Abstract
The strength of conclusions drawn from RNAi-based studies is heavily influenced by the quality of tools used to elicit knockdown. Prior studies have developed algorithms to design siRNAs. However, to date, no established method has emerged to identify effective shRNAs, which have lower intracellular abundance than transfected siRNAs and undergo additional processing steps. We recently developed a multiplexed assay for identifying potent shRNAs and have used this method to generate ~250,000 shRNA efficacy data-points. Using these data, we developed shERWOOD, an algorithm capable of predicting, for any shRNA, the likelihood that it will elicit potent target knockdown. Combined with additional shRNA design strategies, shERWOOD allows the ab initio identification of potent shRNAs that target, specifically, the majority of each gene’s multiple transcripts. We have validated the performance of our shRNA designs using several orthogonal strategies and have constructed genome-wide collections of shRNAs for humans and mice based upon our approach.
Introduction
The discovery of RNAi promised a new era in which the power of genetics could be applied to model organisms for which large-scale studies of gene function were previously inconvenient or impossible (Berns et al., 2004; Brummelkamp et al., 2002; Chuang and Meyerowitz, 2000; Fire et al., 1998; Gupta et al., 2004; Hannon, 2002; Kamath et al., 2003; Kambris et al., 2006; Paddison et al., 2004; Sanchez Alvarado and Newmark, 1999; Svoboda et al., 2000; Timmons and Fire, 1998; Tuschl et al., 1999; Zender et al., 2008). Yet, it quickly became clear that implementing RNAi, especially on a genome-wide scale, could be challenging. This was particularly true for applications in mammalian cells wherein discrete sequences, in the form of siRNAs or shRNAs, were used as silencing triggers (Brummelkamp et al., 2002; Elbashir et al., 2001; Paddison et al., 2002). The overall degree of knockdown achieved was found to vary tremendously, depending upon the precise sequence of the small RNA that is loaded into the RNAi effector complex (RISC) (Chiu and Rana, 2002; Khvorova et al., 2003; Schwarz et al., 2003). Yet, the nature of sequence and structural motifs that favor RISC loading and high turnover target cleavage has yet to be fully revealed (Ameres and Zamore, 2013).
Early studies aimed at optimizing RNAi in mammals used endogenous microRNAs as a guide to the design of effective artificial RNAi triggers (Khvorova et al., 2003; Reynolds et al., 2004; Schwarz et al., 2003; Ui-Tei et al., 2004; Zeng and Cullen, 2003). Canonical microRNAs are processed by a two-step, nucleolytic mechanism (Seitz and Zamore, 2006). The initial cleavage of the primary miRNA transcript in the nucleus by the Microprocessor yields a short, often imperfect, hairpin loop, the pre-miRNA (Denli et al., 2004; Lee et al., 2003). This is exported to the cytoplasm where a second cleavage by Dicer and its associated cofactors yields a short duplex of ~19-20 nucleotides with 2 nucleotide 3′ overhangs (Bernstein et al., 2001; Grishok et al., 2001; Hutvagner et al., 2001; Ketting et al., 2001; Lund et al., 2004; Yi et al., 2003). This duplex serves as a substrate for preferential loading of one strand into Argonaute proteins in the context of RISC (Hammond et al., 2001; Hutvagner and Zamore, 2002; Khvorova et al., 2003; Martinez et al., 2002; Schwarz et al., 2003).
An examination of the sequences of endogenous miRNAs indicated that thermodynamic asymmetry between the two ends of the short duplex was a strong predictor of which strand would be accepted by Argonaute as the “guide” (Khvorova et al., 2003; Schwarz et al., 2003). Applying this insight to artificial triggers, initially in the form of siRNAs, validated the generality of this observation, and thermodynamic asymmetry became a key guiding principle of both siRNA and shRNA design (Reynolds et al., 2004; Silva et al., 2005). Subsequent studies of the structure of the Ago-small RNA complex have also indicated a sequence preference for a 5′ terminal U that fits into a binding pocket in the mid domain of the Argonaute protein (Seitz et al., 2008; Wang et al., 2008).
In many ways, siRNAs gain entry into RISC in mammals by simulating the end product of the two-step miRNA processing pathway. shRNAs, which mimic either the primary miRNA or pre-miRNA must be nucleolytically processed prior to RISC loading (Brummelkamp et al., 2002; Cullen, 2006; Paddison et al., 2002). Therefore, shRNAs are likely subject to additional constraints that lead to efficient recognition by Drosha and Dicer. We do not yet understand the selection rules for effective flux through the miRNA biogenesis pathway and therefore cannot predict ab initio what transcripts will produce small RNAs. However, studies of Drosha, in particular, have implicated patterns of conservation and base pairing in the basal stem, those regions adjacent to the Drosha cleavage site, as determinants of efficient pri-miRNA cleavage (Auyeung et al., 2013; Chen et al., 2004; Han et al., 2006; Seitz and Zamore, 2006). Elements within the hairpin loop have also been shown to have an impact both on Drosha efficiently and its site preference (Han et al., 2006; Zhang and Zeng, 2010).
Several attempts have been made to extract predictive rules for the design of effective small RNAs from endpoint silencing data. The first serious attempt applied Artificial Neural Networks (ANNs) to a set of ~2,000 paired data points associating the sequence of siRNA guides with a corresponding knockdown measurement (established using fluorescent reporters) (Huesken et al., 2005). Experience in the field supported the effectiveness of BIOPREDSi; however, access to the algorithm eventually became impossible. The same dataset was subsequently used to produce a second algorithm, DSIR, which included additional input variables (the frequency of each nucleotide, each 2mer and each 3mer within the guide) (Vert et al., 2006). To accommodate this large number of parameters, linear modeling was performed using Lasso Regression (a form of linear regression that iteratively decreases the use of non-predictive variables in the linear model) (Tibshirani, 1995).
siRNA design algorithms could be applied for the design of shRNAs, and these did inform the design of genome-wide shRNA collections (Berns et al., 2004; Paddison et al., 2004). However, the prognostic power of siRNA design algorithms is compromised for shRNA design. shRNAs, expressed from RNA polII or polIII promoters, reach lower intracellular concentrations than do transfected, synthetic siRNAs (Berns et al., 2004; Paddison et al., 2004). Moreover, shRNAs have additional constraints for effective processing. Therefore, it was imperative that shRNA-specific algorithms be developed.
