Abstract
Translation initiation of alphavirus subgenomic mRNA (sgmRNA) can occur in the absence of several initiation factors (eIFs) in infected cells; however, the precise translation mechanism is still poorly understood. In this study, we have examined the mechanism of initiation and AUG selection in Sindbis virus (SINV) sgmRNA. Our present findings suggest that sgmRNA is translated via a scanning mechanism, since the presence of a hairpin structure before the initiation codon hampers protein synthesis directed by this mRNA. In addition, translation is partially recovered when an in-frame AUG codon is placed upstream of this hairpin. This scanning process takes place without the participation of eIF4A and active eIF2. These results, combined with our findings through modifying the SINV sgmRNA leader sequence, do not support the possibility of a direct initiation from the start codon without previous scanning, or a shunting mechanism. Moreover, studies carried out with sgmRNAs containing two alternative AUG codons within a good context for translation reveal differences in AUG selection which are dependent on the cellular context and the phosphorylation state of eIF2α. Thus, initiation at the additional AUG is strictly dependent on active eIF2, whereas the genuine AUG codon can start translation following eIF2α inactivation. Collectively, our results suggest that SINV sgmRNA is translated by a scanning mechanism without the potential participation of crucial eIFs. A model is presented that explains the mechanism of initiation of mRNAs bearing two alternative initiation codons.
Keywords: AUG selection, initiation factors, scanning mechanism, Sindbis virus translation, viral protein synthesis
INTRODUCTION
Selection of the correct AUG initiation codon is critical for the translation of mRNAs. This selection is accomplished through scanning of the 5′ untranslated region (5′ UTR) by the small ribosomal subunit in conjunction with translation initiation factors (eIFs) (Pestova and Kolupaeva 2002; Asano and Sachs 2007). The initiator Met-tRNAiMet interacts with eIF2, and together with GTP forms the ternary complex. Cap recognition involves the interaction of eIF4E with the methylated structure m7GpppN located at the 5′ end of eukaryotic mRNAs (Gingras et al. 1999). Binding of eIF3 to the eIF4G middle domain promotes the interaction of the preinitiation complex 43S at the 5′ end of mRNAs bearing a cap structure (Lorsch and Dever 2010). Thus, the interaction of the ribosomal subunit 40S, containing several eIFs such as eIF1, eIF1A, eIF3, and eIF2, takes place (Hinnebusch 2011; Valasek 2012). The 40S ribosomal subunit bound to these eIFs is in an “open” conformation, i.e., competent for scanning, a mechanism that involves linear base-by-base inspection of the 5′ UTR (Kozak 1991; Hinnebusch 2011), until an AUG initiation codon is found in a suitable sequence context for initiation (Kozak 1991, 1999). The secondary structure of the 5′ UTR is melted during the scanning process, in part by the helicase activity of eIF4A (Parsyan et al. 2011). Once the preinitiation complex is positioned at the AUG initiation codon, base-pairing takes place with the anticodon present in the initiator tRNA Met-tRNAiMet. Once AUG recognition and codon–anticodon base-pairing has been established, the eIF5 carboxy moiety promotes the dissociation of eIF1, together with inorganic phosphate derived from the GTP hydrolysis of the ternary complex (Cheung et al. 2007; Luna et al. 2012). In addition, the eIF1A carboxy terminus moves closer to the eIF5 amino terminus (Nanda et al. 2013). This movement is coupled to eIF1 exit, which leaves the P site free and allows tighter binding of the initiator tRNA at this site. Concomitant with this rearrangement, eIF5B-GTP can now interact with the 40S subunit. In this manner, eIF5 together with eIF2-GDP are released from the ribosomal subunit, which is now in the “closed” conformation and is committed to continue mRNA translation (Luna et al. 2012; Nanda et al. 2013). This interaction of eIF5B-GTP stimulates the joining of the 60S subunit to form an 80S initiation complex (Pestova et al. 2000). The initiation phase ends with the Met-tRNAiMet accommodated in the P site of the 80S leaving free the A site. Binding of aminoacyl-tRNA-eEF1-GTP to this site starts the elongation phase.
Sindbis virus (SINV) is an archetypical member of the Alphavirus genus, which contains a single-stranded RNA molecule of positive polarity as genome. This genome serves as mRNA (gmRNA) and is translated early during virus infection to produce the nonstructural proteins (nsPs) (Strauss and Strauss 1994). In the late phase of infection, structural proteins are synthesized from the 26S subgenomic mRNA (sgmRNA). Both gmRNA and sgmRNA are capped at their 5′ end and contain a poly(A) tail at their 3′ end (Schlesinger and Schlesinger 1996). The most relevant aspect of sgmRNA translation is that it can take place in the absence of several eIFs. A structural motif located between 27 and 89 nt downstream from the AUG initiation codon (Frolov and Schlesinger 1996), subsequently termed DLP for downstream hairpin loop (Ventoso et al. 2006), is crucial to translate this mRNA when virtually all eIF2α has been phosphorylated (McInerney et al. 2005; Ventoso et al. 2006; Garcia-Moreno et al. 2013). Moreover, eIF4G cleavage by picornavirus proteases, or eIF4A inhibition by hippuristanol, has little effect on protein synthesis directed by sgmRNA (Castello et al. 2006; Garcia-Moreno et al. 2013; Sanz et al. 2013). The lack of an eIF requirement for sgmRNA translation is observed only in SINV-infected cells, but curiously, canonical translation of sgmRNA is evident in transfected cells or in cell-free systems (Sanz et al. 2009; Garcia-Moreno et al. 2013). It is puzzling to envisage the precise mechanism of cap recognition and initiation codon selection in sgmRNA, in the absence of active eIF2 and an intact eIF4F complex. In fact, direct interaction of the 80S ribosome to the initiation codon without scanning has been reported using reconstituted in vitro systems (Skabkin et al. 2010). According to the model proposed, no scanning of the SINV sgmRNA 5′ UTR takes place and only eIF3, DHX29, and eIF2D may suffice for the interaction of the preinitiation complex with the AUG start codon. Contrary to this model, our present findings suggest that the scanning mechanism may be necessary to select the AUG initiation codon and initiate translation. Furthermore, through generation of specific mutant SINV sgmRNAs containing two in-frame AUG start codons, we detected initiation at both AUGs, but each of them was preferentially selected depending on the cell type and the eIF2 phosphorylation state. This suggests that SINV sgmRNA likely initiates translation on the authentic AUG codon by a novel scanning mechanism.
RESULTS
Mechanism of protein synthesis initiation directed by SINV sgmRNA
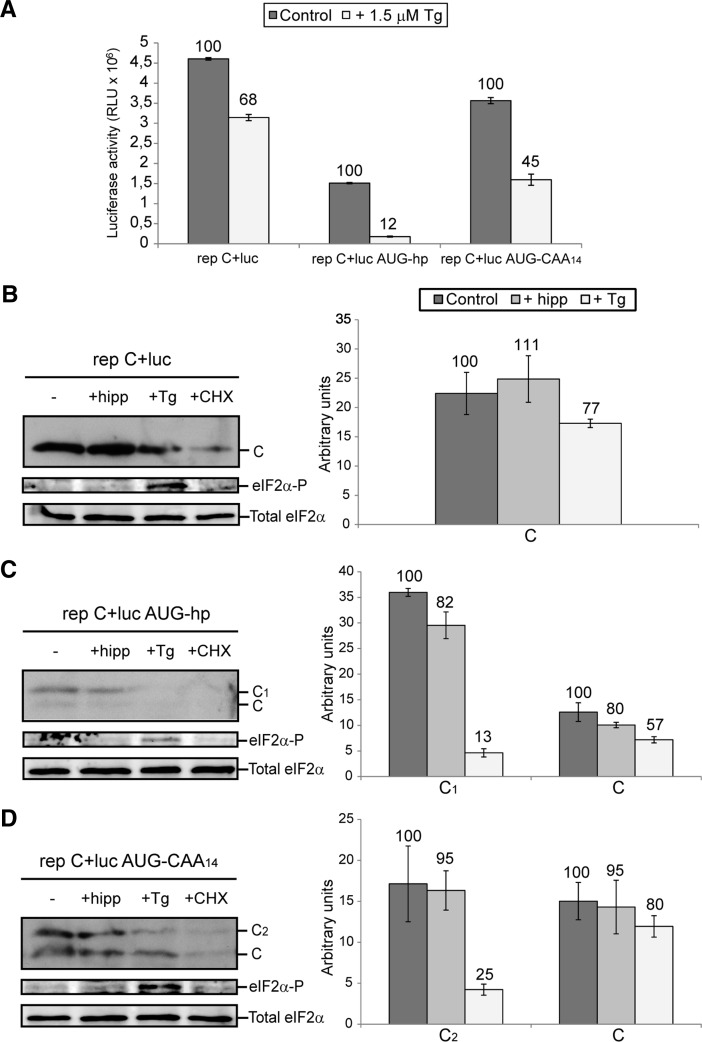
Two different mechanisms can be envisaged for translation initiation of SINV sgmRNA: One is the interaction of the preinitiation complex at the cap structure followed by scanning until the initiator AUG is encountered. Another option is that initiation takes place through the direct interaction of the preinitiation complex with the AUG initiation codon, without scanning. Evidence for this second mechanism has been provided using reconstituted cell-free systems with purified translational components (Skabkin et al. 2010). To determine which of these mechanisms is operational in living cells, several constructs were engineered as depicted in Figure 1A,B (see also Supplemental Table S1 for description). We made use of a SINV replicon containing the luciferase gene as a replacement for the structural proteins PE2, 6K, and E1 (rep C+luc) (Fig. 1B), which is helpful to quantify protein synthesis. Initially, rep C+luc gives rise to the nsPs that are involved in RNA replication and transcription of sgmRNA C+luc from an internal promoter. Therefore, this sgmRNA will be translated in an environment that mimics SINV-infected cells: Cellular translation is shut-off and the requirements for some eIFs are similar (Sanz et al. 2007, 2009; Garcia-Moreno et al. 2013). The construct SINV sgmRNA hp bears a hairpin structure in the midst of the 5′ UTR, located 34 nt from the 5′ end. The other modifications of the sgmRNA leader sequence, represented in Figure 1A, were inserted at this same position. Thus, none of the constructs affect the coding region of nsP4 or its stop codons, which are positioned at the beginning of the sgmRNA sequence. The hairpin hp has a predicted minimum free energy ΔG of −45.30 kcal/mol (RNAfold, University of Vienna) and should not be melted by preinitiation complexes (Kozak 1990, 1991; Babendure et al. 2006). Thus, the placement of this hairpin before the initiation codon hampers translation in the event that scanning occurs (Kozak 1990). Transfection of BHK cells with the control replicon rep C+luc resulted in a robust production of luciferase activity (Fig. 2A, upper panel). In contrast, luciferase synthesis was strongly blocked after rep C+luc hp transfection, suggesting that sgmRNA 5′-UTR scanning is necessary for initiation. As a control, a sgmRNA containing a longer 5′ UTR but without secondary structure was analyzed (rep C+luc CAA14). In this case, no inhibition of luciferase synthesis, directed by sgmRNA, was observed (Fig. 2A, upper panel). Additionally, two further constructs bearing an AUG codon in a good context for initiation (Kozak 1997), located before the hairpin and the CAA14 insert, were constructed: rep C+luc AUG-hp and rep C+luc AUG-CAA14 (Fig. 1A). Transfection of BHK cells with rep C+luc AUG-hp resulted in ∼38-fold stimulation as compared with rep C+luc hp, indicating that after the initiation complex formation, 80S ribosomes can pass through this hairpin. However, luciferase synthesis with this construct was ∼25% to that observed with the control rep C+luc replicon. This result is in good agreement with previous findings showing that insertion of an AUG before the hairpin does not totally rescue translation on the corresponding mRNA (Kozak 1989). On the other hand, rep C+luc AUG-CAA14 translation efficiency was similar to that of rep C+luc and rep C+luc CAA14. These data are consistent with the idea that initiator AUG detection during SINV mRNA translation can occur by a scanning process rather than a direct positioning of the ribosome at the initiation codon.
