Abstract
Fungi in paranasal sinuses are characteristic and considered a major pathogenic factor in a subset of chronic rhinosinusitis (CRS) patients, known as allergic fungal rhinosinusitis (AFRS). CD8+ T cells are enriched in AFRS sinuses but their role in fungal-specific responses is unknown. Alternaria alternata– and Aspergillus fumigatus–specific T lymphocyte responses were investigated in 6 AFRS patients, 10 eosinophilic mucus CRS (EMCRS) patients, 10 CRS with nasal polyps (CRSwNPs) patients, 6 allergic rhinitis with fungal allergy (ARFA) patients, and five controls. Fungal-specific proliferation of human peripheral blood mononuclear cells (PBMCs) was studied prospectively. Proliferating cells were examined for CD3, CD4, CD8, and CD25 expression. Relevant clinical characteristics, fungal allergy, detection of fungi in sinuses, and CD4+ and CD8+ composition of sinus T cells were also examined. CD4+ T-cell division to fungi occurred in all samples, regardless of fungal allergy or CRS. Fungal-specific CD8+ T-cell division occurred in all ARFA and control samples and the majority of CRSwNP patients; however, CD8+ T cells failed to proliferate in AFRS and EMCRS patients. The CD8+ T cells from AFRS patients also did not up-regulate the activation marker, CD25, with fungal antigen exposure. Presence of A. alternata– and A. fumigatus–specific CD4+ and CD8+ T-cell proliferation in healthy individuals, ARFA, and CRSwNP patients suggests that both T-cell subsets may be important in immune responses to these fungi. In AFRS and EMCRS patients, only fungal-specific CD4+ T-cell proliferation occurred; hence, a lack of CD8+ T-cell proliferation and activation in the presence of sinus eosinophilic mucus in these patients, regardless of fungal allergy, is a novel finding. This raises the question whether a dysfunctional CD8+ T-cell response predisposes to ineffective clearance and accumulation of fungi in the sinuses of susceptible patients.
Keywords: Allergic fungal rhinosinusitis, allergic fungal sinusitis, Alternaria, Aspergillus, chronic rhinosinusitis, CD4+, CD8+, dysfunctional, eosinophilic mucus, fungi, human, lymphocytes, mucosa, nasal polyp, proliferation, prospective, sinus, T cells, tissue
Allergic fungal rhinosinusitis (AFRS) refers to a subset of chronic rhinosinusitis with nasal polyp (CRSwNP) patients who have characteristic clinical features and with coexisting fungal allergy and fungi within their sinus eosinophilic mucus.1 AFRS constitutes ∼4–10% of CRS patients and is a recalcitrant NP disease that is difficult to treat and tends to recur.2 An IgE-mediated allergic response to fungi is generally believed to cause AFRS; however, more recent studies have questioned the pathogenic importance of fungal allergy alone.2–4 Fungal hyphae, especially of Alternaria alternata and Aspergillus fumigatus within the sinuses, but without overt tissue invasion is a typical finding in AFRS and also in some nonallergic CRS patients with NPs who have sinus eosinophilic mucus, known as eosinophilic mucus CRS (EMCRS). Currently, it is not understood why fungi accumulate in the sinuses of AFRS and EMCRS patients whereas fungal antigen exposure is common without buildup in healthy individuals, fungal allergic rhinitis patients, and in other forms of CRS diseases with secondary mucociliary dysfunction.5 Although host immune susceptibility is suspected as a contributing factor in AFRS, fungal-specific immune responses have not been thoroughly investigated.
Both CD4+ and CD8+ T cells are important in immunity to fungi.6 A. alternata and A. fumigatus antigens can induce allergic and nonallergic responses. T-cell responses to A. alternata and A. fumigatus antigens in healthy individuals have been described, with both CD4+ and CD8+ peripheral blood (PB) T-cell proliferation shown to several A. fumigatus antigens.7 The presence and magnitude of fungal-specific CD4+ and CD8+ T cells can vary between individuals and is believed to reflect the nature of the local inflammatory response. CD4+ T cells are considered the primary effector cell in protective antifungal immunity but the role of fungal-specific CD8+ T cells is not fully understood.8–10
We hypothesized, based on their clinically distinct phenotype and presence of fungi in their sinuses, that AFRS and EMCRS patients might have a distinctive fungal-specific T-cell response compared with CRSwNPs, non-CRS fungal allergic rhinitis patients, and healthy controls (HCs). Hence, the purpose of this study was to prospectively investigate the magnitude and phenotype of A. alternata– and A. fumigatus-–specific CD4+ and CD8+ T-cell proliferation in PB of AFRS compared with EMCRS and CRSwNP patients. Carboxyfluorescein succinimidyl ester (CFSE) cytometry was used to directly identify the phenotype of T cells that underwent division in response to fungal stimulation.11 PB from HCs and allergic rhinitis with fungal allergy (ARFA) patients, relevant clinical characteristics, fungal allergy, presence of fungi in sinuses, and CD4+ and CD8+ composition of sinus T cells were also examined.
METHODS
The Human Research Ethics Committee for the participating hospitals (The Queen Elizabeth Hospital and Memorial Hospital) approved this study. Voluntary informed consent was obtained from all participants. Patients and controls were recruited prospectively from rhinology and general otolaryngology clinics at the time of surgery.
