Abstract
There is now compelling evidence to indicate a place for heat shock factor 1 (HSF1) in mammary carcinogenesis, tumor progression and metastasis. Here we have investigated a role for HSF1 in regulating the expression of the stem cell renewal factor β-catenin in immortalized human mammary epithelial and carcinoma cells. We found HSF1 to be involved in regulating the translation of β–catenin, by investigating effects of gain and loss of HSF1 on this protein. Interestingly, although HSF1 is a potent transcription factor, it was not directly involved in regulating levels of β-catenin mRNA. Instead, our data suggest a complex role in translational regulation. HSF1 was shown to regulate levels of the RNA binding protein HuR that controlled β-catenin translation. An extra complexity was added to this scenario when it was shown that the long non-coding RNA molecule lincRNA-p21, known to be involved in β-catenin mRNA (CTNNB1) translational regulation, was controlled by HSF1 repression. We have shown previously that HSF1 was positively regulated through phosphorylation by mTOR kinase on a key residue, serine 326 essential for transcriptional activity. In this study we found that mTOR knockdown not only decreased HSF1-S326 phosphorylation in mammary cells, but also decreased β-catenin expression through a mechanism requiring HuR. Our data point to a complex role for HSF1 in the regulation of HuR and β-catenin expression that may be significant in mammary carcinogenesis.
INTRODUCTION
Recent studies indicate a close connection between the transcription of molecular chaperones by Heat Shock Factor 1 (HSF1) and the etiology of cancer (1-4). Molecular chaperones facilitate tumor progression by a range of mechanisms while HSF1 also plays an even more profound role in tumorigenesis(5, 6). HSF1 mediates the carcinogenic effects of a range of agents and plays a versatile role in maintenance of malignant phenotype(3, 7). In breast cancer, HSF1 becomes activated by the cytokine heregulin that activates downstream phosphatidylinositol-3-kinase upon interaction with receptors, HER2 and HER3(8). HSF1 can act as both an activator and inhibitor of gene expression, depending on the regulatory status of the target gene and both effects can contribute to its transforming and carcinogenic properties. Exposure of cells to heregulin leads to HSP synthesis in an HSF1-dependent manner and thus protects tumor cells from apoptosis (8). In addition, heregulin induces the formation of complexes between HSF1 and metastasis associated protein 1 (MTA1) (9). HSF1-MTA1 complexes lead to repression of estrogen-mediated transcription and appear to lead to a proinvasive phenotype in mammary carcinoma cells (9).
Although it is one of the earliest characterized transcription factors, the physiological regulation of HSF1 is still relatively obscure. However, understanding the mechanisms of its physiological control may be a key to discovery of how the factor becomes co-opted and deregulated in cancer. HSF1 is subject to regulation by a range of posttranslational modifications including inhibitory regulation by phosphorylation on serine 303 and positive regulation by phosphoserines 320 and 326 (10-18). The identities of the signaling cascades located upstream of these phosphorylation sites are also beginning to emerge. HSF1-S303 phosphorylation appears to be regulated by a combination of Extracellular Regulated protein Kinase (ERK) and Glycogen Synthase Kinase 3 (GSK3), while S320 is a substrate for protein kinase A (12, 19). In addition, it has recently been shown that S326 is a substrate for the kinase Mammalian Target Of Rapamycin 1 (mTORC1) and that phosphorylation by mTORC1 at this site is critical for HSF1 activation during stress (10). mTORC1 is a protein complex containing a catalytic subunit, the kinase mTOR associated with regulatory proteins such as raptor and LST8. mTORC1 plays a key role in mRNA translation by phosphorylating a number of proteins associated with protein synthesis such as p70 S6kinase (S6K) and 4E-BP (20). Both the activity of mTORC1 and the rate of translation are significantly increased in many cancers (21).
Beta-catenin, the product of the CTNNB1 gene plays major role in development and carcinogenesis both as a structural protein involved in cell adhesion and as a potent transcription factor that functions in combination with proteins such as LEF1 and TCF1 (22-24). In breast cancer β-catenin appears to control renewal of the stem cell fraction through regulation of proteins such as Cyclin D1 (25). There are at least two established modes of β-catenin activation in cancer including: (1) the Wnt pathway, in which β-catenin is regulated by control of its rate of degradation. Destruction is mediated through phosphorylation by the kinase GSK3 on an N-terminal destruction motif, a modification that permits ubiquitinylation by E3 ligase TrCP1 and ultimately targeting to the proteasome. Binding of the ligand Wnt to surface receptors Frizzled and LRP-5/6 triggers Dsh to inhibit GSK3 activity, prevents degradation and permits the influence of β-catenin in stem cell renewal (26). Alternatively, (2) many cancers express β-catenin with mutations in the N-terminal destruction motif and these mutants accumulate through their insensitivity to the degradation pathway (27).
In the present study we have shown HSF1 to regulate β–catenin levels in mammary cells. Activation of HSF1 occurred downstream of mTORC1, through phosphorylation on serine 326 and led to enhanced translation of β-catenin through mechanisms including the decrease in expression of lincRNA-p21, a long non-coding RNA molecule that negatively regulates β-catenin translation. This mechanism was shown to involve upregulation by HSF1 of the RNA binding protein HuR, a molecule previously shown to be involved in antagonizing the effects of lincRNA-p21 on β-catenin translation.
RESULTS
(1) Increased levels of activated, S326-phosphorylated HSF1 are observed in mammary cells with a stem cell surface phenotype
As HSF1 appears to play a key role in development of breast cancer, and the cancer stem cell fraction appears to be essential in tumor initiating component of the tumor cell population, we examined the activity and role of HSF1 in cells with a stem cell phenotype. We first measured HSF1 activity as indicted by phosphorylation status in MDA-MB-231 human mammary carcinoma cells sorted for expression of the stem cell markers CD44 and CD24. The CD44+/CD24− phenotype has been used to identify stem cells in normal mammary epithelial cells as well as malignant cells and in cancer, this population is highly enriched in tumor initiating cells (Suppl. Fig. 1 A)(28). We observed a marked enrichment in HSF1-pS326 in CD44+/CD24− selected cells, with no apparent increase in total HSF1 (Fig. 1A). In addition to being rich in CD44 and low in CD24 these cells excluded the Hoechst 33342 dye and contained over 97% side population, a common stem cell property (Suppl. Fig 1A). (In a previous study we showed that the phospho-antibodies used here could detect HSF1 phosphorylated by purified mTOR in vitro, with phosphorylation at the S326 site validated by Mass Spectrometry (10)). Interestingly the increase in S326 phosphorylation was accompanied by depletion in HSF1 phosphorylated on the inhibitory phosphorylation site serine 303 in CD44+/CD24-selected cells. Thus, the CD44+ / CD24− enriched cell population shows at least two phosphorylation changes associated with HSF1 activation. We next confirmed these experiments in human luminal breast cancer cell line MCF7, in immortalized mammary basal epithelial cells MCF10A as well as in ovarian carcinoma cells (Fig. 1, B, C, Suppl. Fig. 2). The mammary cells enriched in CD44 and low in CD24 (CD44+/ CD24−; Suppl. Fig. 1B) showed marked increase in levels of HSF1-S326-Pi and depletion of HSF1-S303-Pi, as detected by immunoblot with phospho-antibodies (Fig. 1 B, C). Likewise ovarian carcinoma SK-OV-3 cells enriched in CD44 and low in CD24 showed marked accumulation of HSF1-S326-Pi (Suppl Fig. 2). Consonant with increases in HSF1-S326-Pi in mammary carcinoma cells we also observed an increase in p70 S6 kinase (S6K) phosphorylated on T389 in the CD44+/CD24− cells. P70 S6K is a primary substrate of mTORC1 and these data thus suggested constitutively activated mTORC1 in this cell population. In none of the cell lines did we observe significant increases in total HSF1 levels in the CD44+/CD24− subpopulation, suggesting enrichment for activated HSF1 but not total HSF1 in cells with this surface phenotype. HSF1 levels have been shown to be increased in the total cell populations of many clinical mammary cancers (compared to their normal counterparts) and may thus be correspondingly high in both the CSC and bulk population cells (29).
