Abstract
Iron is a key micronutrient for microbial growth but is often present in low concentrations or in biologically unavailable forms. Many microorganisms overcome this challenge by producing siderophores, which are ferric-iron chelating compounds that enable the solubilization and acquisition of iron in a bioactive form. Pantoea stewartii subsp. stewartii, the causal agent of Stewart's wilt of sweet corn, produces a siderophore under iron-limiting conditions. The proteins involved in the biosynthesis and export of this siderophore are encoded by the iucABCD-iutA operon, which is homologous to the aerobactin biosynthetic gene cluster found in a number of enteric pathogens. Mutations in iucA and iutA resulted in a decrease in surface-based motility that P. stewartii utilizes during the early stages of biofilm formation, indicating that active iron acquisition impacts surface motility for P. stewartii. Furthermore, bacterial movement in planta is also dependent on a functional siderophore biosynthesis and uptake pathway. Most notably, siderophore-mediated iron acquisition is required for full virulence in the sweet corn host, indicating that active iron acquisition is essential for pathogenic fitness for this important xylem-dwelling bacterial pathogen.
INTRODUCTION
Pantoea stewartii subsp. stewartii, a Gram-negative bacterial phytopathogen, is the causal agent of a severe seedling wilt in susceptible corn cultivars called Stewart's wilt (1–3). The bacterium is transmitted by an insect vector, the corn flea beetle (Chaetocnema pulicaria), that introduces the pathogen into the plant tissue when it creates scratching wounds on the leaf surface during feeding. There are two phases of Stewart's wilt, a leaf blight and a wilting phase. During the initial infection phase, leaf blight occurs when the bacteria colonize the apoplastic space and cause characteristic water-soaked lesions. As infection progresses, the bacteria preferentially colonize the xylem, where they move systemically through the plant and block water flow, thereby causing the wilting observed in susceptible young seedlings. P. stewartii forms biofilms in the xylem, a process which is regulated in a cell density-dependent manner and requires flagellar-based surface motility and production of an exopolysaccharide (EPS) matrix (4–6). It is these biofilms and the associated EPS that presumably lead to xylem vessel blockage (5, 7). P. stewartii is able to reach very high cell densities and spread quickly within the xylem, in part due to flagellar motility and secretion of a plant cell wall-degrading enzyme (4, 8). However, it is not fully known how the bacterium obtains the suite of required nutrients for rapid growth within the plant or what environmental factors influence the transition from the planktonic to the biofilm state, which is necessary for systemic colonization of the xylem. Iron is commonly a limiting nutrient for microbial growth because it is often insoluble at biological pH within tissues and thus unavailable for utilization. Specifically, in plant tissues the transport of iron involves various chelation and oxidation/reduction steps and iron can also be bound to host ferritin or complexed to phytosiderophores during storage or translocation throughout the plant. In xylem fluid, iron is primarily complexed to citrate and is usually present at concentrations of <10 μM (9–11). In regard to the P. stewartii pathosystem, there is little information about the chemical nature of the iron present in the sweet corn apoplast and xylem niches that P. stewartii colonizes. It is likely that these different microenvironments affect the chemistry of iron and are low in bioavailable iron as in other plant systems (9).
Many species of bacteria secrete iron-chelating compounds, known as siderophores, which enable them to solubilize iron in their environment and transport it back into the cell via specific, high-affinity transport mechanisms (12–14). In terms of pathogenesis, there is often competition for iron between the pathogen and the host. Plants sequester iron for their own metabolic needs leading to a complex set of interactions between the host and invading bacterium involved in iron acquisition. Indeed, siderophores have been implicated as virulence factors for a number of animal pathogens primarily because of their ability to remove bound iron from hemoglobin and other host proteins (15). Some plant-pathogenic bacteria produce siderophores as well, although their importance as virulence factors varies. Production of the siderophore, staphyloferrin B, in Ralstonia solanacearum is not necessary for full virulence in the tomato host, although siderophore mutants are incapable of scavenging iron (14). A siderophore deficient mutant of Xanthomonas oryzae pv. oryzae, a pathogen of rice, was also fully virulent (16). However, Dickeya dadantii, a close relative of P. stewartii, produces two different siderophores, chrysobactin and achromobactin, which are both essential to the infection process and are expressed at different stages of disease progression. These two siderophores likely enable D. dadantii to cope with fluctuating iron levels at distinct phases of the infection process (17).
Siderophores are classified based on their chemical structure as catechol, hydroxamate, or mixed-type siderophores. Aerobactin, a hydroxamate-type siderophore originally isolated from Aerobacter aerogenes (18), is synthesized by a number of bacterial species in the Enterobacteriaceae. In Escherichia coli K-12(pColV-K30) strains, the aerobactin biosynthetic and uptake functions are encoded by an operon found on the virulence plasmid pColV-K30 (19). This operon is induced under iron limiting conditions (20) and consists of five genes. The first four genes, iucA, iucB, iucC, and iucD encode proteins that perform the biosynthetic functions, and a fifth gene, iutA, encodes a specific, high-affinity outer membrane receptor for the ferric-siderophore complex that allows it to be transported back into the cell (19). Aerobactin is not produced by all E. coli strains, but is found in a large number of highly virulent E. coli and Shigella strains (21). The presence of the aerobactin biosynthetic operon is also correlated with high levels of virulence in extraintestinal pathogenic E. coli strains often found in livestock (12). The plant pathogen Pectobacterium carotovorum can synthesize aerobactin as well, but it is only found in a minority of P. carotovorum strains, and its function is dispensable for pathogenicity (22). Analysis of the P. stewartii genome indicates the presence of a conserved iucABCD-iutA operon in P. stewartii (23).
Recent studies have implicated siderophore-mediated active iron acquisition in processes related to bacterial virulence, such as motility, or movement across a solid surface (24–29). The ability to become motile or translocate across a solid surface is an early and essential aspect of microcolony and biofilm formation (30). In the enteric species, Salmonella enterica serovar Typhimurium and E. coli K-12, genes involved in siderophore-mediated iron uptake and iron metabolism were upregulated during the initiation of swarming motility (28, 31). Strains of Pseudomonas putida mutated in either siderophore biosynthetic or receptor genes were compromised in surface motility (24). In the P. stewartii pathosystem, a specialized form of flagellum-based swarming motility is essential during the initial stages of biofilm formation, movement in the xylem and virulence in the plant host (4). In the present study, we demonstrate that siderophore production and uptake are essential for surface motility in vitro, as well as bacterial movement in planta. Furthermore, both are required for virulence and systemic colonization in the host.
MATERIALS AND METHODS
Strains and growth conditions.
All relevant strains and plasmid constructs are listed in Table 1. The P. stewartii strains were grown on Nutrient Agar (Difco Laboratories, Detroit, MI), Luria-Bertani (LB) broth (Difco, Laboratories), or MM9 supplemented with Casamino Acids (0.003%, [wt/vol]) and glucose (0.2%) (32) at 28°C in the presence of nalidixic acid (30 μg/ml), kanamycin (30 μg/ml), ampicillin (100 μg/ml), or chloramphenicol (35 μg/ml) when needed. E. coli strains were grown at 37°C in LB medium in the presence of kanamycin (30 μg/ml), ampicillin (100 μg/ml), or chloramphenicol (35 μg/ml) as needed. E. coli DH10β was used as a cloning host and S17-1λ served as a donor for conjugal transfer of RK-2-based plasmid constructs into P. stewartii. All strains used in the present study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| Pantoea stewartii subsp. stewartii | ||
| DC283 | Wild type; Nalr | 58 |
| ΔiucA mutant | Deletion in iucA; Nalr Kanr | This study |
| ΔiutA mutant | Deletion in iutA; Nalr Kanr | This study |
| Δfur mutant | Deletion in fur; Nalr | This study |
| ΔiucA/iucA+ mutant | Deletion in iucA complemented with pBBR1-MCS4GW:iucA (pLPB44) | This study |
| ΔiutA/iutA+ mutant | Deletion in iutA complemented with pBBR1-MCS4GW:iutA (pLPB43) | This study |
| Δfur/fur+ mutant | Cmr Nalr; deletion in fur complemented by chromosomal integration of wild-type fur into intergenic region downstream of glmS | This study |
| DC283(pHC60) | Wild-type DC283 constitutively expressing GFP | This study |
| ΔiucA mutant(pHC60) | ΔiucA strain constitutively expressing GFP | This study |
| ΔiutA mutant(pHC60) | ΔiutA strain constitutively expressing GFP | This study |
| ΔiucA/iucA+ mutant(pHC60) | ΔiucA/iucA+ strain constitutively expressing GFP | This study |
| ΔiutA/iutA+ mutant(pHC60) | ΔiutA/iutA+ strain constitutively expressing GFP | This study |
| Escherichia coli | ||
| DH10β | F− endA1 recA1 galE15 galK16 nupG rpsL lacX74 80dlacZΔM15 araD139(ara-leu)7697 mcrA(mrr-hsdRMS-mcrBC) | Invitrogen |
| S17-1pir+ | RP4, Mob+; Smr | 59 |
| EC100pir+ | F− mcrA(mrr-hsdRMS-mcrBC) 80dlacZΔM15 lacX74 recA1 endA1 araD139(ara-leu)7697 galU galK-rpsL nupG pir+(DHFR) | Epicentre |
| Plasmids | ||
| pUC18R6K-mini-Tn7-cat | Cmr Apr | 36 |
| pAUC40 | Cmr Smr | 34 |
| pFLP2 | sacB+; FLP recombinase; Apr | 60 |
| pCR8/GW/TOPO | Cloning vector; Spr | Invitrogen |
| pKD4 | Kanr | 35 |
| pBBR1-MCS4GW | Gateway compatible broad-host-range cloning vector | 34 |
| pLPB02 | fur::kan cloned into pCR8/GW/TOPO; Kanr Spr | This study |
| pLPB04 | fur::kan cloned into pAUC40; Kan | This study |
| pLPB29 | fur ORF+promoter cloned into pUC18R6K-mini-Tn7-cat; Cmr Apr | This study |
| pLPB34 | iucA::cloned into pCR8/GW/TOPO | This study |
| pLPB35 | iutA::kan cloned into pCR8/GW/TOPO | This study |
| pLPB36 | iucA::kan cloned into pAUC40 | This study |
| pLPB37 | iutA::kan cloned into pAUC40 | This study |
| pLPB41 | iutA ORF cloned into pCR8/GW/TOPO | This study |
| pLPB42 | iucA ORF+promoter cloned into pCR8/GW/TOPO | This study |
| pLPB43 | iutA ORF cloned into pBBR1-MCS4GW | This study |
| pLPB44 | iucA ORF+promoter cloned into pBBR1-MCS4GW | This study |
| pHC60 | Tetr; constitutively expresses GFP | 38 |
Cmr, chloramphenicol resistance; Tetr, tetracycline resistance; Nalr, nalidixic acid resistance; Apr, ampicillin resistance; Spr, spectinomycin resistance; Smr, streptomycin resistance; Kanr, kanamycin resistance.