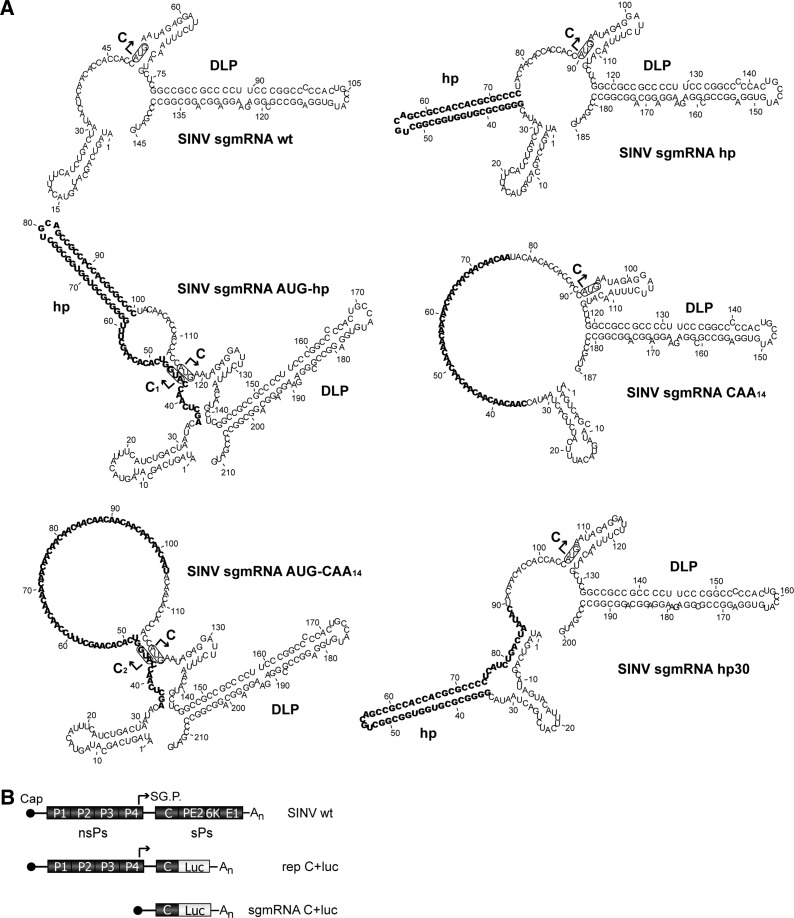
FIGURE 1.
RNA secondary structure models of SINV sgmRNA variants bearing modifications in the leader sequence upstream of the start codon. (A) The RNA secondary structures of the 5′ end of SINV sgmRNA constructs were predicted by RNAfold. The inserts introduced into the leader sequence are highlighted in bold. The initiation AUG codons are shown in boxes; additionally, arrows indicate the start site of protein C from genuine AUG codon and protein variants C1 and C2 from added in-frame AUGs (in AUG-hp and AUG-CAA14 mutants, respectively). (B) Schematic representation of the SINV genome, the rep C+luc replicon, and the subgenomic mRNA C+luc. nsPs, nonstructural proteins; sPs, structural proteins; SG.P., subgenomic promoter (represented by an arrow).
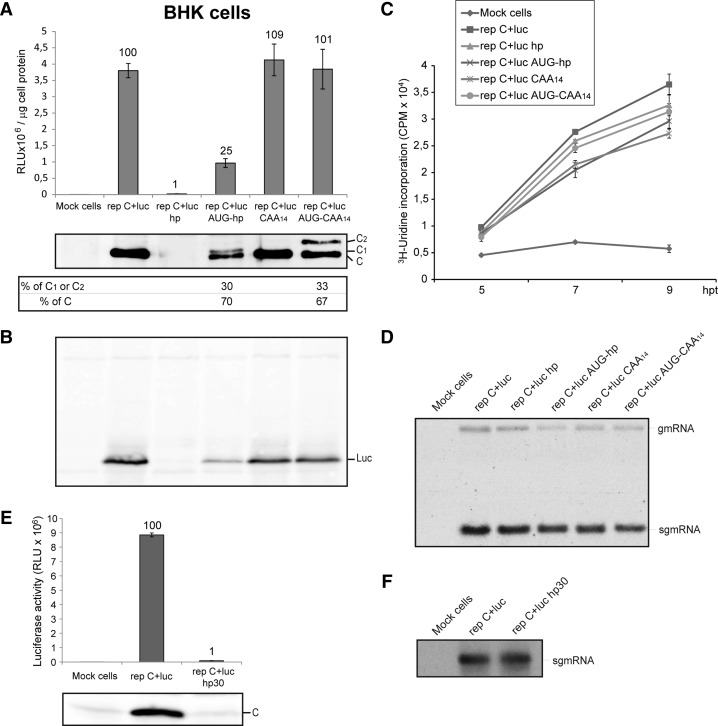
FIGURE 2.
Translation and synthesis of virus-specific RNAs in BHK cells transfected with different SINV replicons. (A,B,E) BHK cells were transfected with Lipofectamine 2000 and the in vitro transcribed SINV replicons depicted in Figure 1. At 7 hpt (hours post-transfection), cells were harvested in luciferase lysis buffer and luciferase activity was measured. Luciferase activity results are displayed in the graph (A and E, upper panels) in RLU per microgram of cell protein and are means ± SD of three representative experiments performed in triplicate. The percentage values obtained from mutant replicons compared with rep C+luc are indicated in the graph. In parallel, SINV C protein and luciferase (Luc) were detected by Western blotting using specific anti-C rabbit polyclonal antibodies (A, middle panel and E, lower panel) and anti-luciferase rabbit polyclonal antibodies (panel B), respectively. The relative percentages of authentic protein C and mutant proteins C1 or C2 were calculated from values obtained by densitometric scanning of the corresponding bands (A, lower panel). (C) BHK cells were transfected as in A, treated with 5 μg/mL actinomycin D from 2 hpt and labeled with 40 μCi/mL [3H]uridine at 4 hpt. Cells were then fixed at the indicated times after transfection, as described in Materials and Methods and harvested to measure [3H]uridine incorporation in a liquid scintillation spectrometer. Cpm values are means ± SD of three representative experiments performed in triplicate. (D,F) BHK cells were transfected as in A. From 2 hpt, cells were treated with 5 μg/mL actinomycin D and from 3 hpt with 40 μCi/mL [3H]uridine. At 7 hpt, total RNA from cells was extracted, separated in 0.7% agarose gels, and then subjected to fluorography and autoradiography.
Translation of sgmRNA from the different replicons described produces a polyprotein, coding for the capsid protein (C) and luciferase. These proteins are released due to the autocatalytic activity of C, which cleaves the polyprotein in the C carboxyl-terminus, immediately after its generation (Strauss and Strauss 1994). To measure the production of C protein in replicating cells, immunoblotting was carried out using a specific rabbit polyclonal antibody. As expected, robust production of C was detected in cells transfected with rep C+luc, whereas virtually no C protein was observed after transfection with rep C+luc hp (Fig. 2A, middle panel). Interestingly, two immunoreactive species with different sizes of C protein were detected when cells were transfected with rep C+luc AUG-hp and rep C+luc AUG-CAA14. The different sequences of extra amino acids at the amino terminus may be the reason for the different mobilities observed in the two C species, C1 and C2. With both constructs, ∼70% of total C protein had the same mobility as wt C and this was consistent with an initiation from the second AUG codon corresponding to the genuine start codon of SINV sgmRNA. The remaining 30% of C was generated from the first upstream AUG added in-frame and within a good context for translation (Fig. 2A, lower panel). Furthermore, synthesis of genuine protein C from rep C+luc AUG-CAA14 was ∼45% less efficient than in rep C+luc CAA14. This result is also supportive of a scanning mechanism model, in which insertion of an upstream AUG in good context for translation should reduce initiation at a downstream AUG. To analyze whether the extra amino acid residues at the C amino terminus may influence the cleavage of the precursor C+luc, we carried out a Western blotting using rabbit polyclonal antibodies against luciferase. Figure 2B shows the production of a single band corresponding to luciferase, whereas the precursor C+luc was not detected.
A possible explanation for the differences observed in protein synthesis with the different replicons tested is that the hairpin, or the sequences, introduced in these replicons, affected the internal promoter which directs transcription of the corresponding sgmRNA. If so, viral RNA synthesis and the quantity of sgmRNA transcribed would be altered in cells transfected with these replicons. To assess this possibility, BHK cells transfected with the different replicons were labeled with [3H]uridine at different times, in the presence of actinomycin D. Under these conditions, cellular RNA synthesis is ablated while viral genome replication, sgmRNA transcription, and translation remain unaffected. Clearly, [3H]uridine incorporation and therefore total viral RNA synthesis was similar in cells transfected with the different replicons (Fig. 2C), suggesting that sgmRNA transcription was not inhibited. To analyze this in more detail, labeled viral RNAs were visualized by electrophoresis on agarose gels. Consistent with the scintillation counting, the quantity of sgmRNA was similar in cells transfected with rep C+luc or with rep C+luc hp (Fig. 2D), supporting the conclusion that inhibition of protein synthesis with rep C+luc hp is not due to viral transcription blockade. The ratio of sgmRNA synthesized versus gmRNA was ∼10:1 in cells transfected with the different replicons.
Further assessment that scanning is the mechanism followed by sgmRNA to initiate translation was obtained with an additional construct, rep C+luc hp30. This construct bears the hp separated 30 nt apart from the genuine initiation codon (see Fig. 1A). The rationale behind this construct was to locate this hairpin separated from the AUG in case it could prevent the hypothetical direct interaction of 80S ribosomes with the initiator AUG. In addition, it must be taken into account that the initial nucleotides after the cap structure must be kept unaltered to maintain the internal promoter sequence and avoid the inhibition of sgmRNA transcription. Transfection of rep C+luc hp30 gave rise to a strong inhibition of luciferase activity (Fig. 2E, upper panel) and C production (Fig. 2E, lower panel), whereas sgmRNA synthesis was not affected (Fig. 2F), suggesting that the hp introduced in the leader sequence hampered translation of this sgmRNA.
In summary, the observations obtained with the different constructs in BHK cells may provide evidence in support of the scanning mechanism in the translation of sgmRNA.
Scanning and AUG selection in SINV-replicating MEFs, PKR−/− MEFs, and mosquito cells
Alphavirus replication induces a strong phosphorylation of eIF2α, mainly mediated through PKR activation by viral dsRNA (Gorchakov et al. 2004; McInerney et al. 2005; Ventoso et al. 2006; Sanz et al. 2009). Activation of other kinases such as GCN2 may be possible (Berlanga et al. 2006), but its contribution to the phosphorylation of eIF2 may not be so important at late times of infection, since this phosphorylation event is absent in mouse cells lacking PKR (PKR−/− MEFs) and in mosquito cells that do not encode for a PKR counterpart (Ventoso 2012). Phosphorylation of eIF2α following SINV replication in the different cell lines used in this work is shown in Figure 3A. In agreement with previous reports, eIF2α became phosphorylated in BHK cells and in MEFs, whereas no induction of eIF2α phosphorylation was found in PKR−/− MEFs and in insect cells. Then, we assayed luciferase synthesis and C production in MEFs transfected with the different replicons described above, but it must be taken into consideration that MEFs are less susceptible for SINV infection (Gorchakov et al. 2004), and transfection with SINV replicons gives rise to a lower expression of C protein or luciferase (Sanz et al. 2013). Indeed, transfection of MEFs with rep C+luc, rep C+luc CAA14 or rep C+luc AUG-CAA14 led to ∼10% production of luciferase as compared with PKR−/− MEFs (Fig. 3B, upper panel). In addition, C production was barely detected by Western blotting and it was only recognized when a high sensitivity reagent was used (Fig. 3B, lower panel). Virtually no expression was found with the replicon rep C+luc hp. Translation was partially restored with rep C+luc AUG-hp but it was still inhibited to a level of 79% as compared with rep C+luc.
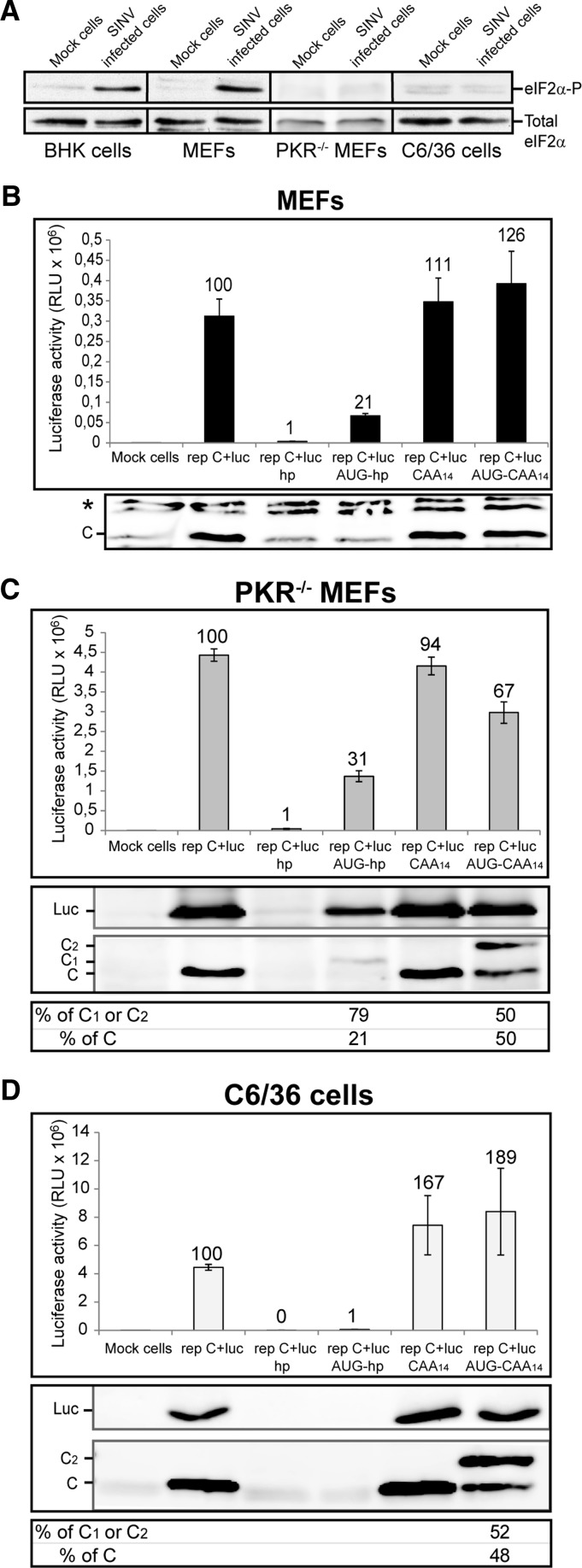
FIGURE 3.
