Abstract
Aberrant angiogenesis plays a large role in pathologies ranging from tumor growth to macular degeneration. Anti-angiogenic proteins have thus come under scrutiny as versatile, potent therapeutics but face problems with purification and tissue retention. We report here on the synthesis of supramolecular nanostructures that mimic the anti-angiogenic activity of maspin, a class II tumor suppressor protein. These maspin-mimetic nanostructures are formed via self-assembly of small peptide amphiphiles containing the g-helix motif of maspin. Using tubulogenesis assays with human umbilical vein endothelial cells, we demonstrate that maspin-mimetic nanostructures show anti-angiogenic activity at concentrations that are significantly lower than those necessary for the g-helix peptide. Furthermore, in vivo assays in the chick chorioallantoic membrane show maspin-mimetic nanostructures to be effective over controls at inhibiting angiogenesis. Thus, the nanostructures investigated here offer an attractive alternative to the use of anti-angiogenic recombinant proteins in the treatment of cancer or other diseases involving abnormal blood vessel formation.
Keywords: peptide amphiphile, self-assembly, maspin, g-helix, anti-angiogenic, cancer therapy
1. Introduction
The growth of a tumor to clinically malignant dimensions requires angiogenesis, the sprouting of new blood vessels from pre-existing vasculature [1–3]. Not only is angiogenesis crucial for tumor growth due to oxygen and nutrient demands, it is also essential for the progression of tumor malignancy. Angiogenesis inhibitors, used either in conjunction with or in place of traditional cytotoxic chemotherapies, have shown promise in restricting tumor growth and have thus become of interest in the development of cancer therapies [4]. Maspin, a 42kDa secreted serine protease inhibitor protein, has been studied due to its potential potency in limiting tumor growth and metastasis [5–7]. Generally regarded as a class II tumor suppressor protein, maspin expression has been shown to be down-regulated during the malignant progression of breast and prostate cancers. Ongoing studies continue to investigate the multifaceted role of maspin in cancer but have suggested that its ability to inhibit angiogenesis is mediated through interactions at the endothelial cell surface with β1 integrin receptor and the urokinase-type plasminogen activator (uPA) and receptor (uPAR) complex. These interactions result in increased adhesion and decreased migration of endothelial cells [8–10]. Recent research has shown that these functions of maspin require its "g-helix" motif [11]. This g-helix, originally identified by the protein crystal structure, is structurally distinct from the reactive serpin loop (RSL) of maspin [12]. However, a 15-mer peptide encompassing the g-helix sequence and two residues on either side (residues 236–250) was able to inhibit migration of multiple cell lines in a β1 integrin-dependent manner similar to full-length maspin, albeit at much higher concentrations [8,11].
Recombinant protein therapeutics such as maspin face challenges relating to high cost, difficult purification, and short retention time in tissue. Peptide-based nanostructures can be clinical alternatives, especially if they offer comparable potency. Rationally designed molecules that conjugate a bioactive peptide epitope to a molecular domain that promotes self-assembly through hydrogen bonding and hydrophobic interactions have shown much promise in mimicking the bioactivity of proteins. These molecules, known as peptide amphiphiles (PAs), typically consist of a hydrophobic alkyl tail, a peptide sequence capable of β-sheet formation, a flexible peptide sequence as linker, and the bioactive peptide epitope at one terminus of the molecule [13–18]. These PAs are known to self-assemble in aqueous solution into high aspect-ratio nanofibers comprised of a hydrophobic core and stabilized by β-sheet formation down their long axis [19,20]. These nanofibers can therefore display on their surfaces a high density of biological signals [21], stabilize epitope secondary structure [22], and improve therapeutic retention in tissue as a result of longer half lives associated with their supramolecular structure [23,24]. For example, protein-mimetic PA supramolecular nanostructures bearing epitopes derived from vascular endothelial growth factor and glucagon-like peptide 1 have shown remarkable bioactivity in stimulating angiogenesis and promoting insulin release, respectively [22,25]. In this work, we investigate a maspin-mimetic PA (MMPA) molecule that incorporates the maspin g-helix motif. Using nanoscale characterization methods, we investigate the supramolecular self-assembly and secondary structure of the PA in the context of its bioactive efficacy. Using in vitro and in vivo assays, we also analyze the anti-angiogenic capabilities of MMPA nanostructures relative to controls.
2. Materials and Methods
2.1 PA Synthesis and Purification
Peptide and PAs were synthesized using standard fluoren-9-ylmethoxycarbonyl (Fmoc) solid-phase peptide synthesis. The maspin-mimetic peptide (MMPep) consists of the maspin g-helix sequence plus two residues on either end (EDESTGLEKIEKQLN) while the maspin-mimetic PA (MMPA) includes a 16-carbon alkylated peptide sequence (C16VVAAGG) on the N-terminus. PA with the same C16VVAAGG region but a scrambled g-helix sequence (ScramPA) of ELQEKLDEITKGENS was additionally synthesized. All molecules were synthesized on rink amide resin and thus display an amide on the C-terminus. Fluorescent versions of the peptide and PAs were synthesized by incorporating a C-terminus lysine with N-methyltrityl side group protection, orthogonally deprotecting the side group, and reacting with fluorescein isothiocyanate (FITC) while on resin. PAs and peptide were dissolved with 20% hexafluoro-2-propanol in acidic 18.2 MOhm pure (MilliQ) water and were purified using reversed-phase high-performance liquid chromatography in an acidic methanol/water gradient containing 0.1% trifluoroacetic acid at a starting condition of 20% methanol. Purified PAs and peptide were lyophilized and stored at −20 °C until use.
Peptide content for non-fluorescent peptide and PAs was analyzed (AIBioTech) in order to establish an accurate conversion between mass concentration and molarity. Stock solutions of peptide and PAs were prepared by dissolving molecules in phosphate buffered saline (PBS) at 1 mM for at least overnight at room temperature prior to use.
2.2 Critical Assembly Concentration Determination by Nile Red Incorporation
The self-assembly of peptide and PAs was assessed by incorporation of hydrophobic solvatochromic fluorophore nile red (NR), which exhibits an emission blueshift in hydrophobic environments. MMPA, MMPep, or ScramPA solutions ranging from 100 nM to 200 µM concentration in PBS were made from stock solutions and NR dissolved in ethanol was diluted 200-fold into solutions to a final concentration of 500 nM. Using a NanoLogHJ spectrofluorometer, samples were excited at 550 nm and spectra were obtained between from 580 nm – 750 nm. The observed blueshift was plotted as a function of concentration to determine the critical assembly concentration.
