Abstract
We previously identified a chlamydial protein designated CPAF (chlamydia protease/proteasome-like activity factor) that is secreted into host cell cytosol for degrading host transcription factors required for major histocompatibility complex antigen expression. Here we report that CPAF, synthesized as a 70-kDa proprotein, is processed into two fragments (designated CPAFn and CPAFc) to form intramolecular dimers that are much more stable than the naïve CPAF. Precipitation with antibodies that recognized CPAF dimers removed the proteolytic activity responsible for degrading host transcription factor RFX5 from chlamydia-infected host cell cytosol, while precipitation with antibodies that recognized free CPAF fragments alone did not remove this activity. Separation of CPAFn from CPAFc resulted in a loss of proteolytic activity. Furthermore, neither expressed full-length CPAF that was not processed nor coexpressed CPAFn and CPAFc fragments that failed to form dimers degraded RFX5. These observations demonstrate that intramolecular dimerization is required for CPAF to degrade host transcription factors, a strategy that is utilized by an obligate intracellular bacterial species to evade host defenses.
Chlamydiae are obligate intracellular bacterial pathogens that must replicate in cytoplasmic vacuoles of eukaryotic cells (11). Despite the severe health problems caused by infection with various chlamydial species (1, 5, 10), the pathogenic mechanisms of chlamydia-induced diseases in humans are still unclear. It is hypothesized that chronic inflammatory responses provoked during chlamydial intravacuolar replication mainly contribute to the chlamydial pathogenesis (1, 3-5, 16, 19). Therefore, investigation of the molecular basis for how chlamydiae maintain long-term intravacuolar residence in infected hosts should improve our understanding of chlamydial pathogenesis.
It has been reported previously that chlamydiae, like viruses (21), have evolved various strategies for evading host defenses in order to achieve long-term survival in infected hosts (8, 26, 27). Chlamydiae have acquired the ability to prevent infected cells from undergoing apoptosis (6, 8, 15), which may allow chlamydiae to avoid immune effector mechanisms mediated by host cell apoptosis. To escape immune detection, chlamydiae suppress major histocompatibility complex (MHC) antigen expression in infected host cells (26, 27). Recently, a chlamydial protein designated CPAF (chlamydia protease/proteasome-like activity factor) that may be responsible for the chlamydial suppression of MHC antigen expression was identified (25). CPAF is secreted from chlamydial vacuoles into host cell cytosol and degrades transcriptional factors required for host MHC gene activation (25). Subsequent studies confirmed that CPAF indeed is the first and only chlamydia-secreted protein for chlamydial manipulation of host cells that has been identified so far (7, 12, 17).
Although CPAF is encoded by a chlamydial open reading frame for a ∼70-kDa protein, it was purified as two shorter fragments with molecular masses of ∼35 kDa (corresponding to the C-terminal half of CPAF; designated CPAFc) and ∼29 kDa (corresponding to the N-terminal portion of CPAF; designated CPAFn) (25). The fact that the CPAFc and CPAFn fragments were coeluted from nondenaturing columns at a molar ratio of almost 1:1 (25) suggests that these two fragments are associated with each other. We hypothesized that CPAF may form intramolecular dimers and that dimerization may be necessary for CPAF activity. In the present study, we tested this hypothesis by using a panel of monoclonal antibodies (MAbs) raised with endogenous CPAF, in combination with various immunoprecipitation schemes. We found that CPAF must form an intramolecular dimer to acquire activity for degrading host transcriptional factors. This finding provided further insight into the molecular mechanism of CPAF-mediated immune evasion, which will no doubt be useful for developing interventional strategies to prevent chlamydiae from evading host immune detection mechanisms.
MATERIALS AND METHODS
Purification of CPAF and generation of MAbs against CPAF.
In order to generate antibodies suitable for detecting CPAF in chlamydia-infected cells, we used endogenous CPAF as the immunogen. About 200 175-cm2 flasks of HeLa cells (American Type Culture Collection, Manassas, Va.) infected with C. trachomatis serovar L2 for 30 to 40 h were harvested to prepare a cytosolic fraction consisting of the supernatant obtained after centrifugation at 100,000 × g (L2S100) (8). CPAF was purified from L2S100 by column chromatography as previously described (25). Purified endogenous CPAF was used to immunize BALB/c mice (The Jackson Laboratory, Bar Harbor, Maine) for generation of antibodies. A fusion protocol described elsewhere was used to make spleen-myeloma (SP20) cell hybridomas (22, 28). Hybridoma cells were screened for antibodies that recognized CPAF by an enzyme-linked immunosorbent assay (29-31), followed by Western blotting (23). A total of six clones were isolated from two independent fusions, and all of the clones were isotyped mouse immunoglobulin G1. Three of the six clones were mapped to the N-terminal fragment (clones 2a, 5, and 54b) and the other three clones were mapped to the C-terminal fragment of CPAF (clones 73, 97, and 100a). The hybridoma culture supernatants were used for Western blotting, radioimmunoprecipitation, and immunofluorescence assays in this study.
Expression of CPAF and CPAF fragments in both bacterial and mammalian cell systems.
