Significance
Factors that influence the virulence of HIV are of direct relevance to ongoing efforts to contain, and ultimately eradicate, the HIV epidemic. We here investigate in Botswana and South Africa, countries severely affected by HIV, the impact on HIV virulence of adaptation of HIV to protective HLA alleles such as HLA-B*57. In Botswana, where the epidemic started earlier and reached higher adult seroprevalence than in South Africa, HIV replication capacity is lower. HIV is also better adapted to HLA-B*57, which in Botswana has no protective effect, in contrast to its impact in South Africa. Modelling studies indicate that increasing antiretroviral therapy access may also contribute to accelerated declines in HIV virulence over the coming decades.
Keywords: HLA, HIV, adaptation, antiretroviral therapy, virulence
Abstract
It is widely believed that epidemics in new hosts diminish in virulence over time, with natural selection favoring pathogens that cause minimal disease. However, a tradeoff frequently exists between high virulence shortening host survival on the one hand but allowing faster transmission on the other. This is the case in HIV infection, where high viral loads increase transmission risk per coital act but reduce host longevity. We here investigate the impact on HIV virulence of HIV adaptation to HLA molecules that protect against disease progression, such as HLA-B*57 and HLA-B*58:01. We analyzed cohorts in Botswana and South Africa, two countries severely affected by the HIV epidemic. In Botswana, where the epidemic started earlier and adult seroprevalence has been higher, HIV adaptation to HLA including HLA-B*57/58:01 is greater compared with South Africa (P = 7 × 10−82), the protective effect of HLA-B*57/58:01 is absent (P = 0.0002), and population viral replicative capacity is lower (P = 0.03). These data suggest that viral evolution is occurring relatively rapidly, and that adaptation of HIV to the most protective HLA alleles may contribute to a lowering of viral replication capacity at the population level, and a consequent reduction in HIV virulence over time. The potential role in this process played by increasing antiretroviral therapy (ART) access is also explored. Models developed here suggest distinct benefits of ART, in addition to reducing HIV disease and transmission, in driving declines in HIV virulence over the course of the epidemic, thereby accelerating the effects of HLA-mediated viral adaptation.
Control of the global HIV pandemic has been focused on prevention of disease and of transmission via antiretroviral therapy (ART) and via attempts to develop HIV vaccines capable of inducing effective immune responses against the virus. Relatively little consideration has been given to the impact of widespread ART and of natural antiviral immunity on the evolution of HIV virulence. Theory predicts, all other things being equal, that infections causing new epidemics will reduce in virulence over time because pathogens require host survival to transmit (1, 2). However, as in HIV infection, a tradeoff typically exists between virulence and transmissibility. In HIV, high viral load is associated with increased transmission risk but more rapid progression to AIDS and death (3–5). It has been estimated that the optimal viral set point for the virus is ∼30,000 copies per mL, which is sufficiently high to provide a reasonable chance of transmission, and sufficiently low to provide long-term survival of the host before progression to disease (5).
We here consider the potential impact of two processes on changing HIV virulence over the course of the epidemic. The first is that of viral evolution in response to HLA-mediated selection pressure. The second is that of widespread use of ART. The term “virulence” refers to a microorganism’s capacity to cause disease (6). It has previously been shown that the viral replicative capacity (VRC) of the transmitted virus predicts both the viral set point and, more strongly, the CD4 decline in the recipient (7, 8). The CD4 decline is the closest marker of rate of progression to disease, and the viral set point predicts that rate of CD4 decline (4). Thus, on this basis, the replicative capacity of HIV at a population level strongly influences the virulence of HIV infection. In turn, virulence is reflected by viral set point and CD4 count. However, it is important to note that, at an individual level, the rate of progression to disease is affected by many additional influences than VRC alone, including host genetic, immunologic, environmental (such as coinfections), and stochastic factors.
Immune control of HIV-1 infection is most strongly influenced by the HLA genes expressed in each individual (9). An important underlying mechanism is the ability of protective HLA molecules to direct cytotoxic T lymphocytes (CTLs) against virus-infected cells, such that HIV can only evade these responses via the selection of escape mutants that also reduce VRC. In this way, even though certain immune responses lose potency, the ability of the virus to replicate can be substantially compromised (10–12).
These CTL escape mutants can be transmitted and accumulate in the population to the point where the virus has successfully adapted to immune responses that were previously protective (13). In Japan, where HLA-B*51 was initially protective in the early part of the epidemic, increase in the frequency of an escape mutant in the critical CTL epitope to 75% of circulating viruses in the Japanese HIV-positive population was associated with loss of the protective effect associated with HLA-B*51. Recent studies (14) indicate that VRC in Japan has been declining over the same timespan, prompting the question of how viral adaptation to HLA-mediated selection might affect population-level VRC.
The rapidly increasing and widespread use of ART worldwide to slow disease progression and reduce transmission has been well documented. However, its potential impact on population-level VRC has received little attention. It is well established that transmission of viruses with high VRC tends to result in high viral set points and rapid CD4 decline in the recipient (7, 8, 15, 16). Thus, the viruses in individuals with the lowest CD4 counts tend to have the highest VRC (14, 17–20). ART initiation along WHO guidelines (21) is in most cases the result of a CD4 count threshold being reached. Successful treatment with ART suppresses viremia to such low levels that the risk of transmissions is effectively eliminated (22). These observations therefore prompt the hypothesis that increasing use of ART would be likely to contribute to a removal from the population of the viruses with the highest VRC, and therefore to decrease the virulence of the HIV epidemic over time.
