Abstract
Persistent gastrointestinal (GI) inflammation, a hallmark of progressive HIV/SIV infection causes disruption of the GI epithelial barrier, microbial translocation and generalized immune activation/inflammation driving AIDS progression. Apart from protein regulators, recent studies strongly suggest critical roles for microRNAs (miRNAs) in regulating and managing certain aspects of the inflammatory process. To examine their immunoregulatory role, we profiled miRNA expression in colon from 12 chronic SIV-infected and 4 control macaques. After applying multiple comparisons correction, 10 (3-up and 7-down) miRNAs showed differential expression. Most notably, miR-34a showed significant upregulation in both epithelial and lamina propria leukocyte (LPL) compartments. Intense γH2AX expression in colonic epithelium and LPL confirmed the contribution of DNA damage response in driving miR-34a upregulation. SIRT1 mRNA and protein decreased significantly in both colonic epithelium and LPL. Luciferase reporter assays validated rhesus macaque SIRT1 as a direct miR-34a target. Decreased SIRT1 expression was associated with constitutively enhanced expression of the transcriptionally active form of the p65 (acetylated on lysine 310) subunit of NFκB exclusively in the LPL compartment. The intensity and number of acetylated-p65+ cells was markedly elevated in LPLs of chronically SIV-infected macaques compared to uninfected controls and localized to increased numbers of IgA+ and IgG+ plasma cells. These findings provide new insights into the potential role of the miR-34a-SIRT1-p65 axis in causing hyperactivation of the intestinal B cell system. Our results point to a possible mechanism where the normal immunosuppressive function of SIRT1 is inhibited by elevated miR-34a expression resulting in constitutive activation of acetylated-p65 (lysine 310).
INTRODUCTION
Regardless of the route of transmission, mucosal tissues, particularly, the gastrointestinal (GI) tract are targeted by HIV/SIV leading to rapid, severe, and sustained depletion of CD4+ T-cells in HIV-infected individuals and SIV-infected rhesus macaques (1-5). As disease progresses GI complications such as anorexia, weight loss and diarrhea become frequent and are being reported in patients despite the extensive use of HAART (6). Histologically, GI disease is characterized by infiltration of the lamina propria by T cells, plasma cells, macrophages and morphologic changes such as villus blunting and crypt hyperplasia. An emerging feature of HIV/SIV pathogenesis is the markedly elevated levels of microbial translocation that occurs in the later stages of infection (7-8). This phenomenon has been proposed to play a key role in driving localized and systemic immune activation, which is a well-recognized correlate of HIV/SIV disease progression. The mechanism(s) leading to increased microbial translocation (MbT) in AIDS patients remains largely unknown. However, the “leaky gut syndrome” is a preferred hypothesis, wherein lumenal bacteria and/or their products enter the intestinal lamina propria through a disrupted epithelial barrier and pass via the portal blood into the systemic circulation.
Viral replication and CD4+ T cell depletion in the LPL compartment is associated with elevated expression of proinflammatory genes and reduced expression of genes involved in maintenance of epithelial barrier, repair, digestive and metabolic functions (9-12). Further, focused longitudinal examination of individual mucosal compartments has revealed deeper insights into the molecular pathological events occurring in the intestinal LPL and epithelial compartments (13-14). While inflammation and immune activation related genes showed marked changes in the LPL compartment, genes regulating enterocyte maturation, differentiation and epithelial barrier function such as Wnt-TCF7L2, Notch signaling proteins, adherens junction, hemidesmosomes and desmosomes were found to be significantly dysregulated in the epithelial compartment following SIV infection (13-14). Overall, these studies demonstrated considerable alterations in enterocyte structure and function that could facilitate microbial translocation.
Although multiple mechanisms involving transcription factors, chromatin modifications and others such as histone modifications are known to regulate gene expression, one important mechanism mediated by small regulatory RNAs called miRNAs has gained a lot of attention recently (15). miRNAs are ~21-23 nts in length and have been described to impact almost all cellular processes by repressing gene expression at the post transcriptional level (15). A growing body of evidence indicates that HIV infection is characterized by dysregulated miRNA expression (16) including direct targeting and crippling of the miRNA biosynthesis machinery by HIV (17). Recent studies performed in SIV-infected rhesus macaques also demonstrated dysregulated miRNA expression in plasma (18), brain (19) and monocyte derived macrophages (20). We recently reported altered miRNA expression in the intestine during acute SIV infection (21). More specifically, we identified miR-190b to be significantly upregulated as early as 7 days post SIV infection and its expression remained elevated throughout SIV infection. Additional studies also suggested that miR-190b could influence disease pathogenesis by directly binding to the 3’ UTR and regulating the expression of MMTR6, a phosphotidylinositol 1-3 bisphosphatase previously shown to inhibit T cell and macrophage activation. In the present study, we performed miRNA profiling in colon during chronic SIV infection and detected marked changes in the expression of miRNAs linked to inflammation, cell cycle arrest and senescence. Among these, the expression of miR-34a, a miRNA shown to regulate apoptosis, cell cycle control and senescence (22-24) was markedly increased in both colonic epithelial and LPL compartments. Further, increased miR-34a expression was associated with reduced expression of SIRT1, a longevity inducing anti-inflammatory, anti-stress and anti-aging class III histone deacetylase in the colonic epithelium and LPL. Most noticeably, reduced SIRT1 expression in the LPL compartment was accompanied by enhanced expression of acetylated p65 (lysine 310) suggesting hyperactivation of the nuclear factor κB (NFκB) signaling pathway. Furthermore, enhanced constitutive expression of acetylated p65 (lysine 310) was localized to lamina propria plasma cells. The results of our study indicate that the miR-34a-SIRT1-acetyl p65 axis represents a potential molecular pathological mechanism that could drive chronic hyperactivation of the intestinal B cell system, leading to their dysfunction.
MATERIALS AND METHODS
Animal ethics statement
All experiments using rhesus macaques were approved by the Tulane University Institutional Animal Care and Use Committee (Protocol No-3574). The Tulane National Primate Research Center (TNPRC) is an Association for Assessment and Accreditation of Laboratory Animal Care International accredited facility (AAALAC #000594). The NIH Office of Laboratory Animal Welfare assurance number for the TNPRC is A3071-01. All clinical procedures, including administration of anesthesia and analgesics, were carried out under the direction of a laboratory animal veterinarian. Animals were anesthetized with ketamine hydrochloride for blood collection procedures. Intestinal resections were performed by laboratory animal veterinarians. Animals were pre-anesthetized with ketamine hydrochloride, acepromazine, and glycopyrolate, intubated and maintained on a mixture of isoflurane and oxygen. Buprenorphine was given intra-operatively and post-operatively for analgesia. All possible measures are taken to minimize discomfort of all the animals used in this study. Tulane University complies with NIH policy on animal welfare, the Animal Welfare Act, and all other applicable federal, state and local laws.
Animals and tissue collection
Colon tissues were collected from a total of sixty one Indian-origin rhesus macaques including forty five animals infected with pathogenic strains of SIV that use CCR5 in vivo and sixteen uninfected macaques. All macaques were infected with SIVmac251. Animal IDs, duration of infection, route of infection, plasma and colonic viral loads and intestinal histopathology in all SIV-infected animals are provided in Table I. For intravenous inoculation, a viral dose of 100 TCID50 was used. However, for intravaginal and intrarectal inoculation, a higher viral dose of 500 and 1000 TCID50, respectively, was used. Twelve chronic SIV-infected animals (Table I) and four control animals (Table I) were used for TLDA miRNA profiling. An additional forty-one animals comprising 5 animals each at 7-10 and 13-14DPI (days post infection), 7 each at 21 and 90DPI, 8 chronic SIV-infected and 9 control macaques (Table I) were included for qRT-PCR confirmation and in situ hybridization studies. Data regarding viral inoculum, duration of infection, plasma and intestinal viral loads for the 24 animals at 7-10, 13-14, 21 and 90DPI have been previously reported (21). In the control group, colon resection segments (~5 cm long) were collected from three macaques (HF54-Pre, HR42-Pre, HD08-Pre) before SIV infection (Table I).
Table 1.
Animal IDs, duration and route of SIV infection, viral loads and intestinal histopathology in chronic SIV-infected macaques.
|
| |||||||
|---|---|---|---|---|---|---|---|
| Animal ID |
Duration of infection (days post infection) |
Route of Infection |
Viral load- Plasma Copies/mL (106) |
Viral load- Colon Copies/mg RNA (106) |
Histopathology (colon) |
Histopathology (Jejunum) |
Colon Histo- pathology score |
|
| |||||||
| Chronic SIV-Infected | |||||||
|
| |||||||
| FE53* | 140 | I/rectal | 0.3 | 5 | Mild colitis | ND | 1 |
| FT11* | 145 | I/v | 500 | 2075 | Moderate colitis | Mild enteritis | 2 |
| GH25* | 148 | I/v | NA | 300 | Mild suppurative colitis | Mild enteritis | 1 |
| CP47* | 151 | I/v | NA | 0.9 | Mild colitis | ND | 1 |
| HI68* | 158 | I/vag | 8 | 3 | Mild colitis | ND | 1 |
| HB31* | 180 | I/v | 3000 | 200 | Lymphoid hyperplasia | Lymphoid hyperplasia | 1 |
| HB48* | 180 | I/v | 20 | 0.8 | ND | Mild enteritis | 0 |
| GK31* | 180 | I/v | 30 | 3 | ND | ND | 0 |
| GA19* | 180 | I/v | 100 | 600 | Lymphoid hyperplasia | ND | 1 |
| DV42* | 226 | I/vag | 0.1 | 1 | Mild colitis | ND | 1 |
| HG45* | 281 | I/vag | 0.9 | 0.3 | ND | ND | 0 |
| HG58* | 286 | I/vag | 0.007 | 0.2 | ND | ND | 0 |
|
| |||||||
| HD08 | 90 | I/v | NA | 300 | Moderate colitis/cryptitis | Moderate enteritis | 2 |
| DE50 | 129 | I/vag | NA | 0.3 | Mild colitis | ND | 1 |
| HN15 | 177 | I/vag | NA | 0.3 | Granulomatous colitis | Granulomatous enteritis | 3 |
| HL01 | 180 | I/vag | 7 | 0.8 | ND | Mild Amyloidosis | 0 |
| HF27 | 180 | I/v | 40 | 0.04 | Lymphoid hyperplasia | Lymphoid hyperplasia | 1 |
| HV95 | 180 | I/v | 10 | 0.1 | Moderate Colitis | ND | 2 |
| FE24 | 271 | I/v | 3.1 | 0.3 | Mild colitis | ND | 1 |
| EI15 | 300 | I/v | 0.4 | 0.9 | Mild colitis | Mild enteritis | 1 |
| R191 | 786 | I/v | NA | NA | Severe colitis | ND | 3 |
|
| |||||||
| Uninfected Controls | |||||||
|
| |||||||
| EL66* | NA | NA | NA | NA | NA | NA | 0 |
| EH70* | NA | NA | NA | NA | NA | NA | 0 |
| EH80* | NA | NA | NA | NA | NA | NA | 0 |
| FK25* | NA | NA | NA | NA | NA | NA | 0 |
|
| |||||||
| GT20 | NA | NA | NA | NA | NA | NA | 0 |
| IC52 | NA | NA | NA | NA | NA | NA | 0 |
| HT74 | NA | NA | NA | NA | NA | NA | 0 |
| IH95 | NA | NA | NA | NA | NA | NA | 0 |
| IT62 | NA | NA | NA | NA | NA | NA | 0 |
| IT13 | NA | NA | NA | NA | NA | NA | 0 |
| EJ34 | NA | NA | NA | NA | NA | NA | 0 |
| HF54-Pre | NA | NA | NA | NA | NA | NA | 0 |
| HR42-Pre | NA | NA | NA | NA | NA | NA | 0 |
| HD08-Pre | NA | NA | NA | NA | NA | NA | 0 |
| HE68 | NA | NA | NA | NA | NA | NA | 0 |
| HP84 | NA | NA | NA | NA | NA | NA | 0 |
|
| |||||||
Colon tissue from 12 chronic SIV-infected (FE53-HG58) and 4 uninfected control (EL66-FK25) macaques shown above the dotted line was used for TLDA miRNA profiling. Colon tissues from all SIV-infected (with exception of CP47, HG58 and R191) and 9 uninfected (GT20-HR42) were used for miRNA qRT-PCR confirmation studies Colon tissues from R191 and three uninfected controls (HD08-Pre, HE68 and HP84) were used for immunofluorescence studies. I/v- Intravenous, I/vag- Intravaginal, I/rectal- Intrarectal , NA- Not applicable for Uninfected controls and Not available for Chronic SIV-infected macaques, ND- None detected
All tissues were collected at necropsy in RNAlater® (Ambion, TX) for total RNA extraction and qRT-PCR. For histopathologic evaluation, colon tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned at 7 μM and stained with hematoxylin and eosin for analysis. Sections were examined in a blinded fashion and histological signs of inflammation was scored semi quantitatively on a scale of 0 to 3 as follows: 0, within normal limits; 1, mild; 2, moderate; 3, severe. In addition, the presence of crypt dilatation/cryptitis, amyloidosis and lymphoid hyperplasia were also recorded (Table I). Mild changes such as lymphoid hyperplasia were assigned a score of 1.