CPAF or CPAF fragments were cloned and expressed in bacterial systems. Primers were designed based on the CPAF sequence of serovar D (CT858; accession number A71461; http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?CMD=search&DB=protein) (20, 25). Both full-length CPAF and various fragments were cloned into a pGEX6p-2 vector (Amersham Biosciences Corp., Piscataway, N.J.) by using serovar L2 genomic DNA as the template. This vector allowed genes of interest to be expressed as fusion proteins with a 26-kDa glutathione S-transferase (GST) as the fusion partner. The following GST-CPAF fusion constructs were produced: 1-283, 1-488, 1-525, 488-525, 1-609, 9-609, 11-609, 60-609, 136-609, 186-609, 242-609, 284-609, 338-609, and 387-609. The potential leader sequence (residues 1 to 24) (http://www.stdgen.lanl.gov/) in CPAF was not excluded during primer design. This is because GST was fused to the N terminus and because there was no difference in the expression level between constructs with the putative leader sequence and constructs without the putative leader sequence. All GST-CPAF constructs were expressed in BL21 with isopropyl-β-d-thiogalactopyranoside (IPTG) as an inducer. GST fusion proteins were purified by using glutathione-conjugated beads (Amersham Biosciences Corp.) as instructed by the manufacturer. For mammalian expression, either the full-length CPAF (residues 1 to 609), the C-terminal fragment (residues 284 to 609), or the N-terminal fragment (residues 1 to 283) was cloned into a pEF vector (Invitrogen, Carlsbad, Calif.) for cytosolic expression with or without a C-terminal fusion myc tag. The CPAF-myc constructs were evaluated for expression in 293T cells after transfection individually or in combinations by using a protocol described elsewhere (24). Proteins encoded by constructs with the putative leader sequence and proteins encoded by constructs without the putative leader sequence exhibited similar levels of solubility and expression.
Western blotting.
The Western blot assays were carried out as described previously (28, 30). Briefly, protein samples (S100 prepared from chlamydia-infected cells, purified CPAF fusion protein, or reaction mixtures from cell-free degradation assays) were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel separation. After proteins were blotted onto nitrocellulose membranes, the primary antibodies were applied. These antibodies included various anti-CPAF mouse MAbs (see above), mouse anti-myc (clone 9E10; American Type Culture Collection), rabbit anti-GST (Amersham Biosciences Corp.), and anti-RFX5 (Rockland Immunochemicals Inc., Gilbertsville, Pa.). Primary antibody binding was examined with corresponding secondary antibodies conjugated with horseradish peroxidase (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.), followed by a standard ECL analysis (Amersham Biosciences Corp.).
Immunoprecipitation.
Precipitation assays were carried out as described elsewhere (24, 25). Briefly, chlamydia-infected cells or transfected cells with or without metabolic labeling with radioactive amino acids ([35S]methionine and [35S]cysteine; ICN Biomedicals Inc., Irvine, Calif.) were lysed with MLB buffer (0.5% IGEPAL [Sigma] in 50 mM Tris [pH 8] with 150 mM NaCl plus various protease inhibitors), and the lysates were mixed with antibodies conjugated to protein A/G agarose beads (Amersham Pharmacia). After unbound material and nonspecifically bound material were washed away with phosphate-balanced salt buffer (pH 7.4), the precipitates were either subjected to direct SDS gel separation and autoradiography or additional treatments. Several immunoprecipitation schemes were designed in the present study in order to address different questions. For sequential precipitation, the supernatants remaining after the first precipitation were subjected to additional precipitation with the same or different antibodies. The goal was to deplete the corresponding antigens or to detect the residual antigens in the remaining supernatants. For serial precipitation, as described previously (25), a pellet precipitated with a given antibody (Io precipitation) was eluted or solubilized, and the solubilized material was subjected to a second precipitation with either the same antibody or a different antibody (IIo precipitation). Solubilization was carried out by soaking antigen-antibody-agarose pellets in an equal volume of SDS buffer (0.5% SDS in 50 mM Tris [pH 7.5]) for 15 min at 37°C with constant rotation. The solubilization reaction was stopped by diluting the mixture with a 100-fold excess of Dulbecco modified Eagle medium containing 10% fetal calf serum. The dilution was often done just prior to the IIo precipitation. It is worth noting that the solubilization conditions used were strong enough to dissociate both the antigen-antibody interactions and the antigen complexes formed via noncovalent bonds. However, the solubilization process was properly controlled so that the conformation of individual antigen subunits was not grossly altered and the subsequent IIo precipitation was not inhibited. The goal of the serial precipitation procedure was to use the IIo precipitation to identify the individual components in the pellets precipitated by the Io precipitation. For pulse-chase experiments, the infected cultures were pulse-labeled with radioactive amino acids for a short time (1 h). After the residual unincorporated radioactive amino acids were washed away, one culture was harvested right away (chasing for 0 h), while parallel cultures were continuously incubated in the presence of a 10-fold excess of cold methionine and cysteine for various periods of time (various chasing periods) prior to harvest.
Cell-free degradation assays.
The cell-free degradation assay was carried out as described previously (27). Briefly, nuclear extracts containing RFX5 were used as substrates, and the enzyme sources included chlamydia-infected cell cytosol (L2S100), as well as the supernatants remaining after antibody precipitation or pellets obtained by antibody precipitation. After incubation of the enzyme-substrate mixtures for 1 h at 37°C, the digestion mixtures were subjected to SDS gel separation, and residual RFX5 was detected by a Western blot assay.