Patients and Controls
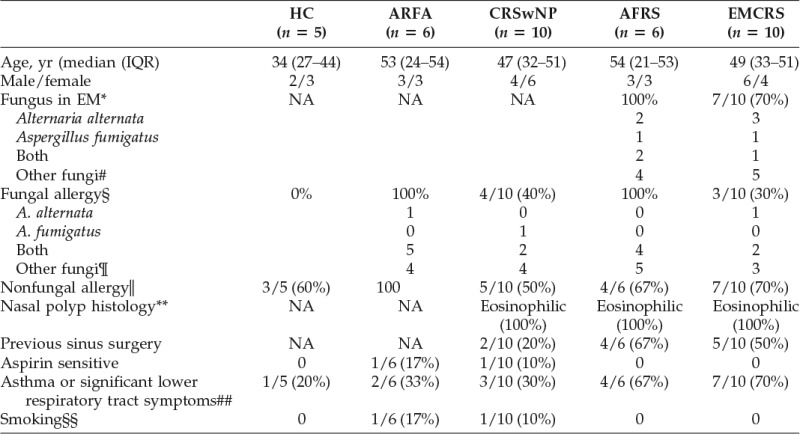
Thirty-two patients and five HCs were recruited. Clinical history, CRS symptom scores, clinical examination, nasal endoscopy, sinus computed tomography (CT) scans, skin-prick tests, serology, relevant laboratory findings, sinus mucus histopathology, fungal culture, and sinus tissue histopathology were available for stratification into the study groups as previously described.12 Twenty-six CRSwNP patients undergoing surgical treatment and six ARFA patients who had fungal allergy and rhinitis but no sinus disease on nasal endoscopy and sinus CT were recruited. HCs did not have sinonasal symptoms (22-item Sino-Nasal Outcome Test score of <1) and had normal nasal endoscopy and a negative skin-prick test or serum IgE to molds.
The CRSwNP patients were defined according to the European Position Paper on Rhinosinusitis and on Nasal Polyps criteria13 and included a >12-week duration of sinonasal symptoms and also endoscopic and sinus CT evidence of sinus mucosal disease and nasal endoscopy evidence of NPs. CRSwNP patients were further subclassified into EMCRS and non-EMCRS patients. EMCRS was defined as CRSwNP patients who had thick, tenacious-colored mucus in the sinuses at surgery, confirmed to be eosinophilic mucus by histology. EMCRS patients with coexisting fungal allergy and a positive sinus EM fungal culture or fungal elements were defined as AFRS patients. CRSwNP patients who did not have sinus eosinophilic mucus are referred to as CRSwNP patients.
All CRS patients undergoing sinus surgical treatment had previously failed medical therapy and intraoperatively had active sinus disease. Medical therapy consisted of intranasal saline, corticosteroid sprays, a 3-week course of a tapering dose of oral prednisolone and in those with endoscopic evidence of purulence, 3 weeks of culture-directed antibiotics.
Exclusion criteria were known coexisting medical problems causing CRS, such as Churg-Strauss syndrome, cystic fibrosis, and immune deficiency; allergic bronchopulmonary aspergillosis; and/or current treatment with systemic corticosteroids or other immunosuppressive therapy at the time of surgery. Patients with a dental etiology for sinusitis, fungus ball, and mucocele were also excluded. Participants had not smoked, had no upper respiratory tract infection, or used topical or systemic corticosteroids, antibiotics, antihistamines, anticholinergics, or homeopathic preparations for a minimum of 4 weeks before collection of tissue and blood samples.
Diagnostic Tests
All CRS and ARFA patients had a sinus CT scan. All participants underwent a nasal endoscopy, had a full blood differential cell count, and allergen-specific serum IgE was tested using the ImmunoCAP system (Uni-CAP-100; Pharmacia Diagnostics AB, Uppsala, Sweden). Skin-prick testing was performed using antigen extracts from Hollister-Stier Laboratories, LLC (Spokane, WA) for the following allergens: mixes of tree pollen, grass pollen, weed pollen, house-dust group, and animal dander and protein. Specific testing for individual molds were performed with skin-prick tests as detailed in the footnote of Table 1.
Table 1.
Patient demographics and clinical characteristics
*By histology or fungal culture.
#Bipolaris, Drechslera, Trichothecium,Candida, Penicillium, Scedosporium, Acremonium, Devriessi, Phialaphora, and Cladosporium species.
§By positive fungal-specific serum IgE or skin-prick test.
¶A. alternata, A. fumigatus, A. nidulans, A. niger, Bipolaris species, Candida albicans, Cladosporium mix (Cladosporium normodendrum, Cladosporium herbarum, and Cladosporium cladosporoides), Epicoccum nigrum, Fusarium vasinsectum, Helminthosporium species, Mucor racemosus, Penicillium mix Penicillium digitatum, Penicillium expansum, and Penicillium notatum), Rhizopus nigricans, Pullularia pullulans, and Trichophyton mix (Trichophyton mentogrophytes, Trichophyton rubrum, and Trichophyton tonsuranis).
‖Mixes of tree pollen, grass pollen, house-dust mite and animal dander.
**Histopathology report from routine diagnostic test.
##Asthma diagnosis inconclusive in 2/4 AFS and 1/7 EMCRS patients with significant lower respiratory tract symptoms.
§§History of cigarette smoking up to 4 weeks before surgery.
AFRS = allergic fungal rhinosinusitis; ARFA = allergic rhinitis with fungal allergy; CRSwNPs = chronic rhinosinusitis with nasal polyps; EM = eosinophilic mucus; EMCRS = eosinophilic mucus chronic rhinosinusitis; HC = healthy controls; IQR = interquartile range (25th–75th percentile); NA = not applicable.
During surgery, tissue samples and mucus were sent fresh for bacterial culture and fixed in 10% buffered formaldehyde for histopathology studies including evaluation for eosinophilic mucus. Eosinophilic mucus was diagnosed when clusters of eosinophils were observed in an amorphous mucus background on hematoxylin and eosin–stained mucus specimens.3
Sinus and PB Samples
Polyps were sampled from the nasal cavity, middle and superior meatus, and ethmoid sinuses from CRSwNP, AFRS, and EMCRS patients. Matched heparinized venous PB samples were obtained from every patient during sinus surgery for comparative analysis. Only PB samples were studied in ARFA and HCs.
Single cell preparation of fresh sinus tissue samples for flow cytometry was performed as previously described, and experiments were performed within a few hours.14 Briefly, whole pieces of tissue were washed to remove macroscopic blood and were minced to <2-mm pieces and passed through an 80-μm nylon mesh (BD Biosciences, San Jose, CA) to isolate single cells without the use of digestive enzymes. Lymphocyte viability was determined by propidium iodide staining and was >92% and viable cells were used for flow cytometry.
PB mononuclear cells (PBMCs) for proliferation and flow cytometry studies were prepared by standard density gradient centrifugation (Lymphoprep; Fresenius Kabi, Bad Homburg, Germany) of heparinized venous PB samples.
