Abstract
Structural disruption of gut microbiota and associated inflammation are considered important etiological factors in high fat diet (HFD)-induced metabolic syndrome (MS). Three candidate probiotic strains, Lactobacillus paracasei CNCM I-4270 (LC), L. rhamnosus I-3690 (LR) and Bifidobacterium animalis subsp. lactis I-2494 (BA), were individually administered to HFD-fed mice (108 cells day−1) for 12 weeks. Each strain attenuated weight gain and macrophage infiltration into epididymal adipose tissue and markedly improved glucose–insulin homeostasis and hepatic steatosis. Weighted UniFrac principal coordinate analysis based on 454 pyrosequencing of fecal bacterial 16S rRNA genes showed that the probiotic strains shifted the overall structure of the HFD-disrupted gut microbiota toward that of lean mice fed a normal (chow) diet. Redundancy analysis revealed that abundances of 83 operational taxonomic units (OTUs) were altered by probiotics. Forty-nine altered OTUs were significantly correlated with one or more host MS parameters and were designated ‘functionally relevant phylotypes'. Thirteen of the 15 functionally relevant OTUs that were negatively correlated with MS phenotypes were promoted, and 26 of the 34 functionally relevant OTUs that were positively correlated with MS were reduced by at least one of the probiotics, but each strain changed a distinct set of functionally relevant OTUs. LC and LR increased cecal acetate but did not affect circulating lipopolysaccharide-binding protein; in contrast, BA did not increase acetate but significantly decreased adipose and hepatic tumor necrosis factor-α gene expression. These results suggest that Lactobacillus and Bifidobacterium differentially attenuate obesity comorbidities in part through strain-specific impacts on MS-associated phylotypes of gut microbiota in mice.
Introduction
Humans are facing a devastating epidemic of metabolic syndrome (MS), symptoms of which include obesity, hyperglycemia, insulin resistance, hyperlipidemia and hypertension (Eckel et al., 2005). Accumulating evidence suggests that dysbiosis of gut microbiota induced by a high fat/high calorie diet has a key role in the development of obesity, insulin resistance and other hallmarks of MS (Ley et al., 2006; Turnbaugh et al., 2006). Reduction in beneficial bacteria (for example, Bifidobacterium, butyrate-producing bacteria) and increases in pro-inflammatory/pathogenic bacteria (for example, Desulfovibrionaceae) are consistently associated with the development of obesity, adipose tissue and systemic inflammation and metabolic comorbidities in both humans (Qin et al., 2012; Vrieze et al., 2012) and rodents (Zhang et al., 2010). Some species of gut bacteria are causally implicated in MS development. For example, the endotoxin-producing strain of Enterobacter cloacae B29 isolated from the gut of an obese human causes obesity in germ-free mice (Fei and Zhao, 2013), while a mucin-degrading strain Akkermansia muciniphila reduces high fat diet (HFD)-induced obesity and comorbidities (Everard et al., 2013; Shin et al., 2014). Taken together, these observations support the concept that targeting HFD-induced gut microbiota dysbiosis is an effective approach to obesity and MS therapy.
Probiotics are defined as ‘live microorganisms that, when administered in adequate amounts, confer a health benefit on the host' (Araya et al., 2002). Oral delivery of probiotic bacteria has been used as a gut microbiota-targeted strategy to combat MS (Delzenne et al., 2011). A variety of beneficial strains, either individually or more often as cocktails, have been shown to alleviate obesity, insulin resistance and/or hepatic steatosis in HFD-fed rodents. These strains include Bifidobacterium spp. (Ma et al., 2008; Amar et al., 2011; An et al., 2011; Chen et al., 2012; Xu et al., 2012), Lactobacillus spp. (Lee et al., 2006; Ma et al., 2008; Aronsson et al., 2010; Xu et al., 2012; Kim et al., 2013; Park et al., 2013b; Yoo et al., 2013), Streptococcus thermophilus (Ma et al., 2008) and Pediococcus pentosaceus (Zhao et al., 2012), as well as the gut commensals, Bacteroides uniformis (Gauffin Cano et al., 2012) and Akkermansia muciniphila (Everard et al., 2013; Shin et al., 2014). Importantly, different probiotic strains have been shown to have dramatically different capacities to modulate metabolic phenotypes (Yin et al., 2010; Fak and Backhed, 2012; Million et al., 2012). Despite these advances, the effects of different probiotics on gut microbiota composition and the relationship between these modulating effects and improvements in host metabolic health is poorly understood.
Most published studies of probiotics in subjects with MS have used culture-based or targeted molecular analyses (for example, quantitative PCR, microarray) to evaluate a limited number of gut microbiota components (for example, Bifidobacterium and Lactobacillus) (An et al., 2011; Chen et al., 2011; Xu et al., 2012; Everard et al., 2013). These studies provide an incomplete profile of probiotic-induced alterations in the microbiota, because bacteria that cannot be cultured or those that share low homology with the specific probes/primers utilized remain unidentified. Recently, using 454 pyrosequencing, Clarke et al. (2013) assessed family-level changes of gut microbiota induced by a bacteriocin-producing probiotic strain L. salivarius UCC118 in HFD-fed mice. However, different bacterial species in the same family or even genus may have contrasting responses to the same intervention (Zhang et al., 2010, 2012b), and it is therefore necessary to investigate the modulating effect of probiotics on the gut microbiota at the species level. Another study by Park et al. (2013b) profiled the gut microbiota of HFD-fed mice in response to a probiotic cocktail of L.plantarum KY1032 and L. curvatus HY7601 by using 454 pyrosequencing and univariate statistical approach. The probiotic cocktail ameliorated MS symptoms while increasing gut Lactobacillus and Bifidobacterium and decreasing Clostridiaceae, Akkermansia and Escherichia coli. Although the two probiotic strains they used showed different capacity in improving lipid metabolism and systemic inflammation when administered individually (Yoo et al., 2013), the effect of each strain on gut microbiota was not assessed separately. Furthermore, it is not clear whether these changed individual bacterial phylotypes were correlated with improvements in MS parameters or not.
The present study elucidates and compares the effects of three individual probiotic strains on host physiology and gut microbial community structure in mice with HFD-induced MS. Each of the three probiotic strains (Lactobacillus paracasei CNCM I-4270 (LC), L. rhamnosus CNCM I-3690 (LR), and Bifidobacterium animalis subsp. lactis CNCM I-2494 (BA)) attenuated weight gain, glucose intolerance and hepatic steatosis. However, the three strains differentially affected host inflammation and gut microbial fermentation. Moreover, although all probiotic strains partially reversed HFD-induced structural changes in the gut microbiota, each strain selectively altered a specific subset of key bacterial species that were significantly associated with one or more features of MS development or progression, and the strain-specific modulating effects of probiotics on these key bacterial phylotypes were partly reflected in strain-specific alleviation of obesity complications. Our results provide novel insight of the role of gut microbiota modulation in probiotic-dependent amelioration of MS.
Materials and methods
Animal trial
After 2 weeks acclimatization, 40 10-week-old male specific pathogen-free C57BL/6J mice were randomly divided into 5 treatment groups (8 mice per group). The eight mice in each group were housed in two cages (four per cage). One group of animals was fed normal chow (NC, containing 10% kcal from fat, 3.85 total kcal g−1, from Research Diets, Inc., New Brunswick, NJ, USA) as healthy controls, one group was fed HFD (containing 60% kcal from fat, 5.24 total kcal g−1, from Research Diets, Inc.) as model controls and received 200 μl de Man–Rogosa–Sharpe broth (OXOID, Basingstoke, UK) as placebo. The other three groups were maintained on HFD with administrations of 200 μl bacterial suspension of each of the three candidate probiotic strains, LC, LR and BA, in de Man–Rogosa–Sharpe broth by gavage at a dose of 108 cells day−1 for 12 weeks. The strain LC was isolated from a vegetable product, and LR and BA were isolated from dairy products. The three strains were selected as probiotics because LC was shown to be anti-inflammatory in vitro (unpublished data), LR was anti-inflammatory in vitro, in a preclinical model of Citrobacter infection (Collins et al., 2014) and in a TNBS-induced colitis model (Grompone et al., 2012), and a fermented milk product containing BA was shown to be anti-inflammatory in a murine model of colitis (Veiga et al., 2010). Details of mouse husbandry and the preparation of bacterial suspensions are described in the Supplementary Materials and Methods. The compositions of the experimental diets are shown in Supplementary Table S1.