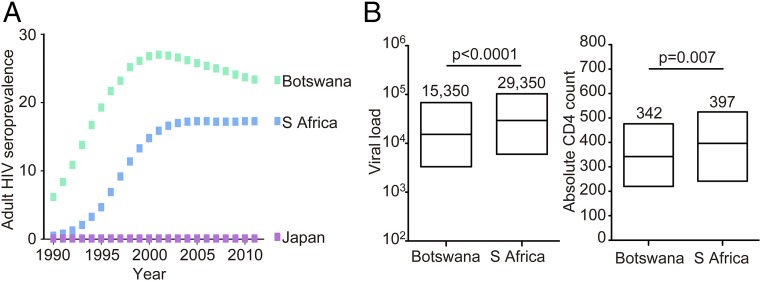
We reasoned here that if the effects of viral adaptation on protective HLA alleles, with subsequent loss of that HLA-associated protection and a measurable decline in VRC, are already evident in the course of the epidemic in Japan, where adult seroprevalence has never exceeded 0.1%, these effects may be at least as evident in southern Africa, where the epidemic has been established for longer and adult seroprevalence rates have been >100-fold higher (23). Although South Africa is the country with the greatest absolute number of HIV infections (6.1 million South Africans currently living with HIV) (24), the epidemic in Botswana preceded it by some years and adult seroprevalence (percentage of the population infected) has been substantially higher (25) (Fig. 1A). Furthermore, the use of ART in South Africa has lagged several years behind that in Botswana (25) (Fig. S1). We hypothesized therefore that, in Botswana compared with South Africa, HIV would be better adapted to protective HLA molecules; that this HLA adaptation would be associated with reduction in or loss of that HLA-linked protective effect; and that the increased frequency of HLA-driven viral variants would be associated with reduced population-level VRC. Finally, we hypothesized that the higher ART coverage in Botswana would have had a corresponding greater impact on reducing population-level HIV virulence in that country.
Fig. 1.
(A) Adult seroprevalence of HIV infection in Botswana, South Africa, and Japan from 1990 to 2011. Data from ref. 25. (B) Viral load and absolute CD4 counts in ART-naïve antenatal cohorts in Gaborone, Botswana and Durban, South Africa. Median viral loads in the Gaborone and Durban cohorts were 15,350 HIV RNA copies per mL and 29,350 copies per mL, respectively (IQR of 3,300 to 68,350 and 6,100 to 103,500). Median absolute CD4 counts in Gaborone and Durban.
Results
Increased HIV Adaptation to HLA in Gaborone Compared with Durban.
Initial observations of two apparently comparable cohorts of ART-naïve antenatal mothers in Gaborone (n = 514) and Durban (n = 328) (mean age 27.5 and 27.3 y) showed somewhat lower viral loads in the Gaborone cohort (Fig. 1B; 15,350 vs. 29,350, P < 0.0001) and also lower absolute CD4 counts (mean 342 cells per mm3 vs. 397 cells per mm3, P = 0.007). The lower CD4 counts suggested that the Gaborone cohort might comprise individuals with more advanced disease, despite the fact that the two cohorts were closely matched in age. In both cohorts viral loads were inversely correlated with CD4 counts (Gaborone: r = −0.36, P < 0.0001; Durban: r = −0.50, P < 0.0001), but for a given CD4 count, viral loads were lower in the Gaborone cohort. Several explanations might underlie such findings, including population differences unrelated to HIV; however, these data were also consistent with the hypothesis that VRC might be lower in Gaborone than in Durban.
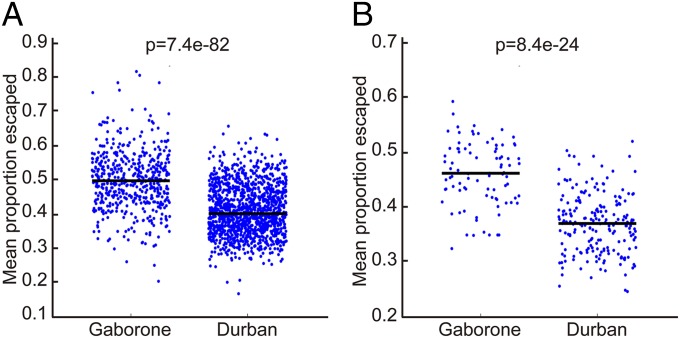
We first addressed the question of whether HIV sequences were better adapted to the HLA molecules expressed in Gaborone compared with Durban. Previous studies of >2,000 HIV-infected subjects in southern Africa—in whom HLA type and autologous Gag, Pol, and Nef HIV-1 sequences had been determined—had identified the escape mutants significantly driven by the HLA molecules expressed in this study group (26). We defined the degree of adaptation as the percentage of HIV amino acid residues that were escape mutants in the autologous virus sequence for the given HLA type of each study subject. We then tested adaptation of all HIV sequences in the cohort against each individual subject’s six class I HLA molecules to estimate the expected level of viral adaptation in that cohort to that individual. Using this approach, it is evident that virus sequences in Gaborone are indeed substantially better adapted to the HLA type of the study subjects than in the Durban cohort (Fig. 2A). This applied both to the cohorts analyzed as a whole and also to the subset of subjects expressing either HLA-B*57 or HLA-B*58:01 (Fig. 2B), the class I molecules previously shown to be most protective against HIV disease progression in Durban (26–28). The statistical significance of the findings was unaltered whether limiting the analysis to the maternal cohort in Durban described above or to an extended Durban cohort of 1,218 ART-naïve subjects (26) that included subjects from outpatient clinics (Methods).
Fig. 2.
Adaptation of Gag, Pol, and Nef sequences in Gaborone and Durban to the HLA repertoire. (A) Viral adaptation to the entire HLA repertoire (all six expressed HLA class I molecules) for the subjects in Gaborone and Durban, comparing all of the sites at which HIV amino acid polymorphisms are associated with any of the HLA molecules expressed, and calculating the average proportion of sites at which viral escape is present. (B) As in A, except restricting to HLA-B*57/58:01–positive individuals.
Reduced Protection from HLA-B*57/58:01 but Not B*39:10/81:01/42:01 in Gaborone.