Standard DNA manipulations.
Genomic DNA was extracted with a DNeasy DNA extraction kit (Qiagen, Valencia, CA). Plasmid DNA was purified with a Zyppy plasmid miniprep kit (Zymo Research Corp., Irvine, CA). DNA fragments were amplified using TaKaRa Ex Taq DNA polymerase (TaKaRa Bio USA, Madison, WI). Synthetic oligonucleotide primers were ordered from Integrated DNA Technologies (Coralville, IA). Restriction enzymes were purchased from New England BioLabs (Ipswich, MA) or Invitrogen, Inc. (Carlsbad, CA).
Mutant construction.
The P. stewartii ΔiucA, ΔiutA, and Δfur mutants were created using a previously described method that is based on Gateway technology (Life Technologies, Inc., Carlsbad, CA) (33). For the ΔiucA mutant, the 5′ and 3′ flanking regions of the target gene and kanamycin cassette (from pKD4) were amplified by using primer pairs iucALFfwd/iucALFrevKan, iucARFfwdKan/iucARFrev, and pKD4fwd/pKD4rev. All primer sequences are included in Table 2. In a second round of PCR, the three PCR products generated from the first round were combined into one tube (without primers) and assembled by PCR overlap extension using optimized PCR conditions. Primer pair iucALFfwd/iucARFrev was added after three rounds of PCR, and then the rest of the PCR was carried out. The assembled PCR product was cloned into vector pCR8/GW/TOPO (Invitrogen) according to the manufacturer's instructions to create plasmid pLPB34. pLPB34 was linearized with XhoI and used to recombine into pAUC40, created from plasmid pKNG101 and modified to contain attL/attR specific recombination sites (34) to create pLPB36. pLPB36 was introduced into P. stewartii DC283 by conjugation, and transconjugants were selected on nutrient agar containing kanamycin at 30 μg/ml and nalidixic acid at 30 μg/ml. DC283 genomic DNA was used as the template for amplification of the flanking regions of the iucA open reading frame (ORF) using optimized PCR conditions. The ΔiutA and Δfur mutants were similarly created using the primer pairs iutALFfwd/iutALFrevKan, iutARFfwdKan/iutARFrev, fur1/fur2, fur3/fur4, and pKD4fwd/pKD4rev that amplify the 5′ and 3′ flanking regions of the iutA gene, the fur gene, and the kanamycin resistance cassette derived from plasmid pKD4 (35), respectively. The resulting overlap PCR products were cloned into vector pCR8/GW/TOPO (Invitrogen) according to the manufacturer's instructions to create plasmids pLPB35 (iutA) and pLPB02 (fur). These plasmids were recombined with pAUC40 to create plasmids pLPB37 (iutA) and pLPB04 (fur). All mutagenesis constructs were separately transformed into E. coli strain S17-1λ for conjugation with wild-type (WT) P. stewartii. For all mutants, the merodiploid state was resolved by sucrose selection. Sucrose resistant colonies were selected for kanamycin resistance (Kanr) and streptomycin resistance (Strs; carried on the pKNG101 plasmid), and double-crossover events were confirmed by using PCR with primers designed to amplify flanking regions and the Kanr marker.
TABLE 2.
Primers used in this studya
All of the primers listed here were from the present study.
Complementation of mutants.
The Δfur mutant strain was complemented by insertion of a WT copy of the respective gene into a neutral region of the chromosome using a mini-Tn7 vector system developed by (36). The fur ORF, with an additional 200 bp upstream to include the native promoter, was amplified by PCR using the primer pair furfwdBamHI/furrevPstI and cloned into the BamHI and PstI restriction sites of the pUC18R6K-mini-Tn7-cat vector (36) to create plasmid pLPB29. The pLPB29 complementation construct was transformed into E. coli strain S17-1λ and subsequently conjugally transferred into strain LB001. The Tn7 construct was expected to integrate into a single conserved chromosomal location in an intergenic region downstream of the glmS gene, as demonstrated in a number of bacterial species (36). Correct insertion was confirmed by DNA sequencing of the insertion site.
ΔiutA and ΔiucA mutants were complemented by providing the WT copy of iutA or iucA on the broad-host-range plasmid vector pBBR1MCS-4 (37). Briefly, the iutA ORF was PCR amplified from WT DC283 genomic DNA using the primer pair iutAfwdORF/iutArev and ligated into vector pCR8/GW/TOPO (Invitrogen) according to the manufacturer's instructions to create pLPB41. For iucA complementation, the iucA ORF was PCR amplified using primer pair iucAFwdBamHI/iucARevKpnI and ligated into pCR8/GW/TOPO to create plasmid pLPB42. pLPB41 and pLB42 were digested with XhoI to linearize them and then recombined with pBBR1MCS4-GW using LR cloning mix (Invitrogen) and according to the manufacturer's protocol to create pLPB43 and pLPB44, respectively. These complementation vectors were transformed into E. coli strain S17-1λ to be used for conjugation with P. stewartii. After conjugation, transformants were screened for resistance to ampicillin and uptake of the correct plasmid was confirmed by plasmid extraction and sequencing.
Siderophore production on CAS plates.
Production of siderophores was detected using colorimetric chromazurol S (CAS) agar plates (32). For each strain, inoculum was taken from a single colony grown on nutrient agar and streaked down the middle of the CAS plates. The zone of color change was photographed after 10 days of incubation at 28°C. For quantitative determination of siderophore production, the zone of color change was measured after 10 days from the edge of colony growth to the edge of the orange halo.
Streptonigrin sensitivity.
Strains were grown overnight in 2 ml of LB medium, diluted 1:20 in fresh LB medium, and divided into untreated and treated samples. Streptonigrin at a final concentration of 1 μM was added to the treated samples. Cell growth was measured by determining the optical density at 600 nm (OD600) over a period of 24 h during incubation at 28°C with shaking at 180 rpm.
Virulence assay.
The scratch inoculation method was designed to mimic the feeding behavior of the corn flea beetle (4). A small amount of bacterial suspension (5 μl) in sterile 1× PBS-T (1× phosphate-buffered saline plus 0.2% Tween 20) containing approximately 5 × 107cells/ml was placed over a wound created by scratching the stem of 7-day-old seedlings with a sterile 20-gauge needle. Plants were observed for symptom development daily, and disease severity was assessed based on an arbitrary rating system modified from Ham et al. (46) between 0 and 4, where 0 = no symptoms, 1 = a few lesions, 2 = spreading lesions that coalesce, 3 = severe lesions plus some wilt, and 4 = severe wilting and death of the plant. The results are shown as the mean disease score of all replicates ± the standard deviations. At least three biological replicates with five technical replicates each (total of 15 plants) were tested for every strain during one experiment and experiments were repeated independently at least three times. Seedlings inoculated with WT DC283 served as positive controls and seedlings inoculated with 1× PBS-T buffer alone served as negative controls.
