Abstract
Eya1 is a conserved critical regulator of organ-specific stem cells. Ectopic Eya1 activities, however, promote transformation of mammary epithelial cells. Signals that instigate Eya1 oncogenic activities remain to be determined. Here, we show that Akt1 kinase physically interacts with Eya1 and phosphorylates a conserved consensus site of the Akt kinase. PI3K/Akt signaling enhances Eya1 transcription activity, which largely attributes to the phosphorylation-induced reduction of Eya1 SUMOylation. Indeed, SUMOylation inhibits Eya1 transcription activity; and pharmacologic and genetic activation of PI3K/Akt robustly reduces Eya1 SUMOylation. Wild type but not Akt phosphorylation site mutant Eya1 variant rescues the cell migratory phenotype of EYA1-silenced breast cancer cells, highlighting the importance of Eya1 phosphorylation. Furthermore, knockdown EYA1 sensitizes breast cancer cells to the PI3K/Akt1 inhibitor and irradiation treatments. Thus, the PI3K/Akt signal pathway activates Eya1. These findings further suggest that regulation of SUMOylation by PI3K/Akt signaling is likely an important aspect of tumorigenesis.
Keywords: Akt, Eya1, Six1, SUMOylation, Breast cancer
Introduction
Eyes absent 1 (Eya1) is a transcriptional coactivator that has intrinsic protein phosphatase activity.1–5 Among four Eya-family genes, Eya1 is essential for proliferation and survival of organ-specific progenitors such as renal and cardiac progenitors.6, 7 Human EYA1 haploinsufficiency causes branchio-oto-renal (BOR) birth defect.7 Under pathological conditions, ectopic EYA1 activity is linked to oncogenic transformation of mammary epithelial cells and breast cancer progression.8, 9 Signals that instigate Eya1 oncogenic activities are unknown.
Eya1 functions as a canonical transcription coactivator of Six1 homeodomain transcription factor.1, 10, 11 The Six1-Eya1 transcription complex regulates expression of a number of downstream target genes that are important for cell proliferation, survival and migration. For instance, c-Myc expression is dramatically diminished in Six1 null mutant embryos.1 Ectopic Six1 expression promotes tumorigenesis by stimulating expression of Cyclin A1.12 Cyclin D1 is also regulated by the Six1-Eya1 transcription complex.13, 14 The intrinsic Eya1 phosphatase activity is in part required to regulate Cyclin D1 gene expression.14 Six1 also increases TGFβ signaling and promotes epithelial-mesenchymal transition (EMT) and metastasis of human mammary carcinoma cells.15 In addition to transcription activities of Eya-family proteins, enzymatic activity of Eya2 and Eya3 controls cytoskeletal organization, and enhance breast cancer cell migration and metastasis independent.8 Upregulation of human SIX1 is linked to advanced stages of breast cancer.13, 15–17 Furthermore, breast cancers with high levels of both EYA (1 or 2) and SIX1 have poor prognosis, including shortened time to relapse, progression to metastasis, and decreased survival rates.9 Given the critical role of Eya-family proteins in regulating cellular behavior and organ development, it is not surprising that aberrant activity of these genes may result in cancers. However, it does raise an important question on how Eya1 protein activity is regulated during cancer development.
Eya1 is post-translational modified by the small ubiquitin-related modifier 1 (SUMO1) protein (SUMOylation) but the potential role of Eya1 SUMOylation is largely unknown.18 Genetic polymorphisms in the SUMO-conjugating enzymes UBC9 and PIAS3 are associated with increased breast cancer grade and reduced DNA-damage repair responses.19–21 UBC9 is aberrantly overexpressed in luminal type of breast cancers.22 Differential expression of SUMO-specific protease 7 (SENP7) isoforms in breast cancer cells regulates tumor progression.23 Thus, losing the balance between substrate SUMOylation and de-SUMOylstion may be involved in Eya1 regulation during cancer development.
The serine/threonine kinase Akt, also known as protein kinase B (PKB), is a central node of the phosphatidylinositol 3-kinase (PI3K) signal pathway. PI3K/AKT signaling is frequently hyperactivated in human cancers, including breast cancers.24–26 In this study, we show that PI3K/Akt signaling enhances transcription activity of Eya1 via repressing its SUMOylation. EYA1 is required for proliferation and migration of a subset of the aggressive triple-negative breast cancer (TNBC) cells. PI3K/Akt signaling represses Eya1 SUMOylation thereby promotes Eya1 oncogenic activity in a phosphorylation-dependent manner. Moreover, genetic silencing of EYA1 significantly increases sensitivity of breast cancer cells to PI3K/Akt1 inhibition and irradiation treatments. Together, results from this study suggest that the PI3K/Akt-mediated repression of substrate SUMOylation, such as Eya1, is an important new aspect of cancer biology.
Results
Eya1 physically interacts with Akt1
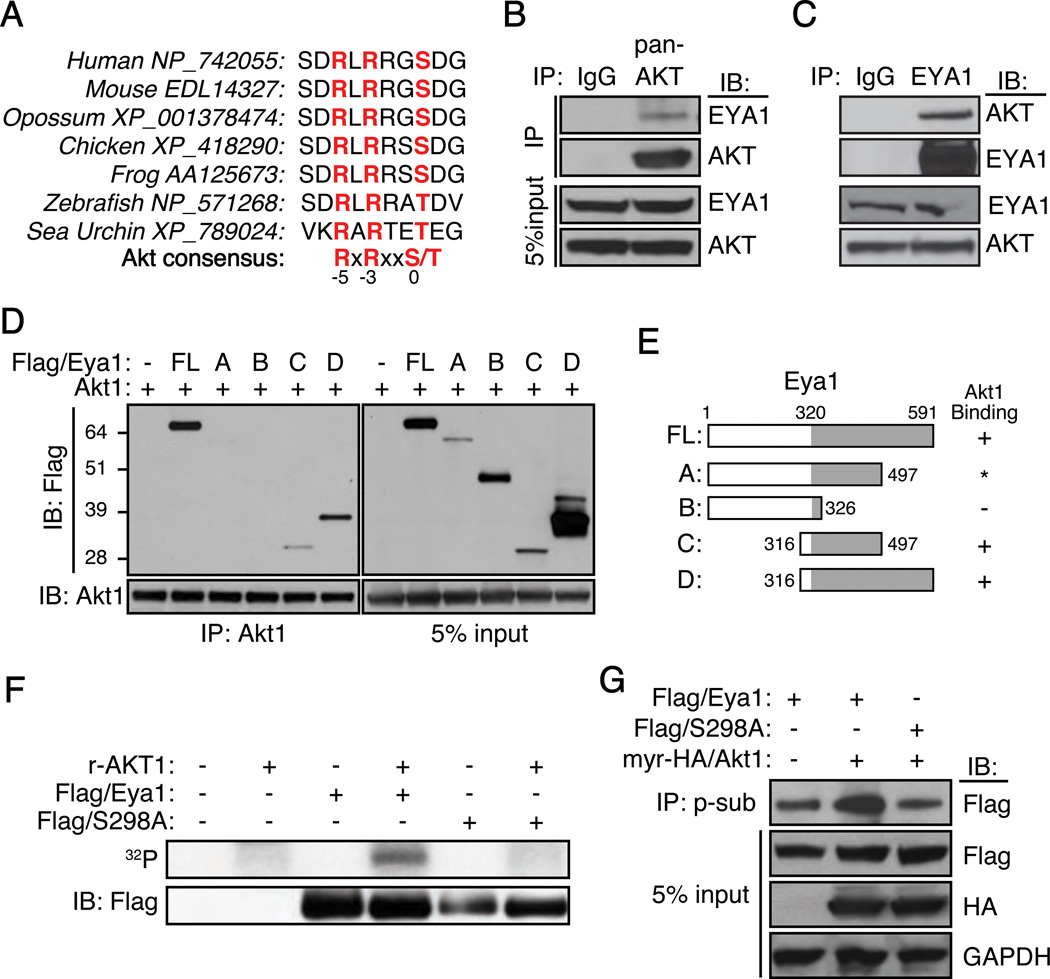
An Akt-substrate consensus motif was identified in Eya1 protein sequence from sea urchin to human (Figure 1A). This high degree sequence conservation suggested that the PI3K/Akt signal pathway is a candidate upstream regulator of Eya1. To test this possibility, we first investigated protein-protein interaction between Akt1 and Eya1 (Figure 1B–E, S1). Endogenous EYA1 was detected in the AKT immunocomplex from human BT549 breast cancer cells (Figure 1B). Conversely, AKT was also detected from the EYA1 protein complex (Figure 1C), indicating an association between these proteins. To further characterize potential interaction domain(s), a series of murine Flag-tagged Eya1 (Flag/Eya1) truncation mutations was created (Figures 1D, E and Figure S1). These variants were co-expressed with wild type murine Akt1 in HEK293 cells, and their interactions were assessed following immunoprecipitation with an Akt1 (Figure 1D) or a Flag-specific antibody (Figure S1). We observed a strong interaction between Akt1 and full-length Eya1 (Figure 1D and S1). Eya1 fragment A containing amino acids 1–497 (aa1-497) was unstable thus precluding a reliable evaluation (Figure 1D and S1). Fragment B (aa1-326) was highly expressed yet no interaction with Akt1 was detected. In contrast, both fragment C (aa316-497) and D (aa316-591) interacted strongly with Akt1. Thus, Akt1 associates with the evolutionarily conserved C-terminal Eya-phosphatase domain.