Figure 1. Accumulation of HSF1 phosphorylated S326 in the CD44+/CD24− cell population of mammary carcinoma cell lines.
(A) Expression levels of HSF1 phospho-S326, HSF1 phospho-S303, total HSF1 and β-catenin was determined by Western blotting in CD44+/CD24− and in the depleted CD44−/CD24+ (Non-CSC) populations of MDA–MB-231 cells. Levels of β-actin expression were also measured as loading controls. Relative levels of phospho-S326 HSF1 and β-catenin, determined by densitometry are indicated. (B) Protein expression levels of HSF1 phospho-S303, HSF1 phospho-S326, total HSF1, β-catenin, S6K phospho-T389, total S6K was determined by Western blotting in MCF7 cells. GAPDH expression level was also measured as loading control. Relative levels of phospho-S326, phospho-S303, β-catenin and phospho-S6K was determined by densitometry as shown. (C) Protein expression levels (as in B) were measured in MCF10A cells. Relative levels of phospho-S326, phospho-S303, β-catenin and phosphor-S6K in the immunoblots were determined by densitometric analyses using Image J software, as shown. Experiments were performed twice with reproducible results.
We also noted that the stem cell self-renewal factor β-catenin was enriched in the CD44+/CD24− fractions of both immortalized and malignant mammary cells that were also abundant in activated HSF1 (Fig. 1 A, B, C). As increases in activated HSF1 accompanied elevated β-catenin, we next examined a potentially causal role for HSF1 in β-catenin expression. (We also aimed to examine the HSF1-β-catenin axis in Her2+/CD44− luminal mammary adenocarcinoma cell line SKBR3. However β-catenin was undetectable by immunoblot in these cells with or without HSF1 knockdown and studies were not continued (Suppl. Fig. 6A).
(2) HSF1 and β-catenin regulation
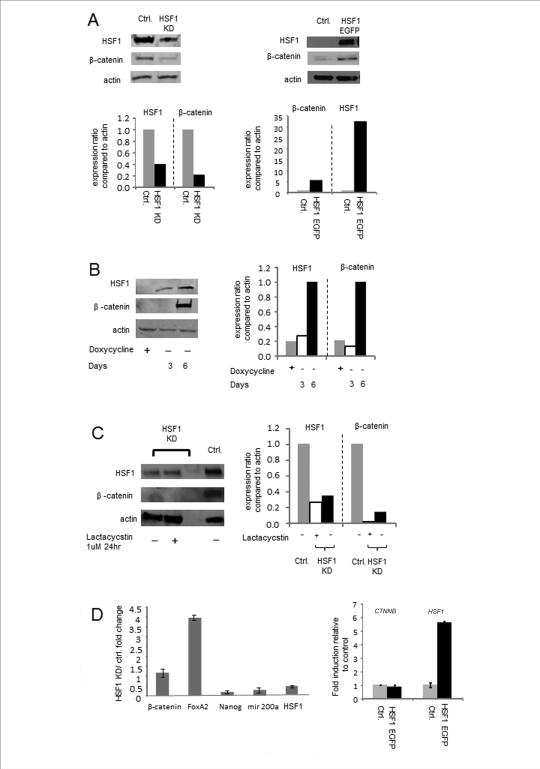
We next examined the consequences for β-catenin expression of reducing intracellular HSF1 levels by RNA interference (Fig. 2). To simplify experiment, these studies were carried out in unsorted cell populations. For most such experiments we used MDA-MB-231 cells as they consist of mostly the CD44+/CD24− sub-population and were able to sustain prolonged (6-20 d) HSF1 knockdown without entering apoptosis, compared to other cancer cells in which only brief (3-4 d) knockdown was feasible before onset of apoptosis (data not shown). MDA-MB-231 cells were stably infected with doxycyclin-inducible lentiviral vector constructs expressing either shRNA targeting HSF1 or control hairpins. As can be seen in Fig. 2 A, 3 days of induction of the HSF1 targeting shRNA with doxycyclin led to partial depletion in HSF1 protein levels, compared with the control hairpins and this reduction in HSF1 was coincident with a decline in β-catenin levels, compared to cells infected with the control siRNA. In addition, HSF1-EGFP overexpression after transfection with an expression vector encoding this protein in the MDA-MB-231 cell line led to increased levels of β-catenin, further suggesting a causal link between HSF1 and β-catenin expression (Fig. 2A, right panel).
Figure 2. β-catenin expression at the protein level correlates with relative levels of HSF1.
(A) Expression levels of HSF1 and β-catenin in MDA-MB-231 cells with either HSF1 knockdown or overexpression were measured by Western blotting. For HSF1 knockdown, cells were treated for three days with doxycycline (2μg/ml) for induction of shRNA expression and knockdown of HSF1. In controls, an irrelevant hairpin was also induced by doxycline. For HSF1 overexpression we used an HSF1-EGFP construct containing the full length HSF1 sequence. Relative levels of HSF1 and β-catenin, determined by densitometry are indicated beneath the immunoblot images. (B) HSF1 knockdown and restoration. Doxycycline treatment was continued for 6 days in cells expressing the inducible shRNA vector (first lane) and then removed from the medium to permit restoration of HSF1 expression. Levels of HSF1, β-catenin and actin expression were measured by Western blotting at day 3 and 6 post removal of doxycycline. (C) Loss of β-catenin expression in HSF1 knockdown cells is not mediated by proteasome degradation. HSF1 knockdown MDA-MB-231 cells were treated with lactacystin for 24hr. β-catenin and HSF1 expression levels were measured by Western blotting. (D) Real time quantitative RT-PCR analysis was performed for analysis of the expression levels of β-catenin, FoxA2, Nanog, miR 200a and HSF1 mRNA in HSF1 knockdown (left panel) and HSF1 overexpressing cells (right panel). Fold change was calculated by normalization to β-actin mRNA levels, followed by comparison with the control untreated sample. Mean values +/− SD are indicated. Each experiment (A-D) was performed at least twice with reproducible findings.