RESULTS
CPAF is synthesized as a 70-kDa proprotein and is processed to form intramolecular dimers.
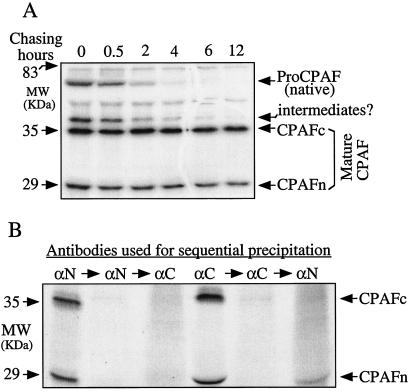
To investigate how CPAF synthesized in chlamydial vacuoles can degrade host cell proteins, we first carried out a series of immunoprecipitation pulse-chase experiments to track the fate of newly synthesized CPAF. As shown in Fig. 1A, a 70-kDa protein was precipitated along with several other proteins from the lysates of cells infected with chlamydiae for 30 h and biosynthetically radiolabeled for 1 h (Fig. 1A, chasing for 0 h). As the parallel pulse-labeled cells were chased, the 70-kDa protein gradually disappeared, while the levels of both the 35-kDa fragment (CPAFc) and the 29-kDa fragment (CPAFn) (25) remained at nearly steady-state levels. By 30 min after chasing, ∼50% of the 70-kDa CPAF had disappeared, while ∼50% of the mature CPAFc and CPAFn fragments remained intact even after chasing for 12 h. The mature CPAFc and CPAFn fragments were apparently more than 10-fold more stable than the 70-kDa CPAF. Based on these observations, the two shorter fragments clearly represented the mature form of CPAF processed from the 70-kDa proprotein (designated proCPAF). The coprecipitated band that migrated just above the mature 35-kDa CPAFc fragment might represent a processing intermediate of CPAF since its half-life was between that of the proCPAF and that of the mature CPAFc and CPAFn fragments. The fact that a large amount of mature CPAFc and CPAFn fragments was present in the sample pulsed for 1 h (Fig. 1A) suggests that processing of proCPAF occurred very quickly. The antibody used for the immunoprecipitation was MAb 73, which was specific for CPAFc (Fig. 2). Coprecipitation of CPAFn with this antibody strongly suggested that CPAFn was in complexes with CPAFc.
FIG. 1.
Identification of CPAFc-CPAFn complexes in chlamydia-infected host cell cytosol. (A). Pulse-chase radioimmunoprecipitation to monitor the fate of newly synthesized CPAF. HeLa cells infected with C. trachomatis serovar L2 for 30 h were metabolically labeled for 1 h (pulse; chasing 0 h), and parallel pulse-labeled cells were chased for various times, as indicated at the top. Cells with or without chasing were harvested and used for antibody precipitation as described in Materials and Methods. Although a CPAFc-specific MAb (MAb 73) was used for the precipitation, CPAFn was coprecipitated. Note that the 70-kDa proCPAF was rapidly processed into two shorter but much more stable fragments, CPAFc (∼35 kDa) and CPAFn (∼29 kDa). The stable CPAFc and CPAFn fragments are designated mature CPAF. (B) Sequential precipitation to determine the extent of complexes formed by CPAFc and CPAFn. Cell lysates prepared as described in Materials and Methods were subjected to precipitation with either MAb 54b (αN) or MAB 73 (αC). After centrifugation, the remaining supernatants were reprecipitated with the same antibodies. The final remaining supernatants after two precipitations were precipitated reciprocally with heterologous antibodies. The pellets brought down during each precipitation were subjected to SDS gel separation and analysis by autoradiography. Note that a small amount of free CPAFn fragment was pulled down (right lane) during the third precipitation from the supernatant that had been precipitated twice with CPAFc-specific MAb 73. MW, molecular mass.
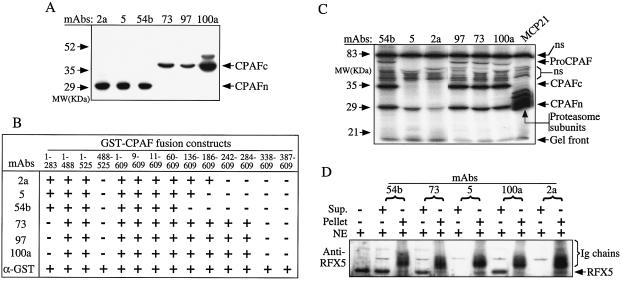
FIG. 2.