Figure 1. Akt1 interacts with and phosphorylates Eya1.
(A) A conserved substrate consensus site of Akt kinase identified in Eya1.
(B and C) Immunoprecipitation (IP) using antibodies against either a pan-AKT (AKT) (B), EYA1 (C) or control IgG from BT549 cell lysates, and resultants were immunoblotted (IB) using indicated antibodies.
(D and E) Co-IP of full-length mouse Akt1 and Flag-tagged Eya1 (Flag/Eya1) fragments from transfected HEK293 cells. Results are summarized in E. *, inconclusive.
(F) Immunoprecipitates (Flag/Eya1 or Flag/S298A) were incubated with recombinant AKT1 (r-AKT1) in an in vitro kinase assay with γ-[32P]-ATP. Top panel, autoradiography; bottom panel, protein immunoblot using a Flag-specific antibody.
(G) Immunoprecipitates from the phospho-specific Akt substrate antibody (p-sub) were probed using indicated antibodies. Flag/S298A, an Eya1 serine 298 to alanine mutation; myr-HA/Akt1, myristoylated HA-tagged Akt1, GAPDH was used as loading control.
To examine whether Eya1 was a substrate of Akt kinase, immunopurified wild type and S298A mutant Eya1 proteins were incubated with a recombinant active AKT1 kinase and 32P-gamma ATP in an in vitro kinase reaction (Figure 1F). Wild type Eya1 was labeled in the IP-kinase reaction. However, mutation of S298 to alanine (S298A), which rendered it incompatible to phosphorylation, was not phosphorylated by AKT1. To study whether this site could be phosphorylated in vivo, we used a phospho-(Ser/Thr) Akt substrate (p-sub) antibody to evaluate Eya1 phosphorylation status. This antibody recognizes a large number of phosphorylated Akt substrates since it preferentially binds to proteins containing phospho-Ser/Thr preceded by Lys/Arg at positions −5 and −3; and the antibody-binding specificity is largely independent of other surrounding sequences.27 Flag/Eya1 was detected by the p-sub antibody (Figure 1G, lane 1 of the top panel). Coexpression of a constitutively active Akt1, which is myristoylated and HA-tagged (myr-HA/Akt1), increased signal of the p-sub immunoreactivity (Figure 1G, compare lane 1 to lane 2, top panel). S298A variant had markedly decreased signal (Figure 1G, compare lane 2 to lane 3, top panel). Together, these observations suggest that S298 is phosphorylated, and activation of Akt1 kinase increases S298 phosphorylation level.
We also noted that the Eya1(S298A) mutant variant remained reactive to the p-sub antibody (Figure 1G, lane 3, top panel). It is therefore possible that endogenous Eya1 could be phosphorylated at multiple sites in addition to S298.
Akt1 enhances Eya1 transcription activity
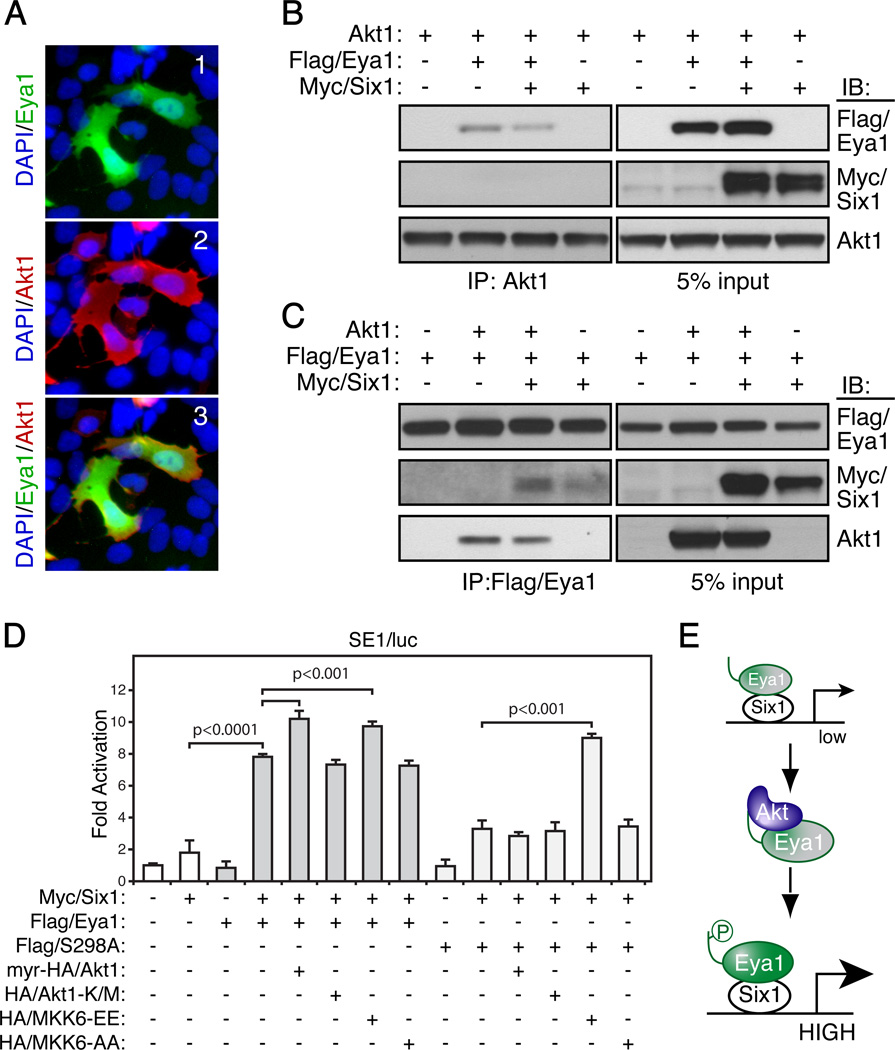
Eya1 is the canonical co-activator of the homeodomain transcription factor Six1. Six1 binds to Eya1 and induces Eya1 translocation from the cytoplasm to the nucleus where the Six1/Eya1 transcription complex activates target gene expression.11 To examine whether Akt1 regulated Eya1 function, we initially examined whether Akt1 influenced Eya1 localization. As shown in Figure 2A, full length Eya1 localized to both cytoplasm and nucleus in the presence of active myr-HA/Akt1 (Figure 2A), which is similar to Eya1 localization pattern in normal HEK293.11 Therefore, Akt1 has no obvious influence on the overall localization pattern of Eya1.
Figure 2. Akt1 induces Eya1 transcription activity.
(A) HEK293 cells were transiently transfected with the Flag/Eya1 and/or HA/Akt1 expression constructs and analyzed by indirect immunocytochemistry using a Flag-antibody (green, 1), HA- antibody (Red, 2) and counter-stained with DAPI (blue). 3, a merged image of 1 and 2.
(B–C) Combinations of Akt1, Flag/Eya1 and Myc/Six1 were co-expressed in HEK293 cells. co-IP and IB were done using indicated antibodies.
(D) A transient transcription reporter assay using a Six1/Eya1-dependent reporter (SE1/luc), which was co-expressed with combinations of indicated expression constructs. Relative fold changes were calculated using basal and Renilla controls (mean ± SEM, n=5).
(E) Schematic diagram of regulation of Eya1 activities. Resting state Eya1 has low transcription activity (low, in a dash box). Akt1 increases Eya1 transcription (HIGH) in part through the conserved S298 phosphorylation (p) site.
We next investigated whether Akt1 was directly associated with the Six1/Eya1 transcription complex. Combinations of wild type Akt1, Flag/Eya1 and Myc-tagged Six1 (Myc/Six1) were co-expressed in HEK293 cells, and immunocomplex were interrogated for the presence of prospective components (Figures 2B and C). Unlike Flag/Eya1, Myc/Six1 was not detected in Akt1 immunocomplex with or without Flag/Eya1 (Figure 2B, the second panel). Instead, co-expression of Myc/Six1 slightly reduced the interaction between Flag/Eya1 and Akt1 (Figure 2B, comparing lanes 2 and 3 of the first panel). A similar phenomenon was observed when the Flag/Eya1 immunocomplex was examined, from which less Akt1 was detected when Myc/Six1 was co-expressed (Figure 2C, compare lanes 2 and 3 of the third panel). Flag/Eya1 interacted with Myc/Six1 with or without co-expression of Akt1 (Figure 2C, the second panel). Thus, Akt1 is unlikely to be a component of the Six1/Eya1 transcription complex.
To determine whether Akt1 is capable of regulating Eya1 transcription activity, we performed a transcription reporter assay using a Six1-binding element-dependent luciferase reporter (SE1/luc).1, 7 The reporter itself had minimal basal activity in HEK293 cells, and coexpression of both Eya1 and Six1 resulted in a significant increase of the reporter activity (Figure 2D). Mutation of the phosphorylation site (S298A) significantly reduced Eya1 transcription activity. A constitutively active myr-HA/Akt1 further enhanced reporter activity mediated by wild type Eya1 but not S298A mutant. MKK6 is a known activator of the Six1/Eya1 transcription reporter.28 The effect of MKK6 was not impaired by the S298A mutation. Thus, activation of Akt1 induces Eya1 transcription activity (Figure 2E), and this in part depends on the conserved S298 residue at the Akt-substrate consensus motif.