Global microRNA profiling using Taqman low density arrays
For TaqMan Low Density Array (TLDA) studies ~350 ng of total RNA from intact colon tissue was first reverse transcribed, pre-amplified and loaded onto TLDA cards and processed according to the manufacturer’s recommended protocol described previously (21).
Quantitative real-time TaqMan stem loop microRNA and SYBR green RT-PCR
Expression of miR-34a and SIRT1 mRNA was further investigated in both colonic epithelium and LPL compartments using qRT-PCR as previously described (21). For SIRT1 qRT-PCR assay, ~1 μg of total RNA was first reverse transcribed using the SuperScript. III First-Strand Synthesis System for RT-PCR kit following the manufacturer’s protocol. Each qRT-PCR reaction (20 μl) contained the following: 2X Power SYBR Green Master Mix without uracil-N-glycosylase (12.5 μl), forward and reverse primer (SIRT1 (For-5’-AAGATGA GCCTGATATTCCAGAGAGAGCTG-3’, Rev-5’-GCCTCTTGATCATCTCCATCAGTCCCAAA-3’), GAPDH (For-5’-CAAGAGAGGCATTCTCACCCTGAA-3’, Rev-5’-TGGTGCCAGA TCTTCTCCATGTC-3’) and Beta-actin (For-5’-CAACAGCCTCAAGATCGTCAGCAA-3’, Rev-5’-GAGTCCTTCCACGATACCAAAGTTGTC-3’) (200 nM) and cDNA (4 μl).
Colonic epithelial and lamina propria cell isolation
In order to determine the mucosal compartment contributing to miR-34a upregulation we separated the intestinal epithelial cells from the underlying LPLs and fibrovascular stroma as described previously (21).
In situ hybridization and immunofluorescence
In situ hybridization for miR-34a was performed using LNA-modified DNA probes (Exiqon Inc, Denmark). Briefly, 7 μm formalin-fixed, paraffin-embedded tissue sections were first deparafinized, rehydrated in descending series of ethanol and pretreated in a microwave with citrate buffer (Antigen unmasking solution; Vector Laboratories, Burlingame, CA) for 20 minutes at high power according to the manufacturer’s instructions. Thereafter, sections were thoroughly washed, placed in a humidified chamber, and hybridized overnight at 50 °C with 60 nM Digoxigenin labeled LNA-modified miR-34a DNA probes (5DigN/ACAACCAGCTAAGACACTGCCA/3Dig_N/, RNA Tm= 85 °C) or scrambled probe (5DigN/GTGTAACACGTCTATACGCCCA/3Dig_N/, RNA Tm= 87 °C) in 1x microRNA in situ hybridization buffer (Exiqon Inc, Denmark). After hybridization slides were washed with 5x SSC (standard saline citrate buffer), 1x SSC, 0.2x SSC, and blocked with Dako protein free blocker (Dako Laboratories) for one hour. Fab fragments of an anti-digoxigenin antibody conjugated with alkaline phosphatase (Roche Diagnostics Corporation, Penzberg, Germany) were used to detect digoxigenin-labeled probes. Positive signals were detected by incubating the sections with permanent red (Dako Laboratories) for seven minutes. Controls included matched tissues hybridized with LNA-modified scrambled probes.
Immunofluorescence for detecting γH2A.X (9F3, 1 in 250) (Abcam), acetyl p65-lysine 310 (1 in 100) (Genetex Incorp), SIRT1 (E104, 19A7AB4, 1F3) (Abcam), SIRT1 (Bioss), SIRT1 (LS-B8356, 1 in 62.5) (LifeSpan Biosciences) and p21 (12D1, 1 in 50) (Cell signaling) was done as described previously (10). Immunophenotyping of acetylated p65 positive cells was done using CD68 (1 in 20) (Dako, Denmark), CD163 (1 in 50) (Serotec, Raleigh, NC), CD3 (1 in 20) (Dako, Denmark), anti-rhesus IgA (1 in 250), IgG (1 in 250) and appropriate Alexa fluor conjugated secondary antibodies.
Quantitative image analysis
Quantitation of cells and regions of interest (ROI) labeled by LNA-modified miR-34a in situ hybridization probes was performed using Volocity 5.5 software (PerkinElmer Inc, MA, USA) after capturing images on a Leica confocal microscope. Several ROI were hand drawn on the epithelial and LPL regions in the images from colon. The data was first graphed and then analyzed using Mann Whitney U test employing the Prism® v5 software (GraphPad software). p< 0.05 was considered statistically significant.
Cloning of 3’-UTR of SIRT1 mRNA and dual-glo luciferase reporter gene assay
The 3’ UTR of the rhesus SIRT1 mRNA contains two predicted miR-34a binding sites (TargetScan 6.2) 31 [Site 1: UUCCACAAGUAUUAAACUGCCAA (nt 891-897)] and [Site 2: CCAGCUAGGACCAUUACUGCCAG (nt 1434-1440)] (Table II). Accordingly, a G-block double stranded DNA sequence (~125 bases long) containing both miR-34a binding sites along with ~30 bases flanking the 5’ and 3’ ends of each site was synthesized (IDTDNA Technologies Inc, IA) for cloning into the pmirGLO dual luciferase vector (Promega Corp, Madison, WI). A second G-block sequence with the exact same flanking UTR sequences but with both miR-34a binding sites (total 14 nucleotides from both sites) deleted was also synthesized to serve as a negative control. Both oligonucleotide sequences were synthesized with a Pme1 site on the 5’ and Xba1 site on the 3’ end for directional cloning. The pmirGLO vector was first cut with Pme1 and Xba1 restriction enzymes, gel purified and ligated with either the wild type sequence containing miR-34a binding sites (SIRT1-WT) or deleted sequence (SIRT1-Del). HEK293 cells were plated at a density of 5x104 cells per well of a 96 well plate. At 60% confluence, cells were co-transfected with ~100 ng of SIRT1-WT or SIRT1-Del UTR miRNA luciferase reporter vector and 100 nM of miR-34a or negative control mimic (CCACAACCUCCUAGAAAGAGUAGA) using the Dharmafect-Duo transfection reagent (ThermoFisher Scientific). In separate wells, cells were also transfected with pmirGLO vector (Promega Corp) as a normalization control. After 72 h, the Dual Glo luciferase assay was performed according to the manufacturer’s recommended protocol using the BioTek H4 Synergy plate reader (BioTek, Winooski, VT). The normalized Firefly to Renila ratio was calculated to determine the relative reporter activity. Experiments were performed in 6 replicates and repeated twice.
Table II.
Schematic representation of SIRT1 3’ UTR depicting predicted binding site for miR-34a. Alignment of SIRT1 mRNA sequence with miR-34a: top strand- SIRT1 mRNA; bottom strand-miR-34a.
|
| |||||
|---|---|---|---|---|---|
| Gene | GenBank Access Number |
Site Conservation | Binding sites on 3’ UTR |
Target Site sequence | Prediction algorithm |
|
| |||||
| SIRT1 (proximal site) |
NM_001142498 | Human/Chimp/Macaque/ Orangutan/Mouse |
888-894 | 5' …UUCCACAAGUAUUAAACUGCCAA… 3' | || || | 3' UGUUGGUCGAUUCUGUGACGGU 5’ |
TargetScan (38) |
|
| |||||
| SIRT1 (distal site) |
NM_001142498 | Human/Chimp/Macaque/ Orangutan/Mouse |
1428-1434 | 5’ ….CCAGCUAGGACCAUUACUGCCAG..3’ || | | | || | || | || 3' UGUUGGUCGAUUCUG--------UGACGGU 5’ |
TargetScan (38) |
|
| |||||
Quantitation of mucosal viral loads
Total RNA samples from all SIV-infected animals were subjected to a quantitative real-time TaqMan two-step RT-PCR analyses to determine the viral load in colon samples. Briefly, primers and probes specific to the SIV LTR sequence were designed and used in the TaqMan real-time RT-PCR assay. Probes were conjugated with a fluorescent reporter dye (FAM) at the 5′ end and a quencher dye at the 3′ end. Fluorescence signal was detected with an ABI Prism 7900 HT sequence detector (Life Technologies Corp). Data were captured and analyzed with Sequence Detector Software (Life Technologies Corp). Viral copy number was determined by plotting CT values obtained from the colon and jejunum samples against a standard curve (y= −3.384x + 39.029) (r2=0.998) generated with in vitro transcribed RNA representing known viral copy numbers.
Data analysis and data availability
TLDA-SDS run files from twelve SIV-infected and four control animals were loaded onto Relative Quantification (RQ) Manager Software v1.2.1 and analyzed using automatic baseline settings and manual threshold of 0.2. One (EL66) of the four control samples was selected as a calibrator and set to 1. The remaining 15 samples were expressed as an n-fold difference relative to the calibrator sample. The results from the RQ manager analysis containing five columns (well, sample, detector, task and CT values) were saved as a tab-delimited text file, imported and analyzed using the DataAssist v3.01 software (Life Technologies, NY) employing global normalization method as this method has been proposed to be more sensitive and accurate for analyzing high throughput TaqMan microRNA qRT-PCR than endogenous controls (25-26). The CT upper limit was set to 33 meaning that all miRNA detectors with a CT value greater than or equal to 33 were excluded. Multiple comparisons correction for TLDA miRNA profiling (simultaneously applied to all 768 miRNA targets on cards A&B) was performed using Benjamini-Hochberg method. p values<0.05 were considered significant. TLDA miRNA data were deposited with GEO (Accession number- GSE60773, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60773).
For miR-34a and SIRT1 qRT-PCR studies, one uninfected control macaque with the highest (for miR-34a) or lowest (for SIRT1) ΔΔCT value served as the calibrator/reference and assigned a value of 1. All differentially expressed miRNAs or mRNAs in SIV-infected and other control macaques are shown as an n-fold difference relative to this macaque. This approach was preferred over calculating miRNA/mRNA fold change using an average of all control animal ΔCT values mainly to facilitate graphing the control samples so that variation within the control group can be displayed. Individual miR-34a and SIRT1 mRNA qRT-PCR data was analyzed by non-parametric Wilcoxon’s rank sum test for independent samples using RealTime STATMINER™ software (Integromics). A Spearman's non-parametric one-tailed correlation analysis was performed to determine the degree of association between colon inflammation scores/tissue viral loads and miR-34a fold expression. Firefly/Renila ratios were analyzed using one-way ANOVA and post-hoc comparisons were performed using Tukey’s multiple comparisons test.