Longer-term reduction in HSF1 levels by 6-day expression of the shRNA construct led to a pronounced decline in HSF1 levels and to major depletion in β-catenin (Fig. 2B, lane 1). Subsequent incubation of cells in the absence of the shRNA-inducing doxycyclin led to recovery in HSF1 levels and renewed intracellular β-catenin expression (Fig. 2B, lanes 2, 3). These data strongly indicated a requirement for HSF1 in the maintenance of β -catenin concentrations in the MBA-MD-231 cells. β-catenin has been shown to be regulated in the canonical Wnt pathway through inhibition of GSK3 and subsequent protection from proteasomal degradation of the dephosphorylated β-catenin (30, 31). We therefore examined the hypothesis that HSF1 may associate with β-catenin and protect it from proteasomal degradation. However, we did not find significant levels of β-catenin in HSF1 immunoprecipitates, suggesting minimal association of the two proteins (data not shown). In addition, treatment of MBA-MD-231 cells with the proteasomal inhibitor lactacystin failed to reverse the effects of HSF1 knockdown and did not lead to significant increases in β-catenin that might have been predicted, if HSF1 acted to reduce proteasomal digestion (Fig. 2C). As HSF1 is a powerful transcription factor controlling the trans-activation of multiple genes, we next examined a potential role for the factor in mediating transcriptional activation of the β-catenin gene (CTNNB1) and stimulation of β-catenin mRNA synthesis. However, although we observed significant changes in a number of factors with activity in mammary cells, including depletion in NANOG mRNA and increases in mRNA encoding forkhead A2 (FOXA2), β-catenin mRNA levels were not significantly altered by HSF1 knockdown (Fig 2D). In repeated experiments, we failed to see a decrease in β-catenin mRNA levels in cells depleted of HSF1 (Fig. 2D, left panel). In addition, when we carried out overexpression of HSF1 we also failed to observe increases in β-catenin/CTNNB1 mRNA levels even though HSF1 mRNA levels were increased markedly (Fig. 2D, right panel). These experiments therefore suggested that regulation of β-catenin levels by HSF1 does not involve major roles for protein stabilization, or direct trans activation of the CTNNB1 gene; regulation at the level of mRNA translation seemed more likely.
(3) Role of HSF1 in translational regulation of β-catenin: potential contributions of microRNA and mRNA binding proteins
The preceding experiments, by process of elimination, suggested a role for HSF1 in maintenance of β-catenin translation. As the translation into proteins of many cancer mRNAs is regulated by their destabilization through small non-coding microRNAs (miRNA) that bind to their 3'-untranslated regions (UTR), we investigated the hypothesis that HSF1 regulates the levels of miRNA targeting CTNNB1 mRNA. Using RT-qPCR we examined expression of specific species of miRNA implicated in β -catenin regulation. These included miR-9a, 200a, 483-3a, 483-5p(32, 33). Among these miRNAs, only miR-200a was significantly reduced by HSF1 KD (Fig. 2D). As miR-200a was reported to be an inhibitor of β-catenin expression, a role for this molecule in HSF1-dependent β-catenin translation seemed unlikely (34, 35). In addition we probed a PCR-based array for an unbiased screen of miRNAs affected by HSF1 knockdown. Twenty seven of the miRNA species in the library were significantly reduced by HSF1 knockdown, indicating a versatile role for this factor in miRNA expression (Suppl. Table 1). In addition, at least 11 miRNA species were increased by HSF1 depletion suggesting a role for HSF1 in repressing expression of some miRNA. MiR-122-5b, miR-1246 and miR-4732-5p were each increased more than three-fold by HSF1 knockdown (Suppl. Table 1). However none of these species were predicted to interact with the 3’ untranslated sequence of the CTNNB1 mRNA as examined by TargetScan analysis.
As there was no compelling miRNA candidate for HSF1 mediation of β-catenin expression, we therefore next examined an alternative mechanism involving the turnover and translation regulatory RNA-binding protein (TTR-RBP) HuR / elavL1 (36-38). HuR can bind to adenine and uracil-rich elements (AU-rich elements, ARE) in the 3’ -UTR or introns of many mRNAs or long non-coding RNA associated with tumor growth and block association of other TTR-RBPs or miRNA, leading to stabilization of the mRNA and/or enhancement of translation (36, 39). In fact previous studies had suggested an interaction between HuR and the CTNNB1 mRNA (40, 41). Our sequence analysis of CTNNB1 indicated a remarkable 38 AU-rich sequences in the 3’UTR, many of which elements are highly conserved, suggesting the potential of interaction between the CTNNB1 3’UTR and HuR (Suppl. Fig. 3; Table 1). We therefore examined the effects of modulating HSF1 on the levels of HuR and β-catenin in MDA-MB-231 cells. Indeed depletion of HSF1 in these cells by knockdown with shRNA led to a profound decrease in the levels of HuR and a concomitant reduction in intracellular β-catenin levels (Fig. 3A). In turn, depletion of HuR by shRNA led to a reduction in β-catenin levels, suggesting by inference the existence of a concerted HSF1>HuR> β-catenin axis (Fig. 3A). In addition, knock down of HuR led to significant loss of HSF1 from the cells possibly indicating another role for HuR in maintaining HSF1 levels in mammary cancer cells through effects on mRNA stability or translation (Fig. 3A). Highly conserved potential ARE sequences, were predicted in the 3'-UTR of HSF1 mRNA by sequence analysis (Suppl. Fig. 4). Similar findings regarding HSF1, HuR and β-catenin were observed in MCF7 and MCF10A cells as HSF1 knockdown by siRNA led to coordinate decreases in expression of β-catenin and HuR, compared to cells exposed to control sequence (Fig. 3B).
Table 1.
AU-rich elements (ARE) in 3’ UTR of HSF1 and CTNNB1 mRNA.
| Gene | Length of 3’-UTR in human | Number of AU-rich elements |
|---|---|---|
| HSF1 | 392 nt | 3 |
| CTNNB1 | 1106 nt (801 nt, 642 nt)* | 37 |
Two shorter variants of 3’-UTR were predicted in CTNNB1 3’-UTR. Detail was shown in supplemental information.
Figure 3. β-catenin expression requires mTOR, HSF1 and HuR expression.
(A) MDAMB-231 HSF1, mTOR and HuR knockdown cells were generated with shRNA expressing lentivirus specific for each targeted mRNA. Expression levels of HuR, β-catenin, HSF1 phospho-S326, total HSF1, phospho S6K p70, total S6K p70 were measured by Western blotting and quantitated by densitometry (right side). Levels of actin were also measured as loading controls. (B) Protein levels of HSF1, β-catenin and HuR were measured in control and HSF1 knocked down MCF7 and MCF10A cells. β-actin was also measured using Western Blotting. Bands in gels were measured by densitometric analyses using Image J software and shown in right side. The experiment was carried out in duplicate with reproducible results.
During these experiments we also noted that HSF1 knockdown reduced the levels of phospho-T389 p70 S6 kinase and its mRNA RPS6KB1 in MCF7 cells (Suppl. Fig. 5 A). These findings therefore suggested that HSF1 is required for the activity of mTORC1, the principle upstream kinase for phospho-T389 p70 S6K phosphorylation. Findings for MCF10A cells were inconsistent reflecting the relatively low expression levels of S6K in these cells (Suppl. Fig 5B). However these findings were confirmed in another mammary cancer cell line SKBR3, indicating regulation of p70 S6K phosphorylation by HSF1 (Suppl. Fig. 5A). Such mechanisms might include direct regulation of mTOR mRNA levels by HSF1 (Suppl. Fig. 5A). These findings are consistent with previous studies using hsf1 knockout cells that showed HSF1 to be required for p70 S6K phosphorylation (7).
