Summary
Prolonged stimulation leads to specific and stable changes in an animal’s behavior. In interneurons, this plasticity requires spatial and temporal control of neuronal protein synthesis. Whether such translational control occurs in sensory neurons is not known. Adaptation of the AWC olfactory sensory neurons of C. elegans requires the cGMP-dependent protein kinase EGL-4. Here we show that the PUF FBF-1 is required in the adult AWC for adaptation and in the odor-adapted animal, increases translation from the egl-4 3’ UTR. Further, the PUF protein may localize translation near the sensory cilia and cell body. Although the RNA-binding PUF proteins have been shown to promote plasticity in development by temporally and spatially repressing translation; this work reveals that in the adult nervous system, they can work in a different way to promote experience-dependent plasticity by activating translation in response to environmental stimulation.
Introduction
Complex behaviors require accurate and efficient sensing of the environment as well as the ability to ignore persistent stimuli that offer little new information. Since sensory neurons provide the information from which most behaviors arise, the ability of these neurons to modulate their excitability, as a function of experience is critical. While brief stimulation elicits rapidly reversible adjustments in excitability that allow the animal to track changes in its environment, prolonged stimulation elicits more enduring homeostatic changes that allow the animal to reset its sensitivity to long-lasting alterations such as day-to-night changes in light intensity (Calvert et al., 2006) or the presence of a persistent odor (Mashukova et al., 2006; Zufall and Leinders-Zufall, 1997). Repeated experiences also elicit changes in behavior that require alterations in the sensitivity of interneurons within neural circuits. Repeated stimulation or prolonged alteration in the pattern of stimulation of specific connections can lead to Long-Term Potentiation (LTP), Long-Term Depression (LTD) or homeostatic synaptic scaling within the synaptic connections between hippocampal cells. These changes have been shown to underlie the animal’s ability to form memories as a function of repeated experience (see Sutton and Schuman, 2005; Sutton and Schuman, 2006 for review).
The nematode C. elegans also adjusts its behavior as a function of experience. Though some odors are inherently attractive to C. elegans, they will begin to ignore such odors after being exposed to them for a prolonged period of time (Bargmann et al., 1993). C. elegans has been shown to respond to over 60 different volatile chemical compounds, many of which stimulate the paired AWA and AWC olfactory sensory neurons which mediate attraction (Bargmann et al., 1993). Remarkably, though each AWC can probably sense tens of odors, sensitivity to individual odors can be decreased independently (Colbert and Bargmann, 1995). Sensitivity to an odor is regulated by the length of prior exposure to that odor (Colbert and Bargmann, 1995; L’Etoile et al., 2002). While initial exposure to an odor gradient elicits chemotaxis to the point source, a 30-minute pre-exposure to the same odor will reversibly decrease chemotaxis (short-term adaptation) and exposures of more than one hour will irreversibly decrease odor-seeking for hours (long-term adaptation) (Colbert and Bargmann 1995). Odor-sensation is thought to occur within the sensory cilia of the AWC neuron where the odor responsive G-protein coupled receptors reside (Sengupta et al., 1996; Troemel et al., 1995). Odor sensation requires the G-alphas ODR-3 and GPA-2 (Roayaie et al., 1998; Lans et al., 2004) and is likely to decrease the amounts of cGMP produced by the membrane guanylyl cyclases DAF-11 and ODR-1 (Birnby et al., 2000; L’Etoile and Bargmann, 2000). Lowered cGMP levels then close the cyclic nucleotide-gated sensory channel TAX-2/4 (Chalasani et al., 2007).
How do these sensory neurons alter their responsiveness as a function of prolonged or repeated experience? Retinal cells within the visual system shuttle previously synthesized adaptation-promoting proteins into the sensory compartment to dampen the incoming signal. Prolonged exposure of rods to bright light induces massive translocation of arrestin from the inner segment to the outer segment where arrestin binds to the GPCR rhodopsin and limits the light-induced signaling (Calvert et al., 2006). Mammalian olfactory neurons adjust their sensitivity by relocating previously synthesized proteins away from possible stimulation: prolonged odor-exposure induces clathrin-mediated endocytosis to remove the olfactory receptor GPCRs from the cell surface (Mashukova et al., 2006). Interneurons, however, use a different strategy to adjust to repeated stimulation: they synthesize new proteins such as CamKII (Miller et al., 2002) and Arc/Arg3.1 (Moga et al., 2004) or sensorin (Lyles et al., 2006) within the vicinity of the stimulated synapse. The net effect of these proteins is to alter channel composition (Chowdhury et al., 2006) or dendritic morphology (Lyles et al., 2006) at the stimulated synapse and thereby adjust the strength of the synaptic connection as a function of experience. Though translational control mechanisms are central to the function of interneurons it is not known whether sensory neurons employ a similar strategy.
In the C. elegans AWC sensory neurons both short-term and long-term adaptation require the cGMP-dependent protein kinase EGL-4 (L’Etoile et al., 2002). Surprisingly, mutations within the egl-4 3’UTR interfere with adaptation (L’Etoile et al., 2002). We found that these residues are part of a highly conserved translation-repressing Nanos Response Element (NRE) (Murata and Wharton, 1995). Thus, translational control within in sensory neurons is crucial for appropriate behavior in the adult animal.
NREs bind the Pumilio/Fem-3 Binding Factor (PUF) family of translational repressors. The founding member, Pumilio, first discovered in Drosophila, was shown to establish the anterior-posterior embryonic axis during development by binding to and repressing translation of maternal hunchback mRNA (Lehmann and Nusslein-Volhard, 1991; Wharton and Struhl, 1991). PUFs have been characterized as translational repressors that seem to work either by reducing translation of the target message or increasing its decay (Wickens et al., 2002). Biochemical evidence suggests that yeast, mammalian, and C. elegans PUFs may accomplish this by interacting with deadenylating and decapping protein complexex; (Goldstrohm et al., 2006; Goldstrohm et al., 2007). However, in oocyte maturation, PUF binding elements in combination with cytoplasmic polyadeylation sites have been shown to increase translation of their targets (Pique et al., 2008) while they seem to repress translation in combination with microRNA binding sites (Nolde et al., 2007). It is not known how Pumilios either by themselves or in combination with other factors act in the context of an adult neuron.
In other systems, PUFs have been shown to focus translation spatially and temporally: in yeast Mtp5p limits HO translation to the bud site (Tadauchi et al., 2001), and in C. elegans, FBF-1/2 restrict the expression of the sperm-promoting factor FEM-3 to the larval period when spermatocytes are produced (Zhang et al., 1997). The ability of PUFs to focus translation in a spatiotemporal fashion makes them attractive candidates for promoting plasticity within neurons. Indeed, in neurons within the developing Drosophila larva, Pumilio’s ability to repress translation is required for regulation of neuronal membrane excitability (Mee et al., 2004); and appropriate synaptic development and plasticity (Menon et al., 2004); as well as proper dendritic branching (Ye et al., 2004). In the adult fly, dPUM is required for olfactory associative learning (Dubnau et al., 2003) and to limit motor neuron excitability (Schweers et al., 2002). In adult mice, PUM2 is found in the neurons of the hippocampus (Rouhana et al., 2005). However, the mechanisms by which PUFs function and the targets they regulate in the adult nervous system are not known.
Here we show that both the PUF binding site within the egl-4 3’UTR and the PUF FBF-1 are required at the time of odor-exposure to promote adaptation to the odors butanone and isoamyl alcohol. The ability of the egl-4 3’ UTR to bind FBF-1 is required both for adaptation and increased expression from the egl-4 3’UTR-dependent Kaede and luciferase transcriptional reporters. Expression of the Kaede reporter in the intact animal reveals that FBF-1 acts via the PUF binding site to perhaps increase translation near the AWC sensory cilia and cell body. The luciferase reporter shows that odor exposure induces a dynamic increase in translation that is dependent upon the NR/FBE. Thus, we describe here a novel function for a PUF binding element. Instead of repressing translation, as was seen in developing tissues, we find that in the adult functioning sensory neurons FBF-1 activates translation of its target in response to stimulation. The mechanism of activation requires a specific sequence within and surrounding the binding site to direct FBF-1 to increase translation and it does this without affecting message levels.