Protein synthesis in MEFs, PKR−/− MEFs, and insect cells transfected with SINV replicons with mutated leader sequences. (A) BHK, MEFs, PKR−/− MEFs, and C6/36 cells were either mock-infected or infected with SINV wt at a MOI of 5 pfu per cell. At 7 (BHK cells and MEFs), 5 (PKR−/− MEFs), or 8 (C6/36 cells) hpi (hours post-infection) cells were collected in sample buffer and analyzed by Western blotting with anti-phospho-eIF2α (eIF2α-P) and anti-eIF2α (Total eIF2α) antibodies. MEFs (B), PKR−/− MEFs (C), and C6/36 cells (D) were transfected with Lipofectamine 2000 and SINV replicons transcribed in vitro. At 7, 5, or 8 hpt, respectively, cells were collected in luciferase lysis buffer and luciferase activity was measured. Luciferase activity results are mean ± SD of three representative experiments performed in triplicate; the percentage values obtained from mutant replicons relative to rep C+luc are indicated (B–D, upper panels). C and luciferase accumulation was analyzed by Western blotting with anti-C and anti-luciferase antibodies, respectively (C and D, middle panels). The percentages of genuine protein C and mutant proteins C1 or C2 relative to the sum of all signals for C species in the lane were estimated by densitometric analysis of the corresponding bands (C and D, lower panels). (*) Unspecific bands.
To analyze sgmRNA translation in replicating cells that do not undergo eIF2α phosphorylation, PKR−/− MEFs were transfected with the same RNA replicons. Luciferase synthesis was strongly inhibited in PKR−/− MEFs transfected with rep C+luc hp, to a level of 99% inhibition with respect to rep C+luc (Fig. 3C, upper panel). In contrast, transfection with rep C+luc AUG-hp increased luciferase synthesis 34-fold, with respect to rep C+luc hp. Similar to that observed in BHK cells (Fig. 2A) there was still an ∼70% inhibition of luciferase synthesis when compared with the rep C+luc replicon. Notably, sgmRNA translation in cells transfected with rep C+luc CAA14 was similar to control rep C+luc, suggesting that increasing the length of the sgmRNA leader sequence has little effect on its translatability. Additionally, insertion of an extra AUG codon within a good context in this leader sequence (rep C+luc AUG-CAA14) diminished luciferase synthesis by only 30%. Therefore, SINV sgmRNA translation initiation may be accomplished by a scanning mechanism in PKR−/− MEFs. Interestingly, analysis of C synthesis by Western blotting revealed that the translation initiation on the first AUG was 80% in rep C+luc AUG-hp, whereas there was ∼50% initiation on each of the two AUG codons in rep C+luc AUG-CAA14 (Fig. 3C, middle and lower panels). This finding is consistent with the concept that the presence of a hairpin located at 14 nt downstream from the first AUG induces initiation on this codon (Kozak 1990). The production of C from the downstream AUG in rep C+luc AUG-CAA14 was reduced by 52% as compared with rep C+luc CAA14 (Fig. 3C, middle panel), which also strongly supports a scanning mechanism. Apart from luciferase activity, we have also analyzed the production of luciferase by immunoblotting (Fig. 3C, middle panel). The results obtained are consistent with those observed with the production of C.
In comparison with our results using PKR−/− MEFs, a number of differences were observed with mosquito C6/36 cells transfected with the above constructs. In agreement with our previous findings, rep C+luc hp was totally inhibited (99.8%) in C6/36 cells (Fig. 3D, upper panel), suggesting that the scanning mechanism is also operative in insect cells that translate alphavirus sgmRNA. Curiously, when a second AUG was placed before the hairpin (rep C+luc AUG-hp), luciferase synthesis was not rescued, and a luciferase synthesis inhibition of 98.8% with respect to rep C+luc was observed (Fig. 3D, upper panel). This result suggests that the mechanism of scanning could be different between mammalian and insect cells. Another possibility is that, perhaps, there are differences in RNA structures (that might depend on different factors, including the host temperature) or in virus replication in the two cell types. Furthermore, luciferase synthesis was increased in C6/36 cells that expressed sgmRNA with a longer leader sequence (rep C+luc CAA14). Finally, a more robust stimulation (189%) was observed with rep C+luc AUG-CAA14. In this situation, translation initiation occurred equally well on both AUGs (Fig. 3D, middle and lower panels). As occurred in PKR−/− MEFs, insertion of the upstream AUG in rep C+luc AUG-CAA14 diminished genuine C production by 55% relative to rep C+luc CAA14, which further supports a scanning mechanism in insect cells. The higher production of luciferase in cells transfected with rep C+luc CAA14 and rep C+luc AUG-CAA14 in comparison with control rep C+luc was also confirmed by Western blotting using anti-luciferase antibodies (Fig. 3D, middle panel).
In conclusion, SINV sgmRNA may be translated by a scanning mechanism in both PKR−/− MEFs and mosquito cells. However, in contrast to BHK cells, the first AUG is preferentially used in rep C+luc AUG-hp in PKR−/− MEFs in which eIF2α is not phosphorylated. The reasons for this difference may relate therefore to the fact that eIF2α is phosphorylated in replicating BHK cells (Sanz et al. 2009) and initiation on the second AUG codon, which is close to the DLP, is resistant to this phosphorylation. Alternatively, it is possible that the translational machinery of BHK cells does not recognize the first AUG codon with high efficiency for unknown reasons. Furthermore, this mRNA is not functional in mosquito cells. Finally, increasing the length of the leader sequence (rep C+luc CAA14) is not detrimental for sgmRNA translation in PKR−/− MEFs, while protein synthesis directed by this sgmRNA is notably increased in mosquito cells.
Translation of SINV sgmRNA variants in the absence of replication
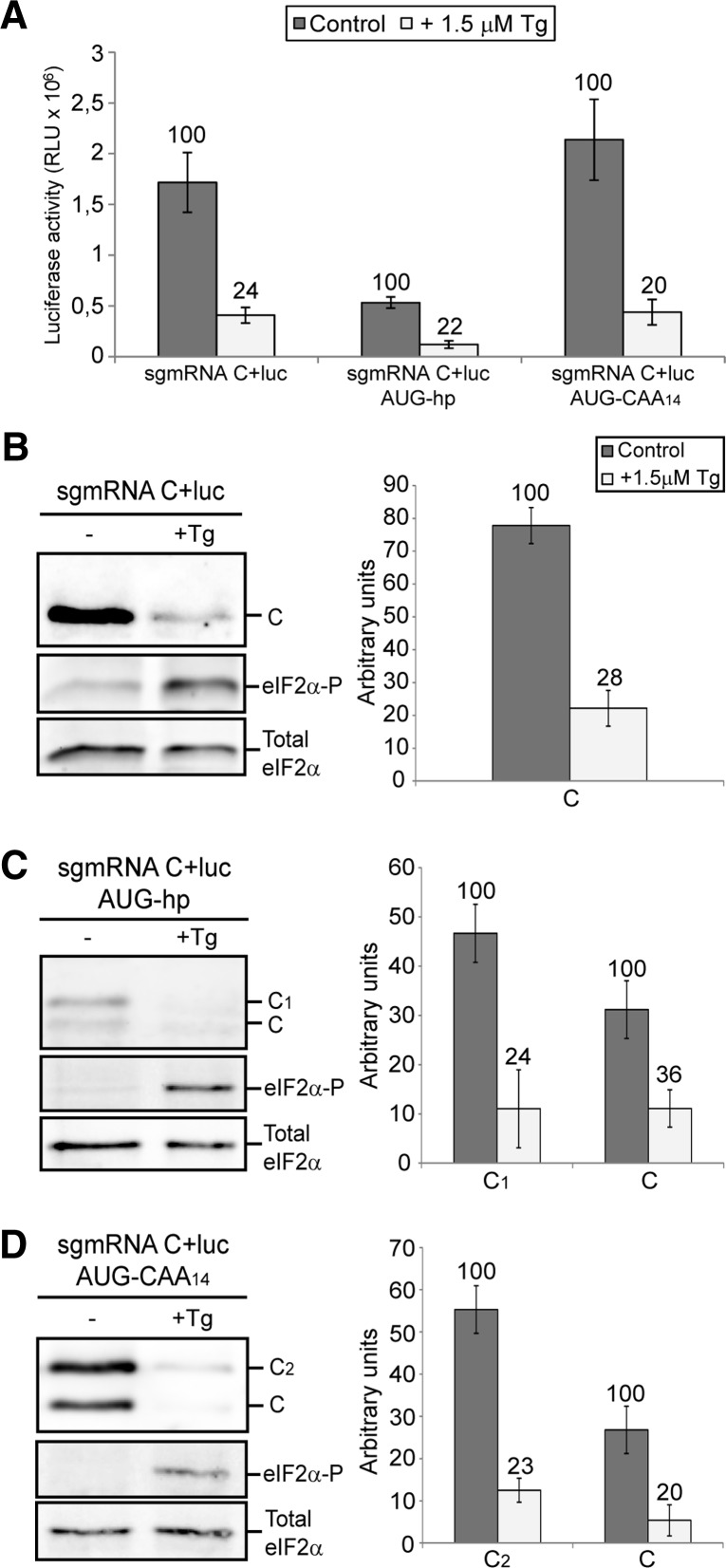
Protein synthesis from SINV sgmRNA is mediated by a dual mechanism of initiation, which is dependent on the context of its translation. Thus, in SINV-replicating cells, sgmRNA is translated in the absence of several eIFs, whereas canonical translation occurs when isolated sgmRNA is transfected in cells or translated in cell-free systems (Sanz et al. 2009; Garcia-Moreno et al. 2013). To analyze the translation of the described sgmRNAs, when detached from the replication context, the corresponding coding regions of the various sgmRNAs were cloned directly under the control of the T7 promoter, without the coding region for the nonstructural proteins. Thereafter, the sgmRNAs were transcribed in vitro and transfected in distinct cells to gauge translation. These sgmRNAs made in vitro by T7 RNA polymerase have exactly the same sequence as those produced in cells from the replicons using the internal promoter. Compared with control transfections, luciferase activity in BHK cells transfected with sgmRNA C+luc hp was abrogated (99% inhibition) (Fig. 4A, upper panel), to a similar degree as the corresponding replicon (rep C+luc hp) (Fig. 2A). This result demonstrates that scanning is a feature of the mechanism used by sgmRNA translation detached from the context of viral replication. This translation was rescued in part with an AUG codon placed upstream of the hairpin (sgmRNA C+luc AUG-hp), leading to a 36-fold stimulation of luciferase synthesis with respect to sgmRNA C+luc hp. However, there was still an 80% inhibition of luciferase synthesis when compared with the control sgmRNA C+luc. The additional sgmRNAs tested, sgmRNA C+luc CAA14 and sgmRNA C+luc AUG-CAA14, were translated to an extent similar to the control (Fig. 4A, upper panel). Protein analysis of C by Western blotting revealed that the degree of initiation from the two AUGs present in sgmRNA C+luc AUG-hp was 36% and 64%, respectively, compared with 45% and 55% for sgmRNA C+luc AUG-CAA14 (Fig. 4A, middle and lower panels). As a control, analysis of sgmRNA levels by qRT-PCR showed that the amount of sgmRNA C+luc variants was very similar in each case after transfection and incubation of BHK cells (Supplemental Fig. S1). These results are consistent with those found for the replicons in BHK cells.
