Summary
Quorum sensing is a process of bacterial cell-cell communication that relies on the production, release, and receptor-driven detection of extracellular signal molecules called autoinducers. The quorum-sensing bacterium Vibrio harveyi exclusively detects the autoinducer N-((R)-3-hydroxybutanoyl)-L-homoserine lactone (3OH-C4 HSL) via the two-component receptor LuxN. To discover the principles underlying the exquisite selectivity LuxN has for its ligand, we identified LuxN mutants with altered specificity. LuxN uses three mechanisms to verify that the bound molecule is the correct ligand: In the context of the overall ligand-binding site, His210 validates the C3 modification, Leu166 surveys the chain-length, and a strong steady-state kinase bias imposes an energetic hurdle for inappropriate ligands to elicit signal transduction. Affinities for the LuxN Kinaseon and Kinaseoff states underpin whether a ligand will act as an antagonist or an agonist. Mutations that bias LuxN to the agonized, Kinaseoff, state are clustered in a region adjacent to the ligand-binding site, suggesting that this region acts as the switch that triggers signal transduction. Together, our analyses illuminate how a histidine sensor kinase differentiates between ligands and exploits those differences to regulate its signaling activity.
Keywords: ligand specificity, quorum sensing, histidine sensor kinase, signal transduction
Introduction
Quorum sensing (QS) is a process of cell-cell communication that bacteria use to control collective behaviors. QS relies on the production, release, and receptor-driven detection of extracellular signal molecules called autoinducers. Strict receptor preferences for cognate autoinducer ligands exist among QS systems, and this feature of QS circuits underpins striking autoinducer detection specificity. Nonetheless, many bacteria harness information garnered from multiple autoinducer-receptor QS pathways to regulate behaviors such as bioluminescence, biofilm formation, sporulation, and virulence. The notion is that bacteria integrate the information encoded in autoinducer mixtures to facilitate intraspecies, intragenus, and interspecies communication, enabling them to precisely execute niche-specific behaviors in accordance with both the cell density and the species composition of the vicinal community (Bassler et al., 1993; Fuqua and Greenberg, 2002; Novick and Geisinger, 2008; Ng and Bassler, 2009; Rutherford and Bassler, 2012). Dissecting QS signal-receptor interactions could enable the understanding of how organisms decode information-containing molecules and inspire strategies to manipulate QS-controlled processes, both harmful and beneficial.
Vibrio harveyi, a Gram-negative marine bacterium that uses QS to control expression of bioluminescence, has been employed as a model to define QS principles. V. harveyi possesses three QS systems, each composed of an autoinducer and a cognate two-component sensor. The LuxM synthase produces Autoinducer-1 (AI-1), an acyl homoserine lactone (AHL) used for intraspecies communication (Cao and Meighen, 1989; Bassler et al., 1993). LuxS synthesizes Autoinducer-2 (AI-2), a furanosyl borate diester involved in interspecies communication (Surette et al., 1999; Schauder et al., 2001; Chen et al., 2002). CqsA produces V. harveyi CAI-1 (Vh-CAI-1), an alpha-enamino-keto fatty acid used for intragenera (i.e., inter-vibrio) communication (Miller et al., 2002; Henke and Bassler, 2004; Higgins et al., 2007; Ng et al., 2011). The three autoinducers are recognized by the LuxN, LuxPQ, and CqsS receptors, respectively (Bassler et al., 1993; Bassler et al., 1994; Miller et al., 2002; Henke and Bassler, 2004). The autoinducers influence whether the receptors act predominately as kinases or predominantly as phosphatases, presumably via autoinducer-triggered conformational changes (Freeman et al., 2000; Neiditch et al., 2005; Neiditch et al., 2006; Wei et al., 2012).
At low cell density (LCD), when little autoinducer is present, the LuxN, LuxPQ, and CqsS receptors are unliganded. Under this condition, the receptors’ autokinase activities dominate. Phosphorelay leads to repression of the QS master transcription factor LuxR (Lenz et al., 2004). At high cell density (HCD), autoinducers accumulate and bind to their cognate receptors, which inhibits the receptors’ autokinase activities (Freeman et al., 2000). Because their constitutive phosphatase activities are unaffected, this step reverses phospho-flow through the QS cascade and allows LuxR to be translated. LuxR regulates hundreds of genes required for group behaviors (Van Kessel et al., 2013), including the luciferase operon (Showalter et al., 1990; Swartzman et al., 1992; Pompeani et al., 2008). Therefore, V. harveyi cells are dark at LCD and bright at HCD. Luciferase, due to its large dynamic range and ease of measurement, has been used as the canonical V. harveyi QS readout.
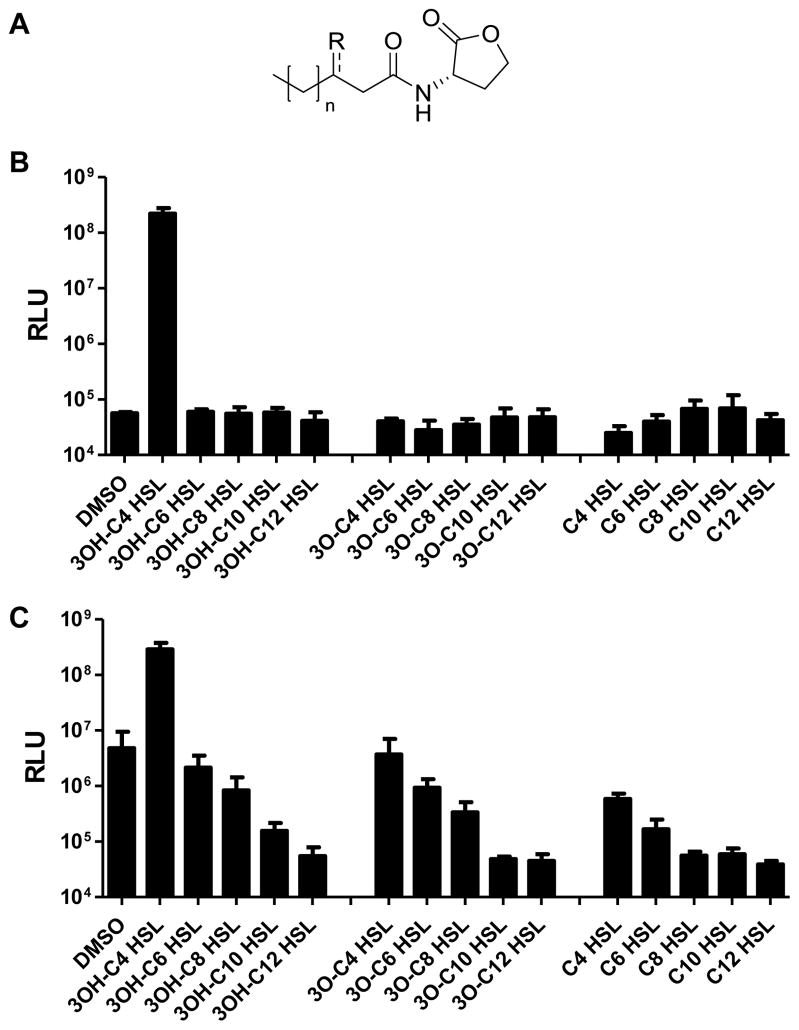
The strongest V. harveyi autoinducer is AI-1. As mentioned, AI-1 belongs to the AHL family, which are often used as autoinducers by Gram-negative bacteria (Fuqua and Greenberg, 2002; Rutherford and Bassler, 2012). AHLs all have a homoserine lactone head group, and they carry different acyl tails (Fig. 1A). Common AHL variations include modifications at the C3 position and differences in acyl chain length (Cao and Meighen, 1989; Pearson et al., 1995; Duerkop et al., 2008). V. harveyi AI-1, N-(3-hydroxybutanoyl)-L-homoserine lactone (designated 3OH-C4 HSL), carries a hydroxyl group at the C3 position (3OH) on a four-carbon long acyl chain (C4) (Fig. 1A). Presumably, bacteria encounter AHL blends in mixed species environments and strict QS receptor specificity must enable Gram-negative bacteria to accurately decode AHL signals to interpret the information encoded in them. In V. harveyi, LuxN detects AI-1. LuxN-type receptors belong to the large family of histidine sensor kinase two-component proteins (Freeman et al., 2000). LuxN is predicted to contain nine transmembrane spanning helices. Previous studies have restricted the AI-1 binding site to membrane-spanning helices 4 to 7 (Swem et al., 2008). To identify residues in LuxN governing ligand specificity, here, we modify AHL structures by chemical synthesis, and we alter the LuxN receptor binding pocket by mutagenesis. We pinpoint His210 as responsible for recognition of the C3 modification, and we show Leu166 determines chain-length preference. We also identify a series of LuxN variants with reduced kinase activity, and find that they harbor mutations in the region immediately adjacent to the AI-1-binding site. We propose that this region acts as the switch that triggers signal transduction. Together, our analyses allowed us to dissect how a histidine sensor kinase differentiates between ligands to regulate its kinase activity.