2.3 Secondary Structure Characterization by Circular Dichroism
The secondary structure of peptide and PAs was probed using circular dichroism (CD) at 22 °C. Peptide and PAs were diluted to 100 µM in 0.1× PBS from stock solution immediately prior to measurement with a JASCO J-715 CD spectrophotometer in a 1 mm pathlength quartz cuvette. From the mean residue ellipticity data, α-helix and β strand content was estimated using the DichroWeb online analysis algorithm [26].
2.4 Imaging by Electron Microscopy
Samples for transmission electron microscopy (TEM) were prepared from 1 mM PA or peptide stock solution by placing 5 µL solution onto a 300 mesh copper grid with an amorphous carbon support film (Electron Microscopy Sciences). The solution was wicked after 2 min, washed twice with MilliQ water, and allowed to dry for 10 min before staining with 2% uranyl acetate. Imaging was performed on a JEOL 1230 TEM with a Hamamatsu ORCA camera at an accelerating voltage of 100 kV.
2.5 Small Angle X-ray Scattering
Small angle x-ray scattering (SAXS) was used to probe the the nanostructure morphology of peptide and PAs at 1mM concentration in PBS. SAXS measurements were performed using beam line 5ID-D in the DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT) Synchrotron Research Center at the Advanced Photon Source of Argonne National Laboratory. Using a double-crystal monochromator to select an energy of 15 keV, scattering data was collected using a CCD detector (MAR) positioned 245 cm behind the sample. Scattering intensity was recorded in the interval 0.008< q < 0.25 A−1 and the wave vector was defined as q = (4 π / λ) sin(θ / 2), where θ is the scattering angle. Solution samples were loaded in 1.5 mm diameter quartz capillaries with exposure times between 2 and 8 s. The 2D SAXS images collected were azimuthally averaged to produce 1D profiles of intensity using the data reduction program Fit2D. Background scattering of the capillary with only PBS was also collected and was subtracted from sample data prior to data analysis.
2.6 Flow Cytometry and Confocal Microscopy to Assess PA Binding
Stock fluorescent PA and peptide solutions were made by dissolving FITC-labeled molecules in PBS at a concentration of 200 µM (assuming peptide content of 90%). Stock fluorescent solutions were then mixed in equal volumes with 1 mM stock solutions of non-fluorescent PA or peptide solution and allowed to sit overnight at room temperature to make working solutions. Subconfluent human umbilical vein endothelial cells (HUVECs) were incubated for 2 hr in starvation media before detaching with 0.05% trypsin/EDTA. Cells were then spun down to remove trypsin/EDTA and were resuspended in starvation media at 250,000 /mL. Working peptide and PA solutions then added to cell suspensions such that the final concentration corresponded to 10 µM non-fluorescent molecule. After incubating at 37 °C for 30 mins, cells were spun down and resuspended in Hank's balanced salt solution (HBSS) twice. Fluorescence signal of each cell was quantified using a LSRFortessa cell analyzer (BD Biosciences). Cells were also plated on poly(d-lysine) coated glass coverslips, fixed with 4% paraformaldehyde, and embedded in mounting media for imaging using an A1R laser scanning confocal microscope (Nikon). Machine parameters were kept constant for all samples in both confocal microscopy and flow cytometry experiments.
2.7 Assessment of Cell Adhesion on Fibronectin
The effect of peptide and PAs on HUVEC adhesion was assessed. Fibronectin from human plasma (Sigma) was dissolved in sterile MilliQ water and was added to tissue culture-treated 96 well plates at 5 ug/cm2, which were then left to dry overnight at room temperature in a sterile laminar flow hood. Coated plates were stored at 4 °C and warmed to 37 °C prior to use. Subconfluent HUVECs were incubated for 2 hr in starvation media (EndoGRO basal media from Millipore supplemented with 0.5% fetal bovine serum and 1× penicillin-streptomycin from Gibco) before detaching with 0.05% trypsin/EDTA. Cells were then spun down to remove trypsin/EDTA and were resuspended in PBS containing peptide or PA molecules. Cells were then incubated for 30 min at 37 °C and then plated at 15,000 cells/well. After 45 mins, wells were washed once with PBS to detach loosely adhered cells. A PicoGreen assay (Invitrogen) was used to quantify number of cells in each well. Cell counts were normalized to that obtained in wells with no treatment. Statistical analysis was performed in GraphPad Prism software using one-way analysis of variance (ANOVA) with a Bonferroni's Multiple Comparisons post-test comparing all pairs of columns. The number of samples for all treatment conditions, n, was equal to 4.
2.8 Assessment of Cell Migration on Fibronectin
Fibronectin from human plasma was coated onto tissue culture-treated 4-chamber glass-bottom petri dishes (In Vitro Scientific) at 7 ug/cm2 as previously described. Subconfluent HUVECs were incubated for 2 hr in starvation media before detaching with 0.05% trypsin/EDTA. Cells were then spun down to remove trypsin/EDTA, re-suspended in starvation media, and plated onto glass-bottom petri dishes at a concentration of 4000 cells per chamber. After cells attached (approximately 1 hr at 37 °C), the media was aspirated and replaced with starvation media containing 80 µg/mL basic fibroblast growth factor (bFGF) and 10 µM MMPA, MMPep, or Scram PA. Live-cell imaging was performed on a Nikon BioStation IM at 37 °C, and cell migration was quantified manually using a plugin to track the location of the cell nucleus every 30 mins in ImageJ software. ANOVA with a Dunnett’s Multiple Comparisons post-test of each treatment condition against control was performed using GraphPad Prism software. For control, MMPA, MMPep, and ScramPA, n = 56, 54, 31, and 42 respectively.
2.9 Assay of Angiogenesis In Vitro by Tube Formation
Wells of a 96-well plate were coated with ECMatrix solution, and 5 × 103 HUVECs were plated in triplicate wells in a volume of 50 µL of endothelial growth medium (EGM) containing 0.5% FBS with 50 ng/mL bFGF for 2 hours. MMPA, MMPep, ScramPA treatments at respective dosages were added to the wells for 5 hours. Tube formation was evaluated by phase-contrast microscopy using an Olympus IX-70 microscope (100× magnification) connected to a Diagnostic Instruments Spot RT Camera. Two images were captured per well. Experiments were repeated independently three times.