FIGURE 4.
Translation directed by in vitro-synthesized SINV sgmRNAs with altered leader sequences. BHK (A), PKR−/− MEFs (B), and C6/36 (C) cells were transfected with Lipofectamine 2000 and in vitro prepared SINV sgmRNAs (described in Fig. 1 and Supplemental Table S1). At 2 hpt, cells were collected in luciferase lysis buffer and luciferase activity was determined (upper panels). In parallel, C products were detected by Western blotting with anti-C antibody (middle panels). (D) One hundred nanograms of SINV sgmRNAs synthesized in vitro by T7 RNA polymerase were added to RRL and incubated for 2 h at 30°C. Luciferase synthesis was estimated by measuring luciferase activity (upper panel). The C proteins synthesized from SINV sgmRNAs were metabolically labeled with [35S]Met/Cys and examined by SDS-PAGE and autoradiography (middle panel). (A–D) Luciferase activity results are means ± SD of three representative experiments performed in triplicate. The percentage values obtained from mutant sgmRNAs compared with sgmRNA C+luc are indicated in the figure. Lower panels represent the relative percentages of authentic protein C and mutant proteins C1 or C2, quantified by densitometric analysis.
In cells lacking PKR protein, such as PKR−/− MEFs or mosquito C6/36, translation of the transfected sgmRNA C+luc hp was also strongly inhibited (Fig. 4B,C, upper panels). Furthermore, significant differences were observed in sgmRNA AUG-hp translation in both cell types. In PKR−/− MEFs, an in-frame AUG located before the hairpin increased translation 30-fold compared with sgmRNA C+luc hp, but there was still a 69% inhibition of translation compared with control sgmRNA C+luc (Fig. 4B). Consistent with the results obtained in replicons, sgmRNA C+luc AUG-hp translation in mosquito cells was hampered, with 97% inhibition in respect to control sgmRNA C+luc (Fig. 4C). Additionally, sgmRNA C+luc CAA14 and sgmRNA C+luc AUG-CAA14 were translated to a greater extent than control mRNA in PKR−/− MEFs (140% and 147% of control, respectively). These two mRNAs were also strongly translated in mosquito C6/36 cells, supporting the conclusion that this modification of the 5′ leader sequence is not detrimental for its translation. The use of the two different AUGs was also observed in PKR−/− MEFs transfected with sgmRNA C+luc AUG-hp or sgmRNA C+luc AUG-CAA14, although the proportion of each AUG used varied depending on the mRNA examined (Fig. 4B, middle and lower panels). Consistent with the observations in replicons (Fig. 3C) the first AUG was preferentially used in PKR−/− MEFs. In mosquito cells, C synthesis could not be detected by Western blotting with sgmRNA C+luc AUG-hp; however, both AUGs were recognized to a similar extent in sgmRNA C+luc AUG-CAA14 (Fig. 4C, middle and lower panels). Comparison of the level of protein C produced from the genuine AUG codon in sgmRNA C+luc CAA14 and sgmRNA C+luc AUG-CAA14 revealed an inhibition of 32%, 62%, and 53% upon insertion of the upstream AUG in BHK, PKR−/− MEFs, and C6/36 cells, respectively, in good agreement with the existence of ribosomal scanning. In conclusion, the scanning mechanism is operative not only in SINV-replicating cells, but also when translation of the isolated mRNAs is examined.
Given that SINV sgmRNA may interact directly with the ribosome in reconstituted in vitro systems (Skabkin et al. 2010), it was of interest to analyze the translation of the different sgmRNA C+luc variants in cell-free systems. For this purpose, RRL was supplemented with each one of the isolated sgmRNAs described above, and luciferase activity, together with radioactive labeling, was measured. As expected, control sgmRNA C+luc efficiently synthesized luciferase as determined by its activity and by [35S]Met/Cys labeling (Fig. 4D, upper and middle panels, respectively). Again, sgmRNA C+luc hp translation was strongly restricted (97% inhibition) in this system, suggesting that scanning is also necessary for SINV sgmRNA translation. However, a partial recovery of luciferase synthesis was obtained with sgmRNA C+luc AUG-hp (13-fold stimulation) compared with sgmRNA C+luc hp, but luciferase synthesis was inhibited 63% with respect to the control (Fig. 4D). Also, in good agreement with the earlier results, luciferase synthesis directed by sgmRNA C+luc CAA14 was increased to 134%, while a similar activity to the control was found with sgmRNA C+luc AUG-CAA14. In this in vitro system, the two initiation codons present in sgmRNAs were recognized with similar efficiencies (∼50% initiation on each AUG), both in sgmRNA C+luc AUG-hp and sgmRNA C+luc AUG-CAA14. Again, the production of protein C from sgmRNA C+luc AUG-CAA14 was diminished by 60% in comparison with sgmRNA C+luc CAA14, which agrees with a translation initiation via scanning in this system.
Participation of eIF4A in SINV sgmRNA translation
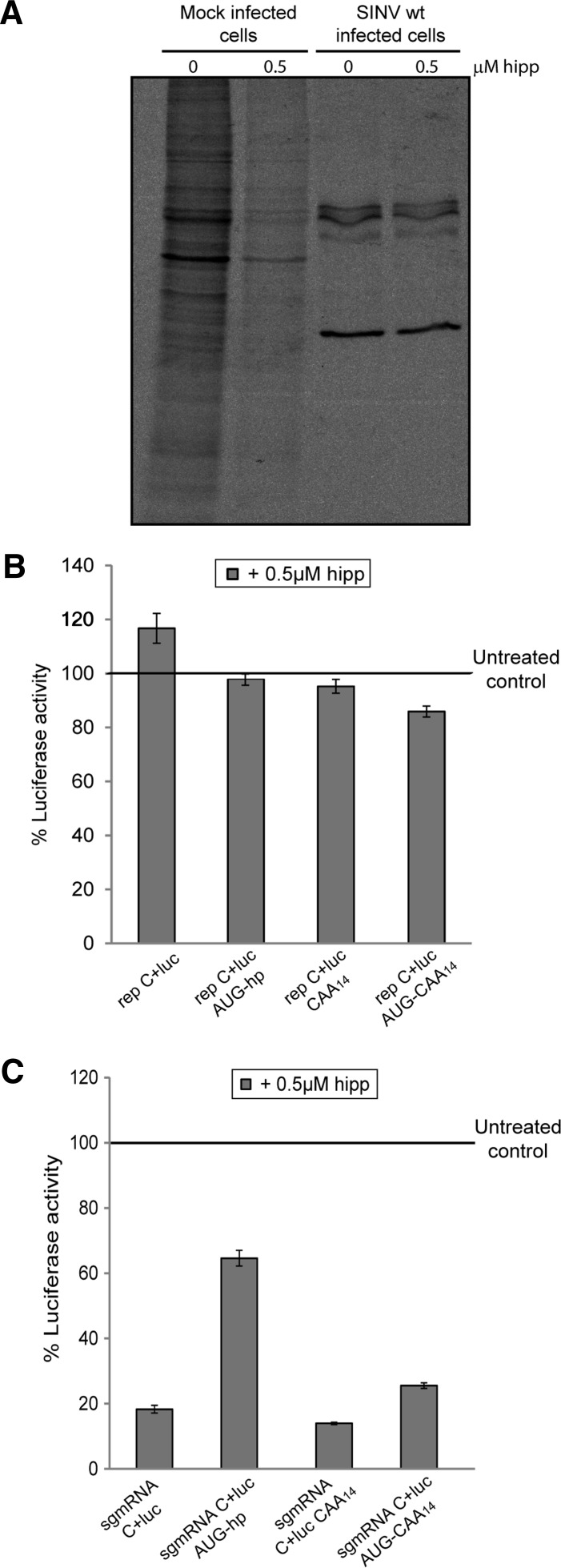
Hippuristanol is a selective inhibitor of eIF4A, a component of the eIF4F complex (Bordeleau et al. 2006; Lindqvist et al. 2008). eIF4A helicase activity in conjunction with eIF4B or eIF4H is necessary to melt the secondary structure within the leader sequence of mRNAs during the scanning process (Parsyan et al. 2011). Our results so far demonstrated that scanning is the mechanism followed by SINV sgmRNA to initiate translation. Thus, it was of interest to determine whether eIF4A participated in scanning of SINV sgmRNA. As a control for hippuristanol activity, uninfected and SINV-infected BHK cells were treated with 0.5 μM hippuristanol and labeled with [35S]Met/Cys. Cellular mRNA translation was drastically inhibited in the presence of the compound, whereas viral protein synthesis was resistant to eIF4A blockade by hippuristanol (Fig. 5A). Luciferase synthesis of replicons in BHK cells was reduced very slightly in the presence of 0.5 μM hippuristanol (Fig. 5B). Similarly, C production from the different AUG codons in rep C+luc AUG-hp and rep C+luc AUG-CAA14 was not affected by hippuristanol treatment in PKR−/− MEFs (Fig. 6B–D). Thus, these results reveal that eIF4A does not participate in the scanning or in the AUG selection of SINV sgmRNA in replicating cells.
FIGURE 5.
Translation of SINV sgmRNAs after eIF4A inhibition with hippuristanol. (A) BHK cells were either mock-infected or infected with SINV wt. At 5.5 hpi, cells were treated with 0.5 μM hippuristanol for 30 min or left untreated. From 6 to 7 hpi, cultures were labeled with [35S]Met/Cys in the presence or absence of the inhibitor. Samples were processed by SDS-PAGE, followed by autoradiography. BHK cells were transfected with Lipofectamine 2000 and SINV replicons transcribed in vitro (B) or in vitro prepared SINV sgmRNAs (C). At 5.5 hpt (B) or 30-min post-transfection (C), 0.5 µM hippuristanol (hipp), 100 µg/mL cycloheximide, or vehicle were added to cells, and the incubation was continued for 90 min before harvesting to analyze luciferase activity. Values obtained from cycloheximide-treated cells were used to subtract the amount of luciferase synthesized prior to hippuristanol addition. The results shown are percentages of the values of hippuristanol-treated cells relative to their respective untreated samples and are the mean ± SD of three independent experiments performed in triplicate.
FIGURE 6.
Selection of the AUG initiation codon after eIF2α phosphorylation induced by Tg. (A) PKR−/− MEFs were transfected with Lipofectamine 2000 and rep C+luc, rep C+luc AUG-hp, or rep C+luc AUG-CAA14. At 3.5 hpt, cells were treated with 1.5 μM Tg or 100 µg/mL cycloheximide (CHX) for 90 min or left untreated. Next, cells were collected in luciferase lysis buffer and luciferase activity was measured. Values obtained from cycloheximide-treated cells were used to subtract the amount of luciferase synthesized prior to Tg addition. (B–D) PKR−/− MEFs were transfected and treated as in (A), including hippuristanol (hipp) at 0.5 μM. Cells were then harvested in sample buffer and analyzed by Western blotting with anti-SINV C, anti-phospho-eIF2α (eIF2α-P), and anti-eIF2α (Total eIF2α) specific antibodies (left panels). The synthesis of protein C and mutant proteins C1 or C2 in the presence of the inhibitors was quantified by densitometric analysis of the corresponding bands (right panels). Values obtained from cycloheximide-treated cells were used to subtract the amount of protein produced prior to the addition of hippuristanol and Tg. The results shown in A–D are the mean ± SD of three independent experiments performed in triplicate. The percentage values of cells treated with hippuristanol or Tg relative to their respective untreated cells are indicated.