Results
The egl-4 3’ UTR contains a novel NRE that is required for olfactory adaptation
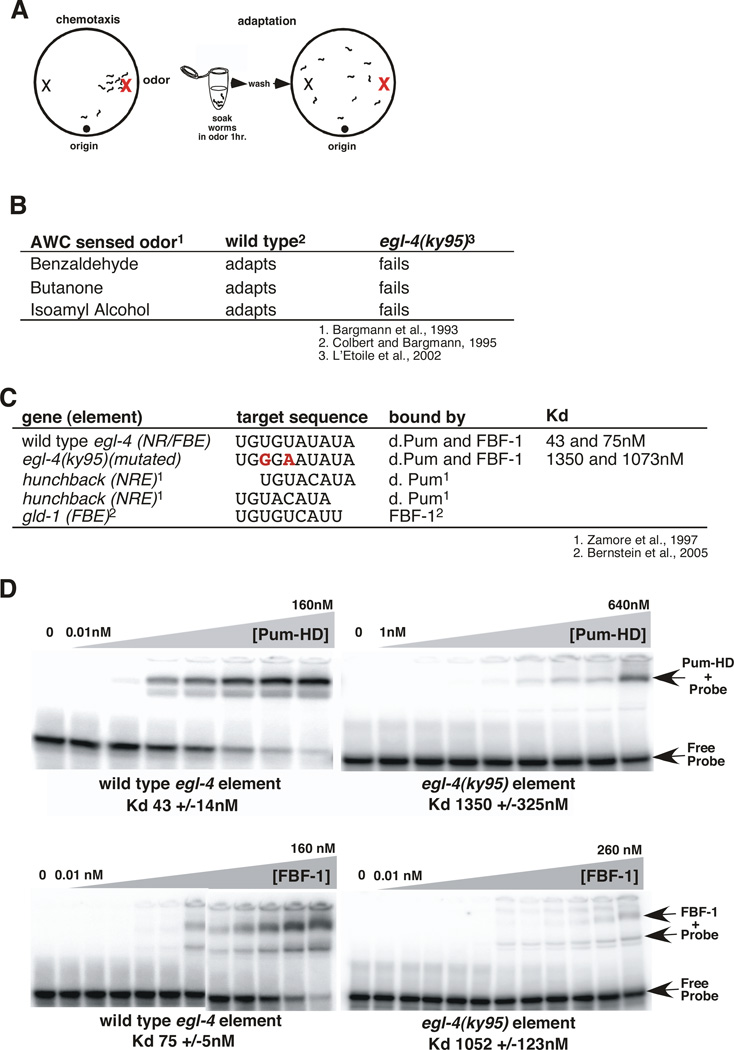
Olfactory adaptation is defined as an animal’s reduced attraction to an odor resulting from prolonged exposure to that odor (Figure 1A; Colbert and Bargmann, 1995). A genetic screen to isolate key players in the adaptation process identified the mutant, egl-4(ky95) (L'Etoile et al., 2002). The egl-4(ky95) mutant strain was defective for adaptation to all AWC-sensed odors we tested (Figure 1B; L'Etoile et al., 2002). Interestingly, the coding region of this allele of egl-4 was intact, instead we found two point mutations in its 3’UTR (L'Etoile et al., 2002) that disrupted a putative NRE (Figure 1C). This indicated that regulation of EGL-4 translation might be important for its function. Points of regulation could include: basal levels of EGL-4, its spatial distribution, or its dynamic expression in response to prolonged odor-stimulation.
Figure 1. The egl-4 3' UTR contains a composite Pumilio and FBF binding site (NR/FBE) required for olfactory adaptation.
(A) Adaptation paradigm. In the chemotaxis assay (left), naïve worms placed on an agar lined Petri dish leave their initial location (origin) to seek an attractive odor source (red X on right side of dish). In the adaptation assay (right), worms have been exposed to the odor diluted into buffer for an hour and they no longer seek its source.
(B) Adaptation to all three AWC sensed odors was impaired by two mutations within the egl-4 3'UTR; egl-4 (ky95) (L'Etoile et al., 2002).
(C) Sequence alignment of Pumilio and FBF-1 binding sites with the egl-4 NR/FBE. Target NREs and FBEs are aligned starting with the UGU and the bases mutated in ky95 are shown in bold red. Both alignments of the egl-4 NR/FBE with the hunchback NRE are shown.
(D) Gel mobility shift assays of the putative response element within wild type (left top and bottom panels) and ky95 mutant (right, top and bottom) 3'UTRs by either purified GST-drosophila Pumilio (PUM-HD) (top panels) or purified GST-FBF-1 (bottom panels). The free probe and shifted complexes are indicated with arrows. 100 fMoles of 32P labeled 30mer RNA probes were incubated with increasing amounts of protein for two hours before being gel resolved. Concentrations of protein incubated with the wild-type element were: 0.01 to 160 nM in 2 fold steps per lane. Concentrations of protein incubated with the ky95 element were 0.01, 0.05, 0.1, 0.5, and 1 nM, then doubled beginning at 5 nM per lane; the 0.01, 0.05, and 0.1 points were omitted for the ky95 probe. The Kds below each gel were generated from three separate gel-shift assays using non-linear regression analysis (GraphPadPrism™) software to fit the % probe bound to sigmoidal dose response (variable slope) curves. Error bars indicate the s.e.m. from at least 3 separate gel shifts.
The egl-4 NRE can bind both FBF-1 and Pumilio proteins
To understand how EGL-4 expression might be regulated, we examined the egl-4 NRE. NREs are defined by their ability to bind the RNA binding Pumilio class of translational repressors (Zhang et al., 1997). The Drosophila genome encodes a single PUF dPum while the C. elegans genome encodes 11 predicted PUFs (Wickens et al., 2002) along with the newly predicted puf-12 (http://www.wormbase.org/db/gene/gene?name=WBGene00014165;class=Gene). The C. elegans PUF family tree has two branches; PUF-8 and PUF-9 are closest to dPUM and the mammalian PUFs, while the other members are on a separate branch (Wickens et al., 2002). Members of each branch bind to distinct RNA sequence elements: dPUM and PUF-8/9 bind an NRE while FBF-1 and FBF-2, two members of the other branch, bind a fem-3 binding element (FBE) (Opperman et al., 2005). Bernstein et al. (2005) showed that dPUM cannot bind an FBE site conversely; FBF-1 does not interact with the hunchback NRE. NREs share sequence similarity with FBEs: both consist of a highly conserved UGUR sequence followed by either one (for the NRE) or two (for the FBE) less conserved nucleotides and then AU (Opperman et al., 2005). The element within the egl-4 3’UTR can be aligned with the hunchback NRE (Zamore et al., 1997) in two registers as well as the FBF-binding gld-1 FBE (Crittenden et al., 2002) (Figure 1C). To determine whether this element was an NRE or FBE, we assessed its ability to bind either GST-tagged protein in a gel mobility shift assay. Both recombinant Drosophila Pumilio (GST-PUM-HD) (Zamore et al., 1997) and C. elegans FBF-1 bound and shifted the electrophoretic mobility of a synthetic 30 nucleotide RNA oligomer containing the wild type egl-4 element (Figure 1D, left). The affinity of drosophila Pumilio for the egl-4 element (Kd=43nM) was comparable to its reported affinity for the canonical NRE (Kd=25nM) (Zamore et al., 1997). Likewise, the affinity of FBF-1 for the egl-4 NRE element (Kd=75nM) was also close to its reported affinity for the gld-1 FBF (Kd = 23 nM) (Bernstein et al., 2005). Neither protein bound appreciably to the corresponding sequence from the mutant ky95 allele (Figure 1D, right). As the egl-4 element bound either the Drosophila Pumilio or the C. elegans FBF-1 it is a verified but novel NRE, which we will refer to as an NR/FBE.
The PUF FBF-1 is required in AWC for adaptation to butanone and isoamyl alcohol
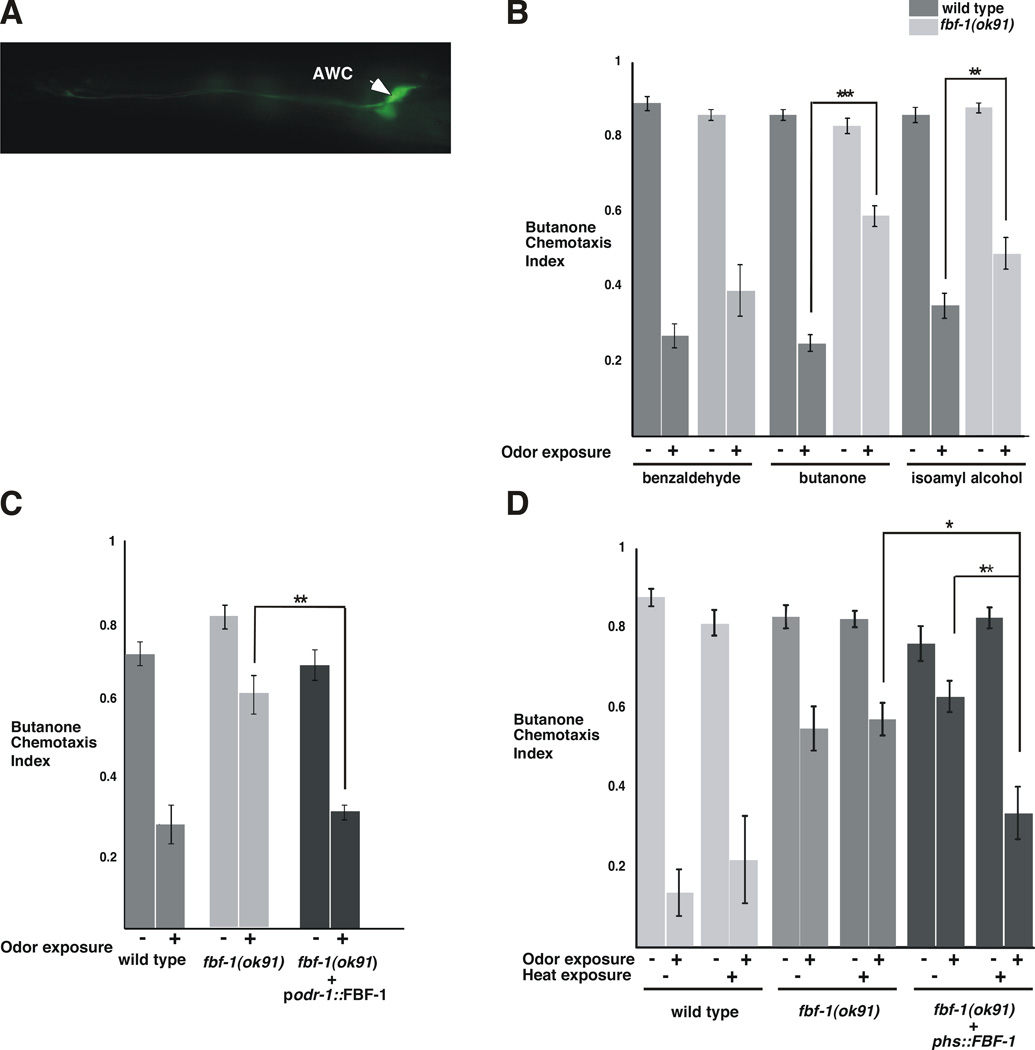
To determine whether FBF-1 is expressed in the same neuron that EGL-4 acts to promote adaptation (L’Etoile et al., 2002), we placed the GFP reporter under the control of sequences upstream of the start site for FBF-1 transcription. GFP was expressed in both AWC neurons (Figure 2A), suggesting that FBF-1 is normally expressed in the same neurons as its proposed target, EGL-4.
