Abstract
The ubiquitous Rho (Ras homology) GTPase Cdc42p can function in different settings to regulate cell polarity and cellular signaling. How Cdc42p and other proteins are directed to function in a particular context remains unclear. We show that the Cdc42p-interacting protein Bem4p regulates the mitogen-activated protein kinase (MAPK) pathway that controls filamentous growth in Saccharomyces cerevisiae. Bem4p controlled the filamentous-growth pathway but not other MAPK pathways (mating or high-osmolarity glycerol response [HOG]) that also require Cdc42p and other shared components. Bem4p associated with the plasma membrane (PM) protein, Sho1p, to regulate MAPK activity and cell polarization under nutrient-limiting conditions that favor filamentous growth. Bem4p also interacted with the major activator of Cdc42p, the guanine nucleotide exchange factor (GEF) Cdc24p, which we show also regulates the filamentous-growth pathway. Bem4p interacted with the pleckstrin homology (PH) domain of Cdc24p, which functions in an autoinhibitory capacity, and was required, along with other pathway regulators, to maintain Cdc24p at polarized sites during filamentous growth. Bem4p also interacted with the MAPK kinase kinase (MAPKKK) Ste11p. Thus, Bem4p is a new regulator of the filamentous-growth MAPK pathway and binds to general proteins, like Cdc42p and Ste11p, to promote a pathway-specific response.
INTRODUCTION
A central problem in the field of signal transduction is to understand how proteins that function in interconnected networks execute a precise response. The Rho-type GTPase Cdc42p is master regulator of cell polarity (1–4) and signal transduction (5, 6). In the budding yeast Saccharomyces cerevisiae, Cdc42p regulates three mitogen-activated protein kinase (MAPK) pathways that sense and respond to different stimuli that comprise an interconnected network (filamentous growth, mating, and high-osmolarity glycerol response [HOG]) (7–9). Cdc42p also has an essential function in polarity establishment (3, 10–15) and exocytosis (16, 17). Therefore, Cdc42p and other general regulators can function in different contexts to regulate cellular responses and control proper growth.
One of the pathways in which Cdc42p operates is the MAPK pathway that controls filamentous (invasive/pseudohyphal) growth. Filamentous growth involves the differentiation to a new cell type in response to nutrient limitation (18–21). In particular, depletion of nitrogen (21) or glucose (22) can induce filamentous growth in yeast. Cdc42p regulates the filamentous-growth pathway by binding to the p21-activated kinase (PAK) Ste20p, which regulates a typical MAPK cascade (Ste11p → Ste7p → Kss1p) (18, 23–29). It is not clear how Cdc42p is activated in the context of the filamentous-growth pathway (Fig. 1A). During normal growth (budding) and mating, Cdc42p is activated by its sole guanine nucleotide exchange factor (GEF), Cdc24p (30–34). At present, it is not known whether Cdc24p regulates the filamentous-growth pathway. It is known that the signaling mucin Msb2p is a plasma membrane regulator of the filamentous-growth pathway that interacts with the active (GTP-bound) form of Cdc42p (26, 35, 36). Thus, Msb2p may stabilize Cdc42p once the GTPase has been activated. Msb2p also associates with Sho1p, another PM regulator of the filamentous-growth pathway (26, 36–38). Sho1p associates with Cdc24p, but the functional significance of this interaction remains unclear (36).
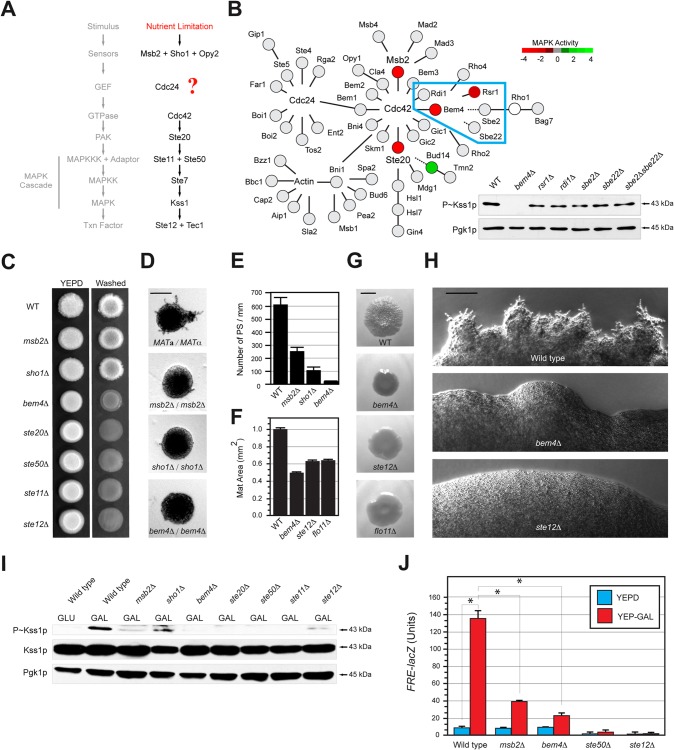
FIG 1.
Bem4p regulates the filamentous-growth pathway. (A) Filamentous-growth pathway. The question mark represents the question of how Cdc42p is activated in the filamentous-growth pathway. (B) Role of the Cdc42p interaction network in regulating the filamentous-growth pathway. Red, positive regulator; green, negative regulator; gray, no phenotype; white, not tested. Color is based on the results of three tests: invasive growth, MAPK growth reporter activity, and Kss1p phosphorylation (see Table S3 in the supplemental material). Dashed line, genetic interaction. (Inset) P∼Kss1p immunoblot from a subset of the network (outlined in blue) that contains Bem4p. Kss1p∼P was detected by anti-phospho-p42/44 antibodies. Pgk1p, loading control. (C) Plate-washing assay. Equal concentrations of cells were spotted onto YEPD medium. Plates were incubated for 4 days and photographed (left). Plates were washed in a stream of water and photographed again (right). (D) Pseudohyphal growth. Diploid strains were grown on low-nitrogen (SLAHD) medium for 4 days. SLAHD is synthetic low-ammonia medium containing glucose as a carbon source and histidine as a nitrogen source (21). Microcolonies were photographed at a magnification of ×5; representative colonies are shown. Bar, 50 μm. (E) Pseudohyphae (PS) were counted at microcolony perimeters and divided by colony perimeters. Microcolonies for separate experiments were examined; error bars represent the standard deviations between trials. Homozygous diploid strains were used for the experiment. (F) Biofilm/mat formation. Strains were spotted on nitrocellulose filters overlaid on YEPD plus 0.3% agar for 4 days at 30°C. Mat expansion was determined by measuring mat areas (in mm2) by ImageJ. Areas were measured at 2 and 4 days; areas for the 4-day time point were normalized to those from the 2-day time point and set to a value of 1 for the wild type. Other values were normalized accordingly. Error bars represent standard deviations between trials. (G) Example of biofilms/mats at 2 days. Bar, 1 cm. (H) Close-up of filamentous mats incubated on YEPD medium for 20d. Bar, 50 μm. (I) P∼Kss1p levels in strains grown to mid-log phase in YEPD (GLU) or YEP-Gal (GAL) medium. Blots were probed with anti-phospho-p42/44 antibodies to detect P∼Kss1p. Blots using antibodies to total Kss1p and Pgk1p are shown as controls. (J) Expression of the FRE-lacZ reporter by β-galactosidase assays (expressed in Miller units). Values are averages from two independent trials; asterisks denote P values of <0.05.
By exploring a protein interaction network surrounding Cdc42p, we identified the Cdc42p-interacting protein Bem4p, which is a general regulator of Rho-type GTPases (39–41), as a major regulator of the filamentous-growth pathway. Bem4p regulated the filamentous-growth pathway but not other MAPK pathways that also require Cdc42p and other common components. We show that Cdc24p regulates the filamentous-growth pathway. Bem4p interacted with Cdc24p, and based on genetic and biochemical evidence, may activate the GEF during filamentous growth. Bem4p also associated with other proteins that regulate the filamentous-growth pathway. Thus, Bem4p is a new regulator of the filamentous-growth pathway that functions through Cdc42p and other general proteins to generate a pathway-specific response.
MATERIALS AND METHODS
Strains, plasmids, and microbiological techniques.
Yeast strains are listed in Table S1 in the supplemental material, and plasmids are listed in Table S2. Yeast and bacterial strains were grown and manipulated by standard methods (42, 43). Gene disruptions and GAL1 promoter fusions were made by PCR-based methods (44, 45), including antibiotic resistance markers (46) and epitope fusions (47). Integrations were confirmed by PCR analysis and phenotype. Unless otherwise stated, all fusion proteins were functional with respect to filamentous-growth pathway activity (by phosphorylated Kss1p [P∼Kss1p] immunoblot analysis) and filamentous/invasive growth (by the plate-washing assay).
Plasmid pIL30-URA3 containing FgTy-lacZ was provided by B. Errede (48). Plasmid pFRE-lacZ was provided by H. Madhani (49). Plasmid reporters containing filamentous-growth pathway targets (KSS1, PGU1, and YLR042c) were provided by C. Boone (50). Plasmid pMSB2-lacZ has been described (51). Overexpression constructs were obtained from an ordered collection from Open Biosystems (52). lacZ gene fusions to the FUS1 promoter (53) were constructed in the V84 vector by in vivo recombination (45). Vector V84 vector (50) was linearized by restriction digest with NotI prior to integration.
Plasmids expressing the BEM4 gene from strong constitutive promoters were constructed (54). The pTEF2 promoter was amplified by PCR and inserted into YEp351 and YEp352 plasmids at SacI and XbaI sites. The BEM4-green fluorescent protein (GFP) fusion was amplified from purified yeast chromosomal DNA and inserted downstream of the pTEF2 promoter at XbaI and SalI sites to create YEp351-pTEF2-BEM4-GFP and YEp352-pTEF2-BEM4-GFP plasmids.
To construct His-tagged version of Cdc42p, the CDC42 open reading frame (ORF) was amplified from PC4368 (pGBDU-Cdc42p) and ligated into pET28b using primers that incorporated 5′ BamHI and 3′ SalI sites. To construct pGST-Cdc24p, the CDC24 gene was subcloned from PC4225 (pGBDU-Cdc24p) and ligated into pGEX4T1 by using BamHI and SalI sites. To construct pMBP-Bem4, the BEM4 gene was amplified from PC4190 (pOAD-Bem4) and introduced into pMAL-C2 with primers that contained 5′ BamHI and 3′ SalI sites. To construct the pMBP-Bem4 truncation alleles, primers were designed to truncate the protein at 100, 200, 300, 400, 500, and 600 amino acid residues downstream of the N terminus. The truncated alleles were amplified from PC4190 with primers that contained 5′ BamHI and 3′ SalI sites.
Plasmids pRS316-CDC24-GFP and YEp351-CDC24-GFP were constructed by amplification of CDC24-GFP from the genomic GFP-tagged strain PC1205 containing 600 bp of the CDC24 promoter using primers that contained 5′ BamHI and 3′ SalI sites. Plasmids pRS315-cdc24-4 and pRS425-cdc24-4 were constructed by amplification of the cdc24-4 allele from PC1436. The PCR fragment included 600 bp of the CDC24 promoter and the entire cdc24-4 ORF. A BamHI site was introduced at the 5′ end of the promoter and a PstI site was introduced at the 3′ end of the cdc24-4 gene. YEp351-Myr-CDC24-GFP was generated in two steps. A PCR fragment containing 600 bp of the CDC24 promoter was amplified with 5′ BamHI and 3′ PstI sites and introduced into YEp351. A second fragment was generated by PCR that introduced a myristoylation site (55, 163) into the N terminus of Cdc24p. This PCR fragment was generated by amplification of CDC24-GFP from strain PC1205. The PCR fragment was ligated into YEp351 that contained the CDC24 promoter using 5′ PstI and 3′ SalI sites.
Alleles of CDC24 were ligated into pGEX4T1 (see Fig. 6D, top). For all CDC24 alleles used, pGEX4T1-CDC24 was used as the template for PCRs. Primers were designed to delete specific regions of Cdc24p corresponding to previously mapped domains. pGEX4T1-CDC24-CHΔ was generated by amplifying the regions of Cdc24p corresponding to amino acids at positions 226 to 854 of the protein. pGEX4T1-CDC24-PB1Δ was generated by amplifying the region of Cdc24p corresponding to amino acid positions 1 to 780. pGEX4T1-CDC24-PHΔPB1Δ was generated by amplifying the region of Cdc24p corresponding to the amino acid positions 1 to 452. pGEX4T1-CDC24-CHΔ-PB1Δ was generated by amplifying the region of Cdc24p corresponding to the amino acids at positions 283 to 681. pGEX4T1-CDC24-PHΔ was generated by amplifying the regions of Cdc24p corresponding to the amino acids at positions 780 to 851. This PCR fragment was then ligated to pGEX4T1-CDC24-PHΔPB1Δ such that a SalI restriction enzyme site served as a linker between the two fragments. The pGEX4T1-CDC24-PH domain only was generated by amplifying the region of Cdc24p corresponding to amino acids 472 to 681. pRS425-CDC24-PHΔ was generated in a fashion similar to that used for YEp351-Myr-CDC24-GFP. The region introduced into pGEX4T1 from the pGEX4T1-CDC24-PHΔ plasmid was amplified and introduced into a pRS425 vector that already contained 600 bp of the promoter.
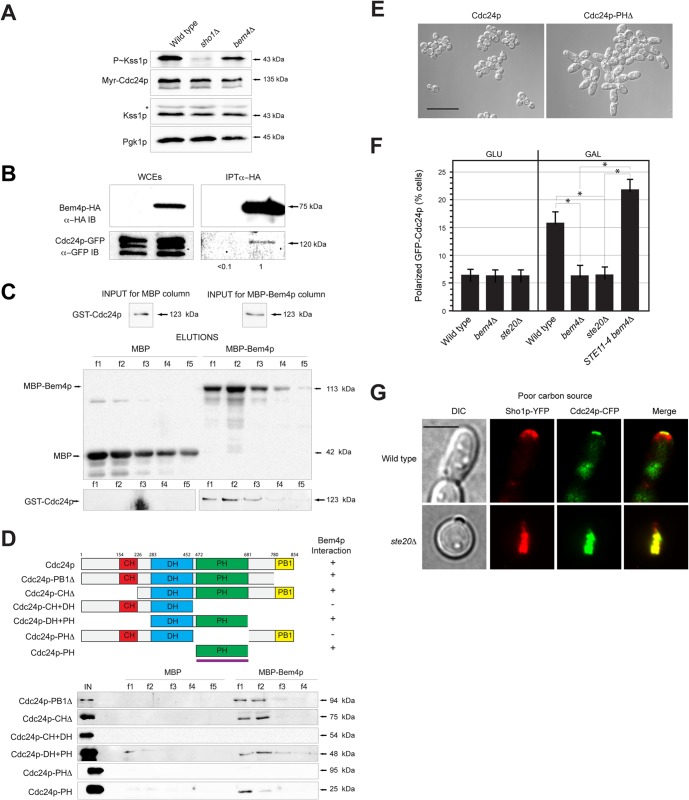
FIG 6.