Proliferation Assays
Fungal Antigens.
Alternaria tenuis (alternata; Cat. No. 5009JF10) and A. fumigatus (Cat. No. 5021JF10), were from Hollister-Stier Laboratories, LLC. The reported value for fungal-specific proliferative responses in an individual corresponds to the highest value obtained for proliferation to either fungal preparation.
CFSE Staining.
5-(6)-Carboxyfluorescein diacetate succinimidyl ester (Mr 557; Molecular Probes, Eugene, OR), was added to 107 mononuclear cells suspended in 1 mL of PBS to final concentration of 10 μM. The suspension was mixed immediately and incubated at 37°C for 15 minutes. Cells were quenched in a fivefold volume of ice-cold RF5 (RPMI-1640 supplemented with 100 U/mL of penicillin, 0.1 mg/mL of streptomycin, 0.3 mg/mL of glutamine, and 5% fetal calf serum) and incubated on ice for 5 minutes. Cells were washed three times in a fivefold volume of RF5 to ensure removal of extracellular 5-(6)-carboxyfluorescein diacetate succinimidyl ester.
Proliferation Assay.
These were conducted as described previously.4 PBMCs were washed twice before suspension at 107 cells/mL in RF5. Two sets of proliferation assays, for tritiated thymidine analysis and for CFSE cytometry were conducted for each individual. The 105 cells in a final volume of 200 μL/well were added in triplicate to flat-bottom, 96-well tissue culture plates (Cell Star, Frickenhausen, Germany) and stimulated with fungal antigens at a final concentration of 7.5 μg/mL The same patient's unstimulated cells were kept in a 37°C 5% CO2 incubator for 36 hours. At 36 hours, tissue culture wells were washed and unstimulated cells were added to adherent antigen-primed cells.
Except for the variables tested, all fungal-specific proliferation assays were conducted under similar conditions. PBMCs from two individuals served as a negative and a positive control for fungal-specific proliferation along with test samples. Internal controls included (a) a negative control where cells were incubated with tissue culture medium alone and (b) a positive control where cells were incubated with phytohemagglutinin (PHA) at a final concentration of 12.5 μg/mL. In some experiments IL-2 was added to their respective wells at a final concentration of 25 U/mL. Percentages of CD4+ and CD8+ T cells, B cells, NK cells, monocyte, and macrophage populations were determined in PBMCs before setting up the proliferation assays.
Tritiated Thymidine Analysis.
Cells were harvested after 96 hours and the amount of thymidine incorporated by cells in response to stimulant was compared with that from unstimulated control cells to yield a stimulation index (SI).
CFSE Proliferation.
CFSE-labeled mononuclear cells were used to set up the fungal-specific proliferation assay described previously. At the end of culture period, cell fluorescence intensity was measured on a FACScan (BD Biosciences) in the 540-nm fluorescence channel 1 parameter. Internal controls for CFSE proliferation assays included (a) CFSE-labeled cells incubated for the same duration without mitogen thereby allowing the position of the undivided cells to be determined and (b) unlabeled cells that were stimulated under the same culture conditions thereby allowing the position of the autofluorescence intensity to be determined. At the end of the culture period, cells were colabeled with combinations of antibodies to CD3, CD4, CD8, and CD25 molecules to determine the phenotype of the dividing cells.
Flow Cytometry: Cell Staining, Acquisition, and Analysis
For cell surface immunostaining, sinus tissue single cell suspension, and PBMC were stained with anti-human monoclonal antibodies against CD3, CD4, CD8, CD14, CD16/56, CD19, CD25, CD45, and CD45RA (BD Biosciences). At least 15,000 gated events were acquired for each sample of PBMC and 10,000 gated events for sinus tissue on a FACS Canto II flow cytometer and analyzed using BD FACSDiva software Version 6.1.3 (BD Biosciences). Depending on the experiment, cell populations were selected using side scatter properties and CD45 or CD3 expression.
For cytometric analysis of CFSE-labeled cells, gates were placed on the blast transformation of lymphocytes represented by increased forward scatter and orthogonal scatter of light.11 Populations of CFSE+, CD4+, CD8+, and CD25+ cells in this gate were examined. Routinely 50,000 gated events were acquired from each sample. Cell viability was assessed by incubating cells with propidium iodide (Invitrogen, Grand Island, NY) for 15 minutes on ice. Nonspecific antibody binding was blocked using human pooled IgG (Intragam P; CSL, Parkville, Australia), and isotype-matched antibody controls (BD Biosciences) were used to define positive and negative staining.
Statistical Analysis
Statistical analysis for independent groups of data was performed with the Mann–Whitney U test for comparison between two groups and the Kruskal–Wallis test with Dunn's post hoc test for comparison between multiple groups using GraphPad Prism software, Version 4.0a for Macintosh (GraphPad, San Diego, CA). For paired groups of data, the Wilcoxon signed-rank test was used to compare between two groups. Data are presented as a median and the 25th–75th percentiles, the limits of the interquartile range (IQR). A value of p < 0.05 was considered significant.
RESULTS
The clinical characteristics of the study groups are shown in Table 1. The age and gender distribution between the study groups were not significantly different. Histopathology reports confirmed eosinophilic mucosal inflammation in all NP samples. A significantly greater proportion of AFRS and EMCRS patients had previous sinus surgeries and asthma compared with other study groups.
CD4+ and CD8+ T-Cell Populations in PB and Sinus Mucosa
The percentage of PB and sinus CD4+ and CD8+ T cells in the patient groups are summarized in Table 2. There was no significant difference in percentage of PB CD4+ and CD8+ T cells between the study groups. In sinus mucosa, AFRS and EMCRS patients had a significantly higher proportion of CD8+ T cells (p = 0.001) and lower CD4+ T cells compared with CRSwNP patients (p = 0.02, Kruskal–Wallis test). There was no significant difference in the percentages of sinus CD4+ and CD8+ T cells between AFRS and EMCRS patients.
Table 2.
