Abstract
The pipid frog Xenopus tropicalis has emerged as a powerful new model system for combining genetic and genomic analysis of tetrapod development with robust embryological, molecular and biochemical assays. Its early development closely resembles that of its well-understood relative X. laevis, from which techniques and reagents can be readily transferred. In contrast to the tetraploid X. laevis, X. tropicalis has a compact diploid genome with strong synteny to those of amniotes. Recently, advances in high-throughput sequencing together with solution-hybridization whole-exome enrichment technology offer powerful strategies for cloning novel mutations as well as reverse genetic identification of sequence lesions in specific genes of interest. Further advantages include the wide range of functional and molecular assays available, the large number of embryos/meioses produced, and the ease of haploid genetics and gynogenesis. The addition of these genetic tools to X. tropicalis provides a uniquely flexible platform for analysis of gene function in vertebrate development.
Keywords: Xenopus, Silurana, tropicalis, genetics, development, organogenesis, gynogenesis, genetic screens, genetic mapping, mutagenesis
1. Introduction
Xenopus embryos have been remarkably productive models for developmental biologists for over 70 years (1, 2). The dominant laboratory species, X. laevis, continues to be an outstanding system for embryological manipulations and gain-of-function gene assays, but its tetraploid genome and long generation time hamper many genetic and genomic approaches. The related diploid species Xenopus (Silurana) tropicalis shares X. laevis’ advantages for experimental embryology while also being ideally suited to genetics and genomics.
Why Xenopus genetics?
Forward genetic screens have the unique ability to identify novel gene functions without bias toward previously known DNA sequence. Our understanding of animal development is founded on genetic studies of invertebrate fly and worm models (3, 4) which identified the transcriptional control networks underpinning the basic animal body plan, but it is expected that many differences may exist in vertebrates. The development of gene targeting for reverse genetic studies in mouse has been especially powerful (5, 6), but forward screens for embryonic mutations in mammals are costly and difficult due to intrauterine development. A large number of informative mutations have been identified in teleosts, particularly zebrafish, where screens have benefited from rapid development of externally fertilized, transparent embryos, high fecundity, and short generation time (7,-10).
Unlike the genome of teleost fish, derived from an ancient duplication (11) or the allopolyploid genomes of other Xenopus, derived from hybridization and genome retention of two separate species (12,13), the genome of X. tropicalis is that of a canonical diploid vertebrate. At ~1.5×109bp it is one of the smallest tetrapod genomes, about the same size as zebrafish, and shows robust synteny with those of amniotes (14), simplifying orthology assignment, functional analysis, and identification of noncoding regulatory elements. Pilot forward genetic screens have already recovered a number of heritable mutants (15-17), several of which have now been mapped to specific genes (18-20). Genetic studies in X. tropicalis are facilitated by the production of up to 9000 embryos from a single mating, sufficient meiotic recombination events to map a mutation or conduct a variety of phenotypic analyses. Extensive genomic resources are available, including a high-quality chromosome-scale draft genome assembly and more than one million ESTs (see Chapter 4 “Navigating the Xenopus tropicalis genome”). Gain-of-function, molecular, and embryological assays are readily transferred from the well-characterized X. laevis system. Transgenic rescue of mutant backgrounds with floxed constructs (21) offers a method for obtaining conditional alleles to delete gene functions in specific tissues or points in development. This uniquely flexible in vivo system now combines the conventional strengths of Xenopus with loss-of-function genetic backgrounds and enhanced genomic tools, multiplying the reach of what has already been described as “perhaps the best vertebrate model organism for functional genomics” (22).
This chapter surveys methods for genetic analysis of X. tropicalis development, including genome manipulations (haploid genetics, gynogenesis and androgenesis), uncovering naturally-occurring mutations, mutagenesis, screening protocols, mapping strategies, sequence-based reverse genetic strategies, and analysis of mutant phenotypes.
2. Materials
2.1 General embryology
0.05 × MMR+ BSA: 1mg/ml Bovine Serum Albumin (BSA) in 0.05 × MMR; Adjust 0.05×MMR to pH 8.3 prior to adding BSA to prevent protein accumulation on pH probe. BSA is slightly acidic and will bring pH down to ~7.7-7.9.
2.2% cysteine: Cysteine hydrochloride (Sigma) in 0.05 × MMR, adjust pH to 7.7-7.9 with 10N NaOH. Use within 2 hrs.
0.4% MS-222: Add 4g of Ethyl 3-aminobenzoate methanesulphonate (MS-222) to 1L 0.05 × MMR. Adjust pH to 7.7 with 1N NaOH. Store at 4°C and reuse up to 10 times.
L15/CS: Leibovitz-15 (L-15) media (GibcoBRL) supplemented with 10% calf serum (CS) (GibcoBRL). Store 10ml aliquots at −20°C.
These materials are used in a variety of different protocols. Materials for specific procedures are indicated below. All chemicals are obtained from Sigma unless otherwise specified.
2.2 Husbandry & obtaining embryos
Human Chorionic Gonadotrophin (HCG) (Chorulon, Intervet). Make stock of 1000 units/ml in sterile H2O and dilute accordingly. Store at 4°C. Sterilize seal with ethanol before and after each use.
Sera Micron Powder (Sera; Heisenberg, Germany) tadpole diet.
Reptomin Sticks (Tetra; Melle, Germany) adult frog diet.
Tropical Fish Flake (Sinclair Animal & Household Care, Gainsborough, UK) or equivalent; diet for metamorphosing tadpoles froglets and supplement for adults.
Sorting tools: manual pipette pump and glass Pasteur pipettes (X. tropicalis embryos tend to stick to plastic transfer pipettes). Notch glass Pasteur pipettes with a diamond pen, break off, and blunt edges with a Bunsen burner flame.
2.3 Karyotyping
27G hypodermic needles
Microscope slides (e.g. positively-charged Superfrost Plus from Fisher) and large coverslips
Paper towels
Distilled H2O
60% acetic acid in distilled H2O
Hoechst 33342 stain (Sigma; Poole, UK), working stock 0.1mg/ml in distilled H2O
70% glycerol in phosphate buffered saline (PBS)
2.4 Mutagenesis
N-nitroso-N-ethylurea (ENU). 1g Isopac (Sigma; Poole, UK)
2-(N-morpholino)ethanesulphonic acid (MES, see Note 1) (Sigma; Poole, UK): Make 2 100mM stocks in dH20. Adjust one to pH 6.0 and one to pH 6.2 with 1N NaOH, store at 4°C.
lab coat, plastic wrist guards, gloves, facemask
Decontamination bath: 10% sodium thiosulphate, 1% sodium hydroxide in H2O
Nutator or roller
2.5 Mapping
Embryo lysis buffer: 50mM Tris-HCl pH 8.8, 1mM EDTA, 0.5% Tween-20, store stock at RT; add 200ug/ml Proteinase K (Roche) immediately prior to use.
PCR-compatible 96 well plates
Standard PCR reagents and equipment
Super Fine Resolution Agarose (SFR) (Amresco; Solon, USA)
2.6 Acrylamide gels and Silver staining
Benchtop sequencing gel apparatus (e.g. Thistle Scientific Model 2) with matching glass plates; 0.4mm spacers & shark’s tooth combs
1 photographic developing dish large enough to fit glass sequencing plate
Denaturing DNA loading buffer: 50ml stock = 49ml formamide, 1ml 0.5M EDTA, 0.1g bromophenol Blue, 0.1g xylene cyanol.
1×Tris Borate EDTA (TBE) Buffer
3-(Trimethoxysily)propyl methacrylate
100% and 70% ethanol
Acrylease (Stratagene)
1L 10% ethanol
1L 1% nitric acid
1L 2g/L silver nitrate
1L Developing solution: 29.6g sodium carbonate, 450μl 37% formaldehyde. Prepare in advance and keep on ice
1L 10% acetic acid
2.7 DNA prep for genotyping from tissue
0.07% MS-222: Add 0.7g of ethyl 3-aminobenzoate methanesulphonate (MS-222) to 1L 0.05 × MMR. Adjust pH to 7.7 with NaOH.
Scalpels/razor blades
Lysis buffer: 100mM Tris-HCl pH8-8.5, 200mM NaCl, 0.2% SDS, 5mM EDTA; 100ug/ml Proteinase K added just before use (Roche)
Isopropanol
70% ethanol
2.8 Sperm Freezing
Cryoprotectant: Disperse one egg yolk (about 15mL) in an equal volume of distilled water; dilute to 20% v/v in 0.4M sucrose, 10mM sodium bicarbonate, 2mM pentoxyfylline solution. Centrifuge for 20 min at 13,000 rpm and use supernatant (store at −20°C for up to one month).
Styrofoam box small enough to fit into −80°C freezer.
3. Methods
3.1. Husbandry & obtaining embryos
Multigeneration genetic studies critically depend on minimizing generation time and maximizing egg quality and fertilization success; diet and husbandry are extremely important, particularly for tadpoles/froglets and egg-producing females. X. tropicalis will not thrive in same conditions as X. laevis, and the two species should never be housed in shared water systems, even with filtration, due to the risk of trans-species infection. See Chapters 2 and 3 for in-depth discussions of husbandry and breeding; short protocols for in vitro fertilization are provided in Section 3.2.1.
3.1.1. Husbandry
Briefly, frogs should be maintained in 24-26°C water, avoiding sudden changes during cleaning. Temperatures as high as 30°C are tolerated, but egg quality from females maintained above 27°C diminishes sharply. Temperatures below 23°C may depress disease resistance; below 20°C is usually lethal for tropicalis. Xenopus tadpoles are filter feeders, so optimal growth is obtained with suspended nutrients, and flow-through recirculation systems should be turned off during feeding. Sera Micron powder, fed frequently, is an excellent diet for small tadpoles and causes minimal fouling of standing water systems. Larger tadpoles and metamorphosing froglets can be supplemented with crushed fish flake daily. Adult frogs do well on a variety of protein-rich diets; we use Tetra Reptomin sticks (aquatic turtle diet) supplemented with fish flake. Reptomin is crushed into fragments for smaller froglets to young adults. (see Note 2).
3.1.2 Xenopus tropicalis Strains
Strains that differ from each other at many sequence loci (polymorphic strains) are valuable for genetic mapping, whether by conventional means or next-generation sequencing approaches. Two strains (IC (originating in the Ivory Coast) and N (Nigeria)) have been inbred for >12 generations and successfully used for mapping mutations(18); a third strain, ICB, is also being inbred (23). An inbred N animal was the basis of the draft genome assembly (14) these animals may thus be more effectively targeted by sequence-based interventions such as morpholino oligonucleotides than the IC strain. Wild caught animals are occasionally available, but extreme care must be taken to prevent disease introduction and taxonomic misidentification, as morphologically-identical non-diploid species (e.g. X. epitropicalis) are known to occur in overlapping range.
3.2 Genome Manipulations: haploid genetics, gynogenesis & androgenesis
The ease with which the entire genome can be manipulated in amphibians is a great advantage for genetic and transgenic applications. Simple and efficient procedures exist for generating both haploid X. tropicalis embryos, which can undergo several days of development, and viable diploid gynogenetic embryos derived solely from the maternal genome. Haploids derived entirely from the paternal genome, or androgenetic haploids, can also be obtained, but in much smaller numbers (24); in this chapter, ‘haploid’ indicates maternally-derived unless otherwise specified. These manipulations are extremely useful for a wide range of applications including high-throughput forward genetic screens, identifying polymorphisms, rapid low-resolution genetic mapping, making transgenes homozygous, and generating completely isogenic strains.