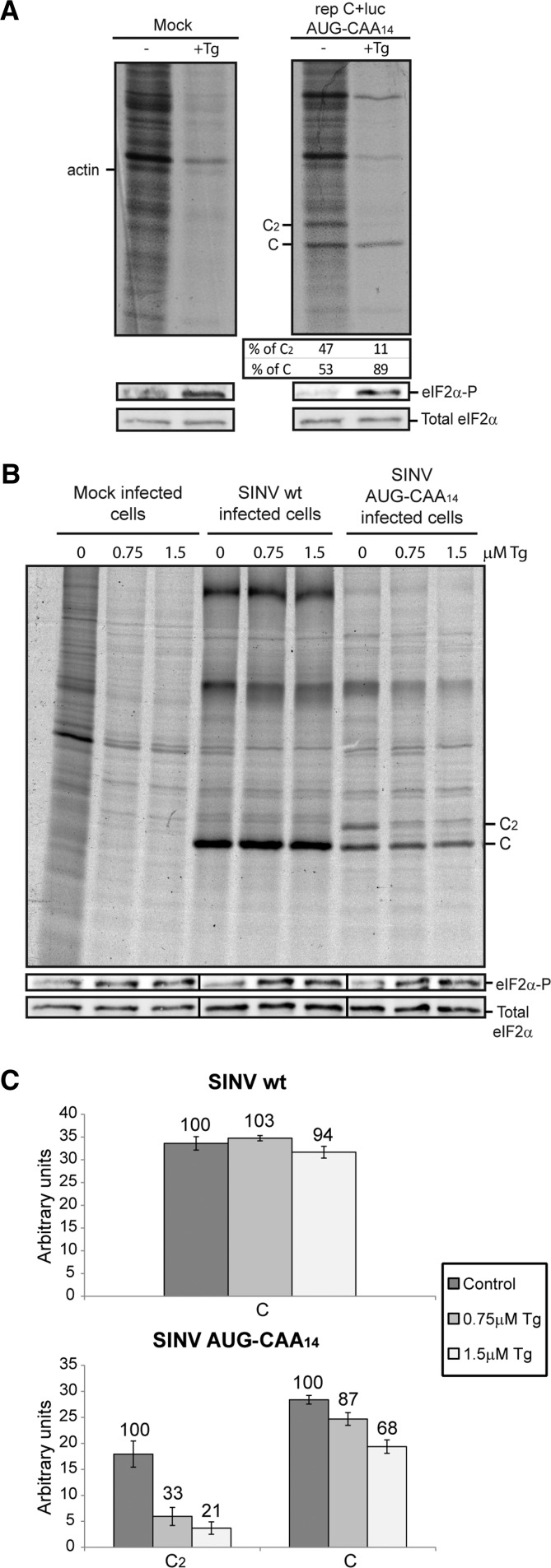
We reported recently that SINV sgmRNA requires eIF4A when translated out of an infection context, for example, when the isolated mRNA is transfected in cells (Garcia-Moreno et al. 2013). When BHK cells were transfected with the isolated sgmRNAs, hippuristanol strongly blocked translation by 70%–80% (Fig. 5C), with the exception of sgmRNA C+luc AUG-hp which was inhibited by ∼40%. These results reinforce the concept that eIF4A participates in the translation of the different sgmRNAs in transfected BHK cells. Therefore, the described SINV sgmRNAs with altered leader sequence manifest a dual mechanism for their translation, i.e., the requirement for eIF4A differs between replicating and nonreplicating cells. In addition, these observations also point to the possibility that scanning may occur without eIF4A in SINV-replicating cells.
Involvement of eIF2 in AUG selection
It is well established that alphavirus sgmRNA can be translated when virtually all eIF2α has been phosphorylated via PKR activation by dsRNA (Gorchakov et al. 2004; McInerney et al. 2005; Ventoso et al. 2006; Sanz et al. 2009). A hairpin structure (DLP) present at 24 nt downstream from the initiation codon is responsible for this eIF2-independent translation mechanism (McInerney et al. 2005; Ventoso et al. 2006). Since two of our constructs give rise to sgmRNAs bearing two functional initiation codons (sgmRNA AUG-hp and sgmRNA AUG-CAA14, see Fig. 1) and codon selection differs between BHK cells and PKR−/− MEFs, it was of interest to know whether this difference was due to eIF2α phosphorylation and if the translation initiation on these two AUGs could occur independently of active eIF2. To this end, PKR−/− MEFs were treated with the inhibitor thapsigargin (Tg), which induces eIF2α phosphorylation through a PKR-independent route (Harding et al. 2000; Linero et al. 2011). Tg treatment had only a modest effect on luciferase synthesis directed by rep C+luc (∼30%), whereas rep C+luc AUG-hp was robustly inhibited by ∼90%, and rep C+luc AUG-CAA14 by 55% (Fig. 6A). Additionally, C synthesis from rep C+luc was marginally decreased upon eIF2α phosphorylation induced by Tg (Fig. 6B). The production of C in the presence of Tg was analyzed by Western blotting and can be estimated by subtraction of the amount of C that appeared in the presence of cycloheximide, which was added at the same time as Tg. In cells transfected with rep C+luc AUG-hp, protein synthesis starting at the first AUG, which is preferentially selected in PKR−/− MEFs (see Figs. 3C, 6C), was profoundly blocked (87% inhibition) (Fig. 6C). Conversely, translation of rep C+luc AUG-CAA14 gave rise to equal amounts of two distinct C proteins (Fig. 6D). In this case, Tg treatment differentially inhibited translation of each C protein as shown in Figure 6D: Protein synthesis starting at the first AUG codon was significantly abrogated (75% inhibition with respect to untreated cells) while translation continued at the authentic AUG codon (20% inhibition). Therefore, the AUG selection pattern in PKR−/− MEFs is modified following eIF2 inactivation. On the other hand, hippuristanol had little effect on C synthesis in this system (Fig. 6B–D), in good agreement with the results described above.
To reinforce the analysis of C synthesis, radioactive labeling was carried out following two different approaches. First, PKR−/− MEFs were transfected with the replicon rep C+luc AUG-CAA14. After 4 h 15 min of incubation, cells were treated with 1.5 μM Tg for 15 min and then radioactively labeled for 30 min in the presence of the inhibitor. The proportion of mutant C2 protein and genuine C protein synthesized in cells transfected with rep C+luc AUG-CAA14 was 47% and 53%, respectively (Fig. 7A). Notably, this proportion drastically changed in the presence of Tg, to 11% and 89%, respectively. The second approach was to obtain recombinant viruses containing the SINV sgmRNA wt and sgmRNA AUG-CAA14 leader sequence (see details in Fig. 1A). PKR−/− MEFs were then infected at a multiplicity of 5 pfu per cell, and at 4 h 15 min post-infection cells were treated for 45 min with 0.75 or 1.5 µM Tg. As shown in Figure 7B, Tg treatment profoundly blocked cellular translation. Moreover, eIF2α phosphorylation resulted in a significant decrease of viral translation at the inserted AUG codon (67% and 79% inhibition in cells treated with 0.75 and 1.5 µM Tg, respectively), whereas protein synthesis from the genuine AUG codon was little affected in SINV AUG-CAA14 and in control SINV wt infected cells (Fig. 7B,C). Induction of eIF2α phosphorylation as well as total eIF2α was tested by Western blotting using specific antibodies (Fig. 7A,B, middle and lower panels). Indeed, eIF2α phosphorylation was apparent in Tg-treated cells.
FIGURE 7.
Influence of eIF2α phosphorylation on SINV sgmRNA AUG-CAA14 codon selection. (A) PKR−/− MEFs were transfected with Lipofectamine 2000 and rep C+luc AUG-CAA14. At 4-h 15-min post-transfection, cells were treated with 1.5 μM Tg for 15 min or left untreated. Cells were then labeled for 30 min with [35S]Met/Cys in the absence or presence of the compound. Radiolabeled proteins were separated by SDS-PAGE followed by autoradiography. The same samples were used to analyze phosphorylated eIF2α and total eIF2α by Western blot. The relative percentages of protein C and mutant protein C2 were calculated by densitometric analysis. (B) PKR−/− MEFs were either mock-infected or infected with SINV wt or SINV AUG-CAA14 at a MOI of 5 pfu per cell. At 4-h 15-min post-infection, cells were pretreated with Tg at the indicated concentrations for 15 min and then metabolically labeled with [35S]Met/Cys for 30 min in the absence or presence of the inhibitor at the same concentrations as before. Whole-cell extracts were analyzed by SDS-PAGE and autoradiography. An aliquot of the same sample was used to analyze eIF2α phosphorylation by Western blotting. (C) Synthesis of C variants was examined by densitometric scanning of the autoradiograph shown in panel B. The percentage values of Tg-treated cells compared with untreated cells are indicated.
We next examined whether eIF2 inactivation would affect translation initiation and AUG selection of the corresponding sgmRNAs out of the replication context. To this end, we transfected PKR−/− MEFs with in vitro-synthesized sgmRNA C+luc, sgmRNA C+luc AUG-hp, and sgmRNA C+luc AUG-CAA14, and induced eIF2α phosphorylation with 1.5 μM Tg. Luciferase activity was reduced by 76%, 78%, and 80%, respectively, in the presence of the inhibitor (Fig. 8A). This result concurs with a previous experiment carried out in BHK cells, in which translation of sgmRNA C+luc was strongly inhibited by arsenite treatment (Sanz et al. 2009). Analysis of C production by Western blotting revealed that the synthesis of genuine protein C was blocked in the three constructs tested following eIF2α phosphorylation (Fig. 8B–D). In addition, synthesis of mutant C1 protein from sgmRNA C+luc AUG-hp and mutant C2 protein from sgmRNA C+luc AUG-CAA14 was also abolished by Tg treatment (Fig. 8C,D, respectively).
FIGURE 8.
Effect of Tg treatment in PKR−/− MEFs transfected with in vitro-synthesized SINV sgmRNAs. (A) sgmRNA C+luc, sgmRNA C+luc AUG-hp, and sgmRNA C+luc AUG-CAA14 synthesized in vitro by T7 RNA polymerase were transfected into PKR−/− MEFs. Thirty minutes later, cells were treated with 1.5 μM Tg or left untreated, and incubated for 90 min before harvesting to measure luciferase activity. (B–D) PKR−/− MEFs were transfected and treated as in A, and then collected in sample buffer. SINV C protein, phosphorylated eIF2α, and total eIF2α were detected by immunoblotting using specific antibodies (left panels). Densitometric quantifications of protein C and mutant proteins C1 or C2 in the presence or absence of Tg are shown in the right panels. The results displayed in A–D are the mean ± SD of three independent experiments. The percentage values of Tg-treated cells relative to untreated cells are indicated.
In conclusion, our results suggest that SINV sgmRNAs containing two initiation codons are translated differently with respect to eIF2 utilization in replicating cells: The first AUG requires active eIF2 for translation, while the second AUG is selected in the presence of phosphorylated eIF2. Strikingly, scanning of the leader sequence and selection of the second AUG initiation codon can be accomplished when eIF2α is highly phosphorylated in a context of viral infection. On the contrary, eIF2 is necessary for SINV sgmRNA scanning and initiation at both AUGs in nonreplicating cells.