2.10 Characterization of Angiogenesis In Vivo by CAM Assay
In vivo assessment of the anti-angiogenic potential of PAs and peptide was performed using the chicken chorioallantoic membrane (CAM). In this well-established CAM assay, angiogenesis of the extraembryonic allantois was measured at several time points after treatment with PAs or peptide. Fertilized chicken eggs (Sunnyside Hatchery) were received and cultured in a temperature controlled, humidified egg incubator. On embryonic day 3, eggs were cracked into 100 mm sterile petri dishes without breaking the yolk sac and were transferred to a water-jacketed humidified CO2 incubator set to 37 °C. On embryonic day 5, 5 µL of treatment (PA, peptide, or scrambled PA dissolved in PBS at 1 mM) or control (PBS) solutions were deposited onto autoclaved filter paper circles 1/4" in diameter. Solution-soaked filter paper circles were placed on top of the CAM and digital images were captured using a Nikon stereomicroscope. ImageJ software was used to extract the green channel from images so that vessels could be more easily seen. Vessel density was quantified by counting the number of vessels intersecting the circumference of filter paper circles and is expressed relative to the initial time point. ANOVA with a Dunnett’s Multiple Comparisons post-test of each treatment condition against control was performed using GraphPad Prism software. A minimum of n = 5 samples was analyzed for each treatment condition at each time point. Chicken embryo assays were conducted following ethical guidelines specified by the Center for Comparative Medicine at Northwestern University.
3. Results and Discussion
3.1 PAs self-assemble at µM concentrations
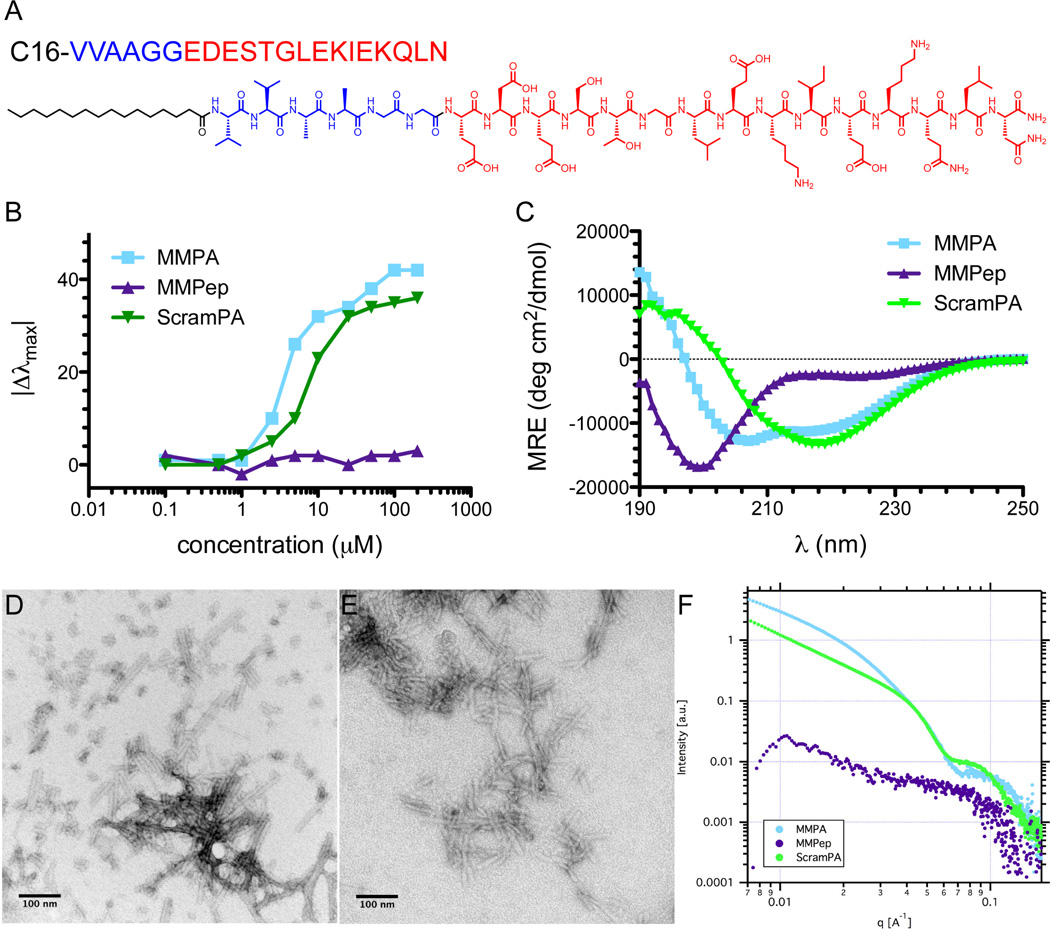
The g-helix of maspin is an 11-residue motif presented on the protein periphery and has been shown to be crucial for decreasing motility and increasing adhesion, two functions which act together on endothelial cells to inhibit angiogenesis [11,12]. The 15-residue peptide encompassing the g-helix plus two additional residues on either side (MMPep) was synthesized as previously reported [11]. MMPA was rationally designed by conjugating MMPep to a C16VVAA self-assembly domain via a flexible glycine linker (Figure 1A). A control PA consisting of the same self-assembly domain and flexible linker but a randomly scrambled version of the MMPep sequence (ScramPA) was also synthesized. The critical assembly concentration (CAC) of MMPA, MMPep, and ScramPA in phosphate buffered saline (PBS) was determined by observing solvatochromic nile red incorporation into hydrophobic domains, which manifests as a blueshift in peak emission. Results indicate that MMPA is capable of forming hydrophobic domains encapsulating nile red at concentrations between 1 µM – 2.5 µM, while ScramPA may begin to encapsulate nile red at concentrations as low as 500 nM (Figure 1B). Similarly to many surfactant molecules, the self-assembly of PAs is partly driven by hydrophobic collapse of alkyl tails, leading to core-shell nanostructures in which the more hydrophilic peptide segments are displayed at the surface. However, in the PA investigated here, the VVAA motif is expected to promote nanofiber formation through aggregation of molecules caused by the high propensity of valine residues to form β-sheets [27]. Furthermore, the presence of valine residues immediately adjacent to the hydrophobic tail is known to promote formation of high aspect ratio nanofibers stabilized by β-sheets parallel to the long axis [28]. As a result, the CAC values of PAs measured here are orders of magnitude lower than those of surfactants such as hexadecyltrimethylammonium bromide [29] and may be even lower in the presence of serum proteins, which could act as nucleation points for aggregation. Thus in contrast to MMPA molecules, MMPep molecules do not show any appreciable aggregation at the concentrations investigated here (up to 200 µM). Circular dichroism (CD) supports the assembly of PA molecules into nanofibers with internal order in the presence of ions, as both MMPA and ScramPA exhibit significantly less random coil signature than MMPep (Figure 1C).
Figure 1.