Body weights and food intake were measured weekly. Stool samples were collected at baseline, second, sixth, eleventh and twelfth week. Serum, epididymal adipose tissue (eAT), liver and jejunum were collected, and oral glucose tolerance tests (OGTT) were conducted at the end of the twelfth week. Details of these procedures are provided in Supplementary Materials and Methods.
Enzyme-linked immunosorbent assay
Enzyme-linked immunosorbent assay kits were used to determine the amount of fasting insulin (Mercodia, Uppsala, Sweden), lipopolysaccharide (LPS)-binding protein (LBP; Cell Sciences, Canton, MA, USA) and adiponectin (R&D Systems, Minneapolis, MN, USA) in serum according to the manufacturer's instructions.
Histomorphology and immunohistochemistry
eAT and liver fixed in paraformaldehyde were embedded in paraffin, sectioned at 5 μm and processed as previously described (Strissel et al., 2007). Hepatic steatosis, adipocyte size, crown-like structures (CLS) and macrophages-expressing matrix metalloproteinase-12 (MMP-12) in eAT were determined as described in the Supplementary Materials and Methods.
Quantification of host gene expression
Total RNA was extracted from eAT, liver and jejunum of each of the 40 animals. For each mouse, transcript levels of GAPDH (glyceraldehyde-3-phosphate dehydrogenase), CD11c, tumor necrosis factor alpha (TNF-α), monocyte chemotactic protein-1, adiponectin and leptin in individual tissues were determined by real-time quantitative PCR as described in Supplementary Materials and Methods. Each qPCR reaction was performed in triplicate, and the expression level was determined as the mean value of the three replicas. Primers are listed in Supplementary Table S2.
454 pyrosequencing of fecal bacterial 16S rRNA gene V3 region
Fecal genomic DNA extraction, amplification of 16S rRNA gene V3 region and pyrosequencing of PCR amplicons were performed as described previously (Zhang et al., 2010, 2012b). PCR primers are listed in Supplementary Table S2.
Bioinformatics and statistical analysis of 454 pyrosequencing data
High-quality sequences were extracted, aligned in Greengenes (release 11_4) to remove sequences with <75% identity with Bacteria and then clustered using Cluster Database at High Identity with Tolerance (CD-HIT) to obtain representative unique sequences. Unique sequences were then subjected to RDP classifier to determine the phylogeny with a bootstrap cutoff of 50% (RDP database version 10.28). Operational taxonomic units (OTUs) were classified with Distance-Based OTU and Richness. Bacterial diversity was assessed with Rarefaction analysis and the Shannon diversity index. Principal component analysis (PCA) and weighted Fast UniFrac principal coordinate analysis (PCoA) based on OTUs were performed to provide an overview of gut microbial dynamics in response to HFD and probiotic treatments. Redundancy analysis models were constructed to identify OTUs that distinguished pairs of treatment groups. Details of these analyses are described in Supplementary Materials and Methods.
Cecal fermentation end products' measurement
The concentrations of the short-chain fatty acids, including acetate, propionate, butyrate and n-valerate, and branched chain fatty acids isobutyrate and isovalerate in the cecal content were determined using an Agilent 6890N gas chromatograph (Agilent Technologies, Wilmington, DE, USA; see Supplementary Materials and Methods).
Statistical analysis of physiological and biochemical data
To test differences in physiological and biochemical values for statistical significance, normally distributed data were analyzed by analysis of variance followed by Tukey post hoc test (SPSS Inc., Chicago, IL, USA). Data that did not meet the assumptions of analysis of variance were analyzed by the Mann–Whitney test (MATLAB R2010a). Differences were considered significant when P<0.05.
Correlations between gut microbial composition (PCoA coordinate scores and OTU abundances) and individual host parameters of obesity, glucose–insulin homeostasis and inflammation were identified using Spearman's correlation (MATLAB R2010a). Correlations between PCoA coordinates and host parameters were considered significant when P<0.05. False discovery rate control was used to account for multiple comparisons when evaluating correlations between OTUs and host parameters in Matlab software, and correlations were deemed significant at false discovery rate<0.25.
Accession number
The sequence information in this paper has been submitted to the GenBank Sequence Read Archive database under accession number SRP020353.
Results
Fecal recovery of the strains in the mouse intestine
One important trait of probiotics is their ability to survive transit through the upper gastrointestinal tract. Accordingly, we determined fecal levels of each of the three probiotic strains using reverse transcription (RT)-quantitative (q) PCR. Fecal levels of each strain increased significantly upon probiotic supplementation (Supplementary Figure S1). The amounts of LC, LR and BA were 107–109 (Supplementary Figure S1a), 106–109 (Supplementary Figure S1b) and 105–107 (Supplementary Figure S1c) cells per gram feces, respectively, during the probiotic administration. These results indicated that active probiotics were continuously present in the mouse gastrointestinal tract throughout the intervention period.
Probiotics attenuate features of the metabolic syndrome in HFD-fed mice
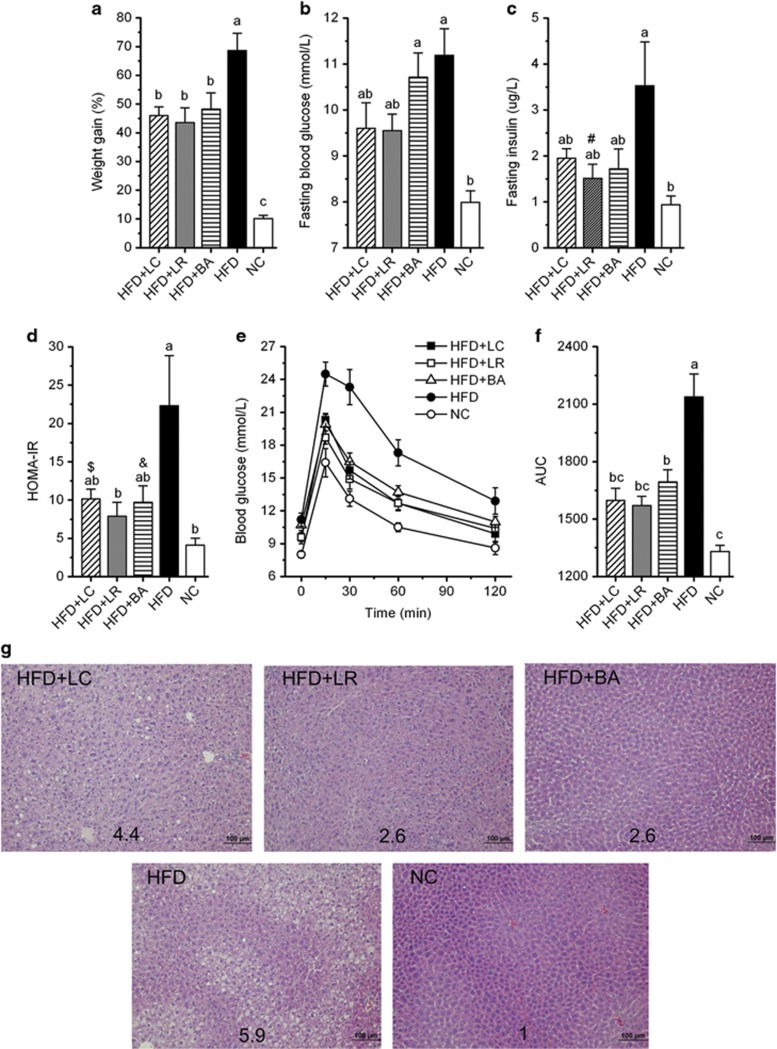
Compared with NC-fed mice, HFD-fed mice gained more weight (Figure 1a, Supplementary Figure S2) and developed hallmark features of metabolic syndrome, including elevated fasting blood glucose (Figure 1b) and fasting insulin (Figure 1c), increased homeostasis model assessment of insulin resistance (HOMA-IR) score (Figure 1d), impaired glucose clearance in the OGTT (Figures 1e and f) and increased hepatic steatosis (Figure 1g). Dietary supplementation with the three probiotic strains significantly attenuated HFD-induced weight gain (Figure 1a, Supplementary Figure S2) despite no reductions in food intake (Supplementary Figure S3). Attenuated weight gain was associated with modestly reduced blood glucose and insulin (Figures 1b and c), resulting in a >50% reduction in HOMA-IR (Figure 1d). In addition, each of the strains significantly increased the clearance of a glucose bolus during the OGTT (Figures 1e and f). Thus each of the probiotic strains enhanced glucose–insulin homeostasis by attenuating HFD-induced hyperinsulinemia, hyperglycemia and glucose intolerance. All probiotic strains also protected against HFD-induced hepatic steatosis (Figure 1g), with LR and BA appearing more protective than LC, based on steatosis score (Figure 1g). Considered together, these results demonstrated the ability of the three probiotic strains to mitigate HFD-induced weight gain and ectopic fat deposition and to enhance glucose–insulin homeostasis.
