Significance
In some cases similar molecular programs (i.e., conserved genes and gene networks) underlie the expression of phenotypic traits that evolve repeatedly across diverse species. We investigated this possibility in the context of social behavioral response, using a comparative genomics approach for three distantly related species: house mouse (Mus musculus), stickleback fish (Gasterosteus aculeatus), and honey bee (Apis mellifera). An experience of territory intrusion modulated similar brain functional processes across species, including hormone-mediated signal transduction, neurodevelopment, chromosome organization, and energy metabolism. Several homologous transcription factors also responded consistently to territory intrusion, suggesting that shared neuronal effects may involve transcriptional cascades of evolutionarily conserved genes. These results indicate that conserved genetic “toolkits” are involved in independent evolutions of social behavior.
Keywords: genetic hotspot, NF-κB signaling, brain metabolism, aggression
Abstract
Certain complex phenotypes appear repeatedly across diverse species due to processes of evolutionary conservation and convergence. In some contexts like developmental body patterning, there is increased appreciation that common molecular mechanisms underlie common phenotypes; these molecular mechanisms include highly conserved genes and networks that may be modified by lineage-specific mutations. However, the existence of deeply conserved mechanisms for social behaviors has not yet been demonstrated. We used a comparative genomics approach to determine whether shared neuromolecular mechanisms could underlie behavioral response to territory intrusion across species spanning a broad phylogenetic range: house mouse (Mus musculus), stickleback fish (Gasterosteus aculeatus), and honey bee (Apis mellifera). Territory intrusion modulated similar brain functional processes in each species, including those associated with hormone-mediated signal transduction and neurodevelopment. Changes in chromosome organization and energy metabolism appear to be core, conserved processes involved in the response to territory intrusion. We also found that several homologous transcription factors that are typically associated with neural development were modulated across all three species, suggesting that shared neuronal effects may involve transcriptional cascades of evolutionarily conserved genes. Furthermore, immunohistochemical analyses of a subset of these transcription factors in mouse again implicated modulation of energy metabolism in the behavioral response. These results provide support for conserved genetic “toolkits” that are used in independent evolutions of the response to social challenge in diverse taxa.
Similar phenotypes can have a shared molecular basis, even among distantly related species (1–3). This phenomenon has been observed for an array of traits, including morphological adaptations like coat color or wing patterning, rapid adaptations like drug resistance, and artificially selected phenological traits like flowering time (reviewed in ref. 2). Shared molecular mechanisms can arise convergently as a result of de novo mutations at genetic hotspots (2) or as a result of conservation. Both of these processes result in “genetic toolkits” or genes that are repeatedly used over evolutionary time to give rise to similar phenotypes (3). The phenomenon of genetic toolkits challenges fundamental notions about evolutionary convergence, conservation, and the origins of biodiversity.
The role of genetic toolkits in shaping behavioral phenotypes is unclear (4, 5). Behaviors are typically polygenic (6) and they show great nuance and plasticity within a species, raising the possibility that cross-species similarities in behavior are superficial. Social behaviors in particular present a challenge to the genetic toolkit concept: these behaviors are critical to survival and reproductive success, but across species there is significant variation in the contexts for and frequency of social interaction, the sensory modalities used to perceive the social landscape, and the structure and function of the brain and endocrine systems that regulate behavior. Nonetheless, due to genetic orthology and striking general similarities in certain social behaviors, it is possible that shared genes, gene networks, or functional processes have been reused to regulate presumably independent evolutions of certain types of social behavior across diverse species.
We used a comparative transcriptomic approach to determine whether shared neuromolecular mechanisms govern the response to an acute social challenge. We focused on a single context, the response to territory intrusion, which is biologically relevant for our focal species and is also generalizable to many other species. We evaluated mechanistic commonalities across a phylogenetic distance that spans ∼650 My of evolution, comparing three species with different ecologies and social organization: the house mouse (male Mus musculus) and stickleback fish (male Gasterosteus aculeatus), both of which are strongly territorial and somewhat social, and the highly social honey bee (female Apis mellifera workers), which exhibits organized collective defensive behavior. This broad comparison enabled us to identify core molecular mechanisms in the brain associated with the response to social challenge.
Results
We sequenced mRNA to compare brain transcriptomes of territory holders exposed to either a conspecific intruder (experimental) or a neutral object (control). We sequenced different brain regions across the three species: honey bee whole brain [which shows a robust transcriptomic signature across multiple aggressive contexts (7)], stickleback diencephalon [which shows intense neural activation in response to intrusion (8)], and mouse ventral hypothalamus (VH) (9). We validated strong involvement of the VH, using brain regional quantitative PCR (qPCR) analysis of several immediate early genes (IEGs) [Fos, Fosl2, Arc, and Egr1 (early growth response protein 1); Fig. S1]. Following sequencing, we ranked all genes within a species based on the degree of differential expression between experimental and control conditions and generated lists of significantly regulated Gene Ontology (GO) terms using a threshold-free rank-based gene set enrichment analysis (GSEA) (10). This approach was applied consistently to the single-species and homologous triplet analyses (SI Materials and Methods and below).
Single-Species Brain Transcriptomic Responses to Territory Intrusion.