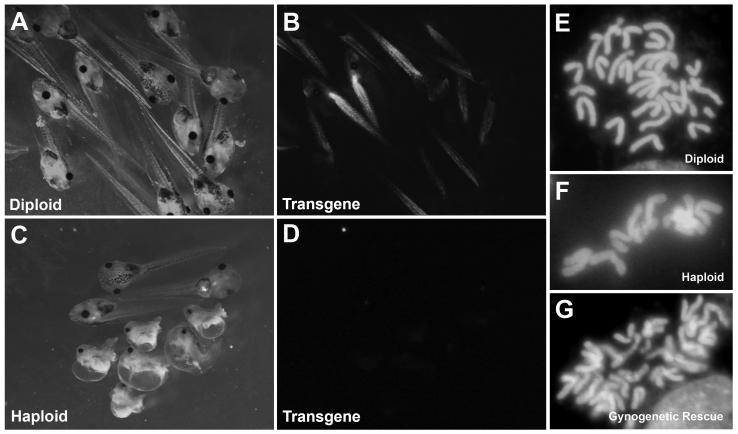
Xenopus eggs, like those of many other lower vertebrates, are deposited prior to completion of second meiosis; extrusion of the second polar body normally begins a few minutes post-activation. If sperm suspensions are UV-irradiated prior to use for in vitro fertilization, egg activation, polar body formation, and cleavage can occur normally, but the crosslinked paternal genome cannot contribute to the zygote, resulting in formation of haploid embryos (Figure 1). Haploid embryos are not viable beyond feeding stages, and show a high level of gastrulation defects and posterior abnormalities. However, they form anterior structures well enough for many phenotypes to be scored, and are also very useful for characterizing parental polymorphisms. If the mother is a heterozygous carrier of a recessive mutation, scorable phenotypes are expected to be uncovered in 50% of the haploid progeny.
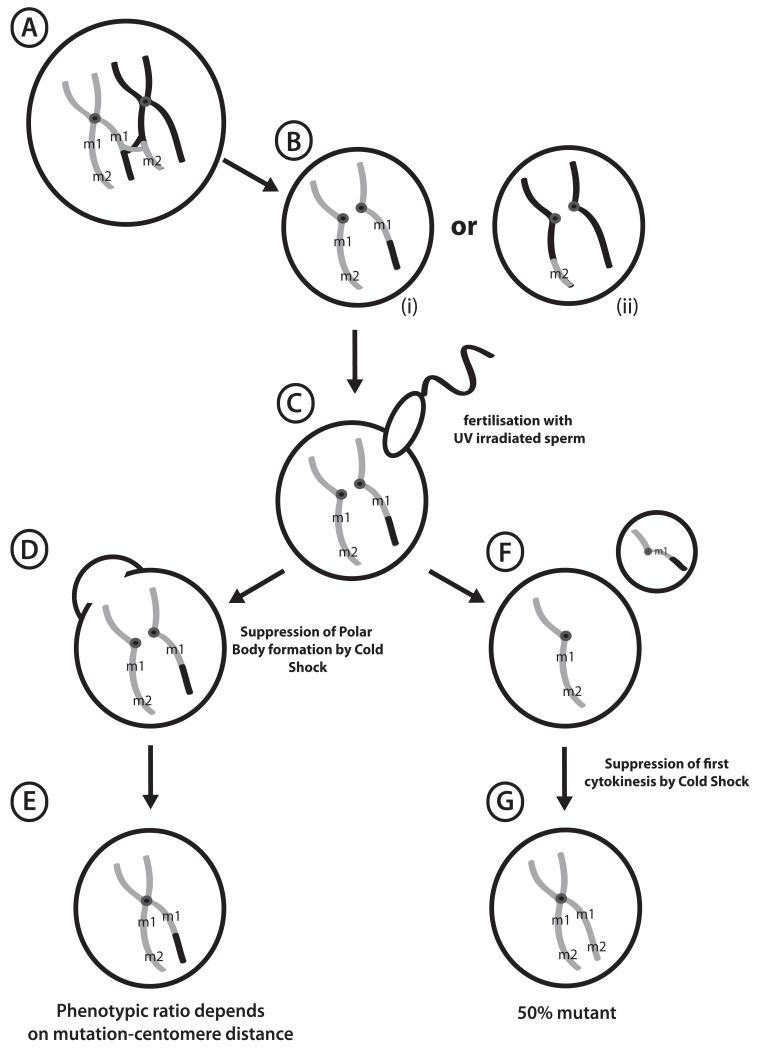
Figure 1. Formation of Haploid and Gynogenetic Embryos.
(A) Diplotene oocyte in a female hybrid for mutagenized gray strain chromosomes and polymorphic black strain, showing a crossover event between mutant loci m1 and m2. (B) Unfertilized eggs showing segregation of sister chromatids after Meiosis I. Note that regions where centromeres hold sister chromatids together are homozygous. (C) UV-irradiated sperm activates development without paternal genetic contribution, forming haploid embryos (F) following polar body extrusion. (D) Early cold shock suppresses formation of the second polar body, with the resulting gynogenote (E) rescued to diploidy and retaining both sets of sister chromatids. Recessive phenotypes at loci closer to centromeres (m1) are more likely to be uncovered than distal loci (m2), where recombination produces heterozygous wild type. (G) Late Cold Shock of haploid embryos following DNA replication prevents first cytokinesis, rescuing haploid to completely homozygous diploids.
In gynogenesis, embryos fertilized with UV-irradiated sperm are rescued to diploidy using one of two basic methods. Polar body formation can be blocked with a simple cold shock shortly after fertilization (early cold shock, ECS) leading to the retention of both sister chromatid products of meiosis II. This method rescues haploids to viable diploidy with high efficiency, and is extremely useful for uncovering recessive phenotypes in the progeny of carrier females. Critically, Mendelian phenotypic ratios are not expected in ECS embryos (gynogenotes) due to recombination during meiosis I, and varies between a maximum of 50% phenotypically recessive mutant (for centromere-linked loci) and a minimum of ~5% for distal loci. The observed ratio can provide useful low-resolution map information (see Figure 1 and section 3.7.4). ECS, which relies on the retention of the maternal polar body, is not applicable to androgenetic haploids.
Alternatively, gynogenetic haploid embryos can be allowed to undergo the first round of DNA duplication, then rescued to diploidy by blocking the first cell division with late cold shock (LCS). This procedure is usually much less efficient than ECS, but can result in completely homozygous isogenic embryos and uncovers recessive phenotypes regardless of chromosomal location in 50% of the progeny of heterozygous carriers. LCS is also theoretically possible using androgenetic haploids, although the number of viable homozygous diploid embryos recovered is very small. These diploidization procedures were originally developed in X. laevis using pressure treatments (25, 26), but have been modified for the simpler cold shock technique in X. tropicalis by Rob Grainger’s group (University of Virginia).
3.2.1 Production of haploid embryos
12-72 hours prior to procedure, prime two or more adult female X. tropicalis with 10u HCG in 100ul sterile water (see Notes 3 and 4).
On day of procedure, boost primed females with 100-200u HCG in 100ul sterile water (see Notes 3 and 4).
~3 hr after boosting females, kill two X. tropicalis males (see Note 5) and dissect testes into L15/CS
Label two 10cm culture dishes ‘haploid’ or ‘control’
Place a few drops of 1×MMR into ‘haploid’ dish for each female. High salt medium prevents premature activation of eggs
Express eggs into media; avoid getting tank water from the frogs onto the eggs. Discard dead, lysing, or stringy eggs (see Note 4).
Transfer small number of eggs with pipette to ‘diploid’ control dish
Place testes in eppendorf tube containing 500μl L15/CS, macerate with eppendorf pestle, add another 500μl L15/CS and mix.
Allow testis fragments to settle, and place sperm suspension onto glass petri dish; save large chunks of testis for diploid control.
UV-irradiate sperm suspension in Stratalinker (Stratagene) or equivalent with 50,000 microjoules (‘energy’ setting 500)
Add irradiated sperm to ‘haploid’ dish (see Note 5).
Add 500μl L15/CS to the non-irradiated testis fragments in eppendorf tube, mix and use to fertilize ‘control’ plate
Gently shake dish to mix eggs and sperm, wait 5 minutes and flood with 0.05×MMR.
Dejelly (see Chapter 2) with 2.2% cysteine in 0.05 × MMR pH 7.7-8.0, rinse, sort evenly-cleaving embryos, and culture overnight at 25°C (see Note 6)
3.2.2 Early Coldshock (gynogenesis by suppression of polar body formation)
As in haploid production (section 3.2.1). In addition:
At least 1 hr before in vitro fertilization, chill ~50ml 0.05 × MMR on ice per female being screened.
For each female, label two 10cm dishes ‘control’, ‘haploid’, plus one 6cm dish ‘ECS’ (Early Coldshock)
Procedure as in haploid protocol (Section 3.2.1) steps 1-9.
-
13.
set timer for 5′, flood embryos with 0.05 × MMR, start timer
-
14.
transfer ~90% of embryos from flooded ‘Haploid’ dish to ECS dish and remove media
-
15.
at 5′, add ice-cold 0.05 × MMR to ECS dish and place in slushy ice bucket for 7′30″.
-
16.
After 7′30″ remove ECS dishes from slush bucket and replace media with RT 0.05 × MMR.
-
17.
Wait >20 before dejellying and sorting. In ECS embryos cleavage will be delayed by 15-20′ relative to haploid and diploid controls (see Notes 5 and 6).
3.2.3 Late cold shock (gynogenesis by suppression of first cleavage)
Suppressing cytokinesis after the first round of DNA replication in the fertilized embryo can also rescue Xenopus haploids to completely homozygous diploids. Time to first cleavage is critical and varies with egg batch as well as temperature. For optimal success, aliquots of each fertilization should be timed. (see protocol below).
As in haploid production (section 3.2.1); in addition
At least 1 hr before squeezing females, chill ~50ml 0.05 × MMR per female in ice bucket as described in early cold shock section.
For each female, label 4 10cm dishes ‘diploid’, ‘haploid’, and ‘LCS’ (late cold shock) and ‘timer’
Procedure as in haploid protocol steps 1-9.
-
13.
set timer to count up, and establish time to first cleavage by removing a small aliquot of sperm & eggs from each haploid dish to ‘timer’ dish and flood with 0.05 × MMR, start timer. Keep all dishes and media at the same temperature.
-
14.
3′ after flooding test fertilization, flood haploid and diploid control dishes.
-
15.
transfer ~90% of embryos from flooded ‘Haploid’ dish to LCS dish
-
16.
after ~45′, remove media from LCS dish.
-
17.
After 45′, observe ‘timer’ dish every one or two minutes for onset of cleavage furrow. First cleavage can take between 45′ and 70′ at 23-25°C
-
18.
At first sign of cleavage furrow in ‘timer’ dish, add ice-cold 0.05 × MMR to LCS dish and place in slushy ice for 5′
-
19.
After 5′ remove LCS dishes from ice bucket and replace media with RT 0.05 × MMR.
-
20
Wait >20′ before dejellying and sorting (see Note 8).
3.2.4 Production of androgenetic haploids
12-72 hours prior to procedure, prime 4-6 adult female X. tropicalis with 10u HCG in 100ul sterile water (see Notes 3 and 4).
On day of procedure, boost primed females with 100-200u HCG in 100ul sterile water (see Notes 3 and 4).
~3 hr after boosting females, kill two X. tropicalis males and dissect testes into L15/CS
Label 6cm culture dishes ‘haploid’ or ‘control’
Prepare sperm solution: place testis from 1 male in a 3cm dish w/ ~400 ul L15+10% FBS and macerate w/ forceps.
- Perform a test fertilization to identify the females producing the best eggs:
- gently squeeze a very small number of eggs (~20-30) from each female into a dry dish; avoid getting tank water from the frogs onto the eggs. Discard batches with dead, lysing, or stringy eggs.
- Add a drop of testis solution to remaining batches of eggs, flood with 1/20× MMR, wait ~10′ and inspect for activation (cortical contraction). Select female(s) showing the best fertilization.
Gently squeeze a very small number of eggs (~30-50) from selected female into a dry 6cm dish; avoid getting tank water from the frogs onto the eggs. Proceed quickly through next steps to avoid drying out the eggs.
Using number 5 watchmaker’s forceps, quickly transfer 10-30 good-looking eggs one by one to a 3cm dish, animal pole up (for irradiation). Avoid eggs that lack a visible germinal vesicle.
Irradiate eggs in Stratalinker 1× 50,000 uJ (‘energy’ setting 500).
Cover eggs w/ testis solution, then remove testis solution for re-use if more eggs are to be treated (step 13 below).
Add a small amount of testis solution to an aliquot of non-irradiated eggs as control.
Flood both irradiated and control eggs with 1/20× MMR
Repeat with several more small squeezes, reusing the testis solution
Fertilization rates in androgenetic haploids under optimal conditions can be nearly as good as with controls, but the total number of cleaving embryos produced is limited by manipulating individual eggs in jelly.
3.2.5 Karyotyping
Karyotying may be used to determine ploidy status of experimentally-manipulated embryos as well as distinguish among X. tropicalis and similar non-diploid species. This protocol was developed by the Grainger lab (University of Virginia) and modified by M. Khokha (Yale University).