Correlation of CPAFc-CPAFn dimers with CPAF degradation of RFX5. (A) Western blot analysis of six MAbs (indicated at the top) with L2S100 as the antigen. MAbs 2a, 5, and 54b recognized the 29-kDa CPAFn band, while MAbs 73, 97, and 100a recognized the 35-kDa CPAFc band. (B) Fine specificity mapping of the MAbs by using a collection of GST-CPAF fusion proteins (indicated at the top) on a Western blot. The numbers for the GST-CPAF constructs indicate the amino acid residues. A plus sign indicates positive binding of an MAb to the corresponding fusion protein, while a minus sign indicates that there was no detectable binding. Proper expression of all fusion proteins was confirmed with a rabbit anti-GST antibody (α-GST). (C) Radioimmunoprecipitation with six anti-CPAF antibodies (MAbs 54b, 5, 2a, 97, 73, and 100a) and one anti-host proteasome antibody (MCP21). The positions of the 70-kDa proCPAF, mature CPAFc and CPAFn fragments, and proteasome subunits are indicated on the right. ns, nonspecifically precipitated bands. Note that all of the antibodies except MAbs 5 and 2a precipitated CPAF dimers consisting of CPAFn and CPAFc fragments. (D) Cell-free degradation of RFX5 by remaining supernatants (Sup.) and pellets after precipitation of L2S100 with the antibodies. The nuclear extracts (NE) containing RFX5 were used as the substrates. The residual RFX5 was detected on a Western blot with rabbit anti-RFX5. The immunoglobulin (Ig) chains of the precipitation antibodies were also visualized on the Western blot. Note that the remaining supernatants after precipitation by MAb 54b, 73, or 100a were not able to degrade RFX5, but the pellets brought down by these MAbs exhibited RFX5 degradation activity. The opposite was true for MAbs 5 and 2a, suggesting that there is a correlation between CPAF activity and CPAF dimerization. MW, molecular mass.
The extent of formation of complexes between CPAFc and CPAFn was determined by a sequential immunoprecipitation assay (Fig. 1B). An MAb recognizing either CPAFn (MAb 54b) (Fig. 2) or CPAFc (MAb 73) precipitated both the 35-kDa CPAFc fragment and the 29-kDa CPAFn fragment, confirming that the CPAFc and CPAFn fragments were physically linked to each other. Reprecipitation of the remaining supernatant with the same antibodies pulled down no additional radioactive signal (Fig. 1B), suggesting that CPAF fragments in complexes were completely depleted during the first precipitation. When the supernatants that remained after depletion by the two consecutive precipitations with the same antibodies were subjected to further precipitation reciprocally with the heterologous antibodies, the CPAFc-specific MAb failed to precipitate any CPAF fragments from the anti-CPAFn-depleted supernatant, while the CPAFn-specific MAb brought down a small amount of free CPAFn from the anti-CPAFc-depleted supernatant. It is worth noting that both the anti-CPAFc and anti-CPAFn MAbs used in this set of experiments were able to recognize the corresponding antigens in either complexes or single chains (Fig. 2). The observations described above clearly demonstrated that most of the mature CPAF fragments were associated with each other in complexes and that that was only a small amount of free CPAFn fragment.
The intramolecular dimers of CPAF are proteolytically active.
A panel of six MAbs was used to determine whether CPAF dimers or free fragments are responsible for CPAF enzymatic activity. The binding sites of the six MAbs (designated MAbs 2a, 5, 54b, 73, 97, and 100a) were mapped by Western blotting, and three of the MAbs bound to CPAFn (MAbs 2a, 5, and 54b) and three bound to CPAFc (MAbs 73, 97, and 100a) (Fig. 2A). These MAbs were further analyzed to determine their fine specificities by using a collection of bacterium-expressed CPAF fragments. As shown in Fig. 2B, the three CPAFc-specific MAbs mapped to a region between residues 284 and 338, while the three CPAFn-specific MAbs displayed different fine specificities; MAb 2a mapped to a region between residues 186 and 242, MAb5 mapped to a region between residues 136 and 186, and MAb 54b mapped to a region between residues 60 and 136. Since these CPAF fragments were expressed as GST fusion proteins, an anti-GST antibody was used to confirm that all CPAF fragments used in the experiment were properly expressed. In a radioimmunoprecipitation assay, four of the six MAbs precipitated both the 35-kDa CPAFc fragment and the 29-kDa CPAFn fragment (Fig. 2C), indicating that the corresponding epitope regions (between residues 283 and 338 recognized by CPAFc-specific MAbs 73, 97, and 100a and between residues 60 and 136 recognized by CPAFn-specific MAb 54b) are surface accessible on CPAF dimers. Although MAbs 2a and 5 specific for CPAFn failed to bring down CPAF dimers, they did precipitate small amounts of free CPAFn fragment (Fig. 2C), confirming that there was excess fragment CPAFn, as shown in Fig. 1B. The failure of MAbs 5 and 2a to precipitate CPAF dimers may have been because the corresponding epitopes (between residues 136 and 242) were no longer accessible to antibody binding when CPAFn dimerized with CPAFc. It is obvious that the conformation of CPAF dimers determines the accessibility of the epitopes to corresponding antibodies.
Importantly, the differential recognition of CPAF by the MAbs allowed us to correlate the CPAF activity with CPAF structure. We evaluated the effects of antibody precipitation on CPAF activity in the chlamydia-infected host cell cytosol. As shown in Fig. 2D, MAbs that were able to precipitate CPAF dimers (including MAbs 54b, 73, and 100a) effectively removed the RFX5 degradation activity from the infected cell cytosol in the antibody pellets, while MAbs 5 and 2a, which precipitated only the CPAFn fragment, did not remove this activity. These observations demonstrated that CPAF activity was associated with CPAF dimers but not with the free CPAFn fragments. However, since the 70-kDa proCPAF was also precipitated along with CPAF dimers, it remains to be determined whether the full-length proCPAF also contributed to the degradation of RFX5, which is discussed below.