Influence of DLP and the leader sequence of SINV sgmRNA in eIF2-independent translation
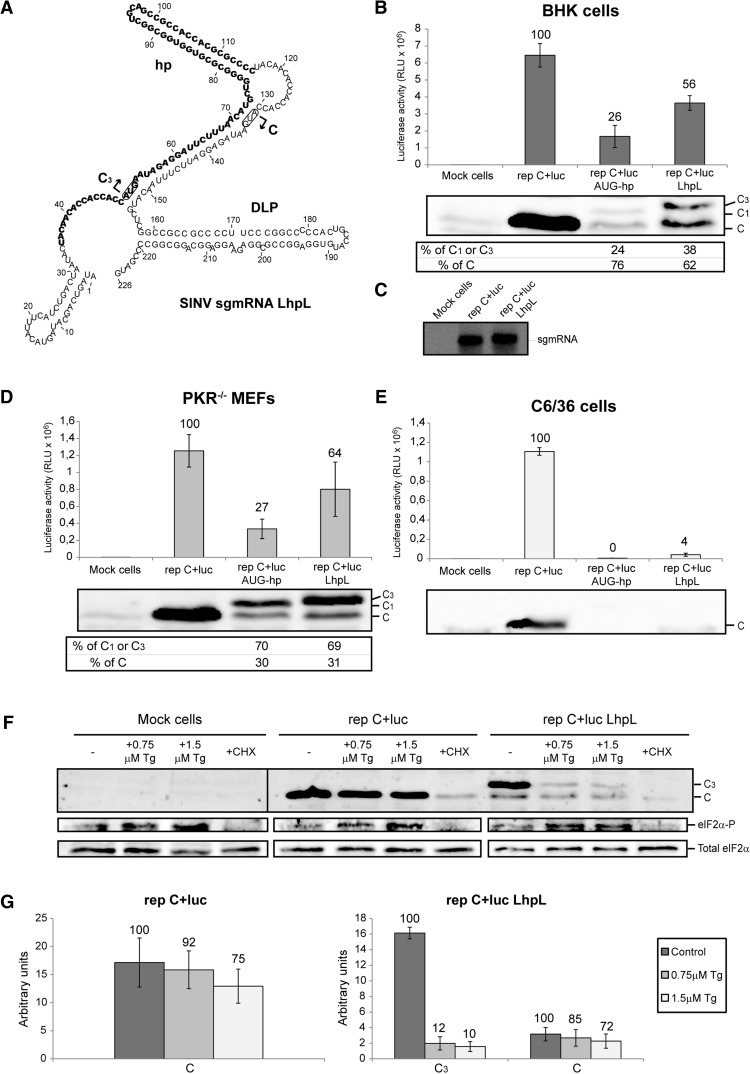
As indicated earlier, the DLP structure leads to eIF2-independent translation of sgmRNA. In agreement with this concept, we have showed that sgmRNAs containing two in-frame initiation codons within a good context were competent to initiate translation at the second AUG upon eIF2α phosphorylation. Previous studies have suggested that the DLP reduces ribosome movement, thus allowing them to initiate translation at limiting amounts of eIF2 (Frolov and Schlesinger 1994, 1996). In such a scenario, the hairpin structure inserted in rep C+luc AUG-hp should confer eIF2 independence also, but instead we observed inhibition of protein synthesis starting at the AUG codon before the hairpin, following eIF2 inactivation. However, the nucleotide sequence surrounding the first AUG in this construct differed from that surrounding the genuine start codon. Therefore, to further analyze the importance of the DLP structure, and also the leader sequence of SINV sgmRNA, in eIF2-independent translation, we generated a new construct named SINV sgmRNA LhpL (Fig. 9A). This construct contains the first 75 nt of SINV sgmRNA, followed by the hairpin hp located at 23 nt downstream from the AUG codon which is now in its authentic sequence context. After the hp, and preceding the DLP structure, there was a duplicate of nucleotides 35–75 with a second AUG codon at a distance of 24 nt to the DLP. BHK, PKR−/− MEFs and insect C6/36 cells were transfected with SINV sgmRNA LhpL, and luciferase activity was measured at the indicated times post-transfection. Luciferase activity measured from rep C+luc LhpL increased 2.2-fold in BHK cells, compared with rep C+luc AUG-hp, but its activity was ∼56% when compared with the control rep C+luc (Fig. 9B). In contrast, sgmRNA production was not affected by the modifications introduced into its leader sequence (Fig. 9C). Similar results were obtained with PKR−/− MEFs cells (2.4-fold and ∼64% when compared with rep C+luc AUG-hp and rep C+luc, respectively) (Fig. 9D). However, C production measured by immunoblotting from collected cells differed between cell lines: In BHK cells, the second AUG codon was preferentially selected, as observed with the other replicons (see Fig. 2A), while in PKR−/− MEFs translation started mostly at the first AUG (as observed with the other constructs) (Figs. 3C, 6C). Notably, the rep C+luc LhpL construct was not operational in C6/36 cells, as luciferase activity was ∼4% to that obtained with control rep C+luc (Fig. 9E). Additionally, to test whether the inserted hp could confer eIF2 independence, PKR−/− MEFs were transfected with the replicon rep C+luc LhpL and treated with Tg to induce eIF2α phosphorylation. As shown in Figure 9F, translation initiation at the first AUG is abolished with Tg treatment (88% and 90% inhibition in cells treated with 0.75 and 1.5 µM Tg, respectively) (Fig. 9G), while translation starting at the genuine AUG codon in control rep C+luc and in rep C+luc LhpL was unaffected by the inhibitor. These results suggest that the inserted hairpin hp does not behave in a similar manner to DLP with respect to AUG selection and eIF2-independent translation. Therefore, the model suggesting that DLP confers eIF2-independence by simply slowing down ribosome movement is not supported by these findings.
FIGURE 9.
Ability of hp and DLP to confer eIF2-independence. (A) Schematic representation of the variants in the 5′ UTR of SINV sgmRNA to obtain mutant LhpL as described in Materials and Methods. RNA secondary structure was predicted and drawn as detailed in Figure 1A. BHK (B,C), PKR−/− MEFs (D), and C6/36 (E) cells were transfected with Lipofectamine 2000 and the indicated SINV replicons transcribed in vitro. At 7, 5, and 8 hpt, respectively, cells were recovered to measure luciferase activity, or processed as described in Figure 2D,F to detect sgmRNA (panel C). Values are represented as means ± SD of three different experiments; the percentage values obtained from mutant replicons relative to control rep C+luc are shown in the chart (B, D, and E, upper panels). In parallel, SINV C production was analyzed in the three cell lines by Western blotting with specific anti-C antibody (B and D, middle panels and E, lower panel). The relative percentages of genuine protein C and mutant proteins C1 or C3 were estimated from values obtained by densitometric analysis (B and D, lower panels). (F) PKR−/− MEFs were transfected with Lipofectamine 2000 and rep C+luc or rep C+luc LhpL. At 3.5 hpt, cells were left untreated or treated with Tg at the indicated concentrations or 100 µg/mL cycloheximide for 90 min. Next, cells were collected in sample buffer and analyzed by SDS-PAGE followed by immunoblotting with anti-SINV C, anti-phospho-eIF2α (eIF2α-P), and anti-eIF2α (Total eIF2α) antibodies. (G) The amount of protein C and mutant protein C3 produced in the presence of Tg was examined by densitometric analysis of the autoradiograph shown in panel F. Values obtained from cycloheximide-treated cells were used to subtract the amount of protein synthesized prior to Tg addition. The percentage values of cells treated with Tg compared with untreated cells are indicated in the graph.
Ribosomal shunting is not operative in SINV sgmRNA AUG-hp
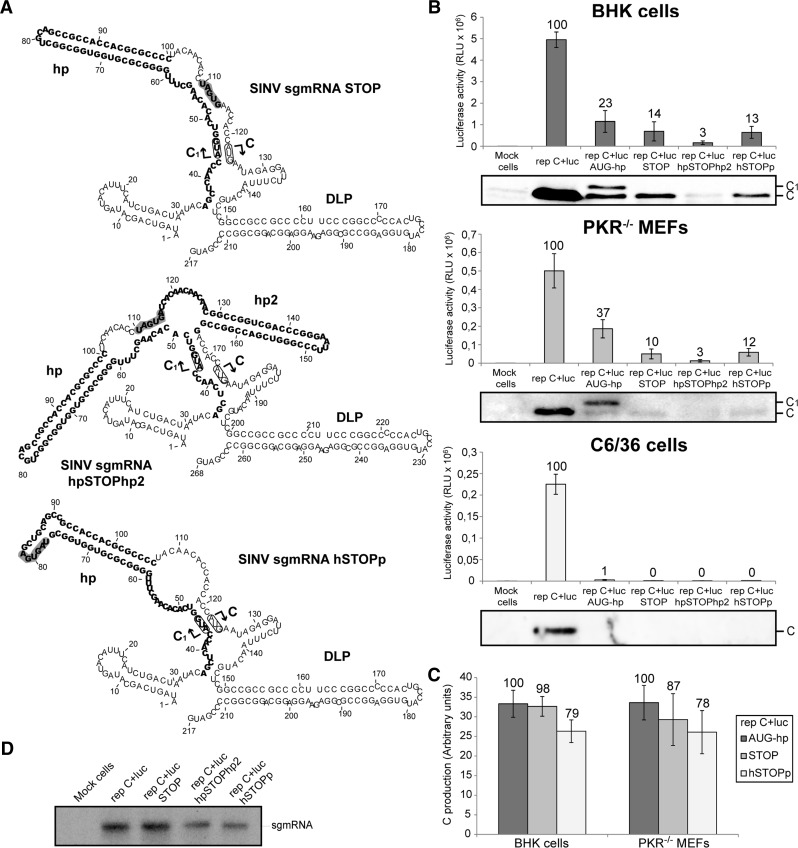
Certain mRNAs can be translated by a mechanism known as “ribosomal shunting.” This process involves the direct translocation of the preinitiation complex from the cap structure to the AUG initiation codon (Yueh and Schneider 1996). Undoubtedly, ribosomal shunting is not the operative mechanism in SINV sgmRNA translation as protein synthesis directed by the construct SINV sgmRNA hp is severely abrogated. However, in sgmRNAs bearing two initiation codons, there are instances where both AUGs are used to initiate translation. In the case of SINV sgmRNA AUG-hp, one possible reason for this is that a proportion of 80S initiation complexes present at the first AUG could translocate directly to the second AUG without scanning. A second possible mechanism could be that elongating 80S ribosomes, which have initiated translation at the first AUG, are able to melt the hairpin during its translation. This process may be used by preinitiation complexes coming behind the 80S ribosomes, to pass through the melted hairpin to initiate protein synthesis at the second genuine AUG. To distinguish between these two possibilities, several additional constructs were engineered by modification of SINV sgmRNA AUG-hp (Fig. 10A). In the first construct, two stop codons were introduced at 9 nt downstream from the hairpin hp (SINV sgmRNA STOP). For the second construction, we inserted a second hairpin hp2 (ΔG of −42.9 kcal/mol) following the stop codons (SINV sgmRNA hpSTOPhp2) that would stall preinitiation complexes coming behind elongating ribosomes, in the case that this event occurs. The third construct contained two stop codons inside the hairpin hp (SINV sgmRNA hSTOPp). Confidence in the structural predictions of these SINV sgmRNAs, as well as all those described above, is shown in Supplemental Figure S2. The three replicons were independently transfected into BHK, PKR−/− MEFs and C6/36 cells followed by measurement of luciferase activity. Results showed that replicons rep C+luc STOP, rep C+luc hpSTOPhp2, and rep C+luc hSTOPp were all potently inhibited in the three cell lines, compared with control rep C+luc: 86%, 97%, and 87% inhibition, respectively, in BHK cells; 90%, 97%, and 88% inhibition in PKR−/− MEFs; 99.6%, 99.7%, and 99.5% inhibition in mosquito cells (Fig. 10B). Additionally, Western blotting showed that only one form of C was detected, albeit to low levels, in the case of the new replicons rep C+luc STOP and rep C+luc hSTOPp in BHK and PKR−/− MEFs, which corresponded to C synthesized from the authentic start codon (Fig. 10B). Larger forms of C produced from the first start codon were not observed, indicating that translation finished at the corresponding termination codons. Also, the amounts of genuine protein C were similar to those produced from the parental rep C+luc AUG-hp construct (only 2%–13% and 21%–22% inhibition in the case of rep C+luc STOP and rep C+luc hSTOPp, respectively) (Fig. 10C). Therefore, the presence of the STOP codons had little effect on initiation at the genuine AUG. On the other hand, the presence of a second hairpin after the stop codons (rep C+luc hpSTOPhp2) further blocked C production (Fig. 10B). This strong reduction in luciferase and C synthesis was not due to variations in the amount of sgmRNA, as observed in Figure 10D. These results discard a shunting mechanism for SINV sgmRNA AUG-hp translation initiation and are consistent with scanning as the mechanism used by this mRNA. In addition, this suggests that the melting of the hairpin hp by elongating 80S ribosomes facilitates preinitiation complexes to reach the authentic AUG codon in the construct SINV sgmRNA AUG-hp.
FIGURE 10.
Translation of SINV sgmRNAs bearing termination codons at different positions. (A) RNA secondary structure of 5′ UTRs predicted by RNAfold (see legend in Fig. 1A). (B) BHK (upper panel), PKR−/− MEFs (middle panel), and C6/36 (lower panel) cells were transfected with Lipofectamine 2000 and the corresponding SINV replicons transcribed in vitro. Seven, 5, and 8 h later, respectively, cells were harvested to measure luciferase activity. Values are plotted as means ± SD of three independent experiments. The percentage values obtained from mutant replicons relative to control rep C+luc are shown in the graph. SINV C accumulation was analyzed in parallel by Western blotting with a specific anti-C antibody. (C) The amounts of genuine protein C were quantified by densitometric scanning of the corresponding autoradiographs. Values are represented as means ± SD of three representative experiments. Numbers above the bars indicate the percentage values obtained from rep C+luc STOP and rep C+luc hSTOPp relative to rep C+luc AUG-hp. (D) Synthesis of sgmRNA in BHK cells transfected with the different SINV replicons and processed as indicated in Figure 2D,F.