Percentage of CD4+ and CD8+ T cells in PB and NPs
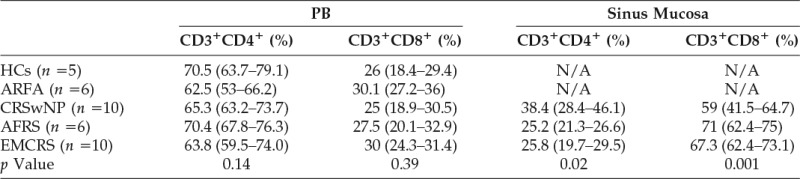
The CD4+ and CD8+ T-cell populations are a percentage of total CD3+ cells. The percentages are shown as the median value and IQR in parenthesis. The p values are derived from Kruskal–Wallis tests between the study groups, with post hoc Dunn test.
AFRS = allergic fungal rhinosinusitis ARFA = allergic rhinitis with fungal allergy; CRSwNPs = chronic rhinosinusitis with nasal polyps; EMCRS = eosinophilic mucus chronic rhinosinusitis; HCs = healthy controls; IQR = interquartile range; NP = nasal polyp; PN = peripheral blood.
Fungal-Specific PBMC Proliferation
The highest proliferative response to either A. alternata or A. fumigatus is plotted as an SI in Fig. 1 A. Compared with HCs, fungal-specific PBMC proliferation was greater in ARFA, CRSwNP, AFRS, and EMCRS (p = 0.007, Kruskal–Wallis test with post hoc Dunn test), with no significant difference between these groups. Interestingly, PBMC proliferation to PHA was much higher in AFRS and EMCRS compared with CRSwNPs, ARFA, and HCs (p = 0.03, Kruskal–Wallis test with post hoc Dunn test; Fig. 1 B).
Figure 1.
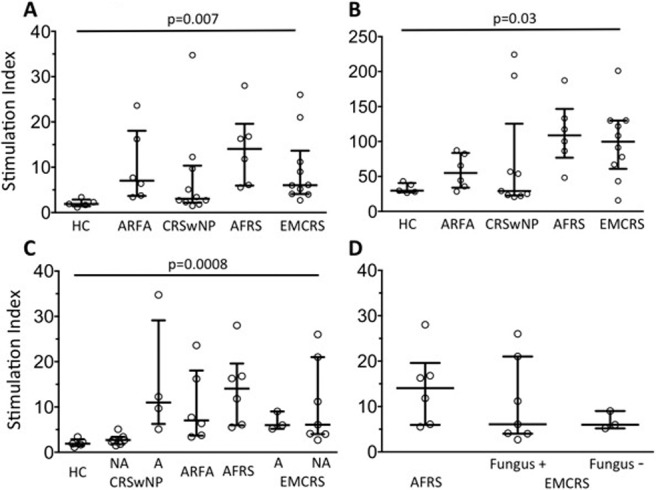
Peripheral blood mononuclear cell (PBMC) proliferation to fungal antigens (A) Alternaria alternata and Aspergillus fumigatus and (B) phytohemagglutinin (PHA) determined by thymidine incorporation expressed as a stimulation index (SI) in healthy controls (HCs; n = 5), allergic rhinitis with fungal allergy (ARFA; n = 6), chronic rhinosinusitis with nasal polyps (CRSwNPs; n = 10), allergic fungal rhinosinusitis (AFRS; n = 6), and eosinophilic mucus chronic rhinosinusitis (EMCRS; n = 10) patients. (C) The fungal antigen–specific SI results based on the presence (allergy [A]) or absence (no allergy [NA]) of fungal allergy in all study groups and (D) based on detection of fungi in sinus eosinophilic mucus of AFRS and EMCRS patients are shown. Each plot refers to a value obtained from one patient (the horizontal bar depicts the mean and vertical bars, the interquartile range [IQR]; p values, Kruskal–Wallis test, post hoc Dunn test).
A significant difference was noted in the fungal-specific proliferation SI between fungal-allergic and non–fungal-allergic groups (p = 0.0008, Kruskal–Wallis test, post hoc Dunn test; Fig. 1 C). In CRSwNPs, an increased response occurred in fungal-allergic individuals (p = 0.001). In contrast, there was no significant difference in the SI between fungal-allergic and nonallergic EMCRS patients (p > 0.05). Overall, the extent of fungal-specific response was similar among fungal-allergic CRSwNPs, ARFA, AFRS, and fungal-allergic and nonallergic EMCRS patients (p > 0.05). There was no significant difference in fungal-specific proliferation between AFRS and sinus fungus-positive and fungus-negative EMCRS patients (p > 0.05; Fig. 1 D).
Phenotype of PB T Cells That Underwent Fungal-Specific Proliferation
A representative gating strategy used in this study is shown in Fig. 2 and a summary of the PB proliferation results is presented in Table 3. In all individuals, PB CD4+ T cells underwent fungal-specific cell division. Interestingly, fungal-specific CD8+ T-cell division only occurred in (1/6) AFRS and (1/10) EMCRS patients compared with 8/10 CRSwNP patients and in all ARFA patients and HCs. PHA-induced cell division occurred in both CD4+ and CD8+ T-cell compartments in all individuals. A representative experiment from a CRSwNP and an AFRS patient is shown in Fig. 3.
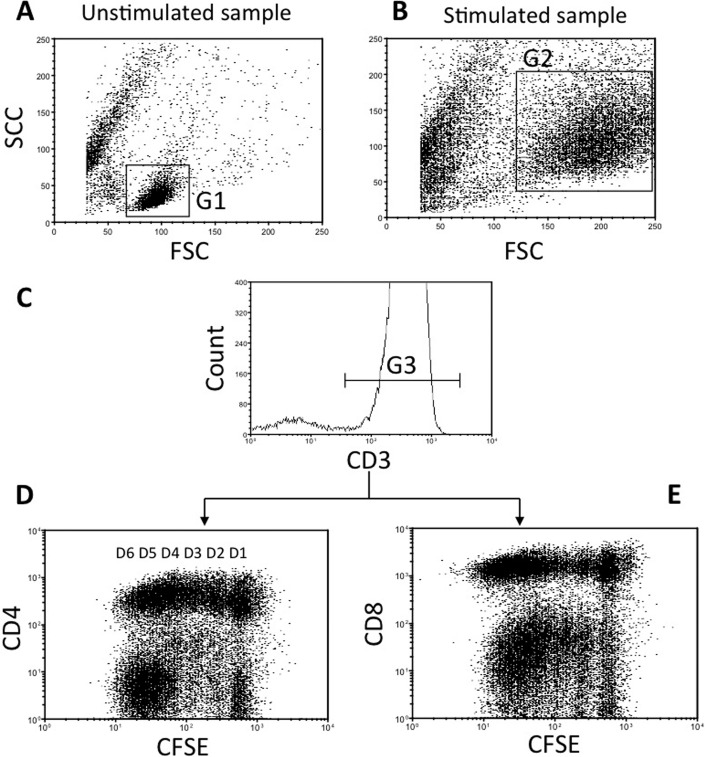
Figure 2.