Intramolecular dimers consisting of the CPAFc and CPAFn fragments are required for CPAF activity.
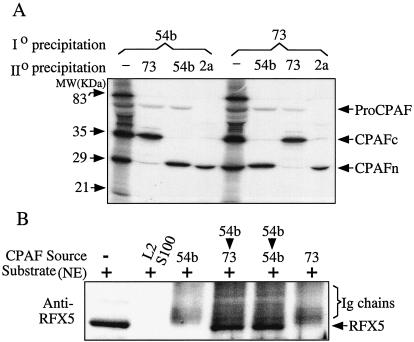
The contributions of various forms of CPAF to the enzymatic activity of CPAF were evaluated by using a serial immunoprecipitation scheme. As shown in Fig. 3A, both CPAFn-specific MAb 54b and CPAFc-specific MAb 73 precipitated the mature CPAFc-CPAFn complexes in addition to the 70-kDa proCPAF (Io precipitation). Three parallel precipitates brought down by each antibody were solubilized to dissociate the CPAFc-CPAFn dimers (see Materials and Methods), and the solubilized material was subjected to reprecipitation with either the same or heterologous antibodies (IIo precipitation) (Fig. 3A). For example, reprecipitation of the solubilized material brought down by MAb 54b with MAb 54b again pulled down only CPAFn and not CPAFc (Fig. 3A), while reprecipitation with CPAFc-specific MAb 73 pulled down only CPAFc and not CPAFn (Fig. 3A), indicating that the solubilization procedure effectively separated CPAFn from CPAFc. Likewise, IIo precipitation of the solubilized material brought down by MAb 73 with either MAb 54b (Fig. 3A) or MAb 73 only pulled down the corresponding single fragments. It is worth noting that regardless of the solubilization treatment, the 70-kDa proCPAF protein was brought down by MAbs 54b and 73 but not by MAb 2a, which is consistent with what is shown in Fig. 2C, suggesting that proCPAF had similar conformations with and without solubilization. Therefore, we concluded that the solubilization procedure used in this experiment was not severe enough to grossly alter the conformation of proteins, yet was strong enough to dissociate protein complexes. We next took advantage of the fact that CPAF dimers that precipitated in the antibody pellets still exhibited measurable RFX5 degradation activity (Fig. 2D), and we compared the RFX5 degradation activities of the antibody pellets brought down during both the Io and IIo precipitations (Fig. 3B). Although the pellets pulled down during Io precipitation by either MAb 54b or MAb 73 degraded RFX5, the pellets brought down during IIo precipitation did not degrade RFX5 regardless of the antibodies used. This is because CPAF dimers were present in the pellets brought down during the Io precipitation but not in the pellets brought down during the IIo precipitation (as shown in Fig. 3A). Since the 70-kDa proCPAF was also brought down by MAbs 54b and 73 during the IIo precipitation (Fig. 3A), the fact that none of the IIo precipitation pellets showed any RFX5 degradation activity (Fig. 3B) allowed us to rule out the possibility that proCPAF contributed to the CPAF proteolytic activity.
FIG. 3.
CPAF dimers are responsible for degradation of host transcription factors. (A) Cell lysates obtained from cultures infected with C. trachomatis serovar L2 and radiolabeled for 1 h were subjected to a first round of precipitation (Io precipitation) with either MAb 54b or MAb 73. The precipitated pellets were solubilized with an SDS buffer in order to dissociate the CPAFc-CPAFn complexes. The solubilized material was subjected to a second round of precipitation (IIo precipitation) with either the same or heterologous antibodies. Note that the CPAFc-CPAFn dimers were precipitated only during the Io precipitation. (B) Cell-free degradation of RFX5 with the precipitated pellets. For details see the legend to Fig. 2. Note that pellets brought down during Io precipitation but not pellets brought down during IIo precipitation degraded RFX5 regardless of the antibodies used. MW, molecular mass.