Place 10 Stage 24-34 tadpoles into a dish of deionized water.
With a scalpel or 27g needle, remove the yolky ventral portion of the tadpole and discard; allow remaining dorsal portions to stand for 20′.
Pipette the dorsal halves with as little water as possible into an Eppendorf tube containing 0.2 ml of 60% acetic acid in water; let stand 5′
Pipette all of the tissue (with minimal acetic acid) and place on a positively charged slide (e.g. Superfrost Plus from Fisher); blot away excess acetic acid.
Place a large coverslip on the slide. Fold a paper towel to the size of the coverslip and place it on top. Apply heavy pressure on top of the paper towel/coverslip for about 5 minutes using a lead brick or by pressing forcefully with a thumb, being careful not to move around (see Note 9).
After five minutes carefully remove the lead brick and paper towel.
Place slide on dry ice for 5 minutes, then remove from dry ice and use a razor blade to gently pry the coverslip from the still-frozen slide.
Place slide on a paper towel and stain chromosomes with Hoechst 33342 (1μl Hoechst 33342 (stock: 0.1mg/ml) in 1 ml distilled water) for 5′. Wear gloves when working with Hoechst.
Tip slide to allow stain to run off onto the paper towel.
Mount in a drop of 70% glycerol/PBS, add large coverslip and seal edges with clear nail polish.
Inspect slide under fluorescent illumination for stained metaphase spreads using a 63× or higher power objective.
3.3 Mutagenesis Strategies
Although naturally occurring mutations may be recovered from untreated animals (16, 17), genetic screens normally begin with mutagenesis. The same mutagenized stocks can often be used for both phenotype-based forward and sequence-driven reverse genetic screening strategies. Methods for inducing mutations in X. tropicalis include X-ray deletions, insertion or mobilization of transgenes (19) and see Chapter 6), gene editing with engineered zinc finger nucleases (27) and see Chapter 7), and chemical mutagenesis (15). With the exception of gene editing, these methods produce randomly distributed genomic lesions to a first approximation.
Chemical mutagenesis is currently the most efficient method for inducing large numbers of simple sequence lesions to obtain a range of specific phenotypes. The combination of next-generation sequencing technology and improved genomic resources has greatly simplified cloning chemically-induced point mutations. The size of deletions produced by X-irradiation is difficult to control, making it hard to associate phenotypes with specific gene functions. Gene disruption by insertional mutagenesis is an attractive strategy for rapid cloning of mutations using inverse PCR-related strategies to isolate endogenous sequences flanking known transgene constructs (28). Insertional mutagenesis by REMI nuclear transfer (see Chapter 11) has already identified a novel limb development gene function (19). Insertional mutagenesis via transposon-mediated transgenesis, which has the potential to greatly increase mutagenesis efficiency, is discussed in Chapter 6, and may eventually make gene trap and related strategies feasible (29). Since transgenesis can induce multiple lesions in the genome, it is important to recognize that identification of a transgene insertion does not supplant linkage analysis and/or independent functional correlation of the putative mutated gene with the phenotype.
3.3.1. Strategies using chemical mutagens
Most extant Xenopus mutations have been induced using chemical mutagens. Positional cloning to identify affected genes (see section 3.7) is greatly simplified by improved genomic resources including the chromosome-scale version 7.1 genome assembly (accessible via www.xenbase.org) and high density meiotic map (30). High-throughput sequencing of whole exomes, either using individual carriers or pooled mutant embryos, is poised to revolutionize identification of mutant lesions both in reverse genetic TILLING strategies and underlying phenotypes identified in forward screens.
Chemical mutagens may be applied in vitro to mature sperm from dissociated testes, or in vivo to target spermatogonia by injection into adult male frogs:
Mutagenesis of dissociated testes followed by in vitro fertilization efficiently induces sequence lesions, and a useful rapid readout of mutagenesis efficiency is provided by the appearance of dominant phenotypes in the resulting embryos. The drawback is that the founding F1 generation is mosaic, because the chemical adducts (usually produced by alkylating agents such as N-nitroso-N-ethylurea (ENU)) on a single strand of the sperm DNA double helix are not repaired and permanently fixed on the complementary strand until the first somatic DNA replication or later. This mosaicism in the F1 generation precludes conventional 3-generation screens, since the likelihood of F2 sibling pairs carrying the same induced allele drops to 1/16 or lower, and the number of matings required to uncover recessive mutations becomes unworkable. Mosaic F1 animals are likewise unsuitable for direct analysis by high-throughput genomic sequencing, since a mutation present in a minority of reads at a given locus is likely to be invisible against the base call error rate. However, in vitro mutagenesis is compatible with haploid or gynogenetic screens using females of either mosaic F1 or non-mosaic F2 generations. The non-mosaic F2 generation is also suitable for tissue sampling for reverse genetic approaches.
In vivo spermatogonial mutagenesis, in contrast, is compatible with either conventional 3-generation or haploid/gynogenetic screens. In practice, it lacks the immediate confirmation of mutagenesis provided by dominant phenotypes following in vitro mutagenesis. Mutagenesis rates can be scored by genomic sequencing in the next generation (see section 3.6).
3.3.2. Insertional mutagenesis
Insertional mutagenesis is an attractive strategy, since known transgene sequences facilitate identification of genomic integration sites and reduce reliance on positional cloning. The first insertional mutation in amphibians, producing a spectacular forelimbless phenotype, was recently described (19). As transgenesis protocols can introduce DNA damage elsewhere in the genome, linkage studies are still useful for confirming association of a phenotype with a specific insertion. A number of protocols have been described for mediating stable transgenesis in Xenopus, including transfer of sperm nuclei (10)(see Chapter 11), various transposable elements (28) and Chapter 14, I-SceI meganuclease (31) and Chapter 12, and phiC31 integrase (32) and Chapter 13. However, the relative inefficiency of transgenesis in X. tropicalis has thus far precluded large-scale screens for insertional mutants. Genetic manipulation of transgenic lines is also potentially powerful. Many reporter lines have been established in X. tropicalis (33), which may be useful substrates for genetic screens focusing on specific tissues or processes, and binary and inducible systems are available for experimental manipulation of gene function (34, 35). Cre recombinase has also been shown to function in Xenopus stable transgenics (36). While targeted knock-in to engineer endogenous loci has not yet been demonstrated in Xenopus, null mutations are becoming increasingly available. Established methods may be used to introduce engineered transgenes and rescue the null background to viability, meanwhile creating extremely useful floxed conditional knockouts, tagged proteins and other informative alleles.
3.4 Chemical Mutagenesis
ENU is highly carcinogenic and must be treated with extreme caution; all manipulations should take place in a fume hood, wearing labcoat, double gloves, and plastic wrist guards. All materials that come into contact with ENU solutions should go into decontamination bath for 24 hrs. ENU solutions are also highly labile, temperature- and pH-sensitive, and biologically effective dosage can be difficult to control. It is recommended that a titration series is performed for each batch prepared, and aliquots frozen at −80°C.
3.4.1 ENU Mutagenesis
3.4.1.1 Preparation of ENU stock solution
Line a fume hood with absorbent benchcoat and place a decontamination bath and waste container/burn bin within.
Prepare 100ml 5mM MES solution from 100mM pH6.0 stock in dH20 (see Note 1).
Remove ENU isopac bottle from protective canister (save can).
Using a 50cc syringe with 18g needle, inject 85.4 ml 5mm MES pH 6.0 into ENU isopac bottle, carefully withdrawing air from bottle into syringe while adding medium to avoid over-pressurizing the bottle.
Return bottle to shipping canister (or cover with aluminum foil) and place on nutator or roller shaker in hood for several hours, occasionally monitoring.
When powder is all (or nearly) in solution, swirl, allow to settle, and freeze 1ml aliquots at −80°C. Retain 20μl for spectrophotometric determination of concentration if desired.
3.4.1.2 Determining ENU concentration by spectrophotometry (optional)
100mM ENU = approx. 11.7 mg/ml (1 g/85.4ml).
Dilute 20 μl ENU solution to 1 ml with 5mm MES pH 6.0 (i.e. 1:50 dilution)
Using a disposable plastic cuvette, determine OD398.
- 1 mg/ml solution of ENU gives OD398 =0.72.
- Therefore, [ENU] mg/ml = (0D398)(50)/0.72
- or = (OD398)(69.4)
3.4.1.3 In vitro ENU mutagenesis of mature sperm
12-72 hours prior to procedure, prime 5 or more adult female X. tropicalis with 10u HCG in 100ul sterile water (see Notes 3 and 4).
On day of procedure, boost primed females with 100-200u HCG in 100ul sterile water (see Notes 3 and 4).
Prepare 10ml of 3mM MES pH6.2 in L15 (without calf serum) (add 0.3ml 100mM MES pH6.2 stock to 9.7ml L15).
Thaw an aliquot of L15/10%CS
Kill 5 males and dissect testes into L15/CS media
-
Prepare 4 15ml tubes with L15/3mM MES (see Note 1) (do not add ENU stock or sperm suspension until the last moment).
Sperm ENU 100mM stock L15/3mM MES6.2 f.c. (mM) 0.1ml 0 0.9ml 0 1ml 0.1 0.9 5mM 1ml 0.15 0.85 7.5 1ml 0.2 ml 0.8 10 Thaw an aliquot of 100mM ENU
Macerate testis in 0.5ml L15/3mM MES pH 6.2 (no CS) using eppendorf and pestle, bring volume to 1.5ml with L15/3mM MES, swirl to mix.
Add 0.1ml sperm solution to 0mM ENU control and 1ml to the 15 ml tubes with L15/3mM MES corresponding to each of the ENU treatments.
Add ENU as indicated in Step 6 and swirl to mix.
Place at 18°C for 1 hour. Swirl to mix every 15′
Add 10ml L15 to each tube and spin down sperm for 5′ 1000 rpm at RT in benchtop centrifuge.
While sperm solutions are spinning, squeeze eggs from females into a drop of 1 × MMR; discard lysing/dead eggs, pool eggs from good females, mix, and split into three 15cm dishes for the ENU doses and a smaller aliquot of eggs in a 10cm dish for no-ENU control.
At conclusion of spin, carefully pipette as much of the supernatant as possible to decontamination bath without disturbing the sperm pellet, then resuspend sperm pellet in residual liquid by flicking. Repeat steps 12 and 14 2×, then gently resuspend in 1ml L15/CS.
Remove remaining MMR from eggs, add treated sperm solution to eggs and mix by shaking briefly.
After 5′, flood with 0.05 × MMR twice (removing first rinse to decontamination bath), dejelly after ~20′ post-fertilization and transfer to fresh dishes. Eggs may now be treated as safe to handle normally.
Important: sort control and ENU-treated dishes at 4-8 cell stage, making sure to make comparable dishes of regularly-cleaving embryos from all doses and controls. It is much simpler to identify regularly-cleaving embryos at 4-8 cells than at later stages; dominant ENU-induced defects will only be scorable if equivalent regularly-cleaving embryos are compared.
-
The following 3 days, sort viable embryos and score samples of control and mutagen-treated for dominant effects on gastrulation, death and other abnormalities.
w.t.(%) gastrul. defect edema other dead control ENU 5 mM ENU 7.5 mM ENU 10mM At feeding stage, select the dose(s) that result in a population of viable embryos, but also show clear dominant effects compared to controls. If desired, expand population of animals treated at this dose.
3.4.2 Spermatogonial mutagenesis
Mitotic spermatogonia, rather than mature sperm, can be targeted for in vivo mutagenesis by injecting adult male frogs with ENU. Replication in the spermatogonial lineage then fixes mutations in the germline, avoiding mosaicism in the F1 generation. Animals usually need several months to recover after an injection series, during which time mutagenized mature sperm (which would contribute to an unwanted mosaic F1) will be cleared. Too much ENU can ablate the testes completely or result in repopulation from a small number of surviving spermatogonia, producing founder effects distorting mutagenesis rates.
Obtain 5 or more adult male frogs
Weigh individual frogs (males are typically between 6-9g).
Record weight and calculate dose of 100mM ENU stock needed for injection (0.1 mg ENU/g frog). Dose per frog (in ml) = [frog weight]×[0.1mg/g]×[1ml/11.7mg ENU], or 0.006333× frog weight.
Immobilize frogs by immersing for 2-5′ in a fresh stock of 0.07% MS222 pH7.8 at RT until they visibly begin to slow down. Immobilizing frogs with anaesthetic during injection reduces risk of serious accidents with ENU-contaminated needles.