Figure 1. LuxN exhibits stringent ligand specificity.
(A) General structure for the AHLs examined in this work (R = OH, O, or H; n = 0, 2, 4, 6, 8). V. harveyi AI-1 is 3OH-C4 HSL (R = OH, n = 0).
(B) Agonist assays were performed by measuring light production from the V. harveyi reporter strain TL25 (ΔluxM ΔluxPQ ΔcqsS) in response to 1 μM of the specified AHLs.
(C) Antagonist assays were performed by measuring light production from V. harveyi TL25 supplied with 20 nM AI-1 and 1 μM of the specified AHLs. For panels B and C, DMSO was used as the control. RLU, Relative Light Units. Error bars represent standard deviations for three replicates.
See also Figs S1 and S2.
Results
LuxN exclusively detects 3OH-C4 HSL
To probe the determinants driving LuxN ligand detection specificity, we measured light production from a V. harveyi bioluminescent reporter strain, TL25 (ΔluxM ΔluxPQ ΔcqsS), that lacks the AI-1 synthase and the AI-2 and Vh-CAI-1 receptors LuxPQ and CqsS, respectively. TL25 makes light only in response to exogenously supplied ligands detected by LuxN. We first tested various AHLs to determine whether or not LuxN is highly specific for AI-1 (Fig. 1B). The V. harveyi TL25 reporter remained dark when 1% v/v DMSO was supplied, whereas addition of 1 μM 3OH-C4 HSL (AI-1) induced an over 1000-fold increase in light production. By contrast, AHLs with longer acyl tails (3OH-C6 HSL – 3OH-C12 HSL) did not induce light production. This result indicates that only AHLs with four carbon acyl chains agonize LuxN. 3O-C4 HSL and C4 HSL also did not induce light production when supplied at 1 μM concentration. Thus, in the context of the required C4 tail, a hydroxyl group at the C3 position is essential for LuxN activation. Consistent with this notion, AHLs carrying longer acyl tails and carbonyl groups at the C3 position (3O-C6 HSL – 3O-C12 HSL), and those lacking functional groups at the C3 position (C6 HSL – C12 HSL) also did not induce light production (Fig. 1B).
The above results demonstrate the exquisite specificity LuxN has for AI-1. The experiment does not, however, provide information concerning whether the non-cognate AHLs cannot bind LuxN or they bind LuxN but cannot convert LuxN from a kinase to a phosphatase. To examine this, we tested the AHLs as antagonists rather than as agonists (Fig. 1C). We added AI-1 at 20 nM, the half maximal effective concentration (EC50), and supplied the other AHLs at a range of concentrations. Our expectation is that a reduction in light production would occur if the non-cognate AHLs bind to LuxN and act as antagonists. As controls, we show that addition of DMSO did not cause any reduction in light output from the half-maximal value, whereas supplying additional AI-1 increased light production thirty-fold, to its maximum level (Fig. 1C). With respect to the test molecules, at 1 μM, 3OH-C6 HSL had little effect on light production, 3OH-C8 HSL caused a five-fold decrease in light production, and 3OH-C10 HSL and 3OH-C12 HSL reduced light production by 30- and 100-fold, respectively. These results suggest that 3OH-C6 HSL cannot compete with AI-1 for LuxN binding while 3OH-C8 HSL and AHLs with longer acyl chains are LuxN antagonists of increasing potency.
To test whether the C3 hydroxyl group is essential for antagonist activity, we examined AHLs carrying other C3 modifications in an experiment analogous to the preceding one (Fig. 1C). 3O-C4 HSL did not decrease light production, whereas all other AHLs tested reduced light production by 5- to 100-fold. Thus, the C3 hydroxyl group is dispensable for LuxN antagonist activity if the AHL harbors a long acyl chain. Indeed, AHL analogs carrying bulky side chains, such as chlorolactone (CL) and phenoxy-thiolactone (PTL) are also potent LuxN antagonists (Fig. S1A, at half maximal inhibitory concentrations (IC50) of 870 nM and 90 nM, respectively).
With regard to AHL chain length, the relative potencies of the antagonists are: C12 HSL > C10 HSL > C8 HSL > C6 HSL (IC50 = 1 nM, 20 nM, 600 nM, 3 μM, respectively, when 20 nM AI-1 was supplied, Fig. S1B). To determine the mechanism of antagonism, we measured IC50 values for 3O-C12 HSL when V. harveyi TL25 was incubated with various concentrations of AI-1 (Fig. S1C). We chose 3O-C12 HSL as the test molecule because it is a naturally occurring autoinducer produced by P. aeruginosa (Pearson et al., 1995) and a potent antagonist of LuxN (IC50 = 5 nM, Figs 1C and S1C). The IC50 increased as AI-1 concentration increased, suggesting a competitive mechanism. We likewise tested a subset of the other AHL antagonists (C8 HSL, C10 HSL, and PTL), and they behaved similarly to 3O-C12 HSL (Fig. S1D). Thus, we infer that all the molecules tested that exhibit antagonist activity act competitively. This finding is not particularly surprising given that the molecules have structures similar to AI-1.
Identification of LuxN mutants with altered ligand specificities
To examine the mechanism underlying the extreme specificity of V. harveyi LuxN for AI-1, we screened for LuxN mutants displaying altered ligand selectivity. To do this, we generated a library of plasmids carrying the luxN gene harboring random mutations in the DNA encoding the transmembrane portion, and we assayed them in V. harveyi XK006 (ΔluxMN ΔluxPQ ΔcqsS), which lacks the AI-1 synthase and all three QS receptors. V. harveyi XK006 is constitutively bright due to the absence of all QS receptor kinase activity. Introduction of wild-type (WT) luxN causes the strain to become dark because the lack of endogenous AI-1 production maintains LuxN in the kinase state (Fig. S2A, DMSO). Supplying exogenous AI-1 converts LuxN to a phosphatase and induces a roughly 500-fold increase in light production. Other AHLs tested (14 total, Fig. S2A) induced less than two-fold increases in light output, and these non-cognate AHLs inhibited AI-1-induced light production (Fig. S2B), proving that this experimental set-up reliably probes the native LuxN ligand detection profile. We reason that luxN mutants with altered ligand specificity should induce light production when non-native AHLs are added. By contrast, null or kinase-defective luxN mutants should be constitutively bright. We screened 5000 luxN mutants for responses to DMSO, C4 HSL, C8 HSL, 3O-C10 HSL, and 3O-C12 HSL. Mutants that were bright in the presence of an AHL, but not when DMSO was added, were isolated, sequenced, and validated. We selected several of them for in-depth analyses and they are the focus of the remainder of this work.
In the ligand-binding site, LuxN His210 determines the preference for the 3-OH modified AHL
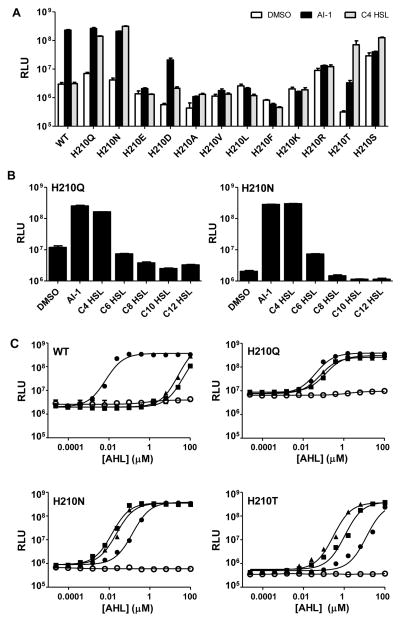
To identify the amino acid residues that determine the LuxN preference for a 3-OH modified ligand, we analyzed the luxN mutant library for those that induced bioluminescence in the presence of C4 HSL (i.e., AI-1 lacking the 3-OH moiety). For reference, V. harveyi XK006 carrying WT luxN produced ~100-times more light following addition of 1 μM 3OH-C4 HSL than when DMSO or 1 μM of C4 HSL was added (Fig. 2A, WT). One mutant, LuxNH210Q, displaying a modestly higher basal level of bioluminescence, induced 50-fold light production in response to 1 μM AI-1 and 30-fold light production in response to 1 μM C4 HSL (Fig. 2A, H210Q). We engineered additional mutations at His210 and found that V. harveyi XK006 carrying luxNH210N behaved similarly to the strain carrying luxNH210Q. Fig. 2A shows that replacing His210 with Glu (H210E) or Asp (H210D) reduced detection of AI-1 and impaired detection of C4 HSL, and hydrophobic or positively charged amino acids eliminated all responses. By contrast, V. harveyi XK006 carrying luxNH210T showed a modest response to AI-1 and a robust response (100-fold induction) to C4 HSL. V. harveyi XK006 carrying luxNH210S behaved similarly, although it exhibited higher basal light production (Fig. 2A). These results establish that His210 is central to the detection of the C3 moiety; substituting His210 with Gln, Asn, Thr, or Ser reduces the stringency for the C3 decoration. AHLs with C6 to C12 chains did not cause induction of light production in V. harveyi XK006 carrying luxNH210Q or luxNH210N (Fig. 2B), with the exception that C6 HSL activated modest light production in V. harveyi XK006 carrying luxNH210N. Thus, LuxN His210 specifies the C3-modification but does not determine the AHL ligand chain-length. We do not exclude the possibility that His210 also interacts with the lactone ring or enhances formation of the LuxN ligand-binding pocket.