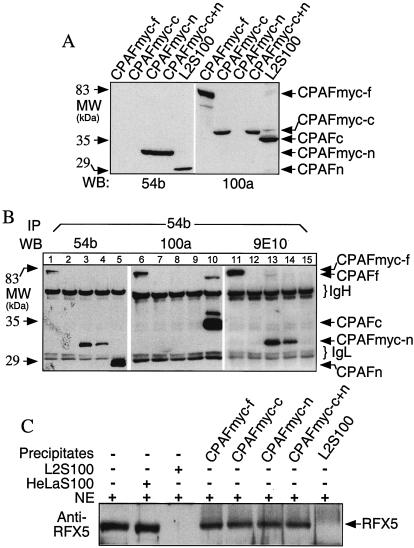
A transfection experiment was used to further confirm the necessity of CPAF dimers for CPAF activity. Three different constructs, full-length CPAF (residues 1 to 609), CPAFn (residues 1 to 283), and CPAFc (residues 284 to 609), were expressed in mammalian cells as fusion proteins with a myc epitope tag at the C terminus. The constructs were designated CPAFmyc-f, CPAFmyc-c, and CPAFmyc-n, respectively. Cells transfected with these constructs either individually or in combination properly expressed the corresponding protein products, as detected with MAbs 54b and 100a on a Western blot (Fig. 4A). Interestingly, the full-length CPAF was not processed when it was expressed via a transgene in mammalian cells. It is worth noting that both CPAFmyc-n and CPAFmyc-c were expressed in cells that were cotransfected with the two plasmids. To test whether coexpression of CPAFc and CPAFn allowed dimer formation, we used the CPAF N-terminus-specific antibody MAb 54b to immunoprecipitate the lysates of the transfectants and detected the CPAF fragments in the precipitates on a Western blot (Fig. 4B). Western blot detection with MAb 54b revealed that both full-length CPAFmyc-f and CPAFmyc-n were precipitated by MAb 54b from cells transfected with the corresponding plasmids, including the cells cotransfected with CPAFmyc-n and CPAFmyc-c (Fig. 4B, lanes 1 to 5). This result was confirmed with the anti-myc MAb 9E10 (Fig. 4B, lanes 11 to 15). As a positive control, MAb 54b successfully precipitated the endogenous CPAFn from the L2S100 sample. However, the CPAF C-terminus-specific antibody MAb 100a failed to detect any CPAFmyc-c in the MAb 54b pellet precipitated from the cotransfected cells, although MAb 100a detected the full-length CPAFmyc-f fusion protein and the endogenous CPAFf and CPAFc in the MAb 54b pellets precipitated from other samples. Since both CPAFmyc-n and CPAFmyc-c were expressed in the cotransfected cells and MAb 100a was able to recognize CPAFmyc-c on a Western blot, the fact that MAb 100a failed to detect any CPAFmyc-c in the MAb 54b pellet precipitated from the CPAFmyc-c- and CPAFmyc-n-cotransfected cell lysates demonstrated that the coexpressed CPAFmyc-n and CPAFmyc-c failed to form dimers. We next correlated dimer formation with proteolytic activity by comparing the abilities of the various MAb 54b precipitates to degrade RFX5 (Fig. 4C). Although the pellet precipitated by MAb 54b from L2S100 degraded RFX5, none of the other MAb 54b pellets precipitated from transfected cells did this, further confirming that CPAFc-CPAFn dimer formation is necessary for the degradation activity. We also tested other conditions for enhancing formation of CPAFc-CPAFn dimers, including transfection with or without tags or the putative CPAF leader in various types of vectors and cell-free systems. None of the conditions tested permitted CPAFc-CPAFn dimer formation or generated detectable proteolytic activity (data not shown). For example, neither dimers nor cleavage activities were found when purified CPAFc and CPAFn fragments were mixed and incubated under various conditions.
FIG. 4.
Transgene-encoded CPAF or CPAF fragments failed to form dimers and to degrade RFX5. 293T cells were transfected with plasmids coding for the full-length protein (CPAFmyc), the C terminus (CPAFmyc-c), or the N terminus (CPAFmyc-n) or were cotransfected with both the C terminus plasmid and the N terminus plasmid (CPAFmyc-c+n). (A) Cell lysates from the transfectants or chlamydia-infected cells (L2S100 was a positive control) were subjected to Western blot analysis with either the N-terminus-specific MAb 54b or the C-terminus-specific MAb 100a. (B) Alternatively, all the cell lysates were precipitated with MAb 54b, and the precipitated pellets were then subjected to Western blot analysis with MAb54b, 100a, or 9E10 (anti-myc tag). Lanes 1, 6, and 11, cells transfected with CPAFmyc-f plasmid; lanes 2, 7, and 12, cells transfected with CPAFmyc-c plasmid; lanes 3, 8, and 13, cells transfected with CPAFmyc-c plasmid; lanes 4, 9, and 14, cells transfected with CPAFmyc-c+n plasmids; lanes 5, 10, and 15, chlamydia-infected cells (L2S100). (C) The precipitates described above were also used to degrade RFX5 in a cell-free degradation assay. Molecular masses (MW) are indicated on the left, while the positions of protein bands are indicated on the right. IgH, immunoglobulin heavy chains; IgL, immunoglobulin light chains; IP, immunoprecipitation; WB, Western blot; NE, nuclear extracts (as the source of RFX5).
DISCUSSION
The identification of CPAF stimulated us to think about how an obligate intracellular bacterium can manipulate the host cell machinery to evade host defenses and improve its intracellular survival. Further elucidation of the molecular mechanisms of CPAF action should ultimately lead to creation of countermeasures for blocking chlamydial intracellular survival. Clearly, many important questions concerning the mechanisms of CPAF action remain to be addressed. For example, how is CPAF secreted from chlamydial vacuoles into the host cells? How is CPAF activity regulated? What is the structural basis for CPAF substrate specificity? In the present study we addressed whether intramolecular dimerization is required for CPAF activity. We obtained convincing evidence which showed that intramolecular dimers consisting of 35-kDa CPAFc fragments and 29-kDa CPAFn fragments are required for CPAF proteolytic activity. First, the 70-kDa proCPAF protein without processing is not enzymatically active and did not contribute to the CPAF activity exhibited by CPAFc-CPAFn dimers (Fig. 3). A transgene-encoded full-length CPAF in mammalian cells was not processed and failed to degrade RFX5 (Fig. 4). Second, neither CPAFc nor CPAFn fragments alone or in a mixture without formation of dimers has any RFX5 degradation activity. Antibodies that precipitated only CPAFn did not precipitate the RFX5 degradation activity (Fig. 2C and D). Neither the free CPAFc fragments nor the free CPAFn fragments released from CPAF dimers exhibited activity (Fig. 3). Transgene-encoded free CPAFc or CPAFn fragments in the absence of dimers did not degrade RFX5 (Fig. 4). Finally, CPAF dimers consisting of CPAFc and CPAFn are responsible for CPAF enzymatic activity. Antibodies able to pull down CPAFc and CPAFn dimers removed the enzymatic activity from the chlamydia-infected cell cytosol and moved it to the antibody pellets (Fig. 2C and D), while the activity disappeared when the dimers were dissociated (Fig. 3).