Figure 1.
Probiotics attenuated HFD-induced obesity, impaired glucose–insulin homeostasis and hepatic steatosis. (a) Body weight gain as the percentage of baseline weight for each mouse. (b) Fasting blood glucose. (c) Fasting insulin. (d) HOMA-IR, calculated by fasting blood glucose (mmol l−1) × fasting insulin (mU l−1)/22.5. (e) Curve of OGTT. (f) Areas under the curve (AUC) of OGTT. (g) Hematoxylin and eosin-stained liver sections, with the average steatosis ‘rank' for each treatment (lowest=1, highest=6) indicated by numbers. Data are shown as means±s.e.m. Values of each group with same letters are not significantly different by analysis of variance followed by Tukey post hoc test. #P=0.055, $P=0.086, &P=0.070 vs HFD group. n=8 mice per group.
Effects of probiotics on inflammation in eAT, liver and jejunum of HFD-fed mice
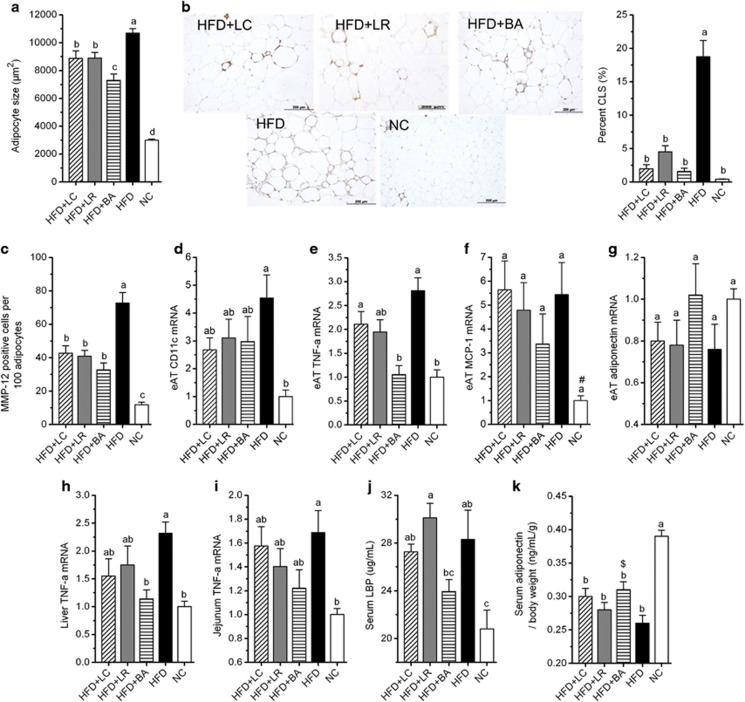
Large increases in adipocyte size (hypertrophy) are associated with recruitment of inflammatory macrophages that express CD11c and MMP-12 and aggregate in inflammatory CLS' around moribund adipocytes (Cinti et al., 2005; Strissel et al., 2007; Shaul et al., 2010). CLS frequency is a predictor of the severity of adipose inflammation and impaired glucose–insulin homeostasis (Strissel et al., 2007; Shaul et al., 2010). In the present study, mean adipocyte size in eAT was increased >250% by HFD feeding (Figure 2a). All probiotic strains reduced mean adipocyte size in response to HFD (Figure 2a), reflecting decreased frequency of larger adipocytes (data not shown). HFD also promoted macrophage infiltration and inflammation in eAT, assessed as increases in CLS and MMP-12-positive macrophages and elevated expression of CD11c, TNF-α and leptin (Figures 2b–e, Supplementary Figure S4). Notably, the three probiotic strains reduced the number of CLS (Figure 2b) and MMP-12-positive cells (Figure 2c) and tended to reduce CD11c expression (Figure 2d) in eAT of HFD-fed mice. In addition, BA (but not LC or LR) significantly reduced TNF-α expression to the level observed in mice fed NC (Figure 2e). Transcript levels of monocyte chemotactic protein-1 (Figure 2f) and adiponectin (Figure 2g) were not significantly altered by diet or probiotics. Neither of the probiotics reduced leptin gene expression compared with the HFD group (Supplementary Figure S4).
Figure 2.
Probiotics mitigated macrophage infiltration in eAT, and BA significantly decreased adipose and hepatic TNF-α gene expression. (a) Mean cell area of adipocyte in eAT (n=8 mice per group). (b) CLS, bar=200 μm; CLS were counted in three sections (⩾600 adipocytes) per mouse (n=5 mice per group); data are expressed as ‘percent CLS' for analysis, data were transformed as ‘arcsin √x'. (c) Quantitation of MMP-12-positive cells in fixed sections of eAT, and data were derived from five ( × 400) microscopic fields from each of 8 mice per diet cohort. (d–g) Gene expression levels of CD11c (d), TNF-α (e), monocyte chemotactic protein-1 (f) and adiponectin (g) in eAT (n=8 mice per group). (h–i) Gene expression levels of TNF-α in the liver (h) and jejunum (i) (n=8 mice per group). All mRNA quantification data were normalized to the housekeeping gene GAPDH. Gene expression levels were expressed as values relative to the NC group. (j) Serum LBP (n=8 mice per group). (k) Serum adiponectin corrected for body weight (n=8 mice per group). Data are shown as means±s.e.m. Values of each group with same letters are not significantly different by analysis of variance followed by Tukey post hoc test. #P=0.078, $P=0.059 vs HFD group.
HFD significantly enhanced TNF-α expression in the liver (Figure 2h) and jejunum (Figure 2i). As in eAT, BA reduced hepatic TNF-α expression to the level measured in the NC group, whereas reductions in TNF-α were not statistically significant in the LC and LR groups (Figure 2h). TNF-α mRNA levels in the jejunum were not significantly reduced by any probiotic strain (Figure 2i).
Taken together, these results indicate that the three probiotic strains significantly mitigated macrophage infiltration in eAT of HFD-fed mice and that BA more robustly attenuated eAT and hepatic TNF-α expression than either LC or LR.
Effects of probiotics on systemic inflammation in HFD-fed mice
We measured circulating LBP (a marker of endotoxin load) and the anti-inflammatory adipokine adiponectin as indices of systemic inflammation (Engeli et al., 2003; Sun et al., 2010). As expected, HFD feeding significantly increased serum LBP (Figure 2j) and lowered serum adiponectin concentrations (corrected for body weight) (Figure 2k). LBP levels of HFD-fed mice receiving probiotic strains LC or LR remained substantially elevated and significantly higher than those of mice fed NC. In contrast, LBP concentrations of HFD-fed mice receiving BA were attenuated to levels intermediate between the HFD and NC groups and were significantly lower than LBP levels in mice receiving probiotic strain LR (P=0.048) (Figure 2j). Adiponectin levels (corrected for body weight) were increased in mice fed HFD+BA vs mice fed HFD alone (P=0.059) but were not significantly increased in HFD-fed mice receiving either LC or LR strains (Figure 2k). These results suggest that BA more effectively attenuated endotoxin load and systemic inflammation than LC and LR.
Overall structural changes of the gut microbiota in response to HFD and probiotic intervention
To profile the effects of diet and probiotics on microbiota structure, we performed 454 pyrosequencing of bacterial 16S rRNA gene V3 region for 80 fecal samples collected from 40 mice at baseline and after 12 weeks. The quality of the 454 run (containing totally 148 samples) that included the 80 fecal samples from our study is shown in Supplementary Table S3. A total of 301 568 usable raw reads were obtained. On average, each fecal sample provided 3770±677 reads (Supplementary Figure S5). A total of 20 785 unique sequences (accounting for 99.9% of usable raw reads) were generated with NAST alignment and CD-HIT clustering and were then binned into OTUs at 98% similarity level, because higher thresholds generated a dramatic increase of OTU numbers, which might represent the microdiversity at the subspecies level (Supplementary Figure S5b), and 2794 OTUs were obtained. At this sequencing depth, rarefaction curves did not plateau (Supplementary Figure S6a), but Shannon diversity indices for all samples were stable (Supplementary Figure S6b). This suggested that although rare new phylotypes would still appear upon further sequencing, most diversity had already been covered.