Figure 2. His210 defines the LuxN preference for the 3-OH modified HSL.
The V. harveyi bioluminescent reporter strain XK006 (ΔluxMN ΔluxPQ ΔcqsS) harboring WT or His210 mutant luxN alleles expressed from pFED343 was assayed for responses to AHLs.
(A) DMSO, white; 1 μM 3OH-C4 HSL, black; 1 μM C4 HSL, grey.
(B) DMSO was used as the control, and AHLs were supplied at 10 μM. For panels A and B, error bars represent standard deviations for three replicates.
(C) Dose-dependent light production from XK006 carrying WT luxN or luxN His210 alleles. DMSO, open circles; AI-1 (3OH-C4 HSL), closed circles; C4 HSL, triangles; 3O-C4 HSL, squares. Representative data from at least three independent experiments are shown. EC50 values are in Table S1.
The above experiments examined ligands with either a hydroxyl group or a hydrogen atom at C3. We wondered whether LuxN His210 variants could accommodate other C3 modified ligands. To examine these possibilities, we measured the dose-dependent responses of WT luxN and the luxN His210 mutants to AHLs carrying different substituents at C3 in the context of a C4 tail. Reinforcing our above results showing that WT LuxN is extremely selective for AI-1, we note that two diastereomers of 3OH-C4 HSL exist, and WT LuxN detected the native (R)-3OH-C4 HSL (EC50 = 9 nM), but not the (S)-3OH-C4 HSL (Fig. 2C and Table S1). This result implies that the (R)-3-OH group is a critical hydrogen-bond donor in the interaction with His210. V. harveyi XK006 with luxNH210Q detected all three C4 AHLs ((R)-3-OH, 3-O and 3-H) with similar sensitivity (Fig. 2C, EC50 values provided in Table S1). LuxNH210N detected all three molecules, with 10-fold less sensitivity for (R)-3OH-C4 HSL than for the other two; LuxNH210T detected, with decreasing sensitivity (>40-fold differences), C4 HSL, 3O-C4 HSL, and (R)-3OH-C4 HSL (Fig. 2C and Table S1). We also measured the above set of LuxN His210 variants’ responses to (S)-3OH-C4 HSL, and to synthetic AHLs carrying a double bond between C2 and C3 (en-C4), and/or a methyl group at the C3 position (3Me). WT LuxN did not respond to any of these synthetic AHLs, whereas the LuxN His210 mutants showed varying degrees of relaxed specificity toward all of them (Table S1). We conclude that LuxN His210 confers specificity for the 3-OH moiety. Asn, Gln, Thr, or Ser at this position reduce the importance of hydrogen bonding with the 3-OH group.
LuxN mutants that detect ligands carrying elongated acyl chains
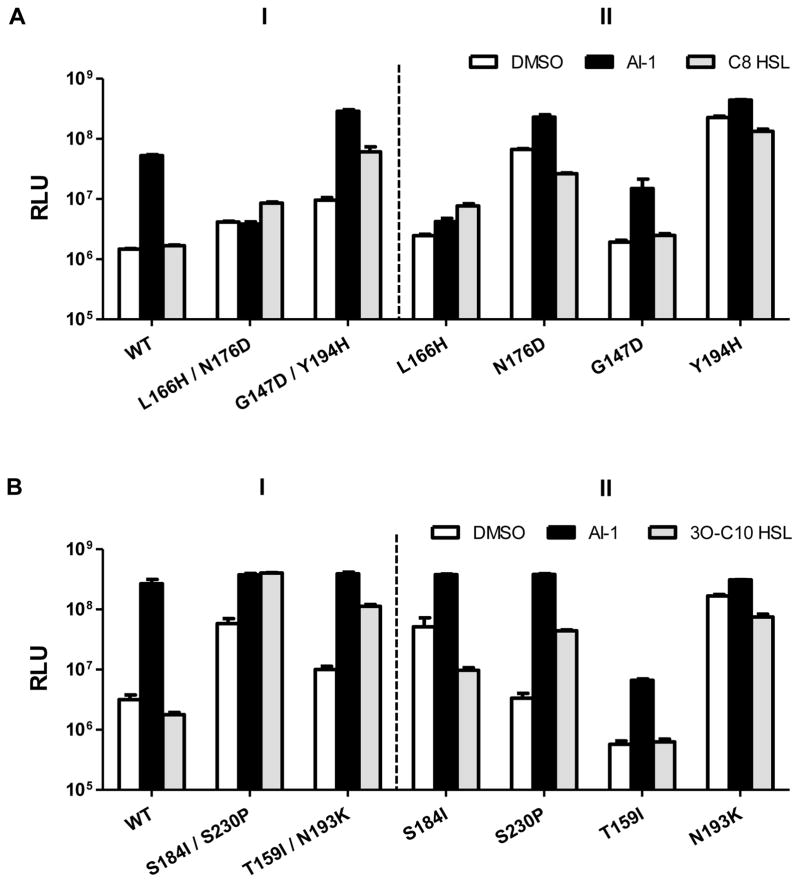
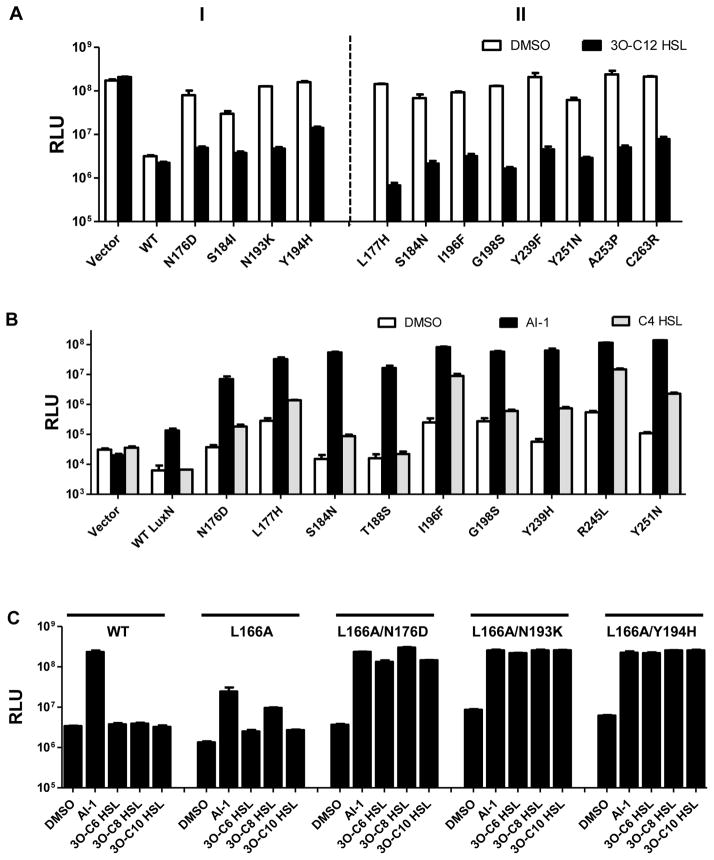
To identify amino acid residues that limit WT LuxN detection to AHLs carrying C4 tails, we isolated luxN mutants from our library that enable V. harveyi XK006 to produce light in response to ligands carrying extended acyl tails. We used C8 HSL as the ligand for the screen and, in secondary assays, examined AHLs with other length tails. We did not conduct a screen with the 3-OH-modified ligand carrying a long tail (e.g., 3OH-C8 HSL) due to limited availability of the molecule. Thus, our strategy did not demand that LuxN be selective for both a 3-OH-moiety and an extended tail, and we address this point explicitly below. Two mutants, LuxNL166H/N176D and LuxNG147D/Y194H detected C8 HSL (panel I of Fig. 3A). Specifically, luxNL166H/N176D induced 3-fold higher light in response to 10 μM C8 HSL compared to when DMSO or 20 nM AI-1 was added. luxNG147D/Y194H exhibited rather high basal light production and induced 30- and 10-fold light production when supplied with AI-1 and C8 HSL, respectively. The mutations were uncoupled and all four single mutants were tested in V. harveyi XK006 (panel II of Fig. 3A). V. harveyi XK006 carrying luxNL166H acted identically to the parent luxNL166H/N176D mutant, whereas the reporter carrying luxNN176D displayed a bright basal phenotype, and addition of C8 HSL did not further enhance light production. Thus, the LuxNL166H mutation is sufficient to confer the response to C8 HSL. By contrast, neither the reporter carrying luxNG147D nor luxNY194H produced increased light in response to C8 HSL. Reconstruction of the double mutants reproduced the parental phenotypes. We conclude that both the LuxNG147D and the LuxNY194H mutations are necessary for detection of C8 HSL. We note that the strains carrying the luxNN176D and luxNY194H single mutants have constitutively bright basal bioluminescence phenotypes. We return to this feature below.