DISCUSSION
Failure to adequately select the AUG initiation codon leads to the synthesis of aberrant proteins or peptides that may be toxic for cells. Therefore, the protein synthesizing machinery has evolved into a sophisticated apparatus to correctly select the initiation codon on mRNAs (Hinnebusch 2011; Valasek 2012). In alphaviruses, the mechanism of initiation codon selection on the sgmRNA is insufficiently understood. Elegant studies using a reconstituted in vitro system with purified components of the translation machinery have determined that eIF3 and DHX29 can promote the formation of 48S initiation complexes on the SINV sgmRNA (Skabkin et al. 2010). In the absence of eIF2, Ligatin (later identified as eIF2D) can recruit the initiator Met-tRNAiMet to the ribosomal P site. The results obtained with this in vitro system were consistent with a placement of the AUG initiation codon directly at the P site, without scanning of the sgmRNA 5′ UTR (Skabkin et al. 2010). However, SINV sgmRNA displays a dual translation mechanism when tested in cell-free systems and virus-infected cells. Thus, the eIF requirements differ when sgmRNA translation is analyzed alone or in replicating cells (Sanz et al. 2009; Garcia-Moreno et al. 2013). In the present work, we provide evidence to support that the initiation mechanism on sgmRNA may involve 5′-UTR scanning. This could represent an example where results obtained in in vitro systems may not reflect the situation observed in virus-infected cells. There are several possibilities for the differences reported by Skabkin et al. (2010) and our present observations. One of them is the different systems used; thus, in vitro systems may contain ionic concentrations that could influence the interaction of 80S ribosomes with SINV sgmRNA. In addition, it is possible that the DLP present in SINV sgmRNA promotes this interaction. Alternatively, it could be possible that the insertion of a hairpin structure into the leader sequence of SINV sgmRNA, such as that described in this work, may affect the potential direct interaction of 80S ribosomes with the genuine initiation codon. However, this possibility is not consistent with the fact that SINV gmRNA does not direct translation from the second cistron, whereas different viral IRES elements placed in substitution for the sgmRNA 5′ UTR can direct the internal translation initiation on gmRNAs (Sanz et al. 2010). In addition, a SINV replicon modified to contain a bicistronic sgmRNA with a duplication of the genuine leader sequence is unable to translate the capsid gene located downstream from the second internal leader sequence (Sanz et al. 2010). These findings supported the idea that the unmodified SINV sgmRNA leader sequence is not promoting internal initiation. Another important observation in support of the scanning mechanism to translate SINV sgmRNA is that the insertion of an upstream AUG in the sgmRNA CAA14 diminishes the utilization of the genuine AUG in the sgmRNA AUG-CAA14. This result indicates that a percentage of preinitiation complexes start translation at the first AUG. This finding was observed in the three different cell lines analyzed and in RRL. One would expect that, if there were internal initiation at the genuine AUG in SINV sgmRNA, the presence of an upstream initiation codon would have no effect on the production of genuine protein C.
A number of structural and sequence-specific features influence AUG selection by the preinitiation complex (Kozak 1991). One of these features is the nucleotide sequence which flanks the AUG; thus a purine present at position −3 and a G at position +4 provide an optimal context. In this respect, SINV sgmRNA does not totally fulfill this requirement since there is an A residue at the position +4. In addition, another important factor for AUG selection is the distance from the cap structure to the initiation codon (Kozak 1991; Hinnebusch 2011). For SINV sgmRNA this distance is 49 nt, which falls within the normal length of leader sequences in vertebrate mRNAs (Kozak 1987). Notably, when this leader sequence is expanded to 91 nt, without introducing secondary structures, the resulting sgmRNA (SINV sgmRNA CAA14) efficiently directs translation both in vertebrate and in mosquito cells. In contrast, the presence of a hairpin structure located between the cap and the AUG codon significantly abolishes translation (SINV sgmRNA hp and SINV sgmRNA hp30). Placement of an AUG within a good context before this hairpin (SINV sgmRNA AUG-hp) partially restores the translation capacity. Interestingly, this mRNA can now initiate translation at either of the two initiation codons, and each one is preferentially chosen depending on the cell type or the degree of eIF2α phosphorylation. Curiously, the presence of this hairpin promotes the initiation at the AUG preceding it in the case of PKR−/− cells but not in BHK cells. In a previous study using BHK cells, van Duijn et al. (1988) expressed mutant Semliki Forest virus sgmRNAs, containing two in-frame AUG codons . They noted that translation started principally at the newly inserted AUG codon. However, the constructs used in those experiments lacked viral replicative proteins, and therefore this was a nonreplication context, and presumably eIF2α remained unphosphorylated. We have analyzed in replicating BHK cells (in which eIF2 has been extensively described to be phosphorylated) the production of C protein from two constructs that contain two initiation codons (rep C+luc AUG-hp and rep C+luc AUG-CAA14). Our results show that the majority of C synthesis started at the authentic AUG codon. This suggests that the cellular context and eIF2α phosphorylation state determines AUG selection.
It is puzzling to understand the mechanism of translation of SINV sgmRNAs bearing two initiation codons, particularly when a hairpin is present between both AUGs, as is the case for SINV sgmRNA AUG-hp. The possibility that ribosomal shunting may take place during the initiation process of alphavirus sgmRNA translation is not supported by the fact that the constructs rep C+luc hp and rep C+luc hp30, which contain a hairpin situated in the leader sequence before the genuine initiation codon, are not functional. Furthermore, these results do not support a direct initiation from the AUG start codon. It could still be formally possible that in sgmRNAs bearing two AUGs a proportion of 80S initiation complexes formed at the first AUG could jump to the second AUG; however, we believe this mechanism is not occurring because a construct bearing two termination codons upstream of the genuine AUG and at 9 nt after the hairpin (rep C+luc STOP) is not operative. The inhibition of luciferase or C synthesis was even stronger in the rep C+luc hpSTOPhp2 construct containing a hairpin between these stop codons and the genuine AUG. These findings do not lend support to the process of ribosomal shunting being important for translation of this mRNA. Thus, the preinitiation complex or even the 80S ribosomes, when they are bound to the cap structure or to the first AUG, might not be able to jump or assemble directly to the second initiation codon without scanning. Instead, a more likely model is depicted in Figure 11, wherein the 80S ribosomes that initiate translation at the first AUG might be able to pass through the hairpin, flattening its structure and permitting the passage of 43S preinitiation complexes that skip the first AUG. These 43S preinitiation complexes would traverse the leader sequence behind the 80S ribosomes until a second AUG is reached. Therefore, initiation on both AUGs could be explained by this model even if a hairpin structure is located between them. According to this model, total suppression of translation at the first AUG would also ablate initiation at the genuine AUG. However, partial inhibition at the first AUG would permit that a few 80S ribosomes could flat the hairpin, and the continuous passage of 43S preinitiation complexes would maintain this hairpin disrupted, leading to initiation at the second AUG. The potential paradox of this model is that reduction of initiation at the first AUG after partial phosphorylation of eIF2α still permits the utilization of the downstream AUG. This paradox can be solved if even a low frequency of initiation events at the upstream AUG is sufficient to melt out the hairpin, allowing the passage of preinitiation complexes until the genuine initiation codon is encountered.
FIGURE 11.
Model for the initiation of translation on SINV sgmRNAs. (A) Model of the scanning mechanism followed by SINV sgmRNA wt to initiate translation. The 40S ribosomal subunit attaches initially to the mRNA at the 5′ cap structure. Then, the 5′ UTR is scanned base-by-base in a 5′–3′ direction until the initiation codon is recognized. (B) Model for translation initiation on SINV sgmRNAs bearing two alternative start codons and a hairpin. Initially, the preinitiation complex containing the 40S ribosomal subunit interacts with the cap structure and scans the leader sequence of sgmRNA until the first AUG is encountered. Then, the 80S initiation complex can be formed and elongation ensues, leading to melting of the hairpin (hp). Another preinitiation complex starts scanning from the cap structure and skips the first AUG start codon. This preinitiation complex bearing 40S subunits can pass through the melted hairpin traveling behind the elongating 80S ribosomes. Once this 40S ribosomal subunit reaches the second AUG, synthesis of authentic C protein starts from this initiation codon.
An important aspect of our work arises from the generation of mRNAs that contain two functional initiation codons; the first one requires active eIF2 whereas the second, which is the authentic AUG, is independent of active eIF2. It could be hypothesized that in SINV-infected cells a small proportion of eIF2 remains unphosphorylated in the foci where viral translation takes place and that this portion of active eIF2 transfers the Met-tRNAiMet to the ribosome. Analysis of translation from mRNAs that contain two AUG initiation codons in replicating cells is inconsistent with such a possibility. Thus, it can be observed that eIF2α phosphorylation blocks the initiation from the first AUG, while the second one, which is close to the DLP, is considerably less affected. Therefore, if eIF2 remained unphosphorylated or if it was dephosphorylated by a viral protein precisely at these translational foci, both AUGs would be used for translation. The fact that, in a context of infection, one of them is inhibited by thapsigargin but the second one is not, further supports the concept that the DLP located 24 nt downstream from this initiation codon provides eIF2 independence for initiation, as previously described in the literature (McInerney et al. 2005; Ventoso et al. 2006).
It has been speculated that the DLP structure confers eIF2 independence for translation, due to partial impediment of the 80S ribosomes to traverse this hairpin (Frolov and Schlesinger 1994, 1996). According to this hypothesis, preinitiation complexes would be stalled at the AUG for some time allowing them to start translation with a low concentration of eIF2. Our present observations do not support such a hypothesis. Thus, in the construct rep C+luc LhpL, the sgmRNA contains the genuine leader sequence and a different hairpin (hp) located at almost the same distance (23 nt) as DLP, but the hairpin hp does not now provide eIF2 independence. Moreover, a similar situation is observed with the construct rep C+luc AUG-hp which bears an AUG codon in a good context and a hairpin situated 14 nt downstream, which in principle is the optimal distance to stall the initiation complexes at the initiation codon (Kozak 1991, 1999). This sgmRNA (SINV sgmRNA AUG-hp) requires active eIF2 to initiate translation at the first AUG. Therefore, the simple mechanical model suggesting that the hairpin slows down ribosome progression and permits initiation under low concentrations of eIF2 is not supported by our present findings. A hypothesis for the functioning of DLP could be the direct interaction of this structure with the ribosomal P site, promoting the interaction of the first aminoacyl-tRNA with the A site. However, further investigation is necessary to unveil the exact participation of DLP during the initiation of SINV sgmRNA translation.
5′-UTR scanning of eukaryotic mRNAs involves the participation of several eIFs including eIF2 as part of the ternary complex. This factor, as well as the initiator tRNA, select the AUG following codon–anticodon base-pairing. If eIF2 does not participate in SINV sgmRNA translation, it is challenging to understand how start codon selection takes place in the absence of this crucial eIF. One possibility, which has been advanced by some laboratories, states that eIF2 is replaced by another cellular factor. With this regard, we suggested previously that eIF2 could be replaced by eIF2A in SINV-infected cells (Ventoso et al. 2006). Another possibility is that eIF2D is used in place of eIF2 (Dmitriev et al. 2010; Skabkin et al. 2010). The exact molecular mechanism used by any of these factors during scanning and AUG selection on the sgmRNA remains to be investigated in alphavirus-infected cells. Another important issue in this field is to understand exactly how scanning occurs in the absence of intact eIF4G and active eIF4A in virus-infected cells (Castello et al. 2006; Garcia-Moreno et al. 2013), because these factors are required to translate sgmRNA in transfected cells and in cell-free systems (Sanz et al. 2009). Further research on the molecular events involved in alphavirus mRNA translation will help to unravel this important process of viral gene expression.
MATERIALS AND METHODS
Cell culture
Baby hamster kidney-21 cells (BHK-21, obtained from ATCC), mouse embryonic fibroblasts (MEFs), PKR−/− MEFs (Yang et al. 1995), and Aedes albopictus C6/36 cells (ATCC) were grown as described (Garcia-Moreno et al. 2013).
RNA structure prediction
Minimum free energy secondary structures of the 5′ end of different SINV sgmRNA constructs were predicted using the default parameters of RNAfold web server (Hofacker 2003): http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi.
Plasmid construction
The plasmids used in this work are listed and described in Supplemental Table S1. The SINV sgmRNA leader sequence in plasmids pT7 rep C+luc hp, pT7 rep C+luc AUG-hp, pT7 rep C+luc CAA14, and pT7 rep C+luc AUG-CAA14 was altered by overlap extension with PCR using the oligonucleotides listed in Supplemental Table S2. Double amplifications were performed as follows: For the first PCR step, specific 5′ primers and 3′Aat were used with pT7 rep C+luc as template, and for the other PCR, specific 3′ primers and 5′Hpa were used with the same template. A mixture of these products, with 5′Hpa and 3′Aat as primers were then used for the second PCR. Next, double PCR products were digested with HpaI and AatII restriction endonucleases (New England Biolabs) and inserted in the same sites of pT7 rep C+luc. To obtain the corresponding sgmRNAs by in vitro transcription, the plasmids pT7 C+luc hp, pT7 C+luc AUG-hp, pT7 C+luc CAA14, and pT7 C+luc AUG-CAA14 were made using the plasmids described above as templates in PCR with oligonucleotides 5′SacI-T7prom and 3′Aat. PCR products were digested with SacI and AatII restriction endonucleases and inserted in the same sites of pT7 rep C+luc. Plasmids pT7 rep C+luc LhpL and pT7 rep C+luc hSTOPp were constructed by inserting the PstI/HpaI-digested PCR product obtained with specific 3′ primers, 5′Hpa and DNA templates pT7 rep C+luc or pT7 rep C+luc AUG-hp, respectively, into the same sites of the shuttle vector phpL26S. Vector phpL26S derives from plasmid pT7 rep C+luc hp and lacks the PstI restriction sites located outside the region delimited by HpaI site in 5′ and AatII site in 3′. Next, the fragment between the HpaI–AatII sites was transferred from the shuttle vector to the same sites in pT7 rep C+luc. Plasmid pT7 SV AUG-CAA14 was obtained by inserting the SpeI/AatII-digested product from pT7 rep C+luc AUG-CAA14 into the same sites of pT7 SVwt. To generate plasmids pT7 rep C+luc STOP and pT7 rep C+luc hpSTOPhp2, a PCR using the specific 5′ primers, 3′Aat and pT7 rep C+luc AUG-hp as template was performed. PCR products were then digested with PstI and AatII enzymes and cloned into the same sites of the shuttle vector pAUGhpL26S. The fragment between the HpaI–AatII sites was subsequently transferred to the same sites in pT7 rep C+luc. The pAUGhpL26S shuttle vector was made by cloning the HpaI/PstI-digested fragment derived from pT7 rep C+luc AUG-hp into the same sites of phpL26S. Plasmid pT7 rep C+luc hp30 was constructed by performing a PCR with primers 5′hp30 and 3′Aat, using pT7 rep C+luc as DNA template. The PCR product was subsequently digested with PstI and AatII enzymes and cloned into the same sites of phpL26S. The fragment between the HpaI–AatII sites was transferred from the shuttle vector to the same sites in pT7 rep C+luc. All generated constructs were sequenced to check the integrity of the cloned fragments.