There are apparent advantages for CPAF to form intramolecular dimers for acquiring proteolytic activity. First, the process of intramolecular dimerization may provide the opportunity to regulate CPAF activity. Since CPAF is mainly used to regulate host cell functions, it may be beneficial for chlamydiae to synthesize CPAF as a zymogen and to make CPAF proteolytically active only when CPAF is secreted into the host cell cytosol. Efforts are under way to test this hypothesis. Second, dimers are usually more stable than single-chain molecules. Not only is the apparent stability of the active CPAF dimers important for chlamydiae to be able to constantly modify host cells during chlamydial intravacuolar persistence, but it also helps CPAF avoid being processed by host proteasomes in the cytosol. Finally, many bacterium-secreted protein toxins, some of which have proteolytic activities, can undergo posttranslational modification, including dimerization or oligomerization, to become active (2, 9, 13, 18). Although CPAF is not a toxin, chlamydiae may have preserved a parallel structure or function for interacting with host cells. Since chlamydiae have become obligate for intracellular growth and intoxication of host cells may no longer be beneficial for intracellular survival, the chlamydiae may have evolved factors such as CPAF to moderate their interactions with host cells, in contrast to bacteria that live in extracellular environments. Chlamydiae may want to minimize toxicity to host cells and protect the infected host cells from destruction by their own defense mechanisms, which is what CPAF can potentially do. Indeed, other intravacuolar bacteria, such as salmonellae, are able to secrete nonexotoxin factors from bacterium-laden vacuoles into the host cell cytosol for modulating host cell actin polymerization to increase bacterial survival (14).
Since neither CPAFc nor CPAFn alone exhibited proteolytic activity, it is difficult to predict which fragment bears the proteolytic domain and which fragment is responsible for substrate recognition. More sophisticated approaches, such as determination of the CPAF tertiary structure, may be required to fully map the functional domains of CPAF and to identify the optimal configuration for CPAF activity. It is reasonable to assume that further oligomerization of intramolecular dimers may further enhance the stability and proteolytic activity of CPAF. Protein folding and/or oligomerization is often an integrated part of protein translocation-secretion pathways. Currently, we are investigating whether CPAF intramolecular dimerization is a critical step in CPAF secretion from the chlamydial vacuole into the host cell cytosol. Determining the mechanisms of CPAF action and understanding CPAF intramolecular dimerization are obvious exciting aspects of the initial identification of CPAF. Clearly, many more questions about how CPAF is utilized by an obligate intracellular bacterial species to evade host defenses will be asked and addressed. An exploration of novel ways to determine the mechanisms of CPAF action on multiple fronts is under way.
Acknowledgments
We thank Joel Baseman and Luzhe Sun for reading the manuscript.
This work was supported in part by grants R01AI47997 and R01HL64883 from the National Institutes of Health to G. Zhong.
Editor: J. B. Bliska
REFERENCES
- 1.Askienazy-Elbhar, M., and J. H. Suchet. 1999. Persistent “silent” Chlamydia trachomatis female genital tract infections. Infect. Dis. Obstet. Gynecol. 7:31-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry, R., S. Moore, A. Alonso, J. Ausio, and J. T. Buckley. 2001. The channel-forming protein proaerolysin remains a dimer at low concentrations in solution. J. Biol. Chem. 276:551-554. [DOI] [PubMed] [Google Scholar]
- 3.Beatty, W. L., T. A. Belanger, A. A. Desai, R. P. Morrison, and G. I. Byrne. 1994. Role of tryptophan in gamma interferon-mediated chlamydial persistence. Ann. N. Y. Acad. Sci. 730:304-306. [DOI] [PubMed] [Google Scholar]
- 4.Beatty, W. L., R. P. Morrison, and G. I. Byrne. 1994. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 58:686-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobo, L. D., N. Novak, B. Munoz, Y. H. Hsieh, T. C. Quinn, and S. West. 1997. Severe disease in children with trachoma is associated with persistent Chlamydia trachomatis infection. J. Infect. Dis. 176:1524-1530. [DOI] [PubMed] [Google Scholar]
- 6.Dean, D., and V. C. Powers. 2001. Persistent Chlamydia trachomatis infections resist apoptotic stimuli. Infect. Immun. 69:2442-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan, P., F. Dong, Y. Huang, and G. Zhong. 2002. Chlamydia pneumoniae secretion of a protease-like activity factor for degrading host cell transcription factors is required for major histocompatibility complex antigen expression. Infect. Immun. 70:345-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan, T., H. Lu, H. Hu, L. Shi, G. A. McClarty, D. M. Nance, A. H. Greenberg, and G. Zhong. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert, R. J., J. L. Jimenez, S. Chen, I. J. Tickle, J. Rossjohn, M. Parker, P. W. Andrew, and H. R. Saibil. 1999. Two structural transitions in membrane pore formation by pneumolysin, the pore-forming toxin of Streptococcus pneumoniae. Cell 97:647-655. [DOI] [PubMed] [Google Scholar]