RDP classifier assigned 98.2% of usable raw reads to 18 different phyla (data not shown). The most abundant phyla included Bacteroidetes (47.4% of usable raw reads), Firmicutes (41.4% of usable raw reads), Proteobacteria (6.4% of usable raw reads), Verrucomicrobia (1.2% of usable raw reads) and Actinobacteria (0.8% of usable raw reads). Twelve weeks of HFD feeding induced widespread changes in gut microbial community structure at the phylum level, with abundances of Firmicutes and Proteobacteria increased and abundances of Bacteroidetes and Actinobacteria decreased (Supplementary Figures S7, S8a, b, d and e). The ratio of Firmicutes to Bacteroidetes was increased upon HFD, and no probiotic strain attenuated this increase (Supplementary Figure S8c). However, LR (but neither BA nor LC) mitigated the HFD-induced increase in Proteobacteria and decrease in Actinobacteria (Supplementary Figures S8d and e).
PCA based on OTU abundance was performed to provide an overview of the gut microbiota composition of five animal groups at the baseline and at the end of the trial. Plotted PCA scores indicated no detectable difference in microbiota composition among groups before the intervention (Supplementary Figure S9). PC1, accounting for 24.3% of total variance, predominantly reflected age-related changes in gut microbiota composition, as PC1 clearly separated samples obtained at baseline from those obtained at week 12. PC2, accounting for 18.4% of total variance, separated the NC group from the four HFD-fed groups at week 12, indicating that PC2 reflects the effect of diet (Supplementary Figure S9).
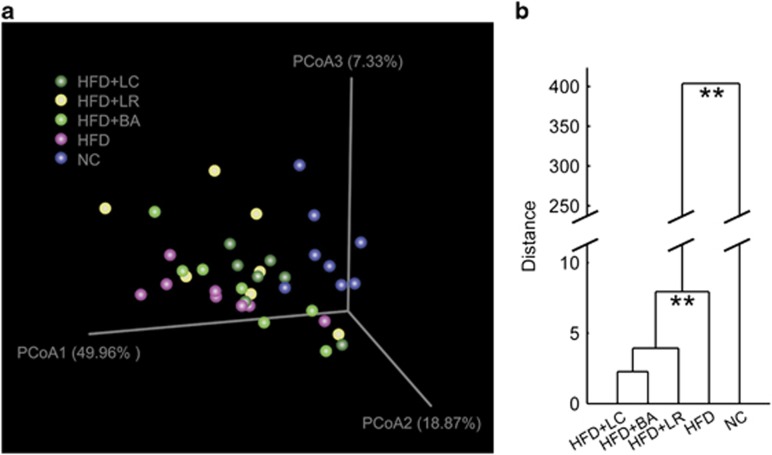
Weighted UniFrac PCoA was then performed on the OTU abundance matrix obtained from the five animal groups at week 12. The PCoA scores clearly separated the HFD group from the NC group with the three probiotic-treated groups distributed in between (Figure 3a). Multivariate analysis of variance derived from PCoA scores confirmed a statistically significant separation between the microbiota of NC- and HFD-fed mice and between the HFD microbiota and the microbiota of the three HFD+probiotic groups (Figure 3b). Thus each of the three probiotic strains shifted the overall structure of the HFD-disrupted gut microbiota toward that of lean mice fed a NC diet.
Figure 3.
Probiotics changed overall structure of gut microbiota. (a) Weighted UniFrac PCoA plot based on OTU abundance. Each point represents the fecal microbiota of a mouse. One mouse from group HFD+LR was detected as an outlier owing to the predominance of OTUs belonging to Erysipelotrichaceae (50.6% of the total bacteria, data not shown) and was thus excluded from the analysis. (b) Clustering of gut microbiota based on mahalanobis distances between different groups calculated with multivariate analysis of variance test, **P<0.01.
Correlation between overall microbiota structure and host MS parameters
We next used Spearman's correlation analysis to determine associations between MS parameters and overall microbiota composition represented by the PCoA coordinates (Table 1). Compositional changes of the gut microbiota along PC1 were significantly associated with obesity (weight gain), glucose–insulin homeostasis (areas under the curve of OGTT, fasting blood glucose, fasting insulin, HOMA-IR) and inflammation (serum LBP, serum adiponectin corrected for body weight, eAT CLS, eAT TNF-α mRNA). In addition, gut microbiota changes along PC2 were significantly correlated with liver TNF-α mRNA.
Table 1. Spearman's correlation between gut microbiota composition represented by the first three coordinates of weighted UniFrac PCoA and host MS parameters.
| MS parameters |
PCoA1 |
PCoA2 |
PCoA3 |
|||
|---|---|---|---|---|---|---|
| r | P | r | P | r | P | |
| Serum adiponectin/body weight | 0.52 | 0.001 | 0.22 | 0.174 | −0.05 | 0.784 |
| eAT adiponectin mRNA | 0.27 | 0.099 | −0.05 | 0.753 | −0.02 | 0.916 |
| Weight gain | −0.57 | <0.001 | −0.36 | 0.023 | 0.02 | 0.917 |
| AUC of OGTT | −0.64 | <0.001 | −0.22 | 0.181 | −0.09 | 0.593 |
| FBG | −0.40 | 0.012 | −0.48 | 0.002 | 0.06 | 0.704 |
| FINS | −0.41 | 0.009 | −0.25 | 0.126 | 0.07 | 0.689 |
| HOMA-IR | −0.43 | 0.006 | −0.32 | 0.044 | 0.05 | 0.752 |
| Serum LBP | −0.42 | 0.008 | −0.04 | 0.806 | 0.35 | 0.028 |
| Adipocyte size | −0.62 | <0.001 | −0.21 | 0.198 | 0.11 | 0.502 |
| eAT CLS | −0.64 | 0.001 | −0.10 | 0.635 | −0.01 | 0.963 |
| eAT CD11c mRNA | −0.57 | <0.001 | −0.14 | 0.403 | −0.09 | 0.567 |
| eAT MMP-12 | −0.47 | 0.002 | −0.42 | 0.007 | 0.02 | 0.904 |
| eAT TNF-α mRNA | −0.39 | 0.014 | −0.23 | 0.156 | −0.04 | 0.819 |
| eAT MCP-1 mRNA | −0.41 | 0.010 | −0.35 | 0.028 | 0.18 | 0.266 |
| Liver TNF-α mRNA | −0.23 | 0.151 | −0.47 | 0.003 | −0.25 | 0.132 |
| Jejunum TNF-α mRNA | −0.33 | 0.043 | −0.26 | 0.107 | −0.04 | 0.796 |
Abbreviations: AUC, area under the curve; CLS, crown-like structures; eAT, epididymal adipose tissue; FBG, fasting blood glucose; FINS, fasting insulin; HOMA-IR, homeostasis model assessment of insulin resistance; LBP, lipopolysaccharide-binding protein; MCP-1, monocyte chemotactic protein 1; MMP-12, matrix metalloproteinase-12; MS, metabolic syndrome; OGTT, oral glucose tolerance tests; PCoA, principal coordinate analysis; TNF-α, tumor necrosis factor-α.
PCoA1–PCoA3 represents the first three principal components of weighted UniFrac PCoA of the gut microbita across all the animal groups at twelfth week of the trial. Significant correlations (P<0.05) are in bold.
Strain-specific modulating effects of probiotic intervention on gut microbiota
Redundancy analysis models were constructed to identify specific bacterial phylotypes whose abundance was changed by HFD feeding (HFD group vs NC group) and by probiotic treatment (HFD group vs each HFD+probiotic group). Gut microbiota was significantly different between the HFD group and each of the other four groups (P=0.002 for all models, Supplementary Figures S10a–d). Specific differences in composition were revealed along the canonical axis, which explained 21.3% (HFD vs NC group), 11.5% (HFD vs HFD+LC group), 11.9% (HFD vs HFD+LR group) and 9.6% (HFD vs HFD+BA group) of the variance in OTU abundance, respectively (Supplementary Figures S10a–d).
