Significance
A major challenge of protein crystallography is to accurately describe protein structure variations. Whereas Bragg peaks only yield a picture in which the variations are superimposed, diffuse X-ray scattering (diffraction between the peaks) reports on the way different atoms move together and can be used to increase the accuracy of models. We have performed a molecular-dynamics (MD) simulation of crystalline staphylococcal nuclease, yielding a rich picture of functionally important motions. This simulation is 100-fold longer than previous studies and yields reproducible calculations of diffuse intensity, overcoming a major challenge in the field. Experimental data support the MD models and indicate that diffuse scattering can be used both to validate and to improve MD simulations.
Keywords: diffuse scattering, protein crystallography, molecular-dynamics simulation, protein dynamics, staphylococcal nuclease
Abstract
X-ray diffraction from protein crystals includes both sharply peaked Bragg reflections and diffuse intensity between the peaks. The information in Bragg scattering is limited to what is available in the mean electron density. The diffuse scattering arises from correlations in the electron density variations and therefore contains information about collective motions in proteins. Previous studies using molecular-dynamics (MD) simulations to model diffuse scattering have been hindered by insufficient sampling of the conformational ensemble. To overcome this issue, we have performed a 1.1-μs MD simulation of crystalline staphylococcal nuclease, providing 100-fold more sampling than previous studies. This simulation enables reproducible calculations of the diffuse intensity and predicts functionally important motions, including transitions among at least eight metastable states with different active-site geometries. The total diffuse intensity calculated using the MD model is highly correlated with the experimental data. In particular, there is excellent agreement for the isotropic component of the diffuse intensity, and substantial but weaker agreement for the anisotropic component. Decomposition of the MD model into protein and solvent components indicates that protein–solvent interactions contribute substantially to the overall diffuse intensity. We conclude that diffuse scattering can be used to validate predictions from MD simulations and can provide information to improve MD models of protein motions.
Proteins explore many conformations while carrying out their functions in biological systems (1–3). X-ray crystallography is the dominant source of information about protein structure; however, crystal structure models usually consist of just a single major conformation and at most a small portion of the model as alternate conformations. Crystal structures therefore are missing many details about the underlying conformational ensemble (4).
Proteins assembled in crystalline arrays, like proteins in solution, exhibit rich conformational diversity (4) and often can perform their native functions (5). Many methods have emerged for using Bragg data to model conformational diversity in protein crystals (6–17). The development of these methods has been important as conformational diversity can lead to inaccuracies in protein structure models (9, 18–20). A key limitation of using the Bragg data, however, is that different models of conformational diversity can yield the same mean electron density.
Whereas the Bragg scattering only contains information about the mean electron density, diffuse scattering (diffraction resulting in intensity between the Bragg peaks) is sensitive to spatial correlations in electron density variations (21–28) and therefore contains information about the way that atomic positions vary together in protein crystals. Because models that yield the same mean electron density can yield different correlations in electron density variations, diffuse scattering provides a means to increase the accuracy of crystallography for determining protein conformational variations (29). Peter Moore (30) and Mark Wilson (31) have argued that diffuse scattering should be used to test models of conformational diversity in X-ray crystallography.
Several pioneering studies used diffuse scattering to reveal insights into correlated motions in proteins (17, 30, 32–49). Some of these studies used diffuse scattering to experimentally validate predictions of correlated motions from molecular-dynamics (MD) simulations (35–37, 40, 42–44). These studies revealed important insights but were limited by inadequate sampling of the conformational ensemble, leading to lack of convergence of the diffuse scattering calculations (35). Microsecond-scale simulations of staphylococcal nuclease were predicted to be adequate for convergence of diffuse scattering calculations (42). Modern simulation algorithms and computer hardware now enable microsecond or longer MD simulations of protein crystals (50).
Here, we present calculations of diffuse X-ray scattering using a 1.1-μs MD simulation of crystalline staphylococcal nuclease. The results demonstrate that we have overcome the past limitation of inadequate sampling. We chose staphylococcal nuclease because the experiments of Wall et al. (49) still represent the only complete, high-quality, 3D diffuse scattering data set from a protein crystal. The calculated diffuse intensity is very similar using two independent halves of the trajectory; the results therefore are reproducible and can be meaningfully compared with the experimental data. The MD simulation provides a rich picture of conformational diversity in the energy landscape of a protein crystal, consisting of at least eight metastable states. Like previous MD studies of crystalline staphylococcal nuclease (42–44), the agreement of the simulation with the total experimental diffuse intensity is excellent, supporting the use of MD simulations to model diffuse scattering data. Unlike previous MD studies, we separately compared the more finely structured, anisotropic component of the diffuse intensity with experimental data. The agreement is substantial but weaker than for the isotropic component, indicating there are inaccuracies in the MD models. Our results therefore point toward using diffuse scattering to improve MD models of protein motions.
Results
Functionally Important Motions of Crystalline Staphylococcal Nuclease.
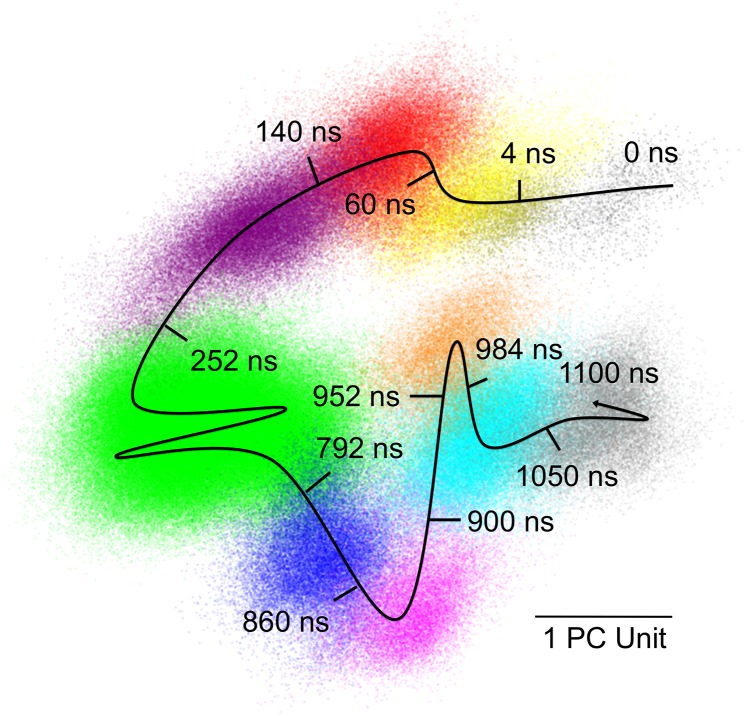
We performed a 1.1-μs MD simulation of a staphylococcal nuclease unit cell (Fig. S1 and Methods), revealing a rich conformational landscape. To analyze the trajectory, we performed principal component analysis (Methods) and gradually added sequential time points to a scatter plot in the space of the two dominant principal components. The points clustered visually into ellipsoidal shaped regions (Fig. 1).