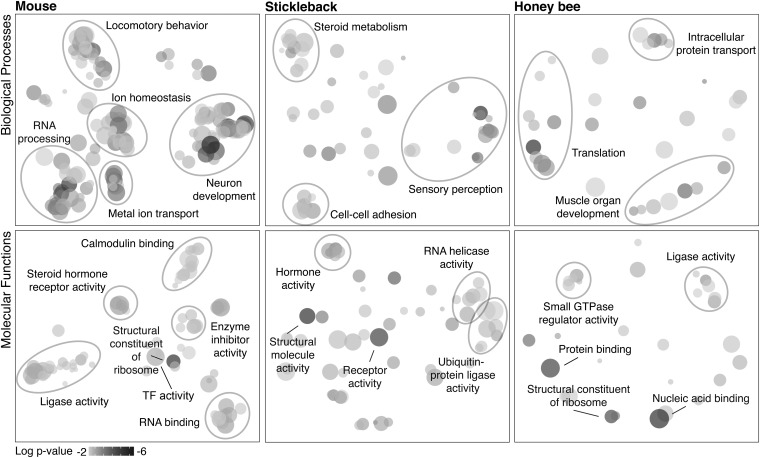
For each species, we derived clusters of related GO terms, using the SimRel function in the program REVIGO (11), which clusters and names groups of GO terms on the basis of semantic similarity, uniqueness, and significance (Datasets S1–S3 and Fig. 1). All species showed a general signature of gene regulatory dynamics, and in mouse and stickleback, several clusters of terms were associated with neural signaling and stimulus response. As expected based on the brain regions assessed and the social context, we found signatures of hormone signaling across all species.
Fig. 1.
Major biological processes (Top) and molecular functions (Bottom) regulated in response to territory intrusion for each species as revealed by Gene Ontology analysis of genes differentially expressed in the brain in response to social challenge. Clustering analyses of single-species GSEA results (adjusted P value < 0.05) are shown. Shading of circles indicates relative significance of the GO term (darker is more significant). Circle size corresponds to the number of genes annotated to the term in the reference database. See Datasets S1–S3 for full lists.
For a subset of significant GO terms, we used post hoc analyses (Materials and Methods and SI Materials and Methods) to determine which genes were most responsible for enrichment of the term. For example, G-protein–coupled receptor (GPCR) activity was significantly enriched across all species and includes proteins integral to the response to hormones and neurotransmitters that modulate behavior (12). Mouse GPCRs included a dopamine receptor (Drd1a) and an adenosine receptor (Adora2a) associated with human panic disorders (13). Honey bee GPCRs included CcapR and hormone receptor EthR, which are responsive to ecdysone (14, 15) (Table S1). Fz2, a Wnt-activated receptor, was another GPCR identified in honey bee. Our GSEA results in mouse also implicated Wnt signaling, suggesting a conserved role for this classic developmental signaling pathway (16) in the response to territory intrusion.
Our analyses also highlighted several other developmental processes, including muscle organ development, which was enriched across all three species. A post hoc analysis showed that differentially expressed genes annotated to muscle organ development in mouse included genes also associated with synapse formation (Col19a1) (17), activity-dependent dendritic outgrowth during development (Mkl2) (18), and glial differentiation in the adult brain (Msx1) (19). Thus, muscle organ development could be indicative of synapse formation, neuronal outgrowth, and glial proliferation. Furthermore, many genes related to muscle contraction in mouse (e.g., Kcnma1, Adora2a, and Adra1d) are involved in vascular muscle function. Though there are structural differences in vertebrate and insect vascular systems, circulatory system development is regulated by evolutionarily conserved gene networks (20), and thus components of this apparent vascular response may represent conserved features of the neural response to territory intrusion.
Cross-Species Core Processes in the Response to Territory Intrusion: Chromosome Organization and Energy Metabolism.
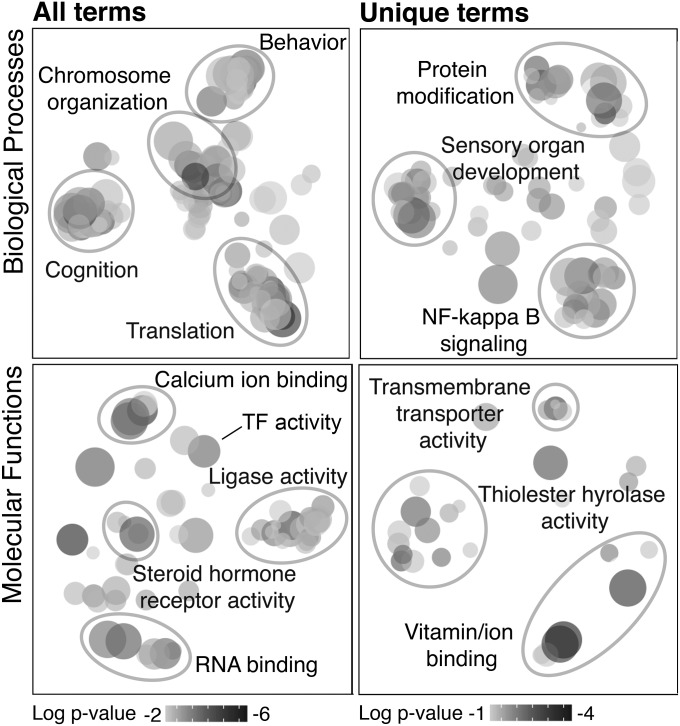
To identify highly conserved (“core”) processes involved in the response to territory intrusion, we analyzed orthologous and paralogous genes (Materials and Methods and SI Materials and Methods). Where possible, we assigned genes to a metazoan evolutionarily orthologous group (EOG) (21) and then filtered each species gene list to contain only EOGs shared across all three species. For each shared EOG, we identified a “homologous triplet” with one gene per species, calculated a combined significance score for the triplet, and ranked triplets on this basis (Dataset S4). Due to design, the GSEA of the ranked triplets identified GO terms enriched within a limited subset of conserved genes.
The GSEA of the triplets (Dataset S5 and Fig. 2) identified a cluster of highly significant GO terms associated with chromosome organization, which was not as prominent in any single-species analysis. Assessing only GO terms unique to the triplet analysis, we also identified a cluster annotated as NF-κB signaling, notable because it is a signal-transduction pathway associated with stress response and energy metabolism in tissues including the brain (22, 23).
Fig. 2.