Functional interaction between Akt1 and Eya1 in vivo
Eya1 is a critical regulator of kidney development and kidney size formation is particularly sensitive to levels of Eya1 activity29. Akt1 is a central regulator of cell proliferation and survival; and is required for growth of all organs including kidney1, 6, 30, 31. To investigate whether Akt1 functionally interacts with Eya1 in vivo, we therefore analyzed the kidney size of compound Akt1−/− and Eya1+/− mutants (Figure S2). The overall body size of newborn Akt1-null mutants was approximately 25% smaller than littermate controls (Figure S2B, n=6), confirming its critical roles in growth and cell proliferation30, 31. The kidney to body weight ratio of Akt1−/− mutants, however, was comparable to that of controls (Figure S2B, F and I), consistent with the finding that Akt1 mutant is proportionally smaller, owing to the global reduction of cell number32. Eya1+/− were grossly normal (Figure S2C, n=9). Compound Akt1−/−;Eya1+/− mutants exhibited overall body sizes comparable to Akt1−/− mutants. Notably, kidneys were significantly smaller in these compound mutants than Akt1−/− or Eya1+/− single mutants, as indicated by a reduced kidney to body weight ratio (Figure S2D, H and I, n=18). Thus, consistent with biochemical observations (Figure 1 and 2), these genetic findings support a functional interaction between Akt1 and Eya1 in vivo.
Eya1 is post-translational modified by SUMO protein
We next wished to understand how PI3K/Akt signaling regulated Eya1 transcription activity. Because of the critical role of SUMOylation in transcription repression;33–37 and Eya1 is a SUMOylated protein,18 we focused on a SUMOylation-dependent mechanism.
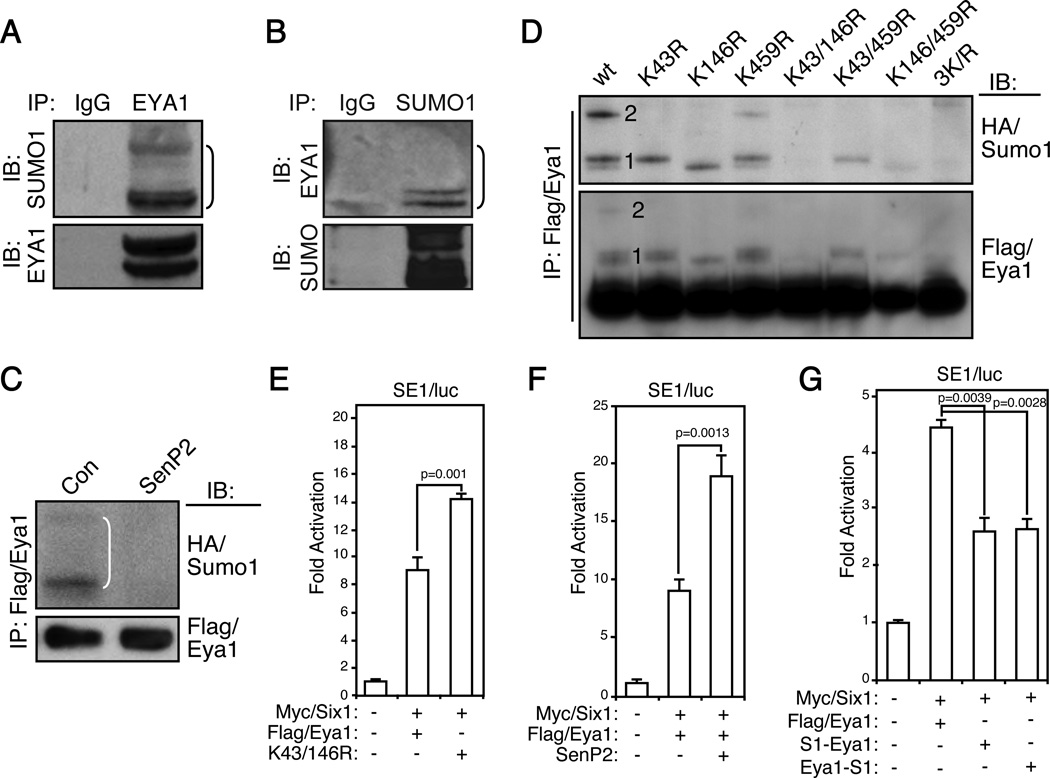
The precise nature of Eya1 SUMOylation and how SUMOylation regulates Eya1 transcription activity remain to be determined. To confirm that Eya1 is indeed SUMOylated, immunopurified endogenous EYA1 proteins were examined using a SUMO1-specific antibody (Figure 3A). Several slow migrating high molecular weight bands were detected, which likely corresponded to SUMOylated EYA1 (Figure 3A). Conversely, examination of SUMO1 immunocomplex using an EYA1 antibody also detected similar high molecular bands (Figure 3B). To ascertain that these slow migration bands were indeed SUMOylated Eya1, Flag/Eya1 and HA-tagged Sumo1 (HA/Sumo1) were coexpressed in HEK293 cells, and SUMOylation pattern of Flag/Eya1 was examined using a HA-specific antibody (the first lanes in Figure 3C and D). We detected three high molecular weight bands, which corresponded to mono- and di-SUMOylation as they had a characteristic incremental size increase of HA/Sumo1 moiety (~12 kDa). Indeed, coexpression of SenP2, a SUMO-specific peptidase, completely eliminated these slow migrating bands (Figure 3C).
Figure 3. SUMOylation represses Eya1 transcription activity.
(A–B) Immunoprecipitation (IP) of endogenous EYA1 (A) or SUMO1 (B) from HEK293 cells and immunobloted (IB) using indicated antibodies. IgG, negative control.
(C) Flag/Eya1 and HA/Sumo1 were transiently expressed in HEK293 cells with SenP2 or a control (con). IP and IB were performed using indicated antibodies.
(D) Combinations of lysine (K) 43, 146 and 459 to arginine (R) Eya1 mutatn variants were used to map Eya1 SUMOylation sites in an IP and IB assay. Number 1 and 2 indicate single and double SUMOylated Eya1, respectively.
(E–G) Reporter assays indicated that SUMOylation represses Eya1 transcription activity. The Six1/Eya1-dependent reporter (SE1/luc) was co-transfected with indicated constructs and the luciferase reporter activity was measure at 48 hours (n=3). S1-Eya1, Eya1-S1: fusion of SUMO1 to Eya1 at N- and C-terminus, respectively.
Eya1 SUMOylation pattern suggested that it was modified at two possible sites. In addition to K43 and K459, sequence analysis revealed another SUMOylation site, K146. To pinpoint which of these three residues were SUMOylated, we generated a series of single, double and triple lysine-to-arginine (K/R) mutations to eliminate SUMOylation on these perspective sites. Wild type Flag/Eya1 immunocomplex exhibited a typical Eya1 SUMOylation pattern, a doublet of mono-SUMOylated and a single di-SUMOylated Eya1 variant (Figure 3D, first lane). K43R and K146R mutants displayed a mono SUMOylation pattern, while K459R showed a similar SUMOylation pattern to wild type control although the overall SUMOylation level was slightly lower than wild type. Mutation of both K43 and K459 residues (K43/459R) had mono-SUMOylation, which was similar to the SUMOylation pattern to K43R variant. In addition, K146/459R and K146R mutants exhibited a similar mono-SUMOylation pattern. Importantly, K43/146R and K43/146/459R mutants were not SUMOylated. Together, these systematic mutagenesis analyses indicate that Eya1 K43 and K146 but not K459 are SUMOylated.
SUMOylation represses Eya1 transcription activity
To determine potential impact of SUMOylation on Eya1 transcription activity, we used both SE1/luc and a native Fgf8 (Fgf8/luc) reporter.1, 7 The de-SUMOylated Eya1 variants, K43/146R (2KR), exhibited a significantly higher transcriptional activity than wild type Eya1 (Figure 3E and S3), raising a possibility that SUMOylation inhibits Eya1 activity. In line with this observation, coexpression of SenP2, which reduced Eya1 SUMOylation (Figure 3C), significantly induced both SE1/luc and Fgf8/luc reporters (Figure 3F and S3B). We also created two fusion proteins, in which Sumo1 was positioned at either C-terminal (Eya1-S1) or N-terminal (S1-Eya1) of Eya1 protein. Previous studies have shown that such fusion protein mimics SUMOylated substrate, particularly with regard to transcription regulators.37, 38 We found that fusion of Sumo1 significantly attenuated Eya1 transcription activities, consistent with the notion that SUMOylation represses Eya1 transcription activity (Figure 3G and S3C).