The generation of accurate siRNA design algorithms was only made possible with the creation of large training datasets. Thus far, a corresponding shRNA dataset has been lacking. Recently, we developed a “sensor” method that allows for the parallel assessment of shRNA potencies on a massive scale (Fellmann et al., 2011). Using the sensor approach, we interrogated ~250,000 shRNAs for their effectiveness in the reporter setting. We have used this dataset to train a machine-learning algorithm for potent shRNA prediction. We have tested this algorithm, which we term, shERWOOD, both at the level of individual shRNAs and at the level of optimized shRNA mini-libraries. We have demonstrated that by applying computational shRNA selection in combination with novel target selection heuristics and with an optimized microRNA scaffold, we are able to create highly potent shRNAs. We have built upon this result to design and construct next-generation shRNA libraries targeting the constitutive exomes of mice and humans. Predictions for other organisms and custom shRNA designs are also made available via a web-based version of shERWOOD.
Results
Neighboring Positions of the Target Sequence are Predictive of ShRNA Strength
As a prelude to creating an shRNA design algorithm, we first developed a large-scale “sensor” dataset in which shRNA potency was measured and associated with sequence information. To perform the assay, we synthesized 12 sets of ~25K constructs that include a doxycycline inducible shRNA and a GFP-tagged shRNA target sequence located downstream of a constitutive promoter (Fellmann et al., 2011). Libraries were packaged and infected (at single copy) into a reporter cell line. In the absence of doxycycline, GFP was detectable in each cell. However, in the presence of doxycycline the shRNAs became expressed and the resultant GFP signal was reduced in a manner proportional to shRNA potency. Using Florescence Activated Cell Sorting (FACS), cells with low GFP levels, in the presence of drug, were gathered and analyzed via NGS to determine which shRNAs became enriched (i.e. which shRNAs have high potency). Operating iterative cycles of this assay has been shown to identify extremely potent constructs (Fellmann et al., 2011).
We next wished to extract what sequence characteristics were most predictive of shRNA efficacy. This subset of characteristics could then be employed as inputs during machine learning. We first developed a method to consolidate the different sensor data points into a single value for each shRNA (Supplementary Materials). These accurately capture the enrichment pattern of individual iterations of the sensor in one single value, thus allowing downstream machine learning to proceed more easily (Figure 1A). Analysis of the coefficients used to consolidate the sensor data shows that information from the final sensor iteration contributes the most to the final potency value, however information from the second iteration is also included, (Figure S1A).
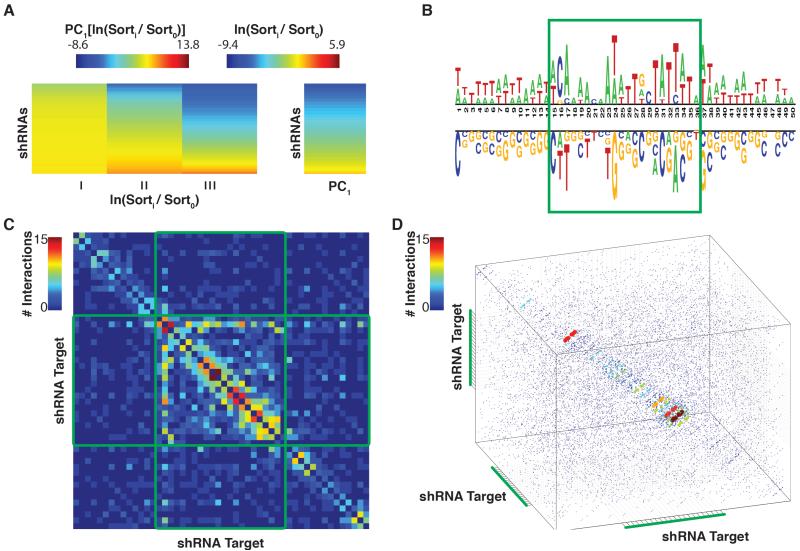
Figure 1. Identification of Sequence Characteristics Predictive of shRNA Efficacy.
A) shRNA score determination via sensor NGS data. On the left is a heatmap representation of normalized shRNA read counts for each on-dox sensor sort. The right panel represents shRNA potencies, calculated by extracting the first principal component of the left panel matrix. B) A nucleotide logo representing enriched (top) and depleted (bottom) nucleotides (p-value < 0.05) in potent shRNAs. C) A heatmap demonstrating the predictive capacity (with respect to shRNA potency) of each pair of positions within the target region. Heatmap cells are colored to represent the number of nucleotide combinations that were significantly predictive (p-value <0.05), at each position-pair. D) The predictive capacity of each triplet of positions within the target region. Data-point colors and sizes represent the number of nucleotide triplets that were significantly predictive (p-value <0.05) at each position-triplet.
To distinguish discretely between strong and weak shRNAs, we applied an Empirical-Bayes Moderated T-Test to the shRNA potency measurements extracted from two biological replicates (Smyth, 2004). Strong and weak shRNAs were those that were enriched or depleted, respectively with an FDR < 0.05.
To test individual nucleotide positions for their predictive capacity, we compared, at each position in the target sequence, each nucleotide’s enrichment and or depletion levels in the potent as compared to the weak shRNAs (Figure 1B and S1B, binomial-test, FDR < 0.05) (Vacic et al., 2006). In general, low GC content is predictive of high efficacy, with the exception of the third nucleotide inside the guide target, which shows a strong selection for cytosine. Also of note is a lack of enrichment for thymidine at the 22nd position of the guide target (corresponding to the first position of the guide). This arose because our input datasets were derived from shRNAs pre-selected by DSIR.
We next tested whether any pairs of positions had predictive capacity for shRNA strength, beyond what was expected based on their individual predictive power. To calculate a measurement for each position pair, we applied linear regression to identify synergistic predictive capacity (p-value < 0.05; see Supplemental Methods). Following this, each position-pair was assigned a value equal to the sum of nucleotide combinations that were predictive of shRNA potency when assessed at the two positions (Figure 1C and S1C). For a given position within the target, the most predictive partner is the neighboring nucleotide. An exception to this trend is observed in the positions corresponding to the shRNA guide seed, where predictive position-pairs are also observed in nucleotides separated by up to four bases.
Finally, we wished to determine if triplets of positions showed a similar trend to that observed in the pair-wise analysis. For this, we performed a modified version of the linear regression tests described above, where triplets instead of pairs of nucleotides were assessed for synergistic predictive capacity. As with the pairwise analysis, neighboring triplets of positions within the target show strong predictive power as compared to triplets of non-neighboring positions (Figure 1D). Further, the distance between predictive triplets is also extended slightly in the guide seed region of the shRNA.