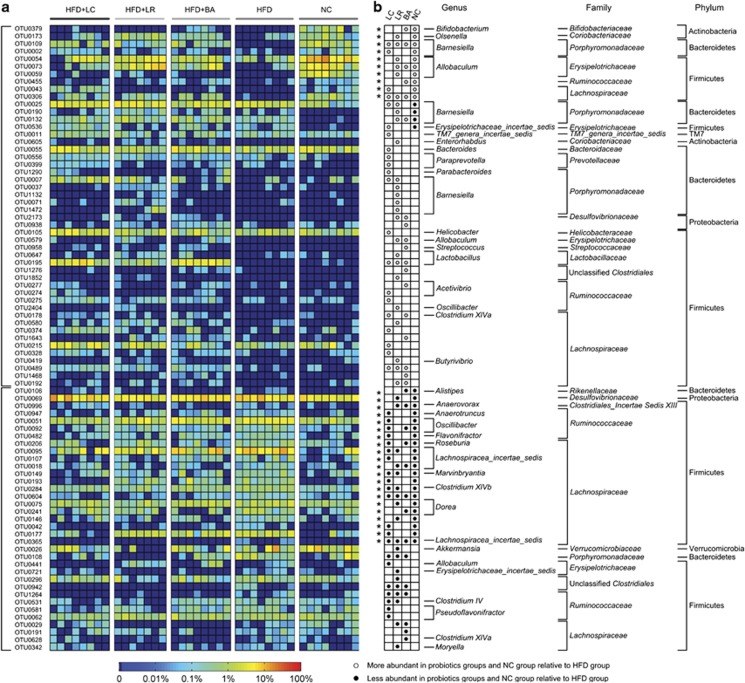
In all, HFD feeding altered the abundance of 118 OTUs (Supplementary Figure S10a). Supplementation with LC, LR and BA enhanced or reduced the abundance of 42, 44 and 32 OTUs, respectively (Supplementary Figures S10b–d), resulting in changes in 83 different OTUs (Figure 4a). OTUs that were changed in abundance by at least two probiotic strains were altered in the same direction (that is, increased or decreased; Figure 4b). Notably, probiotic strains reversed changes in 31 (of 118) OTUs that were altered by HFD (Figure 4b). However, we note that the three probiotic strains altered the abundances of different sets of OTUs (Figure 4, Supplementary Figure S11, Supplementary Table S4). Among the 83 OTUs changed by probiotics, only 9 OTUs were altered by all the three strains, 17 OTUs were altered by two probiotics and 24, 21 and 12 OTUs (totally 57 OTUs) were uniquely altered by LC, LR, and BA, respectively (Supplementary Figure S11, Figure 4). Thus the preponderance of the OTUs was altered by probiotics in a strain-specific manner.
Figure 4.
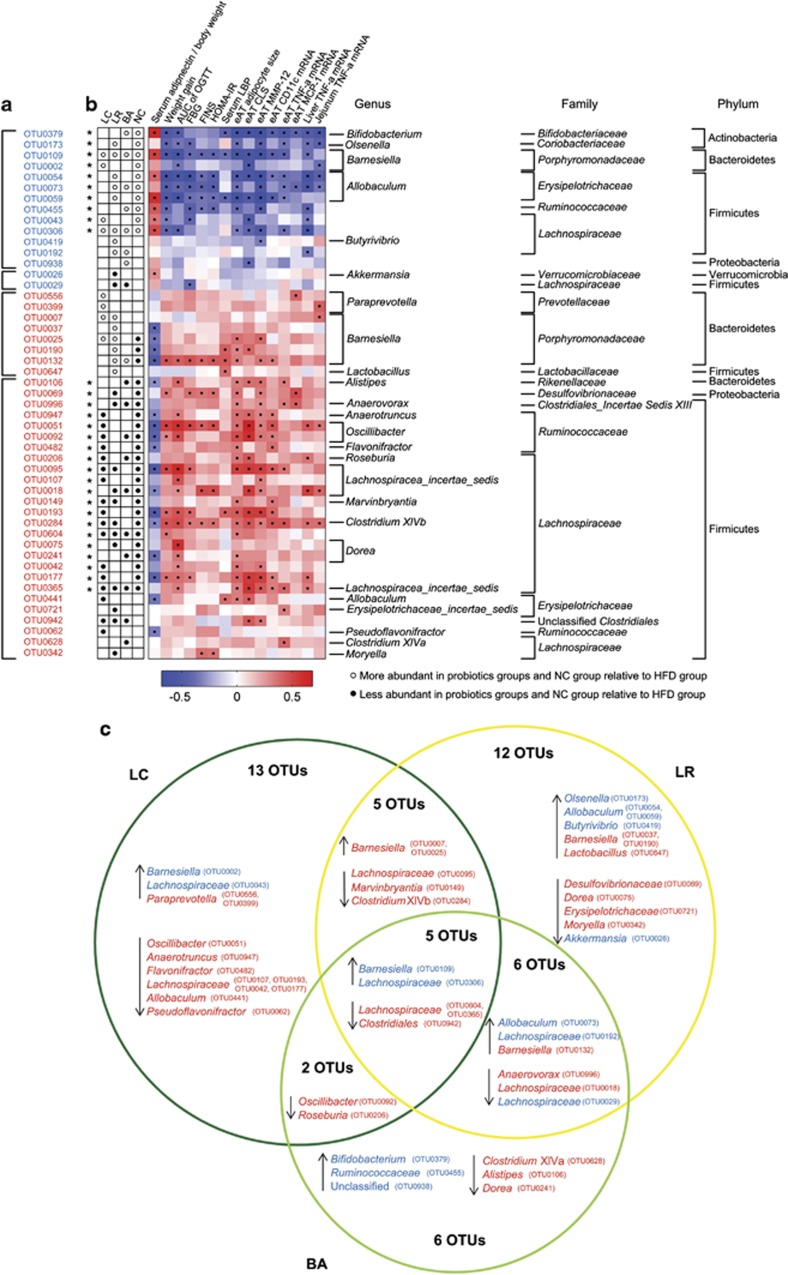
Eighty-three OTUs that were changed in abundance by probiotics according to redundancy analysis (RDA). (a) Heatmap of the abundance of 83 OTUs. Rows correspond to 48 OTUs enriched and 35 OTUs reduced. (b) The changing direction of the 83 OTUs by probiotics according to RDA. Circles and dots indicate the abundance of OTUs that were more and less abundant, respectively, in probiotics groups and NC group relative to HFD group. The taxonomy of the OTUs (genus, family and phylum) is depicted on the right. Asterisk (*) represents the OTU whose abundance was changed by HFD and then the change was reversed by probiotics.
Correlation between 83 OTUs changed by probiotics and host MS phenotypes
Our next goal was to identify specific gut bacteria that potentially mediate the salutary effects of the probiotic strains on HFD-induced MS. Spearman's correlation analysis was performed between the 83 OTUs that were changed by probiotics and specific MS parameters in all the groups of mice. In total, 49 OTUs (herein designated ‘key' OTUs) were significantly correlated with at least one MS parameter. Thirty-four of these key OTUs were correlated with decreased adiponectin and increased weight, inflammation and/or metabolic dysregulation, and thus they were positively correlated with MS disease phenotypes. Fifteen key OTUs were correlated with increased adiponectin and reduced adiposity, inflammation and/or metabolic dysregulation, and thus they were negatively correlated with MS disease phenotypes (Figures 5a and b). Thirty of the 31 OTUs whose abundances were altered by HFD and reversed by probiotics were among the 49 key OTUs (Figure 5; see also Figure 4).
Figure 5.
Forty-nine probiotic-altered OTUs that were significantly correlated with host MS parameters. (a) The changing direction of the 49 OTUs. Circles and dots indicate the OTUs that were more and less abundant, respectively, in the probiotics groups and NC group relative to the HFD group. Asterisk (*) represents OTU whose abundance was changed by HFD and then the change was reversed by probiotics. (b) The correlation between 49 OTUs and host MS parameters. Rows correspond to OTUs with the IDs shown on the left, and columns correspond to MS parameters related to obesity, glucose–insulin homeostasis and inflammation. Colors red and blue denote positive and negative association, respectively. The intensity of the colors represents the degree of association between the OTU abundances and host parameters as assessed by the Spearman's correlations. The black dots in the blue/red cells indicate that the associations were significant (false discovery rate<0.25). The taxonomy of the OTUs is shown on the right. (c) Venn diagrams of 49 OTUs. The OTUs' taxonomy is listed, and blue and red mean that the OTU was negatively and positively, respectively, correlated with MS parameters. ↑ means increased by probiotics, and ↓ represents decreased by probiotics.
Notably, 13 of the 15 OTUs that were negatively correlated with MS disease phenotypes were enriched by at least one probiotic strain (Figures 5a and b), including bacteria belonging to Bifidobacterium, Olsenella, Barnesiella, Allobaculum, Butyrivibrio, unclassified Lachnospiraceae and unclassified Proteobacteria. Similarly, 26 of the 34 OTUs that were positively correlated with MS disease phenotypes were reduced by probiotic treatment (Figures 5a and b), including representatives from Alistipes, Desulfovibrionaceae, Oscillibacter, Clostridium XIVb, Dorea and Clostridium XIVa.