Gating strategy for carboxyfluorescein succinimidyl ester (CFSE) cytometry profiles in peripheral blood mononuclear cells (PBMCs). This represents a patient's PBMC treated with phytohemagglutinin (PHA). (Aand B) The cells represented by dot plots and are gated for forward scatter (FSC) and side scatter (SSC) parameters. Gate (G) 1 is placed on resting PBMC in the unstimulated sample and gate 2 on blasts in the stimulated sample. (C)The histogram is gated (G3) on CD3+ cells in G1 and in G2. Depending on the experiment, cells are then analyzed for cell surface expression of (D) CD4 or (E) CD8. Up to six cell divisions, D1–D6, of CD3+CD4+ and CD3+CD8+ cells can be seen.
Table 3.
Summary of fungal-specific PB CD4+ and CD8+ T-cell proliferation
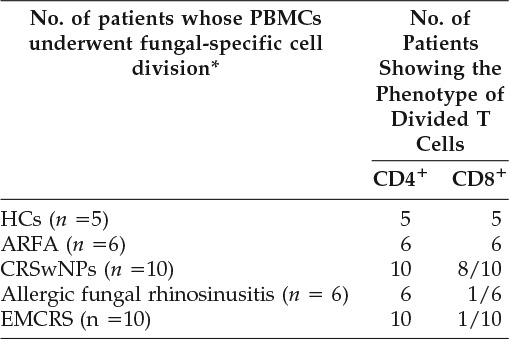
*Cell division in response to stimulation by fungal antigens was determined by CFSE cytometry. A positive fungal-specific CD4+ and/or CD8+ T-cell proliferation result in an individual was considered when there was a greater percentage of CD4+ and/or CD8+ T-cell subset with a lower CFSE fluorescence in fungal-stimulated samples compared with unstimulated samples. Please also refer to an example in Figure 3.
ARFA = allergic rhinitis with fungal allergy; CFSE = carboxyfluorescein succinimidyl ester; CRSwNPs = chronic rhinosinusitis with nasal polyps; EMCRS = eosinophilic mucus chronic rhinosinusitis; HCs = healthy controls; PB = peripheral blood; PBMCs = peripheral blood mononuclear cells.
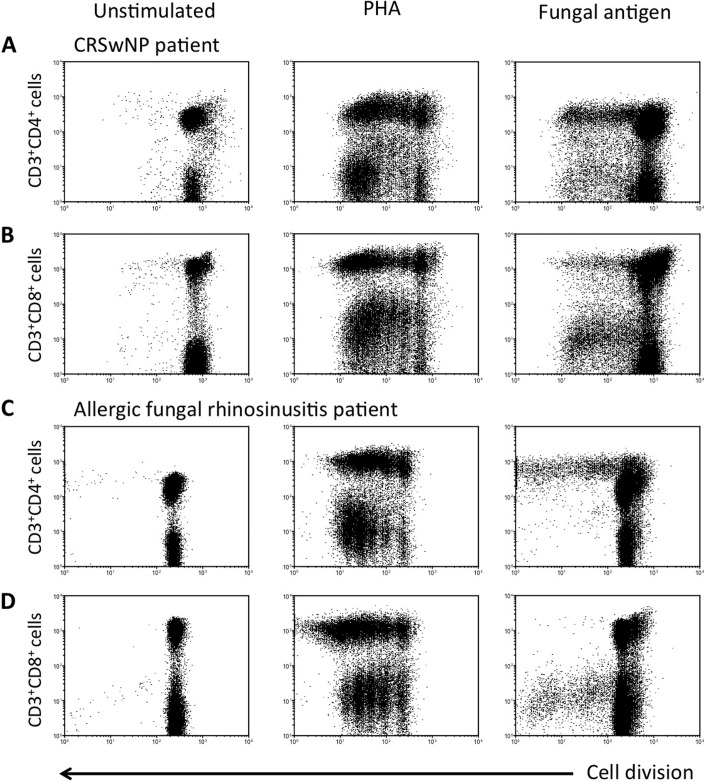
Figure 3.
Fungal-specific blastogenesis of peripheral blood (PB) (A and C) CD3+CD4+ cells and (B and D) CD3+CD8+ cells in a (A and B) chronic rhinosinusitis with nasal polyp (CRSwNP) and (C and D) an allergic fungal rhinosinusitis (AFRS) patient. Carboxyfluorescein succinimidyl ester (CFSE)–labeled peripheral blood mononuclear cells were cultured with no mitogen (unstimulated), phytohemagglutinin (PHA), and Alternaria alternata fungal antigens. Cells were harvested after 6 days of the proliferative period. Dot plots are gated on CD3+ blast cells (please also refer to Fig. 2). Both CD4+ and CD8+ T cells divided in response to fungal antigens in the CRSwNP patient. In contrast, CD8+ T cells did not divide to fungal antigens in the AFRS patient.
IL-2 is required during antigen-specific T-cell activation and proliferation, and we investigated whether the addition of exogenous IL-2 could influence fungal-specific CD8+ T-cell proliferation in AFRS. There was no significant increase in the percentage of dividing CD8+ T cells with fungal antigens and IL-2 (median = 5.8; IQR = 3.3–6.5) compared with IL-2 alone (median = 5.0; IQR = 3.0–7.1; n = 8; p = 0.22, Wilcoxon matched paired test).