Characterization of PA self-assembly. (A) Structure of MMPA, with the bioactive sequence shown in red, the β-sheet and flexible linker sequence shown in blue, and the hydrophobic alkyl tail shown in black. (B) Blueshift in peak nile red emission as a function of concentration, indicating a CAC between 1 – 2.5 µM for MMPA and 0.5 – 2.5 µM for ScramPA in PBS. (C) Circular dichroism of 100 µM PAs and peptide in 0.1× PBS, showing that MMPep exhibits primarily random coil conformation while MMPA and ScramPA have increased α-helix signature. Conventional TEM of (D) MMPA and (E) ScramPA in PBS at 1 mM, showing the existence of short nanofibers. (F) SAXS of PAs and peptides in PBS at 1 mM, indicating that MMPA and ScramPA assemble into a mixture of cylindrical and flat nanofibers while MMPep does not assemble appreciably in solution.
Conventional TEM was performed to investigate the morphology of self-assembled nanostructures in solution. Results show that MMPA and ScramPA form short nanofibers at 1 mM in PBS (Figures 1D and 1E) but do not show any evidence of nanostructure formation in MMPep solutions. SAXS at 1 mM displays relatively low scattering signal for MMPep, which is consistent with lack of nanoscale self-assembly of this peptide as inferred by TEM. SAXS further indicates that the MMPA and ScramPA nanofibers are a mixture of cylindrical and ribbon-like shapes based on a slope of approximately −1.75 in the Guinier region at low q values (Figure 1F). The flatter shape of the Guinier region seen for ScramPA may be due to aggregation of nanostructures in solution. However, it should be emphasized that detailed characterization of PA nanostructure morphology at the micromolar concentrations relevant to biological assays remains an unfortunately difficult challenge and that such nanostructures are likely to vary from those seen at higher concentrations by SAXS and TEM.
3.2 MMPA self-assembly stabilizes g-helix secondary structure
The arrangement of polar (P) and apolar (H) residues in the maspin g-helix can be depicted as PPPXHPPHPPP, where X represents glycine. This periodically hydrophobic sequence is characteristic of amphipathic α-helices and despite the helix-breaking tendency of glycine residues [30], structural data from crystalized maspin confirms that the g-helix exhibits α-helical secondary structure [12]. Thus, CD was further used to examine the secondary structure formation of MMPep, MMPA, and ScramPA (Figure 1C). PAs and peptide were examined at 100 µM in 0.1× PBS. Fitting results obtained with the DichroWeb algorithm (Figure S1) show that MMPep displays primarily random coil conformation with very little α-helical character, while MMPA exhibits significantly enhanced α-helix signature. In comparison, ScramPA also shows increased α-helical character over MMPep but additionally displays higher β-sheet signature. These results suggest that the g-helix sequence alone does not take on the native helical structure found in maspin protein but that PA self-assembly stabilizes and promotes α-helix conformation. Similar stabilization of helical structure has been previously observed in PA systems [22,25,31] and likely results from clustering of helices facilitated by supramolecular self-assembly. Because amphipathic helices such as the g-helix exhibit a hydrophobic face and a polar face, secondary structure stability is greatly improved by coil-coil interactions. Previous studies of helical peptides showing increased helicity with increasing concentration confirm this hypothesis [32,33] and further implicate the role of nanostructure self-assembly in stabilizing the native g-helix conformation. In summary, the assembly of rationally designed MMPA molecules into nanostructures via hydrophobic collapse and β-sheet formation has been confirmed by nile red incorporation, CD, SAXS, and TEM. Furthermore, this supramolecular assembly stabilizes the helical conformation of g-helix epitopes.
3.3 MMPA nanostructures bind to cells and recapitulate multiple functions of maspin
Previous studies have shown that maspin interacts with the β1 integrin receptors of endothelial cells, thereby triggering pathways that inhibit cell migration [9,34,35]. Specifically, the migration inhibiting activity of g-helix peptide alone has been shown to depend on β1 integrin interaction, indicating that the g-helix epitope binds to β1 integrins.11 Therefore, we investigated the ability of PAs and peptide to bind to HUVECs in culture. Fluorescent analogues of MMPA, MMPep, and ScramPA molecules were synthesized by coupling FITC to an additional lysine residue on the C-terminus (Figure S2) and were mixed as a small fraction to the corresponding non-fluorescent molecule for visualization. Confocal microscopy suggests that both MMPA and ScramPA bind to HUVEC membranes and are additionally present in the interior of the cells (Figures 2A–2D). Flow cytometry additionally reveals that cells treated with MMPA or ScramPA have significantly more surface fluorescence than untreated controls or those treated with MMPep (Figure 2E). The median FITC intensity per cell for MMPA, ScramPA, and MMPep was 28.7, 14.1, and 2.6 times that of untreated control, respectively. This result indicates that the binding of MMPA to the surface of HUVECs is enhanced over the binding of g-helix peptide alone. Flow cytometry also indicates that MMPA binds more avidly than ScramPA to HUVEC surfaces, suggesting that the g-helix epitopes displayed by MMPA nanostructures interact specifically with β1 integrins. However, the significant presence of ScramPA on HUVECs compared to untreated controls suggests that binding of PAs to cells is partly non-specific, possibly resulting from ionic interaction between cell surfaces and the highly charged PA nanostructures. Unfortunately, given the degree of non-specific interaction between PA nanostructures and cells, it is difficult to ascertain whether the enhanced binding of MMPA over MMPep results from epitope multivalency or from stabilization of the native g-helix conformation. The significant non-specific binding of PA nanostructures limits the use of standard techniques to assess integrin co-localization studies. In light of the previous work on the mechanisms of g-helix activity, we focus in this work on the mimetic capabilities of MMPA from the perspective of cell behavior and function.
Figure 2.
Binding of PA to cells. Confocal microscopy images of cells treated with (A) PBS control, (B) FITC-labeled MMPep, (C) FITC-labeled MMPA, and (D) FITC-labeled ScramPA show that PAs associate with cell surfaces and are also internalized by cells. (E) FACS analysis of HUVEC fluorescence after treatment with FITC-labeled PAs and peptide. Results show that MMPA binds to cells most effectively but that ScramPA also non-specifically binds to cells, while MMPep binds to cells significantly less effectively than PA nanostructures.
