Abstract
Inflammatory breast cancer (IBC) is the most aggressive type of advanced breast cancer characterized by rapid proliferation, early metastatic development and poor prognosis. Since there are few preclinical models of IBC, there is a general lack of understanding of the complexity of the disease. Recently, we have developed a new model of IBC derived from the pleural effusion of a woman with metastatic secondary IBC. FC-IBC02 cells are triple negative and form clusters (mammospheres) in suspension that are strongly positive for E-cadherin, β-catenin and TSPAN24, all adhesion molecules that play an important role in cell migration and invasion. FC-IBC02 cells expressed stem cell markers and some, but not all of the characteristics of cells undergoing epithelial mesenchymal transition (EMT). Breast tumor FC-IBC02 xenografts developed quickly in SCID mice with the presence of tumor emboli and the development of lymph node and lung metastases. Remarkably, FC-IBC02 cells were able to produce brain metastasis in mice on intracardiac or intraperitoneal injections. Genomic studies of FC-IBC02 and other IBC cell lines showed that IBC cells had important amplification of 8q24 where MYC, ATAD2 and the focal adhesion kinase FAK1 are located. MYC and ATAD2 showed between 2.5 and 7 copies in IBC cells. FAK1, which plays important roles in anoikis resistance and tumor metastasis, showed 6–4 copies in IBC cells. Also, CD44 was amplified in triple-negative IBC cells (10–3 copies). Additionally, FC-IBC02 showed amplification of ALK and NOTCH3. These results indicate that MYC, ATAD2, CD44, NOTCH3, ALK and/or FAK1 may be used as potential targeted therapies against IBC.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-013-2600-4) contains supplementary material, which is available to authorized users.
Keywords: CD44, NOTCH3, MYC, ATAD2, ALK, FAK1/PTK2
Introduction
Inflammatory breast cancer (IBC) is a very aggressive type of advanced breast cancer with a poor prognosis. IBC patients often develop metastasis in brain, bones and soft tissue with variable frequency, being the most aggressive features described in triple-negative (TN) IBC. Although IBC accounts for 1–6 % of all breast cancer cases in the USA and up to 20 % of all breast cancers globally [1], its incidence is dramatically increasing [2, 3]. Due to its propensity to rapidly metastasize, IBC is responsible for a disproportionate number of breast cancer-related deaths [1]. Approximately, 30 % of IBC patients have distant metastases at the time of diagnosis, in contrast to 5 % of patients with non-IBC breast cancers. IBC occurs commonly in patients under the age of 50 years and since it does not present as a lump is often misdiagnosed as an infection [4]. The clinical symptoms of IBC are very distinct from other types of breast cancer and involve the rapid onset of changes in the skin overlying the breast, including edema, redness and swelling, exhibiting a wrinkled and orange-peel appearance of the skin defined as peau d’orange [5]. This peculiar presentation that mimics an inflammation is associated with the invasion of aggregates of tumor cells, defined as tumor emboli into the dermal lymphatics, causing an obstruction of the lymph channels [6]. IBC tumor emboli are non-adherent cell clusters that rapidly spread by a continuous passive dissemination [7], thus favoring both distal metastasis and local recurrence. Although IBC, like non-IBC breast cancers, is a heterogeneous disease and can occur as any of the five molecular subtypes, they are most commonly either ErbB2 overexpressing or triple negative [8].
Few models are currently available to evaluate the peculiar biology of IBC and improve our understanding of the factors associated with early activation of the metastatic process in this disease. The majority of the IBC studies have been performed using the cell lines SUM149 and SUM190 [9]. SUM149 cells are triple negative and SUM190 cells are ER/PR negative and ErbB2 positive [10–13]. Other less studied IBC cell lines are MDA-IBC3, KPL4 and WIBC-9—all ER/PR-negative, ErbB2-positive—and the triple-negative Mary-X xenografts [14–18]. In this work, we report the isolation and characterization of a new human triple-negative IBC model, FC-IBC02. Genomic and expression studies were performed in FC-IBC02 and other IBC cell lines to understand the metastatic process of IBC and determine putative targets for therapy.
Materials and methods
IBC cells and FC-IBC02 breast tumor xenografts
Tumor cells from pleural effusion of IBC patients were grown under non-adherent conditions [19] for a minimum of 14 days and then expanded under adherent conditions. As FC-IBC02, FC-IBC01 (triple negative) [20] and KMO-015 (ER negative, PR negative, ErbB2 positive) were also established in our laboratory. All the cell lines used in these studies were certified by ATCC using (short tandem repeat) profiling.
The tumorigenesis and metastatic capacities of the FC-IBC02 cells were tested in 35- to 40-day-old female SCID mice. After 14 passages, FC-IBC02 cells (105 or 106 cells) were suspended in 150 μl PBS, mixed with 150 μl Matrigel (BD Biosciences, Bedford, MA) and injected into the fourth left inguinal mammary fat pad of SCID mice. Breast tumors and other organs (lungs, heart, liver, spleen, brain, ovaries, kidneys and lymph nodes) were removed, fixed in 10 % neutral buffered formalin and paraffin embedded for histological examination.
Chromosomal and gene expression analyses
The Affymetrix CytoScan™ HD arrays were used to study copy number and loss of heterozygosity (LOH). The intensities of probe hybridization were analyzed by Affymetrix software’s Command Console, and the copy number and genotyping analyses were performed using Affymetrix Chromosome Analysis Suite software with the default settings. Gene expression studies were performed using Affymetrix U133 Plus 2.0 human oligonucleotide microarrays. We evaluated array quality using GCOS, dChip software [21] and R-Bioconductor “affy” package [22]. We calculated the intensities of probe sets using robust multi-chip average algorithm [23]. To identify differentially expressed genes, we conducted two-group comparison using significance analysis of microarrays [24]. Only genes with small false discovery rate (FDR ≤ 0.001) and fold change >fourfold were considered for the analysis.
