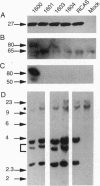
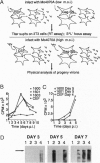
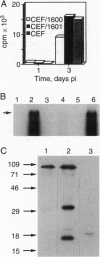
Abstract
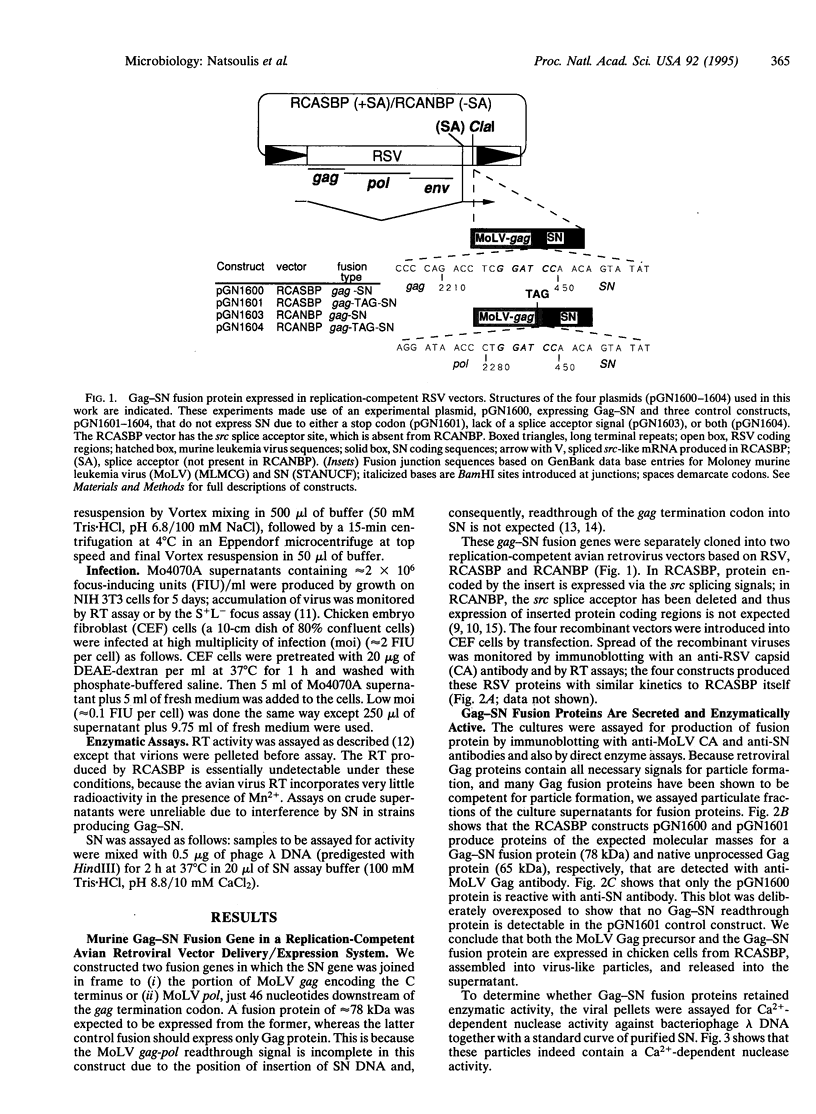
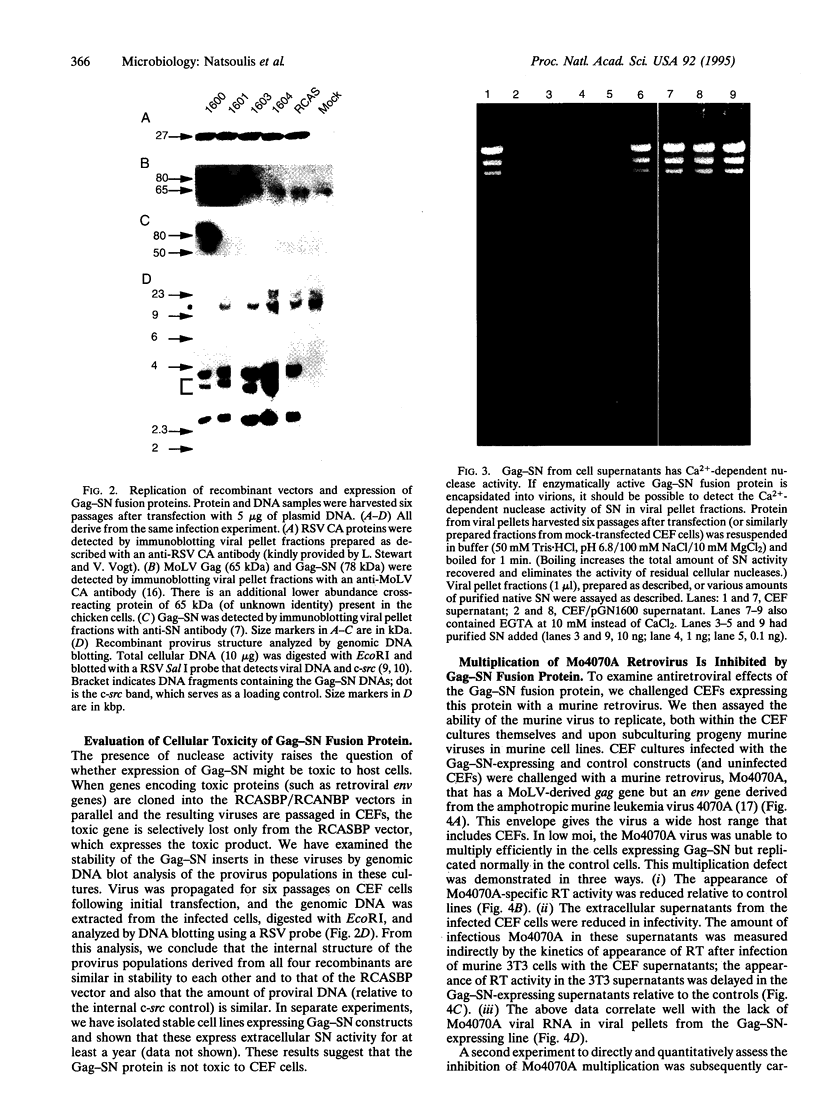
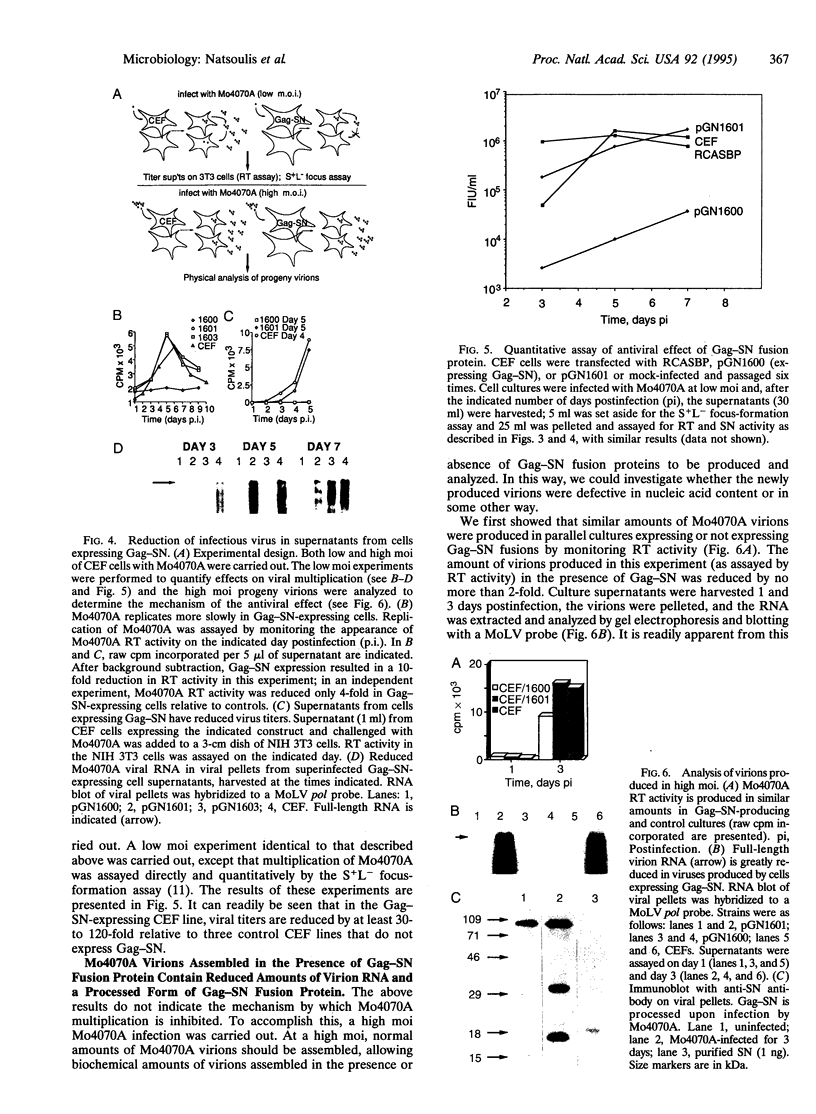
Capsid-targeted viral inactivation is an antiviral strategy in which toxic fusion proteins are targeted to virions, where they inhibit viral multiplication by destroying viral components. These fusion proteins consist of a virion structural protein moiety and an enzymatic moiety such as a nuclease. Such fusion proteins can severely inhibit transposition of yeast retrotransposon Ty1, an element whose transposition mechanistically resembles retroviral multiplication. We demonstrate that expression of a murine retrovirus capsid-staphylococcal nuclease fusion protein inhibits multiplication of the corresponding murine leukemia virus by 30- to 100-fold. Staphylococcal nuclease is apparently inactive intracellularly and hence nontoxic to the host cell, but it is active extracellularly because of its requirement for high concentrations of Ca2+ ions. Virions assembled in and shed from cells expressing the fusion protein contain very small amounts of intact viral RNA, as would be predicted for nuclease-mediated inhibition of viral multiplication.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bassin R. H., Tuttle N., Fischinger P. J. Rapid cell culture assay technic for murine leukaemia viruses. Nature. 1971 Feb 19;229(5286):564–566. doi: 10.1038/229564b0. [DOI] [PubMed] [Google Scholar]