In parallel to reporter assays, endogenous Six1/Eya1 transcription targets were investigated using quantitative real-time PCR (Figure S3D). Consistent with reporter assays, wild type Eya1 significantly increased expression of its endogenous target, Fgf8.1, 7 In comparison, Eya1(K43/146R) mutant variant had a 55% increase of transcription activity than wild type Eya1 (Figure S3D). On the other hand, the hyper-SUMOylated S298A mutant was much less effective (Figure S3D). CYCLIN D1 and c-MYC are two additional Six1-Eya1 target genes.1, 13, 14 Similar to FGF8, de-SUMOylated K43/146R but not hyper-SUMOylated S298A variant significantly induced expression of these genes in HEK293 cells (Figure S3E and S3F). Together, both reporter and endogenous gene expression levels indicate that SUMOylation represses Eya1 transcription activity.
PI3K/Akt promotes transcription activity of Eya1 by repressing its SUMOylation
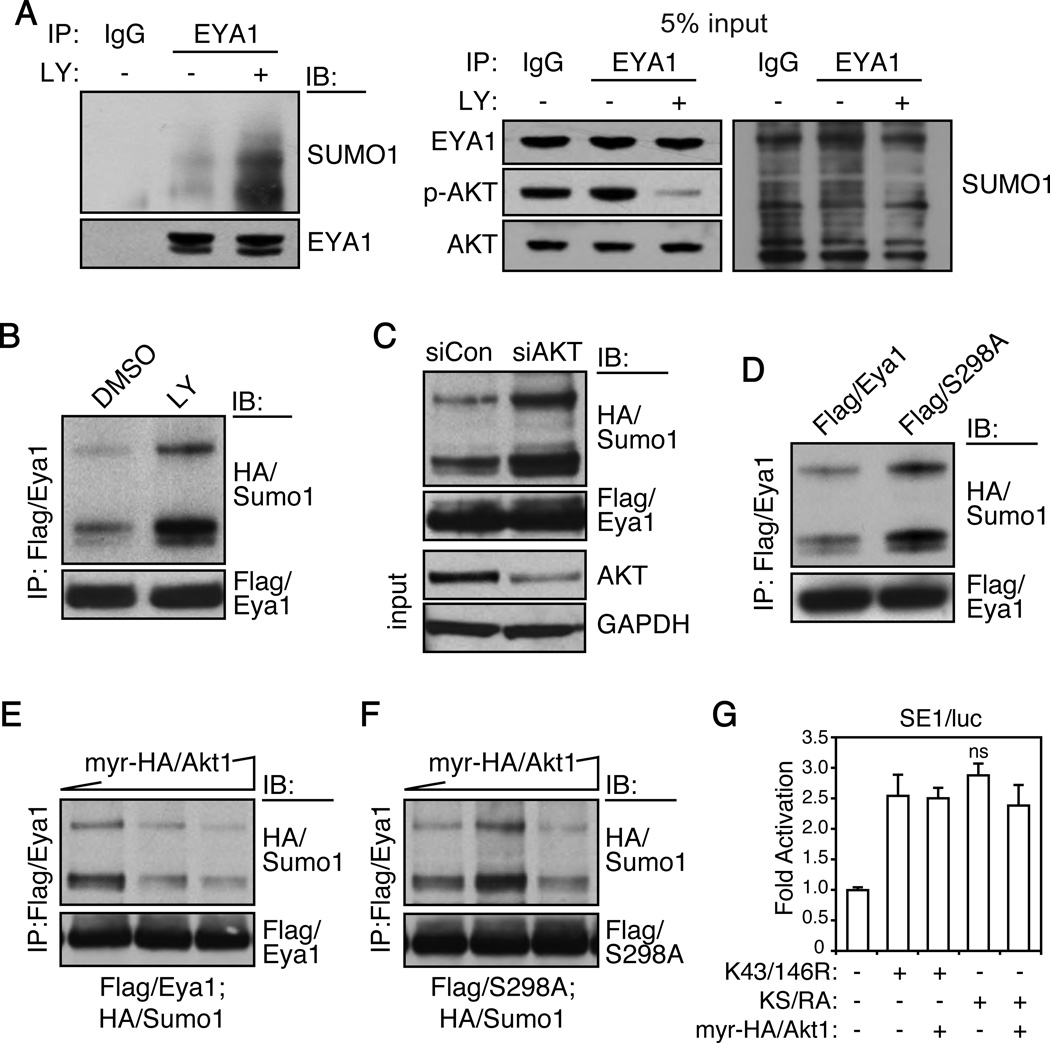
We next examined whether Akt1 regulated Eya1 through a SUMOylation mechanism. Firstly, we manipulated PI3K/AKT signaling using both siRNA and the phosphatidylinositol-3-kinase (PI3K) inhibitor LY294002. To avoid potential compensatory effects among AKT isoforms, all isoforms were targeted by a mixture of siRNAs. Both LY294002 (Figure 4A and B) and siRNA knockdown (Figure 4C) strongly enhanced endogenous EYA1 (Figure 4A) and Flag/Eya1 (Figure 4B and C) SUMOylation levels in HEK293 cells. On the other hand, mutation of the Akt phosphorylation site, S298A, increased Eya1 SUMOylation (Figure 4D). Constitutively active myr-HA/Akt1 attenuated Eya1 SUMOylation in a dose-dependent manner (Figure 4E). However, it had minimal effect on the SUMOylation level of Eya1(S298A) mutant variant (Figure 4F). Thus, PI3K/Akt signaling inhibits Eya1 SUMOylation, and this inhibition is in part mediated by the conserved S298 phosphorylation site.
Figure 4. Akt1 inhibits Eya1 SUMOylation.
(A–C) Inhibition of PI3K/Akt signaling with an inhibitor LY294002 (25 µM, 2 h, A and B) or AKT siRNA oligo sets (C, targeting all three AKT1/2/3 isoforms, Dharmacon) enhanced endogenous EYA1 (A) and Flag/Eya1 (B and C) SUMOylation levels. IP and IB were performed using indicated antibodies.
(D) S298A mutant had enhanced SUMOylation level.
(E and F) Constitutively active myr-HA/Akt1 suppressed SUMOylation of wild type Eya1 (E) but not S298A mutant (F). Flag/Eya1, Flag/S298A and HA/Sumo1 were transiently expressed in HEK293 cells with gradient amount of myr-HA/Akt1.
(G) Akt1 induced Eya1 transcription activity via inhibition of Eya1 SUMOylation. The Six1/Eya1-dependent reporter (SE1/luc) was co-transfected with indicated constructs and the luciferase reporter activity was measure at 48 hours. Relative fold changes were calculated using basal and Renilla controls (mean ± SEM, n=3). ns, not significant; KS/RA, K43R/K164R/S298A triple mutation.
Secondly, we created a compound Eya1 mutant variant, KS/RA, which could not be SUMOylated (K43/146R) or phosphorylated (S298A) at these respective sites, to examine whether Eya1 phosphorylation functions independently to the SUMOylation pathway. Transcription activities of these mutant variants (K43/146R and KS/RA) were compared with or without myr-HA/Akt1. K43R/K146R had a robust transcription activity (Figure 4G). Addition of myr-HA/Akt1 did not further increase K43/146R transcription activity (Figure 4F, compare bar 2 and 3). Transcription activity of Eya1(KS/RA) variant was similar to that of K43/146R; and effect of myr-HA/Akt1 was not significant (Figure 4G, compare bar 4 and 5). Therefore, PI3K/Akt signaling induced Eya1 transcription activity is primarily mediated by the phosphorylation-dependent reduction of Eya1 SUMOylation.