Figure 2. FBF-1 is required in the AWC sensory neuron at the time of odor exposure to promote adaptation of the AWC neuron to butanone.
(A) FBF-1 is expressed in the AWC neurons. Fluorescence micrograph of a worm expressing GFP under the control of 2kB of the fbf-1 promoter region (pfbf-1). Fluorescence was observed in both AWC neurons (marked with an arrowhead), both AWB neurons (not marked), and one unidentified head neuron (not in this plane of focus).
(B) While fbf-1(ok91) null mutant worms were unable to adapt to butanone (***P <0.0001 n=36), and were defective for adaptation to isoamyl alcohol (**P=0.0024 n= 19), they were fully capable of adapting to benzaldehyde (P=0.1758 n=13).
(C) Full-length FBF-1 expressed in AWC and AWB under the odr-1 promoter (L'Etoile and Bargmann, 2000) allowed fbf-1(ok91) mutants to adapt to butanone like wild-type (P=0.6006 n= 4). This was significantly different from the starting fbf-1(ok91) strain (P= 0.0075 n= 4).
(D) Heat shock expression of FBF-1 at the time of odor exposure restored adaptation to fbf-1(ok91) worms. Prior to heat shock, fbf-1(ok91) mutants carrying FBF-1 under the control of the heat shock promoter were as defective for adaptation as the fbf-1 null worms alone (compare 3rd and 5th pair of bars). When the transgenic strain were exposed to ~30° C for 60 minutes, two hours prior to odor exposure, they adapted like wild-type animals (compare 2nd and 6th pair of bars). This adaptation was significantly different from the same strain before heat shock (** P=0.0084) and the heat shocked fbf-1(ok91) (*P=0.0264 n=7). All assays were performed with >50 animals per assay on separate days. P values were obtained using a two-tailed test. Error bars denote s.e.m.
We then used our adaptation paradigm (Figure 1A) (Colbert and Bargmann, 1995) to determine whether FBF-1 is required for adaptation to AWC-sensed odors. The fbf-1(ok91)null strain was proficient at sensing the odors isoamyl alcohol, benzaldehyde and butanone (Figure 2B and Figure S1), but failed to adapt to butanone and was compromised for adaptation to isoamyl alcohol (Figure 2B). This defect was somewhat odor-specific since it was capable of benzaldehyde adaptation (Figure 2C).
Given that EGL-4 acts cell autonomously within the AWCs to promote adaptation (L'Etoile et al., 2002), we asked if FBF-1 does the same. When fbf-1 mutants expressed FBF-1 solely in the pair of AWCs and one other neuron pair (AWBs), they were able to adapt to butanone as well as wild type (Figure 2C). Thus, FBF-1 acts in the same neuron as its proposed translational target, EGL-4 (L’Etoile et al., 2002).
FBF-1 is required during odor-exposure for adaptation
PUF proteins are required for a variety of developmental decisions (Wickens et al., 2002). To determine if FBF-1 mediated translational control is required for development of the AWC neuron or during odor-exposure, we placed the FBF-1 cDNA under the control of the heat shock promoter (Stringham et al., 1992) and expressed this construct in the fbf-1(ok91)strain. When the resulting strain was exposed to heat two hours prior to odor exposure, it adapted as well as wild-type (Figure 2D, dark grey). Heat had no effect on the behavior of the parental fbf-1(ok91) strain (Figure 2D, medium grey). This indicates that FBF-1 is required in the adult neuron, at the time of odor exposure, for adaptation. Furthermore, AWC neuronal cell fate, as assessed by str-2 promoter expression (Troemel et al., 1999), was not altered in fbf-1 defective animals (Table S1).
The Nanos protein, NOS-1 and the poly (A) polymerase GLD-3 are required for adaptation to a wide range of odors
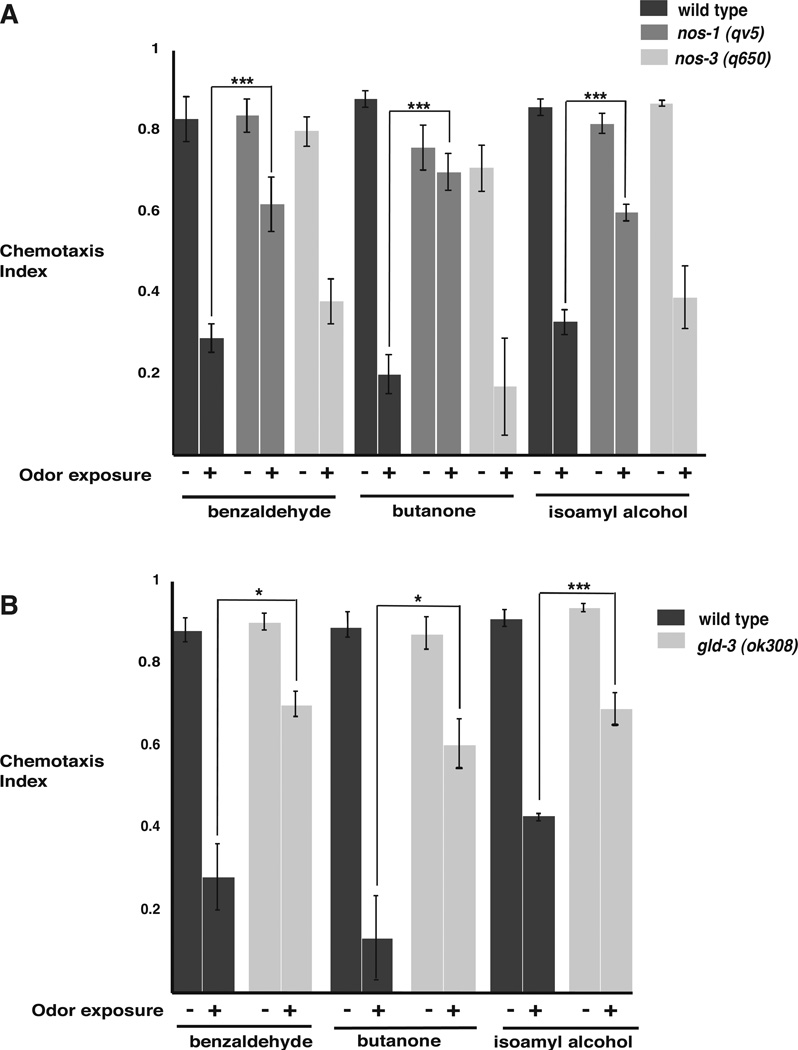
To determine whether proteins that interact with PUFs are required for olfactory adaptation, we examined olfactory adaptation in mutants that lack NANOS. nanos acts with pumilio to repress the Drosophila NRE-containing hunchback message (Wharton and Struhl, 1991). Three nanos-related genes in C. elegans play partially redundant roles in gonad development (Kraemer et al., 1999). Of these, NOS-1 was required for adaptation to all AWC-sensed odors tested (Figure 3A). NOS-3, however, was not required for adaptation to any odor tested (Figure 3A). Thus, though both PUF and NOS proteins were both required to promote adaptation, the nos-1 mutants were defective for adaptation to a wider range of odors than the fbf-1(ok91) null mutants.
Figure 3. The NANOS related molecule NOS-1 and the poly(A)polymerase subunit, GLD-3 are required to promote adaptation to AWC-sensed odors.