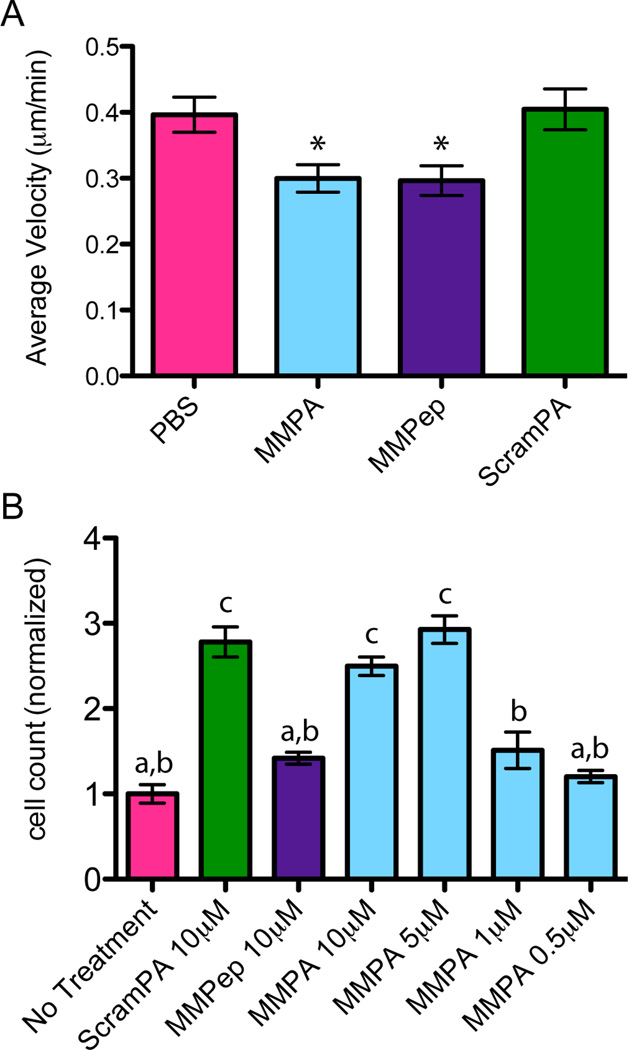
The anti-angiogenic function of maspin relies primarily on two activities: decreasing endothelial cell motility on extracellular matrix (ECM) and increasing endothelial cell adhesion to ECM [9–11]. Live-cell imaging was used to observe HUVEC movement on fibronectin (FN), a common component of ECM, in response to bFGF stimulus. Results demonstrate the ability of both MMPep and MMPA to significantly decrease cell motility over untreated controls (Figure 3A). The average cell speed (measured over at least 2 hrs and up to 8 hrs) on a FN-coated surface was 0.30 µ/min for both MMPA and MMPep treatment conditions compared with 0.40 µ/min for PBS control. The speed of cell movement with ScramPA treatment was not significantly different from control. To investigate HUVEC adhesion to ECM in vitro, PA or peptide-treated cells were allowed to adhere to a surface coated with FN. The number of cells that remained attached to the surface after washing was then quantified. Cells treated with MMPA showed statistically significant increase in adhesion to the FN surface over untreated controls at 5 µM or 10 µM PA concentrations (Figure 3B). The difference between these two higher concentrations was insignificant. However, treatment concentrations that are below the measured CAC of MMPA (i.e. ≤ 1 µM) were not effective at increasing adhesion. Moreover, increased cell adhesion is likely nonspecific, as cells treated with ScramPA showed similar behavior at 10 µM. Because the concentration of PA in solution was extremely low, increased adhesion is not likely to be caused by significant coverage of the FN surface by PAs but is more likely due to the strong non-specific binding of PA nanostructures to the cell surface. This phenomenon covers the cell with attachment points, as PA nanostructures can likely adhere via charge interaction to ECM components as well.
Figure 3.
Effect of PAs on cell mobility and adhesion. (A) Average velocity of HUVECs on FN-coated glass as measured by time-lapse imaging and manual tracking, showing cells treated with MMPA and MMPep to be statistically less migratory (P<0.05) as compared to PBS control. (B) Number of HUVECs, as obtained by PicoGreen assay, that remain adhered to FN-coated tissue culture plastic after treatment with PAs or peptide and repeated washing steps. Results are normalized to the mean of the control group and show that MMPA and ScramPA treatments above their CAC non-specifically increase cell adhesion to FN, while MMPep and unassembled PA do not show this effect. Statistical analysis shows that groups a and b are statistically insignificant, groups a and c are statistically significant (P<0.001), and groups b and c are statistically significant (P<0.01).
Previous studies have shown that the g-helix sequence alone is sufficient and necessary to decrease cell motility in vitro [11]. Thus, it is not surprising that both MMPep and MMPA were able to decrease cell motility on FN while ScramPA did not affect cell motility. However, it is interesting that MMPep was unable to increase cell adhesion on FN. Ravenhill and coworkers similarly showed the inability of the g-helix peptide alone to increase HUVEC adhesion, even though the same function of maspin protein requires the g-helix domain [11]. These findings suggest that even though the ability of maspin to increase cell adhesion and decrease cell motility both rely on having an intact g-helix, only the latter function is directly mediated by activation of β1 integrin by the g-helix. Studies show that this interaction enhances the inactivation rate of β1 integrin and also changes cytoskeletal architecture to a less motile phenotype [11]. These results are in agreement with other previous studies suggesting that activation of β1 integrin by maspin causes reorganization of vinculin and F-actin in HUVECs via inhibition of Rac1 and cdc42, thereby decreasing motility [9]. However, Endsley and coworkers showed that the ability of maspin to increase cell adhesion requires formation of a supercomplex wherein maspin acts as a bridge between between β1 integrin and the uPA/uPAR system [10]. The g-helix motif directly interacts with β1 integrin, while a separate domain of maspin interacts with uPA/uPAR. It is therefore not surprising that the g-helix peptide alone can only recapitulate the anti-motility function but not the pro-adhesion function of maspin. In contrast, MMPA can mimic both functions due to the nature of its supramolecular structure.