PI3K/Akt signaling is required for stress-induced reduction of Eya1 SUMOylation
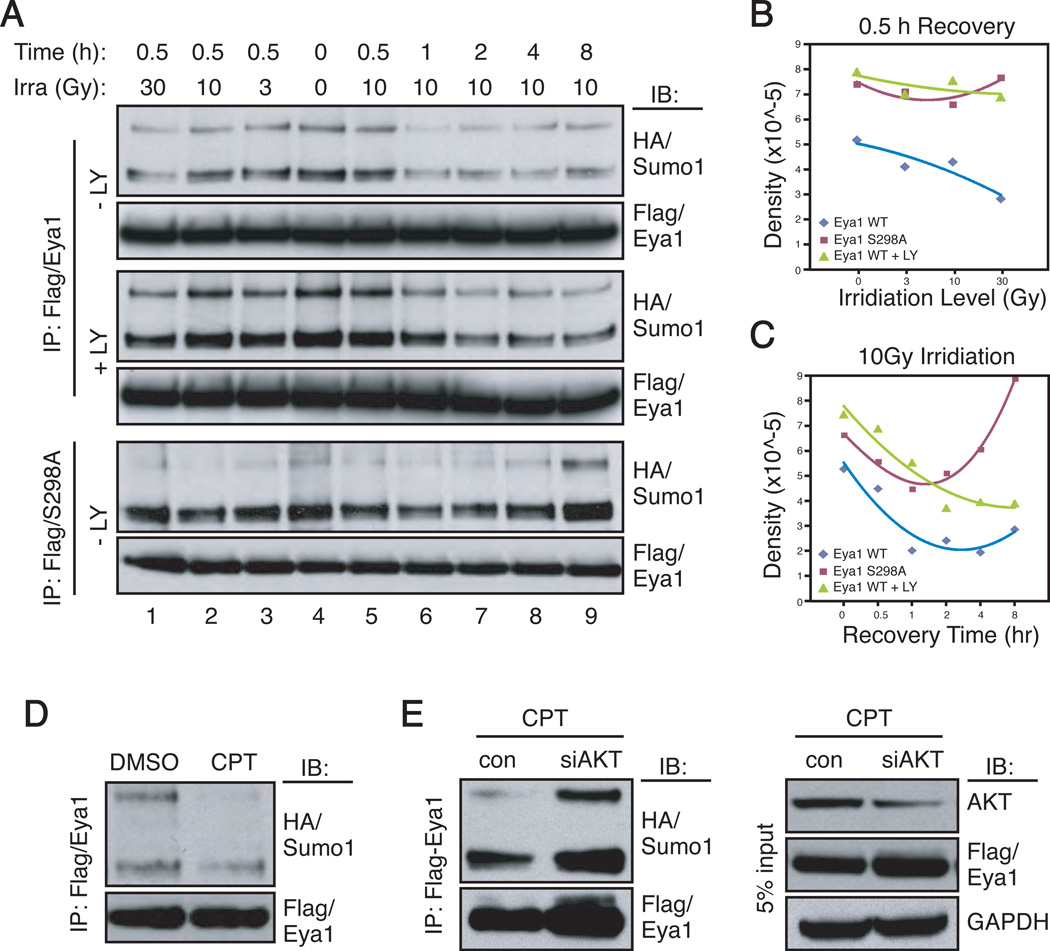
Eya1 is a critical regulator of genotoxic stress response.39 Coincidently, genotoxic stress activates Akt1, which in turn provides a pro-survival signal.40 We therefore investigated whether PI3K/Akt signaling regulated Eya1 SUMOylation during the stress response. Gama-irradiation of HEK293 cells at doses between 5 and 40 Gy resulted in a biphasic activation of AKT, as indicated by AKT phospho-473 immunoreactivity (Figure S4, p-S473/AKT). A similar response was observed using mouse embryonic fibroblasts.40 Coincident to AKT activation, Eya1 SUMOylation decreased after γ-irradiation in a dose-dependent manner (Figure 5A, lanes 1–4). Eya1 SUMOylation reached to its lowest levels within 1 hour after irradiation (10Gy) and persisted at a low level. Inhibition of PI3K/Akt signaling using LY294002 increased Eya1 SUMOylation at each respective time point relative to that of the wild type control (Figure 5A and B). Intriguingly, Eya1 SUMOylation reached its lowest levels after 2–4 hours in the presence of LY294002, which was nearly a 2-hour delay when compared to irradiation induced SUMOylation reduction in the absence of the inhibitor. SUMOylation levels of Eya1(S298A) mutant were high and reached to the lowest level 1 hour after γ-irradiation (Figure 5A–C). Surprisingly, its hypo-SUMOylated state failed to persist, as S298A SUMOylated level increased at 2, 4 and 8 hours of post-irradiation (Figure 5A and C).
Figure 5. Genotoxic stress decreases Eya1 SUMOylation via the PI3K/Akt pathway.
(A–C) HEK293 cells, transfected with Flag/Eya1 and HA/Sumo1, were γ-irradiated with the indicated doses and allowed to recover for indicated time prior to IP and IB assays. Densitometry for quantification were presented in B and C. LY, LY294002.
(D–E) Flag/Eya1- and HA/Sumo1-expressing HEK293 cells were treated with camptothecin (CPT, 10 µM, 30 min) or DMSO (D) in the presence or absence of siAKT (E) prior to the IP and IB analysis using indicated antibodies.
To further examine the dynamics of Eya1 SUMOylation after cellular stress, we treated cells with camptothecin (CPT, 10µM, 30 minutes), a topoisomerase I inhibitor that induces DNA damage and apoptosis. CPT treatment dramatically reduced Eya1 SUMOylation level (Figure 5D). Silencing of all AKT isoforms by siRNA enhanced Eya1 SUMOylation (Figures 4C and 5E), and rendered Eya1 SUMOylation insensitive to CPT treatment (Figure 5E). Taken together, these observations suggest that genotoxic stress represses Eya1 SUMOylation in a PI3K/Akt-dependent manner; and phosphorylation of S298 is required to keep Eya1 at the hypo-SUMOylated state.
Akt1 and Eya1 synergistically regulate breast cancer cell survival and migration
We next wished to investigate the significance of the PI3K/Akt/Eya1 axis in breast cancer cells. Gene expression profile of more than 50 different breast cancer cell lines have been characterized and compared.41, 42 A number of triple negative breast cancer (TNBC) cell lines were among these that had abnormal high levels of EYA1 (Figure S5A). EYA1 expression of four TNBC cell lines, MDA157, BT549, MDA-MB-231 and SUM149, were independently examined and confirmed using a real-time quantitative PCR assay (Figure S5B). MDA157 had the highest of and SUM149 had the lowest of EYA1 expression among four of these cell lines. To investigate a potential role of EYA1 in these TNBC cells, we generated and screened a total of 11 independent lentivirus-based shRNA vectors to target EYA1-specific sequences (Table S1). Among them, five shEYA1 vectors robustly attenuated Six1-Eya1-based reporter activity (shEYA1 1–5, Figure S5C), and significantly silenced endogenous EYA1 transcripts in MDA-MB-231 cells (Figure S5D).
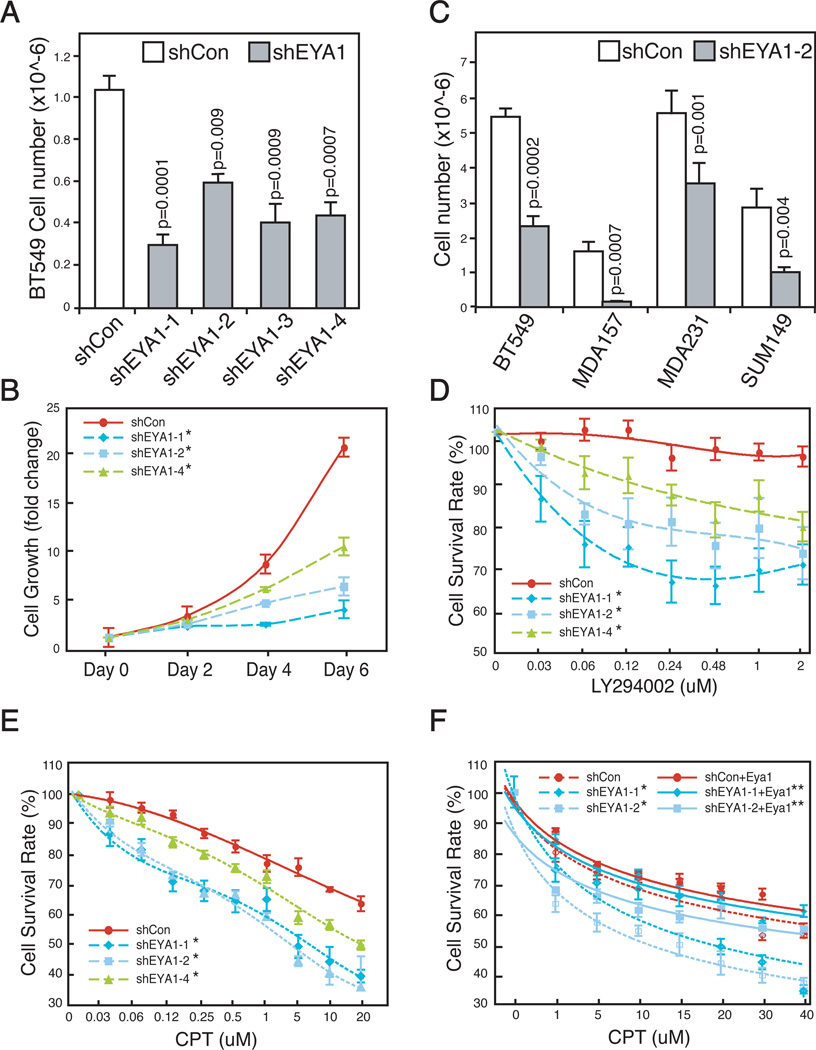
Knockdown of EYA1 using four independent hairpins significantly attenuated BT549 cell growth (Figure 6A and B). Similarly, EYA1 knockdown significantly reduced the growth of three other TNBC cell lines (Figure 6C). We found that MDA157 cells were particularly sensitive to EYA1 level (Figure 6C), suggesting a possibility that they were addicted to high level of EYA1 expression.
Figure 6. EYA1 and PI3K/Akt synergistically promote breast cancer cell survival.
(A–B) Growth rate of stable clones of BT549 breast cancer cells infected with EYA1-specific shRNAs (shEYA1, 1–4) or control (shCon) lentiviral particles. Cell number was quantified after a 3-day culture (A, n=3), or over a 6-day time course (B, n=6). Values are expressed as mean ± SEM. *, p<0.01.
(C) EYA1 knockdown attenuates growth of all four lines of breast cancer cells. The indicated cells were infected with either control (shCon) or EYA1-specific (shEYA1-2) shRNA lentiviral particles, and cell number was quantified after 5 days (BT549, MDA157, SUM149) or 6 days (MDA157).
(D–E) Increased sensitivity of EYA1-silenced BT549 breast cancer cells to genotoxic agent camptothecin (CPT) or PI3K/Akt inhibition (LY294002). Equivalent numbers of control (shCon, solid line) and EYA1-silenced (shEya1-1, 2, 4, dotted lines) cells were treated with either LY294002 or CPT for 24 hours. Relative survival rates were compared with mock transfected cells (DMSO) (mean ± SEM, n=6). *, p<0.01.