Bem4p functions with Cdc24p and regulates its localization. (A) P∼Kss1p levels in cells carrying pMyr-Cdc24p-GFP in the wild type, the sho1Δ mutant, and the bem4Δ mutant. (B) Bem4p-HA interacts with Cdc24p-GFP by co-IP analysis. Band intensity relative to input levels after background subtraction was determined by ImageJ. Input levels are 10% for WCE. (C) MBP-Bem4p interacts with GST-Cdc24p in vitro. Fifty percent of the WCE (input) was used for the pulldown. (D) Bem4p interacts with the PH domain of Cdc24p. Deletion constructs were made at the indicated residues. Fifty percent of the WCE (input) was used for the pulldowns. (E) Cells in mid-log phase expressing wild-type Cdc24p or Cdc24p-PHΔ. Bar, 25 μm. (F) Localization of Cdc24p-GFP in wild-type cells and the indicated mutants. Cells were grown to mid-log phase in SD or S-Gal complete medium. Bar, 5 μm. Approximately 50 cells were counted for each experiment in separate trials; asterisks denote P values of <0.05. (G) Representative cells (from panel F) showing the localization of Sho1p-YFP and Cdc24p-CFP in wild-type cells and the ste20 mutant. Proteins were false colored in red, green, and yellow. Bar, 5 μm.
Two-hybrid constructs in the pOAD and pOBD vectors (56) were provided by S. Fields (University of Washington, Seattle, WA). To construct other two-hybrid plasmids, plasmids pGAD-C1 and pGBDU-C1 were used (57). Complete ORFs were amplified by PCR from genomic DNA isolated from a wild-type yeast strain (PC313). The Ste11p and Cdc42p ORFs were amplified from lab plasmids. For N-terminal and C-terminal truncations of Ste11p, the junction was 1,245 nucleotides from the start of the STE11 ORF. pGAD-SHO1 was subcloned from pGBDU-C1 Sho1p (57). The STE11 gene was subcloned from pGAD-Ste11p (PC4369) and introduced into pGEX4T1 (PC5250) by digestion with BamHI and SalI to create pGST-STE11 (PC6045).
Evaluating Cdc24p function by plasmid shuttle.
CDC24 function was assessed by plasmid shuttle. The CDC24 gene was disrupted by homologous recombination (cdc24::NAT) in a wild-type strain (PC1894) harboring pRS316-CDC24-GFP. LEU2-based plasmids containing versions of CDC24 were introduced into the cdc24 strain. Strains were patched onto 5-fluoroorotic acid (FOA)–Leu medium to force loss of pRS316-CDC24-GFP and retain versions of CDC24 on LEU2-based plasmids. Strains were checked for growth on FOA-Leu and sensitivity on SD-Ura.
Protein purification and in vitro pulldowns.
His-Cdc42p was purified by the following method. Approximately 250 ml of cells (Escherichia coli strain λBL21/DE3) harboring overexpression plasmids (pHis-Cdc42) were grown to exponential phase in 2× yeast Terrific (YT) broth plus kanamycin and induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C. Cells were harvested by centrifugation, and cell pellets were frozen at −80°C. For purification, cells were thawed on ice, resuspended in 10 ml of His column buffer (50 mM sodium phosphate [pH 7.0], 100 mM NaCl) with 1 mM phenylmethylsulfonyl fluoride (PMSF) and incubated at 25°C with 1 mg/ml lysozyme for 30 min. Cells were placed in an ice bath and sonicated for 20 s (2-s pulses with 30 s rest on ice). Cell debris was removed by centrifugation at a relative centrifugal force (RCF) of 10,000 for 10 min at 4°C. Cell lysates were applied to 500 μl prewashed Talon metal affinity resin (product 633502; Clontech) and incubated with end-over-end rotation at 25°C for 30 min. The resin bound to His-tagged proteins was separated from cell lysates by centrifugation at 1,000 rpm for 5 min and washed three times with 5 ml of His column buffer. The resin was then washed in His column buffer with 500 mM NaCl and 10 mM imidazole. The resin was placed in columns, and proteins were eluted with 2.5 ml of His elution buffer (150 mM imidazole, 50 mM sodium phosphate [pH 7.0], 100 mM NaCl). Approximately 30 μl of the elution was saved as the input fraction. Eluted proteins were split into equal concentrations for subsequent pulldown experiments.
To purify glutathione S-transferase (GST)–Cdc24p proteins, an E. coli strain (λBL21/DE3) containing pGST-Cdc24 was grown to mid-exponential phase. Approximately 500 ml of cells harboring GST-Cdc24p were induced with 1.0 mM IPTG for 2 h. Cells were harvested by centrifugation and frozen as cell pellets at −80°C. Cells containing GST-Cdc24p were resuspended in 40 ml of phosphate-buffered saline (PBS) buffer on ice. Cell extracts were prepared as described for His-tagged proteins except that lysates were applied to 400 μl of prewashed glutathione-Sepharose B resin (product 17-0756-01; GE Life Sciences) and incubated with end-over-end rotation at 25°C for 30 min. The resin was harvested by centrifugation at 1,000 rpm for 5 min, washed three times with 4 ml of PBS buffer, and placed in a disposable 20-ml column. GST-Cdc24p proteins were eluted from the columns with 2 ml of GST elution buffer (10 mM glutathione, 50 mM Tris HCl [pH 8]). A 30-μl portion of the elution was saved for the input fraction and elutions were divided into equal portions for pulldown experiments.
To purify Bem4p, cells containing pMBP-Bem4p, pMBP-Bem4p truncation mutants, and pMBP alone were grown to exponential phase in the E. coli strain ER2508 (New England BioLabs). Maltose-binding protein (MBP)–Bem4p (250 ml) and MBP alone (50 ml) were induced with 0.3 mM IPTG for 3 h at 37°C. Cells were harvested by centrifugation and frozen at −80°C. Cells containing MBP-Bem4p were resuspended in 10 ml of MBP buffer (50 mM Tris HCl [pH 7.5], 100 mM NaCl) and 2.5 ml for MBP alone. Cell extracts were prepared as described for His-tagged proteins. A 30-μl portion of the lysates was retained at this step as the input fraction. Lysates were applied to 250 μl of prewashed amylose resin (product E8021S; New England BioLabs) and incubated with end-over-end rotation at 25°C for 30 min. The resins containing MBP-Bem4p and MBP were separated from lysates by gentle centrifugation and washed three times in 2.5 ml of MBP column buffer. The resins were added to disposable 10-ml columns.
To evaluate the interactions between Bem4p and Cdc24p, and Cdc42p, purified His-Cdc42p, GST-Cdc24p, and GST proteins were loaded onto columns that contained MBP-Bem4p or MBP alone, prepared as described above. Columns were washed by addition of 3.75 ml of MBP column buffer. Proteins were eluted in MBP elution buffer (10 mM maltose, 50 mM Tris HCl [pH 7.5], 100 mM NaCl) and evaluated by SDS-PAGE analysis.
For the Bem4p and Ste11p pulldowns, cells containing MBP-Bem4p (PC5807), pGEX4T1 (PC5514), and pGST-STE11 (PC6045) were grown in 200 ml of 2× YT plus 1.3 mM carbenicillin for 3 h to an optical density (OD) of 0.6. IPTG was added to 0.3, 0.1, and 0.6 mM concentrations for PC5807, PC5514, and PC6045, respectively, for 1.5 h. Cells were harvested by centrifugation at 5,000 rpm and stored at −80°C. Cells were thawed on ice in 10 ml of phosphate-buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 11.8 mM KH2PO4 [pH 7.42]) containing 1 mM PMSF and 1 mM dithiothreitol (DTT). Lysozyme was added to 0.75 mg/ml. Individual cultures were sonicated, and unbroken cells and cell debris were removed by centrifugation for 10 min at 10,000 rpm. Clarified lysates containing GST and GST-Ste11p were incubated with 100 μl glutathione-Sepharose beads for 30 min at 4°C using end-over-end rotation. Beads and bound proteins were collected by centrifugation at 2,000 rpm. Clarified lysates containing MBP-Bem4p were split and added to either the GST- or GST-Ste11p-coated beads. Samples were incubated for 30 min at 4°C with end-over-end rotation. Beads and bound proteins were again collected by centrifugation at 2,000 rpm. Samples were loaded onto columns, which were washed twice with 10× bed volumes of PBS. GST and GST-Ste11p were eluted in PBS containing 10 mM reduced glutathione. Elutes were examined by SDS-PAGE for GST, GST-Ste11p, and MBP-Bem4p.
In vitro PAK binding assays.
In vitro pulldowns with the PAK binding domain (PBD-GST, provided by E. Bi) and Cdc42p were performed as described previously (58, 59). Cell lysates were prepared from strains expressing Cdc42p-GFP and a PBD-GST (pGEX-2T-PBD) expressed and purified from Escherichia coli (60). Bacterial cells overproducing pPBD-GST were harvested by centrifugation, resuspended in bacterial cell lysis buffer (1× PBS [pH 7.4], 100 mM NaCl, lysozyme 1 mg/ml, 1× protease inhibitor cocktail [number 11836170001; Roche]), and lysed by sonication (Branson digital sonifier, number 102C). The fusion protein was isolated using immobilized glutathione-agarose beads (number 15160; Thermo Scientific). A 750-μl portion of yeast cell lysate containing Cdc42p-GFP was added to 200 μl of immobilized glutathione-agarose beads with bound PBD-GST or GST alone as a control. A portion of the yeast cell lysate was retained as the whole-cell extract (WCE) control sample. Samples were incubated at 4°C with end-over-end rotation for 1 h. Immobilized beads were harvested by centrifugation at 500 × g for 3 min at 4°C. A portion of the supernatant was retained as the flowthrough (FT) sample. Samples were washed three times by resuspension in 1 ml of bacterial cell lysis buffer containing 0.1% Triton X-100 and centrifuged at 500 × g for 3 min at 4°C. Addition of 200 μl of elution buffer (50 mM Tris HCl [pH 8.0] and 20 mM reduced glutathione [GSH]) followed by incubation at 25°C for 15 min was used to elute associated proteins from the agarose beads and centrifugation 500 × g for 3 min at 25°C. A 200-μl portion of elution buffer was added to the immobilized glutathione beads, and the samples were boiled for 10 min and then centrifuged at 15,800 × g for 3 min at 25°C. Nitrocellulose membranes were incubated for 2 h at 25°C in blocking buffer containing a primary antibodies to detect bound Cdc42p-GFP.
Protein immunoblot analysis.
Immunoblots were performed as described previously (26). Proteins were isolated from cells by bead beating using an MP FastPrep-24 (MP Scientific, Solon, OH) in Thorner buffer (8 M urea, 5% SDS, 40 mM Tris HCl [pH 6.8], 0.1 M EDTA, 0.4 mg/ml bromophenol blue, and 1% β-mercaptoethanol [BME]). Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes (Protran BA85; VWR International Inc., Bridgeport, NJ). After transfer, nitrocellulose membranes were incubated in blocking buffer (5% nonfat dry milk, 10 mM Tris HCl [pH 8], 150 mM NaCl and 0.05% Tween 20) for 16 h at 4°C. Antibodies were used at the manufacturer's concentrations. SuperSignal West Pico (number 1856136; Thermo Scientific) or an ECL Plus Western blotting detection system (RPN2132V1; GE Healthcare) was used to detect secondary antibodies. Precision Plus All Blue protein standards were used as molecular weight markers (Bio-Rad).
Mouse monoclonal antibodies were used to detect green fluorescent protein (clones 7.1 and 13.1; number 11814460001; Roche Diagnostics) and hemagglutinin (HA) (12CA5; number 11583816001; Roche). Rabbit polyclonal anti-HA antibodies (catalog number 600-401-384, lot number 21345; Rockland Immunochemicals Inc., Gilbertsville, PA) were used to detect the HA epitope. Antibodies to phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) (number 4370; Cell Signaling Technology) were used to detect phosphorylated Kss1p and Fus3p. Antibodies against yeast Pgk1p (catalog number 459250, lot number P0660; Life Technologies–Molecular Probes, Grand Island, NY), Kss1p (Santa Cruz, Sc-6775-R), Fus3p (Santa Cruz, Sc-6773), His (catalog number 34660; Qiagen), MBP (E8030S; New England BioLabs), and GST (600-101-200; Rockland Scientific) were also used. Secondary antibodies included goat anti-mouse IgG–horseradish peroxidase (HRP) (170-6516; Bio-Rad), goat anti-rabbit IgG–HRP (111-035-144; Jackson ImmunoResearch Laboratories, Inc.), and donkey anti-goat IgG–HRP (lot number L1010; Santa Cruz).
Colony immunoblots to detect shed Flo11p-HA were performed as described previously (51). Approximately equal numbers of cells were spotted onto yeast extract-peptone-dextrose (YEPD) medium overlaid with a nitrocellulose membrane. Colonies were photographed and washed under a stream of water to remove colonies. The nitrocellulose membrane was removed and placed into blocking buffer for 16 h. The membrane was washed with Tris-buffered saline–Tween (TBST) and incubated with primary and secondary antibodies to detect proteins.
Subcellular fractionation analysis.
Subcellular fractionation was performed as described previously (26, 61) in wild-type strains expressing tagged versions of Cdc42p or Bem4p and in a wild-type strain and a sho1Δ strain expressing a tagged version of Bem4p. Approximately 10 ml of exponentially grown cells were collected by centrifugation at for 5 min at 3,000 × g. Cell pellets were washed with 1 ml of sterile distilled water, harvested by centrifugation, and resuspended in 1 ml DTT buffer (50 mM Tris HCl [pH 8.8], 10 mM DTT). Cells were incubated for 15 min at room temperature. After incubation, the cells were harvested by centrifugation for 2 min at 5,000 × g and resuspended in 1 ml of spheroplast buffer (1.2 M sorbitol, 50 mM potassium phosphate [pH 7.4], 1 mM MgCl2, and 250 mg/ml zymolase). Cells were incubated at 30°C for 1 h, harvested by centrifugation, and washed with 1 ml of 1.2 M sorbitol. Spheroplasts were resuspended in 1 ml of spheroplast lysis buffer (200 mM sorbitol, 50 mM Tris HCl [pH 7.5], 1 mM EDTA, and 1× protease inhibitor cocktail (EDTA free; catalog number 11836170001; Roche Applied Science, Indianapolis, IN) and incubated on ice for 20 min. Following the incubation on ice, samples were harvested by centrifugation at 4°C for 5 min at 500 × g. Supernatants (a portion of which was retained as the S2 fraction) were transferred to sterile tubes and centrifuged at 4°C for 10 min at 15,800 × g. The pellet was retained as the P13 fraction. Supernatants were transferred to ultracentrifuge tubes and centrifuged at 100,000 × g at 30°C for 30 min. Portions of the supernatant and pellet were retained as fractions S100 and P100, respectively. Fractions were immediately processed by SDS-PAGE or frozen at −80°C.
Co-IP analysis.
To evaluate the interaction between Bem4p-HA and GFP-Cdc42p, Cdc24p-GFP, and Ste11p-GFP, Bem4p-HA was precipitated from cell extracts containing GFP-Cdc42p (PC3541), Ste11p-GFP (PC4954), and Cdc24p-GFP (PC5388). Control extracts from wild-type strains expressing GFP-Cdc42p (PC3448), Ste11p-GFP (PC4950), and Cdc24p-GFP (PC5387) were also used. Coimmunoprecipitation (co-IP) analysis was performed in wild-type (PC538) and Bem4p-HA (PC3398) strains containing pSho1-GFP (PC1601) to assess that interaction.