FAK1/PTK2 protein expression studies
To quantify activated (phosphorylated) and total FAK1 (also known as PTK2 or FAK), the FAK FACE (Fast Activated Cell-based ELISA; Active Motif) assays were used. Cells were grown on 96 well plates and incubated with a primary antibody that recognized either phosphorylated FAK1 (Y397) or total FAK1. Subsequently, cells were incubated with a secondary HRP-conjugated antibody and developing solution and the reaction was quantified by colorimetric readout (OD450). The relative number of cells in each well was then determined through use of crystal violet.
Results
FC-IBC02 cell line and breast tumor xenografts
FC-IBC02 was established from the pleural effusion from a 49-year-old Caucasian woman who had developed secondary IBC. The tumor cells from the pleural effusion were selected by growing them a minimum of 14 days under low adherence conditions in serum-free media containing epidermal growth factor (EGF) and fibroblast growth factor, which enrich formation of mammospheres [19]. When cells from the pleural effusion were plated directly into regular culture flasks in DMEM media containing fetal bovine serum, we were unable to isolate tumor cells—indicating that initially culturing in suspension was necessary for their isolation. Following this selection step, the tumor cells were maintained in culture as mammospheres (Fig. 1a) or expanded as adherent cells (Fig. 1d). The clusters of tumor cells growing as mammospheres were approximately ~50 μm in diameter (Fig. 1b, c).
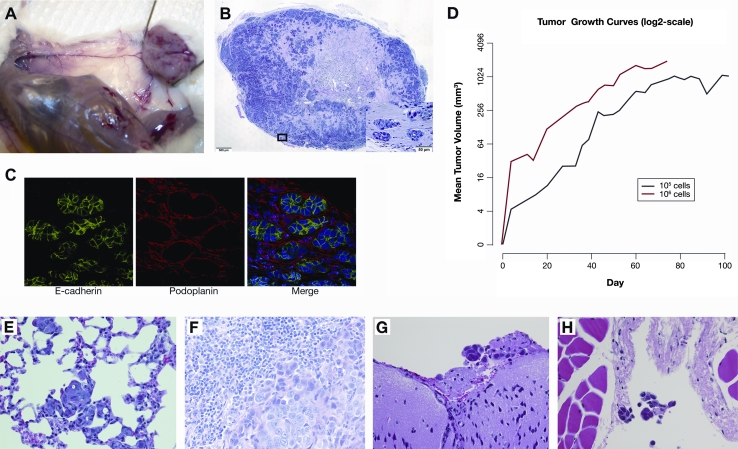
Fig. 1.
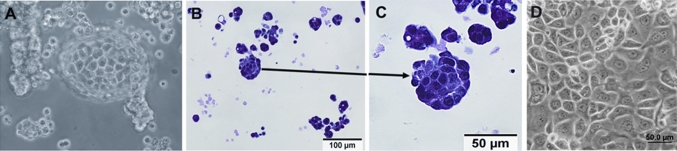
FC-IBC02 cells isolated from a patient with secondary inflammatory breast cancer (IBC). a Morphology of mammospheres after 10 days in culture. b, c Hematoxylin and eosin staining on 4 μm-thick paraffin sections of FC-IBC02 mammospheres (scale bar 100 and 50 μm). d FC-IBC02 growing as adherent cells (passage 30)
For in vivo studies, FC-IBC02 adherent cells were injected into the mammary fat pad of SCID mice, resulting in 100 % tumor growth (Fig. 2a). The xenografts were poorly differentiated adenocarcinomas with frequent massive central tumor necrosis (Fig. 2b). Hematoxylin/eosin staining of paraffin sections of the breast tumor xenografts demonstrated the presence of tumor emboli (Fig. 2b); these tumor emboli expressed E-cadherin and were encircled by podoplanin-positive lymphovascular endothelium indicating their presence in the lymphovascular system (Fig. 2c). FC-IBC02 breast tumor xenografts grew rapidly and a statistically significant increase in tumor volume was observed with time (p value < 1e−06) (Fig. 2d). Breast tumors reached approximately 1 cm3 in diameter at 53 days when 106 cells were injected and at 78 days when 105 cells were injected (Fig. 2d). Although the growth rate of tumors in mice injected with 105 cells was below that in mice injected with 106 cells (p value = 0.017), there was no significant change in tumor volume with time (p value = 0.127) (Fig. 2d). Microscopic examination of the organs of mice bearing FC-IBC02 xenografts demonstrated that all mice developed spontaneous metastases within lungs and lymph nodes (Fig. 2e, f). No brain metastases were found in these animals; as breast tumor FC-IBC02 xenografts grew quickly and the mice had to be killed, there was probably not enough time for the development of spontaneous brain metastasis in the orthotopic xenograft models. Remarkably, injection of FC-IBC02 tumor cells via either the intraperitoneal or intracardiac routes resulted in the formation of brain metastases (Fig. 2g). Examination of tissues revealed the presence of tumor cell aggregates visible within both lymphatic and blood vessels in the orthotopic xenograft models (Fig. 2h).
Fig. 2.