Consistent with the strain-specific effects of the three probiotics on OTU abundances (Supplementary Figure S11, Figure 4), the 49 key bacterial phylotypes were differentially represented among the three probiotic-treated animal groups (Figure 5c). Only 5 OTUs were commonly changed by all three probiotics, whereas 31 of the 49 OTUs (13 by LC, 12 by LR and 6 by BA) were changed by just one individual probiotic strain (Figure 5c). LC and LR increased four and six OTUs affiliated to Barnesiella, respectively, and BA only enhanced two Barnesiella OTUs. Only BA (not LC or LR) increased the abundance of bacteria from Bifidobacterium (B. animalis, data not shown).
Effects of probiotics on cecal end products of fermentation
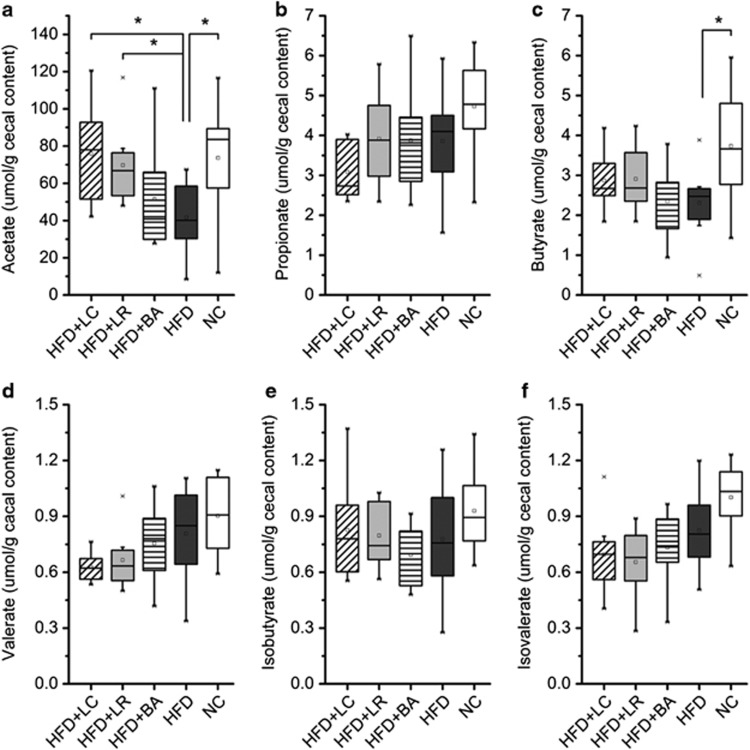
We measured cecal concentrations of the short-chain fatty acids (acetate, propionate, butyrate, valerate) and the branched chain fatty acids (isobutyrate and isovalerate). HFD significantly decreased concentrations of acetate and butyrate as compared with NC (Figures 6a and c). LC and LR administration significantly restored the concentration of acetate in mice fed HFD, whereas BA did not (Figure 6a). The probiotic supplementation did not alter fecal concentrations of propionate, butyrate, valerate, isobutyrate or isovalerate (Figures 6b–f).
Figure 6.
Probiotics LC and LR elevated the concentrations of acetate in cecal content. (a–f) Levels of acetate (a), propionate (b), butyrate (c), valerate (d), isobutyrate (e) and isovalerate (f). In the box plot, the bottom and top are, respectively, the 25th and 75th percentile, a line within the box marks the median and a circle in the box shows the mean. Whiskers above and below the box indicate 1.5 interquartile range of the lower and upper quartile, respectively, and samples beyond are regarded as outliers. *P<0.05 by Mann–Whitney test.
Discussion
Although probiotics are regarded as a gut microbiota-targeted strategy to combat MS, the potential role of gut microbiota modulation in probiotic-dependent amelioration of MS is not clearly understood. The new findings of our study compared with previous related studies are: (1) Not all the gut bacteria changed by probiotics were relevant to the probiotic-mediated improvement of MS; only those that were significantly correlated with disease phenotypes were identified as functionally relevant phylotypes, among which probiotics enriched phylotypes negatively correlated with disease phenotypes and reduced phylotypes positively correlated with disease phenotypes; (2) The strain-specific modulating effects of Lactobacillus and Bifidobacterium probiotics on functionally relevant phylotypes were reflected in differential attenuating impacts of these strains on obesity comorbidities.
In this study, three probiotic strains were tested individually for their capacity to reduce HFD-induced MS. All strains significantly attenuated HFD-induced weight gain, improved glucose–insulin homeostasis and reduced hepatic steatosis, consistent with other studies of Lactobacillus and Bifidobacterium probiotic strains tested individually or as cocktails (Lee et al., 2006; Ma et al., 2008; Aronsson et al., 2010; Amar et al., 2011; An et al., 2011; Chen et al., 2012; Kim et al., 2013; Park et al., 2013b). We additionally demonstrate that each of the three probiotic strains significantly reduced the infiltration of pro-inflammatory (CD11c+, MMP-12+) macrophages into adipose tissue and the formation of CLS. Macrophage infiltration is an underlying cause of chronic adipose inflammation, insulin resistance and other obesity complications (Kanety et al., 1995; Aguirre et al., 2002; Weisberg et al., 2003; Xu et al., 2003; Strissel et al., 2007; Cani et al., 2008). Moreover, reduced numbers of adipose tissue macrophages is a common feature of interventions that attenuate insulin resistance and other obesity complications (Xu et al., 2003; Cancello et al., 2005; Cani et al., 2008). Reducing macrophage infiltration into adipose tissue is therefore one potential mechanism by which probiotics protected HFD-fed mice in the present study. Reductions in macrophage infiltration are consistent with the probiotic-associated reductions in adipocyte size that we observed but may also reflect modulation of gut microbiota, resulting in reduced systemic levels of gut-derived LPS (Caesar et al., 2012) and/or increased gut-derived acetate (Carvalho et al., 2012).
Many environmental perturbations can cause compositional changes of gut microbiota. Indeed, unsupervised multivariate analysis revealed that each of the three probiotic strains induced significant changes in overall gut microbiota structure that partially ameliorated HFD-induced structural dysbiosis. These overall structural changes of gut microbiota (PCs of the UniFrac plot) were correlated with MS parameters in our model. Previous studies showed that different bacterial species in the same genus may respond differentially to the same environmental stressor such as change of diet from high fat to NC (Zhang et al., 2010, 2012a). It is thus important to identify changes of species-level phylotypes in response to probiotic treatment. Park's multi-Lactobacillus-strain cocktail enriched 14 and reduced 8 species-level OTUs (Park et al., 2013b), while our three individual probiotic strains increased 48 and decreased 35 species-level OTUs in total. However, studies have also shown that not all compositional changes induced by an external stressor matter for host health (Robinson et al., 2010). For example, in HFD-fed mice (Cani et al., 2007), the supplementation of oligofructose attenuated endotoxemia, obesity and diabetes and significantly enriched Bifidobacterium spp., Eubacterium rectale–Clostridium coccoides group and decreased Enterobacteriaceae, but only Bifidobacterium spp. was significantly and negatively correlated with plasma endotoxin level in Pearson's correlation analysis and thus was considered as a main gut bacterial group mediating the beneficial effects of the prebiotic treatment; in another study on obese women ingesting inulin-type fructans (Dewulf et al., 2013), correlation analysis between the bacterial groups modified by the prebiotic treatment and host physiological parameters identified Bifidobacterium, Faecalibacterium prausnitzii, Bacteroides intestinalis, B. vulgatus and Propionibacterium as potentially important players in the modulation of host metabolism. In the present study, we applied correlation analysis to further identify phylotypes whose abundances were not only changed by the probiotic treatments but also correlated with at least one host metabolic parameter. Indeed, the results showed that not all but only 49 out of the 83 altered OTUs were significantly correlated with one or more host metabolic indexes.
Among the 49 key phylotypes, 15 and 34 were negatively and positively, respectively, correlated with MS phenotypes (decreased adiponectin and increased weight, inflammation and/or metabolic dysregulation). The key phylotypes that were negatively correlated with MS phenotypes may include potentially beneficial bacteria, and the majority of them (13 out of 15) were increased by at least one of the three probiotic strains. Meanwhile, those key phylotypes that were positively correlated with MS phenotypes may consist of potentially harmful bacteria, and they were, for the most part (26 out of the 34), decreased. For example, the probiotic strains enriched one or more key phylotypes of Bifidobacterium, Olsenella, Barnesiella, Allobaculum and Butyrivibrio that were previously associated with alleviated MS or were shown to prevent dextran sulfate sodium-induced inflammation (Ohkawara et al., 2006; Zhang et al., 2010, 2012b; Le Roy et al., 2012). Conversely, probiotic treatment reduced key phylotypes within Desulfovibrionaceae, Oscillibacter and Clostridium XIVa that were implicated in the development of obesity-related metabolic disorders (Zhang et al., 2010; Lam et al., 2012) or include pro-inflammatory bacterial species (Tuovinen et al., 2013). Thus, these key OTUs were designated as ‘functionally relevant phylotypes' as they may mediate the beneficial effects of the probiotics on the host. Our study is therefore distinguished from previous studies in which associations between individual probiotic-modulated OTUs and improvements in MS were not determined (Park et al., 2013b).