The interaction between Bem4p and Kss1p was assessed using strains containing C-terminally GFP-tagged proteins (Bem4p-GFP, Hog1p-GFP, Fus3p-GFP, Kss1p-GFP, Rsr1p-GFP, Ste20p-GFP, Msb2p-GFP, Ste4p-GFP, Ste5p-GFP, Ste18p-GFP, and Ste2p-GFP from reference 62) that had been transformed with plasmid pGAL-BEM4-HA (52) or a control plasmid, pRS316 (63). Strains were grown to saturation in YEP-Gal and subcultured in YEP-Gal to mid-log phase (7 h). Cells in 100-ml aliquots were harvested by centrifugation and stored at −80°C. Cell extracts were prepared and co-IP analysis was performed as described below. Equal numbers of cells (as determined by OD at 600 nm [OD600]) were harvested by centrifugation, and the pellets were stored at −80°C.
Cell pellets were thawed in ice-cold PBS buffer and transferred to FastPrep tubes (number 5076-200; MP Scientific). Approximately 0.2 ml of glass beads and 1 ml of IP buffer (50 mM Tris [pH 8.0], 1 mM EDTA, 50 mM NaCl, 1.5% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× protease inhibitor cocktail [number 11836170001; Roche]) were added to each sample, followed by two 30-s pulses at 6.5 m/s in a MP FastPrep-24 homogenizer (MP Scientific). Samples were placed on ice for 5 min between cycles. Following FastPrep homogenization, 600 μl of IP buffer was added to each sample, and the samples were vortexed briefly. Samples were centrifuged at 4°C for 10 min at 13,000 rpm. Clarified lysate was then added to 75 μl washed protein G-agarose beads (number 20398; Thermo Scientific, Pittsburgh, PA) and mixed with end-over-end rotation for 30 min at 4°C to preclear lysates. Samples were centrifuged at 4°C for 10 min at 13,000 rpm. Precleared lysates were then transferred to a sterile 1.5-ml tube, and ∼8 mg of anti-HA antibody (Rockland, number 600-401-384, Gilbertsville, PA) was added. Samples were mixed with end-over-end rotation at 4°C for 1 h. Lysate-antibody mixture was then added to 75 μl of washed protein G-agarose beads and mixed with end-over-end rotation at 4°C for 1 h. Samples were then centrifuged (500 × g) for 3 min at 4°C, and the supernatant was discarded. Beads were then washed 3 times with 1 ml IP buffer. Fifty microliters of 2× SDS loading buffer with 2% BME was added to the beads, and they were boiled for 10 min with frequent agitation. Samples were centrifuged at 13,000 rpm, and supernatants were frozen at −80°C.
Two-hybrid analysis.
Two-hybrid constructs were transformed into the PJ69-4a (PC284) strain (57) and assessed by the LYS2::GAL1-HIS3 growth reporter. Approximately equal numbers of cells were spotted on to SD-Ura-Leu or SD-Trp-Leu plates with or without histidine. Plates lacking histidine and containing 5 mM 4-amino-1,2,4-triazole were used to evaluate proteins that showed low levels of autoactivation. Additional evaluation of protein interactions was carried out by assessing β-galactosidase activity using the met2::GAL7-lacZ reporter.
Evaluating MAPK activity for the filamentous growth, mating, and HOG pathways.
Phosphorylation of the MAPK Kss1p (P∼Kss1p) was measured to assess activity of the filamentous-growth pathway (49, 64–66). Phosphorylated Fus3p, Kss1p, and Hog1p were detected following trichloroacetic acid (TCA) precipitation as described previously (67) with suggestions from P. Pryciak and H. Dohlman. Cells were grown to mid-log phase and frozen at −80°C. For phosphorylation assays, wild-type (PC538), msb2Δ (PC948), sho1Δ (PC1531), bem4Δ (PC3016), ste20Δ (PC673), ste50Δ (PC3863), ste11Δ (PC3862), and ste12Δ (PC2382) strains were used. pmi40-101 (PC2148), pmi40-101 sho1Δ (PC448), pmi40-101 bem4Δ (PC3525), pmi40-101 ste20Δ (PC522), pmi40-101 ste50Δ (PC446), and pmi40-101 ste12Δ (PC389) strains were used to examine P∼Kss1p in response to glycosylation deficiency. Cell pellets were thawed by the addition of 300 μl of 10% TCA buffer (10 mM Tris HCl [pH 8.0], 10% trichloroacetic acid, 25 mM ammonium acetate [NH4OAc], 1 mM Na2 EDTA). Approximately 0.2 ml of glass beads was added to each sample, followed by five 1-min pulses at full speed in a multitube vortex mixer. Samples were placed on ice for 3 min between cycles. Cell lysates were transferred to sterile 1.5-ml tubes and centrifuged at 4°C for 10 min at 17,000 × g. Supernatants were transferred to 1.5-ml tubes, and 150 μl of resuspension buffer (0.1 M Tris HCl [pH 11.0], 3% SDS) was added to each sample. Samples were boiled for 5 min and then centrifuged at 17,000 × g for 30 s. Proteins were separated on 12% SDS-PAGE gels and transferred to nitrocellulose membranes (Protran BA85; VWR International Inc., Bridgeport, NJ). After transfer, membranes were incubated with primary (for 16 h) and secondary antibodies (1 h) in TBST containing 5% BSA.
For some experiments, MAPK activity was verified using the FUS1-lacZ growth reporter, which in Σ1278b strains lacking Ste4p shows dependence on Msb2p, Sho1p, Bem4p, Ste20p, Ste11p, Ste7p, Kss1p, and Ste12p (26, 51; this study). FUS1-HIS3 expression was measured by spotting cells onto SD-His with 4-amino-1,2,4-triazole. β-Galactosidase assays were performed as described previously (68). Halo assays were performed as described previously (69).
Filamentous growth and biofilm/mat assays.
The single-cell invasive-growth (22), plate-washing (18), and diploid pseudohyphal-growth assays (21) were performed to evaluate filamentous growth. Biofilm/mat assays were performed as described previously (70). Colony perimeters were examined for filamentous mats as described previously (55). Mats and colonies were photographed using an Evolution MP Color camera (32-0041C-212; MediaCybernetics, Bethesda, MD). Polygalacturonidase activity of secreted Pgu1p was used to evaluate PGU1 expression as described previously (71, 72). Strains were spotted onto synthetic medium containing 1% polygalacturonic acid. Plates were incubated for 2 days prior to addition of 0.1% ruthenium red to detect secreted Pgu1p. The budding pattern was determined by established methodology (73) and was confirmed for some experiments by visual inspection of connected cells (74).
Microscopy.
Differential interference contrast (DIC) and fluorescence microscopy using GFP filter sets were performed using an Axioplan 2 fluorescence microscope (Zeiss) with a Plan-Apochromat 100×/1.4 (oil) objective (numerical aperture [NA], 0.17). Digital images were obtained with the Axiocam MRm camera (Zeiss). Axiovision 4.4 software (Zeiss) was used for image acquisition and analysis. Protein localization was performed as described previously (72) for Bem4p-GFP, Cdc42p-GFP, and GFP-Cdc42p. For microscopic observations of cells exposed to α-factor, strains were grown to mid-log phase and washed three times in water. Cells were incubated in 1 to 5 mM α-factor for 150 min.
Data quantitation and analysis.
Process/function and gene ontology (GO) annotations, fungal homology information, and protein interactions data were assessed using the Saccharomyces Genome Database (http://www.yeastgenome.org/) (75, 76). Protein interactions have been identified by many different studies to create global functional maps (41, 77–81). Immunoblot data were quantitated by measuring band intensities by ImageJ. Specifically, the average background intensity was sampled at multiple positions and subtracted from band intensity values by taking the absolute value of intensity measurements. Normalized values were adjusted to Pgk1p band intensities (after background subtraction) to account for loading differences between samples. Normalized band intensity was set to 1.0 for wild-type samples; other intensities were compared to this value. The lightest exposures for which all bands were visible were used for densitometric analyses. For Kss1p protein blots in which two bands were present, the intensities of both bands were counted. To quantify Flo11p secretion, intensity values were normalized to colony size, and the control was set to 1.0. Samples were analyzed at 1- and 2-day time points. The values were similar, and the 1-day value is shown. Mat/biofilm size and halo size were determined by quantifying the area using the circle tool. A BLAST search was performed to identify BEM4 homologs (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Iterative BLAST searching with Bem4p homologous identified small GTP-binding protein GDP dissociation stimulator (Smg GDS) proteins. Clustal W alignment (http://www.ebi.ac.uk/Tools/msa/clustalw2/) was used to align protein sequences. Alignments are based on reference 82; Armadillo (ARM) repeats were interpreted from reference 83. Statistical significance was determined by the chi-squared test. P values were generated via a two-tailed unpaired Student's t test.
RESULTS
Bem4p Regulates the filamentous growth pathway.
To identify new regulators of the filamentous-growth pathway, genes that comprise an interaction network surrounding Cdc42p, constructed from analysis of global protein interactions screens (41, 84–86), were disrupted and tested for roles in invasive growth and MAPK signaling (Fig. 1B; see Table S3 in the supplemental material). Most of the gene deletions did not induce a phenotype (Fig. 1B, gray circles), but one showed a 4-fold decrease in MAPK activity (Fig. 1B, inset, bem4Δ). Bem4p interacts with Cdc42p (39, 41), the major GTPase that regulates the filamentous-growth pathway (23–26). Bem4p was also uncovered in a genetic screen as a regulator of alcohol-induced filamentous growth (87).
To further resolve the interaction network, proteins with genetic and physical interactions with Bem4p were tested. Bem4p has previously been shown to interact with the Rho-type GTPases Rho1p, Rho2p, Rho4p, and Cdc42p and to function in cell polarity regulation and cell integrity (39, 40). Rho2p and Rho4p did not regulate the filamentous-growth pathway (Fig. 1B and data not shown). Sbe2p and Sbe22p, which control protein trafficking from the Golgi and exhibit genetic interactions with BEM4 (88), did not regulate the filamentous-growth pathway (Fig. 1B, inset; also, see Fig. S1A and B in the supplemental material). Septins, which interact with Bem4p (41, 89) and regulate the formation and constriction of the actomyosin ring (90), showed a normal distribution in the bem4Δ mutant (see Fig. S1C in the supplemental material). Thus, Bem4p may regulate the filamentous-growth pathway independently of its other functions in the cell.
Filamentous growth is a differentiation response that can be quantitatively studied by clear-cut biological and biochemical assays (20, 91, 92). To define the role of Bem4p in regulating filamentous growth, the BEM4 gene was disrupted in haploid (MATa) and diploid (MATa/MATα) strains of the filamentous (Σ1278b) background. Compared to wild-type haploid cells, the bem4Δ mutant was defective for invasive growth by the plate-washing assay (Fig. 1C), which occurs in response to glucose depletion (22). The invasive growth defect of the bem4Δ mutant was more pronounced than that of mutants lacking PM regulators (Fig. 1C, msb2Δ and sho1Δ) and equivalent to that of mutants lacking core regulators (Fig. 1C, ste20Δ, ste50Δ, ste11Δ, and ste12Δ). Homozygous bem4Δ/bem4Δ diploid colonies did not produce pseudohyphae under nitrogen-limiting conditions (Fig. 1D and E; see also Fig. S2A and B in the supplemental material), indicating that Bem4p is required for pseudohyphal growth. The filamentous-growth pathway regulates the formation of colonial mats or biofilms (70). Bem4p was required for the expansion (Fig. 1F) and ruffled appearance (Fig. 1G) of mats. Bem4p was required for the development of filamentous mats (Fig. 1H), which occurs under nutrient-limiting conditions (55). Therefore, Bem4p regulates filamentous growth and biofilm/mat formation in haploid and diploid cells.
The filamentous-growth pathway is activated by glucose limitation, one of the inducers of filamentous growth (22). Bem4p was required for the phosphorylation of the mitogen-activated protein (MAP) kinase Kss1p (P∼Kss1p) during growth in the poor carbon source galactose (Fig. 1I, GAL). The filamentous-growth pathway regulates the response to protein glycosylation deficiency (68, 93, 94). Bem4p was required for the induction of the filamentous-growth pathway in the pmi40-101 mutant (see Fig. S2C and D in the supplemental material), whose conditional glycosylation defect occurs by growth in medium lacking mannose (68). The filamentous-growth pathway controls expression of genes required for differentiation to the filamentous cell type (50, 71, 95–98). Bem4p was required for expression of the FRE-lacZ reporter (Fig. 1J), for production of secreted Pgu1p (see Fig. S2E in the supplemental material), and for production of shed Flo11p (see Fig. S2F in the supplemental material). Thus, Bem4p is a positive regulator of the filamentous-growth pathway.
Bem4p is a specific regulator of the filamentous growth pathway.
Most proteins that function in the filamentous-growth pathway also regulate the mating and/or HOG pathways (35, 99–102). To determine whether Bem4p also regulates these pathways, the bem4Δ mutant was examined for defects in mating or osmotolerance. As shown above, Bem4p was required for filamentous growth. A clear example comes from colony morphology. Wild-type colonies exhibit wrinkled morphology, whereas the bem4Δ mutant, like other pathway mutants (ste12Δ), exhibits smooth morphology (Fig. 2A, Filamentous Growth). By comparison, the bem4Δ mutant showed a normal mating response. The bem4Δ mutant showed growth arrest in response to the pheromone α-factor (Fig. 2A, Mating). The bem4Δ mutant also showed wild-type induction of a mating pathway reporter by α-factor (Fig. 2B), which is different from its role in regulating a filamentous-growth pathway reporter (Fig. 1J). The bem4Δ mutant showed normal rates of shmoo formation (see Fig. S3A in the supplemental material) and diploid formation (see Fig. S3B), concentration-dependent sensitivity to α-factor (see Fig. S3C), and clustering of GFP-Cdc42p at shmoo tips (see Fig. S3D). To determine whether Bem4p regulates the HOG pathway, the BEM4 gene was disrupted in cells lacking the Sln1p branch (ssk1Δ) (103–105). Compared to the ste11Δ ssk1Δ double mutant, which is sensitive to growth in high-salt medium (Fig. 2A, HOG), the bem4Δ ssk1Δ double mutant was not sensitive to osmotic stress (Fig. 2A, HOG). Thus, Bem4p does not play a major role in regulating the mating or HOG pathways.
FIG 2.