Figure 3. LuxN variants detect AHLs carrying long acyl chains.
(A) Bioluminescence from V. harveyi XK006 carrying the specified luxN alleles was measured as in Fig. 2A. Additions were DMSO, white; 20 nM AI-1, black; 10 μM C8 HSL, grey.
(B) As in A with DMSO, white; 20 nM AI-1, black; 10 μM 3O-C10 HSL, grey. Error bars represent standard deviations for three replicates.
See also Fig. S3.
Screening the LuxN mutant library for responses to 3O-C10 HSL yielded two alleles, luxNS184I/S230P and luxNT159I/Y193K (Fig. 3B, panel I), which induced 10-fold and 20-fold increases in light production, respectively. Of the corresponding single mutants, only luxNS230P showed a robust response to 3O-C10 HSL (20-fold induction, panel II of Fig. 3B). LuxNS184I and LuxNN193K conferred a bright basal phenotype. Again, we provide explanation for these phenotypes below.
We measured the double mutants’ dose-dependent light production profiles in response to different AHLs. The LuxNL166H/N176D mutant was severely impaired for response to AI-1, and we assess the ramifications of this result in the next section. By contrast, in the case of LuxNG147D/Y194H, luxNS184I/S230P, and luxNT159I/Y193K, AI-1 remained the most potent agonist. All responded robustly to AHLs possessing C4 to C10 tails, and none responded to C12 HSL (Fig. S3). Thus, these alleles provide LuxN broadened ligand detection capability.
In the ligand-binding site, LuxN Leu166 specifies the AHL tail length
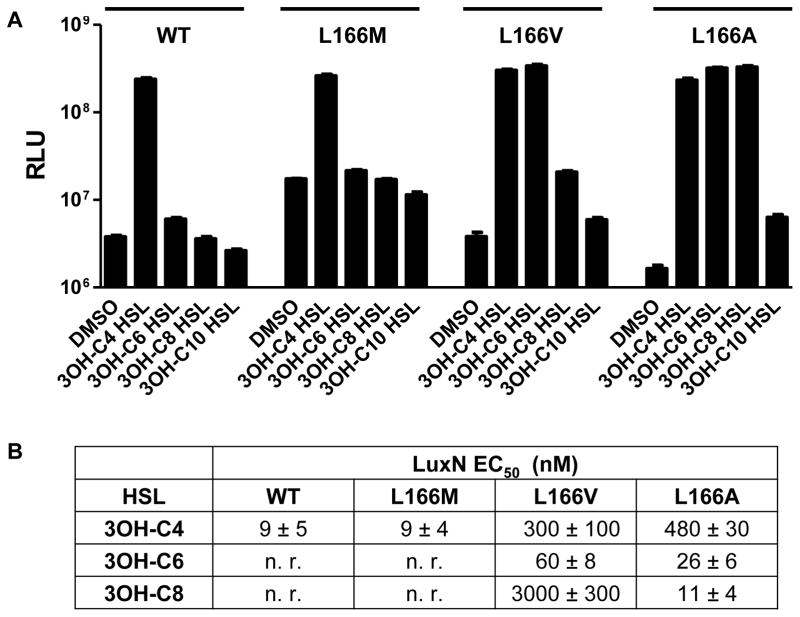
Leu166 is the only residue we identified that, when changed, severely impaired LuxN detection of AI-1 while enabling modest detection of other AHLs (Fig. 3A). Furthermore, of the residues pinpointed by our above analysis, Leu166 displays the most diverse polymorphisms among LuxN homologs (Swem et al., 2008). We speculate that the role LuxN Leu166 has evolved is to specify the C4 AHL tail. To test this idea, we exchanged V. harveyi LuxN Leu166 with Arg, Met, Val, and Ala, and examined these mutants’ responses to AHLs carrying various tail lengths. LuxNL166R did not respond to any AHL in our collection (Fig. S4). LuxNL166M displayed the WT LuxN detection profile (Fig. 4A). LuxNL166V and LuxNL166A responded to 3-OH modified AHLs carrying C4 to C10 length tails (Fig. 4A). Decreasing the size of the residue at position 166 correlated with increasing length of the optimal AHL tail detected (EC50 values in Fig. 4B). Thus, we infer that WT LuxN and LuxNL166M have relatively small binding pockets that restrict detection to C4 HSLs, whereas LuxNL166V and LuxNL166A form expanded ligand-binding pockets that accommodate AHLs carrying correspondingly longer tails (i.e., C6 for LuxNL166V and C8 for LuxNL166A). By contrast, LuxNL166R either eliminates all AHL binding, or it is a locked kinase mutant. We therefore conclude that Leu166 determines the optimal tail length for the LuxN agonist.
Figure 4. Leu166 specifies the LuxN preference for AHL chain-length.
(A) Bioluminescence responses to 1 μM of the specified AHLs were measured using V. harveyi XK006 carrying luxN alleles with mutations at Leu166 expressed from pFED343. Error bars represent standard deviations for three replicates. See also Fig. S4.
(B) EC50 values for WT LuxN and LuxN Leu166 variants. Responses were measured as in Fig. 2C. Numbers are shown as mean ± standard deviation of three replicates, n. r. indicates no response (less than half-maximal induction of light production at 100 μM).
We next tested whether Leu166 has a role in recognizing the C3 modification. We assayed V. harveyi XK006 carrying luxNL166A for responses to elongated AHLs with different moieties at C3. While LuxNL166A detected all of the 3OH modified AHLs tested irrespective of tail length (C4–C10), only modest detection of C8 HSL and 3O-C8 HSL occurred, and only at high concentrations (Table S2). Furthermore, light production was sub-maximal in both cases. Thus, the preference for a 3-OH-moiety is maintained in the LuxNL166A mutant, presumably driven by His210. Because this requirement is not overridden by alteration of Leu166, the role LuxN Leu166 plays in ligand recognition is exclusively in tail length selection.
LuxN His210 and Leu166 act independently to recognize specific moieties on AHL ligands
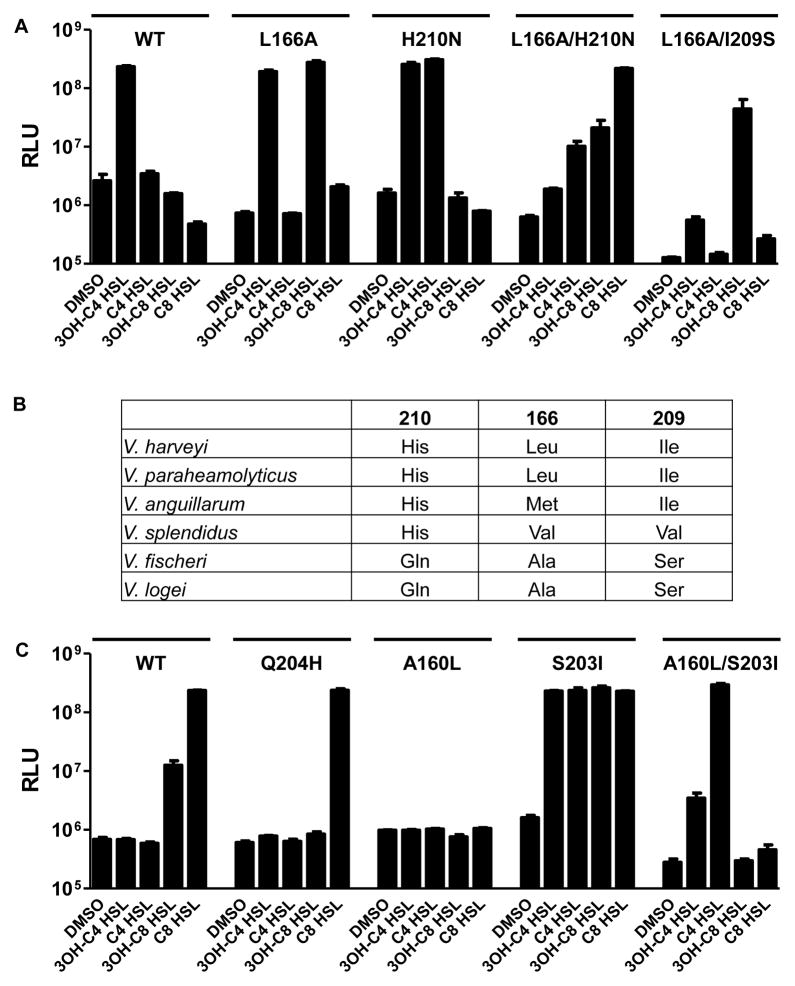
Above we demonstrate that LuxN His210 determines the AHL C3 modification, but it does not play a role in specifying the AHL tail length (Fig. 2B). Conversely, LuxN Leu166 specifies ligand tail length, but it does not select the C3 modification (Table S2). We reason that simultaneous alteration of the His210 and Leu166 residues would produce a LuxN receptor capable of detecting AHL ligands altered in both the C3 substituent and the tail length. We engineered V. harveyi XK006 carrying luxNL166A/H210N, and indeed, it produced maximal light when supplied with C8-HSL or 3O-C8 HSL (Fig. 5A and Table S2). This LuxN variant also detected 3O-C6 HSL and C6 HSL (Table S2). Significantly less light was produced in response to AHLs with C4 tails or to 3OH-C8 HSL (Fig. 5A and Table S2).