Clustering analyses for the homologous triplet GSEA (adjusted P value < 0.05). Triplets were groups of orthologous or paralogous genes as identified by OrthoDB. (Left) Clustering analyses include all significant terms. (Right) Analyses include only terms unique to the homologous triplet GSEA (not identified in any of the single-species analyses). See Dataset S5 for full list.
We identified several other GO terms associated with energy metabolism in the triplet analysis (Dataset S5). Because oxidative phosphorylation, a key energy metabolism pathway, shows a negative relationship with aggression in honey bee (24), we examined the direction of change for all oxidative phosphorylation genes in each species and found they were consistently down-regulated in mouse and honey bee and up-regulated in stickleback (sign test, P < 0.0001; Fig. S2). These results suggest that energy metabolism may be a core mechanism in the response to territory intrusion.
Cross-Species Core Processes in the Response to Territory Intrusion: Transcriptional Regulation.
Transcription factors (TFs) regulate large networks of genes and thus are predicted to play an important role in modulating complex phenotypes like social behavior (7, 23, 25, 26). The term sequence-specific DNA binding transcription factor activity was enriched in mouse (Dataset S1 and Fig. 1, “TF activity”), and a post hoc analysis identified 10 differentially expressed TFs (Table S2). Three of these, Egr2, Foxg1 (forkhead box G1), and Pitx1 (paired-like homeodomain 1), have been implicated in autism spectrum disorders (27–29), and Egr genes are also known to be socially responsive in birds and honey bees (30, 31). The same TF-related GO term was enriched in the homologous triplet analysis (Dataset S5 and Fig. 2), and seven significant homologous TFs were identified (“Tier 1” in Table 1). Four known TFs (32) appeared high on the list of ranked homologous triplets but were not annotated to a significant TF-relevant GO term (“Tier 2”), and 14 other annotated TFs had a combined P < 0.05, but failed to reach the stringent post hoc analysis threshold (“Tier 3”), providing some suggestion of a common role in the response to territory intrusion.
Table 1.
Putative toolkit transcription factors shared across all three species
| Mouse | Stickleback | Bee | Fly |
| Tier 1 | |||
| Nr4a3 | ENSGACG00000010788 | GB49295 | hr38 |
| Lhx1 | ENSGACG00000016592 | GB45975 | lim3 |
| Nr5a1 | ENSGACG00000018317 | GB42142 | ftz-f1 |
| Irx3 | ENSGACG00000016682 | GB55198 | mirr |
| Pou3f4 | ENSGACG00000006709 | GB53100 | pdm2 |
| Emx1 | ENSGACG00000004860 | GB54518 | CG18599 |
| Foxg1 | ENSGACG00000011198 | GB44229 | slp2 |
| Tier 2 | |||
| Gata2 | ENSGACG00000010218 | GB50931 | srp |
| Mycbp | ENSGACG00000013676 | GB51617 | CG17202 |
| Npas4 | ENSGACG00000004132 | GB49843 | dys |
| Chd5 | ENSGACG00000003717 | GB43409 | Various |
| Tier 3 | |||
| Foxp1 | ENSGACG00000018911 | GB40150 | foxP |
| Klf12 | ENSGACG00000020841 | GB52090 | Various |
| Klf15 | ENSGACG00000003717 | GB43409 | Various |
| Meis2 | ENSGACG00000019665 | GB48653 | hth |
| Nfat5 | ENSGACG00000016365 | GB45138 | nfat |
| Nr1h3 | ENSGACG00000004938 | GB48059 | ecR |
| Nr2e1 | ENSGACG00000017060 | GB49738 | hr51 |
| Rax | ENSGACG00000016453 | GB52781 | pph13 |
| Tbx15 | ENSGACG00000012454 | GB55445 | h15 |
| Six6 | ENSGACG00000019038 | GB49751 | optix |
| Bsx | ENSGACG00000020120 | GB49332 | bsh |
| Rfx3 | ENSGACG00000007541 | GB41876 | rfx |
| Nkx2-4 | ENSGACG00000006999 | GB47400 | None |
| Otp | ENSGACG00000015770 | GB51584 | otp |
“Fly” lists the Drosophila melanogaster ortholog to each honey bee gene (annotated using FlyBase; “Various” indicates more than one ortholog). See Results for descriptions of tiers.
We also used a bioinformatics approach to identify TF binding motifs that are significantly enriched in genomic regions near differentially expressed genes (DEGs) [false discovery rate (FDR) < 0.10, experimental vs. control, Dataset S6]. The “cis-Metalysis” tool (33) was used to identify motifs that are consistently associated with DEGs in more than one species (FDR < 0.2, Datasets S7 and S8). One motif, predicted to bind the nuclear receptor protein NR2E1 (a toolkit TF), was enriched in all three species. Several others were enriched in two species, including motifs for TFs involved in neuroendocrine signaling [NR2A1, which was also associated with territorial intrusion in a previous stickleback study (8); LEF1 (34); and DLX1 (35)]. Results also implicated the transcriptional repressor HIC1, which is believed to act via chromatin remodeling (36), and the IEG ZNF354C. We tested for pairs of motifs (in Boolean combinations; Materials and Methods) that are consistently associated with DEGs and identified the pair EGR1 and AP2-A as the only significant combination in all three species (Datasets S8 and S9). This pair of TFs is known to regulate gene expression in the context of synaptic plasticity (37). Other significant pairs involved the toolkit TF FOXG1, as well as other TFs implicated in autism spectrum disorder, e.g., PITX1 (also differentially expressed in mouse, see above), TBX1 [which controls brain vascularization (38)], and TFs involved in neuronal differentiation [OTX1 (39)] and hormone signaling (ESR1).
Regional and Cell Type Specificity of Putative Toolkit TFs.