Swarming motility.
Strains were grown on swarm plates as previously described (4). Swarm plates consist of nutrient soft agar (0.4% agar) plates with added 0.4% glucose. Cells were grown overnight in LB liquid medium with 100 μg of ampicillin/ml when necessary (e.g., for the ΔiucA/iucA+ and ΔiutA/iutA+ complemented strains). Cells were then diluted 1:20 in fresh LB medium and grown to mid-log phase (OD600 = 0.5). The cells were harvested by centrifugation and washed twice with 1× PBS. A 2-μl volume containing ∼107 cells was placed in the center of each swarm plate, followed by incubation at 28°C. Photographs were taken of representative plates for each strain after 16, 24, and 30 h of incubation.
In planta movement assays.
All strains were transformed with plasmid pHC60 (38), which constitutively expresses green fluorescent protein (GFP). Cells were grown overnight in LB medium with tetracycline at 10 μg/ml. The cells were then diluted 1:20 in fresh medium and grown to mid-log phase (OD600 = 0.5). Cells were harvested by centrifugation and washed twice in 1× PBS containing 0.2% Tween 20. Susceptible sweet corn seedlings (cv. Jubilee) were inoculated according to the protocol of Herrera et al. (4). A partial incision was made on the leaf surface and inoculated with 2 μl of bacterial suspension containing ∼107 cells. After 72 h, the distance of movement was quantified by measuring the length of GFP fluorescence using a Leica MZ FLIII fluorescence equipped stereoscope (Leica Microsystems, Wetzlar, Germany). Fluorescent microsphere beads (FluoSpheres; Invitrogen, Carlsbad, CA) were used as a negative control to correct for passive movement within the xylem. The results are expressed as the mean distance from the inoculation point for at least 10 replicates for each strain ± the standard error of the mean.
RESULTS
Siderophore production from the iucABCD-iutA operon.
Genomic analyses revealed that P. stewartii possesses a 7.9-kb gene cluster encoding five ORFs (Fig. 1) with 74% sequence similarity to the aerobactin encoding iucABCD-iutA operon of Escherichia coli S88 (GenBank accession no. NC_011747.1, E value = 0.0) (39). The iucABCD-iutA operon, encoding proteins necessary for aerobactin siderophore biosynthesis and transport, is also found in a number of other pathogenic E. coli strains (12). The first gene, iucA (GenBank accession no. EHT98413), encodes an IucA ortholog with 78% identity to an E. coli IucA/IucC superfamily protein (GenBank accession no. WP_001440858.1, E = 0.0) that is implicated in aerobactin siderophore biosynthesis from N-hydroxylysine and citrate precursors (20, 40). In addition, iutA (GenBank accession no. EHT98409) encodes a putative outer membrane receptor protein essential for transport of ferric-siderophore complexes into the cell. IutA is characterized as a TonB-dependent ligand-gated ion channel protein localized to the outer membrane (40) (Fig. 1). PBLAST analysis of the P. stewartii IutA protein sequence revealed 79% identity to a TonB-dependent siderophore receptor of avian-pathogenic E. coli strain A2363 (GenBank accession no. AY553855, E = 0.0).
FIG 1.
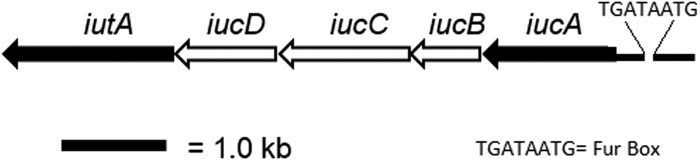
Genomic orientation of the Pantoea stewartii iucABCD-iutA gene cluster. Filled arrows indicate gene deletions created in the present study. The Fur binding sequence is indicated upstream of the iucA ORF.
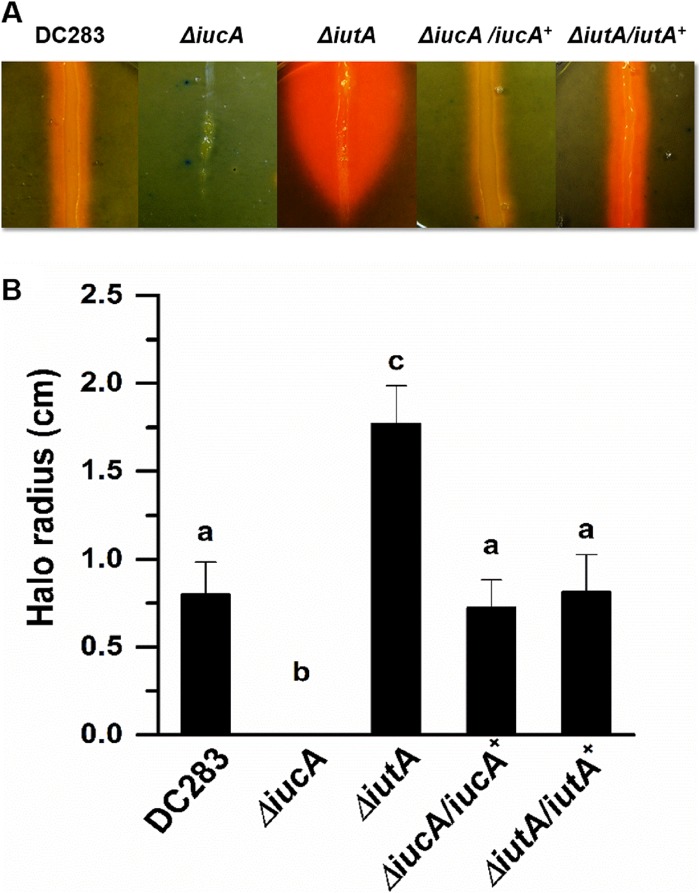
We created nonpolar deletion mutants in the iucA and iutA genes to assess their roles in siderophore production and uptake, respectively, in P. stewartii. CAS medium is a defined medium, which contains 10 μM Fe(III) bound as part of a dye complex, allowing for colorimetric detection of siderophore activity (41). We quantitatively compared the siderophore activity of the different strains by measuring the size of the orange halo at its widest point, and graphically representing the mean of at least 15 replicates after 10 days of incubation (Fig. 2). WT DC283 shows visible siderophore production and had a mean halo radius of 0.79 ± 0.18 cm from the edge of colony growth, whereas the mean radius for the ΔiutA mutant was 1.77 ± 0.2 cm. We reason that this significant increase in siderophore production results from the siderophore biosynthesis proficient ΔiutA mutant continually producing and secreting siderophores, but because it is not able to transport the ferric-siderophore complexes back into the cell siderophore production continues. The mean radius for the ΔiucA knockout mutant was zero since this mutant did not show any visible siderophore activity on CAS medium even after prolonged incubation at 28°C. ΔiucA/iucA+ and ΔiutA/iutA+ complemented strains transformed with iucA or iutA expressed on a broad-host-range plasmid did not differ significantly from the WT (Fig. 2A and B). The differences in the mean halo radius are significant based on Student t test (P ≤ 0.05). Because the ΔiucA strain is compromised in all visible siderophore production, we reason that the iucABCD-iutA operon is the primary locus responsible for siderophore production in P. stewartii.
FIG 2.
Siderophore-mediated iron acquisition requires iucA and iutA. The production of siderophores was detected using colorimetric CAS agar plates (32). (A) For each strain, inoculum was taken from a single colony grown on nutrient agar and streaked down the middle of the CAS plates, followed by incubation for 10 days at 28°C. The radius of the zone of color change was measured at its widest point from the edge of colony growth. (B) The graph represents the mean halo radius from 15 individual plates for each strain. Bars with different letters indicate significance based on a Student t test using P ≤ 0.05. Error bars indicate standard errors of the mean.
Siderophore-mediated active iron acquisition influences swarming motility.
P. stewartii exhibits a unique form of unidirectional swarming motility on soft agar medium (4). After being transferred to swarming medium, cells begin a peripheral colony organization that differentiates into palisade-like ring differentiation after 10 h. This ring ruptures at around 16 h postinoculation and is followed by vigorous unidirectional motility across the agar surface (4).