In vitro transcription and transfection
Plasmid DNA was linearized with XhoI and used as a template for in vitro RNA transcription with T7 RNA polymerase (New England Biolabs) in reactions containing the m7G(5′)ppp(5′)G cap analog (New England Biolabs). Cells were transfected with in vitro-synthesized RNA using Lipofectamine 2000 reagent (Invitrogen), as recommended.
Viral infection
To obtain SINV wt and SINV AUG-CAA14 viruses from the infective cDNA clones, BHK cells were transfected with RNAs synthesized in vitro using linearized plasmids, pT7 SVwt or pT7 SV AUG-CAA14, as DNA template. Viruses were collected from the medium 48 h later. Viral infections of BHK cells, MEFs, and PKR−/− MEFs were performed at a multiplicity of 5 pfu per cell in serum-free Dulbecco's modified Eagle medium (DMEM) at 37°C. Infections of C6/36 cells were carried out at 28°C at the same multiplicity. After 1 h, this medium was replaced with DMEM/10% fetal calf serum (FCS), or M3 medium with 10% FCS in the case of C6/36 cells.
Luciferase activity measurement
Cells were recovered at the indicated post-transfection times in a buffer containing 0.5% Triton X-100, 25 mM glycylglycine (pH 7.8), and 1 mM dithiothreitol. Luciferase activity was measured in standard assays (Luciferase Assay System, Promega) using a Monolight 2010 luminometer (Analytical Luminiscense Laboratory). The amount of total cell protein was determined using the BioRad Protein Assay Kit with bovine serum albumin as a standard, and a GeneQuant Pro Spectrophotometer (Amersham Biosciences).
Western blotting and metabolic labeling
Cells were collected in sample buffer (0.37 M Tris–HCl, pH 6.8, 0.1 M DTT, 2% SDS, 17% glycerol, and 0.024% bromophenol blue), boiled for 5 min, fractionated by SDS-PAGE and transferred to a nitrocellulose membrane for immunoblotting. Specific rabbit polyclonal antibodies raised against SINV C (Sanz et al. 2009), luciferase (Santa Cruz Biotechnology), phospho-eIF2α (Ser 51; Cell Signaling Technology), and total eIF2α (Santa Cruz Biotechnology) were used at 1:1000 dilution. The high sensitivity reagent “SuperSignal West Femto Maximum Sensitivity Substrate” (Thermo Scientific) was used when indicated. Measurement of protein synthesis by radioactive labeling was performed with [35S]Met/Cys (EasyTag Express 35S Protein Labeling mix, PerkinElmer) in methionine/cysteine-free DMEM. Samples were then harvested in sample buffer, boiled, and analyzed by autoradiography of SDS-polyacrylamide gels (15%). Autoradiograms were quantified by densitometry using a GS-800 Calibrated Densitometer (BioRad).
[5-3H]uridine labeling of viral RNAs
[5-3H]uridine was incorporated into cells treated with actinomycin D (5 μg/mL) to enable detection of viral RNA synthesis by agarose gel electrophoresis of labeled RNAs, or by radioactive counting in a scintillation counter. For electrophoretic analysis, total RNA from ∼2.5 × 106 cells treated with actinomycin D from 2 hpt and with [3H]uridine (40 μCi/mL, final concentration) from 3 hpt, was extracted at 7 hpt using the RNAeasy mini Kit (Qiagen) and resuspended in 50 μL water. Samples (25 μL) were denatured with glyoxal and dimethyl sulfoxide and separated by electrophoresis in 0.7% agarose gels containing 10 mM phosphate buffer as described (Sambrook et al. 1989). Gels were then dried and exposed to X-ray film at −70°C. For scintillation counting, ∼6 × 105 cells were treated with actinomycin D from 2 hpt and with [3H]uridine (40 μCi/mL, final concentration) from 4 hpt. At 5, 7, and 9 hpt, medium was discarded and cells were extracted with 5% trichloroacetic acid, washed twice with ethanol, dried, and dissolved in 0.1 N NaOH/1% SDS. Samples were counted in a liquid scintillation spectrometer.
Analysis of sgmRNA levels by quantitative reverse-transcription RT-PCR
Total RNA was extracted at 2 hpt from transfected BHK cells using the RNAeasy mini Kit (Qiagen), and reverse transcribed by using the SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen), according to the manufacturer's recommendations. Samples were then subjected to qRT-PCR using Luc specific primers (Supplemental Table S2) and the SsoFast EvaGreen SuperMix (BioRad). PCR amplification was started by incubation at 95°C for 30 sec followed by 40 cycles at 95°C for 5 sec and 60°C for 5 sec, using the CFX384 Touch Real-Time PCR Detection System (BioRad). The Luc values were normalized with the β-Actin internal control. For this reaction, we used β-Actin specific primers (Supplemental Table S2), TaqMan Fast Universal PCR Master Mix (2×), No AmpErase UNG (Applied Biosystems) and UPL no. 20 probe. PCR amplification was initiated with incubation at 95°C for 20 sec, followed by 40 cycles of 95°C for 3 sec and 60°C for 30 sec. The specificity of the amplification reactions was confirmed by analyzing the corresponding melting curves. Data analysis was carried out using the GenEx software (version 5.3.7).
In vitro translation
Nuclease-treated Rabbit Reticulocyte Lysate system (RRL, Promega) was used for in vitro translation. Reaction mixtures containing 100 ng of in vitro transcribed mRNAs were incubated for 2 h at 30°C. Protein synthesis was estimated by measuring luciferase activity and by radioactive labeling.
Reagents
Hippuristanol (Bordeleau et al. 2006), thapsigargin (Sigma), and cycloheximide (Sigma) were used at the indicated concentrations.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a DGICYT (Dirección General de Investigación Científica y Técnica. Ministerio de Economía y Competitividad, Spain) grant BFU2012-31861. M.G.-M. is holder of a FPI (Formación de Personal Investigador) fellowship. The Institutional Grant awarded to the Centro de Biología Molecular “Severo Ochoa” (CSIC-UAM) by the Fundación Ramón Areces is acknowledged.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.047084.114.
REFERENCES
- Asano K, Sachs MS 2007. Translation factor control of ribosome conformation during start codon selection. Genes Dev 21: 1280–1287. [DOI] [PubMed] [Google Scholar]
- Babendure JR, Babendure JL, Ding JH, Tsien RY 2006. Control of mammalian translation by mRNA structure near caps. RNA 12: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanga JJ, Ventoso I, Harding HP, Deng J, Ron D, Sonenberg N, Carrasco L, de Haro C 2006. Antiviral effect of the mammalian translation initiation factor 2α kinase GCN2 against RNA viruses. EMBO J 25: 1730–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol 2: 213–220. [DOI] [PubMed] [Google Scholar]
- Castello A, Sanz MA, Molina S, Carrasco L 2006. Translation of Sindbis virus 26S mRNA does not require intact eukaryotic initiation factor 4G. J Mol Biol 355: 942–956. [DOI] [PubMed] [Google Scholar]
- Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG 2007. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev 21: 1217–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN 2010. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem 285: 26779–26787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Schlesinger S 1994. Translation of Sindbis virus mRNA: effects of sequences downstream of the initiating codon. J Virol 68: 8111–8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Schlesinger S 1996. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J Virol 70: 1182–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno M, Sanz MA, Pelletier J, Carrasco L 2013. Requirements for eIF4A and eIF2 during translation of Sindbis virus subgenomic mRNA in vertebrate and invertebrate host cells. Cell Microbiol 15: 823–840. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Gorchakov R, Frolova E, Williams BR, Rice CM, Frolov I 2004. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J Virol 78: 8455–8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 5: 897–904. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG 2011. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75: 434–467, first page of table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL 2003. Vienna RNA secondary structure server. Nucleic Acids Res 31: 3429–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15: 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1989. Circumstances and mechanisms of inhibition of translation by secondary structure in eukaryotic mRNAs. Mol Cell Biol 9: 5134–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1990. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci 87: 8301–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem 266: 19867–19870. [PubMed] [Google Scholar]
- Kozak M 1997. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J 16: 2482–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234: 187–208. [DOI] [PubMed] [Google Scholar]
- Lindqvist L, Oberer M, Reibarkh M, Cencic R, Bordeleau ME, Vogt E, Marintchev A, Tanaka J, Fagotto F, Altmann M, et al. 2008. Selective pharmacological targeting of a DEAD box RNA helicase. PLoS One 3: e1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linero FN, Thomas MG, Boccaccio GL, Scolaro LA 2011. Junin virus infection impairs stress-granule formation in Vero cells treated with arsenite via inhibition of eIF2α phosphorylation. J Gen Virol 92: 2889–2899. [DOI] [PubMed] [Google Scholar]
- Lorsch JR, Dever TE 2010. Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. J Biol Chem 285: 21203–21207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna RE, Arthanari H, Hiraishi H, Nanda J, Martin-Marcos P, Markus MA, Akabayov B, Milbradt AG, Luna LE, Seo HC, et al. 2012. The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with eIF1 and eIF2β. Cell Rep 1: 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljeström P 2005. Importance of eIF2α phosphorylation and stress granule assembly in α virus translation regulation. Mol Biol Cell 16: 3753–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda JS, Saini AK, Muñoz AM, Hinnebusch AG, Lorsch JR 2013. Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A, and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal preinitiation complex. J Biol Chem 288: 5316–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, Sonenberg N 2011. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol 12: 235–245. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16: 2906–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335. [DOI] [PubMed] [Google Scholar]
- Sambrook JF, Fritsch EF, Maniatis T 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sanz MA, Castelló A, Carrasco L 2007. Viral translation is coupled to transcription in Sindbis virus-infected cells. J Virol 81: 7061–7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MA, Castelló A, Ventoso I, Berlanga JJ, Carrasco L 2009. Dual mechanism for the translation of subgenomic mRNA from Sindbis virus in infected and uninfected cells. PLoS One 4: e4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MA, Welnowska E, Redondo N, Carrasco L 2010. Translation driven by picornavirus IRES is hampered from Sindbis virus replicons: rescue by poliovirus 2A protease. J Mol Biol 402: 101–117. [DOI] [PubMed] [Google Scholar]
- Sanz MA, Redondo N, García-Moreno M, Carrasco L 2013. Phosphorylation of eIF2α is responsible for the failure of the picornavirus internal ribosome entry site to direct translation from Sindbis virus replicons. J Gen Virol 94: 796–806. [DOI] [PubMed] [Google Scholar]
- Schlesinger S, Schlesinger MJ 1996. Togaviridae and their replication. In Fields virology (ed. Fields BN, et al. ), pp. 825–843 Lippincott-Raven Publishers, Philadelphia. [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV 2010. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev 24: 1787–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG 1994. The αviruses: gene expression, replication, and evolution. Microbiol Rev 58: 491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek LS 2012. ‘Ribozoomin’—translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr Protein Pept Sci 13: 305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijn LP, Holsappel S, Kasperaitis M, Bunschoten H, Konings D, Voorma HO 1988. Secondary structure and expression in vivo and in vitro of messenger RNAs into which upstream AUG codons have been inserted. Eur J Biochem 172: 59–66. [DOI] [PubMed] [Google Scholar]
- Ventoso I 2012. Adaptive changes in αvirus mRNA translation allowed colonization of vertebrate hosts. J Virol 86: 9484–9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M 2006. Translational resistance of late αvirus mRNA to eIF2α phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev 20: 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams BR, Aguet M, Weissmann C 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J 14: 6095–6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yueh A, Schneider RJ 1996. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev 10: 1557–1567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.