RESULTS
Mucosal and plasma viral Loads, CD4+ T cell counts and intestinal histopathology
All chronic SIV-infected macaques obtained at necropsy had substantial viral loads in the colon that ranged from 0.04 × 106 to 2075 × 106 copies/ mg total RNA with a median of 0.9 × 106 copies/ mg total RNA (Table I). Similar to colonic viral loads, plasma viral loads in chronic SIV-infected macaques ranged from 0.3 × 106 to 5000 × 106 copies/mL with a median of 15 × 106 copies/mL. Plasma viral loads were not available for 6 animals (GH25, CP47, HD08, DE50, HN15 and R191).
All chronic SIV-infected animals with the exception of 4 (HF27, HB48, GK31, HG58) progressed to AIDS. CD4+ T cell data were only available for 8 chronic SIV-infected macaques and as shown in supplemental figure 1 all 8 had significant mucosal CD4+ T cell depletion compared to uninfected control macaques. Histologic evaluation of hematoxylin and eosin stained sections of colon (Table I) from all SIV-infected macaques revealed the presence of mild to severe colitis including other intestinal lesions such as crypt abscess/cryptitis (Table I). In contrast to colon, only 9 out of the 21 chronic SIV-infected macaques had mild to moderate enteritis (Table I). Opportunistic pathogens such as Mycobacterium avium intracellulare were not detected in 20 out of 21 chronic SIV-infected macaques.
Chronic SIV infection of the GI tract/immune system is characterized by marked alterations in the expression of inflammation associated miRNAs
We performed global miRNA profiling of colon tissue from 12 chronically SIV-infected and 4 uninfected control macaques using the TLDA cards to determine if miRNA expression was altered in the GI tract/immune system during chronic SIV infection. After applying multiple comparisons correction, 10 miRNAs (3-up and 7-downregulated) were found to be statistically significant (Adjusted p<0.05) and differentially expressed following analysis using DataAssist™ software version 3.01 (Table III).
Table III.
Changes in microRNA expression in colon during chronic SIV infection.
|
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA ID | Chronic SIV (n=12) | Uninfected Controls (n=4) | Fold Change |
Adjusted p value |
||||||||||||||
|
| ||||||||||||||||||
| HB31 | HB48 | GK31 | GA19 | GH25 | FT11 | DV42 | CP47 | HI68 | FE53 | HG45 | HG58 | EL66 | EH70 | EH80 | FK25 | |||
|
| ||||||||||||||||||
| hsa-miR-181-a* | 24.69 | 23.93 | 24.36 | 24.71 | 24.12 | 24.72 | 24.90 | 25.04 | 25.62 | 24.53 | 24.18 | 24.98 | 23.91 | 23.99 | 23.72 | 24.73 | −1.5 | 0.0027 |
| hsa-miR-628-5p | 25.93 | 25.04 | 25.40 | 25.51 | 24.78 | 24.97 | 25.08 | 25.09 | 25.21 | 24.99 | 24.17 | 25.18 | 24.10 | 24.37 | 24.12 | 25.10 | −1.6 | 0.0027 |
| hsa-miR-378 | 25.03 | 24.87 | 25.34 | 25.27 | 24.65 | 25.70 | 25.27 | 25.76 | 26.11 | 24.71 | 25.07 | 25.46 | 24.93 | 24.87 | 24.95 | 25.83 | −1.2 | 0.0174 |
| hsa-miR-424 | 25.92 | 25.16 | 25.54 | 25.32 | 25.13 | 25.27 | 25.97 | 25.92 | 26.52 | 25.18 | 26.09 | 25.92 | 26.07 | 26.06 | 25.84 | 26.94 | 1.4 | 0.0174 |
| hsa-miR-425* | 25.22 | 24.29 | 24.98 | 25.06 | 25.19 | 25.41 | 25.44 | 25.38 | 25.91 | 24.68 | 24.96 | 25.13 | 24.44 | 24.57 | 24.24 | 24.97 | −1.5 | 0.0174 |
| hsa-miR-454* | 29.52 | 28.81 | 29.09 | 29.78 | 29.25 | 29.79 | 29.77 | 29.31 | 29.51 | 29.21 | 29.02 | 29.39 | 28.23 | 28.60 | 28.42 | 28.97 | −1.8 | 0.0174 |
| hsa-miR-223* | 25.99 | 24.89 | 25.66 | 25.48 | 25.57 | 24.65 | 25.35 | 26.13 | 26.65 | 26.33 | 26.53 | 26.75 | 26.54 | 27.23 | 26.67 | 28.97 | 2.7 | 0.019 |
| hsa-miR-24-2* | 25.36 | 24.79 | 25.31 | 24.98 | 25.04 | 24.75 | 24.92 | 25.58 | 25.33 | 24.98 | 24.65 | 25.22 | 24.51 | 24.78 | 24.40 | 25.18 | −1.3 | 0.019 |
| hsa-miR-520c-3p | 32.61 | 29.79 | 32.98 | 31.59 | 31.06 | 31.12 | 31.94 | 32.16 | 31.29 | 31.80 | 31.35 | 31.20 | 30.51 | 30.43 | 30.33 | 31.20 | −1.8 | 0.019 |
| hsa-miR-34a | 21.44 | 20.92 | 20.74 | 20.39 | 19.43 | 19.67 | 20.32 | 21.32 | 21.14 | 20.82 | 19.96 | 21.01 | 21.76 | 21.97 | 20.97 | 22.16 | 2.2 | 0.0306 |
|
| ||||||||||||||||||
| hsa-miR-34a* | 24.17 | 23.40 | 23.32 | 22.79 | 22.21 | 23.14 | 23.90 | 24.83 | 24.38 | 24.37 | 23.53 | 24.67 | 24.99 | 25.56 | 24.60 | 25.11 | 2.6 | 0.0515 |
| hsa-let-7b | 18.06 | 17.44 | 17.62 | 17.68 | 17.00 | 17.77 | 17.98 | 18.00 | 18.51 | 17.24 | 17.51 | 17.87 | 17.98 | 18.21 | 17.72 | 18.68 | 1.3 | 0.0578 |
| hsa-miR-193b* | 27.03 | 26.74 | 26.67 | 26.42 | 25.97 | 26.56 | 27.79 | 28.64 | 28.21 | 28.00 | 26.18 | 28.62 | 27.80 | 28.45 | 28.01 | 29.03 | 2.3 | 0.0589 |
| hsa-miR-146b-5p | 16.11 | 15.98 | 16.81 | 15.35 | 15.29 | 14.79 | 15.72 | 16.44 | 16.67 | 16.20 | 16.47 | 16.59 | 17.21 | 17.05 | 16.23 | 18.16 | 2.1 | 0.0719 |
| hsa-miR-30d | 20.05 | 20.23 | 20.70 | 20.38 | 20.08 | 20.48 | 20.40 | 21.02 | 20.71 | 19.94 | 19.81 | 20.76 | 19.73 | 19.90 | 19.86 | 20.68 | −1.3 | 0.0758 |
| hsa-miR-744* | 26.22 | 25.74 | 25.97 | 26.25 | 25.91 | 27.28 | 26.71 | 26.78 | 27.15 | 25.89 | 25.96 | 26.54 | 25.72 | 25.56 | 25.53 | 26.51 | −1.5 | 0.0796 |
| hsa-miR-18a | 20.89 | 20.28 | 20.64 | 20.25 | 20.09 | 20.69 | 21.24 | 20.44 | 20.70 | 21.05 | 21.14 | 21.00 | 20.87 | 21.18 | 20.79 | 21.97 | 1.4 | 0.0825 |
| hsa-miR-21 | 15.76 | 15.79 | 16.05 | 14.98 | 14.68 | 14.42 | 15.07 | 15.45 | 15.93 | 15.35 | 15.60 | 16.25 | 15.91 | 15.98 | 15.70 | 16.66 | 1.6 | 0.0825 |
| hsa-miR-422a | 24.87 | 24.01 | 24.74 | 24.78 | 24.01 | 24.30 | 23.57 | 24.44 | 24.92 | 23.44 | 23.89 | 23.80 | 23.43 | 23.23 | 22.88 | 24.08 | −1.8 | 0.0825 |
| hsa-miR-656 | 26.98 | 25.27 | 26.07 | 26.86 | 26.39 | 25.95 | 25.89 | 25.98 | 26.35 | 25.02 | 25.63 | 25.58 | 24.64 | 24.96 | 24.64 | 26.02 | −1.9 | 0.0831 |
| hsa-miR-22* | 23.08 | 22.78 | 23.40 | 23.33 | 23.07 | 22.48 | 23.64 | 23.54 | 23.13 | 23.15 | 22.37 | 23.33 | 23.32 | 23.39 | 23.42 | 24.53 | 1.4 | 0.0845 |
| hsa-miR-30c | 15.71 | 14.74 | 14.86 | 14.83 | 14.55 | 14.76 | 14.88 | 15.15 | 15.57 | 14.49 | 14.55 | 14.92 | 14.51 | 14.55 | 14.17 | 15.17 | −1.3 | 0.0947 |
| hsa-miR-129-3p | 26.03 | 26.55 | 25.28 | 25.31 | 26.12 | 25.96 | 26.07 | 25.54 | 26.79 | 24.73 | 24.34 | 24.95 | 24.48 | 25.00 | 24.19 | 25.34 | −1.7 | 0.095 |
|
| ||||||||||||||||||
The table shows raw CT and fold change for all differentially expressed (adjusted p<0.05, above the bar) and miRNAs that showed a tendency to be statistically significant (p<0.1, below the bar) after applying multiple comparisons correction (Benjamini-Hochberg adjusted p values for false discovery rate) in the colon of twelve chronic SIV-infected macaques.
Among the 3 upregulated miRNAs (cards A&B), the expression of miR-223* and miR-34a exceeded 2-fold and miR-424 showed ~1.4 fold increase in colon of chronic SIV-infected macaques (Table III). The expression of 7 miRNAs decreased from 1.2 to 1.8 fold (Table III). At least, 13 other miRNAs that comprised three previously well characterized inflammation associated miRNAs [let-7b (p=0.0578), miR-146b-5p (p=0.0719) and miR-21 (p=0.0825)] showed a tendency to be statistically significant (p < 0.1) (Table III). When multiple comparisons correction was not applied, 68 (31 upregulated and 37 downregulated) miRNAs were found to be differentially expressed (Supplemental Table 1). Among the upregulated miRNAs was miR-190b (3.2 fold increase), which we recently reported to be significantly upregulated throughout the course of SIV infection primarily in response to viral infection/replication and not immune/inflammatory response accompanying viral replication (21). These findings show that chronic SIV infection is characterized by marked changes in the expression of multiple inflammation related miRNAs in the colon and that the profile is dominated by downregulated miRNAs.
miR-34a is an inflammation associated miRNA and its expression is significantly elevated in both colonic epithelial and LPL compartments of chronic SIV-infected macaques
Among the differentially expressed miRNAs, we decided to focus on miR-34a because it is a highly conserved miRNA previously linked to inflammation, apoptosis and cellular senescence /cell cycle arrest (22-24). Further, miR-34a has been directly linked to “inflammaging”, an aging related state characterized by systemic chronic inflammation proposed to promote premature aging of HIV-infected patients (27). Furthermore, miR-34a has also been associated with the induction of cellular senescence in normal colonic epithelial cells in response to treatment with irinotecan, a topoisomerase I inhibiting anti-cancer drug (28). Finally, miR-34a was the most abundant (lowest CT) among the 10 differentially expressed miRNAs whose expression was significantly upregulated in the colon of chronic SIV-infected macaques (Table III).