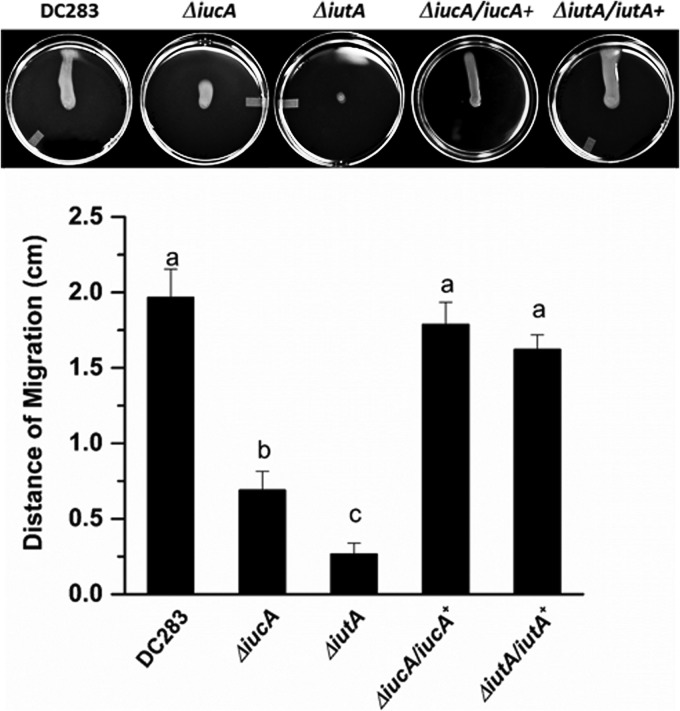
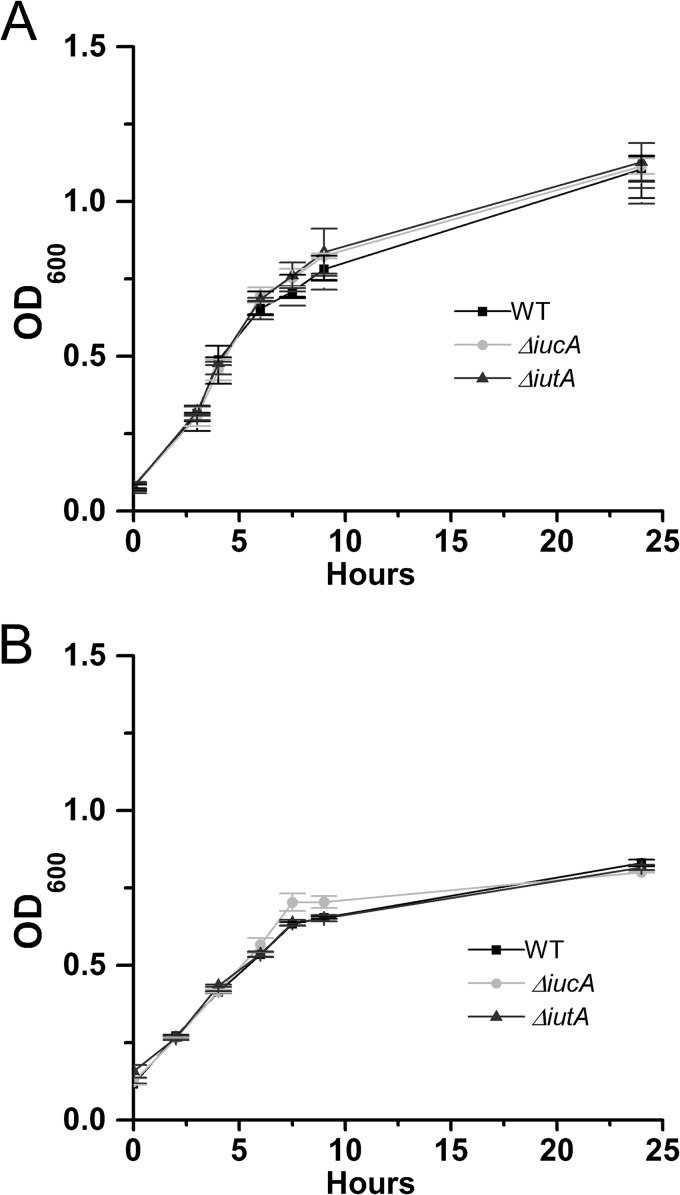
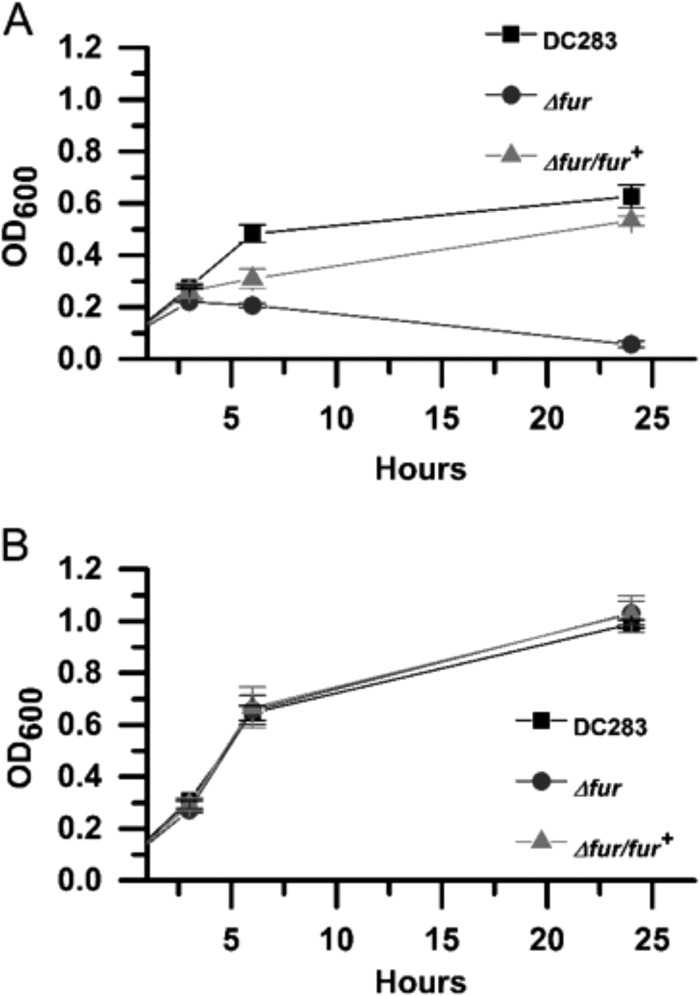
We characterized the behavior of ΔiucA and ΔiutA strains for swarming motility according to the protocol of Herrera et al. (4). The DC283, ΔiucA, ΔiutA, ΔiucA/iucA+, and ΔiutA/iutA+ strains were grown on soft nutrient agar plates (0.4%) supplemented with 0.4% glucose for 24 h. WT DC283 migrates robustly in a single direction across the agar surface, whereas both siderophore mutants showed reduced motility (Fig. 3). The ΔiucA mutant retains unidirectional movement but lags behind and migrates to a much lesser extent compared to the WT. The ΔiutA mutant is completely nonmotile and remains in the center of the plate. The ΔiucA/iucA+ and ΔiutA/iutA+ complemented strains were both restored in swarming motility (Fig. 3). Both siderophore mutants have growth rates equal to the WT in both rich (LB) and minimal MM9 media (Fig. 4). Both mutants also produce similar amounts of stewartan EPS compared to WT, which is required for and facilitates swarming motility in P. stewartii (data not shown). Thus, the deficiency in motility is not attributed to a growth defect or a change in EPS production.
FIG 3.
Siderophore-mediated iron acquisition is required for surface motility. Portions (2 μl) containing ∼107 cells were placed in the centers of motility plates, followed by incubation at 28°C. Images were taken of the same plates after 24 h of incubation. Experiments were performed with at least three biological replicates, with five technical replicates each (total 15 plates per strain). Error bars indicate the standard errors of the mean.
FIG 4.
Siderophore mutants exhibit wild-type growth rates in vitro. Strains were grown in 3 ml of liquid LB medium (A) and MM9 medium (B). The OD600 was recorded over the course of 24 h. The graphs represent the mean of at least three biological replicates plus or minus the standard errors of the mean.
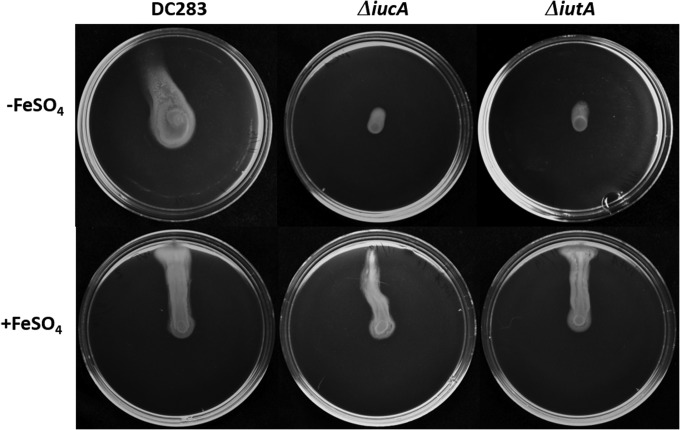
Furthermore, swarming motility in WT DC283 was severely reduced on semisolid medium containing 200 μM 2,2-dipyridyl (DP), a strong iron chelator even after prolonged incubation (48 h) (Fig. 5). This concentration of DP did not reduce the growth rate of WT DC283 in rich LB (data not shown), again indicating that the defect in motility is not attributed to a decrease in growth rate. Moreover, when swarming motility medium was supplemented with 20 μM FeSO4, a readily available source of iron, motility was restored in ΔiucA and ΔiutA strains (Fig. 6). Taken together, these data indicate that iron availability is important for induction of the swarming behavior and that this process is dependent on the siderophore-mediated iron acquisition pathway but can be overridden when sufficient levels of bioavailable iron are supplied.