To examine whether PB fungal-specific CD8+ T-cell activation could be detected in the absence of a proliferative response, cell surface expression of CD25 activation marker was examined in seven patients (four AFRS and three ARFA patients). CD25 (IL-2 α-chain receptor) is up-regulated in activated T cells and its expression is maximal after several days of stimulation.15 These results are summarized in Table 4. Internal controls included unstimulated and PHA-stimulated cells under similar conditions. Fungal antigen–induced CD25 up-regulation was consistently noted in CD4+ T cells in all patients. Although fungal-specific up-regulation of CD25 on CD8+ T cells was significantly increased compared with unstimulated cells in ARFA patients, this was strikingly absent in AFRS. In PHA-treated samples, CD25 up-regulation occurred on PB CD4+ and CD8+ T cells in all patients.
Table 4.
CD25 expression on PB T cells from AFRS and ARFA
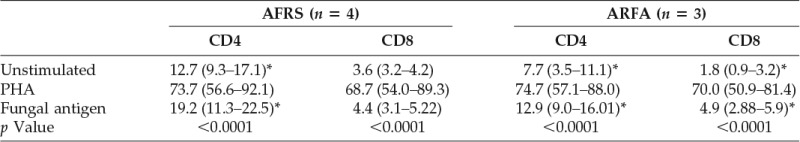
The percentages refer to the proportion of CD4+ and CD8+ T cells expressing CD25 and are presented as the median value and IQR in parenthesis. A significant difference in CD25 expression on T cells between unstimulated and fungal antigen–stimulated cells is indicated by asterisks. The p value, Kruskal–Wallis test followed by post hoc Dunn test.
AFRS = allergic fungal rhinosinusitis; ARFA = allergic rhinitis with fungal allergy; IQR = interquartile range; PB = peripheral blood; PHA = phytohemagglutinin.
To study the influence of fungal allergy and of fungi in sinus eosinophilic mucus on fungal-specific PBMC proliferation, a comparison of the proliferation results (tritiated thymidine and CFSE) in AFRS and EMCRS patients is summarized in Table 5. Increased fungal-specific proliferation was also evident in EMCRS patients who did not have fungal allergy (Table 5; Fig. 1 C).
Table 5.
Alternaria alternata and Aspergillus fumigatus allergy, presence in sinus eosinophilic mucus and PBMC proliferation results in AFRS and EMCRS patients
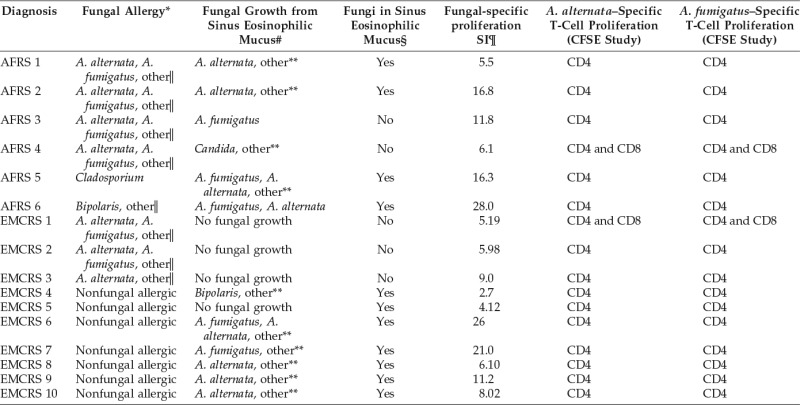
*Fungal-specific serum IgE or skin-prick tests.
#Routine diagnostic test on fresh specimen of sinus eosinophilic mucus for fungal culture.
§Routine diagnostic test on 10% formalin-fixed, hematoxylin and eosin and special fungal-stained specimens of sinus eosinophilic mucus.
¶Determined by tritiated thymidine-based PB fungal proliferation assays. The SI represents the highest proliferation ratio obtained by dividing the value from A. fumigatus– and A. alternata–stimulated tissue culture wells by the value from unstimulated culture wells from the same patient.
‖A. nidulans, A. niger, Bipolaris species, Candida albicans, Cladosporium mix (Cladosporium normodendrum, Cladosporium herbarum, and Cladosporium cladosporoides), Epicoccum nigrum, Fusarium vasinsectum, Helminthosporium species, Mucor racemosus, Penicillium mix (Penicillium digitatum, Penicillium expansum, and Penicillium notatum), Rhizopus nigricans, Pullularia pullulans, and Trichophyton mix (Trichophyton mentogrophytes, Trichophyton rubrum, and Trichophyton tonsuranis).
**Bipolaris, Drechslera, Trichothecium, Candida, Penicillium, Scedosporium, Acremonium, Devriessi, Phialaphora, and Cladosporium species.
AFRS = allergic fungal rhinosinusitis; EMCRS = eosinophilic mucus chronic rhinosinusitis; CFSE = carboxyfluorescein succinimidyl ester; PB = peripheral blood; PBMC = peripheral blood mononuclear cell; SI = stimulation index.
DISCUSSION
The major finding from this study was that A. alternata and A. fumigatus antigens caused proliferation of PB CD4+ and CD8+ T-cell subsets in HCs and ARFA and most CRSwNP patients, whereas CD8+ T-cell response was strikingly absent in AFRS and EMCRS patients. This indicates that in HCs and ARFA and most CRSwNP patients, fungal-specific memory CD4+ and CD8+ T cells were present and implies that both T-cell subsets were important in healthy immune responses generated by these fungi.7
Data from this study also confirm our previous finding that AFRS, EMCRS, CRSwNP, and ARFA patients had elevated fungal-specific PB T-cell response compared with HCs, indicating an increased percentage of fungal-specific T cells in the patient groups.12 Although allergic hypersensitivity was a likely explanation for the increased proliferation in fungal-allergic CRSwNP and ARFA patients, an enhanced response irrespective of fungal allergy in EMCRS suggests stimulation by nonallergenic antigens produced by fungi in their sinuses.7,16 Thus, immune activation by both allergenic and nonallergenic antigens in AFRS could occur, because allergic sensitization and fungi are generally present, and could in part explain their severe clinical phenotype and variable response to fungal desensitization therapy.