Fig. 1.
Scatter plot of structures extracted from the MD trajectory projected on the first two principal components of the α-carbon position covariance matrix. The first component corresponds to the x axis, and the second corresponds to the y axis. Gray regions correspond to the first 10 ns and last 50 ns of the trajectory; colored regions correspond to metastable states (the yellow region overlaps the first gray region). The curved line indicates the rough trajectory of the system; tick marks indicate approximate times at which state transitions occur.
We identified at least eight metastable states by noting when sharp transitions occurred between the regions (Fig. 1, curved black line with tick marks). The time between the transitions varies considerably from 32 ns (Fig. 1, orange basin) to 540 ns (Fig. 1, green basin); notably, these times are longer than the 10-ns duration of the previous MD simulation of a staphylococcal nuclease unit cell (42) (Fig. 1, gray region). The cyan basin is visited twice: the first time for 52 ns, and the second time for 66 ns. The system spends nearly half the duration of the simulation in the green basin (540 ns); visualization in three dimensions revealed that this basin has a fine structure of substates that, compared with the separations in Fig. 1, lie close together in the space of the dominant principal components.
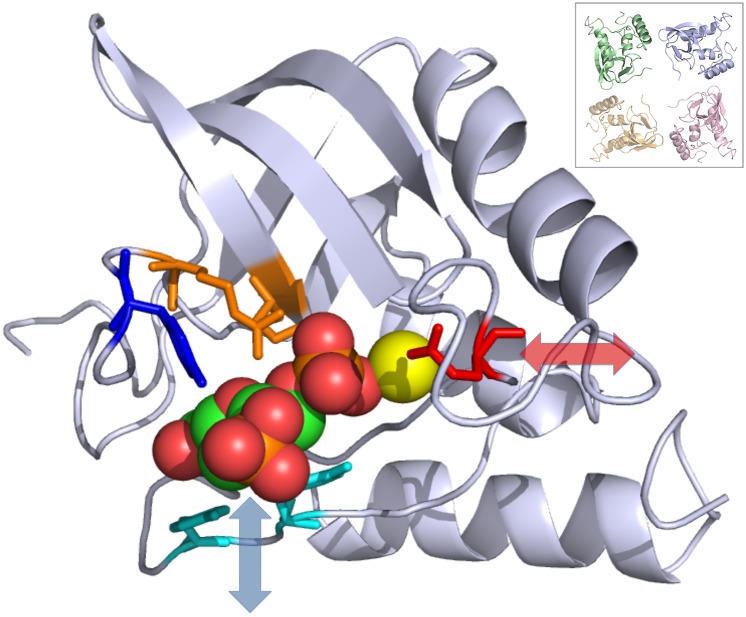
To gain insight into the functional significance of the states in Fig. 1, we created movies to visualize the motions of the two dominant principal components (Movies S1 and S2). Both components are dominated by internal motions rather than overall rotations and translations of the protein. The residues in the active site (51) cluster into localized regions whose motions are highly correlated (Fig. 2): (A) Glu43, at the beginning of the omega loop (Fig. 2, red sticks); (B) Arg35, Tyr85, and Arg87, at the top of the binding pocket (Fig. 2, Arg35 and Arg87 in orange sticks, and Tyr85 in blue sticks); and (C) Tyr113 and Tyr115, at the bottom of the binding pocket (Fig. 2, cyan sticks). Both of the principal components involve substantial motions of regions A and C with little motion in region B. Especially prominent are a clamping motion of region C against region B, which would likely modulate binding interactions (Fig. 2, blue double-headed arrow), and an opening of region A away from region B, modulating the environment of a water molecule putatively involved in the hydrolysis of the 5′-phosphodiester bond (Fig. 2, red double-headed arrow) (52). The motions vary for the four copies of the protein in the unit cell (Fig. 2, Inset): the region C motion is most pronounced for the pink chain; the region A motion is most pronounced for the blue and yellow chains; and the region A and C motions are smallest in the green chain. The states therefore correspond to structures with different active site geometries, resulting in identifiable functional consequences for binding and catalysis.
Fig. 2.
Active-site conformational dynamics in crystalline staphylococcal nuclease. The backbone is rendered using a ribbon, and the P41 unit cell packing along the c axis is shown in the Inset (the screw axis translation is into the page, with the green copy closest and the orange copy farthest away). The residues are shown using sticks, proceeding counterclockwise: Glu43 (red), Arg35 (orange on the β-sheet), Arg87 (orange on the loop), Tyr85 (blue), Tyr115 (cyan), and Tyr113 (cyan). The rest of the protein is rendered as a gray cartoon. The inhibitor thymidine 3′,5′-bisphosphate is shown using spheres and the calcium ion using a yellow sphere. Arrows indicate the direction of motion in the two dominant principal components of the microsecond MD simulation. The loop containing Glu43 (red) moves in the approximate direction indicated by the transparent red double-headed arrow. The loop containing Tyr113 and Tyr115 (cyan) moves in the approximate direction indicated by the transparent blue double-headed arrow. The region containing Tyr85 (blue) and Arg35 and Arg87 (orange) moves much less by comparison. The image was created using PyMOL (66).
MD Model of Diffuse Scattering.