FIG 5.
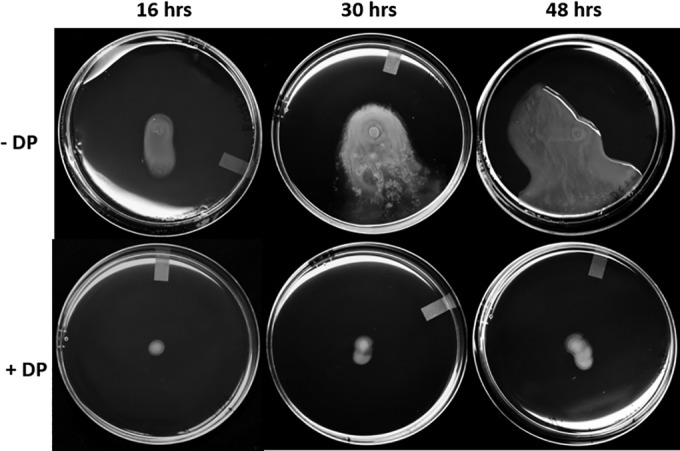
Iron limitation inhibits swarming motility. Portions (2 μl) containing ∼107 cells of WT DC283 were placed in the centers of motility plates with or without 200 μM 2,2-dipyridyl, followed by incubation at 28°C. Images were taken of the same plates after 16, 30, and 48 h of incubation. Experiments were conducted with at least three biological replicates, with five technical replicates each (a total 15 plates per strain).
FIG 6.
Iron supplementation restores motility in ΔiucA and ΔiutA mutants. Portions (2 μl) containing ∼107 WT DC283, ΔiucA, or ΔiutA cells were placed in the centers of motility plates with or without 20 μM FeSO4, followed by incubation at 28°C. Photographs were taken after 24 h of incubation. Experiments were conducted with at least three biological replicates, with five technical replicates each (a total of fifteen plates per strain).
Swarming motility is influenced by intracellular iron.
P. stewartii possesses an ortholog to Fur (ferric uptake regulator), a global repressor that controls iron homeostasis (GenBank accession no. EHU01500). The P. stewartii Fur protein sequence shares 95% identity to the Fur protein found in Salmonella enterica serovar Typhi strain CT18 (GenBank accession no. AL513382.1). Fur typically binds to a conserved sequence in the promoter region of the genes it regulates, acting as an iron-dependent repressor (42, 43). Virtual footprint promoter prediction (44) identified a conserved Fur binding sequence 80 bp upstream of the iucABCD-iutA operon (Fig. 1 and see Fig. S1 in the supplemental material). The P. stewartii Δfur mutant exhibited a significant increase in siderophore activity on CAS plates, a phenotype consistent with loss of repression of the siderophore biosynthetic pathway (Fig. 7 and see Fig. S2 in the supplemental material). Quantitatively, on CAS medium, the Δfur mutant formed a mean halo radius of 1.68 ± 0.2 cm compared to the WT radius of 0.79 ± 0.18 cm (Fig. 7 and see Fig. S2 in the supplemental material). This difference is significant based on Student t test (P ≤ 0.05). Siderophore production in the complemented Δfur/fur+ strain did not differ significantly from the WT.
FIG 7.
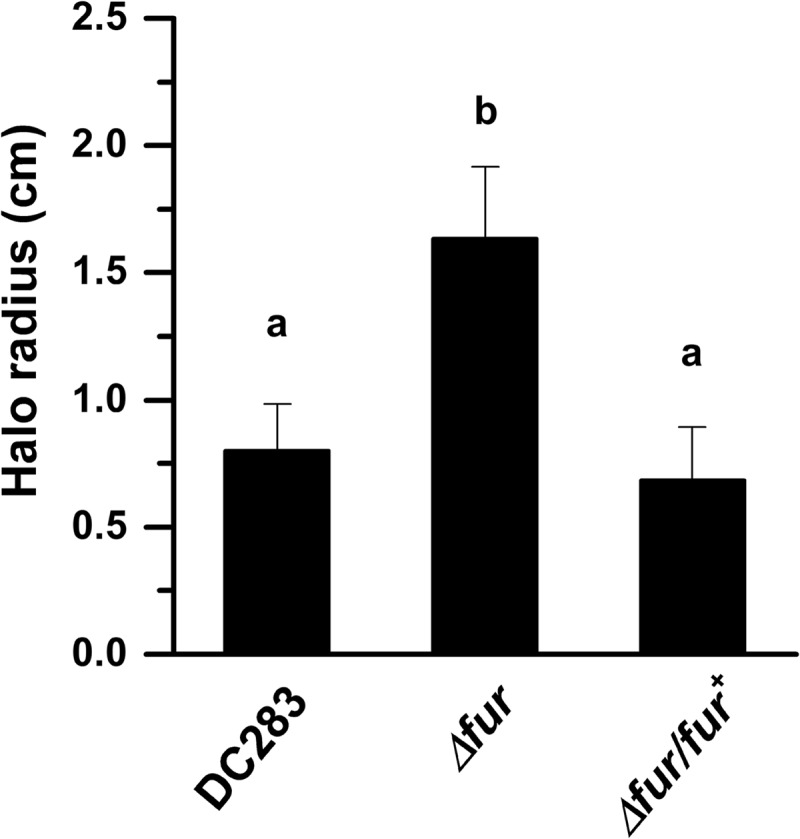
Siderophore production is negatively regulated by Fur. The production of siderophores was detected using colorimetric CAS agar plates (32). For each strain, inoculum was taken from a single colony grown on nutrient agar and streaked down the middle of the CAS plates, followed by incubation for 10 days at 28°C. The halo radius was measured at its widest point from the edge of the colony growth. The graph represents the mean halo radius from 15 plates. Bars with different letters indicate significance based on a Student t test using P ≤ 0.05. Error bars indicate the standard errors of the mean.
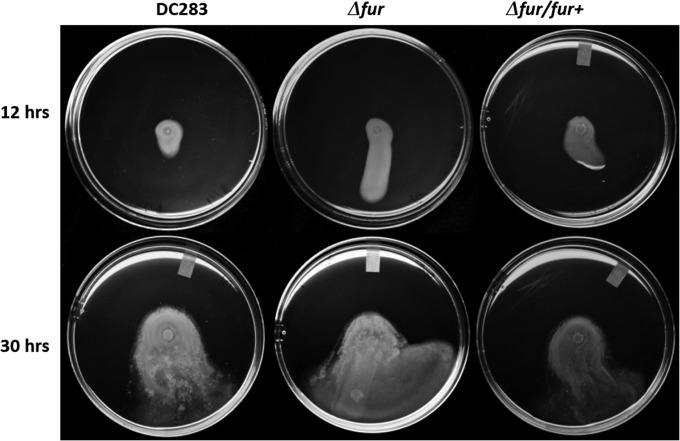
Interestingly, we observed that the Δfur was able to enter a motile state sooner and traveled further than the WT even after 12 h (Fig. 8). The mutation in fur did not result in a change in growth rate in rich medium (LB) compared to the WT (Fig. 9B). Relative amounts of intracellular iron can be determined using the iron-activated antibiotic, streptonigrin, which has increasing bactericidal activity that closely correlates with an increase in intracellular iron levels (45). The Δfur mutant has a severe growth defect in the presence of 1 μM streptonigrin compared to the WT growth rate (Fig. 9A and B), indicating significantly higher intracellular iron levels than the WT, as indicated by a Student t test using P ≤ 0.05. The WT growth rates in the presence of streptonigrin were restored to WT levels in the Δfur/fur+ strain (Fig. 9A).
FIG 8.
Fur regulation influences surface motility. Portions (2 μl) containing ∼107 cells were placed in the centers of swarm plates, followed by incubation at 28°C. Images were taken of the same plates after 12 and 30 h of incubation. Experiments were performed with at least three biological replicates, with five technical replicates each (a total of fifteen plates per strain).
FIG 9.
Δfur mutants accumulate intracellular iron. (A) Growth of streptonigrin-treated cells. (B) Growth of untreated cells. Strains were grown overnight in 2 ml of LB medium, diluted 1:20 in fresh LB medium, and divided into untreated and treated samples. Streptonigrin was added to the treated samples at a final concentration of 1 μM. Cell growth was measured as the OD600 over a period of 24 h, while incubating at 28°C with shaking at 180 rpm. Error bars represent the standard deviations between replicates of each strain.