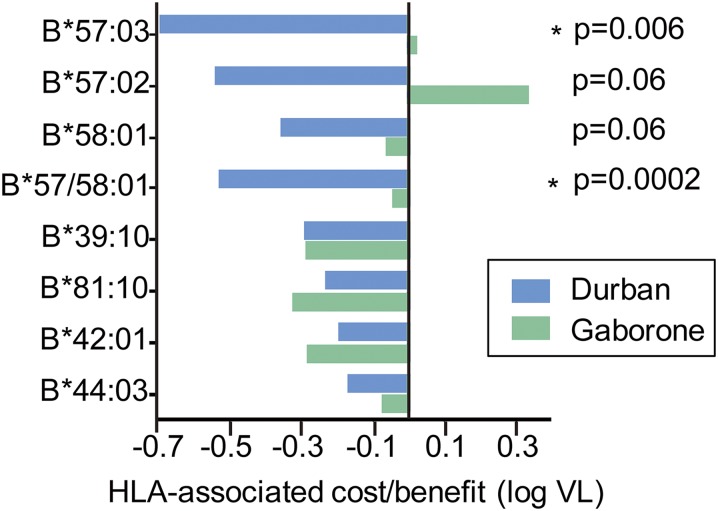
We next investigated whether the increased population-wide viral adaptation in individuals expressing HLA-B*57/58:01 observed in Gaborone was associated with a reduced protective impact of these alleles compared with Durban (Fig. 3). We observed no protective effect of HLA-B*57/58:01 in Botswana, a finding that contrasts significantly with the effect seen in Durban (26–28) (P = 0.0002). This lack of protection conferred by B*57/58:01 in Botswana was consistent for all three closely related alleles; HLA-B*57:03, HLA-B*57:02, and HLA-B*58:01. HLA-B*57 has been associated with the lowest viral set points of all of the HLA alleles that have been studied worldwide (9), and thus this observation in Botswana is unique. However, there was no difference between Gaborone and Durban in the impact on viral set point mediated by the second tier of protective alleles; HLA-B*39:10, HLA-B*81:01, and HLA-B*42:01.
Fig. 3.
Impact of selected HLA-B molecules on viral load in Gaborone and Durban. The relative contributions of HLA-B alleles previously identified as protective in Durban (11–13), to log viral load, in Durban compared with that in Gaborone.
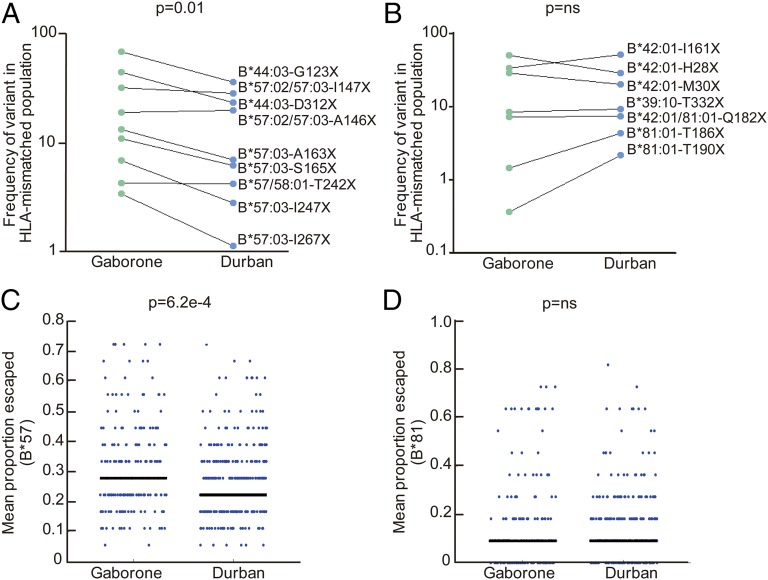
To investigate why there was no difference in the effect of HLA-B*39:10/81:01/42:01 in Gaborone and Durban—in contrast to the differential impact observed for HLA-B*57/58:01 and for HLA-B*44:03 which also carries a significant, albeit modest, protective effect in Durban (26, 28) but not in Gaborone (although the differences here were not significant, P = 0.47 for comparison)—we first compared the frequency of HLA-driven escape mutants in the two groups in Gag (group-specific antigen), which has been proposed as an important CTL target for immune control (29) (Fig. 4 A and B and Table S1). We observed a significant increase in the HLA-B*57/58:01/44:03–driven Gag escape mutants in Gaborone compared with Durban (which remained significant even after exclusion of the HLA-B*44:03 data), but there was no difference in the frequency of the HLA-B*39:10/81:01/42:01–driven escape mutants in these two populations. Extending this analysis to the degree of adaptation in Gag, Pol, and Nef in the two populations showed a significant increase in adaptation to HLA-B*57 in the Gaborone cohort, whereas no difference was observed in adaptation to HLA-B*81 between the populations (Fig. 4 C and D). These findings were not explained by differences in HLA prevalence in the two cohorts (Fig. S2). The only protective HLA allele differing in prevalence between the two cohorts was HLA-B*42:01, which did not in this case affect frequency of HLA-B*42:01–associated escape mutants in the populations.
Fig. 4.
Differential adaptation of HIV to selected HLA-B alleles in Gaborone and Durban. (A) Frequency of HLA-B*57/58:01/44:03–associated Gag mutants in Gaborone vs. Durban (P = 0.01, Student’s t test; P = 0.05 excluding HLA-B*44:03–associated variants). (B) Frequency of HLA-B*39:10/42:01/81:01–associated Gag mutants in Gaborone vs. Durban [P = not significant (ns)]. (C) HLA-B*57–associated adaptation (average proportion of HLA-B*57–associated sites at which escape variants are present per viral sequence) in Durban and Gaborone subjects expressing HLA-B*57. Mann–Whitney test for significance. (D) As in C, except for HLA-B*81.
The impact of HLA alleles associated with high viral set points, such as HLA-B*18:01 and HLA-B*58:02 (refs. 25–28) that do not drive selection of Gag escape mutants (30), did not differ between Gaborone and Durban (P = 0.61 and P = 0.22, respectively).
Lower Population-Level Viral Replicative Capacity in Gaborone.
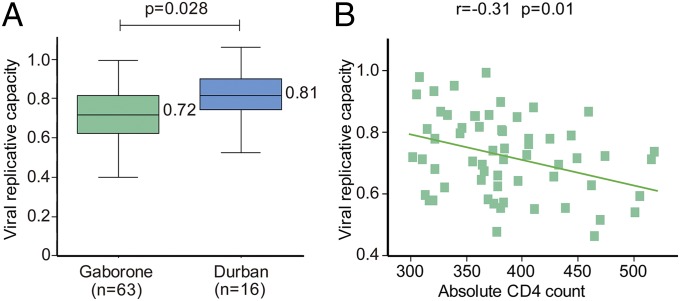
We next addressed the question whether VRC differs between the populations in Gaborone and Durban. Selected samples matched for the donor CD4 count (absolute CD4 count mean: 385 per mm3 and 389 per mm3 in Gaborone and Durban, respectively) were compared in side-by-side assays undertaken in the same laboratory under identical conditions. Consistent with the hypothesis developed above, VRCs were significantly lower in the Gaborone cohort (mean VRC of 0.72 vs. 0.81, P = 0.028; Fig. 5A). Consistent with other studies (17–20), VRC was significantly higher in those with low absolute CD4 counts both in Durban (17) and in Gaborone (Fig. 5B; r = −0.31, P = 0.01).