(A) NOS-1 deficient, nos-1 (gv5) worms failed to adapt to the AWC sensed odors: benzaldehyde (P***= 0.0002 n=9); butanone (P***<0.0001 n= 9); and isoamyl alcohol (P***<0.0001 n=12). In contrast, NOS-3 defective worms (nos-3 (q650)) adapted to all three odors.
(B) Animals that lack the poly (A) polymerase subunit gld-3(ok308) were unable to adapt to the AWC-sensed odors: benzaldehyde (*P= 0.0117 n=5); butanone (*P=0.0127 n=6); and isoamyl alcohol (***P= 0.0007 n=8). Assays were performed on separate days with >50 animals per assay. P values were obtained using a two-tailed test. Error bars denote s.e.m.
Further genetic evidence that translational regulation is required for adaptation came from our observations of mutants that lack the cytoplasmic poly(A) polymerase GLD-3 (Eckmann et al., 2002; Wang et al., 2002). The poly(A) polymerase GLD-2/3 has been proposed to relieve PUF-mediated repression by increasing poly(A) tail length, thereby increasing message stability and/or translation (Eckmann et al., 2002; Suh et al., 2006). We found that the poly(A) polymerase subunit-defective gld-3(ok308) strain failed to adapt to any AWC-sensed odor we tested (Figure 3B). Thus, adaptation may require increased translation and/or RNA stability. Since the egl-4 3’UTR does not contain a cytoplasmic poly-Adenylation element (CPE), egl-4 may be regulated in a novel way by GLD-2/3 and/or, other components of the adaptation machinery may be targeted by GLD-2/3.
Genetic evidence that the egl-4 NRE positively regulates EGL-4
The NR/FBE-mutated egl-4(ky95), the fbf-1 null, the nos-1 null and the gld-3 null strains all fail to adapt to odor exposure. Thus, they may all work in the same direction: to allow EGL-4 to promote adaptation. Since the n479 loss-of-function allele of egl-4 is defective for adaptation (L’Etoile et al., 2002), this suggests that mutations in either the fbf-1 pathway or the NR/FBE decrease levels of EGL-4 relative to wild type. By contrast, previous reports showed that disruption of either puf genes or their targets actually increased the targeted gene’s expression (Gu et al., 2004; Crittenden et al, 2002).
Cell biological evidence that PUF binding may direct the subcellular localization of EGL-4 translation
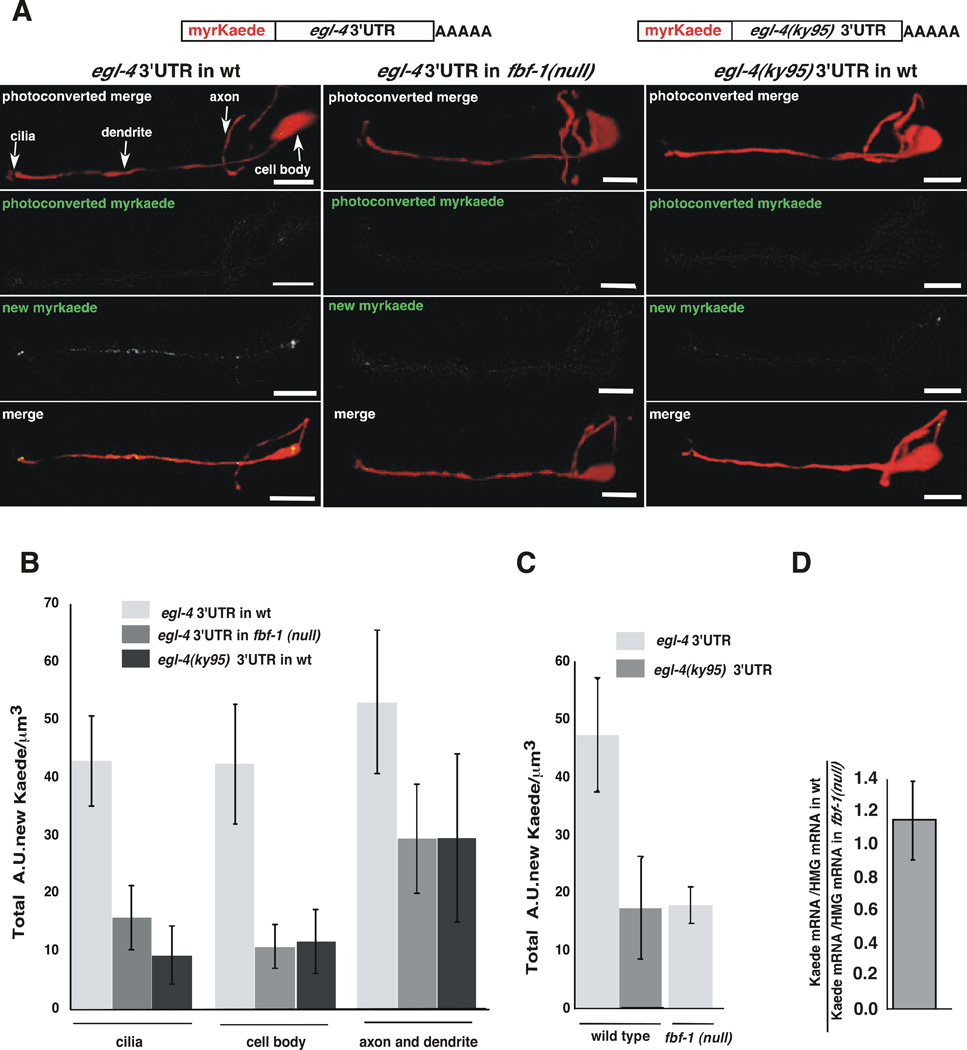
To determine if the NR/FBE control element and FBF-1 regulates where EGL-4 is translated within the AWC of a living animal, we utilized the photoconvertable stony coral protein Kaede (Ando et al., 2002) as a reporter for new protein synthesis. Kaede fluorescence is irreversibly changed from green to red by exposure to U.V. light (Ando et al., 2002). After photoconversion of Kaede to red, only newly synthesized protein will fluoresce green. Kaede coding sequences were placed upstream of either the wild type or ky95-mutated egl-4 3’UTR (Figure 4A). To decrease the potential for diffusion of the newly synthesized Kaede, a myristoylation signal (Adler et al., 2006) was added to it. Myristoylation directs the newly synthesized protein to the cell membrane thus increasing our chance of determining the site of new protein synthesis. Since the coding region of each reporter is identical, changes in the location of the reporter protein should only result from differences in mRNA processing and trafficking. These reporters were expressed from transgenic arrays in either wild-type or fbf-1(ok91) mutant animals. Each line had the same number of reporter transgenes as determined by quantitative real time PCR (Figure S2). Furthermore, the wild-type reporter was examined in populations of siblings that contained the same array but had either wild-type or fbf-1(ok91) genotype.
Figure 4. The egl-4 NR/FBE promotes translation within the AWC neuron.
(A) Top, cartoons of myristoylated Kaede expression reporters under the control of the str-2 promoter were flanked with the wild type egl-4 3’UTR or the egl-4(ky95) 3’UTR. These were expressed in either wild type (left and right columns of images) or fbf-1(ok91) animals (middle column). pstr-2::RFP (red) delimits the AWC neuron (top and bottom rows of images). The parts of the AWC neuron are marked with arrows (upper left). The top row of panels: red and green channels immediately after photoconversion. The second row panels: residual unconverted Kaede in the green channel immediately after photoconversion. The third row of panels: the green channel reveals the subcellular location of accumulated Kaede of the same animal after 3 hours of recovery on food. The fourth row of panels: the merged red and green channels of the same animal.
(B) Kaede synthesis near the sensory cilia and cell body depends on the 3’UTR and FBF-1. Newly synthesized Kaede under the control of the wild type 3’UTR expressed in the fbf-1(ok91) mutant animals was 2.7 fold lower in the cilia (*P=0.0233 n=5) and 3.9 fold lower in the cell body (*P=0.028) than when the same reporter transgene is expressed in wild type animals. In wild type animals expressing Kaede under the control of the egl-4(ky95) 3’UTR, newly synthesized Kaede was 4.6 fold lower in the cilia (**P=0.0032 n=6) and 3.6 fold lower in the cell body (*P=0.0210 n≥6) than in wild type animals expressing the reporter under the control of the wild-type 3’UTR. In the axon and dendrite, the Kaede expression was not significantly different in animals that bear the ky95 mutation (p=0.25) or in animals that lack fbf-1 (p=0.17). The copy number of the wildtype and ky95 3’UTR reporter was the same in each line (Supplemental Materials and Figure S3). P values were obtained using a two-tailed unpaired T test. Error bars denote s.e.m.