Siderophore production influences bacterial dissemination in the xylem.
In addition to in vitro motility, we quantified the dissemination of ΔiucA and ΔiutA in the xylem compared to WT DC283. Sweet corn seedlings were inoculated with DC283, ΔiucA and ΔiutA strains all constitutively expressing GFP encoded on plasmid pHC60 (4, 38). After 72 h postinoculation, both ΔiucA and ΔiutA strains were significantly reduced in the their ability to migrate in the basipetal direction in the xylem compared to the WT based on a Student t test using P ≤ 0.05 (Fig. 10). All strains migrated further than the fluorescent microbeads.
FIG 10.
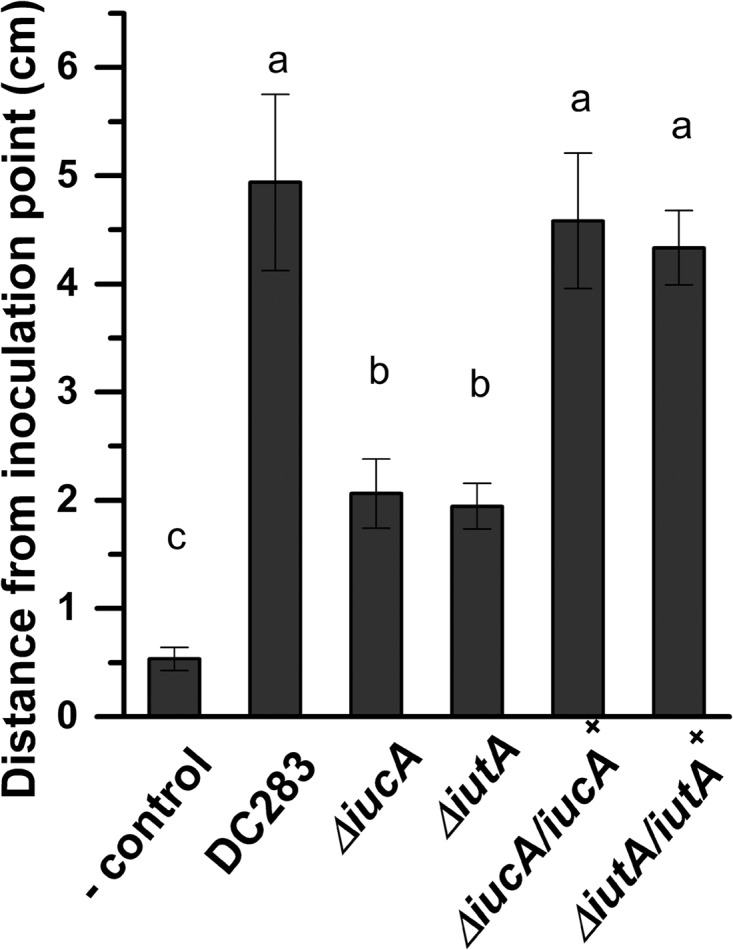
Siderophore production and uptake are necessary for bacterial movement in planta. DC283, ΔiucA, ΔiutA, ΔiucA/iucA+, and ΔiutA/iutA+ strains constitutively expressing GFP from plasmid pHC60 were used to inoculate sweet corn seedlings (cv. Jubilee) in a partial incision on the leaf surface. Plants were inoculated with 2 μl of bacterial suspension containing ∼107 cells in 1× PBS-T. After 72 h, the distance of movement was quantified by measuring the length of GFP fluorescence in the basipetal direction using a Leica MZ FLIII fluorescence equipped stereoscope (Leica Microsystems). Fluorescent microsphere beads (FluoSpheres) were used as a negative control to correct for passive movement. The results are expressed as the mean distance from the inoculation point for at least 10 replicates for each strain ± the standard errors of the mean.
Siderophore production and utilization are necessary for full virulence.
Because mutations in either siderophore biosynthesis or uptake were important for dissemination within the xylem, we hypothesized that the siderophore-mediated iron uptake pathway was necessary for virulence in sweet corn. Ten-day-old sweet corn seedlings were wound-inoculated with either the WT DC283, ΔiucA or ΔiutA or 1× PBS-T buffer. Plants were observed for symptom development daily, and disease severity was assessed based on an arbitrary rating system modified from Ham et al. (46).Both ΔiucA and ΔiutA strains were significantly reduced in their ability to cause symptoms in the plant (Fig. 11). In trans expression of a WT copy of either the iucA and iutA genes on the plasmid pBBR1-MCS4 (pLPB44 and pLPB43) in their respective mutants (ΔiucA/iucA+ and ΔiutA/iutA+) restored symptom development to near WT levels (Fig. 11). The area under the disease progress curve (AUDPC) was calculated using the method of Madden et al. (47). The AUDPC for DC283 was 10.3 after 6 days postinoculation, and for the ΔiucA and ΔiutA mutant strains, the AUDPC were 5.3 and 5, respectively. For the complemented ΔiucA/iucA+ and ΔiutA/iutA+ strains, the AUDPCs were restored to 9.4 and 9.9, respectively. These differences were significant based on a Student t test using P ≤ 0.05. Buffer-inoculated plants did not show disease symptoms.
FIG 11.
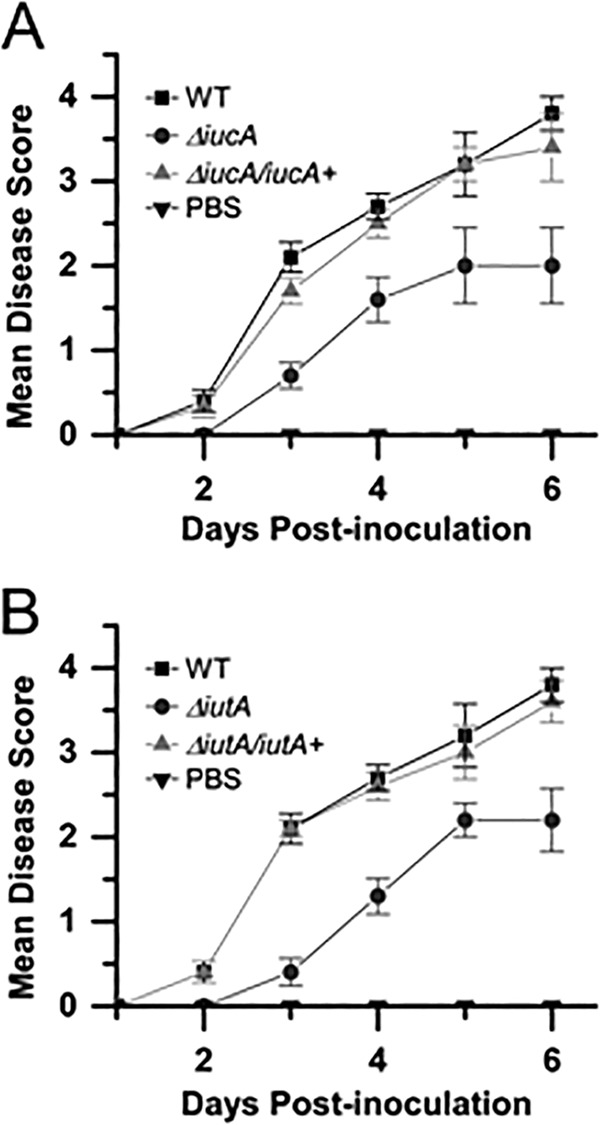
Siderophore production and uptake are required for full virulence. Ten-day-old sweet corn seedlings were inoculated with either a WT DC283, ΔiucA, or ΔiucA/iucA+ strain or 1× PBS buffer (A) or a WT DC283, ΔiutA, or ΔiutA/iutA+ strain or 1× PBS buffer (B). Plants were observed for symptom development daily, and disease severity was assessed based on an arbitrary rating system between 0 and 4. The graph represents the mean of three biological replicates with five technical replicates each (a total 15 plants per strain). Experiments were repeated at least three times with similar results. Error bars indicate the standard errors of the mean.
DISCUSSION
P. stewartii colonizes the xylem in the form of sessile, plant cell wall adherent biofilms. Proper biofilm formation is essential to virulence, and it adheres to a tightly regulated developmental program with spatial and temporal specificity (5). Specifically, during the early stages of biofilm formation, P. stewartii utilizes a specialized form of flagellar-based swarming motility to translocate across a surface and to organize itself into microcolonies that develop into mature, matrix-encased biofilms. Not surprisingly, nonmotile mutants are defective in dissemination in the xylem making flagellar-based surface motility one of the major contributors to pathogenic fitness for P. stewartii (4). Interestingly, mutations in the siderophore-mediated active iron transport system in P. stewartii also led to a decrease in motility both in vitro and in planta. Surface-based motility in P. stewartii is classified as swarming motility (4). In other bacterial systems, cells settle on a surface and experience a lag period before they enter into swarming mode (48). P. stewartii also exhibits a lag period before entering into vigorous swarming mode (4). Interestingly, the ΔiucA and ΔiutA mutants had a longer lag period before swarm expansion initiated, compared to the WT parent, that resulted in a defect in overall motility following 24 h of growth on swarming medium. These mutants continued to have a defective motility phenotype even when incubated for extended periods of up to 48 h (data not shown). Moreover, addition of a strong iron chelator also abrogated motility. It should be noted that neither the mutations in iucA or iutA nor the addition of the iron chelator affected growth rates of P. stewartii in rich medium; thus, the defect in motility is not attributed to a defect in growth on rich swarming medium.