Breast tumor xenograft and metastases in mice. a Breast tumor xenograft in SCID mouse injected with 106 FC-IBC02 cells. b Hematoxylin/eosin staining of breast tumor FC-IBC02 xenograft with tumor emboli. c FC-IBC02 tumor emboli express E-cadherin (green) and are encircled by podoplanin-positive (red) lymphovascular endothelium. d Growth curves of FC-IBC02 breast tumor xenografts after injection of 106 (line in red) or 105 cells (line in black). The growth curves represent the average growth of the breast tumors in five animals per group. Tumor volume measurements were obtained at baseline (Day 0) and at pre-determined time points (measured in days) for each mouse in each group (105 vs. 106 tumor cells injected). e Lung metastasis by orthotopic injection (40× magnification), f lymph node metastasis by orthotopic injection (40× magnification), g brain metastasis after intraperitoneal injection (40× magnification), h cluster of tumor cells within a blood vessel (40× magnification)
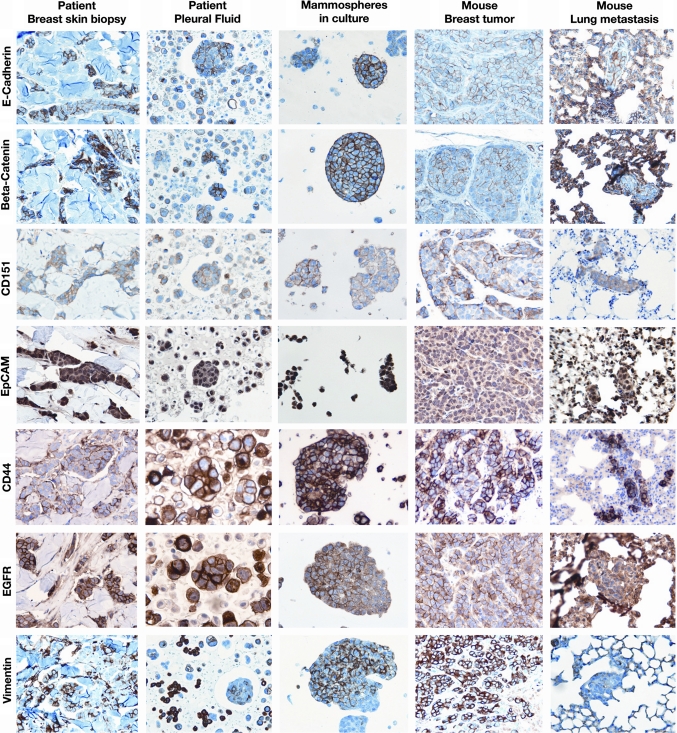
Multiple markers were studied by immunohistochemistry in the skin breast biopsy and original pleural effusion of the patient, from whom the FC-IBC02 cell line was established, and compared to the markers in FC-IBC02 mammospheres in culture and breast tumor xenografts and lung metastasis in SCID mice. The tumor cells present in the breast skin biopsy and original pleural fluid from the patient were ER negative, PR negative and ErbB2 negative (triple negative) (Suppl. Fig. 1). The FC-IBC02 mammospheres in culture, breast tumor xenografts and metastasis in lungs were also triple negative (Suppl. Fig. 1). Tumor cells present in the breast skin biopsy and pleural effusion from the IBC patient as well as FC-IBC02 mammospheres in culture showed strong expression of E-cadherin (CDH1) at the cell membrane (Fig. 3). Mammospheres in culture also showed strong expression of β-catenin (CTNNB1), CD151 (tetraspanin 24) and epithelial cell adhesion molecule (EpCAM) (Fig. 3). The breast tumor xenografts and lung metastases in SCID mice also expressed E-cadherin, β-catenin, CD151 and EpCAM (Fig. 3). Moreover, the FC-IBC02 cells showed strong expression of epidermal growth factor receptor (EGFR) and strong staining for the stem cell marker CD44 (Fig. 3). FC-IBC02 cells showed quite a heterogeneous positive vimentin staining, and this gene was not expressed by these cells in the mice lung metastasis (Fig. 3) probably due to hypermethylation of its promoter. FC-IBC02 cells did not express SMA, S100 or desmin, but showed strong expression of EMA/MUC1 and very weak staining for CK5/6 (data not shown). In the original pleural effusion of the patient, tumor cells were present in clusters or aggregates (Fig. 3 and Suppl Fig. 1).
Fig. 3.
Expression of E-cadherin and other adhesion molecules by FC-IBC02. The FC-IBC02 mammospheres express E-cadherin, β-catenin, CD151 (tetraspanin 24) and EpCAM. Also, these cells are positive for CD44, EGFR (epidermal growth factor receptor) and vimentin. Breast skin biopsy and pleural effusion of the IBC patient, mammospheres in culture, breast tumor xenografts and lungs metastasis in mice are shown for these markers, 20× magnification
Copy number and loss of heterozygosity (LOH) in FC-IBC02 and other IBC cells
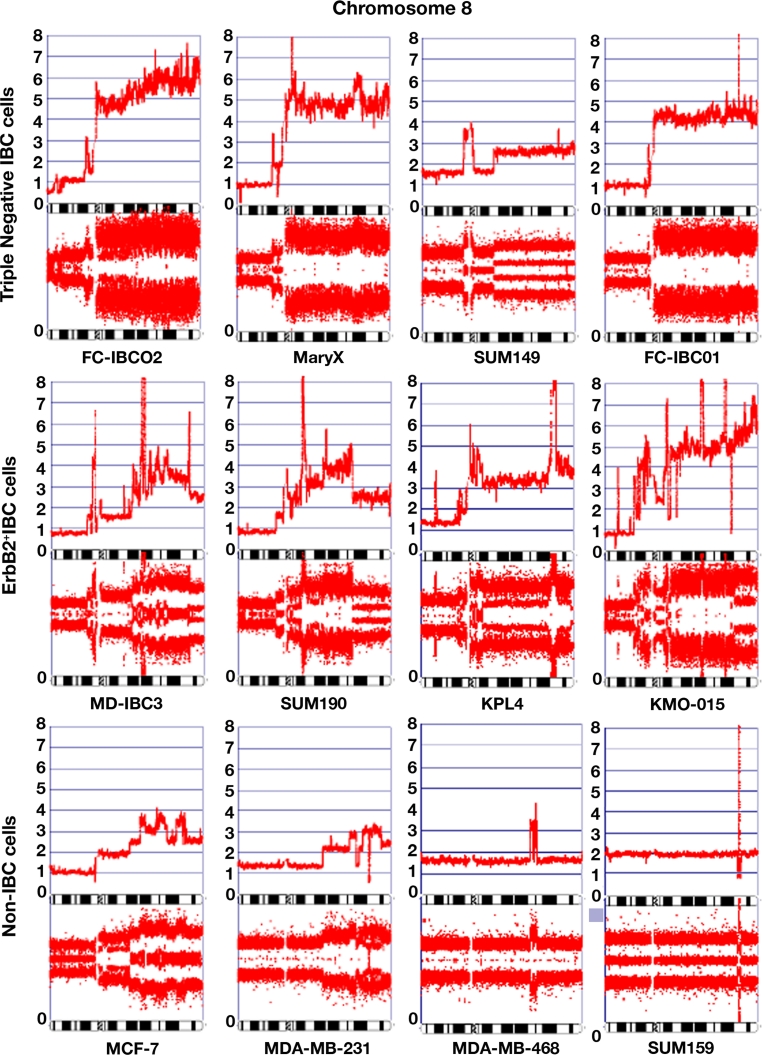