(F) Rescue of EYA1-silenced BT549 cells by wild type mouse Eya1. Equal number of cells was treated with CPT as indicated. Surviving cells were counted 24 h later (n=6). *, EYA1 vs shCon, p<0.01; **, shEYA1 vs shEYA1+Eya1, p< 0.001.
To examine a functional interaction between PI3K/Akt and EYA1 in these cells, we treated and compared survival rate of parental and EYA1-silenced derivative cells with LY294002 inhibitor. MDA-MB-231 cells have relatively low basal level of the PI3K/Akt activity. On the other, BT549 cells have a defective Pten thereby have significantly elevated AKT activity.43 Despite the difference, both cell lines are insensitive to low dose of LY294002 treatment.44 Indeed, suboptimal amount of the drug had no effect on survival of wild type parental BT549 cells (Figure 6D). However, EYA1 knockdown significantly sensitized these cells to drug inhibitions. The enhanced sensitivity was also observed in EYA1-silenced MDA-MB-231 cells (Figure S6A). Similarly, genotoxic stress induced by CPT treatment reduced cell survival of both parental cell lines in a dose dependent manner; and EYA1-silenced cells were significantly more sensitive to CPT treatment (Figure 6E, S6B).
To ascertain the specificity of shRNA targeting constructs, wild type mouse Eya1 was used to rescue effects observed in EYA1-silenced cells. Expression of mouse Eya1, which was insensitive to human shEYA1 targeting vectors, effectively restored drug resistance to Eya1-silenced cells to an extent comparable to that of wild type control cells (Figure 6F), confirming that the observed phenotype was EYA1-specific. Thus, high levels of EYA1 confer to reduced sensitivity to and increased tolerance of breast cancer cells to PI3K/AKT inhibition and genotoxic stress.
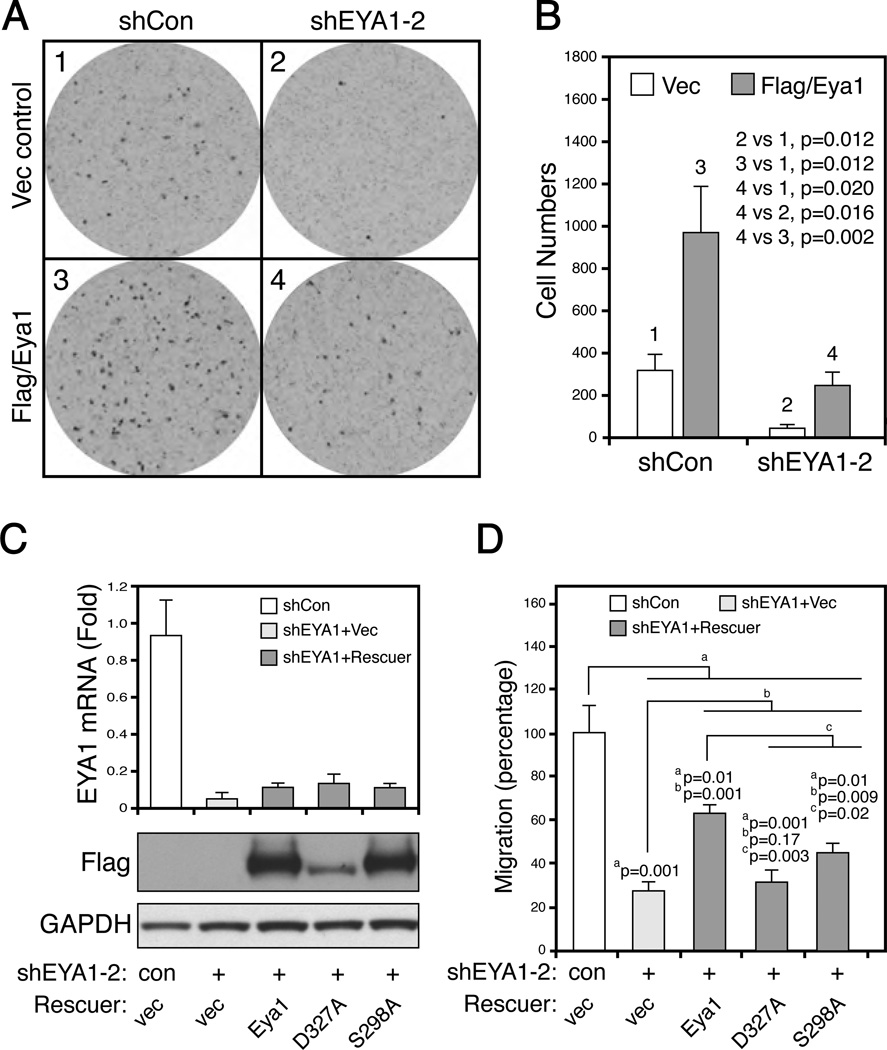
BT549 cells are highly migratory, a hallmark of aggressiveness of invasive cancer cells (Figure 7A and B). Over-expression of wild type mouse Eya1 resulted in a 3-fold increase of cell migration than control cells (Figure 7A and B, compare 1 and 3, n=3, p=0.012). Conversely, silencing of endogenous EYA1 gene significantly reduced cell migration (Figure 7A, compare 1 and 2, n=3, p<0.007). The cell migration defect was partially rescued by wild type mouse Eya1 (Figure 7A and B, compare 1 and 4).
Figure 7. Eya1-mediated cell motility depends on the Akt phosphorylation site.
(A–B) Migration of control BT549 cells (shCon, 1), EYA1-silenced (shEYA1-2), and the corresponding cells rescued with wild type mouse Flag/Eya1, were assayed over a 2h period. Quantified results were shown in B (mean ± SEM, n=3).
(C) Several murine Eya1 variants were stably expressed in EYA1-silenced BT549 cells. The upper bar graph, real-time quantitative PCR assay; the bottom blots, immunoblots of total cell lysate using antibodies specific to Flag (for murine Eya1 variants) and GAPDH (internal loading control). Con, scrambled shRNA; vec, empty expression construct; Eya1, wild type mouse Eya1; D327A, phosphatase-dead mutation; S298A, a phosphorylation-defective mutation. All rescue constructs were Flag tagged.
(D) Migration rate of stable BT549-derived cell lines was examined over a 4 h period. Number of cells was counted and quantitatively analyzed (mean ± SEM, n=3).
Having established the genetic rescue system to assay Eya1 functions, we next aimed to investigate whether phosphorylation was required for endogenous EYA1. Pools of stable EYA1-silenced BT549 cells were established (Figure 7C). Several murine Eya1 variants were subsequently stably expressed in EYA1-silienced cells to examine their ability to rescue the cell migration defect. Wild type and S298A mutant Eya1 variant had a similar level of expression in these stable cell lines (Figure 7C). D327A phosphatase-dead mutant was also expressed but with relatively lower expression level. EYA1 knockdown resulted in more than a 70% reduction of cell migration (Figure 7D, n=3, p=0.001). Expression of wild type mouse Eya1 partially but significantly rescued the migration defect (Figure 7D, n=3, p=0.001). Phosphatase-dead D327A variant was unable to revert the defects, confirming a previous report suggesting that EYA1 phosphatase activity is required for cancer cell motility and invasiveness.8 Interestingly, mutation of Akt phosphorylation site, S298A, significantly attenuated its effectiveness to rescue the migration defect (n=3, p=0.02) (Figure 7D). These results suggest that Eya1-dependent breast cancer motility partially depends on the conserved Akt substrate phosphorylation site, S298.
Discussion
In this study, we provide the first evidence suggesting a functional interaction between the PI3K/Akt signal pathway and Eya1 transcription coactivator. These findings further suggest that the PI3K/Akt-dependent regulation of substrate SUMOylation is an important aspect of tumorigenesis.
The PI3K/Akt/Eya1 axis in breast cancer
PI3K/Akt signaling is often hyperactivated in cancer cells and its hyperactivation promotes tumor progression. Our studies extend this notion by revealing a previously unknown role for the PI3K/Akt/Eya1 axis in breast cancer cell proliferation, migration and stress resistance. Several lines of evidence support the notion that the PI3K/Akt1 signal regulates Eya1. We show that Eya1 is a candidate substrate of Akt1 kinase (Figure 1); and phosphorylation of Eya1 by PI3K/Akt signaling enhances Eya1 transcription activity by inhibiting Eya1 SUMOylation (Figures 2 and 3). It is worth noting that genetic silencing of Eya1 significantly sensitizes breast cancer cells to pharmacological inhibition of PI3K/Akt signaling, suggesting that they may function together to regulate cancer cell behavior (Figure 6). Indeed, Akt1 and Eya1 synergistically regulate kidney growth in vivo (Figure S2); wild type but not the phosphorylation site mutant Eya1 variant is able to rescue a migration phenotype of EYA1-silienced breast cancer cells (Figure 7), indicating that phosphorylation is important for endogenous Eya1 functions.