(C) AWC-wide Kaede expression was determined by integrating Kaede levels over the whole cell using the images analyzed in (B). Over-all Kaede levels were 2.6 fold higher in wild type worms expressing Kaede flanked with the wild-type egl-4 3’UTR than in fbf-1(ok91) mutants expressing the same transgene (*P=0.0165 n=6). Kaede recovery was 2.8 fold higher in wild type worms expressing Kaede flanked with the wild-type egl-4 3’UTR than in wild type worms expressing Kaede with the ky95 3’UTR (*P= 0.0443 n= 6). Newly synthesized Kaede was quantified from confocal images using Volocity software (see Supplemental Materials). (D) Kaede mRNA levels are the same in both wild-type 3’UTR and fbf-1(ok91) mutant worms. Kaede mRNA levels did not differ significantly between these strains (P= 0.416). Real time PCR was performed on cDNA made from worms of each genotype. Units of Kaede mRNA/unit of HMG CoA mRNA were determined for each and the ratio of this value is shown.
In the wild type animal, newly synthesized Kaede under the control of the wild-type egl-4 3’UTR was observed in the sensory cilia as well as throughout the AWC neuron (Figure 4A,B). When we compared the subcellular distribution of newly synthesized Kaede under the control of the ky95 mutant 3’UTR with that of the wild-type 3’UTR, we found that its accumulation in the cilia and in the cell body was 4.6 and 3.6 fold lower respectively while accumulation in either dendrites or axons was not significantly different (Figure 4A compare left to right panels, 4B compare light to dark grey bars). Likewise, when we compared expression from the wild-type 3’UTR reporter in wild types with its expression in fbf-1(ok91) mutants, we found that accumulation of newly synthesized Kaede was significantly lower within the cilia and cell body of the mutants (2.7 and 3.9 fold lower respectively) while accumulation in the axons and dendrites was not significantly different (Figure 4B). These results, however, have enough statistical variation to warrant caution. The fbf-1 mutant background could possibly affect protein transport but it is unlikely that the change in the 3’UTR of the reporter would affect protein trafficking. Thus, accumulation of Kaede in the cell body and cilia probably requires an intact NR/FBE and FBF-1.
Cell biological evidence that FBF-1 enhances EGL-4 translation
When we summed expression of the Kaede reporter throughout the cell, we found that the NR/FBE mutations decreased overall reporter expression appreciably. In wild type animals, Kaede recovery was 2.8 fold higher when the reporter contained the wild type NR/FBE than when it contained the mutant ky95 sequences (Figure 4C). Likewise, loss of the FBF-1 protein reduced translation of the wild-type reporter to the levels seen with the NR/FBE mutant reporter (2.6 fold lower) (Figure 4C). This indicates that FBF-1 acts via binding to the NRE/FBE within the egl-4 3’UTR to keep levels of EGL-4 high presumably to allow for olfactory adaptation. This positive regulation of translation by a PUF and its target-binding site is in contrast to previous reports.
The increased translation we observed could be explained if FBF-1 increases either translation rates or message levels. We asked whether the loss of FBF-1 decreased our reporter’s message levels. Using quantitative real time Reverse Transcriptase-mediated PCR (qRTPCR), we found that the ratio between the reporter’s message level in wild-type as compared to fbf-1(null) mutant animals was ~1.1 (Figure 4D). Thus, the message levels are not appreciably changed by the loss of FBF-1 and the FBF-1-dependent increase in reporter expression is likely a function of an increase in translation rather than in message levels.
The NR/FBE mediates odor-induced translational activation
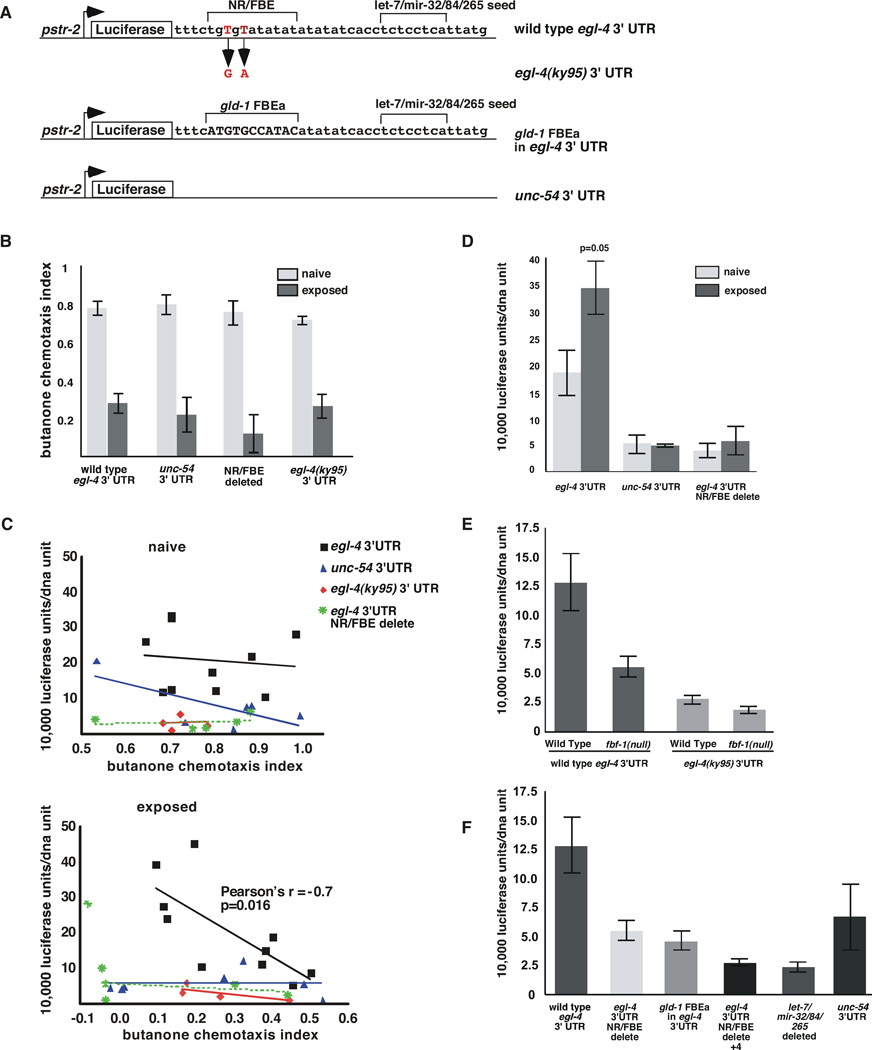
To determine if egl-4’s 3’UTR provides dynamic odor-induced regulation of EGL-4 translation, we developed a luciferase reporter that would allow us to monitor rapid changes in expression. We placed a destabilized luciferase (30’ half-life) (Promega) under the control of an AWCON-specific promoter (pstr-2) and flanked it with the egl-4 5’ and 3’UTRs. To assess the role of the 3’UTR in odor-responsiveness, we altered this reporter by replacing the egl-4 3’UTR with the 3’UTR of the muscle cell-specific myosin unc-54 gene. To elucidate the role of the NR/FBE and its flanking sequences in this process, we mutated the egl-4 3’UTR to mimic the ky95 form or deleted key regulatory elements from this sequence (NR/FBE delete) (Figure 5A). Luciferase values were normalized to numbers of copies of luciferase genes in the population sampled (for expression from each line see Supplemental Materials). A population of animals was exposed to odor for ~one hour. Subsequently, one portion of the population was subjected to a chemotaxis assay while the other portion was subjected to a luciferase assay. Importantly, expression of the transgenes did not alter behavior (Figure 5B).
Figure 5. The egl-4 NR/FBE and a predicted let-7/mir-32/84/265 miRNA seed sequence both promote expression of a Luciferase-based reporter in the AWC neuron.
(A) Diagram of Luciferase-based reporters. All were placed under the control of the str-2 promoter and each contains the indicated 3’ UTR. Upper brackets indicate the nucleotides that were deleted. The longer NR/FBE delete in Figure 5F extends 4 n.t. 3’ of the NR/FBE brackets.
(B) Chemotaxis assays were performed on naïve and odor-exposed animals carrying the indicated transgenes.
(C) Top The chemotaxis behavior (CI) of naive animals (X axis) to butanone was not correlated with luciferase expression (Y axis) in any of the reporter strains examined. When the wild type egl-4 3’ UTR flanks this reporter, Pearson’s r is -0.12 p=0.74, for the unc-54 3’ UTR Pearson’s r is −.71 p=0.11, for the ky95 3’ UTR Pearson’s r is 0.04, p=0.96 and for NR/FBE delete Pearson’s r is 0.17, p=0.78.
Bottom The chemotaxis index of odor-exposed animals correlated well with luciferase expression when the wild type egl-4 3’ UTR flanks this reporter (1 independent line) (Pearson correlation r = −0.7324, *p= 0.016 and Spearman’s correlation r= −0.7939 and **p= 0.0088). Expression was uncorrelated with behavior after odor-exposure when the egl-4 3’UTR (1 line) was replaced by the unc-54 3’UTR (Pearson’s r=0.01, p=0.98), the NR/FBE was mutated to mimic the ky95 allele (2 lines) (red diamonds, Pearson’s r=−0.74, p=0.26), or the NR/FBE was deleted (1 line) (Pearson’s r=−0.33, p=0.58). Each point represents a single assay in which a population was divided and a portion was subjected to a chemotaxis assay and the other portion to luminometry followed by qRT-PCR to obtain units of luciferase per unit of transgenic DNA.