High-resolution arrays were used to evaluate copy number and LOH in FC-IBC02 cells and other triple-negative (Mary X, SUM149, FC-IBC01) and ErbB2-positive (MDA-IBC3, SUM190, KPL4, KMO-015) IBC cell lines. Also, non-IBC cells (MFC-7, MDA-MB231, MDA-MB468, SUM159) were included in these studies to compare the profiles between IBC and non-IBC cells. FC-IBC02 cells showed complicated genomic alterations in all chromosomes with the exception of chromosome 12 (Suppl. Fig. 2). Gains/amplifications were more common than deletions/losses; FC-IBC02 cells showed scattered genomic profile (deletions) on 11q chromosome arm known as chromothripsis; the copy number profile across this chromosome arm shows many positions at which copy number changes between one or two copies (Suppl. Fig. 2). Copy number and LOH studies were also performed for Mary X (Suppl. Fig. 3), SUM149 (Suppl. Fig. 4) and FC-IBC01 (Suppl. Fig. 5). Most of the IBC cells showed multiple amplifications of 8q having 3–10 copies of this chromosomal arm (Fig. 4). The 8q is the location of the oncogene MYC, MTDH1, PVT1, FAK1 (also known as PTK2K) and ATAD2 between other genes. In contrast, the non-IBC cell lines studied showed only 1–4 copies of 8q (Fig. 4). IBC cells showed 2.5–7 copies of the oncogene MYC and, although MYC was also amplified in non-IBC cells, the number of copies was lower in non-IBC (1–3 copies) with the exception of SUM159 cells that have a focal MYC amplification (Table 1). PTK2/FAK1 also showed more copies in IBC (2.5–7 copies) than in non-IBC cells (1–2.5 copies) (Table 1). Metadherin (MTDH1) showed 2.5–6 copies in IBC and 1–3 copies in non-IBC cell lines; ATAD2 showed 2.5–6 copies in IBC and 1–3.5 copies in non-IBC (Table 1). CD44 located in 11p13 showed eight copies in FC-IBC02, ten copies in the triple-negative IBC cells FC-IBC01 and three copies in SUM149; CD44 showed 1–2 copies in the ErbB2 + IBC and non-IBC, except MDA-MB468 (4.5 copies). Additionally, FC-IBC02 cells showed 3.5–4 copies of anaplastic lymphoma kinase (ALK) gene on chromosome 2p23.2 (Table 1).
Fig. 4.
Chromosome 8 in IBC and non-IBC cells. Copy number and loss of heterozygosity (LOH) were studied using Affymetrix CytoScan™ HD arrays. The chromosome 8 in different IBC and non-IBC cells is shown
Table 1.
Copy number of some amplified genes in IBC and non-IBC cells
| Location | Gene | Gene description | Copy number | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IBC | Non-IBC | |||||||||||||
| Triple negative | ER(−) PR(−) ErbB2+ | ER (+) | Triple negative | |||||||||||
| FC-IBC02 | Mary X | SUM 149 | FC-IBC01 | MD-IBC3 | SUM 190 | KPL4 | KMO-015 | MCF-7 | MDA-MB-231 | MDA-MB-468 | SUM-159 | |||
| 2p23.2 | ALK | Anaplastic lymphoma kinase, receptor tyrosine kinase (oncogene) | 3.5–4 | 3 | 1.5 | UPD | 3 | 1 | 3 | 3 hmz | 2 | mos UPD | 3 | 2 |
| 4q11 | c-KIT (CD117) | Receptor tyrosine kinase (oncogene) | UPD | UPD | 1 | 1 | UPD | 6 hmz | 1.8 | 3 | 1 | mos UPD | 1 | 2 |
| 7p12 | EGFR | Epidermal Growth Factor Receptor | 2.5–3 | 2.5 | 2.5–3 | mos UPD | 1 | 4 | 3 | 3 | UPD | mos UPD | 9 | UPD |
| 8p11.22 | FGFR1 | Fibroblast Growth Factor Receptor 1 | 3 | 2 | <2 | 1 | 1 | 1.5 | 2 | 2.5 hmz | 1 | 1 | 1 | 2 |
| 8q24.21 | MYC | MYC (oncogene) | 7 hmz | 4–5 hmz | 2.5 | 4 | 2.5 | 2.5 | 4 | 7 | 3 | 2.5 hmz | 1 | <10 |
| 8q24.3 | PTK2/FAK1 | Focal adhesion kinase 1 | 6 hmz | 6 hmz | 2.5 | 4 | 2.5 | 2.5 | 4 hmz | 7 | 2.5 | 2.5 hmz | 1 | 2 |
| 8q24.13 | ATAD2 | ATPase family, cofactor for MYC | 6 | 4.5 | 2.5 | 4.5 | 3.5 | 2.5 | 4 hmz | 5 hmz | 3.5 | 3 | 1 | 2 |