(4) Role of mTOR upstream of HSF1, HuR and β-catenin
mTOR was also shown to be an essential activator of HSF1 during the stress response and an important upstream kinase that catalyzes HSF1 serine-326 phosphorylation under stress conditions (10). We therefore examined whether mTOR depletion with shRNA could result in a decrease in β-catenin levels as with HSF1 knockdown (Fig. 3A, middle lanes). Indeed mTOR knockdown resulted in a decline in β-catenin levels as well as decreases in HSF1-S326 phosphorylation and HuR concentrations, consistent with a signaling pathway involving a concerted mTOR>HSF1-S326>HuR>β-catenin cascade (Fig. 3A). As a control for the effectiveness in mTOR inactivation, we examined the phosphorylation levels of p70 S6 kinase (Fig. 3A). Levels of phospho-T389 p70 S6 kinase were markedly reduced by mTOR knockdown (Fig. 3A).
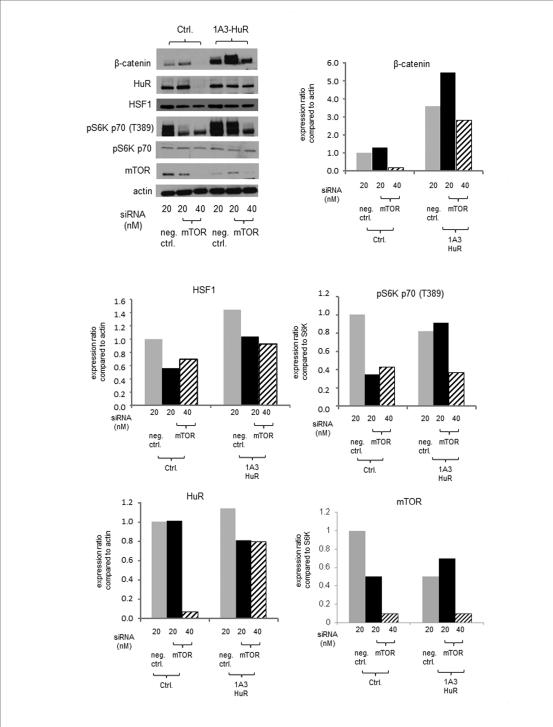
(5) HuR overexpression prevents loss of β-catenin in HSF1 knockdown conditions
We next investigated a potentially causal role for HuR in β-catenin accumulation and assessed the ability of HuR expression from a viral vector to prevent loss of β-catenin in cells exposed to siRNA targeting HSF1. As before, HSF1 knockdown led to loss of β-catenin and HuR (Fig. 4). However, HuR overexpression (IA3-HuR) led to increased β-catenin accumulation and overrode the ability of siRNA targeting HSF1 to reduce the β-catenin levels. As predicted from Fig. 3 and the AREs found in the 3'-UTR of HSF1 (Suppl. Fig. 4), HuR overexpression also led to enhanced HSF1 expression indicating that in addition to β-catenin, HuR targets HSF1 and leads to its increased expression. This suggests the possibility of a positive feedback loop, with HSF1 increasing HuR and elevated HuR in turn enhancing HSF1 levels.
Figure 4. Constitutive expression of HuR overcomes the effects of HSF1 knockdown on β-catenin depletion.
siRNA was used to knock down HSF1 expression in MDA-MB-231 cells (control and 1A3-HuR overexpressing cells). Expression levels of HuR, β-catenin, total HSF1 were measured by Western blotting. Bands in gels were measured by densitometric analyses using Image J software and shown below. Levels of actin expression were also measured as loading controls. The experiment (A-D) was performed twice with reproducible results.
(6) HuR overexpression prevents loss of β-catenin in mTOR knockdown conditions
Another key question was whether HuR expression could compensate for mTOR depletion and spare β-catenin loss in MDA-MB-231 cells. mTOR knockdown led to reduced HuR and β-catenin levels in the cells in an shRNA dose dependent manner (Fig. 5, second and third lanes). The effectiveness of mTOR knockdown is indicated by the western blot analysis of the mTOR catalytic domain as well as mTORC1 substrate phospho-T389 p70 S6K , in which we observed siRNA dose-dependent loss, while total S6K levels were not altered. HuR overexpression, as also shown in Fig 4, increased β-catenin levels and prevented loss due to mTOR knockdown. HSF1 levels were increased by HuR overexpression, and reduced by mTOR knockdown. As mentioned earlier, HSF1 knockdown also reduced the levels of phospho-T389 p70 S6K in mammary cells coordinately with a reduction in HuR (Suppl.Fig. 5, Fig. 3). These data point to a role for the HSF1>HuR axis in mTORC1 activity, p70 S6K phosphorylation and, potentially, translational flux rates in the cell.
Figure 5. Constitutive expression of HuR overcomes the effects of mTOR knockdown on β-catenin depletion.
mTOR concentration was reduced by expression of siRNA in MDA-MB-231 cells (control and 1A3-HuR overexpressing cells). Expression levels of HuR, β-catenin, total HSF1, phospho S6K p70, total S6K p70 and mTOR were measured by Western blotting and quantitated by densitometry (shown below).Actin levels were also measured for loading control. These procedures were performed twice with reproducible results.
Role of HSF1 and lincRNA-p21 in β-catenin translation
It has been reported that HuR can both stabilizes RNA species and enhance mRNA translation and recent studies have shown that the translation of β-catenin could be inhibited by the intergenic long non-coding RNA lincRNA-p21. This process was reversed by increasing levels of HuR and let-7 miRNA, by which lincRNA-p21 was degraded(42). We therefore hypothesized that HSF1 might down-regulate lincRNA-p21 and lead to efficient β-catenin translation. Then we examined the effect of HSF1 depletion on levels of lincRNA-p21 in MDA-MB-231 cells by qRT-PCR. HSF1 knockdown in MDA-MB-231 cells led to significant loss of HSF1 mRNA that was reflected in HuR gene expression but not β-catenin mRNA as observed previously (Fig. 6A). Under these conditions of HSF1 depletion, levels of lincRNA-p21 were increased. We then overexpressed HuR in cells using a viral vector to test if HuR reduced the elevated lincRNA-p21 levels. Indeed, under these conditions of HuR overexpression, HSF1 depletion did not lead to increases in lincRNA-p21 levels. Our experiments are therefore consistent with an HSF1- and HuR-dependent decrease in lincRNA-p21 playing a role in the increase in β-catenin translation observed in the MDA-MB-231 cells.
Figure 6. HSF1 depletion leads to enhanced lincRNA-p21 expression.
(A) Real time quantitative RT-PCR analysis was performed for analysis of the RNA expression levels of lincRNA-p21, CTNNB (β-catenin), HuR and HSF1 mRNA in HuR overexpressing cells with or without HSF1 knockdown. Fold change was calculated by normalization to β-actin levels, followed by comparison with the control untreated sample. (B, C) Real-time quantitative RT-PCR was performed for analysis of the expression levels of HSF1, CTNNB (β-catenin), HuR, and lincRNA-p21 in MCF7 (B) and MCF-10A (C) with or without HSF1 kd. β-actin and 18s rRNA were used as loading controls. Error bars represent the SD of duplicate qPCR. The experiments were performed twice with reproducible results.