(D) Luciferase values in populations of naïve worms that chemotaxed well (CI>0.8, light grey) were compared to those that adapted robustly (CI<0.2, dark grey bars). Luciferase expression was 1.85 fold higher in animals that adapted well than in the naive animals (p=0.05 n=3). Error bars denote the s.e.m.
(E) Luciferase reporter expression in the AWCON neuron in naïve animals was examined as a function of the 3’ UTR. The wild type egl-4 3’UTR (5 independent lines, see Supplemental Materials on details of expression from each line) in wild-type animals conferred 2.4 fold more expression (n=40) than when it was expressed in fbf-1(ok91) mutant animals (3 lines, p=0.0061 n= 15), 4.5 fold higher than the ky95 3’ UTR expressed in wild-type animals (2 lines, p=0.0003 n=15) and 6.4 fold higher than the ky95 3’ UTR expressed in fbf-1(ok91) (2 lines p<0.0001 n=11). fbf-1(ok91) animals expressing the wild-type 3’UTR showed 2.3 fold more luciferase expression than fbf-1(ok91) animals expressing the ky95 3’ UTR (p=0.0025). Expression from the ky95 3’ UTR reporter was not significantly different in wild-type and fbf-1(ok91l) animals (p= 0.1591).
(F) The NR/FBE and flanking sequence elements promote basal levels of expression from the egl-4 3’UTR. In wild-type worms, the egl-4 3’UTR (5 lines) was expressed 2.3 fold more than the NR/FBE deleted 3’ UTR (1 line,** p=0.0077, n=9), 2.8 fold more the gld-1 FBEa substitution (3 lines, **p=0.0028, n=25), 4.8 fold more than the NR/FBE+4 b.p. delete (3 lines, ** p=0.0002 n=9) and 5.2 fold more than the let-7/mir-32/84/265 deletion (2 lines ***p=0.0002, n=8). The unc-54 3’UTR drove 2.4 fold less expression than the wt egl-4 3’UTR 1 line, p=0.0449 n=8). Luciferase/unit DNA values for wild type egl-4 3’ UTR, gld-1-subsitiuted FBE 3’ UTR and unc-54 3’ UTR lines did not follow a normal distribution thus P values were obtained using a Welch-corrected two-tailed T-test using Prism software. Error bars denote s.e.m.
Since EGL-4 is required for both odor-sensation (Daniels et al., 2000) and rapid short-term adaptation (L’Etoile, et al., 2002), we expected that naïve populations should express the reporter at relatively high levels. If robust long-term adaptation requires additional EGL-4 expression, we hypothesized that we might observe an increase in reporter expression above the naïve levels that would correlate with the strength of adaptation. That is, as the animals become less responsive to odor, they might also show increases in EGL-4 and luciferase expression. When we examined populations that expressed luciferase under the control of the wild-type egl-4 3’UTR, we found that luciferase expression correlated inversely with the CI only after animals had been exposed to odor (Figure 5C, bottom). This correlation was not observed in the naïve populations (Figure 5C, top). Thus, a decrease in chemosensation is strongly correlated with increased expression from the egl-4 3’UTR and this relationship is not seen unless the animals have been exposed to odor for a prolonged period.
To determine whether the correlation between luciferase expression and behavior was dependent upon the egl-4 3’ UTR, we examined animals in which luciferase was under the control of the unc-54 3’UTR. Though the behavior of these animals was indistinguishable from those carrying the egl-4 3’UTR (Figure 5B), there was no correlation between CI and luciferase values either in naïve or odor-exposed populations (Figure 5C). Thus, the 3’UTR itself seems to be the target of regulation by prolonged odor-exposure rather than the behavior. We next asked whether the NR/FBE might be required for the odor-responsiveness of the egl-4 3’UTR element. Indeed, changing it to mimic the ky95 3’UTR or deleting it entirely abolished the odor-induced correlation between luciferase expression and behavior (Figure 5C). Thus, correlation between behavior and egl-4 3’UTR reporter expression depends upon an intact NR/FBE. This correlation may be important for long-term adaptation since the egl-4(ky95) mutant strain is unable to adapt to prolonged exposures of odor (L’Etoile et al., 2002). Further, since FBF-1 binding is diminished by the ky95 mutation, this indicates that the odor-responsiveness of the 3’UTR depends upon PUF binding to this element.
Since the strength of adaptation can be measured by the decrease in chemotaxis index (CI) and the longer the odor-exposure, the lower the CI and the more enduring the adaptation (Colbert and Bargmann, 1995; L’Etoile et al., 2002), we decided to compare luciferase reporter expression in populations with high and low CIs. When populations of naïve animals expressing the luciferase reporter under the control of the wild-type egl-4 3’UTR that chemotaxed well (CI>0.8) were compared to identical animals that had adapted robustly (CI<0.2), we saw that luciferase was two fold higher in the adapted populations than in the naive populations (Figure 5D). This odor-regulation was dependent upon the 3’UTR as we failed to see an odor-dependent increase in luciferase levels when the egl-4 3’UTR was replaced with the unc-54 3’UTR or when the NR/FBE was deleted from our reporter (Figure 5D). Thus, though EGL-4 is required in the naïve animal, prolonged odor-exposure acts via the NR/FBE element within the egl-4 3’UTR to increase EGL-4 expression. This indicates that FBF-1 to promotes adaptation by enhancing EGL-4 translation.
The PUF-binding site and its adjacent sequences enhance basal expression
Since both activated and basal levels of expression require PUF control, we wanted to understand how FBF-1 binding to the NR/FBE might affect basal expression of egl-4. We examined luciferase expression in naive animals removed directly from food. Basal levels of expression required FBF-1: expression from the wild type reporter was ~2.4 fold lower in fbf-1(ok91) mutant animals as compared to wild types and loss of FBF-1 binding to the egl-4(ky95) 3’UTR resulted in ~4.5 fold lower expression (Figure 5E). Since expression from the egl-4(ky95) 3’UTR reporter was roughly the same in either fbf-1(ok91) or wild type animals (Figure 5E, compare 3rd and 4th bars) it is unlikely that FBF-1 is acting through additional targets to promote EGL-4 expression.
To understand how the NR/FBE and its surrounding sequences might contribute to basal EGL-4 expression, we engineered specific changes in the 3’UTR (Figure 5A). Selective deletion of the NR/FBE or wholesale replacement of the 3’UTR with that of unc-54 3’ resulted in two-fold lower expression (Figure 5F). This lower expression was not changed by replacement of the NR/FBE with the gld-1 FBEa (Figure 5F, 2nd and 3rd bars) indicating that a PUF-binding site per se may not be sufficient to either promote or repress basal translation. However, FBF-1 may not interact with the hybrid UTR at all since its expression was not affected in the fbf-1 mutant strain (S4). In fact, residues both 5’ and 3’ of the FBEa affected Pumilio binding in vitro (Bernstein et al., 2005). Thus, the context of the PUF-binding site may be critical for the interaction and we may have disturbed this in an unanticipated way. Consistent with this, we were not able to “reprogram” the unc-54 3’UTR by addition of either the NR/FBE or FBEa (S5).
To assess how sequences flanking the NR/FBE might affect PUF-mediated basal translation, we deleted 4 additional bases 3’ of the original NR/FBE deletion (NR/FBE delete+4). The additional 4 base-deletion is predicted to abolish the binding of the microRNA mir-84 (the predicted energy of binding goes from −18.27 kCal/mol to no predicted binding using Miranda, (Enright, et al., 2003)) and to decrease the affinity of the microRNA let-7(−18.27 Kcal/mol to −14.45 kCal/mol). The removal of these residues decreased expression further (compare Figure 5F 2nd and 4th bars), indicating that flanking sequences are crucial for basal translation. Since the Miranda (Enright et al., 2003) predicted seed sequence for let-7/mir-265/mir-32/mir-84 microRNA lies 11–19 nucleotides 3’ of the NR/FBE, we wondered what this site might contribute to basal reporter expression. We found that selective deletion of the seed sequence (Figure 5A) decreased expression by ~6 fold (Figure 5F). Originally, we had hypothesized that microRNA might repress translation by binding to these sites and that FBF-1 might displace the microRNAs and thereby activate translation. Since high levels of basal reporter expression require the predicted microRNA seed sequence, it is formally possible that microRNAs positively affect EGL-4 translation. Such regulation has been observed in cultured cells but thus far, has not been reported in a living, behaving organism.