| 8q22.1 | MTDH1 | Metadherin (oncogene) | 6 | 4.5 | 2.5 | 4.5 hmz | 3.5 | 4 | 3.5 | 4.5 hmz | 3 | UPD | 1 | 2 |
| 8q24.21 | PVT1 | PVT1 (oncogene) | 7 | 5 | 2.5 | 5 | 3 | 2.5 | 10 hmz | 6 | 3.5 | 3 | 1 | 10 |
| 9q34.3 | Notch l | Notch 1 | 2.5 | 2 | 2 | 2.5 | 3 | 2 | 6 | 1 | 2 | 3 | 2 | 2 |
| 19p13 | Notch 3 | Notch 3 | 5–6 hmz | UPD | 2.5 | 2 | 2.5 hmz | 2 | 2 | 2 | 2 | 2 | 3 | 2 |
| 11p13 | CD44 | CD44 | 8 | UPD | 3 | 10 hmz | 1 | 1 | 2 | 1 | 1 | 2 | 4.5 | 2 |
| 17p | TP53 | P53 (tumor suppressor) | 1 | UPD | 2.5 hmz | 1 | 1 | 1 | UPD | 1 | 1 | UPD | 1 | 1 |
Hmz homozygous, UPD uniparental disomy (two copies with LOH), mos mosaic
Expression studies of IBC cells
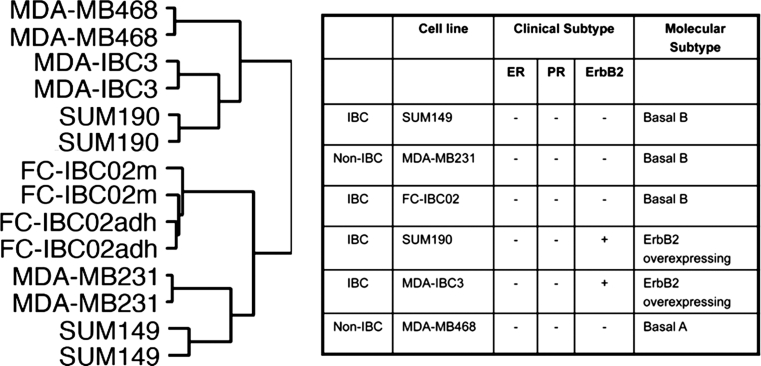
Gene expression studies were performed in IBC triple-negative cells (FC-IBC02, SUM149) and compared to IBC ErbB2-positive (MDA-IBC3, SUM190) and non-IBC triple-negative cells (MDA-MB231 and MDA-MB468). The microarray data have been deposited into the NCBI’s gene expression omnibus (GEO) datasets (GSE40464). Hierarchical clustering of cell lines based on global gene expression showed two cell clusters (Fig. 5) in which FC-IBC02 (mammospheres and adherent cells) clustered together with MDA-MB231 and SUM149, both from the basal B subtype. The second cluster included the triple-negative cell line MDA-MB468 from basal A subtype and the ErbB2-positive IBC cell lines MDA-IBC3 and SUM190 (Fig. 5). The gene expression profiles of FC-IBC02 mammospheres and adherent cells were very similar and only few genes showed significant differences between them.
Fig. 5.
Clustering of IBC and non-IBC cell lines by gene expression profile. A total of 6,575 probe sets passing the criterion of SD (standard deviation) >0.9 and <2 were used in unsupervised hierarchical clustering of samples. The molecular subtypes of the cells used in these studies are described on the right, also ER (estrogen receptor alpha), PR (progesterone receptor) and ErbB2 status of the cell lines are described. FC-IBC02 mammospheres (FC-IBC02 m) and adherent cells (FC-IBC02adh) showed very similar profiles. The triple-negative cell lines FC-IBC02 and SUM149 clustered together with the triple-negative non-IBC MDA-MB231
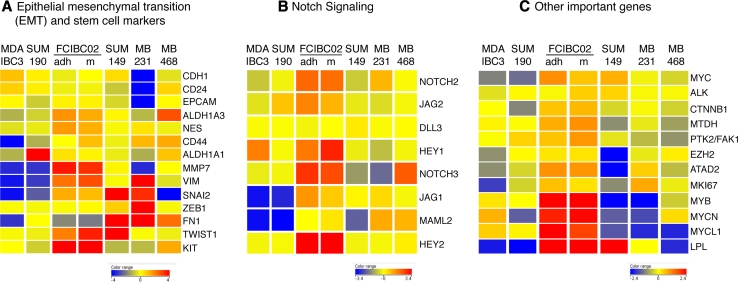
By flow cytometry analyses, FC-IBC02 showed a subpopulation of cells with stem cell characteristics CD44+/CD24low/CD34+ and the number of stem cells was higher in cells growing as mammospheres (15.6 %) than adherent (9.66 %) (Suppl. Figs. 6A and 6B); from these subpopulations of stem cells, higher number of CD133+ cells were seen in mammospheres (20.5 %) than adherent cells (9.47 %) (Suppl. Figs. 6C, 6D). FC-IBC02 also expressed the stem cell markers nestin (NES), ALDH1A3 and high levels of KIT (Fig. 6a). FC-IBC02 showed some characteristics of cells undergoing epithelial–mesenchymal transition (EMT) such as low expression of fibronectin (FN1) and high expression of vimentin (VIM), although these cells expressed genes associated with retention of an epithelial phenotype including E-cadherin (CDH1) and EpCAM (Fig. 3, 6a). All the IBC cell lines expressed low levels of ZEB1 when compared with ZEB1 expression in MDA-MB231; low levels of ZEB1, which is an E-cadherin transcriptional repressor, would explain the expression of E-cadherin in IBC cells (Fig. 6a). Interestingly, expressions of the transcription factors TWIST1 and SNAI2 which are inducers of EMT were high in FC-IBC02 cells (Fig. 6a).