Wall et al. (49) obtained 3D experimental diffuse scattering data by averaging measurements of the continuous intensity at scattering vectors s in the neighborhood of each Bragg peak. This procedure yielded a single diffuse intensity at integer reciprocal lattice points hkl (SI Text); the diffuse lattice showed the expected P4/m Patterson symmetry, which was used to create an eightfold redundant expanded lattice with symmetry averaged observations (49). We similarly computed the diffuse intensity at each reciprocal lattice point and expanded the lattice using the P4/m Patterson symmetry (Methods). To assess the reproducibility of diffuse intensity calculations, we compared calculated using either the first or second half of the following subsections extracted from the full 1.1-μs trajectory: the first 10 ns; the first 100 ns; and the last 1,000 ns. Correlation coefficients using all of the lattice points (∼138,500 in total, of which ∼17,300 are independent, due to symmetry) were calculated between the results obtained using either the first or second half (Methods). Correlations of the total intensity, , were excellent for each of the three subsections of the trajectory: 0.994 for 10 ns, 0.995 for 100 ns, and 0.998 for 1,000 ns.
We performed a more detailed assessment of the reproducibility by first decomposing the diffuse intensity into components that are distributed either isotropically or anisotropically about the origin (Methods). We then calculated correlations for just the anisotropic component, , which corresponds to the striking 3D features visible in the experimental data (see, e.g., figure 3 of ref. 49). The correlations for the anisotropic intensity, , were lower than for the total intensity: 0.646 for 10 ns, 0.654 for 100 ns, and 0.832 for 1,000 ns. The first and second halves of the 1,000-ns trajectory sample a similar range in the dominant principal component (Fig. 1), which is consistent with the high correlation for that trajectory. The difference between the values of and indicates that the total intensity correlation is dominated by the large isotropic component of the signal that varies gradually with resolution. The increase in the anisotropic intensity correlation with simulation time indicates that diffuse scattering calculations become more reproducible for longer simulations, as expected. The fact that = 0.832 for the 1,000-ns trajectory demonstrates that sampling is not a limitation for our validation of MD models of diffuse X-ray scattering.
Meinhold and Smith (42) analyzed diffuse scattering using an approximation in which the variations in atomic positions are normally distributed. In this case, the diffuse intensity in Eq. 1 can be computed using the covariance matrix of atomic displacements between all atom pairs (equation 2 in ref. 42). The reproducibility of diffuse scattering calculations then depends on the reproducibility of these matrix element calculations. Similar to Meinhold and Smith (42), we assessed the reproducibility of the matrix elements by just examining α-carbon coordinates. The covariance matrices were calculated from either the early or late halves of the 1,000-ns trajectory (Methods), and were compared by calculating the Pearson correlation coefficient between all elements. The resulting value of 0.517 is much lower than the values of (0.9975) or (0.832) computed from the diffuse intensity, indicating that calculations of the covariance matrix were not as reproducible as diffuse scattering calculations. A possible explanation is that the diffuse intensity results from the statistically accumulated signal of many atom pairs, whereas the covariance matrix has an element for each individual atom pair.
Experimental Validation of the MD Model.
We compared the MD model to the 64,335 experimental observations collected by Wall et al. (49). The observations were placed on a symmetry expanded diffuse intensity lattice containing 120,845 points (Methods). For each simulation, we multiplied all of the by a single scalar weight w and optimized the value of w to minimize the root-mean-squared deviation (RMSD) of the calculated with respect to the experimental values. Comparisons were performed by calculating correlation coefficients using the lattice points where both calculations and observations were available (∼118,500, of which ∼14,800 are independent due to symmetry). Correlations were computed for four trajectories, either with or without using hydrogen atoms: 0–10, 0–100, 0–1,100, and 100–1,100 ns (Table S1). The correlation of the total intensity ranged from 0.90 to 0.94 for the all-atom calculations, and from 0.85 to 0.92 for the heavy-atom calculations. The correlations for heavy-atom models were in each case lower than for all-atom models (difference of 0.02–0.05), indicating that adding hydrogens somewhat improves the model of total intensity. The correlations increase as the trajectories increase in duration. Excellent agreement was achieved for the 0- to 1,100-ns and 100- to 1,100-ns all-atom simulations, where = 0.94.
We also calculated Meinhold and Smith’s R-factor–like agreement statistic (equation 3 in ref. 42) between and the total computed from the 100- to 1,100-ns simulation. This involved a minimization with respect to an overall weighting factor and baseline shifting of the . We found a value of 0.029, which is much lower than the minimum value of 0.081 found by Meinhold and Smith (table 2 in ref. 42). The agreement factor therefore is substantially improved for our microsecond simulation compared with their 10-ns simulation.
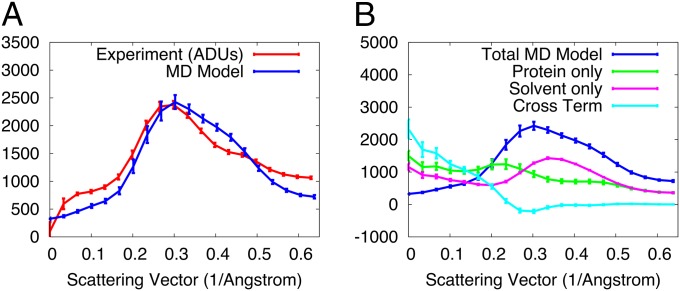
The isotropically averaged diffuse intensity from the MD model and experimental data were very similar (Fig. 3A), which is consistent with the global correlation results. To better understand the origin of the isotropic intensity, we decomposed for the 1,000-ns trajectory into protein (), solvent (), and protein–solvent () terms (Methods). (Note that the contribution of to the sum is negative.) Each component contributes about equally to the total signal (Fig. 3B).
Fig. 3.
Analysis of isotropic diffuse intensity. (A) Comparison of isotropic diffuse intensity for data (red) to the MD model (blue). (B) Decomposition of the total simulated isotropic intensity (blue) into contributions from protein (green), solvent (magenta), and the protein–solvent cross term (cyan). The total intensity is equal to the protein term plus the solvent term minus the cross term (Methods). In both A and B, the SD of the anisotropic diffuse intensity is indicated using error bars.
The protein and solvent curves are similar to those calculated by Meinhold and Smith (42). The cross term, , starts high at small scattering vectors, dips below zero at about 0.3 Å−1, and settles to zero at ∼0.35 Å−1. To gain further insight into the nature of the correlations implied by this behavior, we fit the cross term to a model of correlations that decay exponentially in real space over a length (Methods). A reasonable fit was obtained using Å (Fig. S2), suggesting that the cross term is dominated by short-range correlations between the fluctuations of the protein and solvent (Discussion).