Discussion
This work demonstrates the substantial role that a 3’UTR element can play in determining the behavior of an entire organism. We show that an NRE-containing sequence (the NR/FBE) lying within the 3’UTR of the key adaptation-promoting factor, egl-4 is bound by FBF-1 and regulates EGL-4 expression in adapted animals. This element may also help localize EGL-4 translation near the AWC sensory cilia. To gain mechanistic insight into how the egl-4 3’ UTR enhances expression, we examined both the NR/FBE and its flanking sequences. We found that the NR/FBE itself is required for high levels of reporter expression in naïve worms. In long-term adapted worms, odor increases expression over naïve levels in an NR/FBE-dependent fashion. We provide both genetic and cell biological evidence that in an adult neuron, the normally repressive PUFs may promote plasticity by allowing odor stimulation to activate translation and this may occur near the sensory cilia. These results indicate that 3’UTR elements can act as stimulus-responsive translational enhancers in a sensory neuron. Finally, this PUF-dependent increase in translation is not accompanied by an appreciable increase in message levels, indicating that instead of stabilizing transcripts, FBF-1 is likely to directly activate translation.
Our observation that a PUF and its target site can activate translation is at odds with the way these factors have been shown to work in C. elegans development (Wickens et al., 2002), in the fly motor neurons (Mee et al., 2004), at the fly neuromuscular junction (Menon et al., 2004), or in yeast mating type switching (Gu et al., 2004) where they have been shown to repress translation. However, in meiotic maturation of Xenopus oocytes, PUF binding sites were recently shown to work in conjunction with a cytoplasmic polyAdenylation element (CPE) within the cyclin B1 3’UTR to up-regulate translation (Pique et al., 2008). Since the egl-4 3’UTR does not contain an identifiable CPE, the FBF-1 binding site may be working in a distinct manner to induce translational activation perhaps with the aid of microRNAs.
3’UTR-mediated regulation of translation and behavior
How does a sensory neuron assess the duration or repetition of stimulation in order to alter its excitability? Our studies indicate that prolonged odor exposure regulates new protein synthesis of the EGL-4 kinase via its NR/FBE. This 3’UTR-based strategy is different from the protein translocation and trafficking based strategy used by visual sensory neurons (Calvert et al., 2006). What we see in the C. elegans AWC olfactory sensory neuron may be more akin to what happens in interneurons within a circuit: stimulated synapses exhibit localized increases of plasticity promoting proteins. For example, odor association in flies and long-term memory formation in mice both require regions of the CamKIIα 3’UTR to restrict expression of the kinase spatially and temporally (Miller et al., 2002; Ashraf et al., 2006). Furthermore, long-term facilitation in Aplysia requires highly localized new protein synthesis specifically at the stimulated synapse (Martin, 2002; Martin et al., 1997). How is translation directed to specific regions within the neuron? Mechanisms that limit translation both spatially and temporally have emerged for CamKII: both dendritic transport and activity-dependent degradation of the RISC complex in flies and the Cytoplasmic Poly Adenylation element binding proteins (CPEBs) in mice and Aplysia limit its translation to specific spatiotemporal windows (see Sutton and Schuman, 2006 for review). Our results indicate that a PUF’s interaction with its target may also be able to localize translation within the neuron, whether this localized translation is also increased in response to an odor-stimulus awaits to be seen.
Both the AWC neuron and the interneurons within a circuit respond to multiple stimuli: each AWC neuron probably senses tens of odors. Cells that respond to multiple stimuli might require spatial localization of key plasticity promoting factors; translational control over protein synthesis may provide this precision. PUF-regulated translational control over EGL-4 expression could provide a measure of odor-specificity to the adaptation process by spatially localizing translation. Alternatively, since we have shown that both classes of PUFs bind to the egl-4 3’ UTR and many of these are expressed in AWC (data not shown), unique combinations of PUFs could interact with the NR/FBE in a way that would allow for adaptation to distinct odors. Thus each PUF could provide a unique contribution to odor adaptation. Alternatively, combinations of PUFs could regulate levels of key adaptation-promoting factors. The fact that loss of one of the 11 C. elegans PUF’s, FBF-1, affects fewer odors than loss of NOS-1 or GLD-3, the other members of the PUF translational control pathway, suggests that other PUFs might interact with NOS-1 and GLD-3 to promote adaptation to the additional AWC-sensed odors perhaps via other targets.
Translational activation is directed by the 3’UTR sequence
This work provides an important insight into translation: a 3’UTR element can allow a specific environmental stimulus to dynamically activate translation. This may require an adjacent predicted microRNA binding site. A similar scenario was observed for the repression of the hbl-1 3’UTR transcript: both PUF-9 and the microRNA let-7 were required for proper timing and full repression of the hunchbacklike transcript (Nolde et al., 2007). Thus, there is precedent for Pumilios and microRNAs collaborating to regulate translation. In fact, recent bioinformatic evidence has indicated that PUF and microRNA binding sites are adjacent to each other more frequently than would be predicted if their placement was random, suggesting that these two translational control elements may act together to regulate gene expression (Galango et al., 2008).
Our finding that an otherwise repressive 3’UTR element activates translation is reminiscent of reports on the regulation of Tumor Necrosis Factor alpha (TNFα) (Vasudevan and Steitz, 2007). The TNF alpha 3’UTR contains an AU-rich element (ARE) that can either repress or activate translation depending upon the cellular milieu rather than the ARE sequence itself (Vasudevan et al., 2007). In the postmitotic adult neuron a similar scenario may play out: odor-induced changes in the cellular cohort of PUF-associated proteins and/or RNAs may mediate translational activation. Alternatively, the NR/FBE sequence itself may direct activation. Thus, it will be interesting to determine if the novel NR/FBE element found within other 3’UTRs also activates message translation of other genes. However they work, independent sequence elements within the egl-4 3’UTR such as the PUF and micro-RNA binding sites could generate a “code” for behavior that would enable specific aspects of a behavior to evolve independently as each element is retained or lost.
Finally, in mice, the PUF PUM2 (Xu et al., 2007), NANOS1 (Haraguchi et al., 2003) mammalian GLD2 poly(A) polymerase (Rouhana et al., 2005) are all expressed in the hippocampus suggesting a role for this cohort of proteins in memory formation. It will be interesting to see whether mice that lack these proteins exhibit any behavioral phenotypes and whether the PUF pathway activates translation in the context of mammalian neurons.
Methods
Strains used
Strains used in this work include N2 as wild-type; JK3022 fbf-1(ok91)(9X out crossed); JH1270 nos-1(qv5) (6Xoutcrossed); JK2589 nos-3 (q650 (8X outcrossed)) BS3493 gld-3(ok308)/mIn1[dpy-10(e128) mIs14] II (3X outcrossed). Nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Gel Binding assays
Full-length cDNA encoding FBF-1 (yk322e3 from Yuji Kohara) was PCR amplified and fused to GST sequences in pGex3T using the BamH1 and EcoR1 sites (see Supplemental Materials for details of the purification). Purified GST-FBF-1 was incubated with 100 pM 32P labeled wild type or ky95 30-mer RNA oligomers (Dharmacon) (Bernstein et al., 2005). Gels were resolved for 2 hours, dried and quantified using Image Quant and Kd’s were estimated by non-linear regression curve fitting by GraphPad Prism™ software. GST-tagged Pum HD (generous gift from Phillip Zamore) was purified as previously described (Zamore et al., 1997) and see Supplemental Materials for further details. Binding reactions were performed as described (Bernstein et al., 2005) for FBF-1.
Behavioral Assays
All strains were grown as described (L'Etoile et al., 2002) except gld-3 which was grown at 20 degrees C and shifted to 25 for 4 hours before assaying. Odors were used at the following concentrations: 7 micro liters of Benzaldehyde per 100ml Sbasal, 10–12 micro liters of Butanone per 100 ml S basal (L'Etoile et al., 2002). Isoamyl alcohol adaptation was performed as described (Colbert and Bargmann, 1995). Each chemotaxis assay was performed with a minimum of 50 animals. Statistical tests were performed by GraphPad Prism™ software.
Cell-specific rescue
The full-length cDNA encoding FBF-1 was placed under the control of podr-1::RFP. See Supplemental Materials for details of this construct and generation of the transgenic lines.
Kaede Expression
Transgenic worms expressing either the wild type or ky95 egl-4 3’UTR downstream of luciferase (see Supplemental Materials for the details of the cloning) were immobilized on 2% agarose pads with 1 mM NaN3. Kaede was photoconverted by irradiating with 365 nM U.V. light for 45 seconds using the 63X objective. The irradiated worms were then imaged in the green and red fluorescence channels using a 488- and 543-nM scanning laser. Worms were recovered from pads with S-Basal and allowed to recover on seeded plates for 3 hours prior to reimaging. Worms were imaged on a Zeiss LSM 510 META laser scanning confocal microscope and images were quantified using Volocity software (Supplemental Materials).
Luciferase assays
Transgenic animals containing luciferase upstream of the egl-4 3’ UTR (and the various per-mutations, see Supplemental Materials for details of the generation of these constructs) were grown for 5 days on HB101 at 23–25°C. Worms were collected by washing off the cultured plates with S-Basal and were washed free of E. coli by washing 2 times in S-Basal and 2 times in water. Between washes, animals were spun for 6 seconds in a nanofuge and the final spin was at 12,000 RPM in an Eppendorf centrifuge 5415D for 1 minute. Excess water was removed and worms were frozen immediately in liquid nitrogen and stored at −80°C. Worms were thawed in a water bath at RT and DMSO was added to a final concentration of 10%. Animals were vortexed on maximum for ten seconds, re-frozen in liquid N2 and thawed for 2 minutes in a RT water bath. This was repeated four more times. 40 µl of lysed worm suspension was used in a Turner Biosystems 20/20 luminometer and total luminescence was measured using the Promega Luciferase Assay System as described in technical bulletin number 281. Transgene copy number was obtained using real time PCR using the same supernatant (Supplemental Materials).