A Sensor-Based Computational Algorithm to Predict shRNA Efficacy
Since sequence-based characteristics correlated with shRNA efficiency, we sought to apply machine learning to the sensor-derived efficacy measurements. The goal was to develop a computational algorithm that would predict, for any target sequence, the potency of a corresponding shRNA. We reasoned that the best machine-learning tool to apply to this task was Random Forest Regression Analysis. The reasons for this decision were two-fold. First, there is no decrease in the accuracy of Random Forests when the number of input variables is large. Second, the architecture of the algorithm takes into account increases in accuracy that can be achieved by analyzing combinations of input variables.
Our training dataset was of two distinct types. One comprised an unbiased set of shRNAs that tiled every nucleotide of 9 genes (Fellmann et al., 2011). A second comprised a larger set of shRNAs pre-selected by the DSIR algorithm (described above). We therefore chose to separate data corresponding to each input class and to train separate forests. We also chose to separate data based upon the 5′ nucleotide of the guide. This was done for two reasons. First, previous studies, supported by structural insights, had suggested that the 5′ nucleotide of the guide was a prominent determinant of small RNA potency (Fellmann et al., 2011; Frank et al., 2010; Khvorova et al., 2003; Reynolds et al., 2004). Therefore training forests individually for shRNAs initiating with each base focused the prediction process on additional determinants. Moreover, the DSIR-based predictions were already heavily biased toward U and A at the 5′ position. In fact, the bias was so strong that we did not have sufficient data to train 5′C and 5′G forests for these datasets. This meant that, in the first pass, we trained six independent modules.
In each module, input data were composed of individual base information as well as all neighboring pairs of bases throughout the guide sequence. In addition, the set of triplet-position/nucleotide-combinations found to be predictive, as assessed by linear regression, were also included (Figure 1D). After training each of the modules, we sought to determine which input variables were relied most heavily upon. For each module, each variable was permuted across observations and the resultant reduction in predictive capacity recorded at each regression tree. The resultant changes were then averaged across trees and that mean normalized by their standard deviation. The triplet variables were heavily relied upon (Figure S2A). Particularly the triplet corresponding to shRNA guide positions 2 through 4.
To consolidate these modules, a second-tier random forest was trained using the first tier outputs, the corresponding shRNA-guide base information, and a set of thermodynamic properties extracted from each shRNA (e.g. enthalpy, entropy). We name the compiled algorithm, shERWOOD.
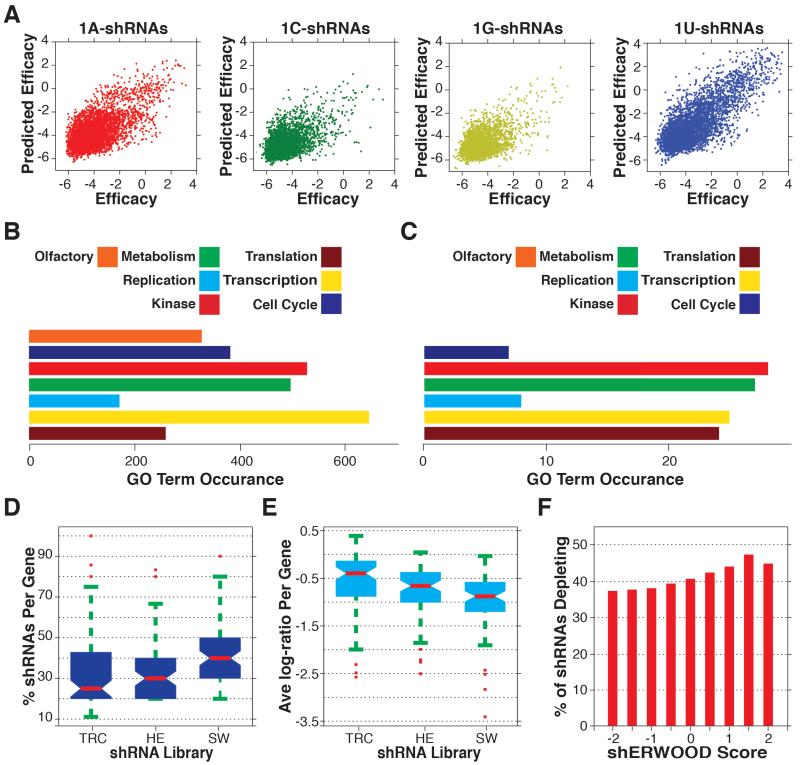
To test the prognostic power of shERWOOD, we took advantage of the unbiased nature of the tiled shRNA sensor data. For each of the 9 genes represented, we independently trained a shERWOOD algorithm without the data corresponding to that gene. We could then test shERWOOD performance against experimental data in a manner that was not skewed by the use of that data for training. We saw an overall Pearson correlation of 0.72 between experimentally derived potency measurements and computational predictions (Figure 2A). For comparison, DSIR achieves a correlation of 0.4 and a prior shRNA prediction algorithm trained on a subset of the sensor data used in this study achieves 0.56 (Matveeva et al., 2012; Vert et al., 2006). This indicates that shERWOOD achieves a roughly 180% increase in performance over currently existing siRNA prediction algorithms and a 126% increase in efficacy over existing shRNA specific prediction algorithms.
Figure 2. Construction and Validation of an shRNA-specific Predictive Algorithm.
A) Consolidated cross validation of predictions vs. sensor-scores for all shRNAs in the Fellmann et al. dataset (shRNAs are separated by the guide 5′ nucleotide). B) GO-term instances associated with the targeted gene set selected for shRNA validation screens. C) GO-term instances associated with genes for which at least two hairpins significantly depleted in each of the TRC, Hannon-Elledge (HE) and shERWOOD (SW) validation screens D) The percentage of shRNAs targeting consensus essential genes that depleted in each of the TRC, HE and shERWOOD shRNA screens. E) Average log-fold change for shRNAs targeting consensus essential genes (per gene) for each of the TRC, EH and shERWOOD validation screens. F) The percentage of shRNAs corresponding to consensus essential genes that, for any given shERWOOD score, depleted in the shERWOOD validation screen.
We have supplemented shERWOOD with additional heuristics to maximize the probability of successfully reducing protein levels in most cell and tissue types. The complex nature of alternative splicing patterns provided a strong motivation for directing shRNAs against constitutive exons. We therefore developed a strategy that iteratively searches for regions within a gene that are shared by at least 80% of transcripts (Supplemental Methods). This algorithm also tests whether high potency shRNAs have the potential to co-suppress paralogous genes. Considered together, these strategies have the potential to maximize the probability of biologically meaningful results from studies using shRNAs.