As for the model , we calculated the anisotropic component of the experimentally observed diffuse intensity, by subtracting the isotropic component (Methods). For both the experimental data and the MD model, at each resolution the SD of the anisotropic intensity is less than 10% of the isotropic intensity (Fig. 3A, error bars). The anisotropic component therefore is weaker than the isotropic component. The anisotropic intensity due the individual protein, solvent, and protein–solvent cross terms show differences in magnitude (Fig. 3B). At scattering vectors greater than 0.1 Å−1, the protein has the strongest anisotropic intensity (largest green error bars). The protein–solvent cross term is weaker (small cyan error bars), and the solvent is weakest (very small magenta error bars). Below 0.1 Å−1, each component has substantial anisotropic intensity; however, different components nearly cancel, summing to a very small total anisotropic intensity (blue bars).
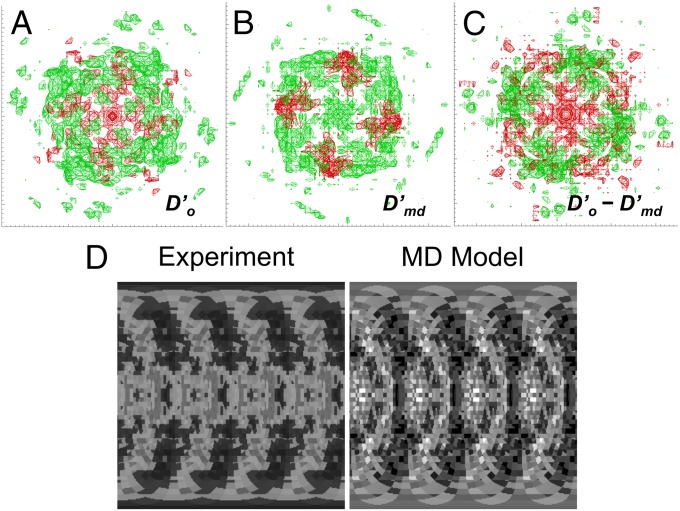
To assess the agreement of the anisotropic component of the MD model () with experimental data (), we visualized level surfaces of (Fig. 4A), (Fig. 4B), and (Fig. 4C). The comparison revealed places where the MD model intensity was either higher (Fig. 4C, green) or lower (Fig. 4C, red) than the experimental intensity. As for the total intensity, to quantify the agreement we calculated the anisotropic correlation coefficient (Methods) between and for each of the trajectories, either with or without hydrogens (Table S1). The correlation of the anisotropic intensity ranged from 0.35 to 0.43 for the all-atom calculations, and from 0.34 to 0.40 for the heavy-atom calculations. The highest anisotropic correlation ( = 0.43) is strongly positive, but is lower than the lowest total intensity correlation ( = 0.85), indicating that the MD model more accurately describes the isotropic than the anisotropic diffuse intensity. The best all-atom correlation is for the 0- to 10-ns trajectory ( = 0.43), and the worst is for the 0- to 100-ns trajectory ( 0.35). The 0- to 1,000-ns and 0- to 1,100-ns trajectories yield intermediate correlations ( = 0.40).
Fig. 4.
Comparison of anisotropic diffuse intensity between models and experimental data. (A–C) Isosurfaces in the (A) experimental (), (B) scaled MD model (), and (C) difference () intensity maps. Positive intensity is shown in green, and negative intensity in red. All isosurfaces including the difference map are displayed at an intensity level equal to the SD of in the solvent ring. The values of were multiplied by a uniform scale factor to yield the same SD in the solvent ring as . The x direction in each panel corresponds to the a* axis, varying from −0.5 Å−1 at the left to 0.5 Å−1 at the right; the y direction corresponds to the b* axis, varying from −0.5 Å−1 at the bottom to 0.5 Å−1 at the top. (D) Visualizations of the experimental (Left) and MD model (Right) anisotropic diffuse intensity in the 0.27-Å−1 resolution shell, for which the agreement is best. The images were constructed as in figure 3 of ref. 49, using the shimlt and seesh routines in LUNUS (63, 72). The y direction corresponds to the polar angle in the shell as measured from the c* axis, varying from 0 at the top to π at the bottom of each image. The x direction corresponds to the azimuthal angle as measured from the a* axis in a right-handed sense, varying from −π at the left to π at the right. Pixel values are displayed as the deviation from the mean on a linear gray scale, with −500 corresponding to black, and 500 corresponding to white.
Correlations for heavy-atom models were lower than for all-atom models by 0.01 or less (Table S1), indicating that adding hydrogens did not substantially improve the model of anisotropic intensity. The correlation for the protein component of the MD model () was = 0.38 (not listed in Table S1), which is very similar to that for the entire MD model. This supports the finding that the protein is the dominant contributor to the anisotropic intensity (Fig. 3A, error bars).
We also calculated the anisotropic correlation within resolution shells, and found the highest value was 0.58 in the 0.27 Å−1 resolution shell. Visualization of the experimental and MD model intensity in this shell shows arcs of diffuse intensity extending between the two poles, and rich large-scale features near the equator (Fig. 4D). A liquid-like motions model showed similar patterns (figure 5 in ref. 49), suggesting that they can be explained largely by the protein structure factor (equation 4 in ref. 49).
Discussion
The present 1.1-μs simulation, which is 100-fold longer than the previous 10-ns simulation of a staphylococcal nuclease unit cell (42), revealed eight metastable states with lifetimes that are longer than the duration of the shorter simulation. Analysis of the motions revealed that the basins correspond to structures with different active-site geometries, resulting in identifiable functional consequences for binding and catalysis.
Diffuse scattering can independently validate the predictions of MD simulations. Conversely, interpretation of the diffuse intensity in terms of a multistate conformational ensemble is now possible (Fig. 1). Although there is room for improvement in modeling the isotropic intensity at resolutions exceeding 0.3 Å−1 (Fig. 3A), the agreement of the MD model with the total experimental diffuse intensity is remarkable considering that no free parameters were adjusted. Moreover, this agreement is robust to sampling (Table S1) and to a model perturbation that includes a change of force field (SI Text).