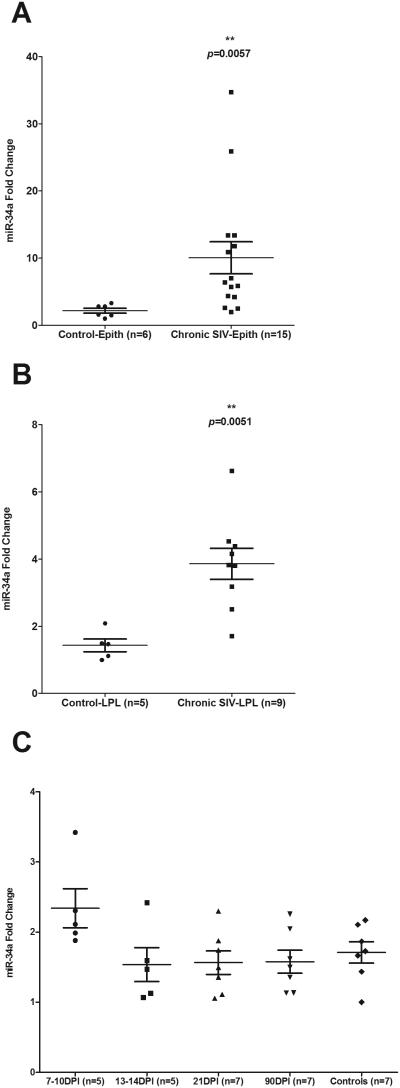
Since miR-34a expression was significantly elevated in intact colons of chronic SIV-infected macaques when using the TLDA cards, we next quantified its expression using miR-34a specific qRT-PCR assay separately in the colonic epithelial and LPL compartments to identify the mucosal compartment contributing to its upregulation. The protocol used for intestinal cell isolation in the present study yields epithelial cells with ~85-90% purity with minimal contamination with IELs (29). Similar purity has also been obtained for LPLs with minimum contamination with epithelial cells (29). The LPLs isolated consists mostly of lymphocytes (50-60%) but also contains substantial number of plasma cells, macrophages and smaller numbers of neutrophils, dendritic cells and eosinophils. Interestingly, miR-34a expression was significantly elevated in both epithelium (~3.8 fold increase; p= 0.0057) (Fig. 1A) and LPL compartments (~2.7 fold increase; p= 0.0051) (Fig. 1B).
FIGURE 1.
qRT-PCR quantification of miR-34a in colon during chronic (A & B) and acute (C) SIV infection. In chronically SIV-infected macaques significant elevation of miR-34a expression was found in both epithelial (A) and lamina propria leukocyte (LPL) (B) compartments of the colon. Data was analyzed using non-parametric Wilcoxon’s rank sum test. The error bars represent standard error of mean fold change within each group. Double asterisks (**) indicate statistical significance (p<0.01) compared to uninfected controls. In contrast, miR-34a expression in the colons of rhesus macaques at 7-10, 13-14, 21 and 90 days post SIV infection (C) did not differ from uninfected control macaques. Data analysis using non-parametric Kruskal-wallis test revealed no differences among the different groups.
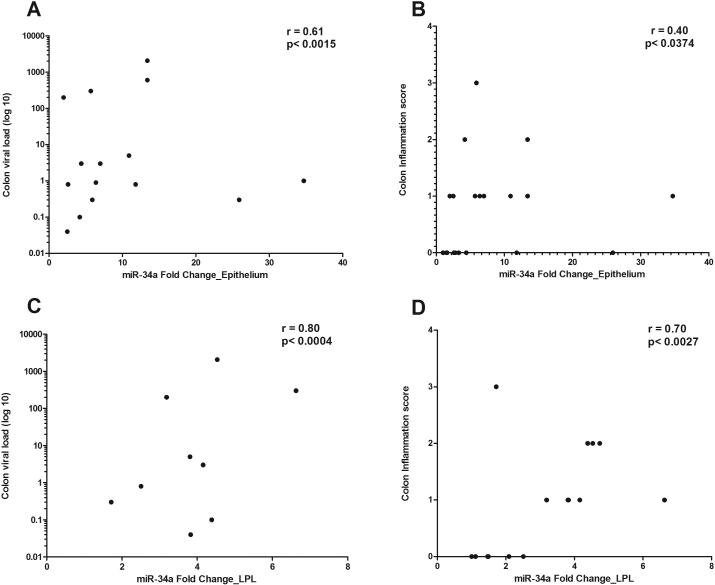
We then sought to determine the timing of miR-34a upregulation during the course of SIV infection by quantifying miR-34a expression in colon samples collected from twenty-four additional macaques at 7-10 (n=5), 13-14 (n=5), 21 (n=7) and 90 (n=7) days post infection. CD4+ T cells status, plasma and intestinal viral loads for all twenty-four animals have been previously reported (21). As shown in figure 1C, no significant increase in miR-34a expression was observed at any of the time points compared to uninfected controls. This suggests that miR-34a upregulation is tied more tightly with inflammation, which is minimal during acute infection than viral load, which is very high acutely, particularly, at 10 to 14 (peak viral load) days post infection. In addition, miR-34a fold change in both colonic epithelium and LPL compartments showed very good positive correlation with tissue viral loads and inflammation scores (Fig. 2A-D). Interestingly, the correlation was higher in the LPL (Fig 2C-D) compared to the epithelial compartment (Fig. 2A-B). These data suggested that miR-34a upregulation had a strong association with inflammation and both epithelial (Fig. 1A) and LPL (Fig. 1B) compartments contribute to its elevated expression detected in intact colon tissue (Table III). Overall, upregulation of the inflammation associated miR-34a in both epithelial and LPL compartments suggests that it could potentially exert distinct pathophysiologic effects in both mucosal compartments through epigenetic regulation of gene expression.
FIGURE 2.
Correlation of miR-34a expression in colonic epithelial (A-B) and lamina propria leukocyte (LPL) (C-D) compartments with tissue viral loads and inflammation scores. miR-34a fold change showed positive statistical correlation with tissue viral load and inflammation scores (p<0.05) in both colonic epithelium and LPLs.
Colonic crypt epithelial cells of chronic SIV-infected macaques strongly express the double stranded DNA damage response (DDR) sensor protein γH2A.X
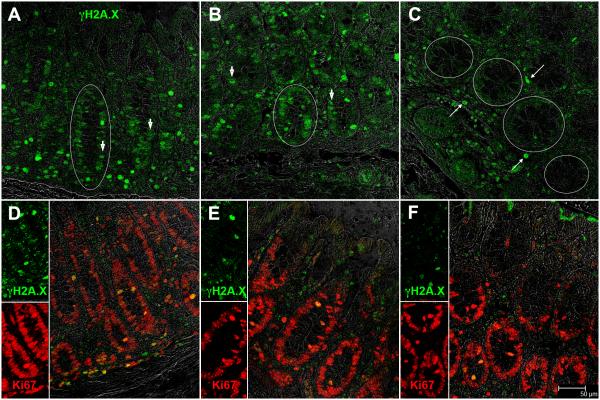
Recent studies show that miR-34a transcription is elevated in response to irradiation or oxidative stress (inflammation) induced DNA damage (30-31). Consequently, we verified the presence of DDR in the colon by determining γH2A.X expression. Both double stranded DNA breaks and telomere uncapping can induce a DDR resulting in the activation and recruitment of ATM/ATR proteins to the site of damage (32) which then phosphorylates histone H2A.X proteins on Ser-139 to facilitate the focal assembly of checkpoint and DNA repair proteins to the DNA damage site (32). Consistent with these findings, enhanced phospho-γH2A.X (Ser-139) expression was detected in colonic crypt epithelial cells of chronic SIV-infected macaques (Fig. 3A&B), an indicator of active DDR signaling. More strikingly, epithelial cells present along the entire circumference of the crypts showed intense nuclear staining for phospho-γH2A.X (white circles in Fig. 3A&B). In contrast, phospho-γH2A.X staining was either extremely weak or completely absent in the crypt epithelial cells of the control macaque (Fig. 3C). In addition, elevated phosphor-γH2A.X expression was also detected in LPLs (Fig. 3A-C).
FIGURE 3.
Colonic epithelial cells of chronic SIV-infected macaques strongly expressed the DNA damage response (DDR) sensor protein phospho-γH2A.X. (A-B). White circles in panels A and B demarcate the periphery of a crypt containing multiple phospho-γH2A.X. positive nuclei. Arrow heads (A-B) point to phospho-γH2A.X. positive crypt epithelial nuclei. In contrast, colonic epithelial cells from the normal uninfected control macaque showed extremely low to undetectable phospho-γH2A.X. expression (C). White circles in panel C demarcate the periphery of four crypts that showed extremely weak to no phospho-γH2A.X. expression in the nuclei. Long thin arrows (C) point to phospho-γH2A.X.+ LPLs. Most of the colonic epithelial cells from both SIV-infected (D-E) and control macaques (F) also stained positive for Ki67, a cell proliferation marker suggesting that these cells did not become senescent. Epithelial and LPL phospho-γH2A.X. (A-C) positive staining cells are shown in green (Alexa fluor-488). Epithelial and LPL Ki67 (D-E) positive staining cells are shown in red (Alexa fluor-568). Panels (D-F) involve double labels with γH2A.X. (green) and Ki67 (red). The individual channels (green for phospho-γH2A.X. and red for Ki67) and grey for differential interference contrast (DIC) to reveal tissue architecture appear on the left with a larger merged image on the right (D-F). Colocalization of green (phospho-γH2A.X.) and red (Ki67) appear light yellow. All panels are shown at 40X magnification.
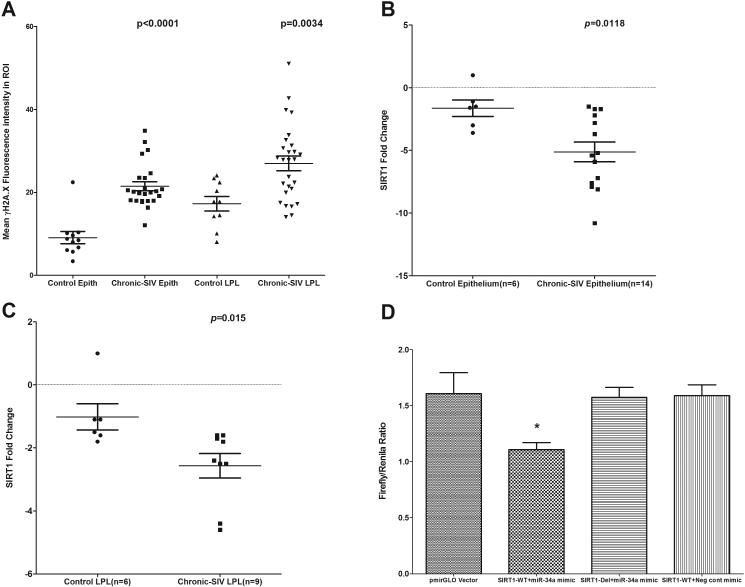
Interestingly, persistent DDR manifested by phospho-γH2A.X accumulation at DNA damage foci is also considered an initiating signal for cellular senescence/aging (33). In this context, elevated miR-34a expression in renal and endothelial cells was well demonstrated to promote cellular senescence (23-24). Accordingly, we used a combination of phosho-γH2A.X and the proliferation marker Ki67 (32) where senescent cells should stain negative for Ki67 and positive for phospho-γH2A.X. Surprisingly, most of the colonic crypt epithelial cells of chronic SIV-infected macaques strongly expressed both phospho-γH2A.X (Fig. 3A-B) and Ki67 (Fig. 3D-F) suggesting that these cells although showed clear evidence of DDR did not undergo senescence and retained the capacity to proliferate. In addition, the absence of cell cycle arrest/cellular senescence was further confirmed based on the expression pattern of cyclin-dependent kinase inhibitor p21, a cell cycle arrest/cellular senescence promoting protein that unlike Ki67 stained differentiated enterocytes near the lumen but not the crypt epithelial cells (Supplemental Figure 2). Quantitative image analysis showed significantly increased expression of phospho-γH2A.X in both colonic epithelial (p<0.0001) and LPL (p=0.0034) compartments (Fig. 4A). Interestingly, between the two mucosal compartments, phospho-γH2A.X expression in the epithelium showed better statistical significance compared to the LPL compartment (Fig. 4A). Collectively, the increased expression of phospho-γH2A.X expression in colonic crypt epithelial cells of chronic SIV-infected macaques provides compelling evidence of oxidative stress mediated DDR secondary to inflammation and represents a potential mechanism driving miR-34a upregulation (34-35). Nevertheless, the positive Ki67 staining suggests that unlike IBD patients (36), colonic crypt epithelial cells in chronic SIV-infected macaques have not been subjected to oxidative stress that is long enough (persistent DDR) to induce changes consistent with cellular senescence.