Benchmarking shERWOOD
To assess the performance of the shERWOOD algorithm, we felt that it was necessary to test a large number of shRNAs for their biological effects, as one can find anecdotal evidence for excellent performance for nearly any algorithm or strategy. We therefore chose ~2,200 genes based upon their enrichment in gene ontology (GO) categories likely to impact the growth and survival of cells in culture (Figure 2B). As controls, particularly for the likelihood of off-target effects, we included 400 olfactory receptor genes. Olfactory receptors are expressed only in olfactory neurons, and even then, they display allelic choice so that only one paralog is expressed per cell. Thus, shRNAs targeting olfactory receptors are highly unlikely to have relevant, on-target biological effects in any cell line screened in vitro. To benchmark the performance of shERWOOD, we compared a focused, mini-library predicted with this algorithm to two widely used genome-wide collections, namely the TRC collection distributed by Sigma-Genosys and the so-called Hannon-Elledge V3 library distributed presently by GE Dharmacon (Chang et al., unpublished). To produce the shERWOOD-based library and a deeper simulation of the V3 library, we used either shERWOOD or DSIR to predict their top 10 scoring shRNAs for our test genes. The sequences of TRC shRNAs are listed on a public web portal and we selected all listed shRNAs for each gene. In the case of TRC shRNAs, it was necessary to adapt them to a 22bp stem for placement into the miR-30 context.
For each test library, we synthesized 27,000 oligonucleotides in solid phase on microarrays (Cleary et al., 2004). These were cleaved, amplified, and cloned directly into a miR-30 scaffold within an MSCV-based retroviral vector without sequence validation. In this arrangement, the primary shRNA was transcribed from the LTR promoter while GFP and Neomycin resistance were separately expressed as a bicistronic transcription unit from the Phosphoglycerate Kinase promoter (PGK; Figure S2D). Pilot sequencing showed that each library was of similar quality and representation.
Each library was infected separately into the pancreatic ductal adenocarcinoma cell line, A385. Two days after infection, cells were collected for a reference time-point, and after ~12 doublings cells were again harvested for a final time-point (Supplemental Methods). shRNA representation was determined following amplification of hairpin inserts from genomic DNA (Sims et al., 2011), and after processing, shRNA read counts were compared between the initial and final time-points (Supplemental Methods, Figure S2E,F and G).
To enable direct comparisons between libraries, we censored the shERWOOD and DSIR-based libraries on a per gene basis to contain the same number of hairpins as were available in the TRC library, keeping those with the best algorithmic scores. We then selected the consensus set of “essential” genes, accepting only those where at least two hairpins in each library passed the statistical threshold (FDR<0.1). As expected, the resulting set of genes that were important for the growth and survival of A385 were depleted of olfactory receptor shRNAs (Figure 2C). In contrast, the set of consensus essential genes was enriched for GO terms associated with translation.
To benchmark shRNA selection strategies against each other, we determined the percentage of shRNAs in each mini-library that scored for each consensus essential gene. For the TRC library, 24% of shRNAs achieved significant depletion, whereas 31% of DSIR-predicted sequences and 40% of shERWOOD-based hairpins scored (Figure 2D). We also considered performance from the perspective of median logfold depletion. For the TRC collection the average log-fold change was −0.4; for DSIR this rose to −0.62, and it increased further to −0.78 for shERWOOD shRNAs (Figure 2E). We note that this type of analysis favors slightly the library with the weakest overall shRNAs, since it will be this collection that sets entry criteria for the consensus essential gene set.
To assess whether shERWOOD scores were a proxy for shRNA potency, we examined the relationship between shERWOOD score and the probability of being significantly depleted for each consensus essential gene. For this, we analyzed all 10 shERWOOD predictions using a sliding scale of shERWOOD score cut-offs (Figure 2F). As an example, considering shRNAs with a score greater than 0.5, the likelihood that an shRNA will be depleted if it targets one of our consensus essential genes is 42%. Again, this underestimates the information content of shERWOOD scores since in the cumulative plot shown, the minimum number of scoring hairpins for a given gene irrespective of scores is 2 (i.e., 20%).
Structure-guided insights expand the shRNA prediction space
Regardless of the accuracy of predictive models, we sometimes found it difficult to identify potent shRNAs due to search space restrictions imposed by sequence constraints (e.g. GC content), gene length, or the complexity of alternative splicing patterns. We therefore sought ways to expand the sequence space to which we could apply the shERWOOD approach. Analysis of miRNA seed sequences as well as other data have suggested that the first base of the small RNA guide does not pair with its target (Lai, 2002; Lewis et al., 2005; Yuan et al., 2006). Structural studies have supported this hypothesis by showing that the first base of the guide is tightly bound within a pocket in the mid domain of Ago proteins (Figure S3A) (Elkayam et al., 2012; Frank et al., 2010; Nakanishi et al., 2012; Wang et al., 2008). Since the first base of the guide is a strong contributor to shRNA efficacy, we reasoned that we could expand the range of possible effective shRNAs by simply changing the first base of all potential guides to a U, promoting their binding to RISC and theoretically not altering target site choice. We will henceforth refer to this as the 1U-strategy. A simulated construction of a human genome-wide shRNA library demonstrates that, when this strategy is implemented, predicted shRNA-potencies increase dramatically, particularly for short GC rich genes (Figure S3B).
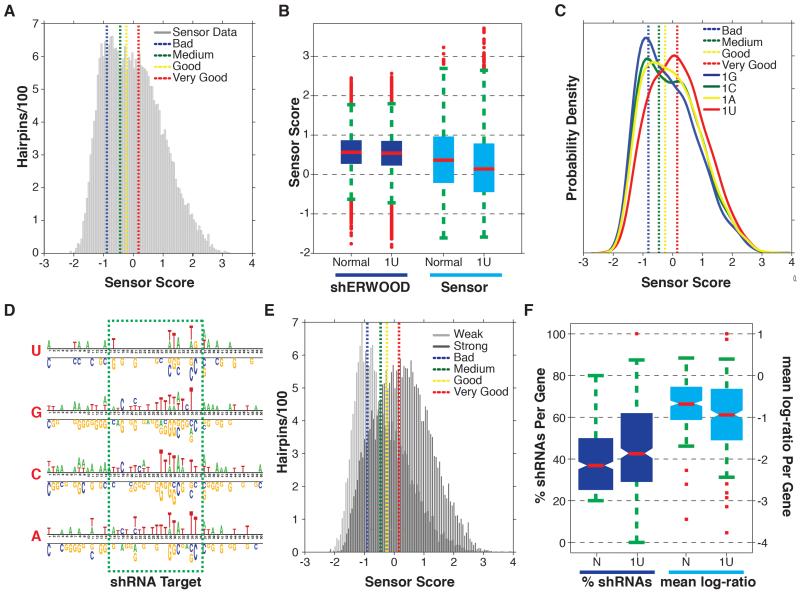
To test the 1U-strategy in a high-throughput manner, we constructed a sensor library where the top 15 shRNAs targeting a set of ~2000 “druggable” genes were predicted using the 1U-strategy. The constructs were designed such that the shRNAs contained the 1U-conversion and the target sites contained the endogenous base. shRNA potencies were extracted as described for Figure 1 (Figure 3A). The distribution indicates that ~50% of the shRNAs were strong or very strong (knockdown efficiency >75%) based on the scores of control shRNAs that were assayed in parallel. When shRNAs were separated into native and artificial 1U sets and the score distributions were plotted, we were surprised to see a significant reduction in the efficacy of the non-native-1U shRNAs (Figure 3B, Wilcoxon ranksum, p-value < 0.01). This was strongly suggestive that RISC interacts not only with the 1U of the guide but also with the first base of the target site.