A predicted phosphorylation site of Eya1 protein is S298, which is located at the highly conserved Akt substrate consensus motif (Figure 1). We show that mutation of this site (S298A) significantly diminished IP-kinase labeling and phospho-specific immunoreactivity (Figure 1). Eya1(S298A) variant becomes insensitive to PI3K/Akt signaling with regard to Eya1 transcription activity (Figure 2) and SUMOylation level (Figure 4); and S298A is unable to substitute wild type Eya1 function in a genetic rescue experiment (Figure 7). Furthermore, our findings suggest that S298 is required to keep Eya1 at hypo-SUMOylated state (Figure 5). We also noted that in addition to S298, Eya1 might have other Akt phosphorylation sites, since S298A mutant is still reactive Akt phospho-substrate specific antibody (Figure 1). Taken together, these observations suggest that S298 is a critical component linking Eya1 to PI3K/Akt signaling.
SUMOylation inhibits Eya1 transcription activity
Results here suggest that SUMOylation inhibits Eya1 transcription. A systemic mutagenesis analysis indicates that Eya1 is SUMOylated at K43 and K146 sites (Figure 3). Eya1 mutant variants that lack these sites are significantly more potent than wild type Eya1 to activate transcription reporters and downstream endogenous target gene expression (Figure 3 and S3). In addition, reduction of Eya1 SUMOylation by Senp2 also potentiates Eya1 transcription activity. Conversely, Eya1 and SUMO1 fused proteins, which mimic SUMOylated Eya1, significantly inhibited transcription reporter gene activities.
Paradoxically, murine Sumo1 heterozygous deletion augments Eya1+/− cleft palate phenotype.18 suggesting that development of the hard palate depends on both Eya1 and SUMOylation. Deletion of Sumo1 may result in a global reduction of SUMOylation level of all substrates. Therefore, it is possible that other SUMOylated proteins in addition to Eya1 contribute to the cleft palate phenotype.18 In contrast to the genetic approach, our study was designed to address specifically the impact of SUMOylation on Eya1 without affecting other SUMO-substrates. These experimental differences may contribute to the discrepancy. It is also worth noting that cleft palate but not renal phenotype was reported in the Sumo1 and Eya1 compound heterozygous.18 This is important because renal development is particularly sensitive to Eya1 level.29 It is therefore possible that the functional relationship between Eya1 and SUMOylation maybe cell type-dependent.
PI3K/Akt signaling inhibits Eya1 SUMOylation
We report here that PI3K/Akt regulates Eya1 activity via a novel SUMOylation mechanism (Figure 4). Activation of the PI3K/Akt signal pathway represses Eya1 SUMOylation level, which is in part mediated through S298 residue. A critical remaining question is how Eya1 phosphorylation inhibits its SUMOylation. Early studies reveal that Dach1 and Six1 are biochemical and genetic partners of Eya1.10, 45 Interestingly, Ubc9, a critical SUMO-conjugating enzyme was identified in the yeast two-hybrid screening that directly interacts with Dach1.46 Many candidate Eya1 interacting partners have since been identified but their function in Eya1 SUMOylation has not been explored yet.47 It is conceivable that SUMOylation machinery might be recruited through Eya1 interacting partners, such as Dach1; and such recruitment could be regulated by PI3K/Akt signaling.
The PI3K/Akt signal pathway plays a prominent role is physiology and diseases. To the best of our knowledge, this is the first example that it hyperactiviates an oncoprotein through a phosphorylation-dependent SUMOyation mechanism. Intriguingly, Akt1 is also SUMOylated; and SUMOylation promotes Akt1 kinase activity.48 Taken together, these observations suggest that crosstalk between the PI3K/Akt signal pathway and SUMOylation mechanism is likely a critical aspect of cancer biology.
Materials and Methods
Mouse strains
All animal studies were performed according to protocols reviewed and approved by the Institutional Animal Care and Use Committee at the Children’s Hospital Boston. Development of heterozygous Eya1 mouse strains was described previously6. Akt1 heterozygous mice were purchased from Jackson Laboratory (Stock number: 004912), donated by Morris Birnbaum31. PCR genotyping was performed as described previously6, 31. Akt1 and Eya1 compound heterozygous mutants, derived from an Eya1+/− and Akt1+/− cross, were used to generate appropriate control and experimental groups. All mice were maintained on C57BL6, 129svj, or CD-1 mixed genetic backgrounds. P0 pups were collected and weighed. Kidneys were dissected and weighed. Bodies and kidneys were imaged on an Olympus DP71 digital camera.
Plasmids and reagents
Six1 reporter constructs, SE1/luc and SE2/luc, were described previously.1, 7 The reporters contained Six1 binding elements (annealing by 5’-TCG AGT AAT CCG AGA CCC TGT AAA TTG ATA CGG AGT AAT TCG ATA TCG A-3’, 5 ’-AGC TCA TTA GGC TCT GGG ACA TTT AAC TAT GCC TCA TTA AGC TAT AGC T-3’) with basal promoters with minimal and high activities, respectively. The Fgf8 promoter reporter was described previously.7 Akt1 expression constructs and siAkt SMARTpool reagents (Thermo Scientific Dharmacon) were obtained from Dr. Rosalyn Adam (Boston Children's Hospital). The lentiviral constructs of Eya1 overexpression and the shRNA were subcloned into pLentiCMV (from Dr. Lan Xu, UMass Med, Worcester) and pLKO.1 vector, respectively.
Cell lines
HEK293, BT549 and MDA157 (from Dr. Joan Brugge, Harvard Medical School) and SUM14949 (from Dr. Charlotte Kuperwasser, Tufts University), were maintained using standard procedures. The stable pools of EYA1 knockdown and Eya1 rescue cells were generated with EYA1 shRNA or Eya1 lentiviral particles and selected with puromycin (2 µg/ml) or blasticidin (10 µg/ml), respectively. Cell proliferation and viability were measured by either cell number counts or using MTT (3-(4,5-Dimethylthiazol-2-yl) - 2,5 - diphenyltetrazolium bromide) assays. For γ-irradiation, cells were treated with a single dose of gamma rays from Cesium-137 (Boston Children’s Hospital), and allowed to recover at 37°C for the indicated times before analysis.
Real-time quantitative PCR
Real-time quantitative PCR was performed using VeriQuest™ SYBR® Green (Affimetrix) on an ABI StepOnePlus (Applied Biosystems). PCR primers were 5’-ACT TAC CAG CTT CAA GAA CCG-3' and 5'-ATT TCC CAT CTG AAC CTC GAC-3' for EYA1; 5’-CAC AGC CCA CTG GTC CTC AAG AGG-3’ and 5’- TGT TTC AAC TGT TCT CGT CGT TTC C-3’ for c-MYC; 5’- GCC CTC TGT GCC ACA GAT GTG AAG-3’ and 5’-GTT CTG CTG GGC CTG GCG CAG GCT-3’ for CYCLIN D1; 5’-CGC TGA GCT GCC TGC TGT TGC-3’ and 5’-GGT AGG TCC GGA TGA GGC GGC-3’ for FGF8; 5’-GCC TCA AGA TCA TCA GCA AT-3’ and 5’-TTC AGC TCA GGG ATG ACC TT-3’ for GAPDH.
Cell migration and soft agar assays
A total of 5×104 cells in 100 µl of serum free medium were placed in the inserts of Costar Transwell® (8.0 µm pore size, Corning). The bottom chamber was filled with complete medium containing 20% fetal bovine serum. After incubation at 37°C, 5% CO2 for 2 – 4 hours, slowly migrating cells on the top layer of the Transwell were removed using cotton swabs. The cells remaining that had migrated and attached to the bottom layer of Transwell inserts were fixed with 100% methanol and stained with 0.1% crystal violet. Total cell number was counted under an Olympus SZX16 microscope and imaged using a DP71 digital camera.
Luciferase assay
Luciferase assays were performed using Dual-Luciferase assay system (Promega) in HEK293 cells) and reporter activities were measured using a Wallac 1420 Multilabel Counter (Perkin Elmer luminometer) and normalized with pRL-TK renilla.
Immunoprecipitation and in vitro kinase assay
The expression plasmids were transfected 293 cells by Lipofectamine 2000 (Invitrogen). The cells were lysed with IP buffer (1.0% Triton X-100, 50 mM Tris-HCl pH 7.6, 300 mM NaCl, 1 mM EDTA, 2.5 mM Sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 5 mM NaF, 1 mM PMSF and protease inhibitor cocktail). Immunoprecipitation were done using standard protocol. For in vitro kinase assay, Flag-tagged proteins were immunoprecipitated by anti-Flag affinity gel (Sigma-Aldrich) and eluted by 3XFlag peptide (150 ng/µl in 50 mM Tris-HCl pH7.6, 150 mM NaCl). Eya1 and S298A mutant proteins and recombinant active AKT1 (R&D Systems) were mixed and incubated in kinase reaction buffer (5 mM MOPS pH 7.2, 2.5 mM beta-glycerophosphate, 5 mM MgCl2, 1mM EGTA, 0.4 mM EDTA, 0.4% glycerol, 5 µM ATP and 5 µCi gamma-[32P]-ATP) at room temperature for 30 min. Reaction products were subjected to SDS-PAGE and transferred to PVDF membrane. The phosphorylation of Eya1 was detected by exposing to X-Ray film. Eya1 protein levels were examined by protein immunoblot using an anti-Flag-HRP antibody.