FIGURE 4.
Quantitation of cells and regions of interest (ROI) labeled by phospho-γH2A.X. was performed using Volocity 5.5 software after capturing images on a Leica confocal microscope. Several ROI were hand drawn on the epithelial and LPL regions in the images from colon of chronic SIV-infected and uninfected macaques (A). Data were analyzed using non-parametric Wilcoxon’s rank sum test. qRT-PCR revealed significantly decreased expression of SIRT1 mRNA in both colonic epithelium (p=0.0118) (B) and LPL (p=0.015) (C) compartments of chronic SIV-infected compared to uninfected control macaques. Data were analyzed using the non-parametric Wilcoxon’s rank sum test. The error bars represent standard error of mean fold change within each group. miR-34a physically associates with the 3’ UTR of rhesus macaque SIRT1 mRNA (D). The SIRT1 wild type and deleted 3′UTR sequences were inserted into the multiple cloning sites situated in the 3′ end of firefly luciferase gene in the pmirGLO vector. HEK293 cells were co-transfected with 100 nM miR-34a or negative control mimic and 100 ng of luciferase reporter constructs containing wild type (WT) or deleted (Del) SIRT1 3′UTR sequences. Firefly and Renila luciferase activities were detected using the Dual-Glo luciferase assay system 72 h after transfection. The ratio of luciferase activities (Firefly/Renilla) was calculated and normalized to the wells transfected with only unmanipulated pmir-GLO vector. Single asterisk (*) indicates statistical significance (p< 0.05) compared to cells transfected with unmanipulated pmirGLO vector.
Enhanced miR-34a expression in the colonic mucosa leads to marked downregulation of SIRT1 mRNA and protein expression
To better understand the biological significance of miR-34a upregulation we focused on SIRT1, a predicted and validated target of miR-34a (34). SIRT1 is a NAD+ -dependent class III histone/protein deacetylase known to deacetylate transcription factors, enzymes, DNA binding histone proteins, etc, and in doing so regulates critical cellular processes such as inflammation, DNA repair, cell senescence/aging, cellular metabolism and stress response (35). Further, decreased expression of SIRT1 has been detected in several chronic inflammatory conditions such as chronic obstructive pulmonary disease, atherosclerosis, obesity and DSS-induced colitis (35,37). Most importantly, SIRT1 inhibits cellular senescence/aging and also functions as a negative regulator of inflammation and immune activation by deacetylating the p65 subunit of NFκB at lysine 310 (35,37).
The 3’ UTR of the rhesus macaque SIRT1 mRNA has two predicted miR-34a binding sites (38) (Table II). While human SIRT1 mRNA has been validated as a direct target of miR-34a (34), it is not known whether mml-miR-34a can bind the 3’ UTR of rhesus macaque SIRT1 mRNA and repress its expression. Since mRNA destabilization has been proposed as a predominant mechanism by which miRNAs repress gene expression (39), we next quantified SIRT1 mRNA separately in colonic epithelium and LPLs. Interestingly, SIRT1 mRNA expression significantly decreased by ~2.4 and ~1.8 fold in colonic epithelium (p=0.0118) and LPLs (p=0.015), respectively (Fig. 4B-C). Further, co-transfection of SIRT1-WT UTR vector with miR-34a mimic resulted in significant reduction (~30%) in Firefly/Renila ratios (p<0.05)). In contrast, co-transfection of SIRT1-Del UTR vector with miR-34a restored the Firefly/Renila ratios to the level observed with unmanipulated pmirGLO vector (Fig. 4D). Interestingly, co-transfection of SIRT1-WT UTR vector with a negative control mimic did not reduce firefly/renila ratios compared to wells transfected with miR-34a mimic (p<0.05) (Fig. 4D). These results clearly show that mml-miR-34a can physically interact with the 3’ UTR of rhesus macaque SIRT1 mRNA and regulate its expression.
We next performed immunofluorescence (IF) on formalin fixed paraffin embedded (FFPE) colon sections using anti-SIRT1 specific antibodies to check if changes in mRNA expression were paralleled at the protein level. After evaluating 5 anti-human SIRT1 specific antibodies one cross-reacted with the rhesus macaque for IF studies. As shown in figure 5, SIRT1 protein expression was detected in the colonic epithelium and LPLs of chronic SIV-infected (Fig 5A-B) and control macaques (Fig. 5D-E). Further, SIRT1 protein localized to both cytoplasm and nuclei of epithelial and LPLs (Fig. 5C). Consistent with decreased SIRT1 mRNA expression, image analysis confirmed significantly (p<0.05) reduced SIRT1 protein expression in the epithelial and LPL compartments of both chronic SIV-infected compared to uninfected control macaques (Fig. 5F). Based on the results of the luciferase assay (Fig. 3C), decreased SIRT1 mRNA and protein expression in the colonic epithelium and LPL may be directly attributed to miR-34a binding and inhibition of target gene expression.
FIGURE 5.
SIRT1 protein expression is significantly reduced in the colon of chronic SIV-infected (A-B) compared to uninfected control macaques (D-E). SIRT1 protein localized to both cytoplasm and nuclei of colonic epithelium and LPLs (C). Panels A-B and D-E contain a single label (SIRT1) in green (Alexa 488). Panel C involves double labels with SIRT1 in green (Alexa 488) and Topro-3 for nuclear labeling in far-red (Alexa 647). Colocalization of green (SIRT1) and far-red (Topro-3) appear light yellow. All panels are shown at 40X magnification. Quantitation of cells and regions of interest (ROI) labeled by anti-SIRT1 antibody using Volocity 5.5 software revealed significantly decreased SIRT1 protein expression in the colonic epithelium and LPLs of chronic SIV-infected compared to uninfected control macaques (F). Several ROI were hand drawn on the epithelial and LPL regions in the images from colon. Data were analyzed using non-parametric Wilcoxon’s rank sum test.
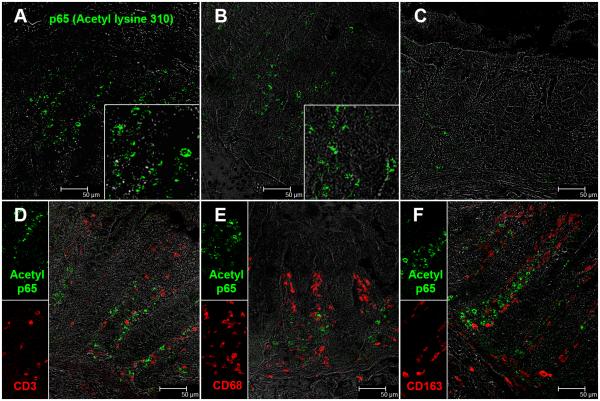
miR-34a mediated downregulation of SIRT1 was associated with constitutive expression of acetylated p65 (lysine 310) in lamina propria plasma cells
Since SIRT1 is known to deacetylate the p65 subunit of NFκB on lysine 310 (35) we hypothesized that miR-34a mediated downregulation of SIRT1 will result in constitutive expression of the transcriptionally active form of p65 (acetylated on lysine 310) in the colon. In agreement with our hypothesis, immunofluorescence studies revealed intense expression of acetylated-p65 exclusively in the LPL compartment of chronic SIV-infected macaques (Fig. 6A-B). The inset image in figure 6A-B shows a magnified view of acetylated p65 positive cells in the colonic lamina propria. This antibody detects p65 only when acetylated on lysine 310 and has been used previously for immunohistochemistry studies (40). In addition, the antibody recognizing the non-acetylated peptide has been removed by chromatography using non-acetylated peptide sequences. In contrast to chronic SIV-infected macaques, occasional acetylated-p65 (lysine 310) positive cells were detected in the colonic lamina propria of the control macaque (Fig. 6C). We next immunophenotyped acetyl p65+ cells using markers specific to macrophages (CD68, CD163) and T lymphocytes (CD3). Surprisingly, none of the markers colocalized with acetylated-p65+ suggesting that neither macrophages nor T lymphocytes were contributing to the elevated acetylated-p65 expression (Fig. 6D-F). Finally, using anti-rhesus IgA and IgG antibodies we successfully identified antibody producing plasma cells as the primary cell type contributing to enhanced acetylated-p65 (lysine 310) expression (Fig. 7A-B). IgA+/acetylated-p65+ and IgG+/acetylated-p65+ cells were rare in the control macaque (Fig. 7C-D). Further, image analysis revealed significantly increased numbers of IgA+/acetylated-p65+ and IgG+/acetylated-p65+ cells in the colonic lamina propria of chronic SIV-infected macaques compared to uninfected control macaques (Figure 8A-D). Image analysis provided additional evidence that plasma cells in the colonic lamina propria of chronic SIV-infected macaques carry a higher percentage of IgG+/acetylated-p65+ cells compared to control macaques (Fig. 8B&D). In contrast, there were very few IgG+/acetylated-p65+ cells in the 3 control macaques (Fig. 8B&D). Similar to the colon, IgA+/acetylated-p65+ cells were also detected in the jejunum but only in macaques that showed histological signs of enteritis (HD08) (Table I) (data not shown). IgA+/acetylated-p65+ cells were extremely rare in the jejunum of chronically SIV-infected macaque (GA19) that did not show histological evidence of enteritis (Table I) (data not shown). Although we could not detect acetylated-p65 in T cells and macrophages, it is possible that the transcriptionally active protein could be present at levels below the detection limit of immunofluorescence.
FIGURE 6.
Presence of increased numbers of acetylated-p65 (lysine 310) expressing cells in the colonic lamina propria of two chronic-SIV infected rhesus macaques (A-B). The inset image shows a close-up magnified view of acetylated-p65+ cells in the colonic lamina propria. Note the fewer acetylated-p65 positive cells in the uninfected control macaque (C). Acetylated-p65 positive staining cells are shown in green (Alexa fluor-488). Lymphocytes [CD3 (D)] and macrophages [CD68 (E), CD163 (F)] are not the predominant cellular source of acetylated-p65 (lysine 310). All panels involve double labels with acetylated-p65 in green and CD3 for lymphocytes and CD68/CD163 for macrophages in red. For each panel the individual channels (green for acetylated-p65, and red for CD3, CD68 and CD163) and grey for differential interference contrast (DIC) to reveal tissue architecture appear on the left with a larger merged image on the right. Note that acetylated-p65+ cells do not colocalize with CD3, CD68 and CD163. All panels are shown at 40X magnification.
FIGURE 7.
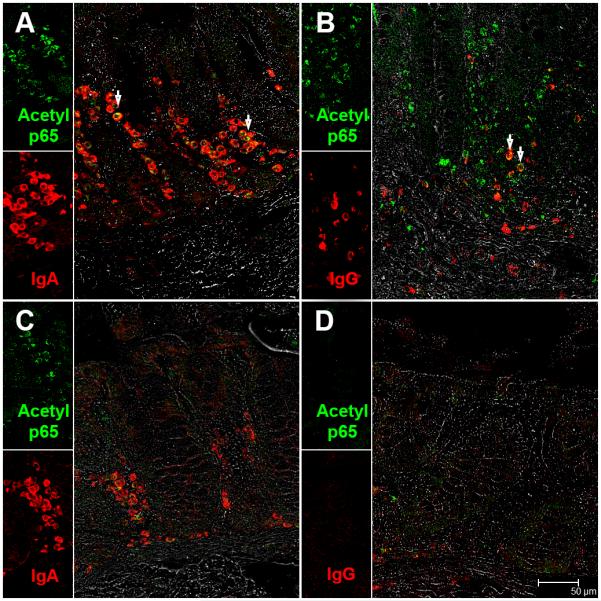
IgA (A) and IgG (B) secreting plasma cells are a major source of acetylated-p65 (lysine 310) in the colonic lamina propria of chronic-SIV infected rhesus macaques. All panels involve double labels with acetylated-p65 (green) and IgA and IgG (red) for plasma cells. For each panel the individual channels (green for acetylated-p65, and red for IgA or IgG and grey for differential interference contrast (DIC) to reveal tissue architecture appear on the left with a larger merged image on the right. Colocalization of green (acetylated-p65) and red (IgA and IgG) appear light yellow and is indicated by white arrow heads. Note the fewer IgA+ and IgA/acetylated-p65++ plasma cells in the uninfected control macaque (C). Also, notice the paucity of IgG+ and IgG/acetylated-p65++ plasma cells in the uninfected control macaque (D).All panels are shown at 40X magnification.