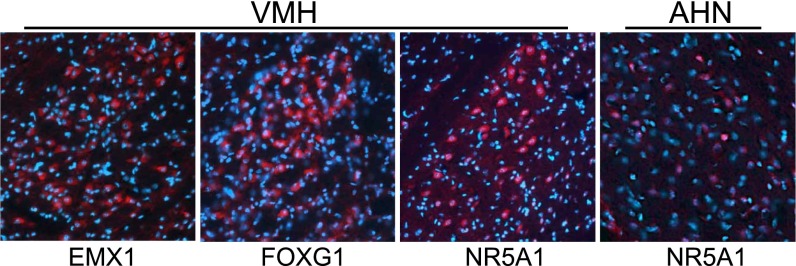
Our analysis highlights a role for developmentally active TFs in adult territory response. To localize and further explore the functional role of the toolkit TFs in the context of intruder response in mouse, we used immunohistochemistry (IHC) to stain adjacent brain sections with antibodies available for three toolkit proteins, EMX1 (a homeobox transcription factor), FOXG1, and NR5A1 (a nuclear receptor protein). These three TFs were colocalized in groups of clustered neurons within the ventromedial hypothalamus (VMH) in experimental animals (Fig. 3); NR5A1 was additionally detected in scattered neurons within the neighboring arcuate hypothalamic nucleus (AHN) (Fig. 3). These results indicate coexpression of these three proteins in adult hypothalamic neurons following territory threat; previously they were considered primarily in the context of embryonic development.
Fig. 3.
Toolkit TF proteins, identified as responsive to social challenge in all three species, localized in mouse brain with immunohistochemistry. Sagittal sections from resident animals were treated with antibodies to EMX1, FOXG1, and NR5A1. Antibodies, stained in red, detected very similar clustered populations of neurons in the ventromedial hypothalamus (VMH); NR5A1 was additionally detected in scattered neurons within the arcuate hypothalamic nucleus (AHN).
Discussion
We present to our knowledge the first comprehensive evaluation of the molecular mechanisms of social challenge that are shared across distantly related taxa. Even though many aspects of the neuromolecular response to territory intrusion are likely taxon or even species specific, we identified cross-species similarities at several biological levels. Our results indicate that chromosome organization and energy metabolism are core processes associated with the response to social challenge.
Changes in chromosome organization may be fundamental to the response to social challenge because they affect DNA accessibility, a prerequisite for transcriptional changes that may be further modified by other mechanisms (40). We observed other types of regulatory mechanisms, such as changes in RNA splicing (mouse), transcription cofactor activity (stickleback), and DNA methyltransferase activity (honey bee), which may add species or context specificity to the response to territory intrusion. Chromosome organization also may be a core mechanism because the processes involved, e.g., histone modifications, are more evolutionarily conserved compared with other epigenetic mechanisms (41).
Brain energy metabolism was the second core process modulated in response to social challenge. A recent study demonstrated a negative causal relationship between brain oxidative phosphorylation activity and territorial aggression in honey bees and fruit flies (24). Consistent with that study, we here specifically identified modulation of NADH dehydrogenase, the oxidative phosphorylation enzyme complex with the best-known relationship to territorial aggression (7, 24). Oxidative phosphorylation genes were generally down-regulated in response to territory intrusion in mouse and honey bee, as in previous studies (7, 24). The stickleback diencephalon showed the opposite pattern, possibly indicative of brain regional heterogeneity or species-level variation in the timing of the metabolic response. A change in energy metabolism might affect a fundamental neural signaling property like excitability and thus play a general and conserved role in modulating behavior across species. Mouse IHC results further implicated metabolism by localizing the molecular response to neurons in the VMH and AHN, regions that interact to regulate food intake and glucose homeostasis (42). Thus, our results associate aggressive social experience and behavioral response with shifts in metabolism at both the cellular and organismal levels. Moreover, AHN neurons are known to regulate metabolism of serotonin (43), and DLX1, which was implicated in cis-motif analysis, is expressed in GABAergic and dopaminergic neurons in the AHN (35). Together these data suggest that metabolic plasticity is also associated with the activity of known aggression-related neurotransmitter systems.
In addition to metabolism and chromosome organization, our study highlighted transcriptional regulation as a significant component of the response to territory threat. In particular, seven homologous TFs were identified as putative “toolkit genes” for social behavior (Tier 1). Several of these have been associated with metabolism or social behavior in previous studies. Nr4a3 (a nuclear receptor protein), Nr5a1, and Irx3 (a homeobox transcription factor) play key roles in energy homeostasis (44–47); Nr5a1, Emx1, and Foxg1 have been associated with behavior and mood disorders (48–50); Nr4a3 is up-regulated in response to novel song in birds (51); and Nr5a1 is linked to social aggression in sticklebacks and mice (8, 48).
Our results also implicated conserved gene networks associated with neuroendocrine signaling. The TF Egr1, implicated in both the GSEA and motif enrichment analysis and known to be involved in social behaviors in other species, is up-regulated by gonadotropins in the brain (52). We identified other neuroendocrine-associated TFs, including three (Rax, Otp, and Nr2e1) that are known to have consistent functions in all three species (53, 54). Mouse and fly Nr2e1/tll proteins share a conserved DNA binding motif, which we identified as significantly enriched in the promoter regions of DEGs in all three species. These results are consistent with previous studies showing that Nr2e1/tll proteins have similar regulatory targets (55, 56) and are known to play a conserved role in aggression (57). Portions of the mouse hypothalamus and the stickleback diencephalon belong to the vertebrate brain social behavior network, a series of connected brain regions that control multiple social behaviors, including aggression (58). Although the bee brain does not include a homologous anatomical correlate to the hypothalamus, the neuroendocrine centers of insects, fish, and mammals function similarly and are thought to share common evolutionary origins (53). Thus, our finding of a conserved role for neuroendocrine signaling suggests that elements of the social behavior network in vertebrates, a network defined by interconnected brain regions, may also be conserved at the molecular level in insects, despite a lack of brain structural homology. The neuromolecular overlap among the three species, despite differences in starting brain material, is striking and suggests the mouse VH captures the evolutionarily conserved response. Additional work is needed to assess the implications of this degree of overlap in terms of the distribution of socially responsive regions in the brain, particularly for honey bee.