Figure 3. Structure-guided Maximization of shRNA-Prediction Space.
A) Histogram of sensor scores for the top fifteen shRNAs, as identified by the shERWOOD-1U strategy, targeting ~2000 “druggable” genes. Overlaid are the mean sensor scores for control shRNAs representing poor, medium, potent and very potent shRNAs (with mean knockdown efficiencies of 25%, 50%, 75% and >90%, respectively). B) The distribution of shERWOOD-1U prediction scores for shRNAs where endogenous 1U-shRNAs are separated from endogenous non-1U-shRNAs. Sensor scores for endogenous 1U- and non-1U-shRNAS are displayed on the left. C) Distribution of sensor scores for shERWOOD-1U-selected shRNAs, separated by endogenous guide 5′ nucleotides. D) A nucleotide logo representing enriched (top) and depleted (bottom) nucleotides (p-value < 0.05) in potent shERWOOD-1U-selected shRNAs (separated by endogenous guide 5′ nucleotides). E) The distribution of sensor scores for shRNAs classified as weak and potent by a random forest classifier trained on the shERWOO-1U sensor data. F) The distributions of the percentage of shERWOOD- and shERWOOD-1U-selected shRNAs targeting consensus essential genes that depleted in validation screens (left). In addition normalized log-fold changes of shRNAs, identified under each selection scheme, are displayed (right).
We therefore stratified 1U shRNAs into four sets based on their endogenous 5′ nucleotide (Figure 3C). This analysis indicated that only a subset of shRNAs perform well when a 1U-switch is made (based on the bi-modal distributions for endogenous 1A, 1C and 1G shRNAs), but the subset that do perform well are predicted to be quite efficacious by the sensor assay. This bimodal distribution is not observed for shERWOOD-selected endogenous 1U shRNAs and we see that the majority of this shRNA class are efficient.
Given these results, we sought to determine whether we could predict those sequences for which a 1U conversion would result in a highly effective shRNA. We fit a Gaussian-mixture model to the sensor scores (Figure S3C) and applied this model to assign shRNAs into one of the two resultant populations (Figure S3D). Following clustering, we applied a binomial test separately for shRNAs where the endogenous base was 1A, 1C, 1G and 1U to determine if any nucleotides were enriched/depleted in the strong shRNAs with respect to weak shRNAs. All sets show a strong enrichment for U in the target region corresponding to the shRNA guide positions 3, 7 and 8 (Figure 3D). There is also a strong selection for Cs in the target region corresponding position 19 of the endogenous 1A, 1C and 1G shRNA guides.
These results prompted us to develop a computational algorithm that could both select the strongest endogenous 1U shRNAs and identify which endogenous 1C, 1G and 1A shRNAs were likely to yield potent 1U-converted molecules. Data points for which the mixed-Gaussian clustering resulting in less than a 70% confidence group assignment were censored (Figure S3E). We trained a random forest using the 22 nucleotides of the endogenous base as well as all neighboring pairs of nucleotides as input and the corresponding 1U-conversion sensor scores as output. The algorithm was able to achieve 80% specificity while maintaining 50% sensitivity. Notably, we were able to increase the specificity to 85% through the supplemental application of previously reported rules for shRNA selection (Figure 3E)(Fellmann et al., 2011; Matveeva et al., 2012).
To validate this addition to the shERWOOD algorithm, we performed an shRNA screen as described above, wherein shRNAs were selected with the 1U-strategy with or without applying the additional filter. We also applied the new variant of the algorithm to shRNA screen described for Figure 2. We found that when additional filters were applied to the 1U strategy, shRNAs targeting our set of consensus essential genes showed a significantly higher percentage of depleted shRNAs per gene (Wilcoxon rank-sum p<0.01) and a stronger mean depletion as measured by log ratio (Wilcoxon rank-sum p<0.01; Figure 3F).
A variant miRNA scaffold increases shRNA potency
Recently completed studies of evolutionarily conserved determinants of Drosha processing raised the possibility that the placement of the EcoRI site in the standard miR-30 scaffold might have reduced the efficiency of pri-miRNA cleavage (Auyeung et al., 2013). Others have reported that alternatively positioning the EcoRI site within the scaffold increases small RNA levels, presumably by improving biogenesis. This led to overall more potent knockdown (Fellmann et al., 2013). We therefore chose to create shRNAs by Gibson assembly, thus removing restriction sites altogether from the shRNA scaffold (Figure S4). We felt that this was the surest way to avoid any unanticipated impacts of altering processing signals. We termed this scaffold, ultramiR.