Moreover, in contrast to studies using a single probiotic strain or a multiple-strain probiotic cocktail, we associate different patterns of probiotic-induced gut microbiota alteration with different impacts on MS pathophysiology. In the present study, each of our three probiotic strains altered a distinct set of functionally relevant gut bacterial phylotypes and also exerted strain-specific alleviation of obesity complications. For example, compared with BA, strains LC and LR increased more phylotypes belonging to Barnesiella, some species of which have been reported to produce acetate (Sakamoto et al., 2007). Indeed, LC and LR (but not BA) elevated acetate production in HFD-fed mice, thereby reversing HFD-induced disruption of gut microbial fermentation. Acetate has been shown to suppress body fat accumulation and inflammation in obese or diabetic rodents by multiple mechanisms (Sakakibara et al., 2006; Yamashita et al., 2007; Kondo et al., 2009; Carvalho et al., 2012). Note also that only BA increased Bifidobacterium animalis, subspecies of which reduced inflammation in vitro or in mice with colitis (Roselli et al., 2006; Veiga et al., 2010). In the present study, BA (but neither LC nor LR) significantly reduced adipose and hepatic TNF-α gene expression and tended to lower circulating LPS load (serum LBP concentration). Together, these observations exemplify how strain-specific modulating effects of probiotics on key gut bacterial phylotypes can be reflected in strain-specific impacts on obesity complications.
The approach used in the present study forms the basis of a discovery pipeline for identifying new probiotics from endogenous gut bacterial species. Specifically, each of the 13 key bacterial phylotypes that were both increased by probiotics and negatively correlated with MS disease parameters will be isolated with a sequence-guided isolation scheme (Fei and Zhao, 2013) and then inoculated into animal models of MS. Bacteria that are cultivable and that attenuate obesity and related disorders in animal models will be considered potential probiotics. The feasibility of this strategy was very recently demonstrated in studies with the endogenous murine commensal A. muciniphila. After its identification as a positive correlate of prebiotic- and pharmacologic-induced improvements in obesity and diabetes, A. muciniphila was administered orally to mice and shown to improve MS symptomology, including excessive fat accumulation, adipose tissue inflammation and glucose intolerance (Everard et al., 2011, 2013; Shin et al., 2014).
Yoo et al. (2013) showed that multiple-strain probiotics were more effective than each of the constituent single-strain probiotics because of the synergistic effects on host health, therefore, it can be inferred that the combination of probiotic strains with complementary effects on gut microbiota and host health would comprehensively improve different symptoms of metabolic syndrome and thus generate optimal synergistic efficacy. Here, our Lactobacillus and Bifidobacterium strains differentially affected host inflammation, gut microbial fermentation and gut microbiota composition; as a result, we hypothesize that multi-strain probiotics consisting of both our Lactobacillus and Bifidobacterium strains would be more effective than probiotics with only Lactobacillus strains. In the future, it would be interesting to compare the effects of different probiotic combinations on MS to formulate multi-strain probiotics with optimum efficacy based on their complementary impacts on gut microbiota and to test the formula at different cellular densities to determine the optimal dosage (Park et al., 2013a).
In conclusion, we demonstrate the utility of two Lactobacillus and one Bifidobacterium strains to individually attenuate HFD-induced obesity, inflammation and MS in mice. One or more probiotic strains beneficially altered the abundance of most ‘functionally relevant' gut bacterial phylotypes that were associated with MS parameters. Most functionally relevant phylotypes were selectively altered by only one probiotic strain, suggesting that the strain-specific salutary effects on MS pathophysiology partially reflect strain-specific impacts on functionally relevant phylotypes. Although translation of these findings to clinical populations remains a significant challenge, ‘gut microbiota-targeted' probiotic intervention strategies may become an important element in diet and lifestyle alterations aiming to prevent or attenuate obesity and related metabolic disorders.
Acknowledgments
We thank Chloé Béal for technical help on RT-qPCR of probiotic strains, M Zhang for statistics expertise and G Wang and J Wang for 454 pyrosequencing data extraction and submission. The Yakult Intestinal Flora-SCAN (YIF-SCAN) technology, developed by Yakult and operated under licence, was used in this study to quantify the live probiotics in feces with RNA-based retro-transcription quantitative PCR. This work was supported by a grant from Danone Research, National Natural Science Foundation of China program grants 31121064 and 81100632.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aguirre V, Werner ED, Giraud J, Lee YH, Shoelson SE, White MF. Phosphorylation of Ser307 in insulin receptor substrate-1 blocks interactions with the insulin receptor and inhibits insulin action. J Biol Chem. 2002;277:1531–1537. doi: 10.1074/jbc.M101521200. [DOI] [PubMed] [Google Scholar]
- Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermudez-Humaran LG, et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med. 2011;3:559–572. doi: 10.1002/emmm.201100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An HM, Park SY, Lee do K, Kim JR, Cha MK, Lee SW, et al. Antiobesity and lipid-lowering effects of Bifidobacterium spp. in high fat diet-induced obese rats. Lipids Health Dis. 2011;10:116. doi: 10.1186/1476-511X-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya MML, Reid G, Sanders ME, Stanton C. Guidelines for the Evaluation of Probiotics in Food. Jt. FAO/WHO Work. Group: London, UK; Ontario, Canada; 2002. [Google Scholar]
- Aronsson L, Huang Y, Parini P, Korach-Andre M, Hakansson J, Gustafsson JA, et al. Decreased fat storage by Lactobacillus paracasei is associated with increased levels of angiopoietin-like 4 protein (ANGPTL4) PLoS One. 2010;5:e13087. doi: 10.1371/journal.pone.0013087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar R, Reigstad CS, Backhed HK, Reinhardt C, Ketonen M, Lunden GO, et al. Gut-derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut. 2012;61:1701–1707. doi: 10.1136/gutjnl-2011-301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancello R, Henegar C, Viguerie N, Taleb S, Poitou C, Rouault C, et al. Reduction of macrophage infiltration and chemoattractant gene expression changes in white adipose tissue of morbidly obese subjects after surgery-induced weight loss. Diabetes. 2005;54:2277–2286. doi: 10.2337/diabetes.54.8.2277. [DOI] [PubMed] [Google Scholar]
- Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- Carvalho BM, Guadagnini D, Tsukumo DM, Schenka AA, Latuf-Filho P, Vassallo J, et al. Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia. 2012;55:2823–2834. doi: 10.1007/s00125-012-2648-4. [DOI] [PubMed] [Google Scholar]
- Chen J, Wang R, Li XF, Wang RL. Bifidobacterium adolescentis supplementation ameliorates visceral fat accumulation and insulin sensitivity in an experimental model of the metabolic syndrome. Br J Nutr. 2012;107:1429–1434. doi: 10.1017/S0007114511004491. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Wang R, Li XF, Wang RL. Bifidobacterium longum supplementation improved high-fat-fed-induced metabolic syndrome and promoted intestinal Reg I gene expression. Exp Biol Med (Maywood) 2011;236:823–831. doi: 10.1258/ebm.2011.010399. [DOI] [PubMed] [Google Scholar]
- Cinti S, Mitchell G, Barbatelli G, Murano I, Ceresi E, Faloia E, et al. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res. 2005;46:2347–2355. doi: 10.1194/jlr.M500294-JLR200. [DOI] [PubMed] [Google Scholar]
- Clarke SF, Murphy EF, O'Sullivan O, Ross RP, O'Toole PW, Shanahan F, et al. Targeting the microbiota to address diet-induced obesity: a time dependent challenge. PLoS One. 2013;8:e65790. doi: 10.1371/journal.pone.0065790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JW, Chervaux C, Raymond B, Derrien M, Brazeilles R, Kosta A, et al. 2014Fermented dairy products modulate C. rodentium induced colonic hyperplasia J Infect Dis; e-pub ahead of print 4 April 2014; doi: 10.1093/infdis/jiu205 [DOI] [PMC free article] [PubMed]
- Delzenne NM, Neyrinck AM, Backhed F, Cani PD. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat Rev Endocrinol. 2011;7:639–646. doi: 10.1038/nrendo.2011.126. [DOI] [PubMed] [Google Scholar]