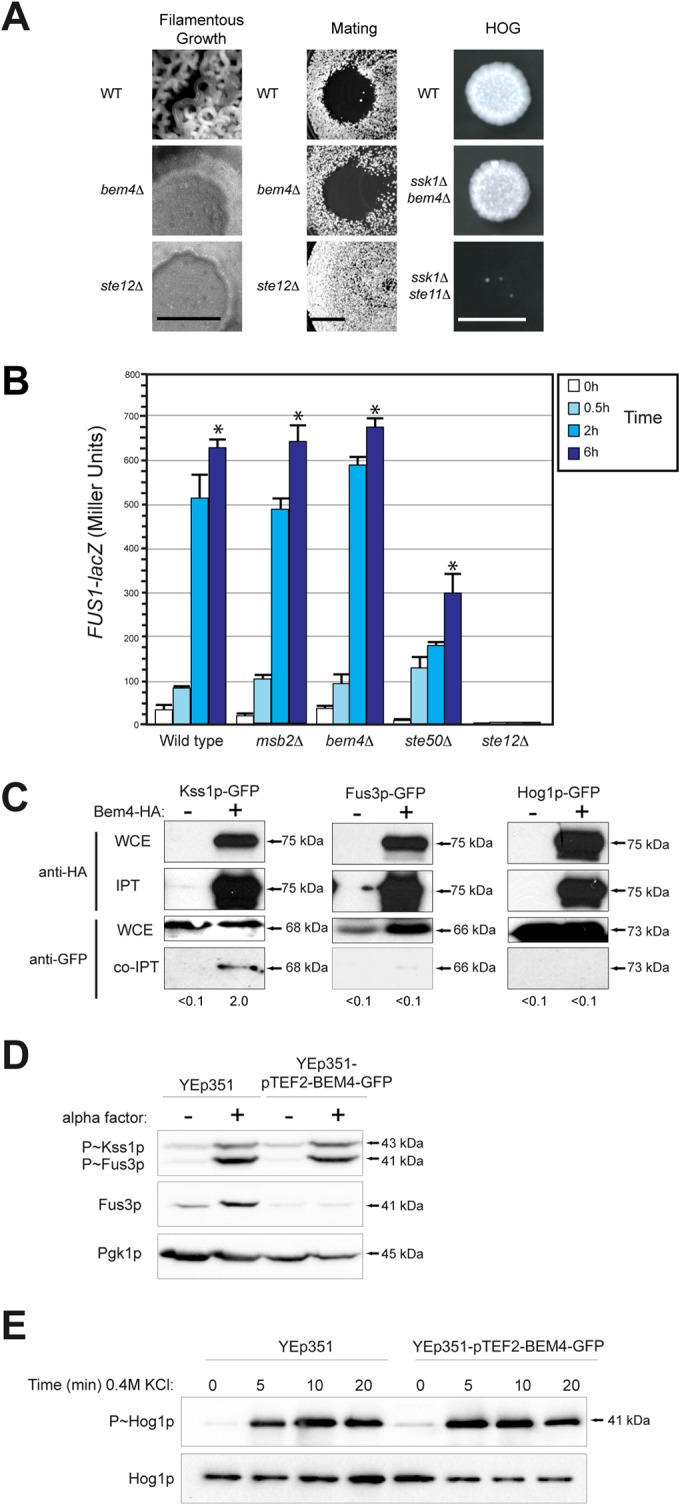
Bem4p is a pathway-specific regulator of the filamentous-growth pathway. (A) (Left) Colony ruffling as an indicator of filamentous growth. Bar, 1 cm. (Middle) Halo assay as a measure of growth arrest in response to α-factor. (Right) Growth of cells in YEPD plus 1 M KCl. Images from the 2-day time point are shown. (B) β-Galactosidase activity of the mating pathway FUS1-lacZ reporter. Cells were grown to mid-log phase in YEPD, washed, and treated with 1 mM α-factor for the indicated times. Values are the averages from two independent trials; error bars show the standard deviations between trials, and asterisks denote P values of <0.05. (C) Co-IP analysis of Bem4p-HA with Kss1p-GFP, Fus3p-GFP, and Hog1p-GFP. Bem4p-HA was overexpressed in cells containing genomic fusions to GFP. Band intensity relative to input levels after background subtraction was determined by ImageJ. Input levels are 10% for WCE. (D) Role of high-copy-number Bem4p-GFP in regulating the mating pathway. Cells were grown to mid-log phase in YEPD and induced by 5 μM pheromone and evaluated by immunoblotting for phosphorylation of Kss1p and Fus3p. (E) Role of high-copy-number Bem4p-GFP in activation of the HOG pathway. Cells were grown to mid-log phase in YEPD and induced by 0.4 M KCl at the indicated times for phosphorylation of Hog1p.
Different MAP kinases regulate the filamentous growth (Kss1p), mating (Fus3p), and HOG (Hog1p) pathways. As determined by coimmunoprecipitation (co-IP) analysis, Bem4p-HA associated with the MAP kinase Kss1p-GFP but not with the MAP kinase Fus3p-GFP or Hog1p-GFP (Fig. 2C). High-copy-number Bem4p, which stimulates the filamentous-growth pathway (see below), did not stimulate the mating pathway in response to a-factor (Fig. 2D, P∼Fus3p and P∼Kss1p) or the HOG pathway in response to salt (Fig. 2E, P∼Hog1p). Thus, Bem4p has a specific function in regulating the filamentous-growth pathway. Besides Kss1p and the transcription factor Tec1p, Bem4p is the only pathway-specific regulator of the filamentous-growth pathway yet identified.
Bem4p was compared to pathway-specific regulators of the mating and HOG pathways. Bem4p is a globular protein that, along with fungal homologs, exhibits similarity to Smg GDS proteins (see Fig. S4 in the supplemental material), which in mammals regulate the activity of Rho GTPases and play diverse roles in development, cell differentiation, and cancer (106–109). By comparison, Pbs2p is a pathway-specific regulator and MAPKK for the HOG pathway (99, 110–112). Ste2p and Ste3p are β-adrenergic-type receptors that connect to a heterotrimeric G protein (Gpa1p, Ste4p, and Ste18p) and the adaptor Ste5p, which is a fungus-specific protein related to E3 ubiquitin ligases, to regulate the mating pathway (113–120). Thus, Bem4p is a unique MAPK regulatory protein that is not related to other pathway-specific regulators. Bem4p may have an evolutionary origin distinct from that of Ste5p and Pbs2p. This conclusion implies that the evolution of pathway-specific regulators may have diversified a core pathway, which can account for why the filamentous growth, mating, and HOG pathways share components, because they arose from a common ancestor (see Fig. S5 in the supplemental material).
Sho1p functions through Bem4p in the filamentous-growth pathway.
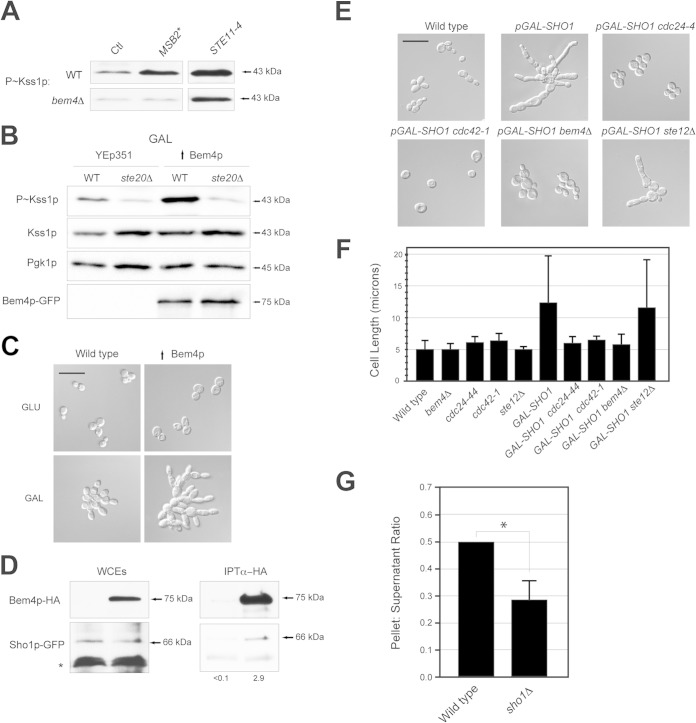
To determine how Bem4p regulates the filamentous-growth pathway, genetic suppression analysis was performed, which allows the ordering of components in a pathway using gain- and loss-of-function alleles. Hyperactive alleles of MSB2, SHO1, and STE11 were tested for their ability to bypass the signaling defect of the bem4Δ mutant. The hyperactive allele MSB2Δ100–181 (26, 36) stimulated Kss1p phosphorylation but did not bypass the signaling defect of the bem4Δ mutant (Fig. 3A; also, see Fig. S3E in the supplemental material). By comparison, the hyperactive allele STE11-4 (121) bypassed the signaling defect of the bem4Δ mutant (Fig. 3A; also, see Fig. S3E in the supplemental material). The increase in Kss1p levels/expression (see Fig. S3E in the supplemental material) is probably due to positive feedback, because KSS1 is a transcriptional target of the filamentous-growth pathway (50). A hyperactive allele of SHO1 gave mixed results (data not shown), perhaps because Sho1p functions at multiple points in the filamentous-growth pathway (100, 112). Thus, Bem4p functions between Msb2p and Ste11p in the filamentous-growth pathway (Fig. 1A).
FIG 3.
Bem4p interacts with Sho1p and is an effector of Sho1p function. (A) P∼Kss1p levels in wild-type cells and the bem4Δ mutant containing the indicated alleles. The total levels of Kss1p and protein (assessed by Pgk1p) were similar between samples. (B) Wild-type cells and the ste20Δ mutant containing an empty plasmid (YEp351) or high-copy-number Bem4p-GFP were grown to mid-log phase in YEP-GAL and evaluated by immunoblot analysis. (C) High-copy-number BEM4 stimulates filamentous growth under nutrient-limiting conditions. Cells containing a control plasmid (YEp351) or high-copy-number Bem4p-GFP were grown to mid-log phase in YEPD (GLU) or YEP-Gal (GAL) medium and examined by microscopy at a magnification of ×100 (bar, 25 μm). (D) Bem4p-HA and Sho1p-GFP associate, as determined by co-IP analysis. Band intensity relative to input levels after background subtraction was determined by ImageJ. Input levels are 10% for WCE. (E) Dependency of Cdc24p, Cdc42p, Bem4p, and Ste12p on the hyperpolarized growth phenotype of Sho1p. Cells were grown for 24 h on YEP-Gal medium. Bar, 25 μm. (F) Quantitation of cell length for the strains used for panel E. More than 100 cells were examined in separate trials for each experiment. With the exception of GAL-SHO1 and GAL-SHO1 ste12 strains, which show a diversity of cell lengths, <10% deviation is seen between trials. (G) Role of Sho1p in regulating the association of Bem4p with the pellet fractions. The experiment was performed in triplicate; the asterisk denotes a P value of <0.05.
A functional Bem4p-GFP fusion expressed from a strong constitutive promoter (pTEF2) on a high-copy-number plasmid (YEp351) induced the filamentous-growth pathway (Fig. 3B, ↑Bem4p). High-copy-number Bem4p also caused hyperfilamentous growth (Fig. 3C). This phenotype was observed only under glucose-limiting conditions (Fig. 3C, compare GLU [glucose] to GAL), which indicates that ↑Bem4p does not constitutively activate the pathway but hyperactivates it under conditions that are permissive for pathway activity. High-copy-number Bem4p failed to bypass the signaling defect of the ste20Δ mutant (Fig. 3B), which indicates that Bem4p functions above Ste20p in the filamentous-growth pathway. Although this result is clear, epistasis analysis with other proteins and conditions suggests that Bem4p impinges on the pathway at multiple levels, including below the Cdc42p-Ste20p module, which is discussed in detail below.
Proteins that function between Msb2p and Ste20p include Sho1p, Opy2p, and Cdc42p (Fig. 1A). Protein interaction tests were used to determine whether Bem4p interacts with these proteins. A functional epitope-tagged version of Bem4p expressed from its endogenous promoter at the BEM4 locus, Bem4p-HA, associated with Sho1p-GFP (also functional and expressed from its own promoter), as determined by co-IP analysis (Fig. 3D). A small amount of Sho1p coprecipitated with a large amount of Bem4p. This may be because Bem4p may associate transiently with Sho1p or with the activated version of the protein, which may comprise only a fraction of the total protein in the cell. Alternatively, the nonionic detergent used to solubilize Sho1p from the PM may affect the conformation of Sho1p or otherwise interfere with the interaction between the proteins. Bem4p did not interact with Msb2p or Opy2p (data not shown). Thus, Bem4p associates with Sho1p and functions at the level of Sho1p in the filamentous-growth pathway.
Sho1p is a PM regulator of the filamentous growth (26, 36, 38, 100, 112) and HOG pathways (35, 99–102). In the filamentous-growth pathway, Sho1p controls the reorganization of cell polarity, which is distinct from its function in the HOG pathway, because osmotic stress does not induce polarized growth. We have previously shown that overexpression of Sho1p by the inducible pGAL1 promoter causes hyperpolarized growth (Fig. 3E, pGAL-SHO1) (36). Cdc42p is the major regulator of cell polarity in yeast (122) and would be expected to mediate this phenotype. The polarized growth induced by pGAL-SHO1 was reduced in the cdc24-1 and cdc42-1 mutants at the semipermissive temperature (Fig. 3E and F). The polarized growth induced by pGAL-SHO1 was also suppressed in the bem4Δ mutant (Fig. 3E and F). This phenotype was not seen in other filamentous-growth pathway mutants (Fig. 3E and F, ste12Δ), which may indicate that Bem4p plays a specific role in mediating the cell polarization induced by overexpression of Sho1p. Subcellular fractionation analysis showed that Bem4p associates with membrane (pellet) fractions (see below). The association of Bem4p with membrane fractions was reduced in the sho1Δ mutant (Fig. 3G), which may indicate that Sho1p contributes to Bem4p localization to the membrane. Therefore, Bem4p functions as an effector for one of the PM regulators of the filamentous-growth pathway and conveys nutrient and polarity signals that emanate at the head of the pathway.
Bem4p regulates Cdc42p activity and can function at the PM.
Given that Sho1p is a PM regulator of the filamentous-growth pathway, Bem4p may also function at the PM. To determine the localization of Bem4p, the Bem4p-GFP fusion protein was examined, which showed a diffuse pattern that was typical of cytosolic proteins but was otherwise uninformative (data not shown). To further define the cellular location of Bem4p, subcellular fractionation analysis was performed (61). By this method, Bem4p-HA partitioned between the membrane fraction (Fig. 4A, P13) and the cytosol (Fig. 4A, S100). Bem4p does not contain a recognizable lipid-binding motif and may associate with the PM through interactions with Sho1p or another protein.
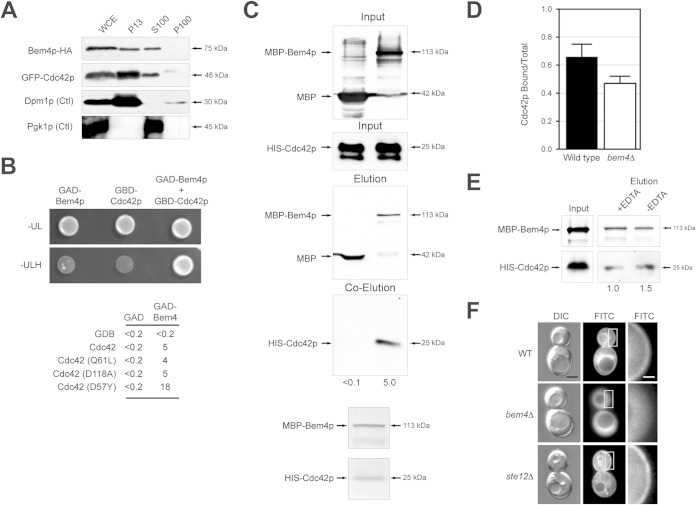
FIG 4.