Statistics
Data analysis was performed with MedCalc software. Paired Student’s t test was used to determine the differences between groups. Data are presented as mean ± SEM (n = 3). P values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgements
We thank S. Morley for suggestions and critical reading of the manuscript. We thank I. Teng, C.S. Guo, X.J. Shi, Z.F. Chai, and K. Pelton for technical supports. We are grateful to R. Adam, G. Gill, J. Brugge, C. Kuperwasser for providing reagents, cell lines and helpful discussion. This work was supported by grant from NIH/NIDCR (1R01DE019823, XL), NIH/NIDDK (R01DK916451 and P50DK65298, XL) and AHA (13GRNT16950006, XL).
Footnotes
Conflict of Interest
Authors declare that they have no conflict interest.
References
- 1.Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 2.Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, et al. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- 3.Rayapureddi JP, Kattamuri C, Steinmetz BD, Frankfort BJ, Ostrin EJ, Mardon G, et al. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003;426:295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- 4.Okabe Y, Sano T, Nagata S. Regulation of the innate immune response by threonine-phosphatase of Eyes absent. Nature. 2009;460:520–524. doi: 10.1038/nature08138. [DOI] [PubMed] [Google Scholar]
- 5.Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 6.Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- 7.Guo C, Sun Y, Zhou B, Adam RM, Li X, Pu WT, et al. A Tbx1-Six1/Eya1-Fgf8 genetic pathway controls mammalian cardiovascular and craniofacial morphogenesis. J Clin Invest. 2011;121:1585–1595. doi: 10.1172/JCI44630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandey RN, Rani R, Yeo EJ, Spencer M, Hu S, Lang RA, et al. The Eyes Absent phosphatase-transactivator proteins promote proliferation, transformation, migration, and invasion of tumor cells. Oncogene. 2010;29:3715–3722. doi: 10.1038/onc.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farabaugh SM, Micalizzi DS, Jedlicka P, Zhao R, Ford HL. Eya2 is required to mediate the pro-metastatic functions of Six1 via the induction of TGF-beta signaling, epithelial-mesenchymal transition, and cancer stem cell properties. Oncogene. 2012;31:552–562. doi: 10.1038/onc.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- 11.Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, et al. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999;19:6815–6824. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coletta RD, Christensen K, Reichenberger KJ, Lamb J, Micomonaco D, Huang L, et al. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci U S A. 2004;101:6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCoy EL, Iwanaga R, Jedlicka P, Abbey NS, Chodosh LA, Heichman KA, et al. Six1 expands the mouse mammary epithelial stem/progenitor cell pool and induces mammary tumors that undergo epithelial-mesenchymal transition. J Clin Invest. 2009;119:2663–2677. doi: 10.1172/JCI37691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu K, Li Z, Cai S, Tian L, Chen K, Wang J, et al. EYA1 Phosphatase Function Is Essential To Drive Breast Cancer Cell Proliferation Through Cyclin D1. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-12-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Micalizzi DS, Christensen KL, Jedlicka P, Coletta RD, Baron AE, Harrell JC, et al. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-beta signaling. J Clin Invest. 2009;119:2678–2690. doi: 10.1172/JCI37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ford HL, Kabingu EN, Bump EA, Mutter GL, Pardee AB. Abrogation of the G2 cell cycle checkpoint associated with overexpression of HSIX1: a possible mechanism of breast carcinogenesis. Proc Natl Acad Sci U S A. 1998;95:12608–12613. doi: 10.1073/pnas.95.21.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reichenberger KJ, Coletta RD, Schulte AP, Varella-Garcia M, Ford HL. Gene amplification is a mechanism of Six1 overexpression in breast cancer. Cancer Res. 2005;65:2668–2675. doi: 10.1158/0008-5472.CAN-04-4286. [DOI] [PubMed] [Google Scholar]
- 18.Alkuraya FS, Saadi I, Lund JJ, Turbe-Doan A, Morton CC, Maas RL. SUMO1 haploinsufficiency leads to cleft lip and palate. Science. 2006;313:1751. doi: 10.1126/science.1128406. [DOI] [PubMed] [Google Scholar]
- 19.Dunnebier T, Bermejo JL, Haas S, Fischer HP, Pierl CB, Justenhoven C, et al. Common variants in the UBC9 gene encoding the SUMO-conjugating enzyme are associated with breast tumor grade. Int J Cancer. 2009;125:596–602. doi: 10.1002/ijc.24286. [DOI] [PubMed] [Google Scholar]
- 20.Dunnebier T, Bermejo JL, Haas S, Fischer HP, Pierl CB, Justenhoven C, et al. Polymorphisms in the UBC9 and PIAS3 genes of the SUMO-conjugating system and breast cancer risk. Breast Cancer Res Treat. 2010;121:185–194. doi: 10.1007/s10549-009-0530-y. [DOI] [PubMed] [Google Scholar]
- 21.Synowiec E, Krupa R, Morawiec Z, Wasylecka M, Dziki L, Morawiec J, et al. Efficacy of DNA double-strand breaks repair in breast cancer is decreased in carriers of the variant allele of the UBC9 gene c.73G>A polymorphism. Mutat Res. 2010;694:31–38. doi: 10.1016/j.mrfmmm.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Moschos SJ, Jukic DM, Athanassiou C, Bhargava R, Dacic S, Wang X, et al. Expression analysis of Ubc9, the single small ubiquitin-like modifier (SUMO) E2 conjugating enzyme, in normal and malignant tissues. Hum Pathol. 2010;41:1286–1298. doi: 10.1016/j.humpath.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Bawa-Khalfe T, Lu LS, Zuo Y, Huang C, Dere R, Lin FM, et al. Differential expression of SUMO-specific protease 7 variants regulates epithelial-mesenchymal transition. Proc Natl Acad Sci U S A. 2012;109:17466–17471. doi: 10.1073/pnas.1209378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellacosa A, Kumar CC, Di Cristofano A, Testa JR. Activation of AKT kinases in cancer: implications for therapeutic targeting. Adv Cancer Res. 2005;94:29–86. doi: 10.1016/S0065-230X(05)94002-5. [DOI] [PubMed] [Google Scholar]
- 25.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller TW, Rexer BN, Garrett JT, Arteaga CL. Mutations in the phosphatidylinositol 3-kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res. 2011;13:224. doi: 10.1186/bcr3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, et al. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J Biol Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 28.Xu Q, Yu L, Liu L, Cheung CF, Li X, Yee SP, et al. p38 Mitogen-activated protein kinase-, calcium-calmodulin-dependent protein kinase-, and calcineurin-mediated signaling pathways transcriptionally regulate myogenin expression. Mol Biol Cell. 2002;13:1940–1952. doi: 10.1091/mbc.02-02-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sajithlal G, Zou D, Silvius D, Xu PX. Eya 1 acts as a critical regulator for specifying the metanephric mesenchyme. Dev Biol. 2005;284:323–336. doi: 10.1016/j.ydbio.2005.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, et al. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 2001;15:2203–2208. doi: 10.1101/gad.913901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001;276:38349–38352. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- 32.Easton RM, Cho H, Roovers K, Shineman DW, Mizrahi M, Forman MS, et al. Role for Akt3/protein kinase Bgamma in attainment of normal brain size. Mol Cell Biol. 2005;25:1869–1878. doi: 10.1128/MCB.25.5.1869-1878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol Cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 35.Yang XJ, Gregoire S. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol Cell. 2006;23:779–786. doi: 10.1016/j.molcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Ouyang J, Shi Y, Valin A, Xuan Y, Gill G. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell. 2009;34:145–154. doi: 10.1016/j.molcel.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross S, Best JL, Zon LI, Gill G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol Cell. 2002;10:831–842. doi: 10.1016/s1097-2765(02)00682-2. [DOI] [PubMed] [Google Scholar]
- 38.Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 39.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell. 2008;30:203–213. doi: 10.1016/j.molcel.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 41.Kao J, Salari K, Bocanegra M, Choi YL, Girard L, Gandhi J, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One. 2009;4:e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.She QB, Chandarlapaty S, Ye Q, Lobo J, Haskell KM, Leander KR, et al. Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to Akt signaling. PLoS One. 2008;3:e3065. doi: 10.1371/journal.pone.0003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torbett NE, Luna-Moran A, Knight ZA, Houk A, Moasser M, Weiss W, et al. A chemical screen in diverse breast cancer cell lines reveals genetic enhancers and suppressors of sensitivity to PI3K isoform-selective inhibition. Biochem J. 2008;415:97–110. doi: 10.1042/BJ20080639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- 46.Machon O, Backman M, Julin K, Krauss S. Yeast two-hybrid system identifies the ubiquitin-conjugating enzyme mUbc9 as a potential partner of mouse Dac. Mech Dev. 2000;97:3–12. doi: 10.1016/s0925-4773(00)00402-0. [DOI] [PubMed] [Google Scholar]
- 47.Tadjuidje E, Hegde RS. The Eyes Absent proteins in development and disease. Cell Mol Life Sci. 2013;70:1897–1913. doi: 10.1007/s00018-012-1144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li R, Wei J, Jiang C, Liu D, Deng L, Zhang K, et al. Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-13-0538. [DOI] [PubMed] [Google Scholar]
- 49.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.