- Dewulf EM, Cani PD, Claus SP, Fuentes S, Puylaert PG, Neyrinck AM, et al. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut. 2013;62:1112–1121. doi: 10.1136/gutjnl-2012-303304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2005;365:1415–1428. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- Engeli S, Feldpausch M, Gorzelniak K, Hartwig F, Heintze U, Janke J, et al. Association between adiponectin and mediators of inflammation in obese women. Diabetes. 2003;52:942–947. doi: 10.2337/diabetes.52.4.942. [DOI] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GG, Neyrinck AM, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes. 2011;60:2775–2786. doi: 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fak F, Backhed F. Lactobacillus reuteri prevents diet-induced obesity, but not atherosclerosis, in a strain dependent fashion in Apoe-/- mice. PLoS One. 2012;7:e46837. doi: 10.1371/journal.pone.0046837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei N, Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013;7:880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauffin Cano P, Santacruz A, Moya A, Sanz Y. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLoS One. 2012;7:e41079. doi: 10.1371/journal.pone.0041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompone G, Martorell P, Llopis S, Gonzalez N, Genoves S, Mulet AP, et al. Anti-inflammatory Lactobacillus rhamnosus CNCM I-3690 strain protects against oxidative stress and increases lifespan in Caenorhabditis elegans. PLoS One. 2012;7:e52493. doi: 10.1371/journal.pone.0052493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanety H, Feinstein R, Papa MZ, Hemi R, Karasik A. Tumor necrosis factor alpha-induced phosphorylation of insulin receptor substrate-1 (IRS-1). Possible mechanism for suppression of insulin-stimulated tyrosine phosphorylation of IRS-1. J Biol Chem. 1995;270:23780–23784. doi: 10.1074/jbc.270.40.23780. [DOI] [PubMed] [Google Scholar]
- Kim SW, Park KY, Kim B, Kim E, Hyun CK. Lactobacillus rhamnosus GG improves insulin sensitivity and reduces adiposity in high-fat diet-fed mice through enhancement of adiponectin production. Biochem Biophys Res Commun. 2013;431:258–263. doi: 10.1016/j.bbrc.2012.12.121. [DOI] [PubMed] [Google Scholar]
- Kondo T, Kishi M, Fushimi T, Kaga T. Acetic acid upregulates the expression of genes for fatty acid oxidation enzymes in liver to suppress body fat accumulation. J Agric Food Chem. 2009;57:5982–5986. doi: 10.1021/jf900470c. [DOI] [PubMed] [Google Scholar]
- Lam YY, Ha CW, Campbell CR, Mitchell AJ, Dinudom A, Oscarsson J, et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS One. 2012;7:e34233. doi: 10.1371/journal.pone.0034233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2012;62:1787–1794. doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- Lee HY, Park JH, Seok SH, Baek MW, Kim DJ, Lee KE, et al. Human originated bacteria, Lactobacillus rhamnosus PL60, produce conjugated linoleic acid and show anti-obesity effects in diet-induced obese mice. Biochim Biophys Acta. 2006;1761:736–744. doi: 10.1016/j.bbalip.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Ma X, Hua J, Li Z. Probiotics improve high fat diet-induced hepatic steatosis and insulin resistance by increasing hepatic NKT cells. J Hepatol. 2008;49:821–830. doi: 10.1016/j.jhep.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Million M, Angelakis E, Paul M, Armougom F, Leibovici L, Raoult D. Comparative meta-analysis of the effect of Lactobacillus species on weight gain in humans and animals. Microb Pathog. 2012;53:100–108. doi: 10.1016/j.micpath.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Ohkawara S, Furuya H, Nagashima K, Asanuma N, Hino T. Effect of oral administration of Butyrivibrio fibrisolvens MDT-1 on experimental enterocolitis in mice. Clin Vaccine Immunol. 2006;13:1231–1236. doi: 10.1128/CVI.00267-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DY, Ahn YT, Huh CS, McGregor RA, Choi MS. Dual probiotic strains suppress high fructose-induced metabolic syndrome. World J Gastroenterol. 2013;19:274–283. doi: 10.3748/wjg.v19.i2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DY, Ahn YT, Park SH, Huh CS, Yoo SR, Yu R, et al. Supplementation of Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032 in diet-induced obese mice is associated with gut microbial changes and reduction in obesity. PLoS One. 2013;8:e59470. doi: 10.1371/journal.pone.0059470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Robinson CJ, Bohannan BJ, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74:453–476. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselli M, Finamore A, Britti MS, Mengheri E. Probiotic bacteria Bifidobacterium animalis MB5 and Lactobacillus rhamnosus GG protect intestinal Caco-2 cells from the inflammation-associated response induced by enterotoxigenic Escherichia coli K88. Br J Nutr. 2006;95:1177–1184. doi: 10.1079/bjn20051681. [DOI] [PubMed] [Google Scholar]
- Sakakibara S, Yamauchi T, Oshima Y, Tsukamoto Y, Kadowaki T. Acetic acid activates hepatic AMPK and reduces hyperglycemia in diabetic KK-A(y) mice. Biochem Biophys Res Commun. 2006;344:597–604. doi: 10.1016/j.bbrc.2006.03.176. [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Lan PT, Benno Y. Barnesiella viscericola gen. nov., sp. nov., a novel member of the family Porphyromonadaceae isolated from chicken caecum. Int J Syst Evol Microbiol. 2007;57:342–346. doi: 10.1099/ijs.0.64709-0. [DOI] [PubMed] [Google Scholar]
- Shaul ME, Bennett G, Strissel KJ, Greenberg AS, Obin MS. Dynamic, M2-like remodeling phenotypes of CD11c+ adipose tissue macrophages during high-fat diet—induced obesity in mice. Diabetes. 2010;59:1171–1181. doi: 10.2337/db09-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63:727–735. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- Strissel KJ, Stancheva Z, Miyoshi H, Perfield JW, 2nd, DeFuria J, Jick Z, et al. Adipocyte death, adipose tissue remodeling, and obesity complications. Diabetes. 2007;56:2910–2918. doi: 10.2337/db07-0767. [DOI] [PubMed] [Google Scholar]
- Sun L, Yu Z, Ye X, Zou S, Li H, Yu D, et al. A marker of endotoxemia is associated with obesity and related metabolic disorders in apparently healthy Chinese. Diabetes Care. 2010;33:1925–1932. doi: 10.2337/dc10-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuovinen E, Keto J, Nikkila J, Matto J, Lahteenmaki K. Cytokine response of human mononuclear cells induced by intestinal Clostridium species. Anaerobe. 2013;19:70–76. doi: 10.1016/j.anaerobe.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Veiga P, Gallini CA, Beal C, Michaud M, Delaney ML, DuBois A, et al. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc Natl Acad Sci USA. 2010;107:18132–18137. doi: 10.1073/pnas.1011737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, et al. 2012Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome Gastroenterology 143913–916.e917. [DOI] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RY, Wan YP, Fang QY, Lu W, Cai W. Supplementation with probiotics modifies gut flora and attenuates liver fat accumulation in rat nonalcoholic fatty liver disease model. J Clin Biochem Nutr. 2012;50:72–77. doi: 10.3164/jcbn.11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H, Fujisawa K, Ito E, Idei S, Kawaguchi N, Kimoto M, et al. Improvement of obesity and glucose tolerance by acetate in Type 2 diabetic Otsuka Long-Evans Tokushima Fatty (OLETF) rats. Biosci Biotechnol Biochem. 2007;71:1236–1243. doi: 10.1271/bbb.60668. [DOI] [PubMed] [Google Scholar]
- Yin YN, Yu QF, Fu N, Liu XW, Lu FG. Effects of four Bifidobacteria on obesity in high-fat diet induced rats. World J Gastroenterol. 2010;16:3394–3401. doi: 10.3748/wjg.v16.i27.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SR, Kim YJ, Park DY, Jung UJ, Jeon SM, Ahn YT, et al. Probiotics L. plantarum and L. curvatus in combination alter hepatic lipid metabolism and suppress diet-induced obesity. Obesity (Silver Spring) 2013;21:2571–2578. doi: 10.1002/oby.20428. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang M, Pang X, Zhao Y, Wang L, Zhao L. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 2012;6:1848–1857. doi: 10.1038/ismej.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4:232–241. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One. 2012;7:e42529. doi: 10.1371/journal.pone.0042529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Higashikawa F, Noda M, Kawamura Y, Matoba Y, Kumagai T, et al. The obesity and fatty liver are reduced by plant-derived Pediococcus pentosaceus LP28 in high fat diet-induced obese mice. PLoS One. 2012;7:e30696. doi: 10.1371/journal.pone.0030696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.