Compared with the total intensity, the MD model had a lower, but still strongly positive, correlation with the anisotropic diffuse intensity. The lower correlation of the anisotropic intensity indicates there are inaccuracies in the MD models. Maximizing the correlation of the anisotropic intensity is therefore a potential strategy for increasing the accuracy of the MD models. For example, both the simulation of a single unit cell with periodic boundary conditions and the assumption of independent unit cells inherent in Eq. 1 might limit the accuracy of the MD model. One way to test this possibility is to simulate larger sections of the crystal with several unit cells (50), as this will enable correlations between unit cells to be included in the model.
Decomposition of the diffuse intensity into protein and solvent components revealed that the protein component dominates the overall magnitude of the anisotropic intensity (Fig. 3A, error bars). In addition, the MD model of anisotropic intensity using the protein alone ( = 0.38) was almost as accurate as using the protein and solvent ( = 0.40). These results support the hypothesis that the anisotropic intensity reports largely on variations in the protein structure (49).
The decomposition also revealed that the protein–solvent interactions contribute a strong negative component to the sum ( is mostly positive in Fig. 3B and enters with a minus sign). Combined with the subatomic correlation length derived from the fit to Eq. 2, these results suggest that the cross term might be mainly due to negative correlations in the density fluctuations that occur in the region between the protein and solvent atoms. One possible explanation for this is that, when atoms move together, the density shifts, resulting in a positive fluctuation () for the leading edge density of one atom (e.g., a protein atom) coupled a negative fluctuation () for the trailing edge density of a neighboring atom (e.g., a solvent atom) (Fig. S2, Inset).
Increasing the duration of the MD trajectory yielded more reproducible calculations of diffuse intensity and better models of the total intensity. Calculations of the covariance matrix of α-carbon displacements, however, were less reproducible than the calculations of diffuse intensity. In addition, only the cyan basin in Fig. 1 is visited more than once. Thus, even though we have achieved reproducible diffuse scattering calculations using an MD model, the sampling of the conformational ensemble is still incomplete. Millisecond all-atom simulations (53), advanced sampling methods such as parallel tempering (54), and acceleration schemes such as Markov state models (55) and parallel replica MD (56, 57) can be used to pursue increased sampling and improved convergence of the calculations.
Our comparisons point to opportunities for improving models of protein motions using diffuse scattering. For example, if the accuracy of the model of anisotropic diffuse intensity can be improved, it might become possible to use diffuse scattering to improve MD force fields. Indeed, use of NMR data for force field improvement provides an example of the potential impact of diffuse scattering. Early comparisons of MD to NMR data for staphylococcal nuclease revealed differences in the residue-by-residue backbone flexibility (58), and more recent force fields now do a much better job of reproducing the NMR data (59–62). Another possibility is to use diffuse scattering for validating results obtained using schemes for increasing sampling (54–57).
There is also room for improvement in integration of experimental diffuse scattering data from diffraction images. For example, our current methods aim to extract the large-scale features (due to short-range correlations within the unit cell) and to ignore the small-scale, streaked features (due to long-range correlations across unit cells) (63). However, these methods can be improved by accounting for better separation of the large-scale from small-scale features, or a finer sampling of all diffuse features (29). Increased diffuse scattering data collection efforts and advances in X-ray detector technology will further this goal (29).
Methods
MD Simulations.
The 48.5 Å × 48.5 Å × 63.5-Å P1 unit cell model was prepared from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) (64) (www.rcsb.org) entry 1STN (65) using the UnitCell code in AmberTools (ambermd.org/#AmberTools). The RMSD of the atomic coordinates [calculated using PyMOL (66)] between 1STN and PDB entry 4WOR (49) (the crystal structure refined against the Bragg data) is 0.70 Å, with backbone deviations concentrated in loops near the ligand binding site (Fig. S3). Waters and counterions were added using genbox and genion in GROMACS (67), resulting in a model with 8,904 protein atoms (four proteins arranged in a P41 configuration), 6,477 TIP3P water atoms, and 40 Cl− ions (Fig. S1). The OPLS-AA force field (68, 69) was selected. An initial model was prepared using energy minimization followed by 100-ps equilibration in a constant-particle number, -volume, and -temperature (NVT) ensemble at 300 K, using harmonic restraints for the protein atom coordinates. The modified Berendsen thermostat with a 0.1-ps time constant was used for temperature equilibration.
The production 1.1-μs simulation was performed in GROMACS (67) on a 128 core 2.00 GHz Intel Xeon X6550 machine using a 2-fs time step with a constant-particle number, -pressure, and -temperature (NPT) ensemble at 300 K and 1 bar. The Parrinello–Rahman barostat with a 2.0-ps time constant was used for pressure equilibration. At a rate of 44.6-ns system time per day of wall clock time, it took roughly 30 d to complete, including periods of down time. Snapshots were recorded every 2 ps in compressed (.xtc) format, yielding a total of 5.5 × 108 frames in a 31.1-GB file.
Upon the switch from the restrained NVT to the unrestrained NPT simulation, the unit cell dimensions began fluctuating (all scaled simultaneously with the volume fluctuations) and the mean values rapidly shrank by 1.3% of their initial values. The structure also drifted away from the crystal structure: the RMSD between the mean coordinates from the simulation and PDB entry 4WOR was 1.8 Å (1.3 Å) for heavy atoms (α-carbons). These values are similar to those obtained by Meinhold and Smith (42) for their 10-ns simulation: they reported deviations of less than 0.8% in unit cell dimensions, and a RMSD between the mean simulation coordinates and PDB entry 2SNS (52) (an experimentally determined crystal structure available at the time) of 1.7 Å (1.3 Å) for heavy atoms (α-carbons).
MD Model Diffuse Scattering Calculations and Reproducibility.
To calculate the diffuse intensity for a given section of the trajectory, ensembles of atomic coordinates were extracted from the full trajectory in 5-ns chunks. This procedure yielded protein and solvent coordinates of the P1 unit cell in a series of separate files (SI Text).