Inject the volume calculated in step 3 to contain 0.1 mg ENU per gram of frog weight subcutaneously into dorsal lymph sac (see Notes 2 and 3).
Allow frogs to recover on wet paper towels in observation tank. They are usually awake and active in~15-20 minutes.
Transfer frogs to observation tank with fresh water. Make sure to discard paper towels and liquid in ENU waste and treat appropriately as ENU waste material.
After several hours, discard frog water in ENU liquid waste and replace with fresh water.
The next day, discard frog water in ENU liquid waste and replace with fresh water. Do this throughout the day for two to three more water changes.
Return frogs to colony.
Re-inject once a week for a total of 3 doses.
Allow frogs to recover for >3 months before breeding.
3.5 Genetic Screens
Forward Genetic Screens
Conventional 3-generation breeding schemes to uncover recessive phenotypes are compatible with spermatogonial mutagenesis (Figure 3). However, in vitro mutagenesis of mature sperm results in a mosaic F1 generation, making recovery of homozygotes by incrossing in subsequent generations prohibitively inefficient.
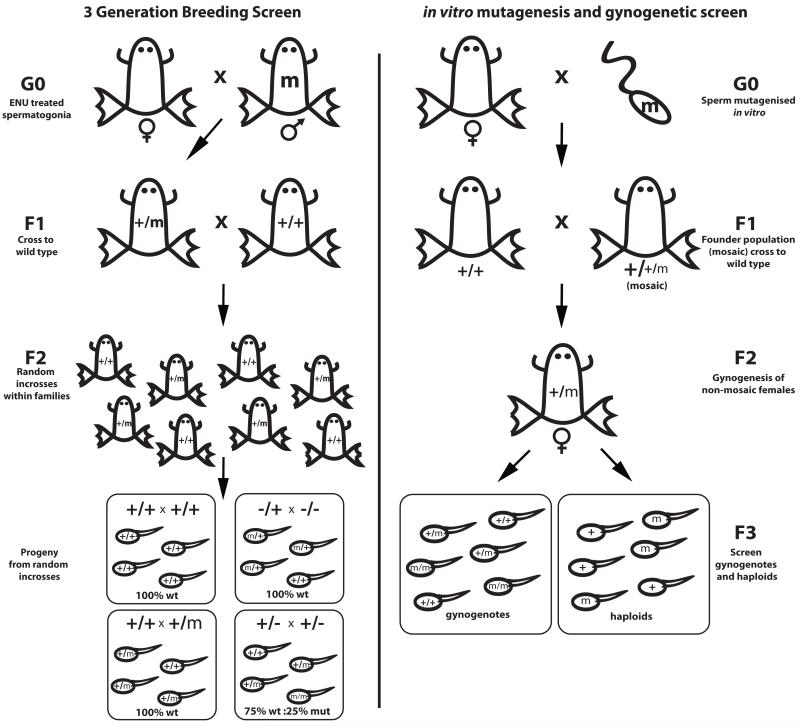
Figure 3. Screening mutagenised populations.
Left: 3-generation breeding screen. In vivo mutagenised males are crossed to wild type females (G0), with the resulting F1 individuals outcrossed again to wild type to create F2 families. Progeny of random incrosses within families are then analysed for mutant phenotypes. Right: in vitro mutagenesis and gynogenetic screen. Eggs are fertilised with mutagenised mature sperm, creating a mosaic F1 population, which is crossed again to create a population of non-mosaic F2 candidate carrier animals. F2 females are then screened by gynogenesis to uncover recessive mutations.
Populations derived from in vitro mutagenesis can still be effectively screened by gynogenesis. Mutations have been successfully recovered by screening the gynogenetic progeny of mosaic F1 females (15), but screens using non-mosaic F2 females are likely to be more efficient. Gynogenesis by polar body suppression (early cold shock) produces some bias towards recovery of centromere-linked alleles, as these loci will be uncovered in a higher proportion (up to 50% of gynogenetic progeny (gynogenotes)) than those produced by more distal loci (as low as ~10% phenotypically mutant for loci unlinked to centromeres, see section 3.7.1). However, females in good condition can produce sufficient numbers of gynogenotes to recover mutations even in telomeric loci.
3.5.1 Early coldshock gynogenetic screen for recessive phenotypes
Gynogenetic screens are primarily used to identify carrier females that are heterozygous for recessive mutations. Ideally, non-mosaic animals are screened, since a greater proportion of progeny will be homozygous for a given mutant locus and more readily detected. Adult females being screened may be housed individually or otherwise marked for the duration of the screen.
Early coldshock gynogenesis (see section 3.2.2) should include haploid and diploid outcross (non-irradiated sperm) controls. The diploid outcross control serves to assess sperm and egg quality, reveals dominant effects, and can be raised as the next generation if desired. Haploid controls help evaluate efficiency of sperm irradiation (see section 3.2.1), and are useful for identifying polymorphisms (section 3.7.2). Some anterior phenotypes may also be scorable on the haploid background, where they would be observed in ~50% of the embryos.
The fraction of ECS gynogenotes in which a given recessive phenotype is observed is not Mendelian. Loci that are close to centromeres will be uncovered in up to 50% of the gynogenetic progeny of heterozygous females. More distal loci will tend to be observed with progressively lower frequency, with a plateau at ~10-15% due to presence of multiple crossovers.
ECS may not result in quantitative rescue of all haploid embryos to diploidy. Background abnormalities from remaining haploid and aneuploid embryos can make it challenging to identify pre-neurulation phenotypes. Post-neurulation phenotypes can be screened efficiently by selecting morphologically perfect wild type embryos from ECS and diploid control dishes on the morning after fertilization (st. 18-22) and monitoring these for subsequent appearance of abnormalities.
3.5.2 Morphological screening checklist
The following screen can be applied to embryos derived either from gynogenesis or from conventional matings; for simplicity, gynogenesis is described.
Compare sibling outcrossed diploid with ECS and haploid embryos for stage-specific developmental processes and to establish a baseline of egg-based non-heritable abnormalities and/or dominant phenotypes. Specific phenotypes that are uncovered in multiple embryos within a clutch are particularly convincing. If you see a phenotype in ECS or haploid dishes, separate those embryos and record specific defect(s) and number of phenotypically mutant and wild type embryos. The phenotype might be lethal; isolating those embryos will make it easier to score the following day. Phenotypes that are scorable in haploids are expected at 50%. Single-gene phenotypes in a clutch of ECS embryos are expected at a maximum of 50% for centromere-linked loci, decreasing to ~10% for distal loci. Record all abnormalities on a scoresheet (example in Figure 4). Collect both mutant and wild type ECS embryos in 96 well plates for use in low-resolution mapping and assignment of linkage group (see section 3.6.1 and 3.7). Also collect a small set (6-12) of haploid embryos for identifying polymorphic markers (section 3.7.2).
Figure 4. Gynogenetic screening checklist.
Sample form for scoring phenotypes during a gynogenetic screen.
Day 0: Gynogenesis & Cleaving Embryo Sorting
Perform Gynogenesis on potential mutant carriers as outlined in section 3.2.2.
Sort regularly-cleaving embryos from unfertilized embryos at 4-16 cells. Irregularly-cleaving embryos will gastrulate poorly, increasing background noise and making it more difficult to detect specific phenotypes. Likewise, treat embryos with optimum care to minimize abnormalities caused by overcrowding or other mistreatment (see Chapter 2).
Day 1: Tailbud stage sort (~16-20 hpf; St. 18-24)
As early as possible, sort normal from dead/abnormal embryos in all dishes, again to obtain a low background of early defects upon which to recognize later-developing phenotypes. Record number of dead embryos and remove them. Remove abnormally developing embryos from ECS dish to fresh plate noting phenotype.
Check ECS dishes for any obvious axial or dorsoventral polarity defects. If these are seen in >50% of embryos, the defect is likely due to imperfect gynogenetic rescue or poor egg quality.
Sort Haploid dishes and discard embryos that fail to develop reasonable heads, then score those with good heads for % of “diploid-looking haploids”. True haploids typically display posterior truncations, failure of blastopore closure, and raised neural folds. Appearance of diploids can be due to either spontaneous polar body failure (in which case both diploid-appearing haploids and ECS can be scored for recessive phenotypes) or failure to inactivate sperm DNA, resulting in diploid embryos in the haploid control and triploid embryos in the ECS dish, reducing the proportion of embryos in which recessive phenotypes may be detected.
Day 2: (48 hpf; St. 35-40)
Check for:-
Axial defects: Embryo shape, truncation/kinking, gross tissue defects.
Mobility: Swirl embryos to centre of dish and gently poke with forcep tip. Wild type embryos respond by twitching or swimming away.
-
Add a few drops of 1:1000 MS-222 to dish, swirl, and repeat until embryos are immobilized.
Score the embryos for the following defects:
Cilia function: do anaesthetized embryos ‘glide’ forwards due to coordinated beating of epidermal cilia?
Heartbeat: does heart beat at normal rate? Note that anaethesia can affect heart rate.
Circulation: Look at the tail above, below and in between the somites for blood movement.
Kidney: is pronephros forming/looping?
Somites: Are somites forming in comparable number, thickness, chevron-shape as wild type?
Pigmentation: Retinal pigment epithelium (RPE) defects? Have melanocytes formed and taken on the spreading star shape? Is there an increase in, or strange patterns of pigmentation?
Edema: Check for edema in unusual or interesting places. Nonspecific edema often forms around the ventral abdomen or heart, but can also be associated with specific phenotypes, e.g. heart, vascular or kidney defects,, and should not necessarily be disregarded.
After screening transfer embryos back to fresh media without anesthetic.
Day 3. (72 hpf; St. 40-43)
Check for:-
Repeat Day 2 checks. Embryos without heartbeat will probably display edema by now. If not, check for blood flow and note any accumulation of blood in the body cavity.
Gut defects: Check for correct coiling of the gut. Does coiling occur, is it always in the same direction? Stage comparison is important.
Otolith/Otic vesicle: Is the size and shape of otic vesicle correct? Are both otoconia present, with normal crystal morphology?
Day 4. (96 hpf; St. 43-46)
Repeat previous checks. Saccular and utricular otoliths will be much clearer on day 4.
Head morphology: By day 5 of development the head will have flattened and cleared. Compare jaw morphology and hindbrain segmentation with diploid controls
3.6 Reverse genetic strategies in X. tropicalis
In contrast to forward genetics, where first mutant phenotypes are described and then the underlying sequence lesion is identified, in reverse genetics the phenotypic outcomes of mutations in previously known sequences are studied. In vertebrates, reverse genetics often refers to mouse knockouts via homologous recombination in ES cells. Equivalent procedures do not currently exist for X. tropicalis, although intriguingly, Xenopus oocytes and extracts efficiently perform extrachromosomal homologous recombination (37) and homology-flanked integration constructs have not been evaluated for targeting. More recently, chimaeric zinc finger nucleases have been shown to be effective for targeted gene disruption in X. tropicalis (27) (and Chapter 7) and zebrafish (38).
Alternatively, reverse genetics can also refer to scanning populations of animals or plants to identify carriers of mutations in particular sequences. One such strategy, Targeting Induced Local Lesions In Genomes (TILLING), shows promise in X. tropicalis (15, 39). Genomic DNA samples are obtained from a population derived from mutagenesis, from which coding regions of target genes are evaluated for induced mutations. This approach was initially developed with non-sequence based methods such as Cel I digestion (refs)(see also Chapter 7) and denaturing HPLC to evaluate a discrete set of amplicons for specific genes from a large mutagenized population, and subsequently adapted for capillary sequencing. However, PCR amplification introduces allele bias and other errors, compromising high-throughput screens (40).
Strategies based on next-generation sequencing technologies can scale up this approach while limiting PCR-based allele bias. One attractive variation is based on directly sequencing the protein-coding space (exome) of genomic sequence. This exon capture involves solution hybridization of sheared genomic DNA with a set of synthetic baits representing the entire exon set (or a subset of specific genes of interest). Non-coding regions are discarded, increasing sequence density in genomic regions likely to give phenotypes and reducing dependence on PCR. Hybridized exomes are then subjected to next-generation sequencing and inspected bioinformatically for heterozygous lesions. Multiple indexed genomes can be run on a single lane of some available sequencing apparatus.