Fig. 6.
Heat maps of the selected marker genes in IBC and non-IBC cells. a Epithelial–mesenchymal transition (EMT) and stem cell markers, b genes involved in Notch signaling pathway and c other selected marker genes are displayed. Red, yellow or blue colors represent expression levels above, at or below the mean level across samples
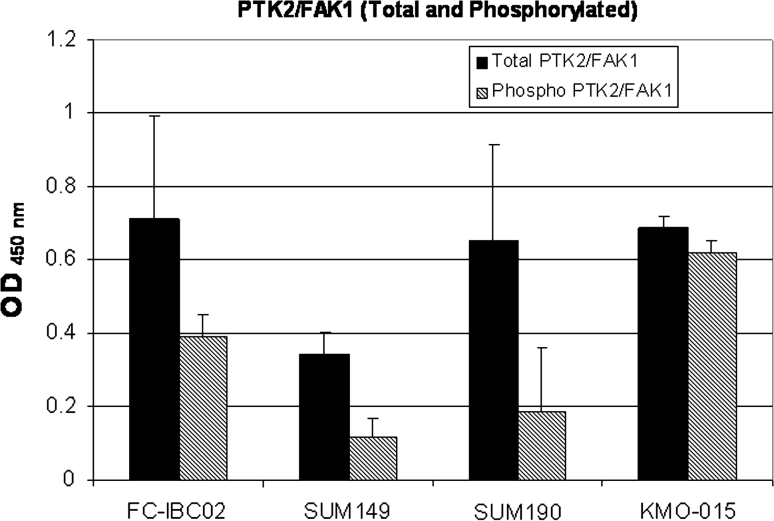
Several genes from the Notch signaling pathway such as NOTCH2, NOTCH3, HEY1, HEY2, JAG1 and JAG2 were upregulated in FC-IBC02 (mammospheres and adherent cells) (Fig. 6b). FC-IBC02 cells expressed high levels of the oncogenes MYCL1, MYB, MYCN and ATAD2 (Fig. 6c). MYC expression was higher in the triple-negative IBC cell lines (FC-IBC02 and SUM149) compared to the triple-negative non-IBC cells (MDA-MB231 and MDA-MB468) and the ErbB2 + IBC cells (MDA-IBC3 and SUM190). All the cells showed ALK (anaplastic lymphoma kinase) expression, although its expression was slightly higher in SUM149 and FC-IBC02 mammospheres (Fig. 6c). PTK2/FAK1 showed higher expression in the IBC cell lines FC-IBC02 and SUM190 and its expression was higher in FC-IBC02 mammospheres compared to adherent cells (Fig. 6c). Total and phosphorylated PTK2/FAK1 protein expression was studied in different IBC cells and all showed phosphorylated PTK2/FAK1 (Fig. 7).
Fig. 7.
Expression of total and phospho-FAK1 (Y397) in IBC cell lines. FAK1 protein expression was studied in the triple-negative IBC cell lines FC-IBC02 and SUM149, and ErbB2-positive IBC cell lines SUM190 and KMO-015. Data were plotted after correction of cell number
Discussion
We have developed a new IBC cell line, FC-IBC02, derived from the pleural effusion of a woman with rapidly progressing secondary IBC. These tumor cells and xenografts in mice showed the same markers as the original pleural effusion in the patient from whom they were isolated. FC-IBC02 breast tumor xenografts grew rapidly and all mice developed spontaneous metastases within lungs and lymph nodes. FC-IBC02 cells were able to recapitulate the peculiar metastatic process observed in IBC patients, as shown by the formation of tumor emboli within lymphatics of SCID mice. Remarkably, injection of FC-IBC02 tumor cells via either the intraperitoneal or intracardiac routes resulted in the formation of brain metastases.
The symptoms of IBC are caused by the invasion of aggregates of tumor cells (tumor emboli) into the dermal lymphatics, causing an obstruction of the lymph channels [6]. Although, tumor emboli are also found in non-IBC tumors, they are more frequent and high in number in IBC [25]. Tumor emboli expressed cell–cell adhesion molecules that maintain the tumor cells together. Approximately, 90 % of human IBCs are associated with increased E-cadherin indicating that the gain of E-cadherin axis contributes to the IBC phenotype [18]. FC-IBC02 cells, breast tumor xenografts and metastatic lesions within lungs in SCID mice showed strong expression of E-cadherin and β-catenin in cell membranes. For most non-IBC tumors, loss of E-cadherin and acquisition of mesenchymal phenotype have been associated with increased invasion and metastatic potential, although in IBC tumor cells gained a survival benefit from the expression of E-cadherin mediating the formation of the characteristic tumor emboli. Moreover, FC-IBC02 cells showed strong expression of the membrane tetraspanin 24 (TSPAN24/CD151). Collectively, these results demonstrate that the FC-IBC02 tumor emboli express E-cadherin, β-catenin and TSPAN24/CD151 suggest that these adhesion molecules may have a functional role in maintaining the tight aggregation of cells within the tumor emboli. The maintenance of cell–cell adhesions allow the migration of tumor cells through the lymphatic and blood vessels as clusters, thus supporting the hypothesis that cohesive or collective migration may provide a survival advantage by protecting cells from immune attack or shear forces during transit through the circulation [26]. The presence of clusters of circulating tumor cells (CTCs) have been observed in the blood of IBC patients (data not shown).