Fig. 5.
Viral replication capacities for Gaborone and Durban populations. (A) Replication capacities, normalized to NL4-3 comparator virus, determined for CD4-matched subjects (CD4 300–500 per mm3) in Gaborone (n = 63) and Durban (n = 16), measured under identical conditions (Methods), were significantly different (P = 0.028, Student’s t test). (B) Correlation between absolute CD4 count and viral replication capacity in the Gaborone study cohort [also previously shown for the Durban cohort (15)].
As expected from the data shown for the entire study cohorts (Figs. 2–4), the frequency of HLA-B*57/58:01–driven Gag mutants in the Botswana subjects whose VRC was determined was higher than in the Durban cohort (mean of 1.3 mutants per subject vs. 1.0, P = 0.045). This is consistent with previous observations that HLA-B*57/58:01–driven Gag mutants reduce VRC (9–12). However, this difference was modest, suggesting the possibility that factors additional to HIV adaptation to HLA-B*57/58:01 may be contributing to the population-level differences in VRC observed (Discussion).
Increased CTL Epitope Variant Frequency over a Decade Within the Same Population.
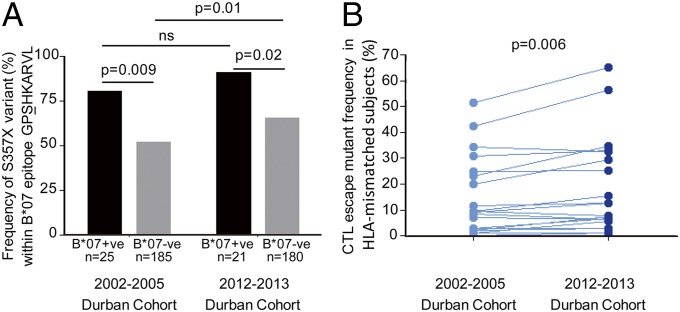
To confirm the increased frequency of CTL escape mutants within a population over time implied by these findings, we first sought to study suitable archived samples from Gaborone, but these were not available. We therefore addressed the question of whether the frequency of CTL mutants would significantly increase over a short time period, such as a decade, by evaluating the same population over time in Durban. To compare data with the maternal cohort in Durban described above that was enrolled between 2002 and 2005, we enrolled a second maternal cohort from the same site in Durban between 2012 and 2013. Gag sequences were determined in HLA-typed subjects and the frequency of variants within Gag CTL epitopes that have been identified in previous studies (13, 26, 30, 31) and confirmed once again in the current study (Table S1), were compared in the two Durban cohorts (Fig. 6 and Table S2). There was no single mutant that was significantly decreased in frequency in the 2012–2013 cohort compared with the 2002–2005 cohort, and there were six mutants that were significantly increased (P < 0.05), as well as 11 other mutants that were also increased in frequency; but individually these increases did not reach statistical significance. Overall, the variant frequency for all of the Gag CTL escape mutants was consistently higher in the 2012–2013 Durban cohort compared with the 2002–2005 Durban cohort (P = 0.006, paired t test) and provides further support for the hypothesis that overall CTL escape mutants increase in frequency over time through the course of the epidemic.
Fig. 6.
Increase in Gag CTL escape mutants within a decade within the same population. (A) Illustrative example of the S357X escape mutant within the HLA-B*07–restricted Gag epitope GPSHKARVL (Gag 355–362). The escape mutant S357X is selected in B*07-positive subjects in both the 2002–2005 Durban cohort and in the 2012–2013 Durban cohort; the frequency of the S357X mutant in the HLA-B*07–negative subjects was significantly higher in the 2012–2013 cohort. (B) Comparison of the frequency of all 23 described Gag CTL escape variants (Table S2) in the 2002–2005 Durban cohort (n = 210) with that in the 2012–2013 Durban cohort (n = 201) (P = 0.001, paired t test).
Discussion
These studies, undertaken in two of the countries worst affected by the HIV epidemic, suggest that here HIV evolution is progressing rapidly. The contrasts between Botswana and South Africa, in the degree of adaptation of HIV to prevailing HLA molecules in the populations and in the protective impact of protective alleles such as HLA-B*57 and HLA-B*58:01, coincide with the substantial differences in duration and magnitude of the epidemic in these two localities. The lack of any HLA-B*57/58:01–associated protective effect in the Botswana cohort that included 510 study subjects—18% of whom expressed one of the HLA-B*57/58:01 alleles—is striking, given that HLA-B*57 subtypes in Caucasian, African, Asian, and Hispanic populations have provided the greatest reduction in viral set points of all of the HLA alleles studied to date (9). However, this process of HIV adaptation to HLA alleles such as HLA-B*57 and HLA-B*58:01 that drive the strongest selection pressure on the virus, with consequent loss of immune protection mediated by those alleles, was anticipated more than a decade ago (32) and similar observations have been made in relation to HLA-B*51 in Japan as described above (13).
The lower VRC observed in the Botswana cohort is consistent with the greater degree of viral adaptation and accumulation of HLA-B*57/58:01–associated mutants, many of which have been shown to reduce VRC, especially in combination (9–12). In vivo, the impact of these escape mutants in reducing VRC is diminished by the presence of compensatory mutants (10–12, 20, 33, 34) and thus one might not necessarily expect increasing number of these mutants invariably to give rise to lower VRC. Nonetheless, the increasing number of HLA-B*57/58:01–associated mutants is correlated with the decreasing VRC in some Southern African cohort studies (Bloemfontein: r = −0.19, P = 0.03; Kimberley: r = −0.25, P = 0.007) (20); and in the current comparison between Gaborone and Durban cohorts, a significantly higher number of HLA-B*57/58:01–associated mutants was observed, both in the Gaborone cohort as a whole (Figs. 2B and 4 A and C) and in the subset selected for the VRC determinations (mean of 1.3 mutants per subject vs. 1.0, P = 0.045). However, the modest increase only in HLA-B*57/58:01–driven mutants in this Botswana subset on which the VRC measurements were undertaken, together with the observation that the number of HLA-B*57/58:01–driven mutations did not correlate significantly with the VRC in the Botswana cohort, suggests that factors additional to mutant number alone may contribute to the lower VRC observed.