X. tropicalis is particularly suited for TILLING studies since mutagenized populations can be archived both as frozen sperm (see section 3.15, (41)) and living stocks. These frogs are significantly longer-lived than other vertebrate genetic models, and can still breed when more than 10 years old. Several X. tropicalis TILLING resources are currently under construction.
In addition to reverse genetic strategies, direct identification of mutations underlying phenotypes recovered from forward screens can be accomplished by exon capture sequencing of tissue from carrier animals as described below. However, given the imperfect annotation of the X. tropicalis genome, not all mutations will be contained in the exon capture set. One very useful variation simultaneously obtains valuable map information. Sequencing exomes harvested from separate pools of ~50 mutant and wild type embryos provides linked SNPs to define the mutation-containing genetic interval as well as possibly identify the mutation directly.
3.6.1 Xenopus tropicalis exon capture sequencing
We have successfully used Agilent Sureselect technology to enrich for the whole X. tropicalis exome, and sequenced the products on the Illumina HiSeq platform. It should also be possible to enrich for specific subsets of genes. Detailed protocols for high-throughput sequencing and solution hybridization kits are available from manufacturers. The following method describes our approach using the genomic resources available at the time.
Design an exome set comprising all X. tropicalis JGI v4.2 gene build Enseml exons, plus cDNAs and ESTs. 2× tile the exome with 120mer oligos in silico, and obtain biotinylated RNA baits corresponding to the 120mer sequences from Agilent. Pool all baits (84Mb, covering the entire known X.tropicalis exome with some duplication due to cDNA/EST overlap with Ensembl exons) into a single tube.
Toe clip live non-mosaic F2 animals derived from in vitro ENU mutagenesis and prepare genomic DNA (see section 3.9.1). F1 animals derived from spermatogonial mutagenesis are also suitable.
Shear DNA using a Covaris sonicator down to 150-300bp.
Prepare a paired end Illumina sequencing library using standard Illumina reagents.
Hybridize the library for 24 hours against the Agilent SureSelect baits using manufacturer’s reagents and conditions.
Isolate hybridizing exome fragments by incubating with streptavidin-coated magnetic beads, wash away non-coding sequences, and digest RNA baits according to manufacturer’s instructions.
Amplify enriched library using Illumina primers, quantify, load onto a single lane of the Illumina HiSeq flowcell, and sequence with paired-end runs (e.g. 76 bp).
Map Illumina HiSeq output back to exome; typically >60% of sequences are found to be on target.
Interrogate sequences that hit the exome with three SNP calling programs (e.g. Qcall, mpileup and GATK), and merge the output from all three programs into a single .vcf file. Note: use of three different SNP-callers is stringent, and may exclude many legitimate SNPs.
Predict consequences of SNPs using the Ensembl genebuild and filter to identify nonsense and essential splice mutations.
Confirm high probability nonsense and essential splice mutations in genes of interest using capillary sequencing of amplicons or another independent SNP detection method (e.g. Kasp (KBioscience) or dCAPS (section 3.7.7.3) to evaluate specific genomic DNA samples. Specific SNP detection methods are also valuable for identifying carriers in the next generation and genotyping embryos during phenotypic analysis.
Outcross animals carrying mutations of interest to obtain an F3 generation for phenotypic analysis. Alternatively, eggs from female F2 carriers can be subjected to gynogenesis to uncover mutations for preliminary studies.
Perform linkage analysis to associate candidate phenotypes with specific identified sequence lesions. Chemical mutagenesis can introduce multiple lesions per genome.
3.7 Mapping mutations
Conventional positional cloning is based on associating a mutant phenotype with nearby naturally occurring differences (polymorphisms) between a mutation-carrying chromosome and a wild type chromosome. To simplify identifying and refining mutation-containing genetic intervals, it is useful to obtain ‘mapcross’ animals that are hybrids between the strain on which the mutation was originally induced and a polymorphic strain which differs at many sequence loci. Many of the mapping strategies developed in other genetic systems (42) can be applied directly to mapping in X. tropicalis. X. tropicalis has several unique advantages for positional cloning. The contiguity of the X. tropicalis genome sequence is steadily increasing (14), and a draft chromosome-scale assembly is accessible at www.xenbase.org. A meiotic map of ~2900 Simple Sequence Length Polymorphisms (SSLPs) has been organized into 10 linkage groups corresponding to the 10 tropicalis chromosomes (30). Many phenotypes uncovered by gynogenesis can be rapidly assigned to one of the 10 chromosomes using a small set of centromere markers (see section 3.7.4 and (7)). Fm, the fraction of phenotypic gynogenotes, also provides an estimate of the gene-centromere distance. Such low-resolution map information is useful for evaluating candidate genes. Higher resolution mapping is accelerated by the large numbers of embryos produced; upwards of 5000 meioses can be scored routinely from a single cross. Figure 5 shows a flowchart with conventional mapping strategies. As the cost of sequencing continues to drop, mapping mutations by direct sequencing of all protein-coding genes using exon capture is also becoming increasingly feasible (see section 3.6).
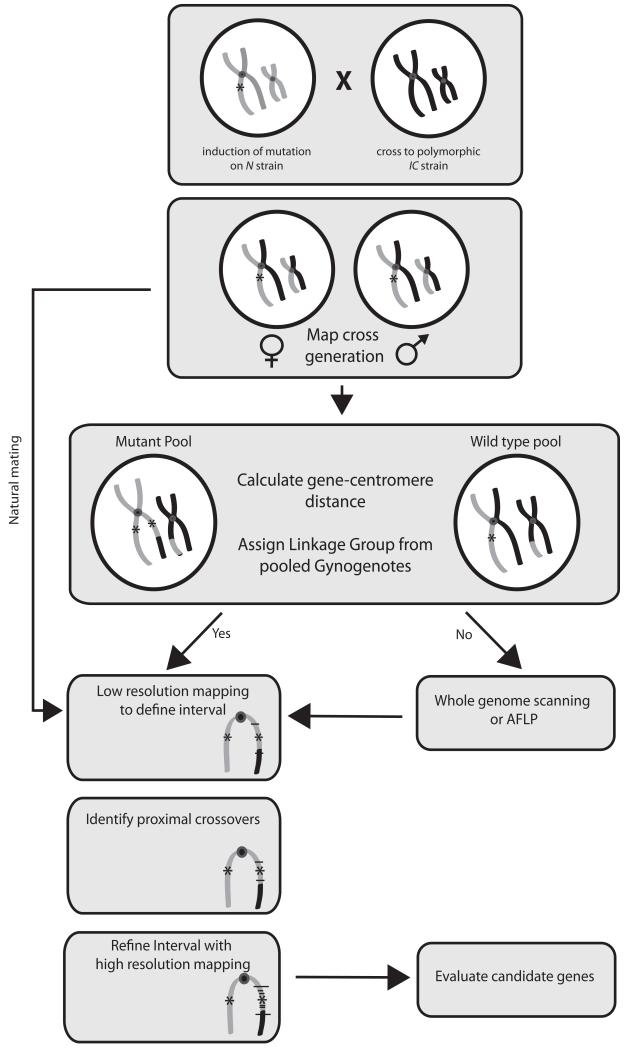
Figure 5. Flowchart for genetic mapping in X. tropicalis.
A recessive mutation induced on one strain (grey) is crossed to a polymorphic mapping strain (black) to obtain hybrid map cross carrier animals. Gynogenetic embryos are obtained from map cross females to calculate gene-centromere distance and for bulk segregant analysis with centromere markers to identify linked chromosome. Conventional crosses between map cross carriers are performed for subsequent analysis. If chromosomal linkage cannot be assigned by bulk segregant analysis, whole genome scanning with polymorphic markers, or Amplified Fragment Length Polymorphism (AFLP) analysis can be used. Low resolution mapping with a small number of mutant embryos is used to identify markers ~3-10cM apart flanking the mutation. These two flanking markers are then used to type large numbers (>500) of mutant embryos to identify those with crossover events between the flanking marker and the mutation. Small sets of recombinants can then be analysed with further markers to refine the interval and number of genes contained within it. Candidate genes are then evaluated by changes in gene expression, spatial expression of transcripts and cDNA sequence. Functional confirmation of any mutation found is accomplished by morpholino phenocopy and rescue with mRNA.
3.7.1 Embryo genomic DNA prep
Genomic DNA for mapping is prepared from whole embryos using a proteinase K based lysis buffer.
When embryos are at least 3 days old, sort phenotypic mutants into a separate dish using a flamed Pasteur pipette.
Place mutant embryos individually in wells of 96 well plates
Collect ~12-24 wild type embryos from the same breeding into clearly marked wells
Remove excess media from each well and freeze at −80°C unless prepping genomic DNA immediately
add 50μl of Lysis buffer w/ proteinase K
Incubate in PCR machine at 56°C for 4 hours followed by 5 minutes at 95°C
Use directly in PCR; no clean up required for most mapping applications (see Note 10)
3.7.2 Identifying polymorphic markers
Differences in the number of short di-, tri- or tetranucleotide repeats, a.k.a. Simple Sequence Length Polymorphisms (SSLPs) are abundant between different strains of frogs (e.g. N and IC), so ideally mapping is conducted on the offspring of N/IC or other hybrid mapcross animals to locate markers linked to the phenotype. However, in all but the most inbred stocks, sufficient polymorphisms for low-resolution mapping are still likely to be present. It may be necessary to test several candidate SSLPs in a genomic region to identify those that are polymorphic in a given cross.
First prepare the following
Genomic DNA extracted from haploid embryos from a mapcross hybrid female (see section 3.7.1)
PCR master mixes with range of potential polymorphic markers
Procedure
Transfer 2μl of DNA from 6 individual haploid embryos to fresh tubes
Set up PCR master mix with primers for marker to test
Add 8μl of Master Mix to each individual Haploid embryo DNA
Run PCR under the following conditions 94°C - 2 minutes, 35 cycles of (94°C - 30 seconds 58°C - 30 seconds 72°C - 1 minute), 72°C for 5 minutes, 4°C Hold.
Run 5μl on 3% Super Fine Resolution (SFR) agarose gel or polyacrylamide gel and silver stain (see section 3.7.3)
If individual haploids produce different molecular weight PCR bands at ~1:1 ratio, the marker is polymorphic in the female parent and can be used for mapping.
Repeat for each candidate marker
3.7.3 Polyacrylamide gels and silver staining
While agarose gels are quick and convenient, resolution is limited and subtle polymorphisms may be missed. Single-base resolution can be obtained using standard sequencing-style denaturing 6% polyacrylamide/8% urea gels, visualizing DNA bands with silver nitrate. This protocol was adapted from (43).
Thoroughly clean and dry both glass plates.
Coat small glass plate with mixture of 5ml 100% EtOH, 75μl 10% acetic acid and 5μl 3-(trimethoxysilyl)propyl methacrylate.
Wash with dH2O followed by 70% EtOH, wipe and allow to dry.
Spray large glass plate with acrylease (Stratagene). Wait 5 minutes then wipe with clean wet tissue.
Pour a 6% acrylamide gel (containing 8M urea) with shark tooth combs.
Pre-run gel in 1×TBE for 30′ at 80W.
Dilute PCR 1:2 with denaturing DNA loading buffer and heat to 95°C for 3 minutes in a thermocycler.
Load 5μl of sample onto gel and run at 55W. Run is complete when the buffer front passes through the bottom of the gel
Split the glass plates apart with a razor blade
Transfer glass plate containing gel to a large photographic developing dish. Gel side up.
Fix gel in 1L 10% EtOH (this can be reused up to 6 times) for 10′
Wash in 1L 1% nitric acid for 3′ (this can be reused twice)
Rinse twice in dH2O 3′ for each wash
Stain for 20′ in 1L silver nitrate
Rinse twice in dH2O 3′ for each wash
Add 1L developing solution and agitate gently until bands appear. This is usually within 5′ depending on temperature of the solution
Stop the reaction in 10% acetic acid for 5′.
Wash gel in dH2O for 10′
Transfer to a light box to photograph with a standard digital camera
3.7.4 Low-resolution mapping with gynogenesis and centromere markers
Placing a mutation on a chromosome, combined with the rough gene-centromere distance provided by phenotype ratio in gynogenetic embryos, allows a genomic region to be inspected for candidate genes. The initial step in positional cloning usually entails defining the chromosome or genetic linkage group that contains the mutation. In many cases this can be accomplished rapidly by analyzing pools of mutant and wild type gynogenetic embryos with polymorphic markers located near each of the ten X. tropicalis centromeres to identify one which segregates with the mutant phenotype (see (23). Examination of DNA from pools of mutant and wild type, know as bulk segregant analysis, simplifies rapid identification of markers linked to the mutant phenotype.