Similar findings were observed in MCF7 and MCF10A cells, with HSF1 knockdown increasing levels of lincRNA-p21 (Fig. 6B and C). However overall levels of lincRNA-p21 were considerably reduced in these two cell lines cells compared to MDA-MB-231 cells. One further difference compared to MDA-MB-231 was that HSF1 knockdown did not markedly alter HuR mRNA levels in MCF7 and MCF10A cells (Fig. 6). The weight of evidence thus suggested that effects of HSF1 on HuR were more likely mediated at the posttranscriptional level in most cells
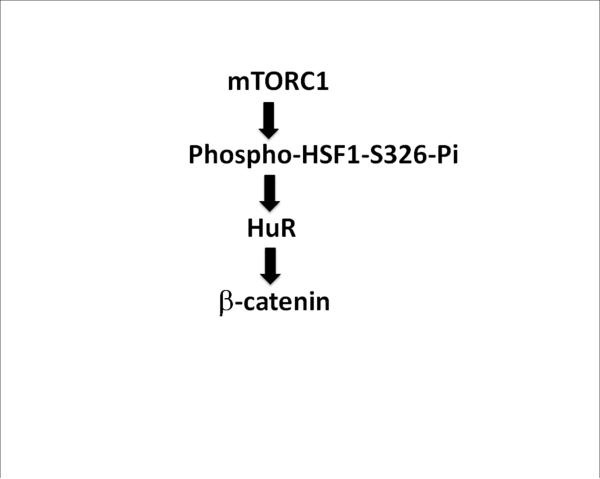
Degradation of lincRNA-p21 was shown to be promoted by the combination of multiple essential factors including HuR and miRNA (42). A potentially complementary mechanism was suggested to us by inspection of the 5’ sequence of lincRNA-p21 for possible miRNA target sites that could be involved regulation by HSF1 (Suppl. Fig. 6). We observed potential interaction sites for miRNA-320 a, b and c, each of which appear to be dependent on HSF1 for expression (Suppl. Table 1, Suppl. Fig. 7). HSF1 regulated HuR and miR-320 or let-7 could thus contribute to control of lincRNA-p21 expression and β-catenin translation. We have summarized some of the basic components of the pathway involved in β-catenin regulation by HSF1 in Fig. 7.
Figure 7. Mechanisms of HSF1-mediated translational control of β-catenin.
HSF1 was activated through S326 phosphorylation by mTOR. Activated HSF1 then led to increased HuR expression, which in turn promoted the translation of CTNNB1 mRNA into β-catenin. A concerted mTOR> HSF1> HuR> pathway appeared to be upstream of β-catenin expression in normal and malignant mammary derived cells.
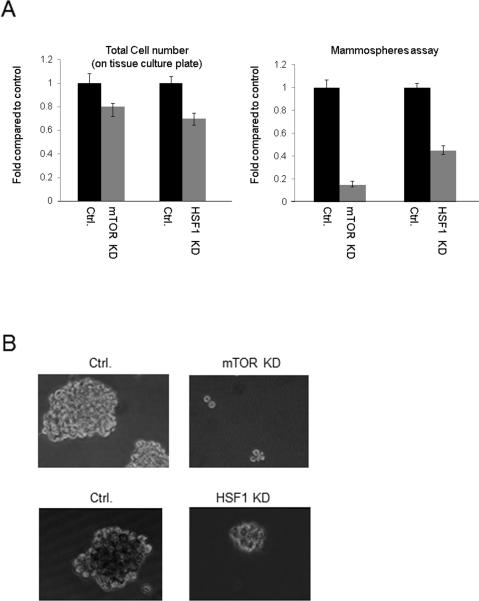
(5) HSF1 knockdown and cell growth in monolayer and mammosphere
In the final experiments, we investigated the role of HSF1 in cell proliferation. MDA-MB-231 cell growth rate under monolayer tissue culture conditions, on a solid matrix was slightly decreased by HSF1 knockdown (Fig 8A). Then, as HSF1 appeared to be activated in cells with a stem cell surface phenotype (Fig. 1) and to positively regulate translation of β-catenin (Fig. 2), a factor involved in stem cell renewal, we next examined the overall effects of HSF1 depletion on growth in mammosphere culture, a property of stem cells. Under these conditions, we observed a marked decrease in growth rate when HSF1 levels were reduced (Fig 8 A, B). HSF1 depletion led to a decrease in both size and number of mammospheres. In addition we investigated the role of mTOR in MDA-MB-231 cell growth in mammosphere, as our studies indicated that mTOR is required for HSF1 activation and accumulation of HuR and β-catenin. Knockdown of mTOR led to an even more pronounced loss of mammosphere formation, which may be related to its upstream role in regulating HSF1, HuR and β-catenin (Fig. 8 A, B).
Figure 8. Depletion of mTOR or HSF1 reduces mammosphere formation in vitro.
Cells were plated at low density in suspension culture (5000/well) on regular or low attachment tissue culture plates.
(A) Number of cells grown on regular tissue culture plates (left panels) and spheroids formed in low attachment plates (right panels) were calculated at day 12 after plating. mTOR control hairpin or shRNA, infected cells were plated at day zero. For HSF1 knockdown, doxycycline (2 ug / ml) was added to cultures every 3 days for continuous induction of the inducible HSF1 control hairpin or HSF1 shRNA expression. Experiments were repeated three times, reproducibly and mean values +/− standard deviation are indicated.
(B) Low magnification image of mammospheres formed by cells expressing control hairpins for mTOR experiment (Ctrl., upper panel), shRNA targeting mTOR (mTOR kd), tripZ ctrl for HSF1 experiment (Ctrl. lower panel) and the tripZ HSF1 knockdown MDAMB-231 cells (HSF1 kd)
DISCUSSION
Our experiments have begun to outline a regulatory pathway that could lead to the increased expression of β-catenin at the level of translation (Fig. 7). The steps that we have deciphered so far include the upstream activation of HSF1 through phosphorylation by mTOR that led to increased HSF1 activity, elevated expression of HuR and an enhanced translation of β-catenin even though CTNNB1 mRNA levels were not markedly altered. HuR could be activated by HSF1 at the transcriptional level in MDA-MB-231 cells and HSF1 knockdown reduced levels of HuR mRNA in this cell line (Fig. 6). Roles for HuR in cell regulation by HSF1 have been shown previously(43, 44). Previous ChIP-sequence experiments carried out in mammary cancer cells indicated occupancy of the HuR gene on chromatin by HSF1(45). However, the reduction in HuR mRNA was only partial in HSF1-depeleted MDA-MB-231 cells and was not observed in MCF7 and MCF10A cells after HSF1 knockdown (Fig. 6). A predominantly posttranscriptional mechanism for HuR regulation by HSF1 was thus suggested by these data. Indeed, HuR has been shown to be positively autoregulatory through binding to AU sequences in its own 3’UTR (36). It has previously been shown that the CTNNB1 mRNA is under negative regulation by lincRNA-p21, a molecule that can hybridize with it and reduce its translation(42). HuR was shown to interact with lincRNAp21 and destabilized it by recruiting a let-7/Ago2 complex, leading to enhanced degradation of the RNA. Our studies indicated that lincRNA-p21 was under control by HSF1 and that this regulation appeared to be mediated by HSF1 dependent increases in HuR (Fig. 6). Overall our data suggested that activated HSF1 could be a potent inducer of gene expression at the posttranscriptional level in mammary cancer. Our experiments suggested that HSF1 might be able to influence translation of a range of proteins through its effects on HuR, lincRNA p21 and microRNAs (Fig. 3, Fig. 6, Suppl. Table 1). Recent studies have emphasized a role for translational flux in regulating HSF1 in cancer, indicating a close interrelationship between the rate of mRNA translation and HSF1(46). In addition the mutual positive interactions between HSF1 and mTORC1 observed in this and previous studies also pointed to a similar conclusion (Suppl. Fig. 6).(10) Stimulation of HuR expression by HSF1 could also suggest a role for the factor in stabilizing mRNA species important in cancer, as well as its effects on translation shown here.