Bem4p binds to and regulates Cdc42p activity. (A) Subcellular fractionation of cells expressing Bem4p-HA and GFP-Cdc42p. Whole-cell extract (WCE), supernatant (S), and pellet (P) fractions from the centrifugation steps (P13, 13,000 × g; P100, 100,000 × g; S100, 100,000 × g) were analyzed by immunoblot analysis. An integral membrane endoplasmic reticulum protein (Dpm1p) and a cytosolic protein (Pgk1p) are shown as controls. (B) (Top) The Gal4p activation domain (GAD)-Bem4p and Gal4p binding domain (GBD)-Cdc42p associate, as determined by two-hybrid analysis. Strains were spotted onto SD-Ura-Leu (-UL) to maintain selection for plasmids and SD-Ura-Leu-His (-ULH) to measure activity of a two-hybrid growth reporter. (Bottom) β-Galactosidase activities (in Miller units) of the two-hybrid reporter GAL7-lacZ. Values are averages from two independent experiments; the standard deviation was less than 10% between trials. (C) MBP-Bem4p and HIS-Cdc42p proteins associate by in vitro pulldown. Fifty percent of the WCE (input) was used for the pulldown. Numbers are band intensities normalized to input protein levels by ImageJ. (D) PAK-binding assay. GFP-Cdc42p was precipitated by beads coated with GST or GST-PAK binding domain. Band intensity was determined by normalizing precipitated GFP-Cdc42p to input levels from wild-type cells or cells lacking BEM4. Equal amounts of GST and GST-PAK binding domain were loaded onto beads. Values are averages from two separate experiments, which showed less than 10% difference between trials. (E) MBP-Bem4p associates with HIS-Cdc42p in buffer with and without 10 mM EDTA. Numbers represent band intensity normalized to input protein levels by ImageJ. (F) Localization of GFP-Cdc42p in wild-type cells and the bem4Δ and ste12Δ mutants. Bar, 2 μm (left and middle) and 0.4 μm (right). Figure S5A in the supplemental material shows additional examples of cells, and quantitation is provided in Fig. S5B and C.
To this point, the data indicate that Bem4p associates with Sho1p and functions at or above Ste20p (Fig. 1A). Cdc42p also regulates the filamentous-growth pathway at the level of Ste20p (23–26) and functions at the PM (123–125). Bem4p and Cdc42p were previously shown to interact and exhibit genetic interactions (39, 41). Specifically, BEM4 (bud emergence) was identified as a high-copy-number suppressor of temperature-sensitive alleles of cdc24, which indicates that Bem4p regulates (in some manner) Cdc42p activity (39). To determine whether Bem4p regulates the activity of Cdc42p in the filamentous-growth pathway, the proteins were examined by biochemical assays. By subcellular fractionation analysis, GFP-Cdc42p showed a profile similar to that of Bem4p (Fig. 4A). By two-hybrid analysis, Bem4p associated with versions of Cdc42p that mimic the GTP-locked (Cdc42pQ61L) and GDP-locked conformations (Cdc42pD118A) and robustly with a dominant negative version of the protein (Cdc42pD57Y) (Fig. 4B). This may indicate that Bem4p is not an effector of Cdc42p, because effectors bind preferentially to the GTP-bound conformation of GTPases.
Using affinity-tagged versions of Bem4p (MBP-Bem4p) and Cdc42p (HIS-Cdc42p) overexpressed in Escherichia coli and purified by affinity chromatography, Bem4p and Cdc42p associated by in vitro pulldown analysis (Fig. 4C). Coomassie staining showed that the proteins interact at a 1:1 ratio (Fig. 4C, bottom). An in vitro PAK binding assay (58) showed that the activity of Cdc42p was dependent on Bem4p (Fig. 4D). This effect was subtle, which is not surprising because Bem4p is not required for growth (budding) and would not be expected to regulate full GTP-Cdc42p activity. Bem4p does not possess a PAK binding domain, which are characteristic of Cdc42p effectors, and purified MBP-Bem4p associated with purified HIS-Cdc42p in buffer with or without EDTA (Fig. 4E), which chelates Mg2+ ions and increases the ratio of nucleotide-free Cdc42p (126). These tests together indicate that Bem4p, rather than functioning as an effector, is a regulator of the activity of Cdc42p in the cell. The in vitro data further indicate that Bem4p can associate with Cdc42p independently of its lipid modification and in the absence of membrane.
Cdc42p is anchored to the PM by a specific lipid modification, geranylgeranylation (11, 127). In the bem4Δ mutant, the localization of GFP-Cdc42p at the PM was reduced (Fig. 4F; also, see Fig. S6 in the supplemental material). This role for Bem4p was not due to its function in regulating the filamentous-growth pathway, as other pathway components did not show this phenotype (ste12Δ) (Fig. 4F; also, see Fig. S6 in the supplemental material). This phenotype was observed under nonfilamentous (high-glucose) conditions and may therefore represent a function for Bem4p that is separate from its role in regulating the filamentous-growth pathway. Bem4p was not required for the localization of Cdc42p to mating projections (see Fig. S3D in the supplemental material), which occurs by an ill-defined mechanism known as clustering (128). Given that Bem4p is not required for viability, the reduced levels of Cdc42p at the PM are sufficient for the protein to perform its essential function in polarity establishment.
Cdc24p regulates the filamentous growth pathway.
The major regulator of Cdc42p during vegetative growth and mating is Cdc24p (30–33). To determine if Cdc24p regulates the filamentous-growth pathway, a plasmid-shuttle approach was employed, because Cdc24p is essential for viability. A high-copy-number plasmid containing CDC24 complemented the viability defect of the cdc24Δ mutant and showed elevated P∼Kss1p levels compared to a CEN-based plasmid expressing normal levels of Cdc24p (Fig. 5A). By comparison, a temperature-sensitive allele, cdc24-4 (129), was defective for filamentous-growth pathway activity at the nonpermissive temperature (Fig. 5A). These results were confirmed at the semipermissive temperature with a growth reporter (FUS1-HIS3), which in cells lacking an intact mating pathway (ste4) provides a readout of filamentous-growth pathway activity (Fig. 5B). Thus, Cdc24p regulates the filamentous-growth pathway.
FIG 5.
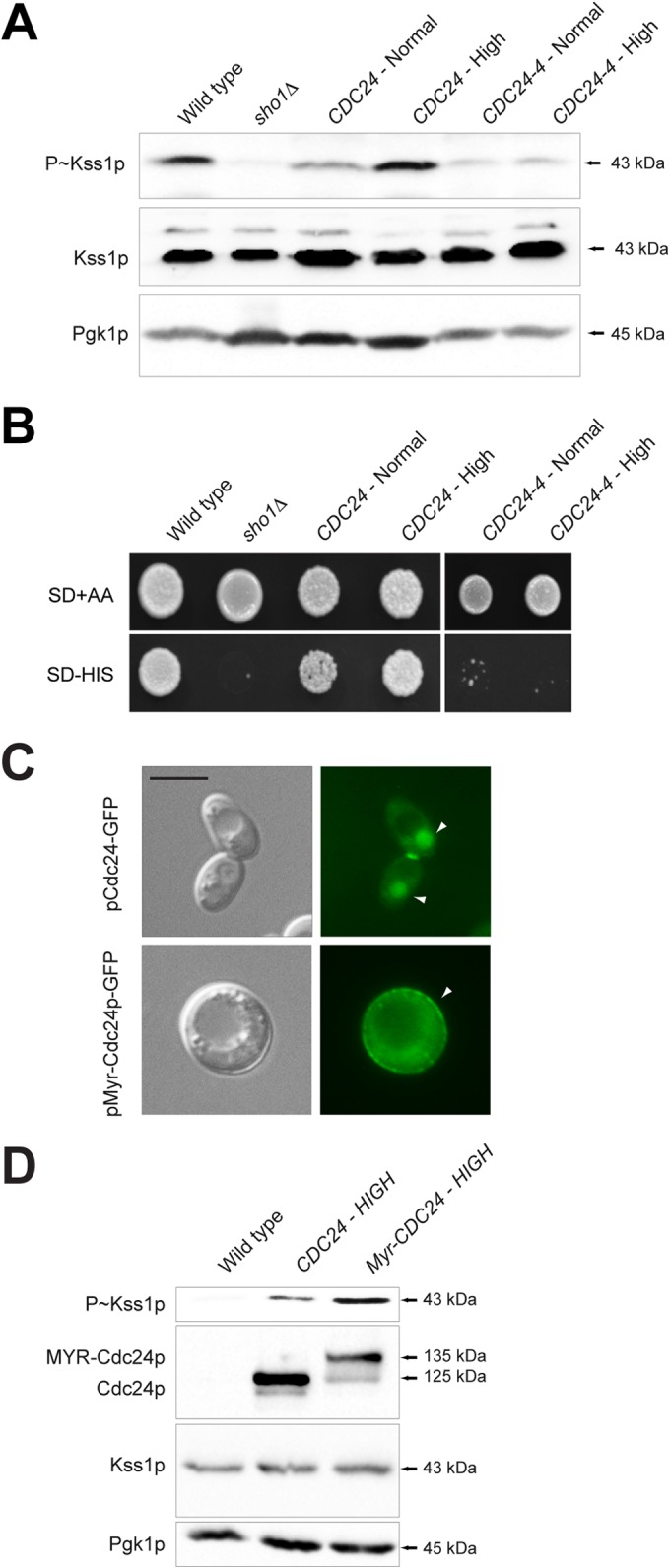
Cdc24p regulates the filamentous-growth pathway. (A) P∼Kss1p levels in cells containing wild-type CDC24 or the cdc24-4 allele. Cells were examined after 2 h of incubation at 37°C. (B) The same strains were assessed at the semipermissive temperature (30°C) with the cross talk growth reporter (ste4 FUS1-HIS3). Growth on SD-His is indicative of filamentous-growth pathway activity. (C) Localization of pCdc24p-GFP and pMyr-Cdc24p-GFP in wild-type cells. Arrow heads indicate Cdc24p-GFP, which is typically seen in the nucleus, at polarized sites, and at the neck for wild-type Cdc24p but mainly at the PM for Myr-Cdc24p-GFP. Bar, 5 μm. (D) P∼Kss1p levels determined by immunoblotting for the wild type, the strain with high-copy-number CDC24, and the strain with high-copy-number Myr-Cdc24p.
Cdc24p is highly regulated in terms of its localization and activity. In the G1 phase of the cell cycle, Cdc24p exits the nucleus and localizes at incipient bud sites at the PM (130–132). During mating, Cdc24p localizes to mating projections (133). To determine whether the localization of Cdc24p at the PM is important for its function in the filamentous-growth pathway, Cdc24p was tethered to the PM by introduction of a myristoylation signal (Myr-Cdc24p). Myr-Cdc42p showed predominately PM localization (Fig. 5C) and showed elevated P∼Kss1p levels (Fig. 5D). Thus, the levels and PM localization of Cdc24p are important determinants in its regulation of the filamentous-growth pathway. This conclusion may be an oversimplification, as MYR-Cdc24p cells are round (not hyperfilamentous), which may result in Cdc42p activation outside its natural context. Nevertheless, these results support the idea that Cdc24p regulates the filamentous-growth pathway.
Bem4p is a regulator of Cdc24p in the filamentous growth pathway.
During vegetative growth, the adaptor Bem1p and the Ras-like GTPase Rsr1p recruit and activate Cdc24p at bud sites (134–136). During mating, Bem1p (3, 137), the heterotrimeric G-protein Ste4p (138), and the cyclin-dependent kinase inhibitor Far1p (129) recruit and activate Cdc24p at shmoo tips. Bem1p, Far1p, Rsr1p, and Ste4p were dispensable for filamentous growth (Fig. 1B). Thus, Cdc24p is likely regulated by a different mechanism in the filamentous-growth pathway. We tested and found that Myr-Cdc24p bypassed the signaling defect of the bem4Δ mutant (Fig. 6A), which indicates that Bem4p functions at or above the level of Cdc24p in the filamentous-growth pathway.
Two-hybrid analysis of cell polarity regulators identified an interaction between Bem4p and Cdc24p (41). Bem4p-HA interacted weakly but reproducibly with Cdc24p-GFP by co-IP analysis (Fig. 6B). Affinity-tagged Cdc24p (GST-Cdc24p) overexpressed and purified from E. coli adhered to an affinity column to which MBP-Bem4p was bound (Fig. 6C, MBP-Bem4p). Thus, Bem4p interacts with Cdc24p in vivo and in vitro.
Cdc24p contains well-established protein-interaction domains that mediate its activation in different contexts. Ste4p, Far1p, and Rsr1p interact with the calponin homology domain (CH, residues 154 to 226). Cdc42p interacts with the Dbl homology domain (DH, residues 284 to 452), and Bem1p interacts with the PB1 domain (residues 781 to 854) (133, 136, 139–142). Cdc24p also contains a pleckstrin homology (PH) domain (residues 477 to 667) (132, 143). The PB1 and PH domains have an autoinhibitory function (136). Binding of Bem1p to the PB1 domain contributes to Cdc24p activation during vegetative growth and mating. To further define the interaction between Bem4p and Cdc24p, in vitro pulldowns were performed with versions of Cdc24p that lack various protein-interaction domains. Bem4p interacted with a version of Cdc24p lacking the PB1 domain (Fig. 6D, Cdc24p-PB1Δ) and a version of Cdc24p lacking the CH domain (Fig. 6D, Cdc24p-CHΔ). Bem4p failed to interact with a version of Cdc24p that contained only the CH and DH domains (Fig. 6D, Cdc24p-CH-DH) but interacted with a version that contained only the DH and PH domains (Fig. 6D, Cdc24p-DH-PH). This analysis indicates that Bem4p interacts with the PH domain of Cdc24p. To further test this possibility, additional derivatives of Cdc24p were constructed and purified. Bem4p was found to interact with a version of Cdc24p that contained only the PH domain (Fig. 6D, Cdc24p-PH) but not a version that lacked only the PH domain (Fig. 6D, Cdc24p-PHΔ). Therefore, Bem4p interacts with the PH domain of Cdc24p.
The interaction between Bem4p and the PH domain of Cdc24p may function to relieve autoinhibition and promote activation of the GEF. In support of this idea, expression of Cdc24p-PHΔ in yeast caused an increase in cell size and cell polarization, which resembled the hyperpolarized growth of cells undergoing filamentous growth (Fig. 6E). PH domains are protein interaction domains, and these domains can also bind to phosphatidylinositide phosphates (PIPs) to selectively direct the association of proteins with membranes (144). The PH domain of Cdc24p does not bind specific PIPs and does not associate with membranes (145). The idea that the binding of Bem4p to the PH domain of Cdc24p may regulate its activity is consistent with the fact that Bem4p is a positive regulator of the MAPK pathway, with the fact that Bem4p functions above Cdc24p in the pathway, and with the role of Bem4p in regulating Cdc42p activity.
The selective recruitment of proteins to the PM increases their local concentration and is a general mechanism for the activation of signaling pathways in a potent and highly selective manner (146, 147). Bem4p may regulate the localization of Cdc24p. To test this possibility, a Cdc24p-GFP fusion protein was examined in wild-type cells and the bem4Δ mutant. Under nutrient-rich (high-glucose) conditions, Cdc24p-GFP was found transiently at polarized sites in emerging buds with equal frequency in wild-type cells and the bem4Δ mutant (Fig. 6F). Cdc24p-GFP was also found in the nuclei of unbudded cells and at the mother bud neck. Under nutrient-limiting (low-glucose) conditions, cells showed an increase in the polarized localization of Cdc24p-CFP, which was reduced in the bem4Δ mutant (Fig. 6F). Other MAPK pathway mutants also showed reduced Cdc24p at polarized sites (Fig. 6F, ste20Δ), and introduction of the hyperactive STE11-4 allele restored polarized Cdc24p-GFP localization to the bem4Δ mutant (Fig. 6F). Thus, Bem4p probably regulates Cdc24p localization through its role in regulating the filamentous-growth pathway. Given that filamentous-growth pathway mutants are viable, it is unlikely that Bem4p and other MAPK regulators are required to recruit Cdc24p to the PM. Rather, the filamentous-growth pathway likely functions to maintain Cdc24p at polarized sites, which may promote polarized growth through a positive-feedback loop (133, 148). Interestingly, Cdc24p-CFP (and Sho1p-YFP) were detected in internal sites in filamentous-growth pathway mutants (Fig. 6G), perhaps because these proteins are internalized in a protein complex from the PM. These findings show that Bem4p is a regulator of Cdc24p in the filamentous-growth pathway and identify a role for the filamentous-growth pathway in maintaining Cdc24p at polarized sites under nutrient-limiting conditions.