Active iron acquisition has been linked to both negative and positive regulation of motility in a number of different bacterial species (24, 26, 48, 49). It has been proposed that iron-mediated, negative regulation of motility is part of a chemotactic response that promotes exploration of new nutrient-rich environments when bacteria find themselves under iron-limited conditions (50). In contrast, motility is positively regulated by iron in P. fluorescens and Campylobacter jejuni (49). Likewise, P. stewartii was less motile when it was either deprived of iron or defective in the ability to actively acquire iron, indicating that motility is positively regulated by iron either directly or indirectly. The xylem niche that P. stewartii predominantly colonizes in the plant is an interconnected network of hollow tubes that conducts water and minerals throughout the plant. In this scenario, the iron limitation that we speculate is occurring in the xylem may causes the bacterium to be nonmotile and settle on the xylem cell wall surface as part of an adaptation strategy to the xylem. This would allow the sessile adherent bacterial population to obtain maximal levels of nutrients from the replenishing sap flow rather than expend energy exploring new areas of the xylem. We speculate that low iron may serve as an environmental cue that promotes biofilm formation for P. stewartii in planta and perhaps other xylem-dwelling bacteria.
Although iron is an essential micronutrient, it can be toxic at high levels, especially when cells are subjected to the oxidative stress of aerobic growth conditions (51). Thus, siderophore secretion and iron uptake pathways are often under tight regulation by transcription factors that respond to iron availability and environmental conditions. Fur (ferric uptake regulator), a highly conserved, iron-responsive transcription factor, often serves as a regulator of siderophore secretion and iron metabolism (17, 52). Fur uses ferrous iron [Fe(II)] as a cofactor, which causes it to bind to a conserved DNA sequence termed a Fur box and act as a repressor of transcription. Under low-iron conditions, Fur is released from the DNA allowing derepression of genes belonging to the Fur regulon, many of which are involved in iron acquisition, including the iucABCD-iutA operon (20). As in many other bacteria, P. stewartii possesses a highly conserved Fur (ferric uptake regulator) protein. The P. stewartii Δfur mutant constitutively overproduced siderophores. Interestingly, the Δfur mutant was hypermotile and accumulated more intracellular iron than the WT parent, highlighting the connection between intracellular iron levels and positive regulation of translocation across a solid surface in P. stewartii. The molecular mechanisms of how intracellular iron levels regulate motility remain obscure (26, 29). In Pseudomonas aeruginosa, the production of rhamnolipid biosurfactants is induced under iron-limiting conditions, leading to an increase in twitching motility (53). Our data also indicate that iron serves as an environmental signal for motility and entry into biofilm development and that Fur is an integral part of that regulatory hierarchy in P. stewartii.
Thus far, there is mixed evidence about the importance of siderophores during bacterial invasion of plants. A number of other plant pathogens produce siderophores that do not have an apparent role in host colonization. For example, there is no distinct correlation between aerobactin production and virulence in Erwinia carotovora where mutants lacking aerobactin biosynthetic functions were still able to cause soft rot symptoms on potato (22). Pseudomonas syringae pv. tomato produces three different types of siderophores, yersiniabactin, pyoverdin, and citrate. All of these siderophores were important for iron-limited growth in vitro, but none of them were necessary for pathogenesis in the plant host (54). In contrast, in Erwinia amylovora, biosynthesis of the siderophore, desferrioxamine, is important for colonization and necrotic symptom development in apple flowers (55). The role of desferrioxamine in virulence was also dependent on the inoculum dose, suggesting that siderophore production is primarily important during the initial stages of infection when E. amylovora is in low numbers during initial blossom infection (55). Siderophore production was also important for colonization of alfalfa sprouts by the human pathogen, Salmonella enterica (56). Interestingly, proteins related to the siderophore-mediated iron uptake pathway were found in the proteome of a virulent isolate of P. stewartii but were conspicuously absent in an avirulent isolate (57). The authors of that study speculated that this active iron transport system is one of the factors that contribute to virulence in P. stewartii. Here, we demonstrate that siderophore production is, indeed, required for full virulence for P. stewartii in susceptible sweet corn seedlings. We attribute the defect in virulence of the siderophore ΔiucA and ΔiutA mutant strains to their deficiency in motility and dissemination in the xylem. However, it cannot be ruled out that the iucA and iutA mutations interfere with bacterial metabolism making them less fit in planta.
The external environmental cues that signal P. stewartii to enter motility mode have, thus far, remained undefined but our data indicate that low iron is one environmental condition sensed by the bacterium that influences motility. Iron-replete conditions favor motility, but iron-limited conditions favor the nonmotile state and initiate the bacterial population to enter into the biofilm maturation phase. These findings highlight the importance of siderophores beyond their primary role in nutrient acquisition and metabolism and present a possible link between plant nutritional status and plant-microbe interactions particularly for xylem-dwelling bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by an award to M.C.R. from the Regents of the University of California, Riverside, and the Agricultural Experiment Station and College of Natural and Agricultural Sciences. This study was also supported by a Hellman fellowship awarded to M.C.R.
We thank James Wells for assistance with statistical analysis.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02503-14.
REFERENCES
- 1.Claflin L. 1999. Compendium of corn diseases, 3rd ed. APS Press, St Paul, MN. [Google Scholar]
- 2.Stewart FC. 1897. A bacterial disease of sweet corn. N Y Agric Exp Station Bull 130:422–439. [Google Scholar]
- 3.Mergaert J, Verdonck L, Kersters K. 1993. Transfer of Erwinia ananas (synonym, Erwinia uredovora) and Erwinia stewartii to the genus Pantoea emend. as Pantoea ananas (Serrano 1928) comb nov. and Pantoea stewartii (Smith 1898) comb nov, respectively, and description of Pantoea stewartii subsp indologenes subsp nov. Int J Syst Bacteriol 43:162–173. doi: 10.1099/00207713-43-1-162. [DOI] [Google Scholar]
- 4.Herrera CM, Koutsoudis MD, Wang X, von Bodman SB. 2008. Pantoea stewartii subsp. stewartii exhibits surface motility, which is a critical aspect of Stewart's Wilt disease development on maize. Mol Plant Microbe Interact 21:1359–1370. doi: 10.1094/MPMI-21-10-1359. [DOI] [PubMed] [Google Scholar]
- 5.Koutsoudis MD, Tsaltas D, Minogue TD, von Bodman SB. 2006. Quorum sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subsp. stewartii. Proc Natl Acad Sci U S A 103:5983–5988. doi: 10.1073/pnas.0509860103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Von Bodman SB, Majerczak DR, Coplin DL. 1998. A negative regulator mediates quorum sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc Natl Acad Sci U S A 95:7687–7692. doi: 10.1073/pnas.95.13.7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun EJ. 1982. Ultrastructural investigation of resistant and susceptible maize inbreds infected with Erwinia stewartii. Phytopathology 72:159–166. doi: 10.1094/Phyto-72-159. [DOI] [Google Scholar]
- 8.Mohammadi M, Burbank L, Roper MC. 2011. Pantoea stewartii subsp. stewartii produces an endoglucanase that is required for full virulence in sweet corn. Mol Plant-Microbe Interact 25:463–470. doi: 10.1094/MPMI-09-11-0226. [DOI] [PubMed] [Google Scholar]
- 9.Briat J-F, Curie C, Gaymard F. 2007. Iron utilization and metabolism in plants. Curr Opin Plant Biol 10:276–282. doi: 10.1016/j.pbi.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Fernández V, Winkelmann G. 2005. The determination of ferric iron in plants by HPLC using the microbial iron chelator desferrioxamine E. Biometals 18:53–62. [DOI] [PubMed] [Google Scholar]