Our experiments also suggested that this pathway could potentially be involved with stem cell regulation. Activated HSF1 and increased β-catenin levels were observed in cells with a stem cell surface phenotype (Fig. 1). In addition, targeting both mTOR and HSF1 with RNA interference approaches reduced ability of cells to form mammospheres, a known property of mammary stem cells (Fig. 7, 8). Beta-catenin is a key factor in stem cell renewal and may thus mediate some of the effects of HSF1 on growth and metastasis. Our recent studies in mouse mammary carcinoma have indicated a profound role for cells with a stem cell phenotype in lung metastasis (47). Our future studies will address a role for HSF1 in breast cancer metastasis in mice using doxycycline-inducible HSF1 knockdown by shRNA as in Fig. 2.
Both HSF1 and mTOR are highly versatile proteins and are likely to influence other aspects of tumor cell metabolism beyond the regulation of β-catenin. HSF1 was shown to perform both transcriptional trans-activating and repressing functions and likely plays a pleiotropic role in cancer. For instance we have shown here the expression of stem cell pluripotency factor nanog to be dependent on HSF1 and previous studies showed that HSF1 can interact with the MTA1-NuRD co-repressor complex known to be involved in stem cell maintenance(9). In addition, HSF1 appeared to regulate a wide range of miRNA targets, many of whose functions are not currently documented, but which might be determinants of cell fate (Suppl. Table 1).
This study and those of others point to HuR as a significant downstream component of HSF1 regulated gene expression. In addition to CTNNB1 / β-catenin as shown here, additional targets for HSF1 and HuR included HIF1 and Cyclin D, also important proteins in mammary cancer (43, 44). As HuR is a potent factor in regulating a broad spectrum of genes at the levels of mRNA stability and translation, this mechanism may play a significant part in the influence of HSF1 in cancer progression and other HSF1>HuR regulated candidates are to be expected in future studies.
MATERIALS and METHODS
Cells, Culture Condition and Reagents
Breast cancer cell lines MCF7, MCF10A, SKBR-3 and MDA-MB-231 and the ovarian cancer cell line SK-OV-3 were purchased from The American Type Culture Collection. MDA-MB-231 cells were maintained in DMEM + high glucose (Invitrogen) and SK-OV-3 and SKBR3 cells in McCoy's 5A (Invitrogen), supplemented with 10 % heat inactivated fetal bovine serum (Invitrogen) and 1000 U of penicillin/streptomycin (Invitrogen) in a 37°C and 5% CO2 humidified incubator. MCF7 cells were maintained in EMEM media supplemented with 10% heat inactivated FBS, Penicillin/streptomycin and 1% insulin. MCF10A cells were maintained in the base medium MEBM along with the additives which was obtained from Lonzae / Clonetics Corporation as a kit: MEGM, Kit Catalog No. CC-3150. To make the complete growth medium, 100 ng/ml cholera toxin was added to the medium. MBA-MD-231 cells were used in most of the experiments as they proved highly resistant to loss of viability seen in some cell types (such as MCF-7, SK-OV3A and SKBR3) on long-term depletion of HSF1 (data not shown). For inhibition of proteasome degradation, cells were treated with 1μM Lactacycstin (Sigma-Aldrich) for 24 h.
CD44+ /CD24− cancer stem cell population cell sorting
MCF7, MCF10A, SK-OV3 and MDA-MB-231 cells were stained with FITC/PE mouse anti-CD44 and PE/FITC mouse anti-CD24 antibody (BD Pharmigen, STEMCELL Technologies) according to standard method for FACScan. Ten million (107) cells from each cell samples were subjected to cell sorting (BIDMC flow cytometry core facility) for the CD44+ /CD24− population. The remaining cell fraction from MDA-MB-231, MCF7, MCF10A and SK-OV3 cells was used as non-CSC. The two sorted cell fractions (CD44+/CD24− and non CD44+/CD24−) from each cell line were collected. The protein concentration in the sorted cell samples was measured and prepared for SDS-PAGE.
Lentiviral shRNA and protein overexpression
To generate the lentiviruses, envelope plasmid, packaging plasmid (Open Biosystems Inc.) and shRNA expressing plasmid were co-transfected into HEK293FT cells (Invitrogen). Lentiviral short hairpin RNA (shRNA) expression vector for mTOR and the control pLKO.1 plasmid were purchased from Addgene. Lentiviral tet-on-driven short hairpin RNA (shRNA) expression vector for HSF1 and the control tripZ plasmid were purchased from Open Biosystems Inc. Lentiviral shRNA expression and control vectors for HuR and the HuR overexpression plasmid 1A3-HuR were kind gifts from Dr. Michael Sherman (Boston University) 13. Virus-containing medium was collected 48 and 72 hr after transfection. MDA-MB-231 cells were infected by incubation with the lentivirus-containing medium and thereafter the cells were treated with puromycin for selection of knockdown cells. Cells overexpressing HuR were positively selected with G418 (Neomycin, 500ug/ml). For induction of shRNA targeting HSF1, MDA-MB-231 cells were treated with 2μg/ml doxycycline. The HSF1-GFP fusion expression plasmid construct used in the study was prepared in pEGFP-N3 vector.
Transient transfection of siRNA
The siRNA or control dsRNA were transfected into cells using lipofectamine RNAi MAX (Invitrogen). Medium was changed 24 hr post-transfection. Total RNA and cell lysate were prepared 72 hr post-tranfection. The siRNA for HSF1 (targeting sequence 5’-CAGGTTGTTCATAGTCAGAAT -3’) and mTOR (targeting sequence 5’-ACTCGCTGATCCAAATGACAA -3') were purchased from Qiagen for transient knockdown. The negative control siRNA (Scr) was purchased from Qiagen (target sequence, AATTCTCCGAACGTGTCACGT).
Western Blot Analysis
Cells were lysed with RIPA buffer containing protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Roche). Protein concentration was quantified with BCA protein assay kit (Pierce) and samples were subjected to SDS-PAGE followed by standard Western blot procedure. Alternatively, sorted cells were directly lysed in SDS reducing sample buffer containing Triton-X100 and βME (beta (2)- Mercaptoethanol) to prepare whole cell lysates. The following antibodies were used for analysis of protein expression: anti-HSF1 (Enzo Life Sciences), p70 S6 Kinase (Cell Signaling), HSF1 phospho-Serine 303 (abcam), HSF1 phospho-Serine 326 (abcam), phospho-S6K Threonine 389 (cell signaling), β-catenin (abcam), β-actin (Sigma-Aldrich), HuR (Abcam), mTOR (Abcam), GAPDH (Santa Cruz). Secondary antibodies used were HRP-goat anti-rat IgG, HRP-goat anti-mouse IgG, HRP-goat anti-rabbit IgG (Santa Cruz).
Densitometric analysis of Western blot bands
Densitometric analysis of the blots was done using Image J software (NIH). Blot images were imported into the Image J software and the contrast is adjusted so that the bands were clearly visible on the blot image. Area around each band was selected and the background intensity was subtracted from the blot image. Values were shown in arbitrary unit and the maximum values were normalized to 1.