3.4 MMPA inhibits tube formation in vitro and blocks angiogenesis in vivo
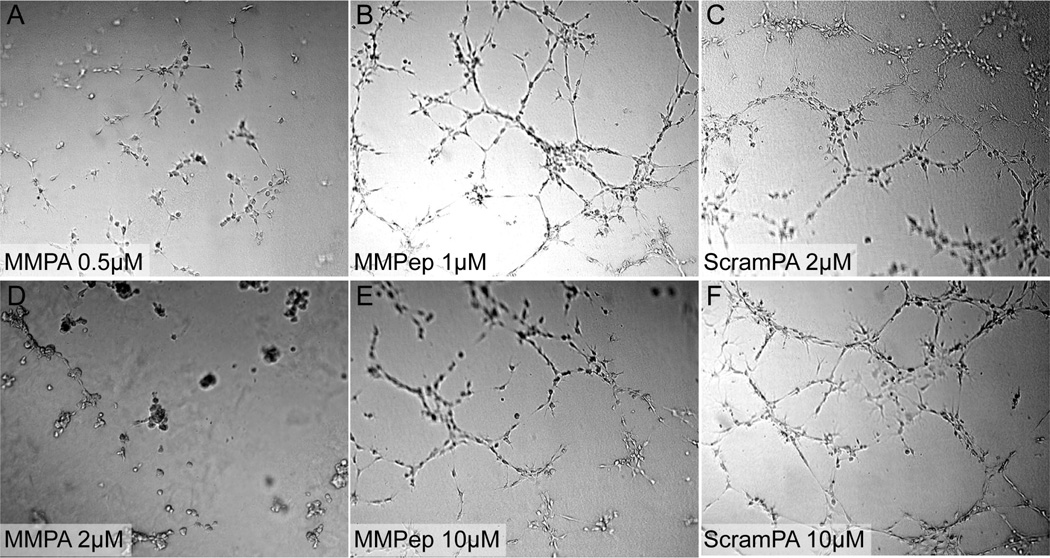
Well-established models exist for assessing angiogenesis in vitro and in vivo [36,37]. Tubulogenesis, the formation of tube-like structures by HUVECs on matrigel, was used to observe the anti-angiogenic potency of PAs and peptide in a quasi-3D system. Based on images obtained 5 hrs after addition of treatments (Figure 4), MMPA was found to be effective in inhibiting tube formation at even sub-micromolar concentrations, which is on par with the bioactivity of native full-length maspin previously reported [8]. The results also show that MMPA is less effective than angiostatin, which can inhibit in vitro tube formation at 10 nM [38], but more effective than small molecule angiogenesis inhibitors such as captopril, which typically require at least 10 µM to inhibit tube formation [39]. In contrast to MMPA, MMPep only partially inhibited tube formation at higher concentrations (10 µM), while ScramPA did not inhibit tube formation even at high concentrations despite non-specific binding to cells. This effect suggests that the g-helix sequence is required for anti-angiogenic activity in vitro. Moreover, PA nanostructures displaying the g-helix show drastically improved efficacy over the g-helix peptide alone.
Figure 4.
Effect of PAs on HUVEC tube formation. Tubulogenesis on Matrigel as imaged 5 hrs after addition of (A, D) MMPA, (B, E) MMPep, and (C, F) ScramPA. Results show that MMPA is capable of inhibiting tube formation at sub-micromolar concentrations while MMPep is slightly effective at concentrations greater than 10 µM. ScramPA does not show efficacy in blocking tube formation at any concentrations investigated.
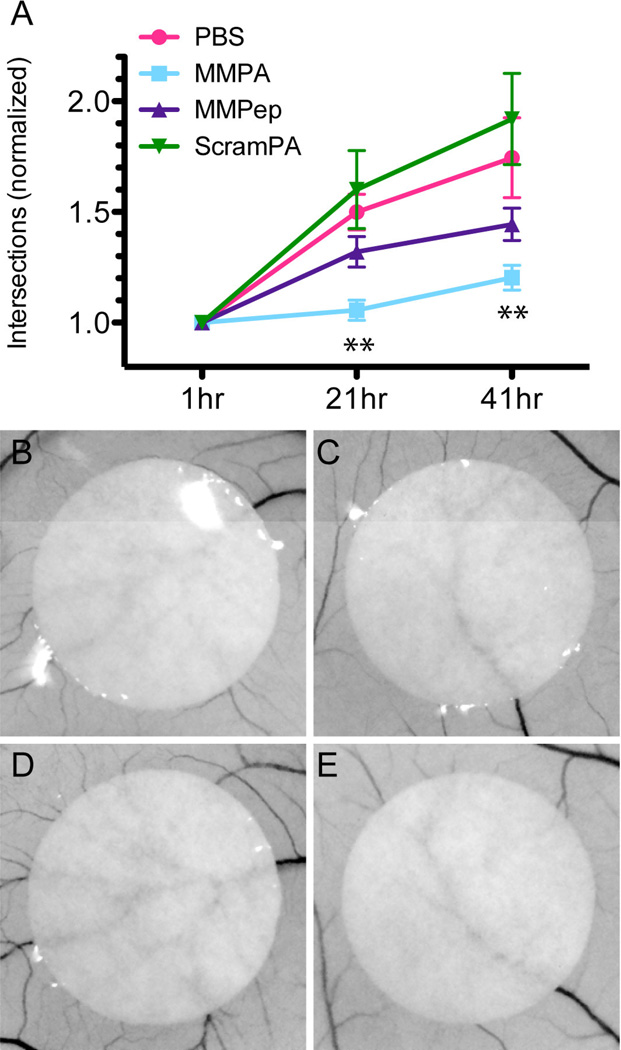
The efficacy of PAs or peptide in blocking angiogenesis in vivo was investigated using a chick embryo chorioallantoic membrane (CAM) assay. Because the CAM is highly angiogenic, especially in stages of early development, PA or peptide treatment was delivered from filter paper circles (each containing 5 µmol treatment) placed onto the shell-less embryo CAM on embryonic day 5. The number of blood vessels intersecting each filter paper perimeter was then counted at 21 hrs and 42 hrs. Figure 5A shows the increase in intersections, normalized to the initial count, for each treatment condition. Only MMPA was found to significantly reduce angiogenesis over PBS controls at both time points, while the anti-angiogenic activity of MMPep was present but is statistically insignificant. Interestingly, ScramPA-treated samples showed a trend towards increased angiogenesis over PBS control. This trend is not statistically significant due to large sample variation but can potentially correlate with increased endothelial cell adhesion in vitro caused by ScramPA. In comparison with native maspin, the data obtained here suggest that MMPA exhibits anti-angiogenic activity in vivo at approximately similar concentrations, as previous reports showed that 3–10 µM maspin inhibits angiogenesis in rat corneal and mouse tumor angiogenesis [8].
Figure 5.
In vivo assay of angiogenesis. (A) Angiogenesis in chick embryo CAM as quantified by counting the number of vessels intersecting the circumference of filter paper circles at 1, 21, and 41 hr after application of treatment. For each sample, counts at later time points are normalized to initial time point (t = 1 hr). Statistical analysis indicates that MMPA significantly decreases angiogenesis at both 21 hr and 41 hr time points (P<0.01). Sample images of PBS treatment at (B) 1 hr and (D) 21 hr as compared to PA treatment at (C) 1 hr and (E) 21 hr.