The PBMC proliferation results was less likely to be a nonspecific mitogenic effect of the fungal antigens17–19 because increased proliferation corresponded with fungal allergy and fungi in sinuses, and not every patient's CD8+ T cells underwent division. Fungal-specific PB CD8+ T-cell division in ARFA and CRSwNP patients and to PHA in all individuals, including AFRS and EMCRS, argues against technical problems with methodology, variations in assay conditions or a generalized T-cell anergy causing fungal-specific CD8+ T-cell inactivity in AFRS and EMCRS.20,21
In the absence of cell division, antigen-specific activation of the CD8+ T cells may be detected by up-regulation of activation markers.22 Accordingly, fungal-induced up-regulation of CD25 on CD8+ T cells in PB was noted in a sample of ARFA patients, but not in AFRS. That CD25 was up-regulated on CD4+ T cells with fungal stimulation, and on CD4+ and CD8+ T cells with PHA in AFRS patients, contends against technical issues and global T-cell anergy accounting for lack of fungal-specific CD8+ T-cell activation. Taken together, these results suggest that in most individuals, both CD4+ and CD8+ T cells were involved in A. alternata– and A. fumigatus–specific responses, whereas in AFRS and EMCRS, the response was mainly caused by CD4+ T cells. Hence, absent fungal-specific CD8+ T-cell activation and proliferation in AFRS and EMCRS patients is a significant finding and raises the possibility of abnormal fungal-specific immune function, where a failure to generate CD8+ T-cell response could contribute toward fungal accumulation in their sinuses.
CD8+ T-cell responders implies presence of MHC I–restricted epitopes, typically associated with intracellular antigen; however, it can also occur because of cross-presentation of extracellular antigen.23–26 Interestingly, defective fungal antigen processing for CD8+ T-cell activation has been proposed as a susceptibility factor for aspergillosis.10 Fungal-specific CD8+ T cells in vivo often display a protective role but the mechanisms controlling their priming, activation, and differentiation are poorly understood.10,22,26–28 Moreover, fungal-specific CD8+ T-cell function and role in CRS inflammatory/anti-inflammatory balance is unknown, in part, because of limited knowledge of the precise antigens of A. alternata and A. fumigatus involved in CRS. Intriguingly, in the presence of defective CD8+ T cells, a protective role for fungal-specific IgG3 has been proposed, the latter which is significantly elevated in AFRS and EMCRS patients.4,29
This study used a fungal antigen mixture commonly used for desensitization therapy. Thus, lack of CD8+ T-cell response in AFRS and EMCRS is significant and also remarkable, given that CD8+ T cells were enriched in their mucosa,12,14,30 hence broaching the potential for a CD8+ T-cell functional abnormality. Our additional studies also showed that CD8+ T cells from AFRS sinus mucosa failed to up-regulate activation markers with fungal antigen exposure, suggesting that the lack of systemic fungal–specific CD8+ T-cell activity was reflected in the mucosal T cells (data not shown). Additional studies of CD8+ T-cell effector activities (cytokine secretion, cytotoxic, and regulatory functions) are needed to understand their role in AFRS and EMCRS.
Absence of fungal-specific CD8+ T-cell response was mainly seen in AFRS and EMCRS patients although NPs were common also to CRSwNP patients. This further supports the notion that sinus eosinophilic mucus may signify a distinct subset of clinically recalcitrant NP disease.1,3,31 AFRS has been historically discerned by fungi in sinus eosinophilic mucus and coexisting fungal allergy,1 but some EMCRS patients who are otherwise clinically indistinguishable from AFRS may not have either fungal allergy or fungi in their sinuses.3 The clinical, sinus cellular infiltrate, fungal-specific humoral, and T-cell characteristics in AFRS and EMCRS subgroups suggest that these diseases have a lot in common and that AFRS may not be a unique entity from EMCRS.3,4,12
In conclusion, data from this pilot study show a significant and novel finding that suggests altered CD8+ T-cell responses to A. alternata and A. fumigatus antigens in AFRS and EMCRS patients and raises the question of dysfunctional host immune mechanisms. In predisposed individuals, failure to control sinus microbial burden and to regulate allergic hypersensitivity, immune tolerance, and eosinophilia could all lead to the clinical phenotype characteristic of AFRS and EMCRS.10,32–34
Footnotes
Funded by Garnett-Passe and Rodney Williams Memorial Foundation
The authors have no conflicts of interest to declare pertaining to this article
REFERENCES
- 1. Bent JP, III, Kuhn FA. Diagnosis of allergic fungal rhinosinusitis. Otolaryngol Head Neck Surg 11:580–588, 1994. [DOI] [PubMed] [Google Scholar]
- 2. Pant H, Schembri MA, Wormald PJ, Macardle PJ. IgE-mediated fungal allergy in allergic fungal sinusitis. Laryngoscope 119:1046–1052, 2009. [DOI] [PubMed] [Google Scholar]
- 3. Pant H, Kette FE, Smith WB, et al. Eosinophilic mucus chronic rhinosinusitis: Clinical subgroups or a homogeneous pathogenic entity? Laryngoscope 116:1241–1247, 2006. [DOI] [PubMed] [Google Scholar]
- 4. Pant H, Kette FE, Smith WB, et al. Fungal-specific humoral response in eosinophilic mucus chronic rhinosinusitis. Laryngoscope 115:601–606, 2005. [DOI] [PubMed] [Google Scholar]
- 5. Ponikau JU, Sherris DA, Kern EB, et al. The diagnosis and incidence of allergic fungal sinusitis. Mayo Clin Proc 74:877–884, 1999. [DOI] [PubMed] [Google Scholar]