Supplementary Material
Acknowledgements
We would like to thank Shih-Yu Chen for his helpful insights, Justine Melo, Damien O'Halloran, Jin Lee, Georgia Woods, Mark Lucanic, Christopher Sullivan, Cori Bargmann, Maria Gallegos and Judith Kimble for critical reading of this manuscript and constructive discussions; Marv Wickens, Laura Opperman for corroborating our findings and sharing unpublished data. We thank Yuji Kohara for cDNA clones and Theresa Stiernagle and the CGC for strains, Pilai Sengupta for pofm-1::GFP, Cori Bargmann and Miri Van Hoven for pstr-2::RFP; Peter Lengyel, Ting Wen Cheng and Danny Rozelle for the reporter constructs and turning the egl-4 3’UTR into a cassette and Philip Zamore for DPUM and advice; Brad Hook for advice on gel shifts and Laurel Beckett for statistical counsel. We are also grateful to, Mehrdad Matloubian and Amanda Kahn-Kirby for help with QRT-PCR and comments on the manuscript; J. Michael Bishop for use of his lab and Robert Bailey for imaging and trouble-shooting and the W.M. Keck Program in Neuroscience Imaging. This work was supported by the NIH Cell and Molecular training grant for JAK; the Sandler Program in Biological Sciences grant for AG and the Joe P. Tupin Intramural Grant Award program administered by the Department of Psychiatry and Behavioral Sciences, UC Davis School of Medicine Health Sciences Award and NSF 0317136 and NIH R01DC5991 to NDL.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler CE, Fetter RD, Bargmann CI. UNC-6/Netrin induces neuronal asymmetry and defines the site of axon formation. Nat Neurosci. 2006;9:511–518. doi: 10.1038/nn1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. Rna. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnby DA, Link EM, Vowels JJ, Tian H, Colacurcio PL, Thomas JH. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in caenorhabditis elegans. Genetics. 2000;155:85–104. doi: 10.1093/genetics/155.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert PD, Strissel KJ, Schiesser WE, Pugh EN, Jr., Arshavsky VY. Lightdriven translocation of signaling proteins in vertebrate photoreceptors. Trends Cell Biol. 2006;16:560–568. doi: 10.1016/j.tcb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert HA, Bargmann CI. Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron. 1995;14:803–812. doi: 10.1016/0896-6273(95)90224-4. [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Cohen J. Statistical power analysis for the behavioral sciences. 2nd ed. Hillsdale, NJ: Lawrence Erlbaum Associates; 1988. [Google Scholar]
- Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- Daniels SA, Ailion M, Thomas JH, Sengupta P. egl-4 acts through a transforming growth factorbeta/SMAD pathway in Caenorhabditis elegans to regulate multiple neuronal circuits in response to sensory cues. Genetics. 2000;156:123–141. doi: 10.1093/genetics/156.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- Eckmann CR, Kraemer B, Wickens M, Kimble J. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev Cell. 2002;3:697–710. doi: 10.1016/s1534-5807(02)00322-2. [DOI] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS ONE. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- Gray NK, Coller JM, Dickson KS, Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. Embo J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Deng Y, Zenklusen D, Singer RH. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi S, Tsuda M, Kitajima S, Sasaoka Y, Nomura-Kitabayashid A, Kurokawa K, Saga Y. nanos1: a mouse nanos gene expressed in the central nervous system is dispensable for normal development. Mech Dev. 2003;120:721–731. doi: 10.1016/s0925-4773(03)00043-1. [DOI] [PubMed] [Google Scholar]
- Kraemer B, Crittenden S, Gallegos M, Moulder G, Barstead R, Kimble J, Wickens M. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr Biol. 1999;9:1009–1018. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- L'Etoile ND, Bargmann CI. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron. 2000;25:575–586. doi: 10.1016/s0896-6273(00)81061-2. [DOI] [PubMed] [Google Scholar]
- L'Etoile ND, Coburn CM, Eastham J, Kistler A, Gallegos G, Bargmann CI. The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron. 2002;36:1079–1089. doi: 10.1016/s0896-6273(02)01066-8. [DOI] [PubMed] [Google Scholar]
- Lans H, Rademakers S, Jansen G. A network of stimulatory and inhibitory Galphasubunits regulates olfaction in Caenorhabditis elegans. Genetics. 2004;167:1677–1687. doi: 10.1534/genetics.103.024786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- Lyles V, Zhao Y, Martin KC. Synapse formation and mRNA localization in cultured Aplysia neurons. Neuron. 2006;49:349–356. doi: 10.1016/j.neuron.2005.12.029. [DOI] [PubMed] [Google Scholar]
- Martin KC. Synaptic tagging during synapse-specific long-term facilitation of Aplysia sensorymotor neurons. Neurobiol Learn Mem. 2002;78:489–497. doi: 10.1006/nlme.2002.4088. [DOI] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- Mashukova A, Spehr M, Hatt H, Neuhaus EM. Beta-arrestin2-mediated internalization of mammalian odorant receptors. J Neurosci. 2006;26:9902–9912. doi: 10.1523/JNEUROSCI.2897-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mee CJ, Pym EC, Moffat KG, Baines RA. Regulation of neuronal excitability through pumilio-dependent control of a sodium channel gene. J Neurosci. 2004;24:8695–8703. doi: 10.1523/JNEUROSCI.2282-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon KP, Sanyal S, Habara Y, Sanchez R, Wharton RP, Ramaswami M, Zinn K. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- Moga DE, Calhoun ME, Chowdhury A, Worley P, Morrison JH, Shapiro ML. Activity-regulated cytoskeletal-associated protein is localized to recently activated excitatory synapses. Neuroscience. 2004;125:7–11. doi: 10.1016/j.neuroscience.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Murata Y, Wharton RP. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- Nolde MJ, Saka N, Reinert KL, Slack FJ. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3'UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev Biol. 2007;305:551–563. doi: 10.1016/j.ydbio.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman L, Hook B, DeFino M, Bernstein DS, Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat Struct Mol Biol. 2005;12:945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- Piqué M, López JM, Foissac S, Guigó R, Méndez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132:434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- Roayaie K, Crump JG, Sagasti A, Bargmann CI. The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron. 1998;20:55–67. doi: 10.1016/s0896-6273(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Rouhana L, Wang L, Buter N, Kwak JE, Schiltz CA, Gonzalez T, Kelley AE, Landry CF, Wickens M. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. Rna. 2005;11:1117–1130. doi: 10.1261/rna.2630205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P, Chou JH, Bargmann CI. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell. 1996;84:899–909. doi: 10.1016/s0092-8674(00)81068-5. [DOI] [PubMed] [Google Scholar]
- Schweers BA, Walters KJ, Stern M. The Drosophila melanogaster translational repressor pumilio regulates neuronal excitability. Genetics. 2002;161:1177–1185. doi: 10.1093/genetics/161.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringham EG, Dixon DK, Jones D, Candido EP. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol Biol Cell. 1992;3:221–233. doi: 10.1091/mbc.3.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Jedamzik B, Eckmann CR, Wickens M, Kimble J. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc Natl Acad Sci U S A. 2006;103:15108–15112. doi: 10.1073/pnas.0607050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Local translational control in dendrites and its role in longterm synaptic plasticity. J Neurobiol. 2005;64:116–131. doi: 10.1002/neu.20152. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Tadauchi T, Matsumoto K, Herskowitz I, Irie K. Post-transcriptional regulation through the HO 3'-UTR by Mpt5, a yeast homolog of Pumilio and FBF. Embo J. 2001;20:552–561. doi: 10.1093/emboj/20.3.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER. Chemosensory signaling in C. elegans. Bioessays. 1999;21:1011–1020. doi: 10.1002/(SICI)1521-1878(199912)22:1<1011::AID-BIES5>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Sagasti A, Bargmann CI. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell. 1999;99:387–398. doi: 10.1016/s0092-8674(00)81525-1. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Wharton RP, Struhl G. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell. 1991;67:955–967. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Xu EY, Chang R, Salmon NA, Reijo Pera RA. A gene trap mutation of a murine homolog of the Drosophila stem cell factor Pumilio results in smaller testes but does not affect litter size or fertility. Mol Reprod Dev. 2007;74:912–921. doi: 10.1002/mrd.20687. [DOI] [PubMed] [Google Scholar]
- Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. Rna. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- Zufall F, Leinders-Zufall T. Identification of a long-lasting form of odor adaptation that depends on the carbon Monoxide/cGMP second-messenger system. J Neurosci. 1997;17:2703–2712. doi: 10.1523/JNEUROSCI.17-08-02703.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.