As outlined in section 3.2.2, gynogenesis prevents second polar body extrusion allowing the post-recombination sister products of meiosis II to be retained. The genome of a gynogenote is therefore completely maternally-derived, but not completely homozygous, analogous to half of a yeast tetrad (see Figure 1 and (44). Polymorphic markers at the centromeres, where each pair of sister chromatids is held together during recombination, will be homozygous, with the different alleles segregating into different individual gynogenotes (see Figure 1). Gynogenetic embryos that are phenotypically mutant for a recessive allele are also by definition homozygous at this mutant locus. If the mutation is located reasonably close to a centromere, a pool of mutant gynogenotes will also appear homozygous for the cognate centromeric marker derived from the mutagenized strain, while the wild type pool will contain the alternative allele (see Figure 6). For the chromosomes that do not contain the mutation, both centromere alleles will contribute equally to mutant and wild type pools. In this fashion, linkage can be established using only the small set of markers corresponding to centromeres of the 10 different chromosomes, and two pools of mutant and wild type gynogenetic DNAs.
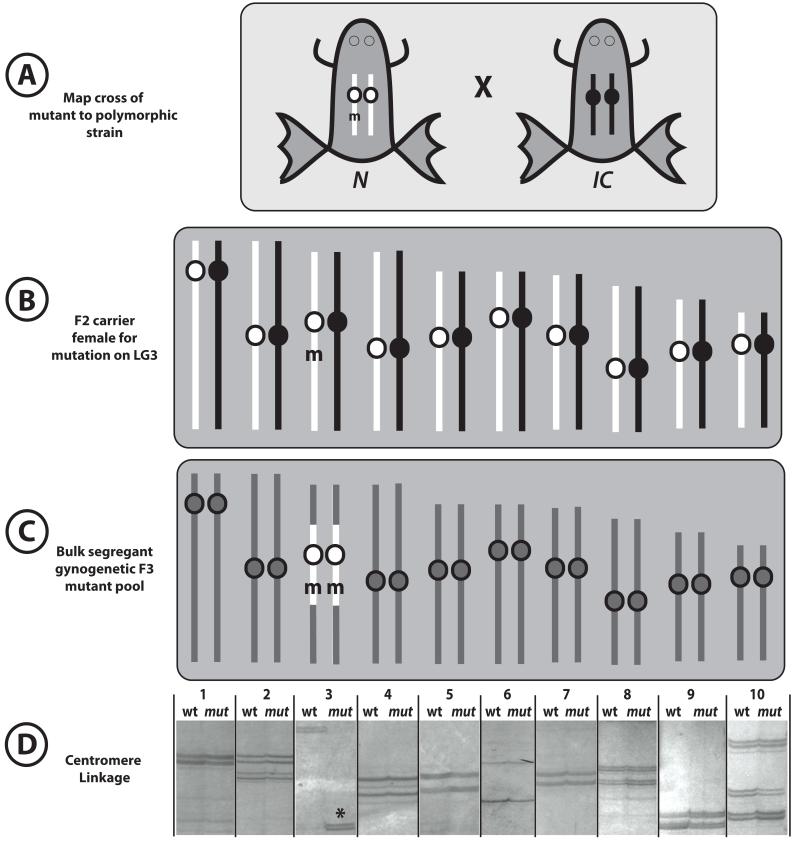
Figure 6. Assigning chromosome linkage by bulk segregant analysis.
(A) A frog carrying a recessive mutation m on the N strain (white chromosomes) is crossed to a polymorphic IC strain (black chromosomes). (B) ‘Mapcross’ hybrid F2 carrier inherits one chromosome from each parent. (C) Pools of ~20 phenotypically mutant and wild type gynogenetic embryos are collected (mutant pool represented). Unlinked chromosomes show equal contribution from white N and black IC alleles (gray chromosomes) in both mutant and wild type pools. However, on the chromosome containing the mutation, the mutant pool is greatly enriched for the white N centromeric allele; the wild type pool may contain either the IC allele or both N and IC . Centromere linkage can often be detected over large genetic distances in gynogenetic embryos. (D) Silver-stained gel showing pools of mutant and wild type embryos scored with polymorphisms at the 10 X. tropicalis chromosomes. Linkage is detected to chromosome 3.
Conveniently, this apparent centromere-mutation linkage extends to more distal mutant loci. Consider a recessive mutation m1 induced on the N background and crossed onto the polymorphic IC strain to create a heterozygous N*/IC carrier female (Figure 1). The gynogenetic offspring of such a hybrid will thus each be homozygous N/N or IC/IC at all centromeres, and the mutant embryos will be Nm1/Nm1 at the mutant locus. For a mutation m2 further from its centromere, recombination events are more likely in the interval, resulting in gradual accumulation of wild type Nm1/IC heterozygotes and decreasing the fraction of mutant gynogenotes. The wild type pool will thus contain both IC/IC (from the original parental allele) and N/N centromeres (from single crossovers producing heterozygotes at the mutant locus). However, the reduced fraction of Nm1/Nm1 mutant embryos are still likely to be homozygous N/N at the corresponding centromere. The exceptions derive from multiple crossover events; half of double crossovers will return linkage to the original centromere allele, while half may switch to the alternative non-parental allele (see (7)). Only when the mutant locus is so distal that the majority of gynogenetic embryos contain multiple intervening crossovers will the mutation no longer appear linked to its centromere.
In addition, a rough estimate of the gene-centromere distance can be obtained from the proportion of mutant gynogenotes observed. If we assume that only single crossovers are present, then
In practice, this formula provides useful information for loci less than ~30cM from centromeres (Fm > 0.2), where single crossovers predominate. If Fm <0.2, the gene-centromere calculation only establishes that the locus is further than 30cM from the centromere, as multiple crossovers are common in the longer chromosome arms. This rough map information can be used to refine candidate gene sets and to select markers for higher-resolution linkage analysis.
3.7.4.1 Assigning linkage group by bulk segregant analysis
First prepare the following
2 pools of DNA from 10-20 phenotypically mutant gynogenotes (5μl from each). Single pools and smaller numbers of embryos can be used, but risk of false positives increases.
2 pools of DNA from 10-20 sibling wild type gynogenotes (5μl from each)
Primer stocks for SSLP markers closely linked to the centromere (see genetic locations of centromeres in (7)).
Procedure
Identify polymorphic SSLP markers within 1.5 cM of X. tropicalis centromeres (see section 3.7.2 and (23)).
Aliquot 2μl of DNA from the two mutant and wild type pools into PCR reaction tubes for each polymorphic centromere marker being tested.
Add 8μl of a standard PCR master mix for polymorphic centromere marker to each tube.
Run PCR under standard conditions for 35 cycles with an annealing temperature of 58°C (for all Wells et al map SSLPs).
Run 5μl on 3% SFR Agarose gel. Some polymorphisms are only scorable using higher resolution 6% polyacrylamide sequencing gels followed by a silver stain (see section 3.7.3).
If a mutation is linked to a given centromere, one band will predominate in mutant lane; the corresponding wild type lane will show either the other band (consistent with a tightly-linked locus) or both species (consistent with a more distal locus). Unlinked centromeres will display identical mutant and wild type bands (see Figure 6).
Repeat until linkage is observed or polymorphisms at all 10 centromeres have been tested (if no linkage is detected, see sections 3.6 and 3.7.5).
Confirm by testing individual embryos with linked centromere marker, and determine linked chromosome arm by testing markers ~5cM either side of centromere.
3.7.5 Alternate strategies
Exon capture for direct sequencing to identify mutations is described in section 3.6; in this section we discuss inexpensive alternative approaches for low-resolution mapping. Distal loci may appear unlinked to centromere markers in bulk segregant analysis, where a small minority of embryos with multiple crossovers can obscure linkage in pools. Weak linkage can sometimes be confirmed by scoring >20 individual mutant gynogenotes for centromere markers.
Alternatively, the larger chromosome arms, or indeed the entire genome, can be scanned using more polymorphisms 10-20 cM apart. Scanning can be performed using bulk segregant analysis with either gynogenetic embryos or the progeny of conventional crosses. Scanning strategies can often be efficiently combined with candidate gene approaches. If similar phenotypes have been characterized in other organisms, X. tropicalis orthologs may be located on the genomic sequence assembly and/or the meiotic map. Nearby SSLP markers may be used for scanning if the scaffolds have been mapped. If not, it is straightforward to identify microsatellite repeat regions and generate homemade ‘bespoke’ markers (see section 3.7.7). Linkage analysis can detect unrelated mutations in the vicinity as well as mutations in the candidate genes themselves.
Amplified Fragment Length Polymorphism (AFLP) analysis can provide a more direct route to obtaining linked sequences, but is somewhat laborious. In brief, mutant and wild type pooled genomic DNAs are digested and randomly amplified in a fashion that allows control over complexity of products (45). The maximum number of bands that can be analyzed on sequencing gels can then be inspected for differences between mutant and wild type. Bands that are present in wild type but not mutant lanes are candidate linked wild type alleles. These can be cut out of the gel, re-amplified, sequenced, and placed on the genome assembly. Additional nearby bespoke SSLPs are then obtained from the identified sequence scaffold and tested to confirm linkage. Convenient AFLP kits are available, to facilitate AFLP analysis, for example e.g. Invitrogen AFLP Analysis System I.
3.7.6 Higher resolution mapping
Most of the considerations for subsequent steps in positional cloning are not specific to X. tropicalis. Gynogenetic embryos, which have fewer crossovers on the centromere side of the mutant locus, are less suitable for fine mapping than those derived from a conventional mating with useful crossover events on both sides. Conventional crosses can also provide larger numbers of embryos than gynogenesis; for successful positional cloning, at least 1000 mutant embryos are often required.
After placing a mutation on a linkage group, the next step involves locating the mutation between two easily-scorable flanking markers <10 cM apart. Initially, the linked chromosome can be scanned with markers spaced at ~10cM intervals, using bulk segregant analysis of mutant and wild type pools of ~20 embryos. Polymorphisms showing strong linkage are then evaluated using ~24-48 individual mutant embryos and 6-12 wild type siblings, and other nearby markers from the meiotic map tested. Markers further from the mutation will yield more recombinants (heterozygotes) than closer markers. Importantly, markers on opposite flanks of the mutation give non-overlapping sets of recombinants, and markers on the same side share recombinants. Flanking markers should be <~10cM apart, and should be relatively easy to score, i.e. simple 2-allele systems, preferable distinguishable on agarose gels, as these will be used to genotype large numbers of embryos.
After flanking markers have been obtained, genotype the available mutant embryos with them to identify those that are recombinant. Mutant embryos that are heterozygous at a flanking marker have informative crossovers closer to the mutation. Once a set of >10-20 recombinants has been obtained, these can be typed with additional markers distributed evenly between the flanking markers to narrow the interval. The available resolution is determined by the number of meioses scored. Genomic regions corresponding to genetic intervals can be inspected for candidate genes easily using Ensembl BioMart (section 3.8.1). intervals can usually be subdivided rapidly with additional markers from the meiotic map, bespoke SSLP markers or SNPs can be generated (see section 3.7.7) to refine the interval.
In some regions, microsatellite repeat polymorphisms may be difficult to find. Single Nucleotide Polymorphisms (SNPs) are abundant in polymorphic crosses. These usually can be identified by sequencing random amplicons from intergenic regions from several mutant and wild type individuals. SNP-containing regions from mutant embryos can be resequenced for linkage analysis, or identified SNPs can be converted into a variety of high-throughput assays. Many SNPs disrupt or create restriction sites, so alleles can be distinguished by digesting an amplification product (Cleaved Amplified Polymorphic Sequence, CAPS). A variation based on introducing a mismatched base in primers, ‘dCAPS’ (derived Cleaved Amplified Polymorphic Sequence) (46) can produce differentially-cleavable alleles starting from any SNP sequence. Kasp and related genotyping techniques combine Fluorescent Resonance Energy Transfer (FRET) with allele-specific PCR to discriminate between SNPs using dye-linked allele-specific primers (www.kbioscience.co.uk). Many other techniques to discriminate between SNPs are continuously being developed.
