhelicity. The four circular templates examined had 0, -50, -110, and -160 superhelical turns under the conditions of the transcription assay. We found that the total amount of RNA synthesis increases as the template acquires more negative superhelical turns. This increased transcription results from more frequent initiation of RNA chains. Transcription of circular DNA with no superhelical turns appears to mimic RNA synthesis in vivo more closely than transcription from either highly superhelical or linear DNA with regard to two criteria: preferential transcription of the region corresponding to early genes and sensitivity to repression by λ cI protein. We suggest that the physical basis for the increased initiation of RNA chains from superhelical DNA is the fact that unwinding events are energetically favored on a DNA molecule with negative superhelical turns. Possible general mechanisms are: (a) RNA polymerase must unwind the DNA duplex as a prelude to initiation; (b) the DNA itself must assume a new conformation at the promoter site which requires an unwinding of the DNA duplex.
Keywords: RNA synthesis, RNA polymerase
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer W., Vinograd J. The interaction of closed circular DNA with intercalative dyes. I. The superhelix density of SV40 DNA in the presence and absence of dye. J Mol Biol. 1968 Apr 14;33(1):141–171. doi: 10.1016/0022-2836(68)90286-6. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Dahlberg J. E. RNA synthesis startpoints in bacteriophage lambda: are the promoter and operator transcribed? Nat New Biol. 1972 Jun 21;237(77):227–232. doi: 10.1038/newbio237227a0. [DOI] [PubMed] [Google Scholar]
- Boedtker H. Dependence of the sedimentation coefficient on molecular weight of RNA after reaction with formaldehyde. J Mol Biol. 1968 Jul 14;35(1):61–70. doi: 10.1016/s0022-2836(68)80036-1. [DOI] [PubMed] [Google Scholar]
- Burgess R. R. A new method for the large scale purification of Escherichia coli deoxyribonucleic acid-dependent ribonucleic acid polymerase. J Biol Chem. 1969 Nov 25;244(22):6160–6167. [PubMed] [Google Scholar]
- Davidson N. Effect of DNA length on the free energy of binding of an unwinding ligand to a supercoiled DNA. J Mol Biol. 1972 May 14;66(2):307–309. doi: 10.1016/0022-2836(72)90482-2. [DOI] [PubMed] [Google Scholar]
- Echols H. Lysogeny: viral repression and site-specific recombination. Annu Rev Biochem. 1971;40:827–854. doi: 10.1146/annurev.bi.40.070171.004143. [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Hayashi M. Template activities of the phi X-174 replicative allomorphic deoxyribonucleic acids. Biochemistry. 1971 Nov;10(23):4212–4218. doi: 10.1021/bi00799a009. [DOI] [PubMed] [Google Scholar]
- Helinski D. R., Clewell D. B. Circular DNA. Annu Rev Biochem. 1971;40:899–942. doi: 10.1146/annurev.bi.40.070171.004343. [DOI] [PubMed] [Google Scholar]
- Losick R. In vitro transcription. Annu Rev Biochem. 1972;41:409–446. doi: 10.1146/annurev.bi.41.070172.002205. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Ptashne M. Multiple repressor binding at the operators in bacteriophage lambda. Proc Natl Acad Sci U S A. 1973 May;70(5):1531–1535. doi: 10.1073/pnas.70.5.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardon J. F., Wilkins M. H. A super-coil model for nucleohistone. J Mol Biol. 1972 Jul 14;68(1):115–124. doi: 10.1016/0022-2836(72)90267-7. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan T., Echols H. Purification and properties of M protein: an accessory factor for RNA polymerase. J Mol Biol. 1973 Aug 25;78(4):675–686. doi: 10.1016/0022-2836(73)90288-x. [DOI] [PubMed] [Google Scholar]
- Richardson J. P. Rates of bacteriophage T4 RNA chain growth in vitro. J Mol Biol. 1970 Apr 14;49(1):235–240. doi: 10.1016/0022-2836(70)90389-x. [DOI] [PubMed] [Google Scholar]
- Saucier J. M., Wang J. C. Angular alteration of the DNA helix by E. coli RNA polymerase. Nat New Biol. 1972 Oct 11;239(93):167–170. doi: 10.1038/newbio239167a0. [DOI] [PubMed] [Google Scholar]
- Schäfer R., Zillig W., Zechel K. A model for the initiation of transcription by DNA-dependent RNA polymerase from Escherichia coli. Eur J Biochem. 1973 Mar 1;33(2):207–214. doi: 10.1111/j.1432-1033.1973.tb02671.x. [DOI] [PubMed] [Google Scholar]
- Steinberg R. A., Ptashne M. In vitro repression of RNA synthesis by purified lambda phage repressor. Nat New Biol. 1971 Mar 17;230(11):76–80. doi: 10.1038/newbio230076a0. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Degree of superhelicity of covalently closed cyclic DNA's from Escherichia coli. J Mol Biol. 1969 Jul 28;43(2):263–272. doi: 10.1016/0022-2836(69)90266-6. [DOI] [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- Wu A. M., Ghosh S., Echols H., Spiegelman W. G. Repression by the cI protein of phage lambda: in vitro inhibition of RNA synthesis. J Mol Biol. 1972 Jun 28;67(3):407–421. doi: 10.1016/0022-2836(72)90459-7. [DOI] [PubMed] [Google Scholar]