FIGURE 8.
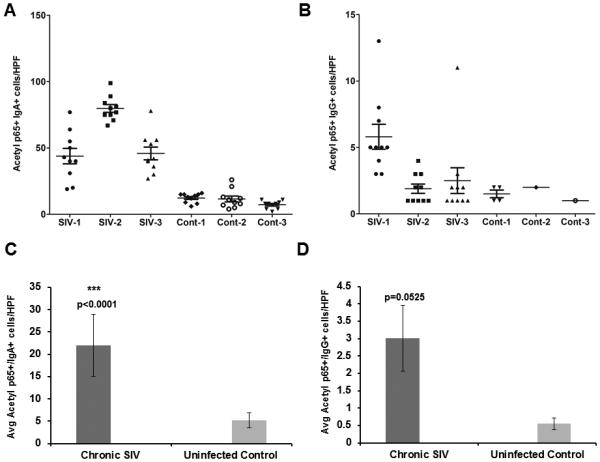
Quantification of the number of IgA/acetylated-p65++ (A) and IgG/acetylated-p65++ (B) in the lamina propria of 3 chronic SIV-infected and 3 control rhesus macaques. Positive cells were counted at an original magnification of 40X in 10 high power fields (HPF). One HPF= 0.2827 mm2. Each data point represents the number of cells in a single HPF; the error bars represent the mean values. Average IgA/acetylated-p65++ (C) and IgG/acetylated-p65++ (D) in the colonic lamina propria of 3 SIV-infected and 3 uninfected control macaques. All slides were analyzed by two investigators in a blinded manner. Data was analyzed using Wilcoxon’s rank sum test. Triple asterisks (***) indicate statistical significance (p< 0.0001) compared to uninfected controls.
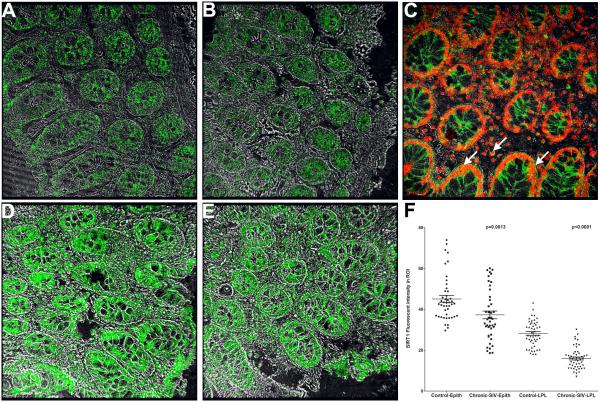
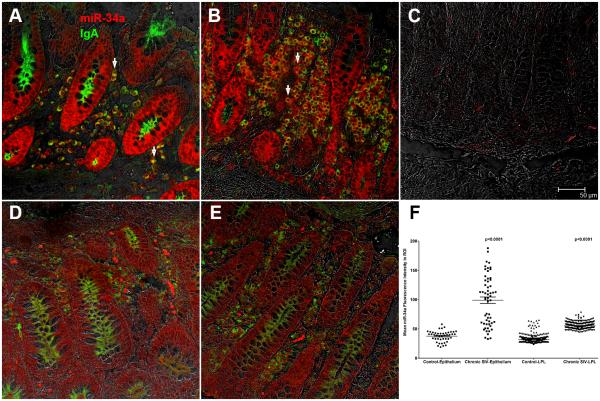
In situ hybridization detects strong miR-34a expression in the colonic epithelium and lamina propria plasma cells
Since acetylated-p65 expression was detected only in plasma cells we next determined if these cells also expressed miR-34a to directly link expression of constitutive acetylated-p65 to elevated miR-34a expression. As shown in figure 9A-B, after 7 minutes incubation with permanent red substrate (Dako Inc) miR-34a expression was strongly detected in colonic epithelium and LPLs of both chronic SIV-infected macaques. In contrast, miR-34a staining was very weak in uninfected control macaques (Fig. 9D-E) examined in the same experiment. Further, the section stained with the scrambled probe did not yield any signal (Fig. 9C). All images were captured using a Leica confocal microscope employing the same laser strength (red and green). These findings are strongly supported by quantitative image analysis showing significantly elevated miR-34a signal intensity in colonic epithelial and LPL compartments of chronic SIV-infected macaques compared to controls (Figure 9F). Double labeling with miR-34a and acetylated-p65 was attempted without success as not all antibodies work well on tissue sections previously subjected to in situ hybridization. The in situ hybridization results were in perfect agreement with the qRT-PCR data showing elevated miR-34a expression in colonic epithelium and LPLs of chronic SIV-infected macaques (Fig. 1A-B). Collectively, the intense miR-34a expression in IgA+ plasma cells (Fig. 9A&B) suggests that enhanced miR-34a expression may promote constitutive activation of acetylated-p65 (lysine 310), possibly, through SIRT1 downregulation.
FIGURE 9.
In situ localization of miR-34a in the colon of two SIV-infected (A-B) and two uninfected control macaques (D-E) with wild type miR-34a or scrambled probe (C). Panels A, B, D and E involve double labels with miR-34a (red) and IgA (green) for plasma cells. The grey channel represents differential interference contrast (DIC) to reveal tissue architecture. Colocalization of green (IgA) and red (miR-34a) appear light yellow. Note the intense miR-34a staining in the epithelium and markedly increased numbers of IgA/miR-34a++ plasma cells (white arrow heads) in the colon of SIV-infected macaques (A-B). In contrast, miR-34a staining is weak in the colonic epithelium and LPL of the control macaques (D-E). The scrambled probe did not yield a signal (C). All panels are shown at 40X magnification. Quantification of cells and regions of interest (ROI) labeled by LNA-modified miR-34a probe was performed using Volocity 5.5 software after capturing images on a Leica confocal microscope. Several ROI were hand drawn on the epithelial and LPL regions in the images from colon (F). Data was analyzed using non-parametric Wilcoxon’s rank sum test.
DISCUSSION
Gastrointestinal inflammation, a major manifestation of HIV/SIV infection that is necessary for pathogen clearance, can, however, result in structural and functional damage when not properly regulated that can promote microbial translocation and systemic immune activation (1-4). In addition to proinflammatory cytokines and transcription factors, recent studies have proposed critical roles for miRNAs in controlling and managing certain aspects of the inflammatory process (41). Moreover, the recent finding that miRNAs primarily regulate gene expression by destabilizing and degrading mRNA targets (39) suggests that the previously reported alterations in intestinal gene expression profiles by us (13-14) and others (11-12) could be directly attributed to the regulatory functions of miRNAs. Therefore, the next logical step is to profile, identify and characterize the specific miRNAs that exert these regulatory effects. To this end, we have reported miRNA dysregulation in colon during acute SIV infection (21). More specifically, we identified miR-190b to be significantly upregulated in both colon and jejunum at all stages of SIV infection primarily in response to viral replication. In the present study, we again focused on the colon as it was found to be more severely impacted than the jejunum/small intestine during chronic SIV infection (Table I). Further and more importantly, the increased bacterial concentration in the colon (1012 bacterial organisms per mL of contents) (42) makes it a major source of bacteria/microbes contributing to microbial translocation during chronic HIV/SIV infection. Here we report the identification of miR-34a, a miRNA previously linked to chronic inflammation, cell cycle arrest, and senescence (22-24,27-28) to be significantly dysregulated in both colonic epithelial and LPL compartments. Further, our findings suggest that the miR-34a-SIRT1-acetylated-p65 axis could potentially play a critical role in promoting immune cell activation, in particular, antibody producing plasma cells.
Among the 10 differentially expressed miRNAs, we further characterized miR-34a in the epithelium and LPL compartments to better understand its role in regulating GI structure and function. Interestingly, miR-34a showed significant upregulation in both colonic epithelium and LPL compartments of chronic SIV-infected macaques. Further, miR-34a is upregulated in response to DNA damage that can be a result of oxidative stress induced by chronic inflammation leading to activation of the p53 mediated DDR (30-31). Therefore, we next probed the expression of the DDR sensor protein γH2A.X and detected elevated nuclear expression of phospho-γH2A.X in the colonic crypt epithelial and LPL compartments. These findings demonstrate activation of DDR signaling and represent a potential mechanism triggering and driving miR-34a upregulation. In addition, the viral protein (HIV/SIV Tat) was also shown to induce the expression of miR-34a (43-44). Therefore, it is likely that Tat protein (direct effects of viral replication) and oxidative stress induced DDR (indirect effects of viral replication) can separately or collectively upregulate miR-34a expression in the colonic epithelium and LPL compartments. The altered expression of miR-34a suggests that it could play a critical role in regulating epithelial cell cycle, survival, lamina propria immune cell activation and inflammatory responses, all of which are dysregulated during chronic HIV/SIV infection.
Since miR-34a expression was markedly elevated in both epithelial and LPL compartments we next investigated the functional relevance of its upregulation by examining the expression of situin1 (SIRT1), a validated target of miR-34a. SIRT1 is a NAD+-dependent class III histone deacetylase that regulates a wide array of cellular processes such as glucose homeostasis, energy metabolism, cell senescence/ageing, apoptosis, double stranded DNA break repair (DSBR) and immune/inflammatory responses (34-35,37). SIRT1 exerts these protective effects by deacetylating transcription factors such as p53, FOXO3, NFκB, DNA repair enzymes and histone proteins (35). Two distinct classes of molecules, namely, RNA binding proteins and non-coding RNAs have been shown to post-transcriptionally regulate SIRT1 expression (45,34). Additionally, the viral protein Tat was shown to activate the expression of miR-34a and miR-217, which then directly targeted and downregulated SIRT1 expression (43-44). This suggests that viral replication in the intestine could contribute significantly to SIRT1 downregulation through enhancing miR-34a and miR-217 expression. More recently, miR-142-5p was also demonstrated to target and downregulate SIRT1 expression in macrophages/microglia from rhesus macaques with SIV induced encephalitis (46). Along similar lines, SIRT1 protein expression decreased in colon of mice with DSS-induced colitis but was restored following treatment with resveratrol, a SIRT1 stimulating polyphenol (37). Consistent with findings in DSS-colitis mice, SIRT1 mRNA and protein expression decreased significantly in both colonic epithelium and LPL compartments of chronically SIV-infected macaques. However, we had to test 5 anti-human SIRT1 antibodies before we found one that cross-reacted with the rhesus macaque for IF studies. This is a challenge facing the non-human primate research community as almost all antibodies are made primarily for use in the human or mouse. The results of the luciferase reporter assay together with the significantly elevated miR-34a expression in both colonic epithelium and LPL provide a potential mechanism, wherein, miR-34a can directly bind to two different sites on the rhesus macaque SIRT1 3’ UTR and inhibit its expression in both compartments.
Enhanced miR-34a expression has been previously associated with cellular senescence in renal (23), endothelial (24) and colonic crypt epithelial cells (28) and thus we were intrigued to determine if similar senescence associated changes occurred in the colonic epithelium during chronic SIV infection. We focused on the colonic epithelium as this compartment is highly proliferative and plays a central role in nutrient absorption and barrier function. Accordingly, senescence induced changes can reduce the proliferation capacity and interfere with restoration of the epithelial barrier. While the accumulation of phospho-γH2A.X foci in response to persistent DDR is considered a major trigger for cellular senescence (33), the strong expression of Ki67, a cell proliferation marker by crypt epithelial cells suggested that these cells did not progress to irreversible cell cycle arrest (senescence) and retained the capacity to proliferate. This may be because SIV-infected rhesus macaques are necropsied when they lose >20% of body weight which supports the argument that the duration of exposure to oxidative stress secondary to inflammation is not long enough to induce cellular senescence unlike in IBD patients who live with the disease for decades (36). Overall, our findings suggest that inflammation induced DDR in the colonic epithelium as evidenced by enhanced γH2A.X expression results in elevated pro-senescent miR-34a and significantly reduced anti-senescent SIRT1 mRNA and protein expression. Nevertheless, low-level viral replication induced chronic intestinal inflammation/immune activation reported to occur in patients receiving long term anti-retroviral therapy (ART) (47) can generate a persistently maintained DDR state similar to IBD patients (36), eventually causing cellular senescence. Future studies are needed to determine whether senescence associated changes occur in long term ART-treated individuals as they can cause accelerated aging of the intestine leading to epithelial barrier dysfunction, microbial translocation and AIDS progression.