One such factor may be the marked diversity of patterns in the frequency of HLA-associated mutations in Gaborone compared with Durban (Fig. 4). Some mutations have higher prevalence in Gaborone, whereas for others the prevalence is similar. For the HLA-B*81:01–associated mutations, prevalence appears lower in Gaborone than in Durban, even though the epidemic is longer established in Gaborone. Within populations the absolute frequency is also very variable (35). Mathematical models of the within-host evolution and between-host spread of CTL-driven escape mutations predict just such diversity. Depending on the rates of escape in HLA-matched hosts and reversion in HLA-mismatched hosts the prevalence of different escape mutations is predicted to display a wide range of dynamics patterns, even including an initial rise followed by a fall (35). These published modeling predictions are consistent with the data presented above, showing an increase in CTL mutant frequency over a decade within the Durban population. Mutants such as the S357X variant within the HLA-B*07–restricted epitope GPSHKARVL (Gag 355–362) accumulate rapidly, as expected for a variant selected at high frequency in acute infection (13) and with a low reversion rate (30); in contrast there was a more modest increase over the same period of the HLA-B*14–associated mutant K302R within the epitope DRFFKTLRA (Gag 298–306), which is selected at lower frequency and reverts rapidly (30).
The expectation from these findings would be that, over the course of the epidemic, HLA adaptation would progressively nullify the direct protective impact of HLA alleles previously associated with slow progression, at the same time mitigating this detrimental effect (to the host) by the concomitant decrease in VRC. For example, the viral loads in HLA-B*57:03–positive subjects were lower in Durban than in Botswana (median 2,630 vs. 9,570), but viral loads in HLA-B*57:03–positive subjects in Botswana were still relatively low given the high degree of viral adaptation to HLA-B*57 in Botswana. The disadvantage to an HLA-B*57–positive host of being infected with a virus preadapted to HLA-B*57 is to some extent compensated for by the low VRC of the transmitted virus which correlates with low early viral set point and high CD4 count (8).
The findings described here in Botswana are consistent with those of a previous study in Amsterdam, where accumulation of HLA-B–associated mutants had accrued over a 20-y period, reducing the number of available epitopes restricted by the most protective HLA molecules in that population, HLA-B*27:05 and HLA-B*57:01 (36). These findings, together with the data presented here and those previously referred to in Japan in relation to HLA-B*51 (13), contrast with the recent study of the North American epidemic in which HIV adaptation was observed to be occurring only slowly, although to a greatest relative extent to the protective alleles (37). The possible reasons for this difference are unknown, but may relate to the heterogeneity of the population structure in North America compared with Botswana, Holland, or Japan, with a consequently diluted and inconsistent selection pressure on the virus driven by the HLA diversity.
An additional factor that may contribute to the lowering of VRC over time in a population is the impact of ART. As described above, transmission of viruses with high replication capacity tends to result in more rapid CD4 decline in the recipient (8) and hence HIV-infected people with low CD4 counts tend to have viruses with high replicative capacity, as observed in this study and several others (14, 17–20). To examine the plausibility of this hypothesis, we developed a mathematical model, first, to investigate how HIV virulence would be expected to change over the course of the epidemic in the absence of ART; and, second, to examine the anticipated impact that ART would have on virulence (Fig. S3 and Table S3). The model that was developed supported the hypothesis that selective treatment of people with low CD4 counts will tend to accelerate the evolution of variants with lower VRC. However, these effects are predicted to be quite subtle over the short term, but would contribute to accelerate the lowering of VRC over the longer term (Fig. S3C).
Several factors may influence the impact of ART on population-level VRC. First, the ART treatment guidelines vary from place to place and change over time. ART treatment programs came into being in South Africa relatively late, government programs being introduced only within the last decade (23, 25, 38, 39). The initial CD4 treatment criterion for ART initiation was an absolute CD4 count of <200 per mm3, and this was changed to <350 per mm3 in 2010. The WHO in 2013 recommended a CD4 count of <500 per mm3 as an appropriate starting point (21). If the ART-naïve 2002–2005 maternal cohort in South Africa is representative, these three CD4 criteria would result in treatment of 19%, 42%, and 72% of the HIV-infected population, respectively. A second factor is the coverage in terms of the percentage of those meeting the CD4 criteria that actually receive ART and maintain viral suppression on ART. A third factor is the proportion of transmissions that occur during acute infection in the donor—that is, before ART can be initiated. This proportion is likely to vary dramatically from one setting to another and estimates range from 1% to 41% (40–43). The consensus figure of ∼25% (40–43) of transmissions occurring in acute or early infection would suggest that the majority of transmissions occur in chronic or late infection, at which times initiation of ART would have an impact.
It is important in these studies of HIV-replicative capacity to draw attention to the caveats that apply with respect to the assay used here. The assay does not take into account the effects of genes other than gag or protease on VRC. Thus, the true VRC of HIV in each study subject is only partially represented by the assay, and also it is possible that mutations outside of Gag-Protease may compensate for mutants within Gag-Protease and vice versa. However, the assay has been adopted and validated by several groups, including the demonstration of a correlation between the replication capacities of Gag-recombinant viruses and complete HIV isolates obtained from the same patient samples (31); and, consistently in these studies, the measured VRC correlates with viral set point and, inversely, with CD4 count (14, 17–20), suggesting that the gag-pro sequence has an important influence on HIV disease outcome, independent of the effects of other genes on VRC. In addition, the CTLs that have been most clearly related to immune control and that dominate the HIV-specific response in subjects expressing the protective HLA alleles HLA-B*57/58:01/27/81:01 are within Gag (9, 44), and the escape mutants within these Gag epitopes have been proposed to play a critical role in immune control via a reduction on VRC (9–12). Thus, the gag-pro region of the viral genome is of particular relevance in evaluating the impact of these escape mutations on VRC that is the subject of this current study.