Each of the files was then processed to calculate the diffuse intensity. The structure factor, , for each unit cell structure n was calculated to 1.6-Å resolution at Miller indices hkl using the iotbx package in the computational crystallography toolbox (CCTBX) (70). The average structure factor, , and the average squared structure factor, , were calculated using a modified version of the Phenix (71) Python script get_struct_fact_from_md.py, which we called get_diffuse_from_md.py. Averages for longer sections of the trajectory were accumulated from averages of the 5-ns chunks. The intensity of the diffuse scattering for the first and second parts of the trajectory were calculated using the following equation:
| [1] |
Eq. 1 appears in the classic text of Guinier (24) and assumes independent unit cell fluctuations. It states that the diffuse intensity is equal to the variance of the unit cell structure factor, and therefore contains information that is distinct from the Bragg peak intensities, which are determined by the square of the mean structure factor (second term of Eq. 1). Eq. 1 is an established method for calculating diffuse scattering from protein MD simulations (35, 42–44).
To decompose the diffuse intensity into isotropic and anisotropic components, reciprocal space was subdivided into concentric spherical shells, each with a thickness equal to the reciprocal unit cell diagonal ( = 0.0336 Å−1). The isotropic intensity was calculated as the mean intensity at scattering vector at the midpoint of each shell n. The anisotropic intensity was then calculated at each lattice point by subtracting the isotropic intensity from the original signal . The value of at scattering vector in the range was obtained by linear interpolation of . The same method was used to obtain isotropic and anisotropic components of the experimentally observed diffuse intensity.
Because the experimental diffuse intensity shows symmetry consistent with the P41 symmetry of the unit cell (49), we enforced the P4/m Patterson symmetry (corresponding to the P41 unit cell symmetry) by replacing each value with the average over all symmetry equivalent hkl positions in the map. The resulting symmetry-expanded map had ∼17,300 independent values on ∼138,500 lattice points with eightfold redundancy. The values were very similar to the original map (Pearson correlation coefficient rsym = 0.996 for total intensity and for anisotropic intensity).
To assess the reproducibility of diffuse scattering calculations in the MD model, we divided the extracted trajectory into first and second halves of equal duration. We then calculated the simulated diffuse scattering from the first and second parts independently, and compared the results quantitatively. A global comparison of and was made using the Pearson correlation coefficients and , respectively.
Covariance Matrices and Principal Components.
To calculate covariance matrices and to perform principal component analysis, the trajectory was adjusted to remove discontinuous jumps of atomic positions to symmetry-related positions during the course of the simulation (trjconv –pbc nojump). Next, the coordinates were adjusted to remove translational drift (trjconv –fit translation) and to preserve the covalent bonding structure of the protein (trjconv –pbc mol). Finally the α-carbon coordinates were selected and used to calculate and diagonalize the covariance matrix (g_covar). Comparisons of covariance matrices and projections of trajectories onto principal components were performed using the tool g_anaeig. Visualization of the energy landscape was performed by displaying the projected coordinates in a 2D scatter plot.
Decomposition into Protein and Solvent Components.
We used Eq. 1 and the equation to decompose into protein , solvent and protein–solvent terms, where and are the structure factors computed from just the protein or solvent component, respectively. Note that positive values of contribute negatively to .
We modeled using the following function (34, 49):
| [2] |
where s is the scattering vector, is the correlation length of atomic displacements, and indicates the 3D Fourier transform of the real-space function , which decays exponentially with distance r. The fitting of to Eq. 2 was performed using the nonlinear-least-squares fitting feature of gnuplot (www.gnuplot.info).
Supplementary Material
Acknowledgments
This work was supported by the US Department of Energy under Contract DE-AC52-06NA25396 through the Laboratory-Directed Research and Development Program at Los Alamos National Laboratory. N.K.S. is supported by NIH Grant GM095887. P.D.A. and T.C.T. are supported by NIH Grant 1P01GM063210. J.S.F. is a Searle Scholar and a Pew Scholar, and is supported by NIH Grants OD009180 and GM110580, and National Science Foundation Grant STC-1231306.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416744111/-/DCSupplemental.
References
- 1.Austin RH, et al. Dynamics of carbon monoxide binding by heme proteins. Science. 1973;181(4099):541–543. doi: 10.1126/science.181.4099.541. [DOI] [PubMed] [Google Scholar]
- 2.Weber G. Ligand binding and internal equilibria in proteins. Biochemistry. 1972;11(5):864–878. doi: 10.1021/bi00755a028. [DOI] [PubMed] [Google Scholar]
- 3.Frauenfelder H, Parak F, Young RD. Conformational substates in proteins. Annu Rev Biophys Biophys Chem. 1988;17:451–479. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- 4.Frauenfelder H, Petsko GA, Tsernoglou D. Temperature-dependent X-ray diffraction as a probe of protein structural dynamics. Nature. 1979;280(5723):558–563. doi: 10.1038/280558a0. [DOI] [PubMed] [Google Scholar]
- 5.Doscher MS, Richards FM. The activity of an enzyme in the crystalline state: Ribonuclease S. J Biol Chem. 1963;238(7):2393–2398. [Google Scholar]
- 6.Burnley BT, Afonine PV, Adams PD, Gros P. 2012. Modelling dynamics in protein crystal structures by ensemble refinement. Elife 1:e00311. [DOI] [PMC free article] [PubMed]