In the LPL compartment, miR-34a expression increased and SIRT1 expression decreased significantly. As the LPL compartment is enriched for immune cells and primarily involved in mediating immune/inflammatory responses, we focused on the RelA/p65 subunit of NFκB, a transcription factor central to immune and inflammatory responses (48). Most notably, post-translational modifications such as phosphorylation, acetylation and sumoylation are necessary for modulating NFκB mediated gene expression (48). Among the different modifications, acetylation of the p65 subunit catalyzed by p300/CBP and PCAF can occur on seven distinct lysine residues (lysine 122, 123, 218, 221, 310, 314, 315) that can either activate or inhibit NFκB signaling depending on which lysine residue is acetylated (48). Among these, acetylation of lysine 310 is of significant interest as it is crucial for full activation of NFκB transcriptional potential (48). Equally important is the finding that SIRT1 (48) and HDAC3 (48) are two major enzymes that can deacetylate lysine 310 on p65 thereby inhibiting its transcriptional activity. In addition, Tat mediated miR-34a upregulation inhibited SIRT1 expression in TZM-bl cells resulting in enhanced acetylated p65 (lysine 310) expression (43). Furthermore, Tat has also been shown to directly bind to the deacetylase domain of SIRT1 and suppress its ability to deacetylate lysine 310 on the p65 subunit of NFκB resulting in enhanced acetylated p65 activity (49). Therefore, we probed the downstream effects of miR- 34a mediated SIRT1 downregulation in the LPL compartment by determining the expression of acetylated p65 (lysine 310). Using IF studies, acetylated p65 (lysine 310) expression was found to be markedly increased solely in the LPL compartment of chronic SIV-infected macaques. Interestingly, acetylated p65+ cells were negative for CD68, CD163 and CD3. Subsequently, using anti-rhesus macaque IgG and IgA antibodies we successfully localized acetylated p65 (lysine 310) to plasma cells. Further, markedly increased numbers of IgA+/Acetyl p65+ and IgG+/Acetyl p65+ cells were detected in the colons of chronic SIV-infected macaques. These findings were not surprising as DDR signaling evidenced by strong phospho-γH2A.X in other lamina propria cells such as lymphocytes and macrophages also activates the JAK-STAT3 pathway for efficient dsDNA repair (50). We previously demonstrated constitutive activation of the phosphorylated form of STAT3 (tyrosine 705) in CD3+ lymphocytes and CD68+ macrophages (10). These together with our previously published findings (10) suggest that while plasma cells activate p65 through acetylation of lysine 310, lymphocytes and macrophages can constitutively activate STAT3 by phosphorylation of tyrosine 705 in response to oxidative stress induced DDR. Moreover, using in situ hybridization we successfully localized miR-34a expression to the colonic epithelium and plasma cells in the lamina propria. These interesting findings are consistent with a previous study that identified miR-34a to be significantly upregulated following B cell activation in vitro with 1 μM CpG (51). More recently, activated B cells isolated from multiple sclerosis patients were found to express significantly high miR-132 and low SIRT1 protein levels (52). Similar to miR-34a, miR-132 has two predicted binding sites on the 3’ UTR of SIRT1 mRNA. Further, SIRT1 downregulation resulted in increased production of lymphotoxin and TNFα. Interestingly, in vitro treatment of B cells from MS patients with resveratrol, a SIRT1 activator, increased SIRT1 protein and normalized lymphotoxin and TNF-α production (52). Although our results do not demonstrate causal relationships, they may partially explain the intestinal B cell hyperactivity reported in HIV-infected individuals characterized by the presence of increased numbers of IgA and IgG producing plasma cells (53). Overall, our findings demonstrate an interesting association between miR-34a upregulation and elevated acetylated-p65 expression and provides a potential mechanism that may cause hyperactivation and possible dysfunction of the intestinal B cell system.
Plasma cells that increase substantially in chronically inflamed tissues (54) can activate the NFκB pathway in response to proinflammatory factors present in their immediate environment leading to the transcription of NFκB responsive genes (55). In this context, the presence of NFκB binding/response elements on the miR-34a promoter may facilitate NFκB proteins to bind and drive miR-34a expression (56). miR-34a may then directly interact and bind to the 3’ UTR of SIRT1 and downregulate its expression. As SIRT1 is required to deacetylate lysine 310 residue on p65, its reduced expression can maintain p65 in its transcriptionally active form and in this way potentially generate a feed forward mechanism involving NFκB, miR-34a and SIRT1 leading to constitutive activation and transcription of NFκB responsive genes. Given that the p65 subunit regulates transcription (57) and assembly of Ig heavy and light chains (58-59) and is indispensable for IgA and IgG production (60), its constitutive activation may partly explain the IgA overproduction reported in the intestine of chronic HIV-infected individuals (53) and, possibly, prolong the survival of plasma cells (long lived plasma cells) in niches like the inflamed colonic lamina propria (53). From the pathogenesis standpoint, IgG producing long-lived plasma cells in the colonic lamina propria of IBD patients were found to produce MMP-3 (54), a NFκB responsive matrix metalloproteinase shown to cause tissue destruction (61). More recently, IgG plasma cells in the colonic mucosa of IBD patients were shown to induce TNF-α and IL-1β production by CD14+ macrophages via IgG-IC-FcγR signaling thereby aggravating colonic inflammation (62). Further, since IgG has strong complement fixing properties, its overproduction may exacerbate intestinal inflammation, structural damage and contribute to disease progression (53). Furthermore, plasma cells have also been demonstrated to produce VEGF, IL-6 and TNFα in various chronic inflammatory conditions (54). Collectively, these findings suggest that intestinal plasma cells may have broader functions in modulating inflammatory responses that can potentially contribute to epithelial barrier disruption and the miR-34a-SIRT1 mediated constitutive NFκB activation may considerably assist in these functions.
To summarize, our results demonstrate marked miRNA dysregulation in the intestine during chronic SIV infection. While miR-34a was significantly upregulated in both colonic epithelium and LPL compartments, SIRT1 mRNA and protein expression significantly decreased in both compartments. Elevated miR-34a and reduced SIRT1 expression was associated with constitutive expression of acetylated-p65 in colonic IgA+ and IgG+ plasma cells. The findings suggest a possible role for the miR-34a-SIRT1-p65 axis in causing hyperactivity and dysfunction of the mucosal B cell system. The recent demonstration that miR-34a expression is elevated following in vitro B cell activation using CpG molecules (51) and its ability to regulate B cell development/differentiation by modulating Foxp1 expression (63) strengthens the possibility. Apart from SIRT1, miR-34a has also been shown to regulate the expression of several cell cycle regulators (NMYC, CCND1, CDK4, CDK6, MET) (22) and future studies involving cell sorting protocols and high throughput proteomics are needed to identify and validate additional targets (~640 predicted) to understand the full functional significance of its upregulation in the colon during HIV/SIV infection. In addition, in vivo miR-34a modulation studies are needed to determine if a causal relationship exists between miR-34a upegulation and constitutive acetylated-p65 expresion. Nevertheless, it is important to note that direct in vivo miRNA inhibition using antagomirs is currently not feasible due to lack of an efficient in vivo delivery system that can selectively target specific segments of the intestine. However, the recent demonstration that a 12-month supplementation of resveratrol, a SIRT1 activating anti-inflammatory polyphenol present in grape extracts, modulated miR-34a expression in peripheral blood mononuclear cells of type 2 diabetes and hypertensive patients (64) is very encouraging as it highlights an alternative approach preferably in combination with anti-retroviral drugs to modulate miR-34a expression and chronic persistent inflammation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Ronald S Veazey, Maurice Duplantis, Yun Te Lin, Faith R. Schiro, Cecily C. Midkiff, Christopher Monjure, Coty Tatum for their technical assistance in the study.