Mapping functional Cdc42p and Cdc24p interaction domains on the Bem4p protein.
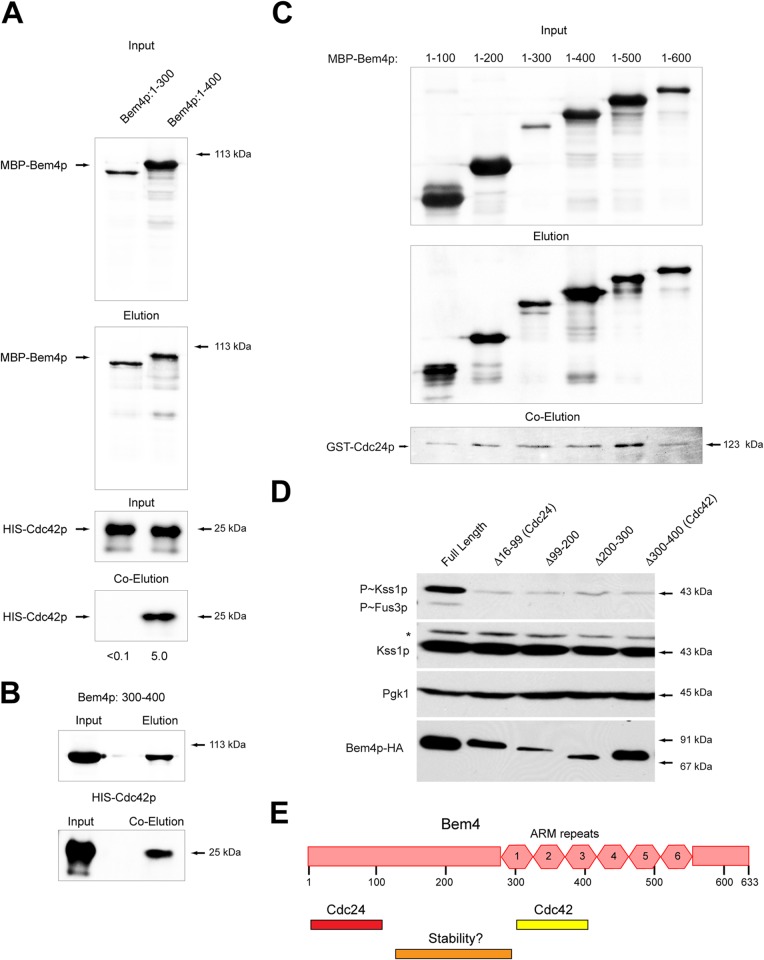
To map the sites of interaction between Bem4p, Cdc24p, and Cdc42p, C-terminal deletions of Bem4p were overexpressed in E. coli and assessed for interaction with Cdc42p and Cdc24p. A version of Bem4p that lacked the last 233 amino acids of the protein (Bem4p1–400) interacted with Cdc42p (Fig. 7A). By comparison, a version of Bem4p that lacked an additional 100 amino acid residues (Bem4p1–300) did not interact with Cdc42p (Fig. 7A). This result indicates that a region of the Bem4p protein (between residues 300 and 400) is required for interaction with Cdc42p. In support of this possibility, a region of the Bem4p protein from 300 to 400 residues interacted with Cdc42p (Fig. 7B). The same truncations were used to map the site of interaction between Bem4p and Cdc24p. All truncations associated with Cdc24p (Fig. 7C), with more or less the same affinity. This result indicates that amino acid residues 1 to 100 of Bem4p are sufficient to mediate that interaction.
FIG 7.
Mapping functional domains on the Bem4p protein. (A) In vitro pulldowns with versions of Bem4p fused to MBP with HIS-Cdc42p. (B) A region of Bem4p from amino acid residues 300 to 400 interacts with HIS-Cdc42p. (C) Versions of MBP-Bem4p that include amino acid residues 1 to 100 interact with GST-Cdc24p, as determined by in vitro pulldown. Controls for the pulldowns can be found in Fig. 6. (D) Impact of internal deletions within the Bem4p protein on the stability of the protein and filamentous-growth pathway activity. (E) Diagram of the Bem4p protein with Cdc24p and Cdc42p sites shown. One region of the protein that affects the stability of the protein is also shown.
To determine the functional significance of these interactions, internal deletions were constructed at the BEM4 locus in the genome, by replacing these regions with a 3×HA epitope. Bem4p-HA 16-99Δ, which encompassed the Cdc24p binding region, was present at the same levels as full-length Bem4p-HA but failed to induce the filamentous-growth pathway (Fig. 7D). Similarly, Bem4p-HA 300-400Δ, which encompasses the Cdc42p binding region, was present at the same levels as Bem4p-HA but failed to induce the filamentous-growth pathway (Fig. 7D). Other deletions were also constructed and tested (99–200Δ and 200–300Δ). These were nonfunctional but affected the stability of the Bem4p protein (Fig. 7E, stability). Although there might be several explanations for why these proteins are not competent to regulate MAPK signaling, a straightforward possibility is that the interaction between Bem4p and Cdc24p and between Bem4p and Cdc42p are required for Bem4p function in the filamentous-growth pathway.
Bem4p interacts with the MAPKKK Ste11p.
Although the major function of Bem4p is at the level of the GTPase module, some data are consistent with a role for Bem4p functioning at other points in the signaling cascade (for an example, see Fig. 2C). To further resolve the roles that Bem4p may play in regulating the filamentous-growth pathway, two-hybrid tests were performed between Bem4p and a panel of pathway regulators (including Bem4p, Ste20p, Ste11p, Ste50p, the cytosolic domain of Msb2p, and the cytosolic domains of Opy2p, Pbs2p, Ste7p, and Ras2p). These tests uncovered a specific interaction between Bem4p and the MAPK kinase kinase (MAPKKK) Ste11p (Fig. 8A). Ste11p regulates the filamentous-growth pathway (Fig. 1A, C, and I) (18, 27) and is a target of the PAK Ste20p (149). Bem4p associated with N-terminal regulatory domain of Ste11p (Fig. 8A). Bem4p also associated to the same degree with wild-type Ste11p and a constitutively active version (Fig. 8A, Ste11-4p). In vitro pulldowns showed that Bem4p-MBP associated with purified GST-Ste11p in vitro (Fig. 8B). Therefore, Bem4p interacts with Ste11p in vivo and in vitro.
FIG 8.

Bem4p interacts with the MAPKKK Ste11p. (A) (Left) Bem4p interacts with Ste11p by two-hybrid analysis but not Ras2p or Pbs2p. (Right) β-Galactosidase activity (in Miller units) of the two-hybrid reporter GAL7-lacZ was measured for the constructs shown. Values are averages from two experiments; the standard deviation was less than 10% between trials. (B) MBP-Bem4p interacts with GST-Ste11p in vitro. Fifty percent of the WCE (input) was used for the pull down. (C) MAPK activity (based on the β-galactosidase activity of the ste4 FUS1-lacZ reporter) in wild-type cells and the bem4Δ mutant containing the indicated versions of Ste50p and Ste11p. Asterisks denote P values of <0.05; the double asterisk denotes a P value of <0.01.
Ste11p is brought to the PM by an adaptor protein, Ste50p, which binds tightly and constitutively to Ste11p (100, 150–157) and the PM regulator Opy2p (37, 158, 159). Given that Bem4p interacts with Ste11p and functions at the PM, Bem4p may recruit or stabilize Ste11p to the PM. To test this possibility, MAPK activity in cells where Ste50p, and consequently Ste11p, was anchored to the PM by a myristoylation signal was recorded. Myr-Ste50p showed elevated filamentous-growth pathway activity, particularly when overexpressed from an inducible promoter (Fig. 8C, Myr-Ste50p). Myr-Ste50p did not bypass the signaling defect of the bem4Δ mutant (Fig. 8C). By comparison, Ste11-4p fully bypassed the signaling defect of the bem4Δ mutant (Fig. 8C). This result indicates that Bem4p plays a role in activating Ste11p at the PM. Activation in this context may be mediated by Bem4p's role in regulating Cdc24p/Cdc42p or by interactions between Bem4p and Ste11p.
DISCUSSION
A critical challenge in the field of cellular signaling is to understand how proteins that function in multiple settings induce a specific response. This is particularly true for small GTPases, which can function in different contexts to execute different types of responses. In this study, we identified a role for a protein which interacts with Cdc42p and which induces a specific differentiation response. Bem4p was first identified as a general regulator of Rho-type GTPases. Our results do not challenge this conclusion. In fact, it is clear that Bem4p has roles in cell polarity regulation and cell integrity (39, 40). In this study, we identified a new and specific function for Bem4p as a regulator of the MAPK pathway that controls filamentous growth. This role for Bem4p was uncovered by exploring a protein interaction network surrounding Cdc42p. Most Cdc42p-interacting proteins did not regulate the filamentous-growth pathway, which is indicative of a highly specific function for Bem4p. The discovery of a new regulator of the filamentous-growth pathway is a key advance in understanding MAPK regulation in this system.
Biochemical and genetic evidence show that Bem4p regulates the filamentous-growth pathway at the level of the Cdc42p GTPase module. Bem4p conveys nutrient and polarity signals that emanate at the Msb2p/Sho1p module (Fig. 9). Bem4p also interacts with the GEF Cdc24p. Cdc24p regulates the filamentous-growth pathway, and we pinpoint a key role for Bem4p in regulating an early step in Cdc24p regulation. We specifically show that Bem4p interacts with the PH domain of Cdc24p. Generally speaking, the PH domains of Rho GEFs can have positive or negative roles in GTPase regulation (160). In yeast, the PH domain of Cdc24p has an autoinhibitory function (136); thus, Bem4p may relieve autoinhibition conferred by that domain (Fig. 9). Bem4p interacts directly with Cdc42p (39; this study). We cannot exclude the possibility that Bem4p facilitates the interaction between Cdc24p and Cdc42p to promote Cdc42p activation (Fig. 9). Likewise, the interaction between Bem4p and the MAPKKK Ste11p connects signaling at the Msb2p/Sho1p module to the MAPKKK through Bem4p. The role of Bem4p in regulating Cdc24p dovetails with the roles of Msb2p and Sho1p in regulating the filamentous-growth pathway. Msb2p interacts with the active (GTP-bound) form of Cdc42p (26) potentially to stabilize activated Cdc42p at complexes containing Sho1p and Bem4p. Sho1p also associates with Cdc24p (36) and may regulate Cdc24p along with Bem4p in the filamentous-growth pathway.
FIG 9.
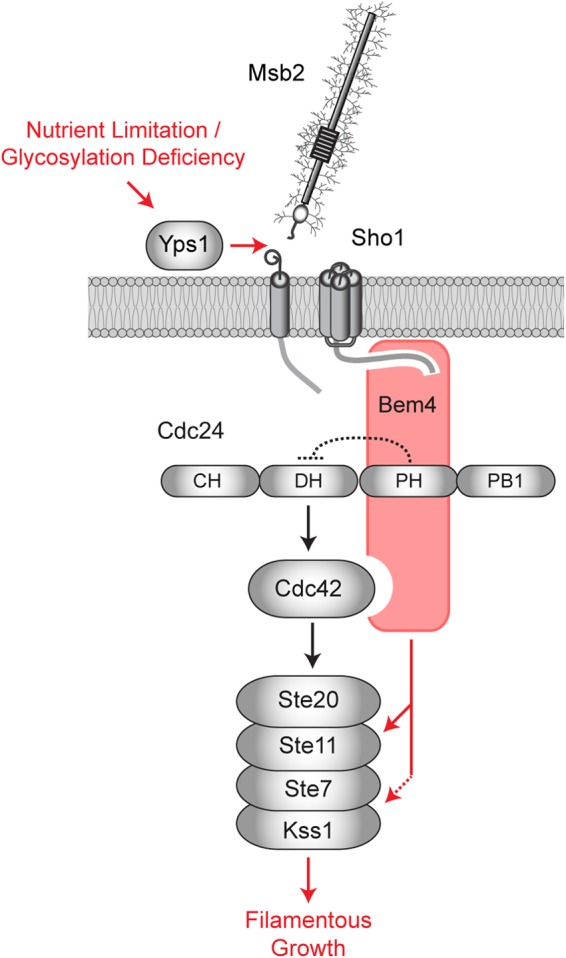
Role of Bem4p in regulating the filamentous-growth pathway. Under nutrient-limiting conditions, Msb2p is activated by processing and release of the inhibitory extracellular domain by the protease Yps1p (36). Activated Msb2p and Sho1p function through Bem4p, which binds to the PH domain of Cdc24p to relieve autoinhibition. Bem4p interacts with Cdc42p and Ste11p and functions in a complex with Kss1p (dashed arrow), to further regulate the filamentous-growth pathway. Although not shown, Cdc42p is at the PM, the PM sensor Opy2p recruits Ste50p-Ste11p to the PM (37, 158, 159), activated Msb2p associates preferentially with GTP-Cdc42p (26), and Sho1p associates with Cdc24p, Ste20p, and Ste11p (100, 112).
It is possible that Bem4p is a GEF for Cdc42p. As is typical for Dbl-type GEFs (161) and Cdc24p (33), Bem4p associates with the inactive (GDP-bound) conformation of Cdc42p. Bem4p also shares limited homology with members of the Smg GDS-type family of GTPase regulators, which regulate Rho-type GTPases in mammals and possess GEF activity (106–109). A previous study failed to uncover GEF activity for Bem4p (40), and the homology between Bem4p and Smg GDS regulators is poorly conserved in the GEF domain. Moreover, Cdc24p is itself required to activate the filamentous-growth pathway. Thus, we favor the possibility that Bem4p regulates Cdc24p-dependent activation of Cdc42p in the filamentous-growth pathway.
The discovery of Bem4p is an important step toward understanding the long-standing mystery of how pathways that share components induce selective responses. Like many pathways, the filamentous-growth pathway is composed of proteins that comprise a broader network that regulates multiple responses (8, 18). A pathway-specific regulator of the filamentous-growth pathway was first postulated in 1994 (18) and continues to be invoked to answer questions surrounding pathway specificity in this system (8). Bem4p is the first pathway-specific regulator to be identified that functions above the MAP kinase Kss1p in the filamentous-growth pathway. Determining how Bem4p executes a pathway-specific response will be an important future challenge. Intriguingly, Smg GDS regulators bind to GTPases and to protein kinases and have been implicated as specificity factors (108, 162).
Supplementary Material
ACKNOWLEDGMENTS
Thanks go to John Pringle (Stanford University, Palo Alto CA), Peter Pryciak (UMASS Medical Center), Henrik Dohlman (University of North Carolina, Chapel Hill, NC), Jeremey Thorner (University of California, Berkeley, CA), Stan Fields and Sven Nelson (University of Washington, Seattle, WA), Hiten Madhani (University of California, San Francisco, CA), Gerald Fink (MIT, Cambridge, MA), Doug Johnson (University of Vermont, Burlington, VT), Scott Emr (Cornell University, Ithaca, NY), Wendell Lim (University of California, San Francisco, CA), Beverly Errede (University of North Carolina, Chapel Hill, NC), Rong Li (Stowers Institute, Kansas City, MO), Erfei Bi (UPENN), Hay-Oak Park (Ohio State University), Mike Yu (SUNY—Buffalo), and Charlie Boone (University of Toronto, ON, Canada) for providing strains, plasmids, reagents, and/or advice. Frank Bright and Ian Horner performed the fluorescence assays with Cdc42p. Lila Balali, Rachel Weigand, and Unnati Dev assisted with experiments.