The ability of MMPA to block angiogenesis both in vitro and in vivo indicates that incorporation of the g-helix motif into self-assembled nanostructures drastically improves its maspin-mimetic functions. Like the g-helix peptide alone, MMPA nanostructures decrease cell motility on FN. Unlike the g-helix peptide alone, MMPA nanostructures also bind aggressively to endothelial cell surfaces and significantly increase cell adhesion as a result of their supramolecular structure. While PA nanostructures bearing a scrambled g-helix sequence also non-specifically increase cell adhesion, they cannot decrease cell motility. It is the combination of specific g-helix-mediated inhibition of cell motility with non-specific promotion of cell adhesion that allows MMPA to mimic multiple functions of full-length maspin and block angiogenesis with high potency.
4. Conclusions
We have developed a supramolecular PA nanostructure that incorporates the maspin g-helix motif and exhibits potent anti-angiogenic bioactivity in vitro and in vivo. These MMPA nanostructures are formed via self-assembly in aqueous environments and display g-helix epitopes in high density as well as stabilize their native helical conformation. In vitro cell assays show that, like maspin protein, MMPA nanostructures inhibit endothelial cell motility via g-helix mediated pathways. Furthermore, MMPA nanostructures can recapitulate the ability of maspin protein to increase endothelial cell adhesion, a function not mediated through the g-helix, via non-specific interactions with cells and substrate. This function results from the supramolecular nature of MMPA. As MMPA nanostructures can mimic multiple functions of the full maspin protein, it is effective in blocking tubulogenesis in vitro at sub-micromolar concentrations and is also effective at inhibiting angiogenesis in vivo in the chicken embryo chorioallantois. Thus, MMPA can form the basis of a versatile therapeutic platform, as it is more accessible synthetically than native maspin and more readily combined with other bioactive molecules (e.g. PAs with other epitopes). Furthermore, MMPA nanostructures can potentially be more resistant to degradation and have prolonged retention time within tissue relative to protein therapeutics.
Supplementary Material
Acknowledgements
This work was supported by a grant from NIH/NCI #5U54CA151880-03. M. Zhang was additionally supported by NCI #CA079736. Experiments utilized the following shared facilities at Northwestern University: the Biological Imaging Facility, the Keck Biophysics Facility, the Cell Imaging Facility, the RHLCC Flow Cytometry Facility, and the Institute for BioNanotechnology in Medicine. The Biological Imaging Facility is generously supported by the NU Office for Research, and electron microscopy was performed on a JEOL 3200 FETEM purchased with the support of NCRR 1S10RR025092. The Cell Imaging Facility is generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. The RHLCC Flow Cytometry Facility is generously supported by a Cancer Center Support Grant NCI CA060553. X-ray scattering studies were performed at the DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT) in Sector 5 of the Advanced Photon Source (APS). DND-CAT is supported by E.I. DuPont de Nemours & Co., The Dow Chemical Company, and Northwestern University. Use of the APS, an Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. Also, the authors wish to thank Dr. Matthew Webber for helpful discussion on the CAM assay, Dr. Ricardo Silva for assistance with using DichroWeb, as well as Dr. Eduard Sleep and Dr. Sushant Tripathy for help with HUVEC cell adhesion assay.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information Available
CD fits by DichroWeb as well as chemical structures for PAs and peptide molecules. This material is available free of charge via the Internet at http://www.sciencedirect.com.
References
- 1.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P, Jain RK. Angiogenesis in Cancer and Other Diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 3.Noonan DM, Benelli R, Albini A. Angiogenesis and Cancer Prevention: A Vision. Recent Results Cancer Res. 2007;174:219–224. doi: 10.1007/978-3-540-37696-5_19. [DOI] [PubMed] [Google Scholar]
- 4.Kerbel R, Folkman J. Clinical Translation of Angiogenesis Inhibitors. Nat Rev Cancer. 2002;2:727–739. doi: 10.1038/nrc905. [DOI] [PubMed] [Google Scholar]
- 5.Zou Z, Anisowicz A, Hendrix MJC, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R. Maspin, A Serpin with Tumor-Suppressing Activity in Human Mammary Epithelial Cells. Science. 1994;263:526–529. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]
- 6.Shi HY, Zhang W, Liang R, Abraham S, Kittrell FS, Medina D, Zhang M. Blocking Tumor Growth, Invasion, and Metastasis by Maspin in a Syngeneic Breast Cancer Model. Cancer Res. 2001;61:6945–6951. [PubMed] [Google Scholar]
- 7.Bailey CM, Khalkhali-Ellis Z, Seftor EA, Hendrix MJC. Biological Functions of Maspin. J Cell Physiol. 2006;209:617–624. doi: 10.1002/jcp.20782. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, Volpert O, Shi YH, Bouck N. Maspin is an Angiogenesis Inhibitor. Nat Med. 2000;6:196–199. doi: 10.1038/72303. [DOI] [PubMed] [Google Scholar]
- 9.Qin L, Zhang M. Maspin Regulates Endothelial Cell Adhesion and Migration Through an Integrin Signaling Pathway. J Biol Chem. 2010;285:32360–32369. doi: 10.1074/jbc.M110.131045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Endsley MP, Hu Y, Deng Y, He X, Warejcka DJ, Twining SS, Gonias SL, Zhang M. Maspin, the Molecular Bridge Between the Plasminogen Activator System and β1 Integrin that Facilitates Cell Adhesion. J Biol Chem. 2011;286:24599–24607. doi: 10.1074/jbc.M111.235788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravenhill L, Wagstaff L, Edwards DR, Ellis V, Bass R. G-Helix of Maspin Mediates Effects on Cell Migration and Adhesion. J Biol Chem. 2010;285:36285–36292. doi: 10.1074/jbc.M110.177253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Law RHP, Irving JA, Buckle AM, Ruzyla K, Buzza M, Bashtannyk-Puhalovich TA, Beddoe TC, Nguyen K, Worrall DM, Bottomley SP, Bird PI, Rossjohn J, Whisstock JC. The High Resolution Crystal Structure of the Human Tumor Suppressor Maspin Reveals a Novel Conformational Switch in the G-Helix. J Biol Chem. 2005;280:22356–22364. doi: 10.1074/jbc.M412043200. [DOI] [PubMed] [Google Scholar]