3.7.7 Bespoke markers for mapping
The X. tropicalis meiotic map (30) currently arranges ~2900 SSLPs in 10 linkage groups corresponding to the 10 X. tropicalis chromosomes. While this provides ample marker density for linkage assignment and rough mapping, higher resolution can require identification of bespoke (custom) markers. SSLP markers are convenient to identify and score, but high-resolution mapping may exhaust SSLP candidates in a region. Single Nucleotide Polymorphisms (SNPs) are abundant in most crosses and also useful for high-resolution mapping.
3.7.7.1 Obtaining bespoke SSLPs
Download FASTA-formatted sequence corresponding to an X. tropicalis genomic region of interest from a genome browser (e.g. JGI, UCSD, Xenbase G-Browse or Ensembl) .
Go to Tandem Repeat finder website (http://tandem.bu.edu/trf/trf.html)
Click “Submit a Sequence for Analysis”
Click “Basic”
Copy and Paste or upload FASTA format sequence to website
Click “Submit Sequence” button (may take a few minutes to load)
On following page click “Tandem Repeats Report”
Look through second column of table (Period Size) for 2, 3 or 4 (di, tri or quad repeat)
Next look through third column (Copy Number) for 2, 3 or 4 period repeats and identify those with 10-30 copies
Click on link for these repeats in first column (Indices)
The following pages give the repeat and sequence flanking
Search the original FASTA format .txt file for the repeat plus 30-50 bp of flanking sequence
Go to Primer3 website (e.g. http://frodo.wi.mit.edu/)
Copy and paste the identified repeat plus flanking sequence into text box on Primer3 (leave parameters unchanged)
Click “Pick Primers” button
Chose primers flanking the repeated sequence
Repeat steps 12-16 for all period sizes of 2, 3 or 4 with a copy number of 10-30
3.7.7.2 Bespoke SNPs
SNPs are abundant in intergenic noncoding regions, and may be identified directly by sequencing PCR products (amplified with a high-fidelity polymerase) from mutant and wild type embryos.
Design PCR primers to amplify ~ 400bp from non-coding regions >5 kb away from exons.
Using a proofreading polymerase, amplify from 3 or more individual wild type and non-recombinant mutant embryos, or >4 unsorted haploids
Sequence all 6 fragments and compare, looking for consistent single nucleotide changes between wild type and mutant embryos.
Analyze by Snip-SNP, dCAPS, or by sequencing amplicons from individual mutant and wild type embryos.
3.7.7.3 Snip-SNP and Derived Cleaved Amplified Polymorphic Sequence Analysis (dCAPS)
Many SNPs identified by sequence result in RFLPs, or Snip-SNPs, that are simpler and cheaper to score than by sequencing multiple embryo DNAs. dCAPS allows virtually any identified SNP to be converted into a snip-SNP for high-throughput analysis (46) by generating PCR primer sequences in which a mismatch is introduced, converting one of the SNPs into a specific cleavable polymorphism. The PCR products can then be digested and compared. The most time-consuming part of this process can be generating mismatched primer sequences to create snip-SNPs; the authors of the dCAPS method have thoughtfully generated an online ‘dCAPS Finder’ at http://helix.wustl.edu/dcaps/dcaps.html. This program also conveniently identifies enzyme choices if conventional snip-SNPs exist between two sequences. SNP-containing genomic regions from mutant and wild type embryos can then be amplified by PCR, digested with the allele-specific restriction enzymes suggested by the dCAPS finder program, and distinguished on agarose gels.
3.8 Evaluation of candidate genes in mapped interval
Strong candidates for gene functions underlying mutant phenotypes can often be identified from larger gene lists based on previously known phenotypes or functions and expression data. Candidate genes are then evaluated in the mutant by expression analysis, cDNA and/or genomic sequence, and functional association (e.g. mRNA rescue or MO phenocopy).
3.8.1 Ensembl BioMart
Mutation-containing intervals can be rapidly inspected for candidate genes by downloading lists of gene models, including GO terms and protein domain information, using the Biomart tool in Ensembl (www.ensembl.org). At the time of writing, this resource references the X. tropicalis v4.1 assembly.
Click on Biomart link (top right)
Choose database (a high number Ensembl or Vega)
Choose dataset ‘X. tropicalis genes’
Under “Filters” in the left hand menu, select “Region” and input scaffold information under ‘Multiple Chromosomal Regions in the format ‘Scaffold_Number:base-base (e.g. ‘scaffold_1 :xxxxx-yyyyy’ to obtain genes on the scaffold between polymorphisms flanking a mutation at base xx,xxx and yy,yyy. GO terms and external references can be added under the “Attributes” section on the left under the “GENE” subsection. Clicking ‘count’ at top left gives the number of Ensembl genes in the set.
Generate a spreadsheet with all the genes and transcripts by clicking ‘Results’ at the top left of the page.
3.8.2 Analysis of candidate cDNAs
Compelling candidate genes in the mutation-containing interval can be evaluated in a number of ways. RT-PCR or in situ hybridization with 3′ probes may be used to detect changes in expression levels in mutant embryos. These are not necessarily the result of changes in transcription level; mutations which introduce stops are frequently degraded by Nonsense-Mediated Decay (47). Likewise, immunostaining or western blot analysis is useful where antibodies are available. Sequencing specific cDNAs from the mutant is an informative and inexpensive option unless the gene is very large.
3.8.3 Confirmation of candidate genes
Many mutagenesis procedures will introduce multiple lesions per genome; induced base changes from chemical mutagenesis are detected as frequently as 1/50,000 bases. Even if a sequence lesion is identified in a coding region within the genetically-defined interval, other mutations may also be present, and independent evidence is usually required to show that one gene is responsible for the mutant phenotype. Ideally, the phenotype can be rescued by a wild type allele delivered by mRNA injection or as a transgene (see Chapters 11-14). Obtaining a specific phenocopy by morpholino oligonucleotide knockdown of the wild type allele is also compelling. Microinjection techniques are similar to those used for laevis, with volumes and dosages adjusted for smaller X. tropicalis embryo. While all mRNA and morpholino oligonucleotides should be titrated, a starting point of 1/10th the dose used for laevis is useful, in an injection volume of up to 2nl in one cell of a 2-cell embryo in filter-sterilised 3% Ficoll/0.05×MMR.
3.8.4 Phenotypic Analysis
Xenopus species are amenable to a broad range of strategies for analysis of gene function and expression. Techniques will vary with each phenotype, and a full review is beyond the scope of this chapter. In general, after effects on external morphology have been described, mutant embryos are processed for histology and fixed for wholemount in situ hybridisation or staining with specific antibodies. Many published protocols for X. laevis are directly transferrable to X. tropicalis.
3.9 Genotyping Adult Frogs
Once a mutation has been identified or mapped to a narrow region, it may be simpler to identify carriers using a PCR or SNP-based approach using genomic DNA from an adult frog (rather than breeding a candidate carrier to known carriers).
3.9.1 Toe clip for genomic DNA
Large quantities of genomic DNA can be harvested non-lethally from adult X. tropicalis toes, which regenerate after about a month.
Anaesthetize frog in 0.09% MS222 for 4 to 6 minutes. Times for this can vary so continually observe until frogs begin to slow their swimming motions, being careful not to over-anaesthetize
Remove frog immediately and briefly rinse in fresh water
Place frog on damp paper towel and remove a single toe with a fresh scalpel/razor blade. Place toe in eppendorf tube containing 400μl lysis buffer, or freeze tissue on dry ice and store at −80°C.
Rinse frog in fresh water and place in clean tank containing wet paper towels. Cover frog in a further damp paper towel. The frog should completely recover in ~1 hour.
To extract DNA, incubate clipped toe in 400μl lysis buffer @ 55°C overnight with agitation
Precipitate DNA by adding 300μl isopropanol
Spin at 4000rpm for 20′
Wash pellet in 500μl 70% ethanol
Resuspend pellet in 100μl nuclease-free water. Usually one toe clip yields ~10μg genomic DNA.
3.9.2 Back swab for genomic DNA
Taking a small sample from the surface the frog is an alternative non-invasive method for obtaining genomic DNA. However, DNA yield is only sufficient for a few reactions, and usually requires amplification with nested primers.
Hold frog with gloved hand (to prevent DNA contamination)
Using a sterile bacterial inoculation stick or pipette tip, wipe across the skin on the frog’s back approximately 10 times
Shake tip or bacterial stick in 500μl of lysis buffer
Incubate at 55°C for 2 hours
Add 1ml 100% EtOH
Spin at 13000 rpm, 4°C for 10′
Wash pellet in 70% EtOH and spin for a further 5′
Resuspend in 14.3μl nuclease-free water ready for PCR
Add a master mix of primary PCR to this tube and cycle in a thermocycler under primer specific conditions for 35 cycles.
Use 0.2μl of this PCR as template for a second reaction with nested primers
3.10 X. tropicalis sperm freezing
Storage of frozen sperm at −80°C facilitates archiving specific stocks and strains; shipping frozen sperm on dry ice is often simpler and more reliable than shipping adult animals. These protocols have been adapted from (41).
3.10.1 Cryopreservation of sperm
Prepare cryoprotectant.
Inject males with 100u HCG 12-24 hours before harvesting testes.
The next day, kill males and dissect out testes, rolling on clean paper towel to remove traces of blood.
Macerate both testes with eppendorf pestle in a single 1.5 ml Eppendorf tube with 500μl L15/CS
Add 500μl cryoprotectant.
Aliquot into 4-10 tubes.
Place tubes in small styrofoam box, wrap lid with aluminum foil.
Place the box in −80°C for at least 24 hours, then transfer tubes to rack or box for long-term storage at −80°C or in liquid nitrogen (see Note 11).
3.10.2 In vitro fertilization using frozen sperm
Prepare 25°C water bath and express eggs from females into a dry dish, discarding poor-quality eggs.
Thaw frozen sperm in 25°C water bath. Remove immediately when thawed (<30”).
Dilute sperm with 2ml distilled water and add to eggs.
Gently mix sperm and eggs with a pipette tip.
After 2 minutes, flood with distilled water.
Fertilization rates of 10-15% are typically obtained with a ½ testis frozen aliquot on ~1000 eggs.
4. Notes
MES buffer (unlike many other common buffers such as Tris) does not contain amine groups that react with ENU.
A variety of diets work well for adults as long as they are relatively protein-rich and small enough for frogs of a given size to swallow easily, but large enough to grasp with their forelimbs. Live foods such as insect larvae or unprocessed offal carry some risk of transmitting parasites. Powdered fish flake is useful to supplement the diet of larger tadpoles in flow-through systems, but rapidly fouls standing water.
Injections are made into the dorsal lymph sac by inserting a 27G needle subcutaneously between the dorsal lateral line stripes. For optimal egg quality, females may be re-ovulated every 6 weeks to 6 months. Some groups re-use males as often as once a week, and females once a month, although it is likely that ovulating females more than 5-6 times per year may be stressful in the long term.
Always handle amphibians with wet hands to prevent damage to their skin. To squeeze eggs from frogs, grasp the female dorsally, with her left leg between the index and middle finger of your right hand, and her left leg in your left hand. Gently massage her abdomen in an anterior-to-posterior direction with your thumb to express eggs. Eggs will activate prematurely if they contact low-salt solutions, so avoid dripping water from the frog while squeezing. Good quality eggs will be spherical, surrounded by clear jelly, with uniform pigmentation in the animal hemisphere except for a lighter polar patch overlying the germinal vesicle; poor quality eggs may be lysing or stringy. X. tropicalis in vitro fertilization is typically less efficient than with laevis; testes are smaller, contain less sperm, and appear to be more salt-sensitive. X. tropicalis embryos do not consistently undergo upward reorientation of the animal hemisphere as laevis do upon activation; cortical pigment contraction and germinal vesicle breakdown are more reliable indicators. When sacrificing frogs, use RT anaesthetic solution; chilled solutions may immobilize animals without anaesthetic effect.
If available, testes from males bearing fluorescent transgenes can be used to assess haploid formation; efficient UV-irradiation will block paternal transgene transmission. Development of haploid embryos is strongly affected both by egg quality and genetic background. Haploid development typically includes deficits in axis elongation and posterior structures, incomplete blastopore closure and other gastrulation defects, but anterior structures are often well-formed (compare Figure 2 A & C). Diploid-appearing embryos are also observed in some batches of haploids. These may result from incomplete UV-irradiation of sperm, but spontaneous diploidization of haploids has also been observed, probably due to failure of polar body formation (see Figure 2 A-D).