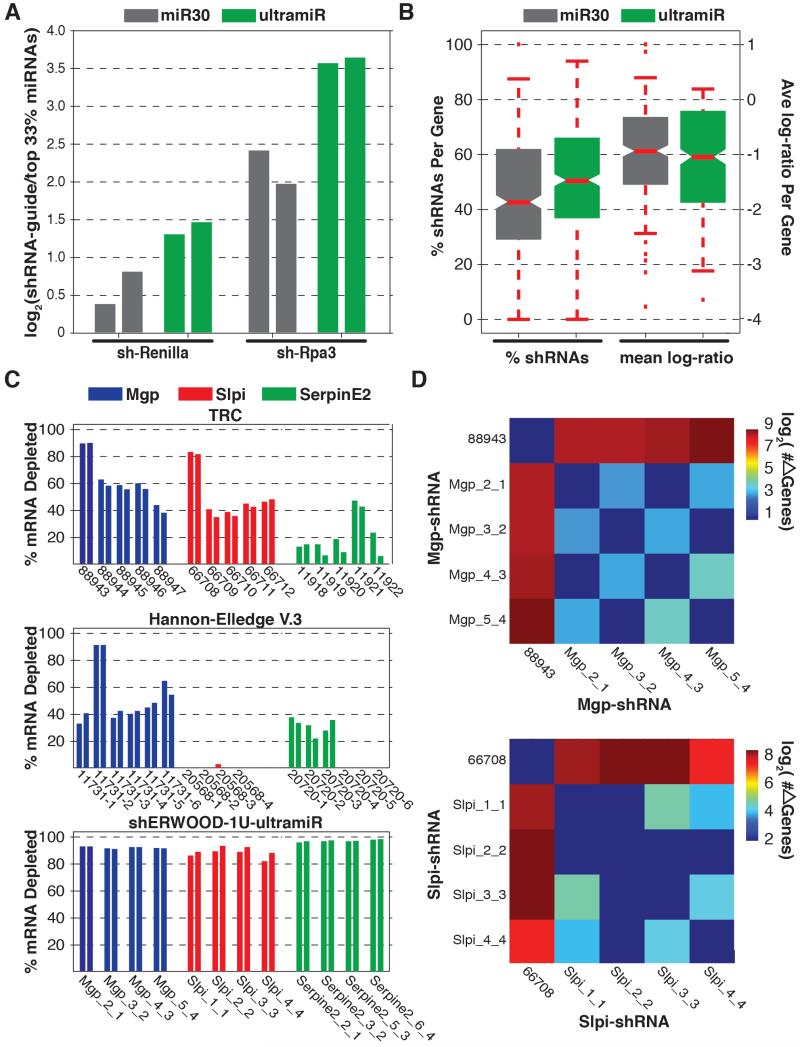
To test ultramiR performance, we inserted two shRNAs, targeting luciferase or mouse RPA3, into the standard scaffold and into ultramiR. These constructs were packaged and infected in duplicate (MOI < 0.3) into HEK293T cells and the modified DF1 reporter line used for the sensor screen, respectively (Fellmann et al., 2011). Following selection for singly infected cells, we analyzed levels of mature shRNAs by small RNA sequencing (Malone et al., 2012). shRNA guide counts were normalized across libraries by determining their log-fold enrichment relative to the 66th quantile of endogenous microRNA levels. A comparison of the normalized shRNA values indicated that, when shRNAs were placed into the ultramiR scaffold, mature small RNA levels were significantly increased relative to levels observed using the standard miR-30 scaffold (Figure 4A). Notably, the performance of ultramiR and the previously described alternate scaffold, miR-E, were indistinguishable (not shown).
Figure 4. Validation of an Alternative Mir Scaffold.
A) Relative abundances of processed guide sequences for two shRNAs (as determined via small RNA cloning + NGS analysis) when cloned into traditional miR30 and ultramiR scaffolds. Values represent the log-fold enrichment of shRNA guides with respect to sequences corresponding to the ten most abundant microRNAs. B) Distributions of the percentage of shHERWOOD-1U-selected shRNAs targeting consensus essential genes that depleted in validation screens when shRNAs were placed into miR30 and ultramiR scaffolds. Log-fold changes for the same constructs are displayed on the left. C) Knockdown efficiencies for shRNAs targeting mouse genes Mgp, Slpi and Mgp. shRNAs assessed were those contained within the TRC collection, those initially designed for the Hannon-Elledge V.3 library and those designed using the current strategies. the TRC and Hannon-Elledge V.3 shRNAs are housed within each libraries lentiviral vectors, while the shERWOOD-1U selected shRNAs are housed within an ultramiR scaffold in a retroviral vector. Ultramir is constitutively expressed from the LTR. D) The number of differentially expressed genes (> 2-fold change and FDR < 0.05) identified through pairwise comparisons of the cell lines corresponding to Mgp and Slpi knockdown by the shERWOOD-1U selected shRNAs and the TRC shRNAs 88943 and 66708.
To provide a more rigorous test of ultramiR performance, we created a variant of shERWOOD-selected 1U-strategy shRNA library, and compared its performance to that of the same library in the standard scaffold. Considering the consensus essential gene set, over half of all shRNAs in the library were significantly depleted (Figure 4B). This substantial improvement (from 42% to 51%, Wilcoxon rank-sum p<0.01) was accompanied by a greater degree of mean log-fold depletion for each construct (from −0.95 to −1.05, Wilcoxon rank-sum p<0.01).
We also tested a limited number of individual shRNAs for their potency by measuring reductions in target mRNA levels. We selected the four shRNAs with the highest shERWOOD scores for mouse Mgp, Serpine2 and Slpi. These were cloned into an MSCV-based ultramiR vector wherein hygromycin resistance and mCherry were also expressed as a bicistronic transcript from the PGK promoter. We also chose to compare these shRNAs to those developed using previous library construction strategies. For this, we obtained the current TRC (5 shRNAs per gene) and V.3 Hannon-Elledge (6 shRNAs per gene) library constructs targeting these genes. For the Hannon-Elledge library, because there were not 4 pre-cloned shRNAs for each gene, we assembled the remaining shRNAs that were designed as part of that library but never constructed. We failed to clone two constructs (both targeting Slpi) after multiple attempts, meaning that only 4 V3 constructs were tested for that gene. Mouse 4T1 cells were infected at single copy and knockdown was tested following selection of infected cells. The TRC library is carried within a vector lacking a fluorescent marker. We therefore calibrated infection levels to achieve single copy by comparison to parallel infections and selections with V3 constructs. The knockdown efficiency of each shRNA was assessed by comparing transcript levels (via qPCR) to those in cells infected with corresponding empty vectors. The TRC shRNAs showed modest knockdown in most cases, with only two shRNAs showing greater than 80% transcript reduction (88943 and 66708, Figure 4C). The Hannon-Elledge V.3 shRNAs produced relatively modest levels of knockdown. In comparison the majority of shRNAs designed using the strategies outlined here reduced target mRNA levels by over 80%, with most reducing target mRNA levels by more than 90% (Figure 4D). Considered together, our data indicate that the combined use of shERWOOD and the ultramiR scaffold consistently produces highly potent shRNAs.