- 7.Chaudhry C, Horwich AL, Brunger AT, Adams PD. Exploring the structural dynamics of the E. coli chaperonin GroEL using translation-libration-screw crystallographic refinement of intermediate states. J Mol Biol. 2004;342(1):229–245. doi: 10.1016/j.jmb.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z, Chapman MS. Conformational disorder of proteins assessed by real-space molecular dynamics refinement. Biophys J. 2001;80(3):1466–1472. doi: 10.1016/S0006-3495(01)76118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DePristo MA, de Bakker PI, Blundell TL. Heterogeneity and inaccuracy in protein structures solved by X-ray crystallography. Structure. 2004;12(5):831–838. doi: 10.1016/j.str.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 10.Fraser JS, et al. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462(7273):669–673. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser JS, et al. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc Natl Acad Sci USA. 2011;108(39):16247–16252. doi: 10.1073/pnas.1111325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuriyan J, et al. Exploration of disorder in protein structures by X-ray restrained molecular dynamics. Proteins. 1991;10(4):340–358. doi: 10.1002/prot.340100407. [DOI] [PubMed] [Google Scholar]
- 13.Kuriyan J, Weis WI. Rigid protein motion as a model for crystallographic temperature factors. Proc Natl Acad Sci USA. 1991;88(7):2773–2777. doi: 10.1073/pnas.88.7.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pellegrini M, Grønbech-Jensen N, Kelly JA, Pfluegl GM, Yeates TO. Highly constrained multiple-copy refinement of protein crystal structures. Proteins. 1997;29(4):426–432. doi: 10.1002/(sici)1097-0134(199712)29:4<426::aid-prot3>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 15.Sternberg MJ, Grace DE, Phillips DC. Dynamic information from protein crystallography. An analysis of temperature factors from refinement of the hen egg-white lysozyme structure. J Mol Biol. 1979;130(3):231–252. doi: 10.1016/0022-2836(79)90539-4. [DOI] [PubMed] [Google Scholar]
- 16.van den Bedem H, Bhabha G, Yang K, Wright PE, Fraser JS. Automated identification of functional dynamic contact networks from X-ray crystallography. Nat Methods. 2013;10(9):896–902. doi: 10.1038/nmeth.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wall ME, Clarage JB, Phillips GN. Motions of calmodulin characterized using both Bragg and diffuse X-ray scattering. Structure. 1997;5(12):1599–1612. doi: 10.1016/s0969-2126(97)00308-0. [DOI] [PubMed] [Google Scholar]
- 18.García AE, Krumhansl JA, Frauenfelder H. Variations on a theme by Debye and Waller: From simple crystals to proteins. Proteins. 1997;29(2):153–160. [PubMed] [Google Scholar]
- 19.Furnham N, Blundell TL, DePristo MA, Terwilliger TC. Is one solution good enough? Nat Struct Mol Biol. 2006;13(3):184–185. doi: 10.1038/nsmb0306-184. , discussion 185. [DOI] [PubMed] [Google Scholar]
- 20.Kuzmanic A, Pannu NS, Zagrovic B. X-ray refinement significantly underestimates the level of microscopic heterogeneity in biomolecular crystals. Nat Commun. 2014;5:3220. doi: 10.1038/ncomms4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zachariasen W. Theory of X-Ray Diffraction in Crystals. Wiley; New York: 1945. [Google Scholar]
- 22.James R. The Optical Principles of the Diffraction of X-Rays. Bell; London: 1948. [Google Scholar]
- 23.Wooster WA. Diffuse X-Ray Reflections from Crystals. Oxford Univ Press; Oxford: 1962. [Google Scholar]
- 24.Guinier A. X-ray Diffraction in Crystals, Imperfect Crystals, and Amorphous Bodies. W. H. Freeman and Company; San Francisco: 1963. [Google Scholar]
- 25.Amorós JL, Amorós M. Molecular Crystals; Their Transforms and Diffuse Scattering. Wiley; New York: 1968. [Google Scholar]
- 26.Warren BE. X-Ray Diffraction. Addison-Wesley; Reading, MA: 1969. [Google Scholar]
- 27.Willis BTM, Pryor AW. Thermal Vibrations in Crystallography. Cambridge Univ Press; Cambridge, UK: 1975. [Google Scholar]
- 28.Welberry TR. Diffuse X-Ray Scattering and Models of Disorder. Oxford Univ Press; Oxford: 2004. [Google Scholar]
- 29.Wall ME, Adams PD, Fraser JS, Sauter NK. Diffuse X-ray scattering to model protein motions. Structure. 2014;22(2):182–184. doi: 10.1016/j.str.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore PB. On the relationship between diffraction patterns and motions in macromolecular crystals. Structure. 2009;17(10):1307–1315. doi: 10.1016/j.str.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 31.Wilson MA. Visualizing networks of mobility in proteins. Nat Methods. 2013;10(9):835–837. doi: 10.1038/nmeth.2606. [DOI] [PubMed] [Google Scholar]
- 32.Caspar DL, Clarage J, Salunke DM, Clarage M. Liquid-like movements in crystalline insulin. Nature. 1988;332(6165):659–662. doi: 10.1038/332659a0. [DOI] [PubMed] [Google Scholar]
- 33.Chacko S, Phillips GN., Jr Diffuse x-ray scattering from tropomyosin crystals. Biophys J. 1992;61(5):1256–1266. doi: 10.1016/S0006-3495(92)81934-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarage JB, Clarage MS, Phillips WC, Sweet RM, Caspar DL. Correlations of atomic movements in lysozyme crystals. Proteins. 1992;12(2):145–157. doi: 10.1002/prot.340120208. [DOI] [PubMed] [Google Scholar]
- 35.Clarage JB, Romo T, Andrews BK, Pettitt BM, Phillips GN., Jr A sampling problem in molecular dynamics simulations of macromolecules. Proc Natl Acad Sci USA. 1995;92(8):3288–3292. doi: 10.1073/pnas.92.8.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doucet J, Benoit JP. Molecular dynamics studied by analysis of the X-ray diffuse scattering from lysozyme crystals. Nature. 1987;325(6105):643–646. doi: 10.1038/325643a0. [DOI] [PubMed] [Google Scholar]
- 37.Faure P, et al. Correlated intramolecular motions and diffuse X-ray scattering in lysozyme. Nat Struct Biol. 1994;1(2):124–128. doi: 10.1038/nsb0294-124. [DOI] [PubMed] [Google Scholar]
- 38.Glover ID, Harris GW, Helliwell JR, Moss DS. The variety of X-ray diffuse-scattering from macromolecular crystals and its respective components. Acta Crystallogr B Struct Sci. 1991;47(Pt 6):960–968. [Google Scholar]
- 39.Helliwell JR, Glover ID, Jones A, Pantos E, Moss DS. Protein dynamics—use of computer-graphics and protein crystal diffuse-scattering recorded with synchrotron X-radiation. Biochem Soc Trans. 1986;14(3):653–655. [Google Scholar]
- 40.Héry S, Genest D, Smith JC. X-ray diffuse scattering and rigid-body motion in crystalline lysozyme probed by molecular dynamics simulation. J Mol Biol. 1998;279(1):303–319. doi: 10.1006/jmbi.1998.1754. [DOI] [PubMed] [Google Scholar]