P.J.C. is supported by the U.S. Public Health Service (GM number 098629).
This work is dedicated to Jon Widom.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00850-14.
REFERENCES
- 1.Bendezu FO, Martin SG. 2013. Cdc42 explores the cell periphery for mate selection in fission yeast. Curr Biol 23:42–47. doi: 10.1016/j.cub.2012.10.042. [DOI] [PubMed] [Google Scholar]
- 2.Das M, Drake T, Wiley DJ, Buchwald P, Vavylonis D, Verde F. 2012. Oscillatory dynamics of Cdc42 GTPase in the control of polarized growth. Science 337:239–243. doi: 10.1126/science.1218377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howell AS, Jin M, Wu CF, Zyla TR, Elston TC, Lew DJ. 2012. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell 149:322–333. doi: 10.1016/j.cell.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howell AS, Savage NS, Johnson SA, Bose I, Wagner AW, Zyla TR, Nijhout HF, Reed MC, Goryachev AB, Lew DJ. 2009. Singularity in polarization: rewiring yeast cells to make two buds. Cell 139:731–743. doi: 10.1016/j.cell.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melendez J, Grogg M, Zheng Y. 2011. Signaling role of Cdc42 in regulating mammalian physiology. J Biol Chem 286:2375–2381. doi: 10.1074/jbc.R110.200329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson KF, Erickson JW, Antonyak MA, Cerione RA. 2013. Rho GTPases and their roles in cancer metabolism. Trends Mol Med 19:74–82. doi: 10.1016/j.molmed.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz MA, Madhani HD. 2004. Principles of map kinase signaling specificity in Saccharomyces cerevisiae. Annu Rev Genet 38:725–748. doi: 10.1146/annurev.genet.39.073003.112634. [DOI] [PubMed] [Google Scholar]
- 8.Saito H. 2010. Regulation of cross-talk in yeast MAPK signaling pathways. Curr Opin Microbiol 13:677–683. doi: 10.1016/j.mib.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Bardwell L. 2006. Mechanisms of MAPK signalling specificity. Biochem Soc Trans 34:837–841. doi: 10.1042/BST0340837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park HO, Bi E. 2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev 71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams AE, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. 1990. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol 111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. 1997. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 13.Wedlich-Soldner R, Altschuler S, Wu L, Li R. 2003. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299:1231–1235. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- 14.Atkins BD, Yoshida S, Saito K, Wu CF, Lew DJ, Pellman D. 2013. Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J Cell Biol 202:231–240. doi: 10.1083/jcb.201301090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyer JM, Savage NS, Jin M, Zyla TR, Elston TC, Lew DJ. 2013. Tracking shallow chemical gradients by actin-driven wandering of the polarization site. Curr Biol 23:32–41. doi: 10.1016/j.cub.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Bi E, Novick P, Du L, Kozminski KG, Lipschutz JH, Guo W. 2001. Cdc42 interacts with the exocyst and regulates polarized secretion. J Biol Chem 276:46745–46750. doi: 10.1074/jbc.M107464200. [DOI] [PubMed] [Google Scholar]
- 17.Okada S, Leda M, Hanna J, Savage NS, Bi E, Goryachev AB. 2013. Daughter cell identity emerges from the interplay of Cdc42, septins, and exocytosis. Dev Cell 26:148–161. doi: 10.1016/j.devcel.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts RL, Fink GR. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev 8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 19.Ryan O, Shapiro RS, Kurat CF, Mayhew D, Baryshnikova A, Chin B, Lin ZY, Cox MJ, Vizeacoumar F, Cheung D, Bahr S, Tsui K, Tebbji F, Sellam A, Istel F, Schwarzmuller T, Reynolds TB, Kuchler K, Gifford DK, Whiteway M, Giaever G, Nislow C, Costanzo M, Gingras AC, Mitra RD, Andrews B, Fink GR, Cowen LE, Boone C. 2012. Global gene deletion analysis exploring yeast filamentous growth. Science 337:1353–1356. doi: 10.1126/science.1224339. [DOI] [PubMed] [Google Scholar]
- 20.Cullen PJ, Sprague GF Jr. 2012. The regulation of filamentous growth in yeast. Genetics 190:23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077–1090. doi: 10.1016/0092-8674(92)90079-R. [DOI] [PubMed] [Google Scholar]
- 22.Cullen PJ, Sprague GF Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc Natl Acad Sci U S A 97:13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall JE, Thomas DY. 1997. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J 16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosch HU, Roberts RL, Fink GR. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peter M, Neiman AM, Park HO, van Lohuizen M, Herskowitz I. 1996. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J 15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 26.Cullen PJ, Sabbagh W Jr, Graham E, Irick MM, van Olden EK, Neal C, Delrow J, Bardwell L, Sprague GF Jr. 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous-growth pathway in yeast. Genes Dev 18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Styles CA, Fink GR. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 28.Bao MZ, Schwartz MA, Cantin GT, Yates JR III, Madhani HD. 2004. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell 119:991–1000. doi: 10.1016/j.cell.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 29.Chou S, Huang L, Liu H. 2004. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell 119:981–990. doi: 10.1016/j.cell.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 30.Sloat BF, Adams A, Pringle JR. 1981. Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J Cell Biol 89:395–405. doi: 10.1083/jcb.89.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bender A, Pringle JR. 1989. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc Natl Acad Sci U S A 86:9976–9980. doi: 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bender A, Pringle JR. 1992. A Ser/Thr-rich multicopy suppressor of a cdc24 bud emergence defect. Yeast 8:315–323. doi: 10.1002/yea.320080409. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Y, Cerione R, Bender A. 1994. Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem 269:2369–2372. [PubMed] [Google Scholar]
- 34.Gulli MP, Peter M. 2001. Temporal and spatial regulation of Rho-type guanine-nucleotide exchange factors: the yeast perspective. Genes Dev 15:365–379. doi: 10.1101/gad.876901. [DOI] [PubMed] [Google Scholar]
- 35.Tatebayashi K, Tanaka K, Yang HY, Yamamoto K, Matsushita Y, Tomida T, Imai M, Saito H. 2007. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J 26:3521–3533. doi: 10.1038/sj.emboj.7601796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ. 2008. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J Cell Biol 181:1073–1081. doi: 10.1083/jcb.200704079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto K, Tatebayashi K, Tanaka K, Saito H. 2010. Dynamic control of yeast MAP kinase network by induced association and dissociation between the Ste50 scaffold and the Opy2 membrane anchor. Mol Cell 40:87–98. doi: 10.1016/j.molcel.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 38.O'Rourke SM, Herskowitz I. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev 12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mack D, Nishimura K, Dennehey BK, Arbogast T, Parkinson J, Toh-eA, Pringle JR, Bender A, Matsui Y. 1996. Identification of the bud emergence gene BEM4 and its interactions with Rho-type GTPases in Saccharomyces cerevisiae. Mol Cell Biol 16:4387–4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirano H, Tanaka K, Ozaki K, Imamura H, Kohno H, Hihara T, Kameyama T, Hotta K, Arisawa M, Watanabe T, Qadota H, Ohya Y, Takai Y. 1996. ROM7/BEM4 encodes a novel protein that interacts with the Rho1p small GTP-binding protein in Saccharomyces cerevisiae. Mol Cell Biol 16:4396–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drees BL, Sundin B, Brazeau E, Caviston JP, Chen GC, Guo W, Kozminski KG, Lau MW, Moskow JJ, Tong A, Schenkman LR, McKenzie A III, Brennwald P, Longtine M, Bi E, Chan C, Novick P, Boone C, Pringle JR, Davis TN, Fields S, Drubin DG. 2001. A protein interaction map for cell polarity development. J Cell Biol 154:549–571. doi: 10.1083/jcb.200104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 43.Rose MD, Winston F, Hieter P. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 44.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res 21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. doi:. [DOI] [PubMed] [Google Scholar]
- 46.Goldstein AL, McCusker JH. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541–1553. doi:. [DOI] [PubMed] [Google Scholar]
- 47.Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 48.Laloux I, Jacobs E, Dubois E. 1994. Involvement of SRE element of Ty1 transposon in TEC1-dependent transcriptional activation. Nucleic Acids Res 22:999–1005. doi: 10.1093/nar/22.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madhani HD, Styles CA, Fink GR. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91:673–684. doi: 10.1016/S0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 50.Roberts CJ, Nelson B, Marton MJ, Stoughton R, Meyer MR, Bennett HA, He YD, Dai H, Walker WL, Hughes TR, Tyers M, Boone C, Friend SH. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 51.Chavel CA, Dionne HM, Birkaya B, Joshi J, Cullen PJ. 2010. Multiple signals converge on a differentiation MAPK pathway. PLoS Genet 6:e1000883. doi: 10.1371/journal.pgen.1000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, Gerstein M, Dumont ME, Phizicky EM, Snyder M, Grayhack EJ. 2005. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev 19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagen DC, McCaffrey G, Sprague GF Jr. 1991. Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol Cell Biol 11:2952–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mumberg D, Muller R, Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 55.Karunanithi S, Joshi J, Chavel C, Birkaya B, Grell L, Cullen PJ. 2012. Regulation of mat responses by a differentiation MAPK pathway in Saccharomyces cerevisiae. PLoS One 7:e32294. doi: 10.1371/journal.pone.0032294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCraith S, Holtzman T, Moss B, Fields S. 2000. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc Natl Acad Sci U S A 97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.James P, Halladay J, Craig EA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caviston JP, Tcheperegine SE, Bi E. 2002. Singularity in budding: a role for the evolutionarily conserved small GTPase Cdc42p. Proc Natl Acad Sci U S A 99:12185–12190. doi: 10.1073/pnas.182370299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benard V, Bohl BP, Bokoch GM. 1999. Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J Biol Chem 274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- 60.Otsuki Y, Tanaka M, Yoshii S, Kawazoe N, Nakaya K, Sugimura H. 2001. Tumor metastasis suppressor nm23H1 regulates Rac1 GTPase by interaction with Tiam1. Proc Natl Acad Sci U S A 98:4385–4390. doi: 10.1073/pnas.071411598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horazdovsky BF, Emr SD. 1993. The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J Biol Chem 268:4953–4962. [PubMed] [Google Scholar]
- 62.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 63.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Courchesne WE, Kunisawa R, Thorner J. 1989. A putative protein kinase overcomes pheromone-induced arrest of cell cycling in S. cerevisiae. Cell 58:1107–1119. doi: 10.1016/0092-8674(89)90509-6. [DOI] [PubMed] [Google Scholar]
- 65.Ma D, Cook JG, Thorner J. 1995. Phosphorylation and localization of Kss1, a MAP kinase of the Saccharomyces cerevisiae pheromone response pathway. Mol Biol Cell 6:889–909. doi: 10.1091/mbc.6.7.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cook JG, Bardwell L, Thorner J. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 67.Lee MJ, Dohlman HG. 2008. Coactivation of G protein signaling by cell-surface receptors and an intracellular exchange factor. Curr Biol 18:211–215. doi: 10.1016/j.cub.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cullen PJ, Schultz J, Horecka J, Stevenson BJ, Jigami Y, Sprague GF Jr. 2000. Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics 155:1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cullen PJ, Sprague GF Jr. 2002. The Glc7p-interacting protein Bud14p attenuates polarized growth, pheromone response, and filamentous growth in Saccharomyces cerevisiae. Eukaryot Cell 1:884–894. doi: 10.1128/EC.1.6.884-894.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reynolds TB, Fink GR. 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- 71.Madhani HD, Galitski T, Lander ES, Fink GR. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc Natl Acad Sci U S A 96:12530–12535. doi: 10.1073/pnas.96.22.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pitoniak A, Birkaya B, Dionne HS, Vadiae N, Cullen PJ. 2009. The signaling mucins Msb2 and Hkr1 differentially regulate the filamentation MAPK pathway and contribute to a multimodal response. Mol Biol Cell 20:3101–3114. doi: 10.1091/mbc.E08-07-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chant J, Pringle JR. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J Cell Biol 129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cullen PJ, Sprague GF Jr. 2002. The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol Biol Cell 13:2990–3004. doi: 10.1091/mbc.E02-03-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, Weng S, Botstein D. 1998. SGD: Saccharomyces Genome Database. Nucleic Acids Res 26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Krieger CJ, Livstone MS, Miyasato SR, Nash RS, Oughtred R, Skrzypek MS, Weng S, Wong ED, Zhu KK, Dolinski K, Botstein D, Cherry JM. 2008. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res 36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Washburn MP, Wolters D, Yates JR III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 78.Ito T, Chiba T, Yoshida M. 2001. Exploring the protein interactome using comprehensive two-hybrid projects. Trends Biotechnol 19:S23–27. doi: 10.1016/S0167-7799(01)01790-5. [DOI] [PubMed] [Google Scholar]
- 79.Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S, Quondam M, Zucconi A, Hogue CW, Fields S, Boone C, Cesareni G. 2002. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295:321–324. doi: 10.1126/science.1064987. [DOI] [PubMed] [Google Scholar]
- 80.Gene Ontology Consortium. 2001. Creating the gene ontology resource: design and implementation. Genome Res 11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weng S, Dong Q, Balakrishnan R, Christie K, Costanzo M, Dolinski K, Dwight SS, Engel S, Fisk DG, Hong E, Issel-Tarver L, Sethuraman A, Theesfeld C, Andrada R, Binkley G, Lane C, Schroeder M, Botstein D, Cherry JM. 2003. Saccharomyces Genome Database (SGD) provides biochemical and structural information for budding yeast proteins. Nucleic Acids Res 31:216–218. doi: 10.1093/nar/gkg054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mody A, Weiner J, Ramanathan S. 2009. Modularity of MAP kinases allows deformation of their signalling pathways. Nat Cell Biol 11:484–491. doi: 10.1038/ncb1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peifer M, Berg S, Reynolds AB. 1994. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell 76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 84.Miller JP, Lo RS, Ben-Hur A, Desmarais C, Stagljar I, Noble WS, Fields S. 2005. Large-scale identification of yeast integral membrane protein interactions. Proc Natl Acad Sci U S A 102:12123–12128. doi: 10.1073/pnas.0505482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 86.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 87.Lorenz MC, Cutler NS, Heitman J. 2000. Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol Biol Cell 11:183–199. doi: 10.1091/mbc.11.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Santos B, Snyder M. 2000. Sbe2p and sbe22p, two homologous Golgi proteins involved in yeast cell wall formation. Mol Biol Cell 11:435–452. doi: 10.1091/mbc.11.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Casamayor A, Snyder M. 2003. Molecular dissection of a yeast septin: distinct domains are required for septin interaction, localization, and function. Mol Cell Biol 23:2762–2777. doi: 10.1128/MCB.23.8.2762-2777.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hall PA, Russell SEH, Pringle JR (ed). 2008. The spetins. Wiley Publishers, New York, NY. [Google Scholar]