- 10.Grayston, J. T., and S. Wang. 1975. New knowledge of chlamydiae and the diseases they cause. J. Infect. Dis. 132:87-105. [DOI] [PubMed] [Google Scholar]
- 11.Hackstadt, T., E. R. Fischer, M. A. Scidmore, D. D. Rockey, and R. A. Heinzen. 1997. Origins and functions of the chlamydial inclusion. Trends Microbiol. 5:288-293. [DOI] [PubMed] [Google Scholar]
- 12.Heuer, D., V. Brinkmann, T. F. Meyer, and A. J. Szczepek. 2003. Expression and translocation of chlamydial protease during acute and persistent infection of the epithelial HEp-2 cells with Chlamydophila (Chlamydia) pneumoniae. Cell Microbiol. 5:315-322. [DOI] [PubMed] [Google Scholar]
- 13.Malghani, M. S., Y. Fang, S. Cheley, H. Bayley, and J. Yang. 1999. Heptameric structures of two alpha-hemolysin mutants imaged with in situ atomic force microscopy. Microsc. Res. Tech. 44:353-356. [DOI] [PubMed] [Google Scholar]
- 14.Miao, E. A., M. Brittnacher, A. Haraga, R. L. Jeng, M. D. Welch, and S. I. Miller. 2003. Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol. Microbiol. 48:401-415. [DOI] [PubMed] [Google Scholar]
- 15.Rajalingam, K., H. Al-Younes, A. Muller, T. F. Meyer, A. J. Szczepek, and T. Rudel. 2001. Epithelial cells infected with Chlamydophila pneumoniae (Chlamydia pneumoniae) are resistant to apoptosis. Infect. Immun. 69:7880-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen, S. J., L. Eckmann, A. J. Quayle, L. Shen, Y. X. Zhang, D. J. Anderson, J. Fierer, R. S. Stephens, and M. F. Kagnoff. 1997. Secretion of proinflammatory cytokines by epithelial cells in response to chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J. Clin. Investig. 99:77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw, A. C., B. B. Vandahl, M. R. Larsen, P. Roepstorff, K. Gevaert, J. Vandekerckhove, G. Christiansen, and S. Birkelund. 2002. Characterization of a secreted chlamydia protease. Cell Microbiol. 4:411-424. [DOI] [PubMed] [Google Scholar]
- 18.Stein, P. E., A. Boodhoo, G. D. Armstrong, S. A. Cockle, M. H. Klein, and R. J. Read. 1994. The crystal structure of pertussis toxin. Structure. 2:45-57. [DOI] [PubMed] [Google Scholar]
- 19.Stephens, R. S. 2003. The cellular paradigm of chlamydial pathogenesis. Trends Microbiol. 11:44-51. [DOI] [PubMed] [Google Scholar]
- 20.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 21.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 22.Zhong, G., J. Berry, and R. C. Brunham. 1994. Antibody recognition of a neutralization epitope on the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 62:1576-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong, G., and R. C. Brunham. 1992. Antibody responses to the chlamydial heat shock proteins hsp60 and hsp70 are H-2 linked. Infect. Immun. 60:3143-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong, G., F. Castellino, P. Romagnoli, and R. N. Germain. 1996. Evidence that binding site occupancy is necessary and sufficient for effective major histocompatibility complex (MHC) class II transport through the secretory pathway redefines the primary function of class II-associated invariant chain peptides (CLIP). J. Exp. Med. 184:2061-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong, G., P. Fan, H. Ji, F. Dong, and Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193:935-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong, G., T. Fan, and L. Liu. 1999. Chlamydia inhibits interferon gamma-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J. Exp. Med. 189:1931-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong, G., L. Liu, T. Fan, P. Fan, and H. Ji. 2000. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in chlamydia-infected cells. J. Exp. Med. 191:1525-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong, G., C. Reis e Sousa, and R. N. Germain. 1997. Production, specificity, and functionality of monoclonal antibodies to specific peptide-major histocompatibility complex class II complexes formed by processing of exogenous protein. Proc. Natl. Acad. Sci. USA 94:13856-13861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong, G., G. P. Smith, J. Berry, and R. C. Brunham. 1994. Conformational mimicry of a chlamydial neutralization epitope on filamentous phage. J. Biol. Chem. 269:24183-24188. [PubMed] [Google Scholar]
- 30.Zhong, G., I. Toth, R. Reid, and R. C. Brunham. 1993. Immunogenicity evaluation of a lipidic amino acid-based synthetic peptide vaccine for Chlamydia trachomatis. J. Immunol. 151:3728-3736. [PubMed] [Google Scholar]
- 31.Zhong, G. M., and R. C. Brunham. 1991. Antigenic determinants of the chlamydial major outer membrane protein resolved at a single amino acid level. Infect. Immun. 59:1141-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]