The expression of epithelial markers by IBC cells is paradoxical to the current hypothesis that metastasis occurs as part of the process of epithelial–mesenchymal transition (EMT). Through the EMT process, epithelial cells lose cell–cell contact and cell polarity, downregulate epithelial-associated genes and acquire a mesenchymal phenotype; this cellular process culminates with single cells having increased motility, invasiveness and metastatic capacity. FC-IBC02 cells showed some, but not all the characteristic expression patterns of EMT; these cells expressed vimentin (VIM) and the EMT-transcription factors SNAI2 and TWIST1, although they showed strong expression of the epithelial markers EpCAM, MUC1 and E-cadherin. It has been suggested that SNAI2 and TWIST1 are probably responsible for maintaining the stem cell state rather than inducing mesenchymal marker expression [27, 28]. Recent studies suggest that EMT is not a pre-requisite for tumor cell invasion and cells can move in a collective manner dependent on maintenance of cell–cell adhesion molecules [29].
At the genomic level, FC-IBC02 cells and other IBC cells showed extensive chromosomal copy number changes indicating a higher degree of genomic instability in IBC that is in agreement with their high grade. NOTCH3 was amplified in FC-IBC02, and several genes from the Notch signalling pathway were upregulated in this cell line. Recently, it was shown that Notch 3 was also activated in Mary X [30]. Furthermore, IBC cells showed amplification of the 8q chromosomal arm where the oncogene MYC and ATAD2, a cofactor for MYC, are located. MYC showed 2.5–7 copies in IBC cells. MYC amplification is one of the most consistent markers of adverse prognosis for cancer and is more often amplified in IBC than non-IBC tumors [31]. MYC expression was higher in the triple-negative IBC cell lines FC-IBC02 and SUM149 when compared with the triple-negative non-IBC MDA-MB-231 and MDA-MB468, and the ErbB2 + IBC cells SUM190 and MDA-IBC3. MYC has a large number of targeted genes and it upregulates genes involved in cell growth and proliferation [32]. ATAD2 was amplified and upregulated in FC-IBC02; it was shown that this gene is usually overexpressed and amplified in aggressive tumors [33]. ATAD2 associates through its bromodomain (BRD) with histone H3 acetylated at Lys 14 during late mitosis, regulating the expression of genes required for cell cycle progression [33]; recently, specific inhibitors to BRD-containing proteins have been developed [34–36]. Targeting ATAD2 through the use of specific inhibitors to its BRD domain will be useful as therapeutic targets in MYC-driven tumors.
Metadherin (MTDH), a gene related to anoikis resistance, located at 8q, was also amplified and upregulated in FC-IBC02 cells. Anchorage-independent survival or anoikis resistance is characteristic of tumor cells and contributes to metastasis [37]. All tumor cells that eventually form metastases at distant sites must survive in circulation and then in metastatic organs, and all of these microenvironments are quite distinct from that of the breast. The ability of tumor cells to survive in these altered matrix conditions is known as anoikis resistance (adhesion-independent growth) and is in contrast to normal epithelial cells, which undergo cell death (anoikis) when deprived of signals from the normal extracellular matrix [37]. MTDH functions as a downstream mediator of the transforming activity of oncogenic Ha-Ras and c-MYC and promotes lung, bone and brain metastasis [38]. Interestingly, another gene previously reported to contribute to metastasis via regulation of anoikis resistance is the focal adhesion kinase FAK1 (or PTK2) [37], which shows 2.5–7 copies in IBC. FAK1 is a non-receptor tyrosine kinase that localizes to focal adhesions and controls a number of cell pathways including proliferation, viability and survival. Its overexpression was linked to anoikis resistance [39]. FAK1 mRNA and protein levels are absent or low in normal tissue and benign neoplasms, and are upregulated in invasive and metastatic tumors [40]. Elevated FAK1 levels have been reported in many tumors such as epithelial tumor of the breast [39]. We have studied FAK1 in the triple-negative IBC cell lines FC-IBC02 and SUM149, and ErbB2-positive cell lines SUM190 and KMO-015, and this protein showed to be activated in these cells (phosphorylated in Y397). Studies for evaluating a new dual ALK-FAK1 inhibitor in IBC cells and xenograft models are ongoing in our laboratory.
In summary, the gene signature and phenotypic characteristics of FC-IBC02 and other IBC cell lines suggest that IBC exhibits characteristics of epithelial plasticity, where tumor cells retain an epithelial phenotype through E-cadherin and EpCAM expressions, while simultaneously expressing markers of cancer stem cells. FC-IBC02 xenografts represent an ideal model for studying spontaneous metastases in lungs and lymph nodes and brain metastases by the intracardiac route. Moreover, FC-IBC02 and other IBC cells demonstrated amplification of MYC, ATAD2, CD44, NOTCH3, ALK and FAK1 indicating that these genes could be used as potential target therapies against IBC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by the American Airlines-Komen for the Cure Foundation Promise Grant KGO81287 (FMR, MC), NIH NCI 1R01 CA 138239 (MC) and the Inflammatory Breast Cancer Foundation. We thank Maria Florencia Arisi for the corrections of this manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Sandra V. Fernandez, Phone: +1-215-5037783, FAX: +1-215-5039334, Email: Sandra.Fernandez@jefferson.edu
Massimo Cristofanilli, Phone: +1-215-5035098, FAX: +1-215-5033408, Email: Massimo.Cristofanilli@jefferson.edu.