RNA isolation, cDNA synthesis and quantitative PCR
Total RNA was isolated using the RNeasy mini Kit (Qiagen) and reverse transcription (RT) was carried out using a High capacity cDNA synthesis Kit (Applied Biosystems) following instructions provided by the manufacturers. For analysis of HSF1, HUR/ERAVL1, NANOG, CTNNB1, ACTB mRNA and miR-200a expression, TaqMan assay was performed using TaqMan Universal Master Mix (Life technologies), TaqMan Gene Expression Assays (Life Technologies), primer pairs and probes specific for the amplification of target genes. Alternatively, HSF1, CTNNB1, HUR/ERAVL1, MTOR, RPS6KB1 (encoding S6K) mRNA and 18s rRNA and lincRNAp21 levels were quantified using specific primer pairs and SYBR green in the realtime PCR system. All reaction was performed on ABI 7300 Realtime PCR System. Thermocycling condition includes a 15 min at 95°C followed by 40 cycles of 15 sec denaturation at 95°C and 1 min of annealing and extension at 60. Sample for each treatment group were performed in triplicates and experiments repeated three times. Data analysis was done by using the comparative Ct method with β-actin as normalization control. Alternatively, relative expression levels were quantified as ratio to 18s rRNA levels.
PCR array and microRNA array
RT2 Profiler PCR Array (Qiagen) for human stem cells was performed using cDNA from tripZ control and tripZ HSF1 knockdown MDA-MB-231 cells for gene expression profiling. For analysis of miRNA expression, mRNA collected from the tripZ ctrl and tripZ HSF1-knockdown MDA-MB-231 cells were sent to Phalanx Bio Inc. for microRNA OneArray microarray profiling.
Mammosphere formation in vitro
To generate mammospheres, cells were plated at low density (5,000 cells/well) in 6-well ultra-low attachment plates. Cells were grown in serum-free DMEM/F12 medium supplemented with 20ng/ml bFGF, 20ng/ml EGF and B27 (Life Technologies). Doxycycline (2ug/ml) was added for induction of shRNA targeting HSF1. Suspension cultures were incubated for 12 days and numbers of spheroid formed were counted at three individual fields.
In silico analysis of putative HuR-binding sequences
AU-rich elements in 3' UTR of CTNNB1 and HSF1 mRNA were searched utilizing the TargetScan program. Successive 5 or more A or U nucleotide sequences were counted as AU-rich elements.
Supplementary Material
Acknowledgements
We thank the Department of Radiation Oncology, Beth Israel Deaconess Medical Center for support and encouragement. We are very grateful for help with construct preparation from Dr Tom Prince.
# This work was supported by NIH research grants RO-1CA047407, R01CA119045 and RO-1CA094397
Footnotes
& Conflict of interest.
The authors declare no conflict of interest.
References
- 1.Ciocca DR, Arrigo AP, Calderwood SK. Heat shock proteins and heat shock factor 1 in carcinogenesis and tumor development: an update. Arch Toxicol. 2012 Aug 11; doi: 10.1007/s00204-012-0918-z. PubMed PMID: 22885793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calderwood SK, Gong J. Molecular chaperones in mammary cancer growth and breast tumor therapy. J Cell Biochem. 2011 Apr;113(4):1096–103. doi: 10.1002/jcb.23461. PubMed PMID: 22105880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Min JN, Huang L, Zimonjic DB, Moskophidis D, Mivechi NF. Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene. 2007 Aug 2;26(35):5086–97. doi: 10.1038/sj.onc.1210317. PubMed PMID: 17310987. [DOI] [PubMed] [Google Scholar]
- 4.Jin X, Moskophidis D, Mivechi NF. Heat shock transcription factor 1 is a key determinant of HCC development by regulating hepatic steatosis and metabolic syndrome. Cell Metab. 2011 Jul 6;14(1):91–103. doi: 10.1016/j.cmet.2011.03.025. PubMed PMID: 21723507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderwood SK. HSF1, a versatile factor in tumorogenesis. Curr Mol Med. 2012 Jul 17; doi: 10.2174/156652412803306675. PubMed PMID: 22804234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell stress & chaperones. 2005;10(2):86–103. doi: 10.1379/CSC-99r.1. Summer PubMed PMID: 16038406. Pubmed Central PMCID: 1176476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenisis. Cell. 2007;130:1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khaleque MA, Bharti A, Sawyer D, Gong J, Benjamin IJ, Stevenson MA, et al. Induction of heat shock proteins by heregulin beta1 leads to protection from apoptosis and anchorage-independent growth. Oncogene. 2005 Sep 29;24(43):6564–73. doi: 10.1038/sj.onc.1208798. PubMed PMID: 16007186. [DOI] [PubMed] [Google Scholar]
- 9.Khaleque MA, Bharti A, Gong J, Gray PJ, Sachdev V, Ciocca DR, et al. Heat shock factor 1 represses estrogen-dependent transcription through association with MTA1. Oncogene. 2008 Mar 20;27(13):1886–93. doi: 10.1038/sj.onc.1210834. PubMed PMID: 17922035. [DOI] [PubMed] [Google Scholar]
- 10.Chou SD, Prince T, Gong J, Calderwood SK. mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PloS one. 2012;7(6):e39679. doi: 10.1371/journal.pone.0039679. PubMed PMID: 22768106. Pubmed Central PMCID: 3387249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Murshid A, Prince T, Calderwood SK. Protein kinase A regulates molecular chaperone transcription and protein aggregation. PloS one. 2011;6(12):e28950. doi: 10.1371/journal.pone.0028950. PubMed PMID: 22216146. Pubmed Central PMCID: 3245242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murshid A, Chou SD, Prince T, Zhang Y, Bharti A, Calderwood SK. Protein kinase A binds and activates heat shock factor 1. PloS one. 2010;5(11):e13830. doi: 10.1371/journal.pone.0013830. PubMed PMID: 21085490 Pubmed Central PMCID: 2976705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Grammatikakis N, Siganou A, Stevenson MA, Calderwood SK. Interactions between extracellular signal-regulated protein kinase 1, 14-3-3epsilon, and heat shock factor 1 during stress. The Journal of biological chemistry. 2004 Nov 19;279(47):49460–9. doi: 10.1074/jbc.M406059200. PubMed PMID: 15364926. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Grammatikakis N, Siganou A, Calderwood SK. Regulation of molecular chaperone gene transcription involves the serine phosphorylation, 14-3-3 epsilon binding, and cytoplasmic sequestration of heat shock factor 1. Molecular and cellular biology. 2003 Sep;23(17):6013–26. doi: 10.1128/MCB.23.17.6013-6026.2003. PubMed PMID: 12917326. Pubmed Central PMCID: 180972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu B, Soncin F, Price BD, Stevenson MA, Calderwood SK. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. The Journal of biological chemistry. 1996 Nov 29;271(48):30847–57. doi: 10.1074/jbc.271.48.30847. PubMed PMID: 8940068. [DOI] [PubMed] [Google Scholar]
- 16.Kline MP, Morimoto RI. Repression of the heat shock factor1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol Cell Biol. 1997;17:2107–15. doi: 10.1128/mcb.17.4.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knauf U, Newton EM, Kyriakis J, Kingston RE. Repression of heat shock factor 1 activity at control temperature by phosphorylation. Genes Dev. 1996;10:2782–93. doi: 10.1101/gad.10.21.2782. [DOI] [PubMed] [Google Scholar]