In conclusion, these data suggest that viral adaptation to protective HLA alleles such as HLA-B*57 and HLA-B*58:01, together with the increasing use of ART, are both forces driving the virulence of transmitted viruses down. If this proves to be the case, this process would substantially accelerate the success of current prevention and treatment programs that are designed ultimately to bring about population-level eradication of HIV.
Methods
Study Cohorts.
We studied antenatal women with chronic, ART-naïve HIV-1 infection recruited from two cohorts as follows: (i) Durban, South Africa (n = 328), enrolled between 2002 and 2005; and (ii) Gaborone, Botswana (n = 514), enrolled between 2007–2008, from the Mma Bana Study as previously described (10, 44–46). A third maternal cohort was enrolled from the same site in Durban, South Africa (n = 264) in 2012–2013, comprising 38 ART-naïve antenatal mothers and 226 postnatal mothers attending a baby immunization clinic [enrolled a median of 186 d postdelivery, interquartile range (IQR) of 71 to 375 d]. These mothers had received AZT from booking to delivery and then single dose of nevirapine during labor according to the South African national guidelines, and no ART postdelivery. Analyses were also undertaken using data from an extended Durban cohort (n = 1,218) that included subjects from outpatient clinics enrolled up to 2007 as previously described (25). Viral load was measured from plasma using the identical Roche Amplicor Version 1.5 assay in both cohorts. Samples from study subjects were HLA-A–, -B–, and -C–sequenced based typed in the Clinical Laboratory Improvement Amendments/American Society of Histocompatibility and Immunogenetics-accredited laboratory of William Hildebrand (University of Oklahoma Health Sciences Center, Oklahoma City) using a locus-specific PCR amplification strategy and a heterozygous DNA-sequencing methodology for exons 2 and 3 of the class I PCR amplicon. Relevant ambiguities (47) were resolved by homozygous sequencing. DNA sequence analysis and HLA allele assignment were performed with the software Assign-SBT Version 3.5.1 (Conexio Genomics). Ethics approval was given by the Office of Human Research Administration, Harvard School of Public Health and the Health Research Development Committee, Botswana Ministry of Health, University of KwaZulu-Natal Review Board, and Oxford Research Ethics Committee. Written informed consent was obtained from all individuals.
Amplification and Sequencing of Proviral DNA.
Gag, Pol, and Nef sequences were generated from genomic DNA extracted from peripheral blood mononuclear cells, amplified by nested PCR using previously published primers to obtain population sequences, as previously described (48). Sequencing was undertaken using Big Dye Ready Reaction Terminator Mix Version 3 (Applied Biosystems). Sequences were analyzed using Sequencher Version 4.8 (Gene Codes Corporation).
Statistics: Identification of HLA-Associated Viral Polymorphisms and Codon Covariation.
HLA-associated viral polymorphisms and viral amino acid covariations were identified from proviral DNA using a previously described method that corrects for phylogeny, HLA-linkage disequilibrium, and codon covariation (29, 49). Briefly, a maximum likelihood phylogenetic tree was constructed for each gene and for every HLA allele and amino acid, two generative models of the observed presence or absence of the amino acid in each sequence were created—one representing the null hypothesis that the observations are generated by the phylogenetic tree alone and the other representing the alternative hypothesis that additional escape or reversion takes place due to HLA pressure, as estimated using a modified logistic regression model (25).
The likelihood of the observations was then maximized over the parameters of both models with an expectation maximization algorithm, and a P value was computed with a likelihood ratio test based on those likelihoods. To increase power, the tests were made binary such that the presence or absence of a given HLA allele was correlated with the presence or absence of a given amino acid. In addition, HLA polymorphism pairs were analyzed only when both the amino acid and the HLA were independently observed in at least 10 individuals. For every amino acid at each position, the HLA allele with the strongest association was added to the model and the analysis was repeated to identify the next most significant HLA, conditioned on those previously added to the model. This procedure was iterated until no HLA allele yielded an association with a P value of less than 0.05. A q value statistic, estimating the proportion of false positives among the associations identified at a given P value threshold, was estimated using the method of Storey and Tibshirani (50). Statistical significance was reported using q values of ≤ 0.05 (5% false-discovery rate) for each P value threshold.
Associations were learned using a previously defined, multicohort set of 2,066 individuals from various regions in Southern Africa, including South Africa (n = 1,254), Botswana (n = 326), Zambia (n = 326), and n = 66 individuals of Southern African descent who enrolled in the Thames Valley clinic in the United Kingdom (26). High-resolution HLA typing was available for each individual, as were sequences for Gag (n = 1,897; n = 1,135 for p15), Pol (n = 1365; n = 1,315, 1,364, and 698 for Pr, RT, and Int, respectively), and Nef (n = 1,336). Gag, Pol, and Nef alignments were constructed using HIVAlign (51), then hand edited. Maximum likelihood trees were constructed separately for Gag, Pol, and Nef using PHYML (52). The extent to which an HIV sequence was adapted to a given HLA allele was measured as the proportion of sites associated with that allele that were adapted, defined as the presence of a polymorphism (either by itself or as a called mixture) that is positively correlated with the HLA or the presence of a polymorphism that is different from the polymorphism that is negatively associated with the HLA (if such a polymorphism exists). Sites containing indel characters are treated as missing data. In Fig. 2, for example, each point represents the HLA repertoire of each study subject (i.e., the six HLA class I molecules expressed) and the average, over all sequences in the cohort, of the number of sites in Gag, Pol, and Nef that are adapted to those HLA class I molecules, divided by the total number of sites associated with selection pressure mediated by those HLA class I molecules. This calculation therefore represents the average degree of adaptation of all sequences in that cohort to that individual HLA repertoire.
Viral Replication Assay.
Patient gag-protease isolated from plasma RNA was inserted into a NL4-3 gag-protease–deleted plasmid to generate recombinant viruses, as previously described (17, 18).