To assess the specificity of shRNA knockdown, we performed RNAseq on all cell lines expressing shERWOOD-ultramiR shRNAs targeting Slpi and Mgp and the two cell lines harboring TRC constructs 88943 and 66708, which target Mgp and Slpi, respectively. Even in the absence of off-target effects, the silencing of a gene through RNAi will likely elicit biological effects that result in changes in the abundance of other mRNAs. Unlike so-called “off-target” effects, phenotypic effects that emanate from on-target silencing should be consistent for all efficacious shRNAs. Thus, by comparing the expression profiles of cells harboring different shRNAs corresponding to a single gene, one should be able to infer the scope of off-target effects for each construct. Those shRNAs, which show the greatest propensity to off-targets, will be those, which create expression profiles most dissimilar to the mean profile.
When either Mgp or Slpi were silenced using the strategies outline here, the expression profiles in the resultant lines were found to be highly similar. Less than 25 genes were altered in their expression (DESeq, fold-change > 2 and FDR < 0.05) between any pair of corresponding lines. However, when these were compared to lines that had Mgp or Slpi silenced using potent TRC constructs, a significant difference in expression profiles is observed. Over 500 genes are altered in the line where Mgp has been silenced using the TRC constructs, and approximately 250 are altered in the line expressing the TRC Slpi-shRNA (Figure 4D).
These results could reflect our current strategies for reducing off-targeting or to our use of a microRNA-based scaffold. Recently, others have observed strong phenotypic changes, related to microRNA dysregulation, when U6 driven stem-loop shRNAs were expressed in cells where the target gene had been deleted (Baek et al., 2014). In contrast, when these same shRNAs were expressed from a microRNA scaffold, the phenotype was not observed. Overall, the aforementioned analysis indicates that shRNAs produced using the strategies outlined in this report, when expressed in an ultramiR scaffold, show strong knockdown capacity and limited off-target effects.
Discussion
The application of RNAi in mammalian cells promised a revolution in understanding gene function and in the discovery and validation of therapeutic targets. While the impact of RNAi has been enormous, there have also been substantial frustrations in attempts to fully realize the potential of this technology. Many different sequences often need to be tested in order to obtain one that potently suppresses expression, a problem that is particularly acute with shRNAs expressed from single-copy transgenes. This, and the resulting variability in the quality of publicly available genome-wide shRNA collections, has caused consternation, particularly when very similar shRNA screens carried out by different investigators yield largely non-overlapping results (Babij et al., 2011; Luo et al., 2009; Scholl et al., 2009). We have tried to address problems with current shRNA technologies both by optimizing target sequence choice and by optimizing small RNA production.
We have leveraged our prior development of a high-throughput assay for testing shRNA potency to develop a computational algorithm capable of accurately predicting the outcome of the sensor screen and in turn predicting potentially potent shRNAs. Though iterative cycles of training and refinement, we have produced a tool that permits highly efficacious shRNAs to be generated for nearly any gene.
We have validated the performance of our approach and benchmarked it against current tools using non-sequence verified, focused shRNA libraries. Based upon our analyses, we can now generate shRNA libraries where nearly 60% of all hairpins targeting essential genes are strongly depleted in multiplexed screens. This means that for any library containing on average 4 hairpins per gene, most bona fide hits will be identified by multiple hairpins, greatly reducing the probability of false-positive calls. Since our libraries were used in their raw form, we feel that this is a lower boundary of performance, since sequence-validated and arrayed collections will not contain a mixture of shRNA variants generated by synthesis and PCR errors.
Given the promise of our approach, we have undertaken the construction of fourth- and fifth-generation, sequence-verified shRNA libraries targeting the mouse and human genomes. The fourth generation toolkit takes advantage of shERWOOD in a canonical miR-30 scaffold and currently comprises over 75,000 shRNAs targeting human genes and 40,000 shRNAs targeting mouse genes. The fifth generation toolkit places shERWOOD shRNAs in the ultramiR scaffold and is presently ~50% complete.
We have predicted shERWOOD shRNAs targeting constitutive exons of annotated human, mouse and rat protein coding genes, and these are available via a web portal (http://sherwood.cshl.edu:8080/sherwood/). We have additionally made shERWOOD available as a web-based tool for custom shRNA prediction, for example for the design of shRNAs for other model organisms or for specific mRNA isoforms or non-coding RNAs.
Overall, we feel that the combination of improvements to shRNA technologies described herein creates a next-generation RNAi toolkit that will produce more reliable outcomes for investigators, whether applied on a gene-by-gene basis or in the context of unbiased, genome-wide screens.
Experimental Procedures
Cell Lines
The sensor algorithm was performed using ERC cells (derived from DF-1 chicken embryonic fibroblasts (Fellmann et al., 2011). All shRNA screens were performed in the pancreatic adenocarcinoma cell line A385 (Cui et al., 2012). small RNA analysis for RPA2 shRNAs was performed in the ERC cell line (Fellmann et al., 2011) and in HEK293Ts for the Renilla shRNAs. Individual shRNA knockdown experiments were performed in the 4T1 murine mammary cancer cell line (Dexter et al., 1978).
Vectors
All RNAi screens and small RNA cloning experiments were performed with an MSCV-based retroviral vector harboring a bi-cistronic transcript (eGFP-IRES-Neomycin) downstream of the PGK promoter (Figure S2D). Single target knockdown experiments for shERWOOD-ultramiR shRNAs were performed with a similar vector where Neomycin is replaced with Hygromycin and eGFP is replaced with mCHERRY. Single target knockdown experiments for the Hannon-Elledge V3 and TRC shRNAs were performed with the GIPZ and pLKO.1 vectors, respectively (GE Dharmacon).
shRNA Library Construction
To ensure high complexity end products, all shRNA libraries were amplified from raw chip material using 16 separate 1 ul 100 uM aliquots with 22 PCR cycles. All transformations were performed with Invitrogen’s MegaX DH10B T1 Electrocompetent cells using a Biorad Gene Pulser Xcell and Biorad Gene Pulser 1mm cuvettes for electroporation. For each library, a minimum of 25M successfully transformed cells were obtained.
shRNA Library Screening
shRNA libraries were packaged using the Platinum-A retrovirus packaging cell (Cell Biolabs). Cells were co-transfected with VSVG and siRNAs targeting the shRNA processing protein Pasha (Qiagen). Viral infections were performed at a multiplicity of infection (MOI) 0.3 to ensure a maximum of one shRNA infection per cell. shRNA representation in the infected cell population was maintained at a minimum of 1000 infected cells per shRNA on each passage. All screens were performed in triplicate. Two days after infection, cells were collected for a reference time-point, and after ~12 doublings cells were again harvested for a final time-point. Neomycin selection began after the initial time-point and continued throughout the screens.
shRNA Library Processing and Analysis
Following cell harvests, DNA was extracted with the Qiagen QIAamp DNA Blood Maxi Kit. For each sample, shRNA molecules were extracted from genomic DNA in 96 separate 25-cycle PCR reactions where 2 ug of input DNA was included in each reaction. Following this initial PCR, illumina adapters were added via PCR and samples were processed on the Illumina Hi-Seq-2.0 platform (read depth was maintained at ~1000 short-reads per shRNA). Following sequencing shRNA counts were extracted with the bowtie algorithm (allowing zero mismatches) and normalized by their total counts. Log-fold changes demonstrated a GC-bias in the control shRNA population (Figure S2E). To remove this bias a one-degree polynomial was fit to each screen replicate’s log-fold change vs. GC content data, and this curve was then subtracted from each data-point (Figure S2F). Following this, values were further normalized such that the control population had population variance of one. shRNAs were classified as depleted with an FDR cutoff of 0.1 using an Empirical-Bayes Moderated T-Test (Figure S2G)(Smyth, 2004).