NF-κB signaling also was part of the conserved response to territory intrusion. Altered NF-κB signaling in white blood cells has been implicated in the human response to social stress (59), and this mechanism has been proposed to link chronic stress to increased prevalence of diseases, including mental health disorders, cardiac illnesses, and cancer (60). A model of a honey bee brain transcription regulatory network identified NF-κB as a global regulator of behaviorally related brain gene expression in contexts including aggression (23). A conserved role of immune and stress responses across multiple social contexts would suggest that these pathways play a general role in social behavioral modulation. The elements of these processes and the contexts that contribute to adverse health outcomes remain to be investigated.
Evidence for a shared neuromolecular response to territory intrusion spans multiple biological levels. We have identified evidence for shared core processes, components of gene networks, and single genes. Understanding the regulation and evolution of social behavior across diverse species requires a comparative regulatory genomics approach at the network (and subnetwork) level that goes beyond single genes. For instance, one toolkit TF, Pou3f4 (honey bee GB53100), is the ortholog to the Drosophila melanogaster gene Pdm2, a target of the iconic insect transcription factor fruitless, a master regulatory gene involved in social behaviors including aggression (61). Because there is no vertebrate ortholog to fruitless, one implication of this finding is that some of the gene networks regulated by fruitless may be involved in social interactions in other species, despite the absence of the specific upstream regulator gene.
Most of the behavioral toolkit TFs identified in our study (Nr4a3, Lhx1, Nr5a1, Irx3, Emx1, and Foxg1) are involved in brain or neural development, including postnatal neurogenesis (62–64), and the involvement of developmental processes also was revealed by both the GSEA and cis-motif analyses. Previous studies have associated developmental gene expression in the adult brain with plasticity in neural connectivity and the maintenance of regional boundaries (65, 66); other examples of genetic toolkits also involve genes associated with developmental patterning (3). Development genes may be ideal toolkit candidates because their critical roles in the embryonic stage lead to a high degree of conservation over evolutionary time. Our results demonstrate an additional property of these genes, the ability to show both highly canalized and highly plastic expression at different points in an organism’s life. Elucidating the regulatory features that alternately constrain and facilitate plasticity in gene expression within the lifespan of the organism will help explain why some genes function as components of genetic toolkits, ultimately improving our understanding of the evolution of complex traits like social behavior.
Materials and Methods
See SI Materials and Methods for details.
Behavioral Paradigms.
Focal animals were allowed to establish a territory in a closed container in the laboratory. Experimental individuals were exposed to an intruder, which elicited an aggressive response from the focal animal (confirmed by observation). Control individuals were exposed to an inanimate object, which provoked exploration but did not elicit aggression. Animals were exposed to the intruder or object for 5 min (honey bee and stickleback) or 10 min (mouse) and then the intruder or object was removed. Animals were killed after an additional 15 min (mouse) or 25 min (honey bee and stickleback). These slight variations in timelines were required to keep mouse and stickleback experiments consistent with previously published protocols (8, 43, 67, 68). The protocol for honey bee was novel, so we followed the stickleback timeline because expression differences for IEGs and other socially responsive genes are generally best detected 30 min poststimulus (69, 70). We sampled three individuals per treatment for mouse and stickleback and six groups per treatment for honey bees.
RNA Sequencing.
Brains were dissected and RNA was extracted and quantified following standard procedures. Poly-A RNA was enriched from total RNA, using Dynabeads Oligo(dT)25 (Life Technologies), and RNA-seq libraries were prepared using the NEXTflex Directional RNA-seq Kit with Illumina-compatible adaptors (Bioo Scientific). Single-end sequencing was performed on an Illumina HiSEq 2500 instrument by the W. M. Keck Center (University of Illinois).
Data Processing and Enrichment Analyses.
RNA-seq reads were aligned to reference genomes (mouse, NCBI build 37.2; stickleback, Ensembl release 70; honey bee, ref. 71), using TopHat and Bowtie, and HTSeq was used to calculate read counts per gene. Within-species clustering analyses identified one mouse control sample and two bee control samples as outliers, and these samples were excluded from further analyses (final sample sizes were 3/2, 3/3, 6/4 experimental/control for mouse, stickleback, and honey bee, respectively). For each species, we filtered out genes with extremely low expression levels. We assessed differential expression between experimental and control, using the “exactTest” function in EdgeR (72). We ranked all genes on the basis of raw P value. Ranked lists were assessed for Gene Ontology enrichment, using the logistic regression function in Babelomics 4.3 (73). This threshold-free analysis determines Gene Ontology categories that are represented by genes near the top of a ranked list (10).
Homologous Triplets.
EOGs were obtained from OrthoDB (21) (accessed October 15, 2013). To use a broad criterion for homology, we retained all paralogs (74), ultimately selecting the paralog showing the highest degree of differential expression between experimental and control conditions as the species representative for each homologous triplet. Homologous triplet combined significance scores were calculated using Fisher’s method [R package MADAM (75)].
Gene Annotations.
We used mouse GO annotations curated by the database Babelomics 4.3 (73). For stickleback and honey bee, we derived Gene Ontology assignments, using protein family annotations from the database PANTHER (76). For the homologous triplets analysis, we assigned functional enrichment on the basis of the mouse gene representative.
GSEA Post Hoc Analyses.