- 6. Syme RM, Wood CJ, Wong H, Mody CH. Both CD4+ and CD8+ human lymphocytes are activated and proliferate in response to Cryptococcus neoformans. Immunology 92:194–200, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaudhary N, Staab JF, Marr KA. Healthy human T-cell responses to Aspergillus fumigatus antigens. PLoS One 5:e9036, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramadan G, Konings S, Kurup VP, Keever-Taylor CA. Generation of Aspergillus- and CMV-specific T-cell responses using autologous fast DC. Cytotherapy 6:223–234, 2004. [DOI] [PubMed] [Google Scholar]
- 9. Beck O, Topp MS, Koehl U, et al. Generation of highly purified and functionally active human TH1 cells against Aspergillus fumigatus. Blood 107:2562–2569, 2006. [DOI] [PubMed] [Google Scholar]
- 10. Carvalho A, De Luca A, Bozza S, et al. TLR3 essentially promotes protective class I-restricted memory CD8+ T-cell responses to Aspergillus fumigatus in hematopoietic transplanted patients. Blood 119:967–977, 2012. [DOI] [PubMed] [Google Scholar]
- 11. Fulcher D, Wong S. Carboxyfluorescein succinimidyl ester-based proliferative assays for assessment of T cell function in the diagnostic laboratory. Immunol Cell Biol 77:559–564, 1999. [DOI] [PubMed] [Google Scholar]
- 12. Pant H, Beroukas D, Kette FE, et al. Nasal polyp cell populations and fungal-specific peripheral blood lymphocyte proliferation in allergic fungal sinusitis. Am J Rhinol Allergy 23:453–460, 2009. [DOI] [PubMed] [Google Scholar]
- 13. Fokkens WJ, Lund VJ, Mullol J, et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2012. Rhinol Suppl 23:1–298, 2012. [PubMed] [Google Scholar]
- 14. Pant H, Hughes A, Miljkovic D, et al. Accumulation of effector memory CD8+ T cells in nasal polyps. Am J Rhinol Allergy 27:e117–e126, 2013. [DOI] [PubMed] [Google Scholar]
- 15. Biselli R, Matricardi PM, D'Amelio R, Fattorossi A. Multiparameteric flow cytometric analysis of the kinetics of surface molecule expression after polyclonal activation of human peripheral blood T lymphocytes. Scand J Immunol 35:439–447, 1992. [DOI] [PubMed] [Google Scholar]
- 16. Templeton SP, Buskirk AD, Law B, et al. Role of germination in murine airway CD8+ T-cell responses to Aspergillus conidia. PLoS One 6:e18777, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Syme RM, Spurrell JC, Ma LL, et al. Phagocytosis and protein processing are required for presentation of Cryptococcus neoformans mitogen to T lymphocytes. Infect Immun 68:6147–6153, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mody CH, Wood CJ, Syme RM, Spurrell JC. The cell wall and membrane of Cryptococcus neoformans possess a mitogen for human T lymphocytes. Infect Immun 67:936–941, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Graybill JR, Alford RH. Cell mediated immunity in cryptococcosis. Cell Immunol 14:12–21, 1974. [DOI] [PubMed] [Google Scholar]
- 20. Brenchley JM, Karandikar NJ, Betts MR, et al. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood 101:2711–2720, 2003. [DOI] [PubMed] [Google Scholar]
- 21. Horiuchi N, Aiba S, Ozawa H, et al. Peripheral blood lymphocytes from psoriatic patients are hyporesponsive to beta-streptococcal superantigens. Br J Dermatol 138:229–235, 1998. [DOI] [PubMed] [Google Scholar]
- 22. Heintel T, Breinig F, Schmitt MJ, Meyerhans A. Extensive MHC class I-restricted CD8 T lymphocyte responses against various yeast genera in humans. FEMS Immunol Med Microbiol 39:279–286, 2003. [DOI] [PubMed] [Google Scholar]
- 23. Hon H, Oran A, Brocker T, Jacob J. B lymphocytes participate in cross-presentation of antigen following gene gun vaccination. J Immunol 174:5233–5242, 2005. [DOI] [PubMed] [Google Scholar]
- 24. Randolph GJ, Jakubzick C, Qu C. Antigen presentation by monocytes and monocyte-derived cells. Curr Opin Immunol 20:52–60, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brode S, Macary PA. Cross-presentation: Dendritic cells and macrophages bite off more than they can chew! Immunology 112:345–351, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin JS, Yang CW, Wang DW, Wu-Hsieh BA. Dendritic cells cross-present exogenous fungal antigens to stimulate a protective CD8 T cell response in infection by Histoplasma capsulatum. J Immunol 174:6282–6291, 2005. [DOI] [PubMed] [Google Scholar]
- 27. Beno D, Stover AG, Mathews HL. Growth inhibition of Candida albicans hyphae by CD8+ lymphocytes. J Immunol 154:5273–5281, 1995. [PubMed] [Google Scholar]
- 28. Levitz SM, Dupont MP, Smail EH. Direct activity of human T lymphocytes and natural killer cells against Cryptococcus neoformans. Infect Immun 62:194–202, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yuan RR, Casadevall A, Oh J, Scharff MD. T cells cooperate with passive antibody to modify Cryptococcus neoformans infection in mice. Proc Natl Acad Sci U S A 94:2483–2488, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ragab A, Samaka RM. Immunohistochemical dissimilarity between allergic fungal and nonfungal chronic rhinosinusitis. Am J Rhinol Allergy 27:168–176, 2013. [DOI] [PubMed] [Google Scholar]
- 31. Ferguson BJ. Eosinophilic mucin rhinosinusitis: A distinct clinicopathological entity. Laryngoscope 110:799–813, 2000. [DOI] [PubMed] [Google Scholar]
- 32. Enomoto N, Hyde E, Ma JZ, et al. Allergen-specific CTL require perforin expression to suppress allergic airway inflammation. J Immunol 188:1734–1741, 2012. [DOI] [PubMed] [Google Scholar]
- 33. Tang Y, Guan SP, Chua BY, et al. Antigen-specific effector CD8 T cells regulate allergic responses via IFN-gamma and dendritic cell function. J Allergy Clin Immunol 129:1611–1620, 2012. [DOI] [PubMed] [Google Scholar]
- 34. Ahn CN, Wise SK, Lathers DM, et al. Local production of antigen-specific IgE in different anatomic subsites of allergic fungal rhinosinusitis patients. Otolaryngol Head Neck Surg 141:97–103, 2009. [DOI] [PubMed] [Google Scholar]