For further details on the Experimental Procedures, please see Supplemental Information
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute as well as grants from the NIH (G.J.H.) and a kind gift from Kathryn W. Davis. S.R.V.K is supported by a fellowship from The Hope Funds For Cancer Research. E.W. is supported by a fellowship from the Boehringer Ingelheim Foundation. We thank all members of the Hannon lab for help with initial library construction. We also like to thank Blake Simmons (Transomics Technologies) for help in constructing sequence verified versions of the libraries. Finally, we thank Vasily Vagin for helpful comments on this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability
All raw and processed data is available through NCBI under the accession number GSE62189.
Author Contributions
S.R.V.K and G.J.H designed the experiments and wrote the manuscript. S.R.V.K designed the algorithm. A.M performed all shRNA screens. A.M. and X.Z. performed the 1U-sensor. N.E. performed all small RNA cloning. S.R.V.K, A.M. and N.E. constructed the sequence verified libraries. K.C. and K.M. performed the DSIR-sensors. S.R.V.K and A.G. developed and implemented the exom-inclusion and off-target minimization strategies. A.G. and O.E.D designed the shERWOOD website. E.W. and S.K. performed the individual knockdown experiments. S.R.V.K and S.K. performed the RNAseq experiments.
References
- Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nature reviews. Molecular cell biology. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babij C, Zhang Y, Kurzeja RJ, Munzli A, Shehabeldin A, Fernando M, Quon K, Kassner PD, Ruefli-Brasse AA, Watson VJ, et al. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer research. 2011;71:5818–5826. doi: 10.1158/0008-5472.CAN-11-0778. [DOI] [PubMed] [Google Scholar]
- Baek ST, Kerjan G, Bielas SL, Lee JE, Fenstermaker AG, Novarino G, Gleeson JG. Off-target effect of doublecortin family shRNA on neuronal migration associated with endogenous microRNA dysregulation. Neuron. 2014;82:1255–1262. doi: 10.1016/j.neuron.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science (New York, N.Y.) 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science (New York, N.Y.) 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Rana TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Molecular cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- Chuang CF, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary MA, Kilian K, Wang Y, Bradshaw J, Cavet G, Ge W, Kulkarni A, Paddison PJ, Chang K, Sheth N, et al. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nature methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- Cui Y, Brosnan JA, Blackford AL, Sur S, Hruban RH, Kinzler KW, Vogelstein B, Maitra A, Diaz LA, Jr., Iacobuzio-Donahue CA, et al. Genetically defined subsets of human pancreatic cancer show unique in vitro chemosensitivity. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:6519–6530. doi: 10.1158/1078-0432.CCR-12-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Induction of stable RNA interference in mammalian cells. Gene therapy. 2006;13:503–508. doi: 10.1038/sj.gt.3302656. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Dexter DL, Kowalski HM, Blazar BA, Fligiel Z, Vogel R, Heppner GH. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer research. 1978;38:3174–3181. [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell reports. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, Elledge SJ, Hannon GJ, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Molecular cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Gupta S, Schoer RA, Egan JE, Hannon GJ, Mittal V. Inducible, reversible, and stable RNA interference in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1927–1932. doi: 10.1073/pnas.0306111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science (New York, N.Y.) 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J, Meloon B, Engel S, Rosenberg A, Cohen D, et al. Design of a genome-wide siRNA library using an artificial neural network. Nature biotechnology. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science (New York, N.Y.) 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science (New York, N.Y.) 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, Lee WJ, Ueda R, Lemaitre B. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Current biology: CB. 2006;16:808–813. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & development. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nature genetics. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science (New York, N.Y.) 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C, Brennecke J, Czech B, Aravin A, Hannon GJ. Preparation of small RNA libraries for high-throughput sequencing. Cold Spring Harbor protocols. 2012;2012:1067–1077. doi: 10.1101/pdb.prot071431. [DOI] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Matveeva OV, Nazipova NN, Ogurtsov AY, Shabalina SA. Optimized models for design of efficient miR30-based shRNAs. Frontiers in genetics. 2012;3:163. doi: 10.3389/fgene.2012.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & development. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O’Shaughnessy A, Gnoj L, Scobie K, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nature biotechnology. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Sanchez Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, Silver SJ, Tamayo P, Wadlow RC, Ramaswamy S, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5′ precision of both MicroRNAs and their miRNA* strands in flies. Current biology: CB. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Zamore PD. Rethinking the microprocessor. Cell. 2006;125:827–829. doi: 10.1016/j.cell.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nature genetics. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Sims D, Mendes-Pereira AM, Frankum J, Burgess D, Cerone MA, Lombardelli C, Mitsopoulos C, Hakas J, Murugaesu N, Isacke CM, et al. High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome biology. 2011;12:R104. doi: 10.1186/gb-2011-12-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development (Cambridge, England) 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. Regression Shrinkage and Selection via the Lasso. Journal of the Royal Statistical Society. 1995;58:267–288. [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & development. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic acids research. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, Iakoucheva LM, Radivojac P. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics (Oxford, England) 2006;22:1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]
- Vert JP, Foveau N, Lajaunie C, Vandenbrouck Y. An accurate and interpretable model for siRNA efficacy prediction. BMC bioinformatics. 2006;7:520. doi: 10.1186/1471-2105-7-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & development. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Chen HY, Tuschl T, Patel DJ. A potential protein-RNA recognition event along the RISC-loading pathway from the structure of A. aeolicus Argonaute with externally bound siRNA. Structure (London, England: 1993) 2006;14:1557–1565. doi: 10.1016/j.str.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA (New York, N.Y.) 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic acids research. 2010;38:7689–7697. doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.