Because the GSEA approach is threshold-free, it did not identify specific genes that account for significant enrichment of a particular GO term. To identify these genes for a subset of significantly enriched terms of interest, we performed a post hoc analysis consisting of iterative hypergeometric tests to determine the threshold cutoff point within the ranked list of genes that corresponded to the strongest significance for enrichment of the GO term of interest.
cis-Motif Analysis.
We used the Stubb algorithm (77) for genome-wide scanning of motif matches in conjunction with cis-Metalysis to identify TF motifs or pairs of motifs enriched in noncoding regions around up- and down-regulated socially responsive genes. We used a collection of 368 motifs from JASPAR (78) and Jolma et al. (79). For “meta-associations,” i.e., motifs (or combinations of two motifs) enriched for DEGs across two or three species, we corrected “meta P values” reported by cis-Metalysis for multiple hypothesis testing, using an empirical FDR estimation.
Quantitative PCR.
We generated cDNA from pools of RNA dissected from the VH, the bed nucleus of the stria terminalis (BNST), and cortex of experimental or control mice. Gene expression levels determined by qPCR were normalized to the average expression of mouse 18S rRNA (primers are in Table S3).
IHC.
We performed IHC on adjacent brain sections, using antibodies against ARC, EMX1, NR5A1, and FOXG1. Alexa-Fluor goat-α-mouse or mouse-α-rabbit (Molecular Probes, Invitrogen) was used as secondary antibody. Tissue was counterstained using 10 µg/mL Hoechst 33342 nuclear staining (Invitrogen; no. H3570). Fluorescent images were reviewed using an Olympus BX60 microscope and images were captured using a NanoZoomer high-resolution scanner (Hamamatsu) and an ApoTome Structured Illumination Optical Sectioning System (Zeiss) incorporated into an Axiovert 200M microscope (Zeiss).
Supplementary Material
Acknowledgments
We thank K. M. Kapheim, M. M. Wheeler, A. T. Magis, and M. S. McNeill for RNA-seq consultation and C. Blatti for cis-motif analysis consultation. This work was funded by Grant SFLIFE 291812 from the Simons Foundation (to L.S., G.E.R., A.M.B., S.S., and J.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420369111/-/DCSupplemental.
References
- 1.Stern DL. The genetic causes of convergent evolution. Nat Rev Genet. 2013;14(11):751–764. doi: 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]
- 2.Martin A, Orgogozo V. The loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67(5):1235–1250. doi: 10.1111/evo.12081. [DOI] [PubMed] [Google Scholar]
- 3.Gellon G, McGinnis W. Shaping animal body plans in development and evolution by modulation of Hox expression patterns. BioEssays. 1998;20(2):116–125. doi: 10.1002/(SICI)1521-1878(199802)20:2<116::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 4.Toth AL, Robinson GE. Evo-devo and the evolution of social behavior. Trends Genet. 2007;23(7):334–341. doi: 10.1016/j.tig.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Sumner S. The importance of genomic novelty in social evolution. Mol Ecol. 2014;23(1):26–28. doi: 10.1111/mec.12580. [DOI] [PubMed] [Google Scholar]
- 6.Kültz D, et al. New frontiers for organismal biology. Bioscience. 2013;63(6):464–471. [Google Scholar]
- 7.Alaux C, et al. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc Natl Acad Sci USA. 2009;106(36):15400–15405. doi: 10.1073/pnas.0907043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanogo YO, Band M, Blatti C, Sinha S, Bell AM. Transcriptional regulation of brain gene expression in response to a territorial intrusion. Proc Biol Sci. 2012;279(1749):4929–4938. doi: 10.1098/rspb.2012.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin D, et al. Functional identification of an aggression locus in the mouse hypothalamus. Nature. 2011;470(7333):221–226. doi: 10.1038/nature09736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sartor MA, Leikauf GD, Medvedovic M. LRpath: A logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics. 2009;25(2):211–217. doi: 10.1093/bioinformatics/btn592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459(7245):356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton SP, et al. Evidence for genetic linkage between a polymorphism in the adenosine 2A receptor and panic disorder. Neuropsychopharmacology. 2004;29(3):558–565. doi: 10.1038/sj.npp.1300311. [DOI] [PubMed] [Google Scholar]
- 14.Lahr EC, Dean D, Ewer J. Genetic analysis of ecdysis behavior in Drosophila reveals partially overlapping functions of two unrelated neuropeptides. J Neurosci. 2012;32(20):6819–6829. doi: 10.1523/JNEUROSCI.5301-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park Y, Kim YJ, Dupriez V, Adams ME. Two subtypes of ecdysis-triggering hormone receptor in Drosophila melanogaster. J Biol Chem. 2003;278(20):17710–17715. doi: 10.1074/jbc.M301119200. [DOI] [PubMed] [Google Scholar]
- 16.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 17.Su J, Gorse K, Ramirez F, Fox MA. Collagen XIX is expressed by interneurons and contributes to the formation of hippocampal synapses. J Comp Neurol. 2010;518(2):229–253. doi: 10.1002/cne.22228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalita K, Kuzniewska B, Kaczmarek L. MKLs: Co-factors of serum response factor (SRF) in neuronal responses. Int J Biochem Cell Biol. 2012;44(9):1444–1447. doi: 10.1016/j.biocel.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Ramos C, Martinez A, Robert B, Soriano E. Msx1 expression in the adult mouse brain: Characterization of populations of beta-galactosidase-positive cells in the hippocampus and fimbria. Neuroscience. 2004;127(4):893–900. doi: 10.1016/j.neuroscience.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313(5795):1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. OrthoDB: A hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 2013;41(Database issue):D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mauro C, et al. NF-κB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat Cell Biol. 2011;13(10):1272–1279. doi: 10.1038/ncb2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandrasekaran S, et al. Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proc Natl Acad Sci USA. 2011;108(44):18020–18025. doi: 10.1073/pnas.1114093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li-Byarlay H, Rittschof CC, Massey JH, Pittendrigh BR, Robinson GE. Socially responsive effects of brain oxidative metabolism on aggression. Proc Natl Acad Sci USA. 2014;111(34):12533–12537. doi: 10.1073/pnas.1412306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell P, Reep RL, Stoll ML, Ophir AG, Phelps SM. Conservation and diversity of Foxp2 expression in muroid rodents: Functional implications. J Comp Neurol. 2009;512(1):84–100. doi: 10.1002/cne.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson GE, Fernald RD, Clayton DF. Genes and social behavior. Science. 2008;322(5903):896–900. doi: 10.1126/science.1159277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swanberg SE, Nagarajan RP, Peddada S, Yasui DH, LaSalle JM. Reciprocal co-regulation of EGR2 and MECP2 is disrupted in Rett syndrome and autism. Hum Mol Genet. 2009;18(3):525–534. doi: 10.1093/hmg/ddn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Philippi A, et al. Association of autism with polymorphisms in the paired-like homeodomain transcription factor 1 (PITX1) on chromosome 5q31: A candidate gene analysis. BMC Med Genet. 2007;8:74. doi: 10.1186/1471-2350-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ariani F, et al. FOXG1 is responsible for the congenital variant of Rett syndrome. Am J Hum Genet. 2008;83(1):89–93. doi: 10.1016/j.ajhg.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clayton DF. The genomics of memory and learning in songbirds. Annu Rev Genomics Hum Genet. 2013;14:45–65. doi: 10.1146/annurev-genom-090711-163809. [DOI] [PubMed] [Google Scholar]
- 31.Lutz CC, Robinson GE. Activity-dependent gene expression in honey bee mushroom bodies in response to orientation flight. J Exp Biol. 2013;216(pt 11):2031–2038. doi: 10.1242/jeb.084905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fulton DL, et al. TFCat: The curated catalog of mouse and human transcription factors. Genome Biol. 2009;10(3):R29. doi: 10.1186/gb-2009-10-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ament SA, et al. New meta-analysis tools reveal common transcriptional regulatory basis for multiple determinants of behavior. Proc Natl Acad Sci USA. 2012;109(26):E1801–E1810. doi: 10.1073/pnas.1205283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coyle-Rink J, Del Valle L, Sweet T, Khalili K, Amini S. Developmental expression of Wnt signaling factors in mouse brain. Cancer Biol Ther. 2002;1(6):640–645. doi: 10.4161/cbt.313. [DOI] [PubMed] [Google Scholar]
- 35.Yee CL, Wang Y, Anderson S, Ekker M, Rubenstein JL. Arcuate nucleus expression of NKX2.1 and DLX and lineages expressing these transcription factors in neuropeptide Y(+), proopiomelanocortin(+), and tyrosine hydroxylase(+) neurons in neonatal and adult mice. J Comp Neurol. 2009;517(1):37–50. doi: 10.1002/cne.22132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B, Chambers KJ, Leprince D, Faller DV, Wang S. Requirement for chromatin-remodeling complex in novel tumor suppressor HIC1-mediated transcriptional repression and growth control. Oncogene. 2009;28(5):651–661. doi: 10.1038/onc.2008.419. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Petersohn D, Schoch S, Brinkmann DR, Thiel G. The human synapsin II gene promoter. Possible role for the transcription factor zif268/egr-1, polyoma enhancer activator 3, and AP2. J Biol Chem. 1995;270(41):24361–24369. doi: 10.1074/jbc.270.41.24361. [DOI] [PubMed] [Google Scholar]
- 38.Cioffi S, et al. Tbx1 regulates brain vascularization. Hum Mol Genet. 2014;23(1):78–89. doi: 10.1093/hmg/ddt400. [DOI] [PubMed] [Google Scholar]
- 39.Larsen KB, Lutterodt MC, Møllgård K, Møller M. Expression of the homeobox genes OTX2 and OTX1 in the early developing human brain. J Histochem Cytochem. 2010;58(7):669–678. doi: 10.1369/jhc.2010.955757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 41.Fuchs J, Demidov D, Houben A, Schubert I. Chromosomal histone modification patterns—from conservation to diversity. Trends Plant Sci. 2006;11(4):199–208. doi: 10.1016/j.tplants.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 42.Ramírez-Amaya V, et al. Spatial exploration-induced Arc mRNA and protein expression: Evidence for selective, network-specific reactivation. J Neurosci. 2005;25(7):1761–1768. doi: 10.1523/JNEUROSCI.4342-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karl T, et al. Y1 receptors regulate aggressive behavior by modulating serotonin pathways. Proc Natl Acad Sci USA. 2004;101(34):12742–12747. doi: 10.1073/pnas.0404085101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smemo S, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baba T, et al. Glycolytic genes are targets of the nuclear receptor Ad4BP/SF-1. Nat Commun. 2014;5:3634. doi: 10.1038/ncomms4634. [DOI] [PubMed] [Google Scholar]
- 46.Büdefeld T, Tobet SA, Majdic G. Steroidogenic factor 1 and the central nervous system. J Neuroendocrinol. 2012;24(1):225–235. doi: 10.1111/j.1365-2826.2011.02174.x. [DOI] [PubMed] [Google Scholar]
- 47.Nonogaki K, et al. Serotonin 5-HT2C receptor-independent expression of hypothalamic NOR1, a novel modulator of food intake and energy balance, in mice. Biochem Biophys Res Commun. 2009;386(2):311–315. doi: 10.1016/j.bbrc.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 48.Silva BA, et al. Independent hypothalamic circuits for social and predator fear. Nat Neurosci. 2013;16(12):1731–1733. doi: 10.1038/nn.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kinsler R, Taylor MM, Flores NM, Leffert JJ, Beech RD. Altered response to antidepressant treatment in FoxG1 heterozygous knockout mice. Synapse. 2010;64(2):169–171. doi: 10.1002/syn.20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao BJ, Li Y. Reduced anxiety— and depression-like behaviors in Emx1 homozygous mutant mice. Brain Res. 2002;937(1-2):32–40. doi: 10.1016/s0006-8993(02)02461-7. [DOI] [PubMed] [Google Scholar]