Figure 5. LuxN His210 and Leu166 function independently to recognize AHL moieties.
(A) V. harveyi XK006 carrying pFED343 harboring WT or mutant luxN alleles were assayed for responses to the specified AHLs. See also Table S2.
(B) Sequence alignments of vibrio LuxN homologs showing the amino acid residues at key positions.
(C) V. harveyi XK006 harboring WT and mutant V. fischeri ainR alleles on pFED343 were assayed for responses to the specified AHLs.
In A and C, bioluminescence assays were performed as in Fig. 1 and AHLs were added at 1 μM. Error bars represent standard deviations for three replicates. See also Fig. S5 and Table S3.
V. fischeri AinR uses a mechanism analogous to that of LuxN for AHL tail length determination
Bioinformatic analyses of sequences of vibrio LuxN homologs show that different amino acid residues exist at positions corresponding to V. harveyi LuxN His210 and Leu166 (Fig. 5B). We wondered if those receptors’ abilities to detect particular AHL autoinducers stem from polymorphisms at these two critical residues. We examined this idea using the Vibrio fischeri LuxN (named AinR) that contains Gln204 and Ala160 at His210 and Leu166, respectively. V. fischeri AinR detects C8 HSL (Gilson et al., 1995; Kimbrough and Stabb, 2013). We cloned AinR and then replaced Gln204 and Ala160 with His and Leu, respectively. These constructs were introduced into V. harveyi XK006 (Fig. 5C). No endogenous light production occurred when WT AinR was expressed in XK006 suggesting that AinR is a kinase in the absence of ligand. Light production also did not occur when C4 AHLs were added, but light production was induced when 3OH-C8 HSL and C8 HSL were supplied (Fig. 5C). The reporter was most sensitive to C8 HSL (Table S3), indicating that our heterologous system faithfully reproduces the AinR ligand detection specificity.
First, we examined the role of Gln204 in C3 moiety specification. The AinRQ204H mutant did not detect AHLs carrying C4 tails. Furthermore, it lost the ability to detect 3OH-C8 HSL, but retained the capability to detect C8 HSL (Fig. 5C and Table S3). AinRQ204A and AinRQ204T displayed phenotypes similar to WT AinR (Fig. S5A). Thus, AinR Gln204 is unlikely to be responsible for the preferred detection of an undecorated C3 on C8 HSL. We likewise investigated if AinR Ala160 determines the AHL tail-length. Replacing AinR Ala160 with Leu abolished detection of AHLs carrying C4 to C12 tails (Figs 5C and S5B and Table S3). These results show that AinR Ala160 is necessary for C8 tail length detection, however, the single AinRA160L alteration is not sufficient to convert the receptor to a C4 HSL detector. We mutated other amino acid residues in this region of AinR to those present at the corresponding sites in LuxN. Replacing AinR Ser203 by Ile enabled detection of AHLs carrying C4 to C10 tails (Figs 5C and S5B) with C8 HSL remaining the most potent agonist (Table S3). Thus, AinR Ser203 is responsible for discrimination against short chain AHLs. When we also replaced Ala160 with Leu, we found that AinRA160L/S203I detected only AHLs carrying C4 and C6 tails (Figs 5C and S5B and Table S3). We interpret these results to mean that, relative to LuxN, WT AinR harbors an enlarged ligand-binding pocket via Ala160, and possibly other residues, and this pocket accommodates AHLs carrying long and short tails. Ser203 restricts detection to only AHLs with the optimal C8 tail. We suggest that an enlarged AinR binding pocket poses a problem: it must simultaneously enable detection of long tailed AHLs while excluding short tailed AHLs. Thus, the AinR receptor uses a mechanism analogous to LuxN for detecting AHLs of particular tail length with an additional selectivity feature that excludes shorter non-cognate signals. Indeed, when we incorporated Ser209 into the LuxNL166A mutant, ligand detection was restricted to 3OH-C8 HSL (Fig. 5A, compare to L166A), showing that a Ser at this position can discriminate against shorter chain ligands in both AinR and in LuxN.
Mutants with broadened ligand specificity bias LuxN to the Kinoff state
Our previous work suggests that LuxN exists in two states: the kinase on (Kinon) state that is favored when no autoinducer is bound, and the kinase off (Kinoff) state when bound by autoinducer. Above we noted that several LuxN variants (N176D, S184I, N193K, and Y194H) cause higher light production than does WT LuxN when no autoinducer is present (Fig. 3). One possible interpretation is that these mutations bias the unliganded LuxN receptor towards the Kinoff state. Another possibility is that these mutations are simply null alleles. To distinguish between these possibilities, we assayed whether these LuxN mutants are expressed and produced as stable proteins and if their kinase activities could be re-activated by the antagonist 3O-C12 HSL. This set of LuxN proteins are all expressed (as judged by mRNA) and produced (as judged by Western Blot) (Their levels range from 30%–500% of WT LuxN, Fig. S6A, B). In addition, 3O-C12 HSL inhibited light production to basal levels in all cases (panel I of Fig. 6A). Together these results demonstrate that the kinase activities of this set of LuxN mutants can indeed be reactivated and thus, they are not null mutants. From here forward, we refer to them as Kinoff LuxN mutants. Since 3O-C12 HSL activates LuxN kinase activity whereas AI-1 inhibits LuxN kinase activity, we conclude that 3O-C12 HSL acts as an “inverse agonist” (Geisinger et al., 2009).
Figure 6. Kinoff biased LuxN mutants are antagonized by 3O-C12 HSL and display increased sensitivity to AI-1 and non-cognate AHLs.
(A) Panel I, V. harveyi XK006 harboring WT luxN or the Kinoff biased luxN alleles (from Fig. 3) were assayed for bioluminescence with DMSO (white) and in response to 10 μM 3O-C12 HSL (black). Additional LuxN mutants exhibiting high basal light production are shown in Panel II and Fig. S6D.
(B) V. harveyi XK847 harboring WT luxN and the specified Kinoff luxN alleles on pFED343 were assayed for responses to the specified AHLs. DMSO, white; 20 nM AI-1 (3OH-C4 HSL), black; 1 μM C4 HSL, grey.
(C) V. harveyi XK006 harboring luxNL166A and additional mutations were assayed for responses to the specified AHLs. Bioluminescence assays were performed as in Fig. 1 and AHLs were added at 1 μM. Error bars represent standard deviations for three replicates.
See also Figs S6, S7 and Table S4.
The above findings inspired us to identify additional Kinoff biased LuxN mutants. To do this, we screened for luxN mutants that are constitutively bright in V. harveyi XK006 when no ligand is present, but become dark when 3O-C12 HSL is added. We isolated and sequenced eight such mutants (Fig. S6C). All point mutants were reengineered to verify phenotypes. V. harveyi XK006 carrying these luxN alleles produced 20- to 50-fold more light than V. harveyi carrying WT luxN when no AI-1 was supplied. In all cases, addition of 3O-C12 HSL reduced light production to basal levels (panel II of Fig. 6A). We also examined previously engineered luxN mutants in our collection for this phenotype (Swem et al., 2008 and unpublished results), and identified several more Kinoff mutants with the same phenotype (Fig. S6D). Expression and protein production levels of a representative subset of these mutants were verified (Fig. S6A, B).
In the absence of ligand, V. harveyi XK006 carrying the Kinoff luxN mutants produced varying levels of bioluminescence, indicating that the Kinoff LuxN receptors have differing kinase activities which results in them having varying abilities to drain phosphate from the QS pathway (Figs 6A and S6C, D). To quantify the extent of Kinoff bias harbored by each LuxN mutant, we introduced additional phosphate flow through the QS pathway using V. harveyi XK847 (ΔluxMN ΔluxS ΔcqsS), which contains the WT LuxPQ receptor. This strain lacks the AI-2 synthase LuxS, and thus, LuxPQ is a constitutive kinase. Over-expression of WT luxN (Kinon) in V. harveyi XK847 resulted in a completely dark phenotype when DMSO or 3O-C12 HSL was added (Fig. S7A). Addition of 100 nM AI-1 increased light production by 1000-fold. This result shows that, when WT LuxN is converted to the Kinoff state, its phosphatase is capable of overpowering the endogenous LuxPQ kinase activity. All of the Kinoff luxN mutants produced more light than the strain carrying WT luxN in the absence of ligand, they all produced maximal light when AI-1 was supplied, and with the exception of LuxNG238A, they all became dark when 3O-C12 HSL was provided (Fig. S7A). Their sensitivities to 3O-C12 HSL ranged from IC50 15 nM to 640 nM, (Table S4). These results indicate that the kinase activity in the Kinoff LuxN mutants can be fully activated and deactivated by appropriate ligand supplementation. Thus, Kinoff LuxN mutants are adjustable protein switches, each distinctly biased toward the Kinoff state. The LuxNG238A mutant is an exception, which appears to be locked in the Kinoff state. It is not, however, impaired for phosphatase activity because it is capable of overriding the LuxPQ kinase activity.