- Bolognesi D. P., Montelaro R. C., Frank H., Schäfer W. Assembly of type C oncornaviruses: a model. Science. 1978 Jan 13;199(4325):183–186. doi: 10.1126/science.202022. [DOI] [PubMed] [Google Scholar]
- Feng Y. X., Yuan H., Rein A., Levin J. G. Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J Virol. 1992 Aug;66(8):5127–5132. doi: 10.1128/jvi.66.8.5127-5132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel A., Boeke J. D. Reverse transcriptase encoded by a retrotransposon from the trypanosomatid Crithidia fasciculata. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9794–9798. doi: 10.1073/pnas.88.21.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S., Traktman P., Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981 Apr;38(1):239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhouse J. J., Petropoulos C. J., Crittenden L. B., Hughes S. H. Helper-independent retrovirus vectors with Rous-associated virus type O long terminal repeats. J Virol. 1988 Dec;62(12):4809–4812. doi: 10.1128/jvi.62.12.4809-4812.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Jelinek L., Whiting S., Barklis E. Transport and assembly of gag proteins into Moloney murine leukemia virus. J Virol. 1990 Nov;64(11):5306–5316. doi: 10.1128/jvi.64.11.5306-5316.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. H., Greenhouse J. J., Petropoulos C. J., Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol. 1987 Oct;61(10):3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S., Kosik E. Mutagenesis of the region between env and src of the SR-A strain of Rous sarcoma virus for the purpose of constructing helper-independent vectors. Virology. 1984 Jul 15;136(1):89–99. doi: 10.1016/0042-6822(84)90250-2. [DOI] [PubMed] [Google Scholar]