- 51.Dong S, et al. Discrete molecular states in the brain accompany changing responses to a vocal signal. Proc Natl Acad Sci USA. 2009;106(27):11364–11369. doi: 10.1073/pnas.0812998106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang EJ, Nasipak BT, Kelley DB. Direct action of gonadotropin in brain integrates behavioral and reproductive functions. Proc Natl Acad Sci USA. 2007;104(7):2477–2482. doi: 10.1073/pnas.0608391104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tessmar-Raible K, et al. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: Insights into hypothalamus evolution. Cell. 2007;129(7):1389–1400. doi: 10.1016/j.cell.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 54.de Velasco B, et al. Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev Biol. 2007;302(1):309–323. doi: 10.1016/j.ydbio.2006.09.035. [DOI] [PubMed] [Google Scholar]
- 55.Yu RT, McKeown M, Evans RM, Umesono K. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature. 1994;370(6488):375–379. doi: 10.1038/370375a0. [DOI] [PubMed] [Google Scholar]
- 56.Yu RT, et al. The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proc Natl Acad Sci USA. 2000;97(6):2621–2625. doi: 10.1073/pnas.050566897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis SM, Thomas AL, Nomie KJ, Huang L, Dierick HA. Tailless and Atrophin control Drosophila aggression by regulating neuropeptide signalling in the pars intercerebralis. Nat Commun. 2014;5:3177. doi: 10.1038/ncomms4177. [DOI] [PubMed] [Google Scholar]
- 58.Goodson JL. The vertebrate social behavior network: Evolutionary themes and variations. Horm Behav. 2005;48(1):11–22. doi: 10.1016/j.yhbeh.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller GE, et al. A functional genomic fingerprint of chronic stress in humans: blunted glucocorticoid and increased NF-kappaB signaling. Biol Psychiatry. 2008;64(4):266–272. doi: 10.1016/j.biopsych.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cole SW. Elevating the perspective on human stress genomics. Psychoneuroendocrinology. 2010;35(7):955–962. doi: 10.1016/j.psyneuen.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neville MC, et al. Male-specific fruitless isoforms target neurodevelopmental genes to specify a sexually dimorphic nervous system. Curr Biol. 2014;24(3):229–241. doi: 10.1016/j.cub.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiecker C, Lumsden A. Compartments and their boundaries in vertebrate brain development. Nat Rev Neurosci. 2005;6(7):553–564. doi: 10.1038/nrn1702. [DOI] [PubMed] [Google Scholar]
- 63.Abellán A, Vernier B, Rétaux S, Medina L. Similarities and differences in the forebrain expression of Lhx1 and Lhx5 between chicken and mouse: Insights for understanding telencephalic development and evolution. J Comp Neurol. 2010;518(17):3512–3528. doi: 10.1002/cne.22410. [DOI] [PubMed] [Google Scholar]
- 64.Shen L, Nam HS, Song P, Moore H, Anderson SA. FoxG1 haploinsufficiency results in impaired neurogenesis in the postnatal hippocampus and contextual memory deficits. Hippocampus. 2006;16(10):875–890. doi: 10.1002/hipo.20218. [DOI] [PubMed] [Google Scholar]
- 65.Zapala MA, et al. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci USA. 2005;102(29):10357–10362. doi: 10.1073/pnas.0503357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.French L, Pavlidis P. Relationships between gene expression and brain wiring in the adult rodent brain. PLoS Comput Biol. 2011;7(1):e1001049. doi: 10.1371/journal.pcbi.1001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bell AM, Backström T, Huntingford FA, Pottinger TG, Winberg S. Variable neuroendocrine responses to ecologically-relevant challenges in sticklebacks. Physiol Behav. 2007;91(1):15–25. doi: 10.1016/j.physbeh.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 68.Raskin K, et al. Conditional inactivation of androgen receptor gene in the nervous system: Effects on male behavioral and neuroendocrine responses. J Neurosci. 2009;29(14):4461–4470. doi: 10.1523/JNEUROSCI.0296-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ellis LL, Carney GE. Socially-responsive gene expression in male Drosophila melanogaster is influenced by the sex of the interacting partner. Genetics. 2011;187(1):157–169. doi: 10.1534/genetics.110.122754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cummings ME, et al. Sexual and social stimuli elicit rapid and contrasting genomic responses. Proc Biol Sci. 2008;275(1633):393–402. doi: 10.1098/rspb.2007.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elsik CG, et al. HGSC production teams Honey Bee Genome Sequencing Consortium Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genomics. 2014;15:86. doi: 10.1186/1471-2164-15-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Medina I, et al. Babelomics: An integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 2010;38(web server issue):W210–W213. doi: 10.1093/nar/gkq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kristiansson E, et al. A novel method for cross-species gene expression analysis. BMC Bioinformatics. 2013;14(70):70. doi: 10.1186/1471-2105-14-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kugler KG, Mueller LA, Graber A. MADAM - An open source meta-analysis toolbox for R and Bioconductor. Source Code Biol Med. 2010;5:3. doi: 10.1186/1751-0473-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8(8):1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sinha S, van Nimwegen E, Siggia ED. A probabilistic method to detect regulatory modules. Bioinformatics. 2003;19(suppl 1):i292–i301. doi: 10.1093/bioinformatics/btg1040. [DOI] [PubMed] [Google Scholar]
- 78.Portales-Casamar E, et al. JASPAR 2010: The greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38(database issue):D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jolma A, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152(1-2):327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.