- 11.Lopez-Millan AF, Morales F, Abadia A, Abadia J. 2000. Effects of iron deficiency on the composition of the leaf apoplastic fluid and xylem sap in sugar beet: implications for iron and carbon transport. Plant Physiol 124:873–884. doi: 10.1104/pp.124.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garénaux A, Caza M, Dozois CM. 2011. The ins and outs of siderophore mediated iron uptake by extra-intestinal pathogenic Escherichia coli. Vet Microbiol 153:89–98. doi: 10.1016/j.vetmic.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 13.Enard C, Diolez A, Expert D. 1988. Systemic virulence of Erwinia chrysanthemi 3937 requires a functional iron assimilation system. J Bacteriol 170:2419–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhatt G, Denny TP. 2004. Ralstonia solanacearum iron scavenging by the siderophore staphyloferrin B is controlled by PhcA, the global virulence regulator. J Bacteriol 186:7896–7904. doi: 10.1128/JB.186.23.7896-7904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu B, Garcia-Herrero A, Johanson T, Krewulak K, Lau C, Peacock RS, Slavinskaya Z, Vogel H. 2010. Siderophore uptake in bacteria and the battle for iron with the host; a bird's eye view. Biometals 23:601–611. doi: 10.1007/s10534-010-9361-x. [DOI] [PubMed] [Google Scholar]
- 16.Pandey A, Sonti RV. 2010. Role of the FeoB protein and siderophore in promoting virulence of Xanthomonas oryzae pv. oryzae on rice. J Bacteriol 192:3187–3203. doi: 10.1128/JB.01558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franza T, Mahé B, Expert D. 2005. Erwinia chrysanthemi requires a second iron transport route dependent of the siderophore achromobactin for extracellular growth and plant infection. Mol Microbiol 55:261–275. [DOI] [PubMed] [Google Scholar]
- 18.Gibson F, Magrath DI. 1969. The isolation and characterization of a hydroxamic acid (aerobactin) formed by Aerobacter aerogenes 62-1. Biochim Biophys Acta 192:175–184. doi: 10.1016/0304-4165(69)90353-5. [DOI] [PubMed] [Google Scholar]
- 19.Lorezo V, Binereif A, Paw BH, Neilands JB. 1986. Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J Bacteriol 165:570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neilands JB. 1992. Mechanism and regulation of synthesis of aerobactin in Escherichia coli K-12 (pColV-K30). Can J Microbiol 38:728–733. doi: 10.1139/m92-119. [DOI] [PubMed] [Google Scholar]
- 21.Lorenzo V, Martinez JL. 1988. Aerobactin production as a virulence factor: a reevaluation. Eur J Clin Microbiol Infect Dis 7:621–629. doi: 10.1007/BF01964239. [DOI] [PubMed] [Google Scholar]
- 22.Ishimaru CA, Loper JE. 1992. High-affinity iron uptake systems present in Erwinia carotovora subsp. carotovora include the hydroxamate siderophore aerobactin. J Bacteriol 174:2993–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glasner JD, Rusch M, Liss P, Plunkett G, Cabot EL, Darling A, Infield-Harm P, Gilson MC, Perna NT. 2006. ASAP: a resource for annotating, curating, comparing and disseminating genomic data. Nucleic Acids Res 34:D41–D45. doi: 10.1093/nar/gkj164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matilla MA, Ramos JL, Duque E, de Dios Alché J, Espinosa-Urgel M, Ramos-González MI. 2007. Temperature and pyoverdine-mediated iron acquisition control surface motility of Pseudomonas putida. Environ Microbiol 9:1842–1850. doi: 10.1111/j.1462-2920.2007.01286.x. [DOI] [PubMed] [Google Scholar]
- 25.Ojha A, Hatfull GF. 2007. The role of iron in Mycobacterium smegmatis biofilm formation: the exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth. Mol Microbiol 66:468–483. doi: 10.1111/j.1365-2958.2007.05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banin E, Vasil ML, Greenberg EP. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci U S A 102:11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. 2011. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol 193:3606–3617. doi: 10.1128/JB.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Frye JG, McClelland M, Harshey RM. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol Microbiol 52:169–187. doi: 10.1111/j.1365-2958.2003.03977.x. [DOI] [PubMed] [Google Scholar]
- 29.Singh PKP MR, Greenberg EP, Welsh MJ. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 30.López D, Vlamakis H, Kolter R. 2010. Biofilms. Cold Spring Harbor Perspect Biol 2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue T, Shingaki R, Hirose S, Waki K, Mori H, Fukui K. 2007. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J Bacteriol 189:950–957. doi: 10.1128/JB.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Louden BC, Haarmann D, Lynne A. 2011. Use of blue agar CAS assay for siderophore detection. J Microbiol Biol Educ 12:51–53. doi: 10.1128/jmbe.v12i1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol 5:30. doi: 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlier A, Burbank L, Von Bodman SB. 2009. Identification and characterization of three novel EsaI/EsaR quorum-sensing controlled stewartan exopolysaccharide biosynthetic genes in Pantoea stewartii ssp. stewartii. Mol Microbiol 74:903–913. doi: 10.1111/j.1365-2958.2009.06906.x. [DOI] [PubMed] [Google Scholar]
- 35.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi K-H, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 37.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM. 1995. Four new derivatives of the broad host range cloning vector pBBR1-MCS carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 38.Cheng H-P, Walker GC. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol 180:5183–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peigne C, Bidet P, Mahjoub-Messai F, Plainvert C, Barbe V, Médigue C, Frapy E, Nassif X, Denamur E, Bingen E, Bonacorsi S. 2009. The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect Immun 77:2272–2284. doi: 10.1128/IAI.01333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. 2013. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41:348–352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 42.Bagg A, Neilands JB. 1987. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 43.De Lorenzo V, Giovannini F, Herrero M, Neilands JB. 1988. Metal ion regulation of gene expression: Fur repressor-operator interaction at the promoter region of the aerobactin system of pCoIV-K30. J Mol Biol 203:875–884. doi: 10.1016/0022-2836(88)90113-1. [DOI] [PubMed] [Google Scholar]
- 44.Münch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- 45.Yeowell HN, White JR. 1982. Iron requirement in the bactericidal mechanism of streptonigrin. Antimicrobial Agents Chemother 22:961–968. doi: 10.1128/AAC.22.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ham JH, Majerczak DR, Arroyo-Rodriquez AS, Mackey DM, Coplin DL. 2006. WtsE, an AvrE-family effector protein from Pantoea stewartii subsp. stewartii, causes disease-associated cell death in corn and requires a chaperone protein for stability. Mol Plant Microbe Interact 19:1092–1102. doi: 10.1094/MPMI-19-1092. [DOI] [PubMed] [Google Scholar]
- 47.Madden LV, Hughs G, van den Bosch F. 2007. The study of plant disease epidemics. APS Press, St Paul, MN. [Google Scholar]
- 48.Patrick JE, Kearns DB. 2012. Swarming motility and the control of master regulators of flagellar biosynthesis. Mol Microbiol 83:14–23. doi: 10.1111/j.1365-2958.2011.07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butcher J, Stintzi A. 2013. The transcriptional landscape of Campylobacter jejuni under iron replete and iron limited growth conditions. PLoS One 8:e79475. doi: 10.1371/journal.pone.0079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Déziel E, Lépine F, Milot S, Villemur R. 2003. RhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005–2013. doi: 10.1099/mic.0.26154-0. [DOI] [PubMed] [Google Scholar]
- 51.Zheng M, Doan B, Schneider TD, Storz G. 1999. OxyR and SoxRS regulation of Fur. J Bacteriol 181:4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bagg A, Neilands JB. 1987. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev 51:509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glick R, Gilmour C, Tremblay J, Satanower S, Avidan O, Déziel E, Greenberg PE, Poole K, Banin E. 2010. Increase in rhamnolipid synthesis under iron-limiting conditions influences surface motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol 192:2973–2980. doi: 10.1128/JB.01601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones AM, Wildermuth MC. 2011. The phytopathogen Pseudomonas syringae pv. tomato DC3000 has three high-affinity iron-scavenging systems functional under iron limitation conditions but dispensable for pathogenesis. J Bacteriol 193:2767–2775. doi: 10.1128/JB.00069-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dellagi A, Brisset M-N, Paulin J-P, Expert D. 1998. Dual role of desferrioxamine in Erwinia amylovora pathogenicity. Mol Plant Microbe Interact 11:734–742. doi: 10.1094/MPMI.1998.11.8.734. [DOI] [PubMed] [Google Scholar]
- 56.Hao l-y, Willis DK, Andrews-Polymenis H, McClelland M, Barak JD. 2012. Requirement of siderophore biosynthesis for plant colonization by Salmonella enterica. Appl Environ Microbiol 78:4561–4570. doi: 10.1128/AEM.07867-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Q, Jiang Z, Liao J, Chen Z, Li H, Mei M, Zhang l-H. 2007. Identification of genetic markers to distinguish the virulent and avirulent subspecies of Pantoea stewartii by comparative proteomics and genetic analysis. Appl Microbiol Biotechnol 74:186–193. doi: 10.1007/s00253-006-0656-3. [DOI] [PubMed] [Google Scholar]
- 58.Coplin DL, Frederick RD, Majerczak DR, Haas ES. 1986. Molecular cloning of virulence genes from Erwinia stewartii. J Bacteriol 168:619–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 60.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.