Kinoff LuxN mutants represent a spectrum of biases towards the Kinoff state
Each of the above Kinoff LuxN mutants displays an intrinsic level of bias. We wonder if these mutations act independently, and if so, whether mutations could be combined to increase the bias toward the Kinoff state. To test this idea, we engineered three combinations of luxN Kinoff mutations, introduced them into V. harveyi XK847, and examined their phenotypes. All strains carrying luxN Kinoff double mutants produced higher bioluminescence than the strain carrying WT luxN and the single Kinoff biased parents (Fig. S7B). For instance, V. harveyi XK847 carrying luxNN176D or luxNS184N produced 10-fold more light than the strain carrying WT luxN, whereas V. harveyi XK847 carrying luxNN176D/S184N produced 5000-fold more light than the strain carrying WT luxN (panel I of Fig. S7B). All double mutants produced close to the maximal level of light in the presence of AI-1, and light production could be inhibited by 3O-C12 HSL (Fig. S7B). We interpret these results to mean that the Kinoff LuxN single mutants represent states on a continuum of biases towards the full Kinoff state, and the mutations act additively. If this is correct, we predict that the bias towards the Kinoff state should be reversed by incorporation of a mutation we previously characterized, F163A, that causes LuxN to be biased toward the Kinon state (Swem et al., 2008). Indeed, V. harveyi XK847 carrying luxNF163A/N176D produced 5-fold less light than the strain carrying luxNN176D, and it responded only modestly to AI-1 (Fig. S7C). V. harveyi XK847 carrying luxNF163A/K191A behaved similarly (Fig. S7C). Finally, introducing the F163A mutation into the LuxNG238A mutant reduced basal light production (Fig. S7C, white bar), showing again that LuxNG238A is not a null mutant but, rather, strongly biased to Kinoff.
Kinoff LuxN mutants exhibit reduced AHL selectivity
A previous report indicates that LuxN Kinoff biased mutants (S184N, R245L) are more sensitive to AI-1 than WT LuxN (Swem et al., 2008). We found that this is also the case with our newly discovered Kinoff mutants: all of them display increased sensitivity to AI-1 compared to WT LuxN (Fig. 6B). Additionally, most of the Kinoff biased LuxN mutants also detected C4 HSL, whereas WT LuxN does not (Fig. 6B). We thus wondered if Kinoff biased mutations could be combined with ligand specificity altering mutations to further enhance detection of non-cognate AHLs. As shown above, V. harveyi XK006 carrying luxNL166A robustly detects 3OH-C8 HSL (Figs 4 and 5). When the luxN L166A mutation was combined with the Kinoff mutations N176D, N193K, or Y194H, the double mutant receptors became more sensitive to AI-1, 3O-C6 HSL, 3O-C8 HSL, and 3O-C10 HSL, producing 3- to 10-fold more light than the strain carrying luxNL166A alone (Fig. 6C). We reason that the LuxN L166A mutation accommodates AHLs carrying extended acyl tails, and introduction of a Kinoff biased mutation enables AHLs in general to more effectively convert the receptor from Kinon to Kinoff. We conclude that the Kinoff mutations override the requirement for specific moieties on AHL ligands, and thus cause the LuxN receptor to become less selective overall in AHL detection. This could explain why we isolated this type of mutant when we screened for LuxN receptors capable of responding to C8 HSL and 3O-C10 HSL (Fig. 3).
Discussion
To understand QS receptor specificity, we examined the responses of the V. harveyi LuxN receptor to a panel of AHLs. The LuxN N-terminal domain contains 9 transmembrane helices connected by intervening loops (Fig. 7A). Previous mutant analyses indicate that AI-1 binds to a region composed of helices 4 to 7, and the intervening periplasmic loops 2 and 3 (Fig. 7A, required amino acids shown in red). WT LuxN exclusively detects AI-1 ((R)-3OH-C4 HSL). AI-1 homologs lacking the C3-hydroxy group (3O-C4 HSL and C4 HSL) are weak LuxN agonists, and they have no antagonist activity. AHLs carrying extended acyl tails (C6 – C12) are competitive antagonists of LuxN and potency increases with acyl tail length (Fig. 1). LuxN His210 in helix 6 specifies the C3 modification, and LuxN Leu166 in helix 5 defines the ligand tail length (Figs 2, 4, and 7A, green). These two residues act independently as “gatekeepers” in the ligand-binding pocket to ensure the proper moieties are present on the autoinducer (Fig. 5). Mutations that modestly broaden ligand preference coupled with Kinoff mutations also relax ligand specificity (Figs 3 and S3). Consistent with this idea, Kinoff mutants are more sensitive to AI-1 than WT LuxN, and combining a Kinoff mutation with a mutation at the Leu166 gatekeeper site further enhances detection of non-cognate AHLs (Fig. 6).
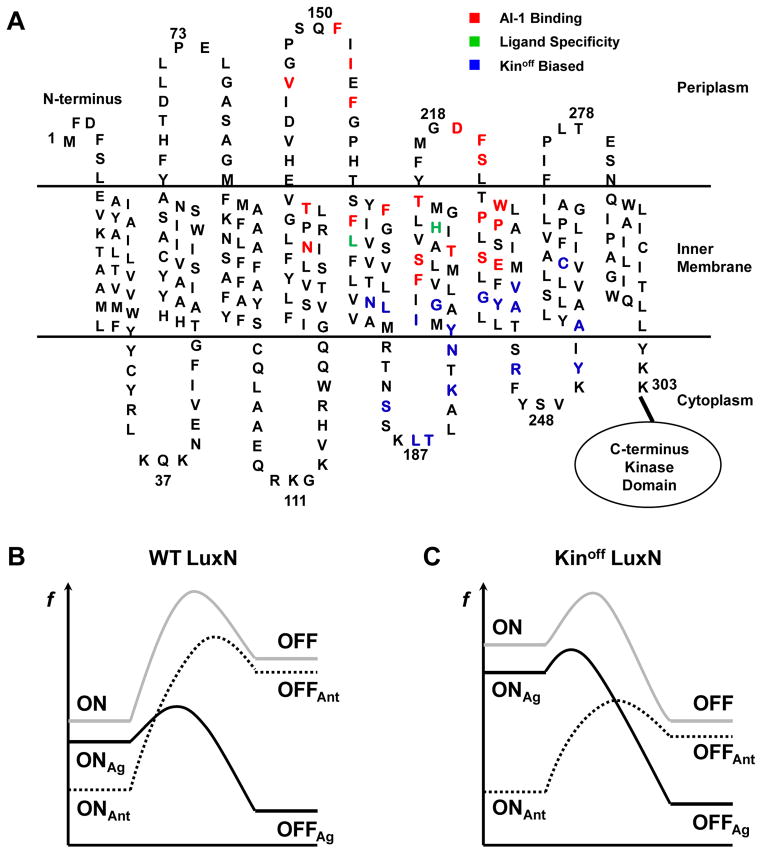
Figure 7. Sites of LuxN mutants with altered ligand specificity and a proposed model for LuxN agonism and antagonism.
(A) Predicted topology of the LuxN transmembrane domain. Residues required for AI-1 ((R)-3OH-C4 HSL) detection are shown in red. Leu166 and His210 that determine ligand specificity are shown in green. Kinoff biased mutation sites are shown in blue.
(B) An energy profile illustrating theoretical transitions of the WT LuxN receptor between the Kinon and the Kinoff states upon ligand binding. Notations are: f (free energy), ON (unliganded Kinon state), OFF (unliganded Kinoff state), ONAg (agonist-bound Kinon state), OFFAg (agonist-bound Kinoff state), ONAnt (antagonist-bound Kinon state), OFFAnt (antagonist-bound Kinoff state). Grey solid, unliganded; black solid, agonists bound; dotted black, antagonists bound.
(C) An energy profile illustrating the analogous transitions for the Kinoff biased LuxN receptor. Notations are as in B.