Titration of virus stocks and replication assays were performed as described (12, 13), using a multiplicity of infection of 0.003. The mean slope of exponential growth from days 2 to 7 was calculated in Excel (Microsoft) using the LOGEST function and converted to natural logs. This was then divided by the slope of growth of the WT NL4-3 control included in each replication assay, to generate a normalized replication capacity for comparison between different assays. Replication assays were performed in duplicate and the results averaged.
The samples used for the VRC assays were selected at random from subjects in each cohort whose absolute CD4 counts were matched in the range of 300 to 500 cells per mm3 and retrospectively compared and confirmed to be not significantly different (385 per mm3 and mean of 389 per mm3 in Gaborone and Durban, respectively). The VRC had previously been evaluated in >400 HIV-infected study subjects in Durban (17). The subset of viruses selected at random from the Durban cohort were established as representative of the entire sample group of 119 Durban subjects whose absolute CD4 counts were in the range of 300 to 500 per mm, in having the same mean VRC: The mean VRC as determined in Durban in this subset of 16 viruses was 0.62 (SD 0.09), compared with a mean VRC of 0.62 (SD 0.1) for the whole group. Viral titres and replication capacities for these 16 viruses were determined at the University of Oxford as described above and normalized to the same viral stock of the NL4-3 control used for the 63 Gaborone samples. The VRC determined at the University of Oxford for the 16 Durban Gag-protease recombinant viruses was strongly correlated with the VRCs determined for these same viruses in Durban in the previously published study (12) (r2 = 0.94 P < 0.0001; Pearson’s correlation).
Viral Replication Assay Validation.
Gag-Protease recombinant viruses generated by this method have been shown to be representative of original plasma quasispecies (17, 18). In this study, Gag p17+p24 was sequenced from 27 randomly selected recombinant viruses from the Gaborone study cohort and compared with the original plasma HIV RNA sequence. Viral RNA was isolated from recombinant virus supernatants using a QIAamp Viral RNA mini kit (Qiagen). RT-PCR was performed and the product sequenced along with the gag-protease PCR product from plasma viral RNA, as previously described (17, 18), to obtain gag p17 and p24 population sequences from both plasma viral RNA- and recombinant virus RNA-derived cDNA. Sequences were analyzed and edited in Sequencher 4.8 and aligned to the HXB2 reference sequence (GenBank accession no. K03455) in Se-Al Version 2.0a11. Plasma viral RNA-derived cDNA sequences and recombinant virus RNA-derived cDNA sequences from the Gaborone subjects were directly compared with ascertain similarity. Twenty-seven pairs of sequences were randomly chosen and percentage pairwise distances calculated using HyPhy Version 1.0β (53). The median number of nucleotide differences between the compared sequences was 2.9 (IQR 0.9–5.2), with mixed bases included as differences, resulting in an average nucleotide similarity of 0.97 between pairs, in support of previous findings (12, 13).
Mathematical Model.
The model shown diagrammatically in Fig. S3 is formally described by the following set of coupled, first order, ordinary differential equations:
These equations were solved numerically using the deSolve package in R and the lsoda method (54). The initial values given were
and time steps of size 0.01 y were used. For the first 35 y ν was set to 0 and then this was increased to 0.4 from t = 35 onward, so that from this point 40% of new infections end up being treated when they pass the treatment threshold. All parameters and their given values are detailed in Table S3.
Supplementary Material
Acknowledgments
This work is funded by grants from the National Institutes of Health [R01AI46995 (to P.J.R.G.)], the Wellcome Trust (to P.J.R.G.), and the Medical Research Council UK. Z.B. is supported by a New Investigator Award (Canadian Institutes of Health) and the Michael Smith Foundation for Health Research. M.B. holds a Canada Research Chair, Tier 2, in Viral Pathogenesis and Immunity.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KP208181–KP208313).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413339111/-/DCSupplemental.
References
- 1.Weiss RA. The Leeuwenhoek Lecture 2001. Animal origins of human infectious disease. Philos Trans R Soc Lond B Biol Sci. 2001;356(1410):957–977. doi: 10.1098/rstb.2001.0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control. Oxford University Press; Oxford: 1992. [Google Scholar]
- 3.Quinn TC, et al. Rakai Project Study Group Viral load and heterosexual transmission of human immunodeficiency virus type 1. N Engl J Med. 2000;342(13):921–929. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 4.Mellors JW, et al. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272(5265):1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 5.Fraser C, Hollingsworth TD, Chapman R, de Wolf F, Hanage WP. Variation in HIV-1 set-point viral load: Epidemiological analysis and an evolutionary hypothesis. Proc Natl Acad Sci USA. 2007;104(44):17441–17446. doi: 10.1073/pnas.0708559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herbeck JT, et al. Is the virulence of HIV changing? A meta-analysis of trends in prognostic markers of HIV disease progression and transmission. AIDS. 2012;26(2):193–205. doi: 10.1097/QAD.0b013e32834db418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goepfert PA, et al. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J Exp Med. 2008;205(5):1009–1017. doi: 10.1084/jem.20072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prince JL, et al. Role of transmitted Gag CTL polymorphisms in defining replicative capacity and early HIV-1 pathogenesis. PLoS Pathog. 2012;8(11):e1003041. doi: 10.1371/journal.ppat.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goulder PJR, Walker BD. HIV and HLA class I: An evolving relationship. Immunity. 2012;37(3):426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Picado J, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80(7):3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford H, et al. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J Exp Med. 2009;206(4):909–921. doi: 10.1084/jem.20081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boutwell CL, et al. Frequent and variable cytotoxic-T-lymphocyte escape-associated fitness costs in the human immunodeficiency virus type 1 subtype B Gag proteins. J Virol. 2013;87(7):3952–3965. doi: 10.1128/JVI.03233-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawashima Y, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458(7238):641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nomura S, et al. Significant reductions in Gag-protease-mediated HIV-1 replication capacity during the course of the epidemic in Japan. J Virol. 2013;87(3):1465–1476. doi: 10.1128/JVI.02122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollingsworth TD, et al. HIV-1 transmitting couples have similar viral load set-points in Rakai, Uganda. PLoS Pathog. 2010;6(5):e1000876. doi: 10.1371/journal.ppat.1000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hecht FM, et al. HIV RNA level in early infection is predicted by viral load in the transmission source. AIDS. 2010;24(7):941–945. doi: 10.1097/QAD.0b013e328337b12e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright JK, et al. Gag-protease-mediated replication capacity in HIV-1 subtype C chronic infection: Associations with HLA type and clinical parameters. J Virol. 2010;84(20):10820–10831. doi: 10.1128/JVI.01084-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miura T, et al. HLA-associated alterations in replication capacity of chimeric NL4-3 viruses carrying gag-protease from elite controllers of human immunodeficiency virus type 1. J Virol. 2009;83(1):140–149. doi: 10.1128/JVI.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brockman MA, et al. Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity that may be largely compensated for in chronic infection. J Virol. 2010;84(22):11937–11949. doi: 10.1128/JVI.01086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang KH, et al. Bloemfontein-Oxford Collaborative Group Progression to AIDS in South Africa is associated with both reverting and compensatory viral mutations. PLoS ONE. 2011;6(4):e19018. doi: 10.1371/journal.pone.0019018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. World Health Organization (2014) Consolidated ARV guidelines 2013. Available at www.who.int/hiv/pub/guidelines/arv2013/art/statartadolescents_rationale/en/index.htm. Accessed July 1, 2014.