- 91.Granek JA, Kayikci O, Magwene PM. 2011. Pleiotropic signaling pathways orchestrate yeast development. Curr Opin Microbiol 14:676–681. doi: 10.1016/j.mib.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arkowitz RA, Bassilana M. 2011. Polarized growth in fungi: symmetry breaking and hyphal formation. Semin Cell Dev Biol 22:806–815. doi: 10.1016/j.semcdb.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 93.Yang HY, Tatebayashi K, Yamamoto K, Saito H. 2009. Glycosylation defects activate filamentous growth Kss1 MAPK and inhibit osmoregulatory Hog1 MAPK. EMBO J 28:1380–1391. doi: 10.1038/emboj.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee BN, Elion EA. 1999. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc Natl Acad Sci U S A 96:12679–12684. doi: 10.1073/pnas.96.22.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Madhani HD, Fink GR. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 96.Guo B, Styles CA, Feng Q, Fink GR. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci U S A 97:12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lo WS, Dranginis AM. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell 9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lambrechts MG, Bauer FF, Marmur J, Pretorius IS. 1996. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc Natl Acad Sci U S A 93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maeda T, Takekawa M, Saito H. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 100.Tatebayashi K, Yamamoto K, Tanaka K, Tomida T, Maruoka T, Kasukawa E, Saito H. 2006. Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J 25:3033–3044. doi: 10.1038/sj.emboj.7601192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hao N, Behar M, Parnell SC, Torres MP, Borchers CH, Elston TC, Dohlman HG. 2007. A systems-biology analysis of feedback inhibition in the Sho1 osmotic-stress-response pathway. Curr Biol 17:659–667. doi: 10.1016/j.cub.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 102.Marles JA, Dahesh S, Haynes J, Andrews BJ, Davidson AR. 2004. Protein-protein interaction affinity plays a crucial role in controlling the Sho1p-mediated signal transduction pathway in yeast. Mol Cell 14:813–823. doi: 10.1016/j.molcel.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 103.Fassler JS, Gray WM, Malone CL, Tao W, Lin H, Deschenes RJ. 1997. Activated alleles of yeast SLN1 increase Mcm1-dependent reporter gene expression and diminish signaling through the Hog1 osmosensing pathway. J Biol Chem 272:13365–13371. doi: 10.1074/jbc.272.20.13365. [DOI] [PubMed] [Google Scholar]
- 104.Reiser V, Raitt DC, Saito H. 2003. Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J Cell Biol 161:1035–1040. doi: 10.1083/jcb.200301099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Singh KK. 2000. The Saccharomyces cerevisiae Sln1p-Ssk1p two-component system mediates response to oxidative stress and in an oxidant-specific fashion. Free Radic Biol Med 29:1043–1050. doi: 10.1016/S0891-5849(00)00432-9. [DOI] [PubMed] [Google Scholar]
- 106.Tew GW, Lorimer EL, Berg TJ, Zhi H, Li R, Williams CL. 2008. SmgGDS regulates cell proliferation, migration, and NF-kappaB transcriptional activity in non-small cell lung carcinoma. J Biol Chem 283:963–976. doi: 10.1074/jbc.M707526200. [DOI] [PubMed] [Google Scholar]
- 107.Zhi H, Yang XJ, Kuhnmuench J, Berg T, Thill R, Yang H, See WA, Becker CG, Williams CL, Li R. 2009. SmgGDS is up-regulated in prostate carcinoma and promotes tumour phenotypes in prostate cancer cells. J Pathol 217:389–397. doi: 10.1002/path.2456. [DOI] [PubMed] [Google Scholar]
- 108.Ferro E, Trabalzini L. 2010. RalGDS family members couple Ras to Ral signalling and that's not all. Cell Signal 22:1804–1810. doi: 10.1016/j.cellsig.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 109.Ntantie E, Gonyo P, Lorimer EL, Hauser AD, Schuld N, McAllister D, Kalyanaraman B, Dwinell MB, Auchampach JA, Williams CL. 2013. An adenosine-mediated signaling pathway suppresses prenylation of the GTPase Rap1B and promotes cell scattering. Sci Signal 6:ra39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 111.Posas F, Saito H. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 112.Zarrinpar A, Bhattacharyya RP, Nittler MP, Lim WA. 2004. Sho1 and Pbs2 act as coscaffolds linking components in the yeast high osmolarity MAP kinase pathway. Mol Cell 14:825–832. doi: 10.1016/j.molcel.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 113.Pryciak PM. 2008. Systems biology. Customized signaling circuits. Science 319:1489–1490. doi: 10.1126/science.1156414. [DOI] [PubMed] [Google Scholar]
- 114.Takahashi S, Pryciak PM. 2008. Membrane localization of scaffold proteins promotes graded signaling in the yeast MAP kinase cascade. Curr Biol 18:1184–1191. doi: 10.1016/j.cub.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Good M, Tang G, Singleton J, Remenyi A, Lim WA. 2009. The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell 136:1085–1097. doi: 10.1016/j.cell.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zalatan JG, Coyle SM, Rajan S, Sidhu SS, Lim WA. 2012. Conformational control of the Ste5 scaffold protein insulates against MAP kinase misactivation. Science 337:1218–1222. doi: 10.1126/science.1220683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Choi KY, Satterberg B, Lyons DM, Elion EA. 1994. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell 78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 118.Printen JA, Sprague GF Jr. 1994. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 138:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Marcus S, Polverino A, Barr M, Wigler M. 1994. Complexes between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc Natl Acad Sci U S A 91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cote P, Sulea T, Dignard D, Wu C, Whiteway M. 2011. Evolutionary reshaping of fungal mating pathway scaffold proteins mBio 2:e00230–10. doi: 10.1128/mBio.00230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stevenson BJ, Rhodes N, Errede B, Sprague GF Jr. 1992. Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev 6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 122.Bi E, Park HO. 2012. Cell polarization and cytokinesis in budding yeast. Genetics 191:347–387. doi: 10.1534/genetics.111.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Richman TJ, Toenjes KA, Morales SE, Cole KC, Wasserman BT, Taylor CM, Koster JA, Whelihan MF, Johnson DI. 2004. Analysis of cell-cycle specific localization of the Rdi1p RhoGDI and the structural determinants required for Cdc42p membrane localization and clustering at sites of polarized growth. Curr Genet 45:339–349. doi: 10.1007/s00294-004-0505-9. [DOI] [PubMed] [Google Scholar]
- 124.Das A, Slaughter BD, Unruh JR, Bradford WD, Alexander R, Rubinstein B, Li R. 2012. Flippase-mediated phospholipid asymmetry promotes fast Cdc42 recycling in dynamic maintenance of cell polarity. Nat Cell Biol 14:304–310. doi: 10.1038/ncb2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Smith SE, Rubinstein B, Mendes Pinto I, Slaughter BD, Unruh JR, Li R. 2013. Independence of symmetry breaking on Bem1-mediated autocatalytic activation of Cdc42. J Cell Biol 202:1091–1106. doi: 10.1083/jcb.201304180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hamel B, Monaghan-Benson E, Rojas RJ, Temple BR, Marston DJ, Burridge K, Sondek J. 2011. SmgGDS is a guanine nucleotide exchange factor that specifically activates RhoA and RhoC. J Biol Chem 286:12141–12148. doi: 10.1074/jbc.M110.191122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ohya Y, Qadota H, Anraku Y, Pringle JR, Botstein D. 1993. Suppression of yeast geranylgeranyl transferase I defect by alternative prenylation of two target GTPases, Rho1p and Cdc42p. Mol Biol Cell 4:1017–1025. doi: 10.1091/mbc.4.10.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Richman TJ, Sawyer MM, Johnson DI. 2002. Saccharomyces cerevisiae Cdc42p localizes to cellular membranes and clusters at sites of polarized growth. Eukaryot Cell 1:458–468. doi: 10.1128/EC.1.3.458-468.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Butty AC, Pryciak PM, Huang LS, Herskowitz I, Peter M. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282:1511–1516. doi: 10.1126/science.282.5393.1511. [DOI] [PubMed] [Google Scholar]
- 130.Nern A, Arkowitz RA. 2000. Nucleocytoplasmic shuttling of the Cdc42p exchange factor Cdc24p. J Cell Biol 148:1115–1122. doi: 10.1083/jcb.148.6.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shimada Y, Gulli MP, Peter M. 2000. Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nat Cell Biol 2:117–124. doi: 10.1038/35000073. [DOI] [PubMed] [Google Scholar]
- 132.Toenjes KA, Sawyer MM, Johnson DI. 1999. The guanine-nucleotide-exchange factor Cdc24p is targeted to the nucleus and polarized growth sites. Curr Biol 9:1183–1186. doi: 10.1016/S0960-9822(00)80022-6. [DOI] [PubMed] [Google Scholar]
- 133.Butty AC, Perrinjaquet N, Petit A, Jaquenoud M, Segall JE, Hofmann K, Zwahlen C, Peter M. 2002. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. EMBO J 21:1565–1576. doi: 10.1093/emboj/21.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Park HO, Bi E, Pringle JR, Herskowitz I. 1997. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc Natl Acad Sci U S A 94:4463–4468. doi: 10.1073/pnas.94.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Park HO, Kang PJ, Rachfal AW. 2002. Localization of the Rsr1/Bud1 GTPase involved in selection of a proper growth site in yeast. J Biol Chem 277:26721–26724. doi: 10.1074/jbc.C200245200. [DOI] [PubMed] [Google Scholar]
- 136.Shimada Y, Wiget P, Gulli MP, Bi E, Peter M. 2004. The nucleotide exchange factor Cdc24p may be regulated by auto-inhibition. EMBO J 23:1051–1062. doi: 10.1038/sj.emboj.7600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Leeuw T, Fourest-Lieuvin A, Wu C, Chenevert J, Clark K, Whiteway M, Thomas DY, Leberer E. 1995. Pheromone response in yeast: association of Bem1p with proteins of the MAP kinase cascade and actin. Science 270:1210–1213. doi: 10.1126/science.270.5239.1210. [DOI] [PubMed] [Google Scholar]
- 138.Simon MN, De Virgilio C, Souza B, Pringle JR, Abo A, Reed SI. 1995. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature 376:702–705. doi: 10.1038/376702a0. [DOI] [PubMed] [Google Scholar]
- 139.Wiget P, Shimada Y, Butty AC, Bi E, Peter M. 2004. Site-specific regulation of the GEF Cdc24p by the scaffold protein Far1p during yeast mating. EMBO J 23:1063–1074. doi: 10.1038/sj.emboj.7600123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. 1994. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J Cell Biol 127:1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ogura K, Tandai T, Yoshinaga S, Kobashigawa Y, Kumeta H, Ito T, Sumimoto H, Inagaki F. 2009. NMR structure of the heterodimer of Bem1 and Cdc24 PB1 domains from Saccharomyces cerevisiae. J Biochem 146:317–325. doi: 10.1093/jb/mvp075. [DOI] [PubMed] [Google Scholar]
- 142.Ito T, Matsui Y, Ago T, Ota K, Sumimoto H. 2001. Novel modular domain PB1 recognizes PC motif to mediate functional protein-protein interactions. EMBO J 20:3938–3946. doi: 10.1093/emboj/20.15.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zheng Y. 2001. Dbl family guanine nucleotide exchange factors. Trends Biochem Sci 26:724–732. doi: 10.1016/S0968-0004(01)01973-9. [DOI] [PubMed] [Google Scholar]
- 144.Sato TK, Overduin M, Emr SD. 2001. Location, location, location: membrane targeting directed by PX domains. Science 294:1881–1885. doi: 10.1126/science.1065763. [DOI] [PubMed] [Google Scholar]
- 145.Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. 2004. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell 13:677–688. doi: 10.1016/S1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 146.Pryciak PM, Huntress FA. 1998. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev 12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM. 2007. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell 128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gulli MP, Jaquenoud M, Shimada Y, Niederhauser G, Wiget P, Peter M. 2000. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol Cell 6:1155–1167. doi: 10.1016/S1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- 149.Drogen F, O'Rourke SM, Stucke VM, Jaquenoud M, Neiman AM, Peter M. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr Biol 10:630–639. doi: 10.1016/S0960-9822(00)00511-X. [DOI] [PubMed] [Google Scholar]
- 150.Rad MR, Xu G, Hollenberg CP. 1992. STE50, a novel gene required for activation of conjugation at an early step in mating in Saccharomyces cerevisiae. Mol Gen Genet 236:145–154. [DOI] [PubMed] [Google Scholar]
- 151.Xu G, Jansen G, Thomas DY, Hollenberg CP, Ramezani Rad M. 1996. Ste50p sustains mating pheromone-induced signal transduction in the yeast Saccharomyces cerevisiae. Mol Microbiol 20:773–783. doi: 10.1111/j.1365-2958.1996.tb02516.x. [DOI] [PubMed] [Google Scholar]
- 152.Posas F, Witten EA, Saito H. 1998. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol Cell Biol 18:5788–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ramezani Rad M, Jansen G, Buhring F, Hollenberg CP. 1998. Ste50p is involved in regulating filamentous growth in the yeast Saccharomyces cerevisiae and associates with Ste11p. Mol Gen Genet 259:29–38. doi: 10.1007/s004380050785. [DOI] [PubMed] [Google Scholar]
- 154.Jansen G, Buhring F, Hollenberg CP, Ramezani Rad M. 2001. Mutations in the SAM domain of STE50 differentially influence the MAPK-mediated pathways for mating, filamentous growth and osmotolerance in Saccharomyces cerevisiae. Mol Genet Genomics 265:102–117. doi: 10.1007/s004380000394. [DOI] [PubMed] [Google Scholar]
- 155.Ramezani-Rad M. 2003. The role of adaptor protein Ste50-dependent regulation of the MAPKKK Ste11 in multiple signalling pathways of yeast. Curr Genet 43:161–170. doi: 10.1007/s00294-003-0383-6. [DOI] [PubMed] [Google Scholar]
- 156.Truckses DM, Bloomekatz JE, Thorner J. 2006. The RA domain of Ste50 adaptor protein is required for delivery of Ste11 to the plasma membrane in the filamentous growth signaling pathway of the yeast Saccharomyces cerevisiae. Mol Cell Biol 26:912–928. doi: 10.1128/MCB.26.3.912-928.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hao N, Zeng Y, Elston TC, Dohlman HG. 2008. Control of MAPK specificity by feedback phosphorylation of shared adaptor protein Ste50. J Biol Chem 283:33798–33802. doi: 10.1074/jbc.C800179200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ekiel I, Sulea T, Jansen G, Kowalik M, Minailiuc O, Cheng J, Harcus D, Cygler M, Whiteway M, Wu C. 2009. Binding the atypical RA domain of Ste50p to the unfolded Opy2p cytoplasmic tail is essential for the high-osmolarity glycerol pathway. Mol Biol Cell 20:5117–5126. doi: 10.1091/mbc.E09-07-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wu C, Jansen G, Zhang J, Thomas DY, Whiteway M. 2006. Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev 20:734–746. doi: 10.1101/gad.1375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rossman KL, Worthylake DK, Snyder JT, Siderovski DP, Campbell SL, Sondek J. 2002. A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange. EMBO J 21:1315–1326. doi: 10.1093/emboj/21.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hart MJ, Eva A, Zangrilli D, Aaronson SA, Evans T, Cerione RA, Zheng Y. 1994. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J Biol Chem 269:62–65. [PubMed] [Google Scholar]
- 162.Hao Y, Wong R, Feig LA. 2008. RalGDS couples growth factor signaling to Akt activation. Mol Cell Biol 28:2851–2859. doi: 10.1128/MCB.01917-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Karunanithi S, Cullen PJ. 2012. The filamentous growth MAPK pathway responds to glucose starvation through the Mig1/2 transcriptional repressors in Saccharomyces cerevisiae. Genetics 192:869–887. doi: 10.1534/genetics.112.142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.