References
- 1.Hance KW, Anderson WF, Devesa SS, Young HA, Levine PH. Trends in inflammatory breast carcinoma incidence and survival: the surveillance, epidemiology, and end results program at the National Cancer Institute. J Natl Cancer Inst. 2005;97:966–975. doi: 10.1093/jnci/dji172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chia S, Swain SM, Byrd DR, Mankoff DA. Locally advanced and inflammatory breast cancer. J Clin Oncol. 2008;26:786–790. doi: 10.1200/JCO.2008.15.0243. [DOI] [PubMed] [Google Scholar]
- 3.Dawood S, et al. Differences in survival among women with stage III inflammatory and noninflammatory locally advanced breast cancer appear early: a large population-based study. Cancer. 2011;117:1819–1826. doi: 10.1002/cncr.25682. [DOI] [PubMed] [Google Scholar]
- 4.Robertson FM, et al. Inflammatory breast cancer: the disease, the biology, the treatment. CA Cancer J Clin. 2010;60:351–375. doi: 10.3322/caac.20082. [DOI] [PubMed] [Google Scholar]
- 5.Cristofanilli M, et al. Inflammatory breast cancer (IBC) and patterns of recurrence: understanding the biology of a unique disease. Cancer. 2007;110:1436–1444. doi: 10.1002/cncr.22927. [DOI] [PubMed] [Google Scholar]
- 6.Molckovsky A, Fitzgerald B, Freedman O, Heisey R, Clemons M. Approach to inflammatory breast cancer. Can Fam Physician. 2009;55:25–31. [PMC free article] [PubMed] [Google Scholar]
- 7.Alpaugh ML, Tomlinson JS, Kasraeian S, Barsky SH. Cooperative role of E-cadherin and sialyl-Lewis X/A-deficient MUC1 in the passive dissemination of tumor emboli in inflammatory breast carcinoma. Oncogene. 2002;21:3631–3643. doi: 10.1038/sj.onc.1205389. [DOI] [PubMed] [Google Scholar]
- 8.Bertucci F, Finetti P, Birnbaum D, Viens P. Gene expression profiling of inflammatory breast cancer. Cancer. 2010;116:2783–2793. doi: 10.1002/cncr.25165. [DOI] [PubMed] [Google Scholar]
- 9.Van Laere SJ, et al. Identification of cell-of-origin breast tumor subtypes in inflammatory breast cancer by gene expression profiling. Breast Cancer Res Treat. 2006;95:243–255. doi: 10.1007/s10549-005-9015-9. [DOI] [PubMed] [Google Scholar]
- 10.Willmarth NE, Ethier SP. Autocrine and juxtacrine effects of amphiregulin on the proliferative, invasive, and migratory properties of normal and neoplastic human mammary epithelial cells. J Biol Chem. 2006;281:37728–37737. doi: 10.1074/jbc.M606532200. [DOI] [PubMed] [Google Scholar]
- 11.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forozan F, et al. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer. 1999;81:1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charafe-Jauffret E, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–2284. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 14.Kurebayashi J, et al. Isolation and characterization of a new human breast cancer cell line, KPL-4, expressing the Erb B family receptors and interleukin-6. Br J Cancer. 1999;79:707–717. doi: 10.1038/sj.bjc.6690114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alpaugh ML, Tomlinson JS, Shao ZM, Barsky SH. A novel human xenograft model of inflammatory breast cancer. Cancer Res. 1999;59:5079–5084. [PubMed] [Google Scholar]
- 16.Shirakawa K, et al. Absence of endothelial cells, central necrosis, and fibrosis are associated with aggressive inflammatory breast cancer. Cancer Res. 2001;61:445–451. [PubMed] [Google Scholar]
- 17.Xiao Y, Ye Y, Yearsley K, Jones S, Barsky SH. The lymphovascular embolus of inflammatory breast cancer expresses a stem cell-like phenotype. Am J Pathol. 2008;173:561–574. doi: 10.2353/ajpath.2008.071214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomlinson JS, Alpaugh ML, Barsky SH. An intact overexpressed E-cadherin/alpha, beta-catenin axis characterizes the lymphovascular emboli of inflammatory breast carcinoma. Cancer Res. 2001;61:5231–5241. [PubMed] [Google Scholar]
- 19.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson F et al (2012) Genomic profiling of pre-clinical models of inflammatory breast cancer identifies a signature of epithelial plasticity and suppression of TGF B signalling. J Clin Exp Pathol 2:1–11
- 21.Li C, Hung Wong W (2001) Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2: RESEARCH 0032 [DOI] [PMC free article] [PubMed]
- 22.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 23.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 24.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo AC, et al. Analysis of RhoC expression and lymphovascular emboli in inflammatory vs non-inflammatory breast cancers in Egyptian patients. Breast. 2009;18:55–59. doi: 10.1016/j.breast.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 27.Chui MH. Insights into cancer metastasis from a clinicopathologic perspective: Epithelial mesenchymal transition is not a necessary step. Int J Cancer. 2012;132(7):1487–1495. doi: 10.1002/ijc.27745. [DOI] [PubMed] [Google Scholar]
- 28.Mani SA, et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 30.Xiao Y, et al. The lymphovascular embolus of inflammatory breast cancer exhibits a Notch 3 addiction. Oncogene. 2011;30:287–300. doi: 10.1038/onc.2010.405. [DOI] [PubMed] [Google Scholar]
- 31.Bekhouche I, et al. High-resolution comparative genomic hybridization of inflammatory breast cancer and identification of candidate genes. PLoS ONE. 2011;6:e16950. doi: 10.1371/journal.pone.0016950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciro M, et al. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 2009;69:8491–8498. doi: 10.1158/0008-5472.CAN-09-2131. [DOI] [PubMed] [Google Scholar]
- 34.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller S, Filippakopoulos P, Knapp S. Bromodomains as therapeutic targets. Expert Rev Mol Med. 2011;13:e29. doi: 10.1017/S1462399411001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frisch SM, Screaton RA. Anoikis mechanisms. Curr Opin Cell Biol. 2001;13:555–562. doi: 10.1016/S0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- 38.Hu G, et al. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gabarra-Niecko V, Schaller MD, Dunty JM. FAK regulates biological processes important for the pathogenesis of cancer. Cancer Metastasis Rev. 2003;22:359–374. doi: 10.1023/A:1023725029589. [DOI] [PubMed] [Google Scholar]
- 40.Cance WG, et al. Immunohistochemical analyses of focal adhesion kinase expression in benign and malignant human breast and colon tissues: correlation with preinvasive and invasive phenotypes. Clin Cancer Res. 2000;6:2417–2423. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.