- 22.Cohen MS, et al. HPTN 052 Study Team Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365(6):493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.WHO Report in partnership with UNICEF and UNAIDS (2013) Global update on HIV treatment 2013. Available at apps.who.int/iris/bitstream/10665/85326/1/9789241505734_eng.pdf. Accessed July 1, 2014.
- 24. AVERT (2014) HIV & AIDS in South Africa. Available at www.avert.org/hiv-aids-south-africa.htm. Accessed July 1, 2014.
- 25. The World Bank (2014) Data. Available at data.worldbank.org. Accessed July 1, 2014.
- 26.Carlson JM, et al. Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. J Virol. 2012;86(9):5230–5243. doi: 10.1128/JVI.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiepiela P, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432(7018):769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 28.Leslie A, et al. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J Virol. 2010;84(19):9879–9888. doi: 10.1128/JVI.00320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goulder PJR, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8(8):619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews PC, et al. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J Virol. 2008;82(17):8548–8559. doi: 10.1128/JVI.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prado JG, et al. Replicative capacity of human immunodeficiency virus type 1 transmitted from mother to child is associated with pediatric disease progression rate. J Virol. 2010;84(1):492–502. doi: 10.1128/JVI.01743-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brander C, Walker BD. Gradual adaptation of HIV to human host populations: Good or bad news? Nat Med. 2003;9(11):1359–1362. doi: 10.1038/nm941. [DOI] [PubMed] [Google Scholar]
- 33.Schneidewind A, et al. Transmission and long-term stability of compensated CD8 escape mutations. J Virol. 2009;83(8):3993–3997. doi: 10.1128/JVI.01108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crawford H, et al. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J Virol. 2007;81(15):8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fryer HR, et al. SPARTAC Trial Investigators Modelling the evolution and spread of HIV immune escape mutants. PLoS Pathog. 2010;6(11):e1001196. doi: 10.1371/journal.ppat.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schellens IM, et al. Loss of HIV-1-derived cytotoxic T lymphocyte epitopes restricted by protective HLA-B alleles during the HIV-1 epidemic. AIDS. 2011;25(14):1691–1700. doi: 10.1097/QAD.0b013e32834981b3. [DOI] [PubMed] [Google Scholar]
- 37.Cotton LA, et al. Genotypic and functional impact of HIV-1 adaptation to its host population during the North American epidemic. PLoS Genet. 2014;10(4):e1004295. doi: 10.1371/journal.pgen.1004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. UNAIDS (2013) Global Report: UNAIDS report on the global AIDS epidemic 2013. Available at www.unaids.org/en/media/unaids/contentassets/documents/epidemiology/2013/gr2013/UNAIDS_Global_Report_2013_en.pdf. Accessed July 1, 2014.
- 39.Johnson LF. Access to antiretroviral treatment in South Africa, 2004-2011. South Afr J HIV Med. 2012;13(1):22–26. doi: 10.4102/sajhivmed.v18i1.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wawer MJ, et al. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J Infect Dis. 2005;191(9):1403–1409. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- 41.Goodreau SM, et al. Concurrent partnerships, acute infection and HIV epidemic dynamics among young adults in Zimbabwe. AIDS Behav. 2012;16(2):312–322. doi: 10.1007/s10461-010-9858-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powers KA, et al. Role of acute infection in HIV transmission - Authors’ reply. Lancet. 2011;378:1914. doi: 10.1016/S0140-6736(11)61832-1. [DOI] [PubMed] [Google Scholar]
- 43.Gray RH, et al. Rakai Project Team Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet. 2001;357(9263):1149–1153. doi: 10.1016/S0140-6736(00)04331-2. [DOI] [PubMed] [Google Scholar]
- 44.Kiepiela P, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13(1):46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 45.Matthews PC, et al. HLA-A*7401-mediated control of HIV viremia is independent of its linkage disequilibrium with HLA-B*5703. J Immunol. 2011;186(10):5675–5686. doi: 10.4049/jimmunol.1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shapiro RL, et al. Antiretroviral regimens in pregnancy and breast-feeding in Botswana. N Engl J Med. 2010;362(24):2282–2294. doi: 10.1056/NEJMoa0907736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cano P, et al. Common and well-documented HLA alleles: Report of the Ad-Hoc committee of the american society for histocompatiblity and immunogenetics. Hum Immunol. 2007;68(5):392–417. doi: 10.1016/j.humimm.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 48.Leslie A, et al. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J Exp Med. 2005;201(6):891–902. doi: 10.1084/jem.20041455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlson JM, et al. Phylogenetic dependency networks: Inferring patterns of CTL escape and codon covariation in HIV-1 Gag. PLOS Comput Biol. 2008;4(11):e1000225. doi: 10.1371/journal.pcbi.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Los Alamos National Laboratory (2014) HIV databases. Available at www.hiv.lanl.gov. Accessed July 1, 2014.
- 52.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 53.Pond SL, Frost SD, Muse SV. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 54.Soetaert K, Petzoldt T, Setzer R. 2010 Solving differential equations in R: Package deSolve. J Stat Softw 33(9):1–25. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.