- Mathias S. L., Scott A. F., Kazazian H. H., Jr, Boeke J. D., Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991 Dec 20;254(5039):1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Natsoulis G., Boeke J. D. New antiviral strategy using capsid-nuclease fusion proteins. Nature. 1991 Aug 15;352(6336):632–635. doi: 10.1038/352632a0. [DOI] [PubMed] [Google Scholar]
- Ott D. E., Keller J., Sill K., Rein A. Phenotypes of murine leukemia virus-induced tumors: influence of 3' viral coding sequences. J Virol. 1992 Oct;66(10):6107–6116. doi: 10.1128/jvi.66.10.6107-6116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos C. J., Hughes S. H. Replication-competent retrovirus vectors for the transfer and expression of gene cassettes in avian cells. J Virol. 1991 Jul;65(7):3728–3737. doi: 10.1128/jvi.65.7.3728-3737.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteegen R. J., Oroszlan S. Effect of chemical modification and fragmentation on antigenic determinants of internal protein p30 and surface glycoprotein gp70 of type C retroviruses. J Virol. 1980 Mar;33(3):983–992. doi: 10.1128/jvi.33.3.983-992.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon R. A., Jr, Erdie C. R., Oliver M. G., Wills J. W. Incorporation of chimeric gag protein into retroviral particles. J Virol. 1990 Sep;64(9):4169–4179. doi: 10.1128/jvi.64.9.4169-4179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills J. W., Craven R. C. Form, function, and use of retroviral gag proteins. AIDS. 1991 Jun;5(6):639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- Wills N. M., Gesteland R. F., Atkins J. F. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]