- 18.Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, et al. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol Cell Biol. 2003 Apr;23(8):2953–68. doi: 10.1128/MCB.23.8.2953-2968.2003. PubMed PMID: 12665592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu B, Zhong R, Soncin F, Stevenson MA, Calderwood SK. Transcriptional activity of heat shock factor 1 at 37 degrees C is repressed through phosphorylation on two distinct serine residues by glycogen synthase kinase 3 and protein kinases Calpha and Czeta. The Journal of biological chemistry. 1998 Jul 17;273(29):18640–6. doi: 10.1074/jbc.273.29.18640. PubMed PMID: 9660838. [DOI] [PubMed] [Google Scholar]
- 20.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nature reviews Molecular cell biology. 2009 May;10(5):307–18. doi: 10.1038/nrm2672. PubMed PMID: 19339977. [DOI] [PubMed] [Google Scholar]
- 21.Yecies JL, Manning BD. mTOR links oncogenic signaling to tumor cell metabolism. Journal of molecular medicine. 2011 Mar;89(3):221–8. doi: 10.1007/s00109-011-0726-6. PubMed PMID: 21301797. [DOI] [PubMed] [Google Scholar]
- 22.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Developmental cell. 2009 Jul;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. PubMed PMID: 19619488 Pubmed Central PMCID: 2861485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemler R. From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends in genetics : TIG. 1993 Sep;9(9):317–21. doi: 10.1016/0168-9525(93)90250-l. PubMed PMID: 8236461. [DOI] [PubMed] [Google Scholar]
- 24.Vleminckx K, Kemler R, Hecht A. The C-terminal transactivation domain of beta-catenin is necessary and sufficient for signaling by the LEF-1/beta-catenin complex in Xenopus laevis. Mechanisms of development. 1999 Mar;81(1-2):65–74. doi: 10.1016/s0925-4773(98)00225-1. PubMed PMID: 10330485. [DOI] [PubMed] [Google Scholar]
- 25.Gotoh J, Obata M, Yoshie M, Kasai S, Ogawa K. Cyclin D1 over-expression correlates with beta-catenin activation, but not with H-ras mutations, and phosphorylation of Akt, GSK3 beta and ERK1/2 in mouse hepatic carcinogenesis. Carcinogenesis. 2003 Mar;24(3):435–42. doi: 10.1093/carcin/24.3.435. PubMed PMID: 12663502. [DOI] [PubMed] [Google Scholar]
- 26.Metcalfe C, Bienz M. Inhibition of GSK3 by Wnt signalling--two contrasting models. Journal of cell science. 2011 Nov 1;124(Pt 21):3537–44. doi: 10.1242/jcs.091991. PubMed PMID: 22083140. [DOI] [PubMed] [Google Scholar]
- 27.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research. 2011 Jan;39(Database issue):D945–50. doi: 10.1093/nar/gkq929. PubMed PMID: 20952405. Pubmed Central PMCID: 3013785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charafe-Jauffret E, Ginestier C, Birnbaum D. Breast cancer stem cells: tools and models to rely on. BMC Cancer. 2009;9:202. doi: 10.1186/1471-2407-9-202. PubMed PMID: 19555472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE, et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011 Nov 8;108(45):18378–83. doi: 10.1073/pnas.1115031108. PubMed PMID: 22042860. Pubmed Central PMCID: 3215027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindvall C, Bu W, Williams BO, Li Y. Wnt signaling, stem cells, and the cellular origin of breast cancer. Stem Cell Rev. 2007 Jun;3(2):157–68. doi: 10.1007/s12015-007-0025-3. PubMed PMID: 17873348. [DOI] [PubMed] [Google Scholar]
- 31.Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003 May 22;423(6938):409–14. doi: 10.1038/nature01593. PubMed PMID: 12717450. [DOI] [PubMed] [Google Scholar]
- 32.Veronese A, Visone R, Consiglio J, Acunzo M, Lupini L, Kim T, et al. Mutated beta-catenin evades a microRNA-dependent regulatory loop. Proc Natl Acad Sci U S A. 2010 Mar 22;108(12):4840–5. doi: 10.1073/pnas.1101734108. PubMed PMID: 21383185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010 Mar;12(3):247–56. doi: 10.1038/ncb2024. PubMed PMID: 20173740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saydam O, Shen Y, Wurdinger T, Senol O, Boke E, James MF, et al. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/beta-catenin signaling pathway. Mol Cell Biol. 2009 Nov;29(21):5923–40. doi: 10.1128/MCB.00332-09. PubMed PMID: 19703993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su J, Zhang A, Shi Z, Ma F, Pu P, Wang T, et al. MicroRNA-200a suppresses the Wnt/beta-catenin signaling pathway by interacting with beta-catenin. Int J Oncol. 2012 Apr;40(4):1162–70. doi: 10.3892/ijo.2011.1322. PubMed PMID: 22211245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srikantan S, Gorospe M. HuR function in disease. Front Biosci. 2012;17:189–205. doi: 10.2741/3921. PubMed PMID: 22201738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nabors LB, Gillespie GY, Harkins L, King PH. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 2001 Mar 1;61(5):2154–61. PubMed PMID: 11280780. [PubMed] [Google Scholar]
- 38.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, et al. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001 Dec;108(11):1657–65. doi: 10.1172/JCI12973. PubMed PMID: 11733561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic acids research. 2005;33(22):7138–50. doi: 10.1093/nar/gki1012. PubMed PMID: 16391004 Pubmed Central PMCID: 1325018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Molecular cell. 2011 Aug 5;43(3):340–52. doi: 10.1016/j.molcel.2011.06.008. PubMed PMID: 21723171. [DOI] [PubMed] [Google Scholar]
- 41.Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003 Oct 16;22(46):7146–54. doi: 10.1038/sj.onc.1206862. PubMed PMID: 14562043. [DOI] [PubMed] [Google Scholar]
- 42.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Molecular cell. 2012 Aug 24;47(4):648–55. doi: 10.1016/j.molcel.2012.06.027. PubMed PMID: 22841487. Pubmed Central PMCID: 3509343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gabai VL, Meng L, Kim G, Mills TA, Benjamin IJ, Sherman MY. Heat shock transcription factor Hsf1 is involved in tumor progression via regulation of hypoxia inducible factor 1 and RNA-binding protein HuR. Molecular and cellular biology. 2012 Mar;32(5):929–40. doi: 10.1128/MCB.05921-11. PubMed PMID: 22215620. Pubmed Central PMCID: 3295200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim G, Meriin AB, Gabai VL, Christians E, Benjamin I, Wilson A, et al. The heat shock transcription factor Hsf1 is downregulated in DNA damage-associated senescence, contributing to the maintenance of senescence phenotype. Aging cell. 2012 Aug;11(4):617–27. doi: 10.1111/j.1474-9726.2012.00827.x. PubMed PMID: 22510478. Pubmed Central PMCID: 3433748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012 Aug 3;150(3):549–62. doi: 10.1016/j.cell.2012.06.031. PubMed PMID: 22863008. Pubmed Central PMCID: 3438889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santagata S, Mendillo ML, Tang YC, Subramanian A, Perley CC, Roche SP, et al. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science. 2013 Jul 19;341(6143):1238303. doi: 10.1126/science.1238303. PubMed PMID: 23869022. Pubmed Central PMCID: 3959726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weng D, Penzner JH, Song B, Koido S, Calderwood SK, Gong J. Metastasis is an early event in mouse mammary carcinomas and is associated with cells bearing stem cell markers. Breast cancer research : BCR. 2012;14(1):R18. doi: 10.1186/bcr3102. PubMed PMID: 22277639. Pubmed Central PMCID: 3496135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.