- 13.Hartgerink JD, Beniash E, Stupp SI. Self-Assembly and Mineralization of Self-Assembling Nanofibers. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 14.Hartgerink JD, Beniash E, Stupp SI. Peptide-Amphiphile Nanofibers: A Versatile Scaffold for the Preparation of Self-Assembling Materials. Proc Natl Acad Sci USA. 2002;99:5133–5138. doi: 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui H, Webber MJ, Stupp SI. Self-Assembly of Peptide Amphiphiles: From Molecules to Nanostructures to Biomaterials. Biopolymers. 2010;94:1–18. doi: 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webber MJ, Kessler JA, Stupp SI. Emerging Peptide Nanomedicine to Regenerate Tissues and Organs. J Intern Med. 2010;267:71–88. doi: 10.1111/j.1365-2796.2009.02184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matson JB, Zha RH, Stupp SI. Peptide Self-Assembly for Crafting Functional Biological Materials. Curr Opin Solid State Mater Sci. 2011;15:225–235. doi: 10.1016/j.cossms.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webber MJ, Berns EJ, Stupp SI. Supramolecular Nanofibers of Peptide Amphiphiles for Medicine. Isr J Chem. 2013;53:530–554. doi: 10.1002/ijch.201300046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang H, Guler MO, Stupp SI. The Internal Structure of Self-Assembled Peptide Amphiphiles Nanofibers. Soft Matter. 2007;3:454–462. doi: 10.1039/b614426h. [DOI] [PubMed] [Google Scholar]
- 20.Velichko YS, Stupp SI, Olvera de la Cruz M. Molecular Simulation Study of Peptide Amphiphile Self-Assembly. J Phys Chem B. 2008;112:2326–2334. doi: 10.1021/jp074420n. [DOI] [PubMed] [Google Scholar]
- 21.Storrie H, Guler MO, Abu-Amara SN, Volberg T, Rao M, Geiger B, Stupp SI. Supramolecular Crafting of Cell Adhesion. Biomaterials. 2007;28:4608–4618. doi: 10.1016/j.biomaterials.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 22.Webber MJ, Tongers J, Newcomb CJ, Marquardt KT, Bauersachs J, Losordo DW, Stupp SI. Supramolecular Nanostructures that Mimic VEGF as a Strategy for Ischemic Tissue Repair. Proc Natl Acad Sci USA. 2011;108:13438–13443. doi: 10.1073/pnas.1016546108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghanaati S, Webber MJ, Unger RE, Orth C, Hulvat JF, Kiehna SE, Barbeck M, Rasic A, Stupp SI, Kirkpatrick CJ. Dynamic In Vivo Biocompatibility of Angiogenic Peptide Amphiphile Nanofibers. Biomaterials. 2009;30:6202–6212. doi: 10.1016/j.biomaterials.2009.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Shape Effects of Filaments Versus Spherical Particles in Flow and Drug Delivery. Nat Nanotech. 2007;2:249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan S, Sur S, Newcomb CJ, Appelt EA, Stupp SI. Self-Assembling Glucagon-Like Peptide 1-Mimetic Peptide Amphiphiles for Enhanced Activity and Proliferation of Insulin-Secreting Cells. Acta Biomater. 2012;8:1685–1692. doi: 10.1016/j.actbio.2012.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitmore L, Wallace BA. Protein Secondary Structure Analyses from Circular Dichroism Spectroscopy: Methods and Reference Databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 27.Koehl P, Levitt M. Structure-Based Conformational Preferences of Amino Acids. Proc Natl Acad Sci USA. 1999;96:12524–12529. doi: 10.1073/pnas.96.22.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paramonov SE, Jun HW, Hartgerink JD. Self-Assembly of Peptide–Amphiphile Nanofibers: The Roles of Hydrogen Bonding and Amphiphilic Packing. J Am Chem Soc. 2006;128:7291–7298. doi: 10.1021/ja060573x. [DOI] [PubMed] [Google Scholar]
- 29.Fuguet E, Ràfols C, Rosés M, Bosch E. Critical Micelle Concentration of Surfactants in Aqueous Buffered and Unbuffered Systems. Anal Chim Acta. 2005;548:95–100. [Google Scholar]
- 30.MacPhee CE, Woolfson DN. Engineered and Designed Peptide-Based Fibrous Biomaterials. Curr Opin Solid State Mater Sci. 2004;8:141–149. [Google Scholar]
- 31.Forns P, Lauer-Fields JL, Gao S, Fields GB. Induction of Protein-Like Molecular Architecture by Monoalkyl Hydrocarbon Chains. Biopolymers. 2000;54:531–546. doi: 10.1002/1097-0282(200012)54:7<531::AID-BIP60>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 32.Eisenberg D, Wilcox W, Eshita SM, Pryciak PM, Ho SP, DeGrado WF. The Design, Synthesis, and Crystallization of an Alpha•Helical Peptide. Proteins: Struct, Funct, Genet. 1986;1:16–22. doi: 10.1002/prot.340010105. [DOI] [PubMed] [Google Scholar]
- 33.Ho SP, Degrado WF. Design of a 4-Helix Bundle Protein: Synthesis of Peptides which Self-Associate into a Helical Protein. J Am Chem Soc. 1987;109:6751–6758. [Google Scholar]
- 34.Cella N, Contreras A, Latha K, Rosen JM, Zhang M. Maspin is Physically Associated with β1 Integrin Regulating Cell Adhesion in Mammary Epithelial Cells. FASEB J. 2006;20:1510–1512. doi: 10.1096/fj.05-5500fje. [DOI] [PubMed] [Google Scholar]
- 35.Bass R, Wagstaff L, Ravenhill L, Ellis V. Binding of Extracellular Maspin to β1 Integrins Inhibits Vascular Smooth Muscle Cell Migration. J Biol Chem. 2009;284:27712–27720. doi: 10.1074/jbc.M109.038919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnaoutova I, Kleinman HK. In Vitro Angiogenesis: Endothelial Cell Tube Formation on Gelled Basement Membrane Extract. Nat Protoc. 2010;5:628–635. doi: 10.1038/nprot.2010.6. [DOI] [PubMed] [Google Scholar]
- 37.West DC, Thompson WD, Sells PG, Burbridge MF. Angiogenesis Assays Using Chick Chorioallantoic Membrane. Methods Mol Med. 2001;46:107–129. doi: 10.1385/1-59259-143-4:107. [DOI] [PubMed] [Google Scholar]
- 38.O'Reilly MS, Holmgren L, Shing Y, Chen C, Rosenthal RA, Cao Y, Moses M, Lane WS, Sage ES, Folkman J. Angiostatin: A Circulating Endothelial Cell Inhibitor That Suppresses Angiogenesis and Tumor Growth. Cold Spring Harb Symp Quant Biol. 1994;59:471–482. doi: 10.1101/sqb.1994.059.01.052. [DOI] [PubMed] [Google Scholar]
- 39.Volpert OV, Ward WF, Lingen MW, Chesler L, Solt DB, Johnson MD, Molteni A, Polverini PJ, Bouck NP. Captopril inhibits angiogenesis and slows the growth of experimental tumors in rats. J Clin Invest. 1996;98:671–679. doi: 10.1172/JCI118838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.