Wait at least 15′ after activation to allow completion of cortical rotation; dejellying during this early period can produce axial defects. BSA-coating dishes prevents embryos from sticking to the plates, which can affect gastrulation.
Depending on egg quality and genetic background, this procedure typically rescues 25% or more of the cleaving haploid embryos to viable diploids.
Haploid and outcrossed diploid controls should be reaching the 4-cell stage when LCS embryos undergo the first cell division; LCS embryos cleaving in synch with controls should be discarded. Rescue efficiency is strongly dependent on egg quality and genetic background. Use of transgenic sperm is recommended to control for efficiency of UV-inactivation.
Heavy compression is important for getting good spreads, but the coverslip should not slide around. This technique can produce hundreds of stained nuclei, but finding those with complete countable spreads requires patience.
Providing the Proteinase K is adequately denatured (by the 5 minute, 95°C step), this DNA can be immediately used in PCR reactions without the need for precipitation or cleanup. For mapping purposes, collect as many mutant embryos as are available. Assignment of mutant loci to specific chromosomes by centromere linkage can be accomplished with as few as 12 mutant and wild type gynogenotes (see 3.7.4). Analysis of 1000 mutant embryos from conventional crosses (~2000 parental meioses) provides a theoretical resolution of 0.05cM (see 3.7.5), which is usually sufficient to define the mutation-containing interval on a single sequence scaffold.
It is advisable to test-fertilize some eggs with aliquot of fresh sperm from each male; if fresh sperm is not capable of fertilization, frozen testis from that male should be discarded. This freezing method seems to work as well as more elaborate methods described in (41).
Figure 2. Haploid and gynogenetic embryos.
Wild type eggs were fertilized using homozygous cardiac actin-RFP transgenic sperm, either untreated to form conventional diploid embryos (A) or UV-irradiated to form haploids (C). Paternal transgene transmission is visible in the diploid clutch (B) but absent in the haploids (D). Haploids can form anterior structures well, but posterior structures are truncated. Note that three spontaneously diploidized embryos without paternal transgene appear in the haploid clutch. Panels E-G show the karyotype of outcrossed diploid (E), haploid (F) and early cold shock gynogenetic diploid embryos (G).
5. Summary/Discussion
We have tried to outline some useful strategies and protocols for developmental genetics using Xenopus tropicalis. Amphibian embryos have been a valuable system for understanding vertebrate development using a broad range of embryological, molecular and, more recently, genomic tools. With its compact genome and short generation time, X. tropicalis offers the prospect of combining genomics and precision loss-of-function genetic approaches with the conventional Xenopus toolkit for a unique range of analysis of gene function in a single in vivo vertebrate model system.
Acknowledgments
Many colleagues contributed to these protocols. In addition to past and present members of the Zimmerman and Stemple laboratories, the authors would particularly like to thank Rob Grainger and Takuya Nakayama (University of Virginia), Richard Harland (UC Berkeley), and Mustafa Khokha (Yale). T.J.G. and L.B.Z. are funded by UK Medical Research Council U117560482; DLS by the Wellcome Trust WT 077047/Z/05/Z.
References
- 1.Gurdon JB, Hopwood N. The introduction of Xenopus laevis into developmental biology: of empire, pregnancy testing and ribosomal genes. Int J Dev Biol. 2000;44:43–50. [PubMed] [Google Scholar]
- 2.Harland RM, Grainger RM. Xenopus research: metamorphosed by genetics and genomics. Trends Genet. 2011 doi: 10.1016/j.tig.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 4.Wang BB, Muller-Immergluck MM, Austin J, Robinson NT, Chisholm A, Kenyon C. A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell. 1993;74:29–42. doi: 10.1016/0092-8674(93)90292-x. [DOI] [PubMed] [Google Scholar]
- 5.Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, Smithies O. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 6.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 7.Streisinger G, Walker C, Dower N, Knauber D, Singer F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio) Nature. 1981;291:293–296. doi: 10.1038/291293a0. [DOI] [PubMed] [Google Scholar]
- 8.Kimmel CB. Genetics and early development of zebrafish. Trends Genet. 1989;5:283–288. doi: 10.1016/0168-9525(89)90103-0. [DOI] [PubMed] [Google Scholar]
- 9.Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 10.Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nusslein-Volhard C. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- 12.Bewick AJ, Anderson DW, Evans BJ. Evolution of the closely related, sex-related genes DM-W and DMRT1 in African clawed frogs (Xenopus) Evolution; international journal of organic evolution. 2010;65:698–712. doi: 10.1111/j.1558-5646.2010.01163.x. [DOI] [PubMed] [Google Scholar]
- 13.Tinsley RCK, H.R. The Biology Of Xenopus. 1996. [Google Scholar]
- 14.Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichmann DS, Dubchak I, Amaya E, Detter JC, Fletcher R, Gerhard DS, Goodstein D, Graves T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Wills A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goda T, Abu-Daya A, Carruthers S, Clark MD, Stemple DL, Zimmerman LB. Genetic screens for mutations affecting development of Xenopus tropicalis. PLoS Genet. 2006;2:e91. doi: 10.1371/journal.pgen.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grammer TC, Khokha MK, Lane MA, Lam K, Harland RM. Identification of mutants in inbred Xenopus tropicalis. Mech Dev. 2005;122:263–272. doi: 10.1016/j.mod.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Noramly S, Zimmerman L, Cox A, Aloise R, Fisher M, Grainger RM. A gynogenetic screen to isolate naturally occurring recessive mutations in Xenopus tropicalis. Mech Dev. 2005;122:273–287. doi: 10.1016/j.mod.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Abu-Daya A, Sater AK, Wells DE, Mohun TJ, Zimmerman LB. Absence of heartbeat in the Xenopus tropicalis mutation muzak is caused by a nonsense mutation in cardiac myosin myh6. Dev Biol. 2009;336:20–29. doi: 10.1016/j.ydbio.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abu-Daya A, Nishimoto S, Fairclough L, Mohun TJ, Logan MP, Zimmerman LB. The secreted integrin ligand nephronectin is necessary for forelimb formation in Xenopus tropicalis. Dev Biol. 2010;349:204–212. doi: 10.1016/j.ydbio.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geach TJ, Zimmerman LB. Paralysis and delayed Z-disc formation in the Xenopus tropicalis unc45b mutant dicky ticker. BMC Dev Biol. 2010;10:75. doi: 10.1186/1471-213X-10-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waldner C, Sakamaki K, Ueno N, Turan G, Ryffel GU. Transgenic Xenopus laevis strain expressing cre recombinase in muscle cells. Dev Dyn. 2006;235:2220–2228. doi: 10.1002/dvdy.20880. [DOI] [PubMed] [Google Scholar]
- 22.Amaya E. Xenomics. Genome Res. 2005;15:1683–1691. doi: 10.1101/gr.3801805. [DOI] [PubMed] [Google Scholar]
- 23.Khokha MK, Krylov V, Reilly MJ, Gall JG, Bhattacharya D, Cheung CY, Kaufman S, Lam DK, Macha J, Ngo C, Prakash N, Schmidt P, Tlapakova T, Trivedi T, Tumova L, Abu-Daya A, Geach T, Vendrell E, Ironfield H, Sinzelle L, Sater AK, Wells DE, Harland RM, Zimmerman LB. Rapid gynogenetic mapping of Xenopus tropicalis mutations to chromosomes. Dev Dyn. 2009;238:1398–1346. doi: 10.1002/dvdy.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elsdale TR, Gurdon JB, Fischberg M. A description of the technique for nuclear transplantation in Xenopus laevis. J Embryol Exp Morphol. 1960;8:437–444. [PubMed] [Google Scholar]
- 25.Reinschmidt D, Friedman J, Hauth J, Ratner E, Cohen M, Miller M, Krotoski D, Tompkins R. Gene-centromere mapping in Xenopus laevis. J Hered. 1985;76:345–347. [PubMed] [Google Scholar]
- 26.Tompkins R, Reinschmidt D. Experimentally induced homozygosity in Xenopus laevis. Methods Cell Biol. 1991;36:35–44. doi: 10.1016/s0091-679x(08)60271-x. [DOI] [PubMed] [Google Scholar]
- 27.Young JJ, Cherone JM, Doyon Y, Ankoudinova I, Faraji FM, Lee AH, Ngo C, Guschin DY, Paschon DE, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Harland RM, Zeitler B. Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A. 2011;108:7052–7057. doi: 10.1073/pnas.1102030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yergeau DA, Kuliyev E, Mead PE. Injection-mediated transposon transgenesis in Xenopus tropicalis and the identification of integration sites by modified extension primer tag selection (EPTS) linker-mediated PCR. Nat Protoc. 2007;2:2975–2986. doi: 10.1038/nprot.2007.428. [DOI] [PubMed] [Google Scholar]
- 29.Yergeau DA, Kelley CM, Kuliyev E, Zhu H, Sater AK, Wells DE, Mead PE. Remobilization of Tol2 transposons in Xenopus tropicalis. BMC Dev Biol. 2010;10:11. doi: 10.1186/1471-213X-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wells D, Gutierrez L, Xu Z, Krylov V, Macha J, Blankenburg K, Hitchens M, Bellot L, Spivey M, Kowis A, Ye Y, Pasternak S, Owen J, Tran T, Slavikova R, Tumova L, Tlapakova T, Seifertova E, Scherer S, Sater A. A Genetic Map of Xenopus tropicalis. Dev Biol. 2010 doi: 10.1016/j.ydbio.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogino H, McConnell WB, Grainger RM. Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease. Mech Dev. 2006;123:103–113. doi: 10.1016/j.mod.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Allen BG, Weeks DL. Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat Methods. 2005;2:975–979. doi: 10.1038/nmeth814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirsch N, Zimmerman LB, Gray J, Chae J, Curran KL, Fisher M, Ogino H, Grainger RM. Xenopus tropicalis transgenic lines and their use in the study of embryonic induction. Dev Dyn. 2002;225:522–535. doi: 10.1002/dvdy.10188. [DOI] [PubMed] [Google Scholar]
- 34.Chae J, Zimmerman LB, Grainger RM. Inducible control of tissue-specific transgene expression in Xenopus tropicalis transgenic lines. Mech Dev. 2002;117:235–241. doi: 10.1016/s0925-4773(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 35.Hartley KO, Nutt SL, Amaya E. Targeted gene expression in transgenic Xenopus using the binary Gal4-UAS system. Proc Natl Acad Sci U S A. 2002;99:1377–1382. doi: 10.1073/pnas.022646899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryffel GU, Werdien D, Turan G, Gerhards A, Goosses S, Senkel S. Tagging muscle cell lineages in development and tail regeneration using Cre recombinase in transgenic Xenopus. Nucleic Acids Res. 2003;31:e44. doi: 10.1093/nar/gng044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehman CW, Carroll D. Homologous recombination catalyzed by a nuclear extract from Xenopus oocytes. Proc Natl Acad Sci U S A. 1991;88:10840–10844. doi: 10.1073/pnas.88.23.10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carruthers S, Stemple DL. Genetic and genomic prospects for Xenopus tropicalis research. Semin Cell Dev Biol. 2006;17:146–153. doi: 10.1016/j.semcdb.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 40.Showell C, Carruthers S, Hall A, Pardo-Manuel de Villena F, Stemple D, Conlon FL. A Comparative Survey of the Frequency and Distribution of Polymorphism in the Genome of Xenopus tropicalis. PloS one. 2011;6:e22392. doi: 10.1371/journal.pone.0022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sargent MG, Mohun TJ. Cryopreservation of sperm of Xenopus laevis and Xenopus tropicalis. Genesis. 2005;41:41–46. doi: 10.1002/gene.20092. [DOI] [PubMed] [Google Scholar]
- 42.Bahary N, Davidson A, Ransom D, Shepard J, Stern H, Trede N, Zhou Y, Barut B, Zon LI. The Zon laboratory guide to positional cloning in zebrafish. Methods Cell Biol. 2004;77:305–329. doi: 10.1016/s0091-679x(04)77017-x. [DOI] [PubMed] [Google Scholar]
- 43.Bassam BJ, Caetano-Anolles G, Gresshoff PM. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- 44.Streisinger G, Singer F, Walker C, Knauber D, Dower N. Segregation analyses and gene-centromere distances in zebrafish. Genetics. 1986;112:311–319. doi: 10.1093/genetics/112.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neff MM, Neff JD, Chory J, Pepper AE. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 47.Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]