The Kinoff biased LuxN mutants provide insight into how signal transduction occurs following agonist binding. Kinoff LuxN mutants cluster in a region composed of helices 5 to 8, and the associated cytoplasmic loops (Fig. 7A, blue). This region is highly conserved among LuxN-type receptors (Swem et al., 2008), with many residues showing complete conservation (Asn176, Leu187, Lys191, Tyr194, Gly198, Gly238, Tyr239, Arg245, and Tyr251). This region is located immediately downstream of the putative AI-1 binding pocket and it connects directly to the cytoplasmic kinase domain. We suspect that this “linker” region acts as the switch that regulates LuxN kinase activity in response to ligand binding. Mutants in this region display various degrees of Kinoff bias (Figs. 6 and S7), and we suggest they mimic naturally occurring states explored by the receptor during transitions between its Kinon and Kinoff states. We imagine that AI-1 binding to WT LuxN induces movement in this region to promote signal transduction. Piston-like or rotational movement of the TM helices, as in the Che/MCP and LuxQ receptors, could disrupt the auto-kinase activity (Ottemann et al., 1999; Neiditch et al., 2006; Stock, 2006). Alternatively, AI-1 binding could enforce a large scale rearrangement of the LuxN dimers, perhaps similar to the scissor blade-like closing movement of the Bacteroides thetaiotaomicron BT4633 receptor (Lowe et al., 2012). The transmembrane and kinase domains of receptor histidine kinases are often linked by cytoplasmic coiled-coil HAMP domains (Gao and Stock, 2009). Alternatively, a short cytoplasmic helical region in AgrC connects the sensor and the kinase domains and exercises rheostat-like control over kinase activity (Wang et al., 2014). No obvious linker domain is present in LuxN. Instead, we have identified a region within the transmembrane domain that seemingly harbors the linker function. Analogous regions could exist in other histidine kinases that contain multipass transmembrane domains and that lack canonical cytoplasmic linker domains.
We propose that LuxN uses at least three verification mechanisms to ensure proper ligand interactions prior to eliciting signal transduction. First, LuxN surveys the ligand structure for the presence of the hydroxyl group at C3. Indeed, 3O-C4 HSL and C4 HSL are significantly less potent than AI-1 in agonizing LuxN because their access to the LuxN binding pocket is gated by His210 (Fig. 2). The C3-OH likely acts as a hydrogen-bond donor for the imidazole side chain on His210, and this putative interaction presumably fosters productive AI-1 binding but not 3O-C4 HSL or C4 HSL binding. Mutations at His210 alter the preference for the C3 decoration, and, for example, we can make LuxN prefer 3-Me-en-C4 HSL by replacing His210 with Thr (Table S1). Presumably, hydrophobic interactions mediated by Thr foster 3-Me-en-C4 HSL binding over AI-1. Analogous mechanisms exist in CqsS recognition of CAI-1. The preference for amino-CAI-1 over hydroxy-CAI-1 by V. cholerae CqsS is governed by Trp104 and Ser107 (Ng et al., 2010). We conclude that two-component QS receptors fine-tune their binding to ligands through specific interactions between a few key residues in their binding pockets and signature chemical moieties on the corresponding ligands. These interactions can be harnessed to drive ligand specificity.
The second test for the presence of the correct ligand occurs through LuxN control over the bound ligand’s ability to elicit signal transduction. Many AHL molecules carrying extended acyl tails bind LuxN with high affinity, but only AI-1 agonizes the receptor (Fig. 1). We know other ligands bind at the LuxN binding site because they act as competitive antagonists (Fig. S1). We interpret these findings in light of our previous postulate that LuxN transitions between two reversible conformations, Kinon and Kinoff in which AI-I binds to the Kinoff state and stabilizes that state (Swem et al., 2008). In our model, unliganded WT LuxN is strongly biased toward the Kinon state, and this bias imposes an energetic hurdle that must be overcome to switch the receptor to the Kinoff state (Fig. 7B, grey curve). Agonists stabilize the receptor Kinoff state (Fig. 7B, from OFF to OFFAg) and reduce the free-energy barrier making the transition to the Kinoff state favorable (Fig. 7B, black curve). An antagonist, by contrast, cannot stabilize the Kinoff state to overcome the energy barrier for transition. Possibly, antagonists further stabilize the Kinon state (Fig. 7B, from ON to ONAnt), enlarging the energy barrier, and in turn, inhibiting the Kinon to Kinoff transition (Fig. 7B, dotted curve). Under this framework, we rationalize that WT LuxN, when in the Kinoff state, has a restricted ligand binding pocket and only AI-1 can stabilize this conformation. AI-1 binding does not, however, stabilize the Kinon state. Rather, the LuxN Kinon conformation has affinity for AHL antagonists carrying extended acyl tails. Binding of an antagonist maintains the LuxN receptor in the Kinon state. Thus, LuxN in the Kinon and Kinoff states harbor different preferences for AHL tail length. We hypothesize that transitions between the LuxN Kinon and LuxN Kinoff states are accompanied by significant changes in the ligand binding pocket. AI-1 and antagonist AHLs with long tails stabilize one conformation but not the other (induced fit model). By contrast, we propose that AHLs carrying C6 tails do not stabilize either conformation or they stabilize both conformations to the same extent. Such an architectural design could fortify the barrier segregating AI-1 binding and signal relay from binding of a non-cognate molecule, and in so doing, prevent inaccurate signal transduction. V. harveyi LuxN uses Leu166 to set the C4 tail length preference, and this restriction can be adjusted to C6 or C8 by replacing Leu166 with Val or Ala (Fig. 4). We reason that incorporation of Val or Ala at position 166 forms an enlarged ligand binding pocket, enabling AHLs with longer tails to bind, and importantly, to stabilize the LuxN Kinoff state. In V. fischeri AinR, Ala160 specifies restriction to a C8 HSL tail. Additionally, AinR employs Ser203 to reduce AinR overall sensitivity to AHLs, thus excluding detection of AHLs carrying shorter tails (Fig. 5C). LuxN does not require such a signal-dampening residue presumably because harboring a small binding pocket naturally excludes AHL carrying long tails. LuxN and AinR signaling can be easily adjusted to accommodate other ligands by alteration of a few key residues, suggesting impressive plasticity that could enable adaptation of emerging species to new environments.
A third mechanism we propose LuxN uses to enhance ligand stringency is the imposition of overall reduced sensitivity to ligands due to the strong bias to the Kinon state. This mechanism ensures that weak agonists cannot elicit a signaling response. LuxN mutants biased to the Kinoff state display higher sensitivity to AI-1, and they readily respond to non-cognate AHLs (Fig. 6). We again interpret these results using our two-state model framework. As mentioned, the Kinon bias of unliganded WT LuxN imposes a large energetic threshold that needs to be overcome for AHLs to act as agonists. By contrast, LuxN mutants biased to the Kinoff state harbor a reduced energy barrier for the transition from Kinon to Kinoff (Fig. 7C, grey curve), and so there is a lower threshold for AHLs to cross to act as agonists (Fig. 7C, from OFF to OFFAg). This interpretation could explain why Kinoff biased mutations magnify the LuxN response to AI-1, and allow non-optimal agonists to stabilize the Kinoff state and induce the QS response (Fig. 6B and C). Regarding AHL antagonists and Kinoff biased LuxN mutants, increased stabilization of the Kinon state is required for inhibition compared to WT LuxN (Fig. 7B and C, compare the difference from ON to ONAnt), explaining why 3O-C12 HSL is a less potent antagonist of the most highly Kinoff biased LuxN mutants than it is for WT LuxN (Fig. S7 and Table S4). Had the native LuxN receptor evolved to be biased to the Kinoff state, it would harbor increased sensitivity to AI-1, but would be vulnerable to non-cognate ligands functioning as efficient agonists. The endogenous LuxN Kinon bias mechanism probably also reduces signaling noise at LCD by enabling the LuxN receptor to be impervious to fluctuations in autoinducer concentrations. This design guarantees that V. harveyi executes its QS response only at HCD when sufficient AI-1 signal is present. If so, this feature of the LuxN receptor guarantees that the QS response is all-or-none, and this mode-changing action is only triggered by the cognate signal.
In conclusion, only an AHL with high affinity for the LuxN ligand binding site that is capable of stabilizing the LuxN Kinoff state but not the Kinon state, and also capable of overcoming the endogenous bias to the Kinon state can elicit signal transduction. This arrangement likely makes the LuxN receptor ultra-sensitive to AI-1 while remaining impervious to similar AHLs including those carrying C4 tails. This feature allows signaling by AI-1 in the presence of other highly similar C4 HSLs. Importantly, the LuxN kinase is potently activated by AHLs carrying extended tails, eliminating the possibility of signaling. Antagonism potency follows the order C6 < C8 < C10 < C12. Thus, molecules that are most divergent from the native V. harveyi autoinducer are the most potent antagonists. We suggest that, in its native environment, V. harveyi encounters mixtures of signals produced by other species occupying the same niche. Thus, V. harveyi needs a mechanism to gauge the relative numbers of self versus non-self in the vicinity. Possibly, V. harveyi uses relative antagonism potency to measure its relationships to others in the community: the stronger the antagonist the more distance the species-relatedness. Finally, becoming “locked” into the Kinon state by long chain AHLs, desensitizes LuxN to AI-1. It is curious why LuxN remains susceptible to antagonists since our results show that antagonism can be prevented by altering a few amino acid residues. We suggest this “vulnerability” ensures that V. harveyi does not respond to its own signal in the face of fierce competition, which prevents the leakage of benefits of QS-controlled public goods to non-kin.
Experimental Procedures
Bacterial strains and culture conditions
All V. harveyi strains were derived from V. harveyi BB120 (Bassler et al., 1997) and grown aerobically at 30°C in Luria-Marine (LM) or Autoinducer Bioassay (AB) medium (Greenberg et al., 1979). Plasmids were maintained in E. coli strain XL10-Gold (Agilent) at 37°C in LB. Genotypes of strains and plasmids are provided in the Supplemental Data (Table S5). Unless otherwise specified, chloramphenicol (Cm) was added to a final concentration of 10 μg ml−1 and Isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM.