- 41.Kolatkar AR, Clarage JB, Phillips GN., Jr Analysis of diffuse scattering from yeast initiator tRNA crystals. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 2):210–218. doi: 10.1107/S0907444993011692. [DOI] [PubMed] [Google Scholar]
- 42.Meinhold L, Smith JC. Fluctuations and correlations in crystalline protein dynamics: A simulation analysis of staphylococcal nuclease. Biophys J. 2005;88(4):2554–2563. doi: 10.1529/biophysj.104.056101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meinhold L, Smith JC. Correlated dynamics determining x-ray diffuse scattering from a crystalline protein revealed by molecular dynamics simulation. Phys Rev Lett. 2005;95(21):218103. doi: 10.1103/PhysRevLett.95.218103. [DOI] [PubMed] [Google Scholar]
- 44.Meinhold L, Smith JC. Protein dynamics from X-ray crystallography: Anisotropic, global motion in diffuse scattering patterns. Proteins. 2007;66(4):941–953. doi: 10.1002/prot.21246. [DOI] [PubMed] [Google Scholar]
- 45.Meinhold L, Merzel F, Smith JC. Lattice dynamics of a protein crystal. Phys Rev Lett. 2007;99(13):138101. doi: 10.1103/PhysRevLett.99.138101. [DOI] [PubMed] [Google Scholar]
- 46.Mizuguchi K, Kidera A, Gō N. Collective motions in proteins investigated by X-ray diffuse scattering. Proteins. 1994;18(1):34–48. doi: 10.1002/prot.340180106. [DOI] [PubMed] [Google Scholar]
- 47.Phillips GN, Jr, Fillers JP, Cohen C. Motions of tropomyosin. Crystal as metaphor. Biophys J. 1980;32(1):485–502. doi: 10.1016/S0006-3495(80)84985-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riccardi D, Cui Q, Phillips GN., Jr Evaluating elastic network models of crystalline biological molecules with temperature factors, correlated motions, and diffuse x-ray scattering. Biophys J. 2010;99(8):2616–2625. doi: 10.1016/j.bpj.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wall ME, Ealick SE, Gruner SM. Three-dimensional diffuse x-ray scattering from crystals of Staphylococcal nuclease. Proc Natl Acad Sci USA. 1997;94(12):6180–6184. doi: 10.1073/pnas.94.12.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janowski PA, Cerutti DS, Holton J, Case DA. Peptide crystal simulations reveal hidden dynamics. J Am Chem Soc. 2013;135(21):7938–7948. doi: 10.1021/ja401382y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grissom CB, Markley JL. Staphylococcal nuclease active-site amino acids: pH dependence of tyrosines and arginines by 13C NMR and correlation with kinetic studies. Biochemistry. 1989;28(5):2116–2124. doi: 10.1021/bi00431a023. [DOI] [PubMed] [Google Scholar]
- 52.Cotton FA, Hazen EE, Jr, Legg MJ. Staphylococcal nuclease: Proposed mechanism of action based on structure of enzyme-thymidine 3′,5′-bisphosphate-calcium ion complex at 1.5-Å resolution. Proc Natl Acad Sci USA. 1979;76(6):2551–2555. doi: 10.1073/pnas.76.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dror RO, Dirks RM, Grossman JP, Xu H, Shaw DE. Biomolecular simulation: A computational microscope for molecular biology. Annu Rev Biophys. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- 54.Earl DJ, Deem MW. Parallel tempering: Theory, applications, and new perspectives. Phys Chem Chem Phys. 2005;7(23):3910–3916. doi: 10.1039/b509983h. [DOI] [PubMed] [Google Scholar]
- 55.Pande VS, Beauchamp K, Bowman GR. Everything you wanted to know about Markov state models but were afraid to ask. Methods. 2010;52(1):99–105. doi: 10.1016/j.ymeth.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martínez E, Uberuaga BP, Voter AF. Sublattice parallel replica dynamics. Phys Rev E Stat Nonlin Soft Matter Phys. 2014;89(6):063308. doi: 10.1103/PhysRevE.89.063308. [DOI] [PubMed] [Google Scholar]
- 57.Voter AF. Parallel replica method for dynamics of infrequent events. Phys Rev B Condens Matter. 1998;57(22):13985–13988. [Google Scholar]
- 58.Chatfield DC, Szabo A, Brooks BR. Molecular dynamics of staphylococcal nuclease: Comparison of simulation with 15N and 13C NMR relaxation data. J Am Chem Soc. 1998;120(21):5301–5311. [Google Scholar]
- 59.Showalter SA, Brüschweiler R. Validation of molecular dynamics simulations of biomolecules using NMR spin relaxation as benchmarks: Application to the AMBER99SB force field. J Chem Theory Comput. 2007;3(3):961–975. doi: 10.1021/ct7000045. [DOI] [PubMed] [Google Scholar]
- 60.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lindorff-Larsen K, et al. Systematic validation of protein force fields against experimental data. PLoS One. 2012;7(2):e32131. doi: 10.1371/journal.pone.0032131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beauchamp KA, Lin YS, Das R, Pande VS. Are protein force fields getting better? A systematic benchmark on 524 diverse NMR measurements. J Chem Theory Comput. 2012;8(4):1409–1414. doi: 10.1021/ct2007814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wall ME. Methods and software for diffuse X-ray scattering from protein crystals. Methods Mol Biol. 2009;544:269–279. doi: 10.1007/978-1-59745-483-4_17. [DOI] [PubMed] [Google Scholar]
- 64.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hynes TR, Fox RO. The crystal structure of staphylococcal nuclease refined at 1.7 Å resolution. Proteins. 1991;10(2):92–105. doi: 10.1002/prot.340100203. [DOI] [PubMed] [Google Scholar]
- 66. DeLano WL, The PyMOL Molecular Graphics System (Schrödinger, LLC, New York), Version 1.7.1.1.
- 67.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel modecular dynamics implementation. Comput Phys Commun. 1995;91(1-3):43–56. [Google Scholar]
- 68.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118(45):11225–11236. [Google Scholar]
- 69.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B. 2001;105(28):6474–6487. [Google Scholar]
- 70.Grosse-Kunstleve RW, Sauter NK, Moriarty NW, Adams PD. The Computational Crystallography Toolbox: Crystallographic algorithms in a reusable software framework. J Appl Cryst. 2002;35(Pt 1):126–136. [Google Scholar]
- 71.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wall ME. 1996. Diffuse features in X-ray diffraction from protein crystals. PhD thesis (Princeton University, Princeton)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.