Supported by NIH grants: R01DK083929 to MM, AI084793 and OD011104 (formerly RR00164)
REFERENCES
- 1.Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 2.Smit-McBride Z, Mattapallil JJ, McChesney M, Ferrick D, Dandekar S. Gastrointestinal T lymphocytes retain high potential for cytokine responses but have severe CD4(+) T-cell depletion at all stages of simian immunodeficiency virus infection compared to peripheral lymphocytes. J Virol. 1998;72:6646–6656. doi: 10.1128/jvi.72.8.6646-6656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- 4.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, Boden D, Racz P, Markowitz M. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacArthur RD, DuPont HL. Etiology and pharmacologic management of noninfectious diarrhea in HIV-infected individuals in the highly active antiretroviral therapy era. Clin. Infect. Dis. 2012;55(6):860–867. doi: 10.1093/cid/cis544. [DOI] [PubMed] [Google Scholar]
- 7.Klatt NR, Funderburg NT, Brenchley JM. Microbial translocation, immune activation, and HIV disease. Trends Microbiol. 2013;21(1):6–13. doi: 10.1016/j.tim.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchetti G, Tincati C, Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev. 2013;26(1):2–18. doi: 10.1128/CMR.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGowan I, Elliott J, Fuerst M, Taing P, Boscardin J, Poles M, Anton P. Increased HIV-1 mucosal replication is associated with generalized mucosal cytokine activation. J. Acquir. Immune. Defic. Syndr. 2004;37:1228–1236. doi: 10.1097/01.qai.0000131846.12453.29. [DOI] [PubMed] [Google Scholar]
- 10.Mohan M, Aye PP, Borda JT, Alvarez X, Lackner AA. Gastrointestinal disease in simian immunodeficiency virus-infected rhesus macaques is characterized by proinflammatory dysregulation of the interleukin-6-janus kinase/signal transducer and activator of transcription-3 pathway. Am J Pathol. 2007;171:1952–1965. doi: 10.2353/ajpath.2007.070017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George MD, Sankaran S, Reay E, Gelli AC, Dandekar S. High-throughput gene expression profiling indicates dysregulation of intestinal cell cycle mediators and growth factors during primary simian immunodeficiency virus infection. J. Virology. 2003;312(1):84–94. doi: 10.1016/s0042-6822(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 12.Sankaran S, George MD, Reay E, Guadalupe M, Flamm J, Prindiville T, Dandekar S. Rapid onset of intestinal epithelial barrier dysfunction in primary human immunodeficiency virus infection is driven by an imbalance between immune response and mucosal repair and regeneration. J Virol. 2008;82(1):538–545. doi: 10.1128/JVI.01449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohan M, Kaushal D, Aye PP, Alvarez X, Veazey RS, Lackner AA. Focused examination of the intestinal lamina propria yields greater molecular insight into mechanisms underlying SIV induced immune dysfunction. PLoS One. 2012;7(4):e34561. doi: 10.1371/journal.pone.0034561. D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohan M, Kaushal D, Aye PP, Alvarez X, Veazey RS, Lackner AA. Focused examination of the intestinal epithelium reveals transcriptional signatures consistent with disturbances in enterocyte maturation and differentiation during the course of SIV infection. PLoS One. 2013;8(4):e60122. doi: 10.1371/journal.pone.0060122. D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 16.Swaminathan S, Murray DD, Kelleher AD. miRNAs and HIV: unforeseen determinants of host-pathogen interaction. Immunol. Rev. 2013;254(1):265–280. doi: 10.1111/imr.12077. [DOI] [PubMed] [Google Scholar]
- 17.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 18.Witwer KW, Sarbanes SL, Liu J, Clements JE. A plasma microRNA signature of acute lentiviral infection: biomarkers of central nervous system disease. AIDS. 2011;25(17):2057–2067. doi: 10.1097/QAD.0b013e32834b95bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yelamanchili SV, Chaudhuri AD, Chen LN, Xiong H, Fox HS. MicroRNA-21 dysregulates the expression of MEF2C in neurons in monkey and human SIV/HIV neurological disease. Cell Death Dis. 2010;1:e77. doi: 10.1038/cddis.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sisk JM, Witwer KW, Tarwater PM, Clements JE. SIV replication is directly downregulated by four antiviral miRNAs. Retrovirology. 2013;10(1):95. doi: 10.1186/1742-4690-10-95. JE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohan M, Chandra LC, Torben W, Aye PP, Alvarez X, Lackner AA. miR-190b is markedly upregulated in the intestine in response to SIV replication and partly regulates myotubularin related protein-6 expression. J. Immunol. doi: 10.4049/jimmunol.1303479. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen F, Hu SJ. Effect of microRNA-34a in cell cycle, differentiation, and apoptosis: a review. J. Biochem. Mol. Toxicol. 2012;26(2):79–86. doi: 10.1002/jbt.20412. [DOI] [PubMed] [Google Scholar]
- 23.Bai XY, Ma Y, Ding R, Fu B, Shi S, Chen XM. miR-335 and miR-34a Promote renal senescence by suppressing mitochondrial antioxidative enzymes. J. Am. Soc. Nephrol. 2011;22(7):1252–1261. doi: 10.1681/ASN.2010040367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin B, Yang H, Xiao B. Role of microRNAs in endothelial inflammation and senescence. Mol Biol Rep. 2012;39(4):4509–1458. doi: 10.1007/s11033-011-1241-0. [DOI] [PubMed] [Google Scholar]
- 25.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'haene B, Mestdagh P, Hellemans J, Vandesompele J. miRNA expression profiling: from reference genes to global mean normalization. Methods Mol Biol. 2012;822:261–272. doi: 10.1007/978-1-61779-427-8_18. [DOI] [PubMed] [Google Scholar]
- 27.Rippo MR, Olivieri F, Monsurró V, Prattichizzo F, Albertini MC, Procopio AD. MitomiRs in human inflamm-aging: A hypothesis involving miR-181a, miR-34a and miR-146a. Exp Gerontol. 2014 Mar 7; doi: 10.1016/j.exger.2014.03.002. pii: S0531-5565(14)00074-6. [DOI] [PubMed] [Google Scholar]
- 28.Rudolf E, John S, Cervinka M. Irinotecan induces senescence and apoptosis in colonic cells in vitro. Toxicol Lett. 2012;214(1):1–8. doi: 10.1016/j.toxlet.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Pan D, Das A, Liu D, Veazey RS, Pahar B. Isolation and characterization of intestinal epithelial cells from normal and SIV-infected rhesus macaques. PLoS One. 2012;7(1):e30247. doi: 10.1371/journal.pone.0030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato M, Paranjape T, Muller RU, Nallur S, Gillespie E, Keane K, Esquela-Kerscher A, Weidhaas JB, Slack FJ. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu H, Gatti RA. MicroRNAs: new players in the DNA damage response. J. Mol. Cell. Biol. 2011;3(3):151–158. doi: 10.1093/jmcb/mjq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawless C, Wang C, Jurk D, Merz A, von Zglinicki T, Passos JF. Quantitative assessment of markers for cell senescence. Exp. Gerontol. 2010;45(10):772–778. doi: 10.1016/j.exger.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 33.Wang C, Jurk D, Maddick M, Nelson G, Martin-Ruiz C, von Zglinicki T. DNA damage response and cellular senescence in tissues of aging mice. Aging. Cell. 2009;8(3):311–323. doi: 10.1111/j.1474-9726.2009.00481.x. [DOI] [PubMed] [Google Scholar]
- 34.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U S A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kauppinen A, Suuronen T, Ojala J, Kaarniranta K, Salminen A. Antagonistic crosstalk between NF-κB and SIRT1 in the regulation of inflammation and metabolic disorders. Cell Signal. 2013;25(10):1939–1948. doi: 10.1016/j.cellsig.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Risques RA, Lai LA, Brentnall TA, Li L, Feng Z, Gallaher J, Mandelson MT, Potter JD, Bronner MP, Rabinovitch PS. Ulcerative colitis is a disease of accelerated colon aging: evidence from telomere attrition and DNA damage. Gastroenterology. 2008;135(2):410–418. doi: 10.1053/j.gastro.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh UP, Singh NP, Singh B, Hofseth LJ, Price RL, Nagarkatti M, Nagarkatti PS. Resveratrol (trans-3,5,4'-trihydroxystilbene) induces silent mating type information regulation-1 and down-regulates nuclear transcription factor-kappaB activation to abrogate dextran sulfate sodium-induced colitis. J. Pharmacol. Exp. Ther. 2010;332(3):829–839. doi: 10.1124/jpet.109.160838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis BP, Burge CB, Bartel DP. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 39.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su BH, Tseng YL, Shieh GS, Chen YC, Shiang YC, Wu P, Li KJ, Yen TH, Shiau AL, Wu CL. Prothymosin α overexpression contributes to the development of pulmonary emphysema. Nat. Commun. 2013;4:1906. doi: 10.1038/ncomms2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 42.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8(6):411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 43.Zhang HS, Chen XY, Wu TC, Sang WW, Ruan Z. MiR-34a is involved in Tat-induced HIV-1 long terminal repeat (LTR) transactivation through the SIRT1/NFκB pathway. FEBS. Lett. 2012;586(23):4203–4207. doi: 10.1016/j.febslet.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 44.Chang JR, Mukerjee R, Bagashev A, Del Valle L, Chabrashvili T, Hawkins BJ, He JJ, Sawaya BE. HIV-1 Tat protein promotes neuronal dysfunction through disruption of microRNAs. J. Biol. Chem. 2011;286(47):41125–41134. doi: 10.1074/jbc.M111.268466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Z, Fan D, Gui B, Shi L, Xuan C, Shan L, Wang Q, Shang Y, Wang Y. Neurodegeneration-associated TDP-43 interacts with fragile X mental retardation protein (FMRP)/Staufen (STAU1) and regulates SIRT1 expression in neuronal cells. J. Biol. Chem. 2012;287(27):22560–22572. doi: 10.1074/jbc.M112.357582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaudhuri AD, Yelamanchili SV, Marcondes MC, Fox HS. Up-regulation of microRNA-142 in simian immunodeficiency virus encephalitis leads to repression of sirtuin1. FASEB. J. 2013;27(9):3720–3729. doi: 10.1096/fj.13-232678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chun TW, Nickle DC, Justement JS, Meyers JH, Roby G, Hallahan CW, Kottilil S, Moir S, Mican JM, Mullins JI, Ward DJ, Kovacs JA, Mannon PJ, Fauci AS. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis. 2008;197(5):714–720. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- 48.Huang B, Yang XD, Lamb A, Chen LF. Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cell Signal. 2010;22(9):1282–1290. doi: 10.1016/j.cellsig.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon HS, Brent MM, Getachew R, Jayakumar P, Chen LF, Schnolzer M, McBurney MW, Marmorstein R, Greene WC, Ott M. Human immunodeficiency virus type 1 Tat protein inhibits the SIRT1 deacetylase and induces T cell hyperactivation. Cell. Host. Microbe. 2008;3(3):158–167. doi: 10.1016/j.chom.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barry SP, Townsend PA, Knight RA, Scarabelli TM, Latchman DS, Stephanou A. STAT3 modulates the DNA damage response pathway. Int J Exp Pathol. 2010;91(6):506–514. doi: 10.1111/j.1365-2613.2010.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S, Moffett HF, Lu J, Werner L, Zhang H, Ritz J, Neuberg D, Wucherpfennig KW, Brown JR, Novina CD. MicroRNA expression profiling identifies activated B cell status in chronic lymphocytic leukemia cells. PLoS One. 2011;6(3):e16956. doi: 10.1371/journal.pone.0016956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyazaki Y, Li R, Rezk A, Misirliyan H, Moore C, Farooqi N, Solis M, Goiry LG, de Faria Junior O, Dang VD, Colman D, Dhaunchak AS, Antel J, Gommerman J, Prat A, Fillatreau S, Bar-Or A, CIHR/MSSC New Emerging Team Grant in Clinical Autoimmunity and the MSSRF Canadian B cells in MS Team A Novel MicroRNA-132-Surtuin-1 Axis Underlies Aberrant B-cell Cytokine Regulation in Patients with Relapsing-Remitting Multiple Sclerosis. PLoS One. 2014;9(8):e105421. doi: 10.1371/journal.pone.0105421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nilssen DE, Øktedalen O, Brandtzaeg P. Intestinal B cell hyperactivity in AIDS is controlled by highly active antiretroviral therapy. Gut. 2004;53(4):487–493. doi: 10.1136/gut.2003.027854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gordon JN, Pickard KM, Di Sabatino A, Prothero JD, Pender SL, Goggin PM, MacDonald TT. Matrix metalloproteinase-3 production by gut IgG plasma cells in chronic inflammatory bowel disease. Inflamm Bowel Dis. 2008;14(2):195–203. doi: 10.1002/ibd.20302. [DOI] [PubMed] [Google Scholar]
- 55.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2(6):a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Wang K, Chen X, Meng H, Song M, Wang Y, Xu X, Bai Y. Transcriptional activation of microRNA-34a by NF-kappa B in human esophageal cancer cells. BMC. Mol. Biol. 2012;13:4. doi: 10.1186/1471-2199-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lefranc G, Lefranc MP. Regulation of the immunoglobulin gene transcription. Biochimie. 1990;72:7–17. doi: 10.1016/0300-9084(90)90167-f. M.P. [DOI] [PubMed] [Google Scholar]
- 58.Oltz EM. Regulation of antigen receptor gene assembly in lymphocytes. Immunol. Res. 2003;23:121–133. doi: 10.1385/IR:23:2-3:121. [DOI] [PubMed] [Google Scholar]
- 59.Durandy A. Hyper-IgM syndromes: a model for studying the regulation of class switch recombination and somatic hypermutation generation. Biochem. Soc. Trans. 2002;30:815–818. doi: 10.1042/bst0300815. [DOI] [PubMed] [Google Scholar]
- 60.Doi TS, Takahashi T, Taguchi O, Azuma T, Obata Y. NF-kappa B RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J. Exp. Med. 1997;185(5):953–961. doi: 10.1084/jem.185.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roman-Blas JA, Jimenez SA. NF-kappaB as a potential therapeutic target in osteoarthritis and rheumatoid arthritis. Osteoarthritis. Cartilage. 2006;14(9):839–848. doi: 10.1016/j.joca.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 62.Uo M, Hisamatsu T, Miyoshi J, Kaito D, Yoneno K, Kitazume MT, Mori M, Sugita A, Koganei K, Matsuoka K, Kanai T, Hibi T. Mucosal CXCR4+ IgG plasma cells contribute to the pathogenesis of human ulcerative colitis through FcγR-mediated CD14 macrophage activation. Gut. 2013;62(12):1734–1744. doi: 10.1136/gutjnl-2012-303063. [DOI] [PubMed] [Google Scholar]
- 63.Rao DS, O'Connell RM, Chaudhuri AA, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33(1):48–59. doi: 10.1016/j.immuni.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tomé-Carneiro J, Larrosa M, Yáñez-Gascón MJ, Dávalos A, Gil-Zamorano J, Gonzálvez M, García-Almagro FJ, Ruiz Ros JA, Tomás-Barberán FA, Espín JC, García-Conesa MT. One-year supplementation with a grape extract containing resveratrol modulates inflammatory-related microRNAs and cytokines expression in peripheral blood mononuclear cells of type 2 diabetes and hypertensive patients with coronary artery disease. Pharmacol Res. 2013;72:69–82. doi: 10.1016/j.phrs.2013.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.