DNA manipulation
Standard procedures were used for DNA manipulation (Sambrook et al, 1989). PCR reactions were performed with iProof High-Fidelity DNA polymerase (Bio-Rad). Restriction endonucleases, dNTPs, and T4 ligase were purchased from New England Biolabs. The IBI High-Speed Plasmid Mini kit (IBI) was used for plasmid preparations, and Zymoclean Gel DNA Recovery Kit (Zymogen) was used for PCR purifications. Oligonucleotide sequences used for PCR, site-directed mutagenesis, and sequencing reactions will be provided upon request.
V. harveyi strain and luxN mutant library construction
V. harveyi XK006 (ΔluxMN, ΔluxPQ, ΔcqsS) was constructed by deleting luxMN from strain TL25 (ΔluxM, ΔluxPQ, ΔcqsS) using a previously described method (Swem et al., 2008). Likewise, V. harveyi XK847 (ΔluxMN, ΔluxS, ΔcqsS) was derived from strain TL26 (ΔluxN, ΔluxS, ΔcqsS). The luxN gene was cloned into pFED343 and mutagenized as previously described (Swem et al., 2008). The luxN mutant library was conjugated into V. harveyi XK006 with the helper plasmid pRK2013 (Ditta et al., 1980). Approximately 50,000 exconjugants were screened for bioluminescence phenotypes in response to selected AHLs. Plasmids from mutants were isolated and backcrossed into V. harveyi XK006 to confirm phenotypes. Mutant luxN genes were sequenced and all mutations were re-engineered via Quikchange II site-directed mutagenesis (Agilent). luxN single mutant alleles were conjugated into V. harveyi XK006 and when specified, into V. harveyi XK847. Sequences encoding the FLAG tag were introduced at the 3′ end of luxN (QuikChange II) for LuxN protein detection by Western blot.
Bioluminescence assays
V. harveyi strains were grown overnight in LM medium and diluted 1:1000 in AB medium containing 0.5 mM IPTG in 96-well microtiter plates. Antibiotics were added when appropriate. DMSO or AHLs were added at specified concentrations. For dose-response measurements, AHLs were added at 100 μM, and serial 4-fold dilutions were made to final AHL concentrations of 24 pM. The cultures were allowed to grow for ~9 h, at which time, bioluminescence and optical density were measured with an Envision Multilabel plate reader (Perkin Elmer). RLU (Relative Light Units) are defined as counts min−1 ml−1/OD600.
Quantitative real-time PCR analysis
V. harveyi XK006 carrying WT or luxN mutant alleles was grown overnight in LM medium and diluted 1:1000 in AB medium containing 0.5 mM IPTG and shaken for 9 h, after which cell pellets were isolated and flash frozen with liquid nitrogen. Pellets were stored at −80°C prior to RNA isolation using the RNeasy Mini Kit (Qiagen). RNA was quantified and 2 μg of RNA was converted to cDNA with Superscript III reverse transcriptase (Invitrogen). Quantitative real-time PCR analysis was performed with primers for luxN and hfq, where hfq served as an internal control.
Western blot analyses
V. harveyi XK006 carrying WT luxN-FLAG or mutant luxN-FLAG constructs were grown overnight in LM medium and diluted 1:1000 in AB medium containing 50 μM IPTG and grown with shaking for additional 9 h. 1 ml of cells were collected and resuspended in 100 μl of Bugbuster (Novagen) with 50 μg ml−1 lysozyme (Sigma), 10 U ml−1 Benzonase Nuclease (Novagen), and diluted into 4x SDS-PAGE protein loading buffer. Samples were electrophoresed on 4–15% Mini-Protein Gels (Bio-Rad), and subsequently transferred to PVDP membranes. Membranes were blotted with anti-LuxS antibody produced in rabbit and with monoclonal anti-FLAG M2-Peroxidase (HRP) antibody produced in mouse (Sigma). Chemiluminescence was visualized by ImageQuant LAS 4000 (GE Healthcare).
AHL molecules and synthesis
Commercial sources and synthesis protocols are provided in the Supplementary Experimental Procedures.
Supplementary Material
Acknowledgments
We are indebted to members of the Bassler laboratories and to Dr. Ned Wingreen for insightful discussions and suggestions. This work was supported by the Howard Hughes Medical Institute, National Institutes of Health (NIH) Grant 5R01GM065859 and National Science Foundation (NSF) Grant MCB-0343821 to BLB. LCM is supported by the Air Force Office of Scientific Research (Grant No. FA9550-12-1-0367). XK is supported by HHMI International Student Research fellowship.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Author Contributions
X.K. designed all experiments, constructed all strains, and conducted all biological assays. L.C.M. designed and synthesized all chemical molecules. B.L.B. initiated and supervised the project.
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- Cao JG, Meighen EA. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Ditta G, Stanfield S, Corbin D, Helinski DR. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980;77:7347–51. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop BA, Herman JP, Ulrich RL, Churchill MEA, Greenberg EP. The Burkholderia mallei BmaR3-BmaI3 quorum-sensing system produces and responds to N-3-hydroxy-octanoyl homoserine lactone. J Bacteriol. 2008;190:5137–41. doi: 10.1128/JB.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JA, Lilley BN, Bassler BL. A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol Microbiol. 2000;35:139–149. doi: 10.1046/j.1365-2958.2000.01684.x. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Greenberg EP. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol. 2002;3:685–95. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–54. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, Muir TW, Novick RP. agr receptor mutants reveal distinct modes of inhibition by staphylococcal autoinducing peptides. Proc Natl Acad Sci U S A. 2009;106:1216–21. doi: 10.1073/pnas.0807760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson L, Kuo A, Dunlap PV. AinS and a new family of autoinducer synthesis proteins. J Bacteriol. 1995;177:6946–51. doi: 10.1128/jb.177.23.6946-6951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg EP, Hastings JW, Ulitzur S. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch Microbiol. 1979;120:87–91. [Google Scholar]
- Henke JM, Bassler BL. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol. 2004;186:6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- van Kessel JC, Rutherford ST, Shao Y, Utria AF, Bassler BL. Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J Bacteriol. 2013;195:436–443. doi: 10.1128/JB.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbrough JH, Stabb EV. Substrate specificity and function of the pheromone receptor AinR in Vibrio fischeri ES114. J Bacteriol. 2013;195:5223–32. doi: 10.1128/JB.00913-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Lowe EC, Baslé A, Czjzek M, Firbank SJ, Bolam DN. A scissor blade-like closing mechanism implicated in transmembrane signaling in a Bacteroides hybrid two-component system. Proc Natl Acad Sci U S A. 2012;109:7298–303. doi: 10.1073/pnas.1200479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell. 2005;18:507–518. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, et al. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Perez LJ, Wei Y, Kraml C, Semmelhack MF, Bassler BL. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol Microbiol. 2011;79:1407–1417. doi: 10.1111/j.1365-2958.2011.07548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Wei Y, Perez LJ, Cong J, Long T, Koch M, et al. Probing bacterial transmembrane histidine kinase receptor-ligand interactions with natural and synthetic molecules. Proc Natl Acad Sci U S A. 2010;107:5575–5580. doi: 10.1073/pnas.1001392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- Ottemann KM, Xiao W, Shin YK, Koshland DE. A piston model for transmembrane signaling of the aspartate receptor. Science. 1999;285:1751–4. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1995;92:1490–4. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompeani AJ, Irgon JJ, Berger MF, Bulyk ML, Wingreen NS, Bassler BL. The Vibrio harveyi master quorum-sensing regulator, LuxR, a TetR-type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol Microbiol. 2008;70:76–88. doi: 10.1111/j.1365-2958.2008.06389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- Showalter RE, Martin MO, Silverman MR. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J Bacteriol. 1990;172:2946–54. doi: 10.1128/jb.172.6.2946-2954.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM. Transmembrane signaling by asymmetry. Nat Struct Mol Biol. 2006;13:862–3. doi: 10.1038/nsmb1006-862. [DOI] [PubMed] [Google Scholar]
- Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci U S A. 1999;96:1639–44. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzman E, Silverman M, Meighen EA. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J Bacteriol. 1992;174:7490–3. doi: 10.1128/jb.174.22.7490-7493.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem LR, Swem DL, Wingreen NS, Bassler BL. Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell. 2008;134:461–473. doi: 10.1016/j.cell.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhao A, Novick RP, Muir TW. Activation and inhibition of the receptor histidine kinase AgrC occurs through opposite helical transduction motions. Mol Cell. 2014;53:929–40. doi: 10.1016/j.molcel.2014.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Ng WL, Cong J, Bassler BL. Ligand and antagonist driven regulation of the Vibrio cholerae quorum-sensing receptor CqsS. Mol Microbiol. 2012;83:1095–108. doi: 10.1111/j.1365-2958.2012.07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.