Abstract
ATAD2, a remarkably conserved, yet poorly characterized factor is found upregulated and associated with poor prognosis in a variety of independent cancers in human. Studies conducted on the yeast Saccharomyces cerevisiae ATAD2 homologue, Yta7, are now indicating that the members of this family may primarily be regulators of chromatin dynamics and that their action on gene expression could only be one facet of their general activity. In this review, we present an overview of the literature on Yta7 and discuss the possibility of translating these findings into other organisms to further define the involvement of ATAD2 and other members of its family in regulating chromatin structure and function both in normal and pathological situations.
Keywords: AAA ATPase, ATAD2, chromatin, histone chaperone, transcription, yeast
INTRODUCTION
With the exception of mammals, the function of the members of the ATAD2 protein family remains mostly undefined in multicellular eukaryotes. The first reported activity of ATAD2 (ATPase family AAA+ Domain-containing protein 2) is its ability to act as a co-activator of estrogen and androgen receptors (Zou et al., 2007; 2009). Other studies later defined ATAD2 as an E2F and MLL co-factor stimulating E2F-dependent cell proliferation (Revenko et al., 2010) and also as a MYC partner (Ciro et al., 2009). These transcriptional activities of ATAD2 have been, at least partly, attributed to its two conserved domains (Revenko et al., 2010; Zou et al., 2007). Indeed, a conserved feature of ATAD2 and of its homologues across species is the presence of a two partite AAA+ ATPase (ATPases Associated with diverse cellular Activities) domain that mediates protein multimerization and of a bromodomain that is responsible for its binding to his-tones (Boussouar et al., 2013). This high conservation of the domain organization of the protein in the vast majority of eukaryotes, including unicellular organisms such as yeasts, is also indicative of its conserved functional features.
Functional studies of ATAD2 family members carried out in mammals were mostly in cancer settings and have recently been reviewed (Boussouar et al., 2013). However, in addition to these studies, there is a relatively rich literature on Yta7 (Yeast Tat-binding Analog 7), the unique ATAD2 homologue expressed in the budding yeast Saccharomyces cerevisiae. This review considers all the molecular data available on Yta7 in the light of our knowledge on ATAD2. The data reported highlight the implication of Yta7 not only in gene expression, as expected, but also in genome organization, as a possible histone chaperone acting at boundary sites and regulating transcription. Altogether, these data point to conserved functional features of the Yta7/ATAD2 family.
ATAD2-LIKE PROTEINS ARE CONSERVED FROM YEAST TO HUMAN
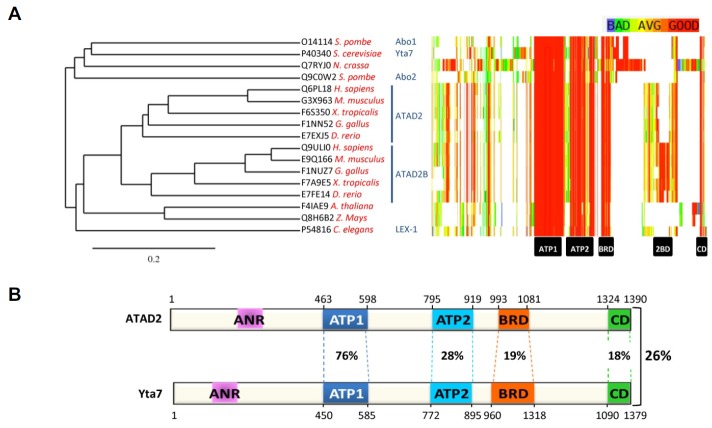
A phylogenetic tree of ATAD2-like proteins, shown in Fig. 1A (left), illustrates the strong conservation of this protein family within the eukaryotic kingdom, including yeast. These proteins all share a two-partite AAA+ ATPase domain, which is found N-terminal to a bromodomain.
Fig. 1.
Members of the ATAD2 protein family are conserved from yeast to human. (A). Left part of the panel is a graphical representation of a phylogenetic tree performed with TreeDyn (v198.3) (Dereeper et al., 2008; 2010) from the amino acid sequences of ATAD2-like proteins (UniProt, 2014) aligned by CLUSTALO (McWilliam et al., 2013). Scale bar: 0.2 changes per site. Right part of the panel shows a multiple sequence alignment performed using PSI-coffee (Di Tommaso et al., 2011; Kemena and Notredame, 2009). (B) Schematic representation of human ATAD2 and S. cerevisiae Yta7 drawn to scale. The percentages of identity are indicated both for the overall length of the two proteins (on the right side of the panel) and for each conserved domain (Alignment done with CLUSTALO). ANR, Acid N-terminal Region; ATP1 and ATP2, AAA+ ATPase domain 1 and 2; BRD, BRomoDomain; 2BD, ATAD2B specific Domain; CD, C-terminal Domain.
This tree also shows that in several organisms the genome encodes two ATAD2 paralogs. These include Chordata and the fission yeast Schizosaccharomyces pombe that possess two ATAD2-like proteins known as ATAD2A and B and Abo1 and 2, respectively. It is of note that the vast majority of the published functional studies concerns the ATAD2A (also termed ATAD2) member in both human and mouse. The high sequence similarity between these paralogs argues in favor of a possible functional redundancy across all species.
Other eukaryotes such as plants (Arabidopsis thaliana, Zea mays), the worm Caenorhabditis elegans, and the budding yeast S. cerevisiae possess only one gene encoding an ATAD2-like protein. Intriguingly, in Drosophila melanogaster and Tetrahymena thermophila no gene encoding a homologue of ATAD2 has been identified. In these two organisms, the closest relative proteins belong to the Valosin-Containing Protein/Transitional Endoplasmic Reticulum ATPase (VCP/TERA family), which is also a conserved protein family that, to a certain extent, shares a similar domain organization with ATAD2 family members, although the amino-acid sequence aside the AAA+ ATPase domain is poorly conserved. Xenopus laevis is quite peculiar, since it possesses a gene expressing a shorter version of ATAD2-like protein. This protein contains only the first AAA+ ATPase domain and the N-terminal part of the second one. The existence of organisms that have lost part or the totality of ATAD2-like proteins might indicate that a concomitant functional adaptation has occurred to compensate for the lack of ATAD2 functions in these organisms. This observation also points to the existence of possible redundant functional pathways in various eukaryotic cells that express ATAD2 family members.
ATAD2-like proteins have their highest sequence similarity within the two AAA+ ATPase domains and the bromodomain (Fig. 1A, right). The AAA+ ATPase domain is found in all kingdoms of living organisms, in proteins possessing many different cellular functions. ATAD2-like proteins contain two AAA+ ATPase domains both located in their N-terminal part. The first domain appears more conserved across evolution (Fig. 1A, right and 1B). ATP binding and ATPase activities were demonstrated for ATAD2 (Zou et al., 2007). Additionally, mutations in the first AAA+ ATPase domain that affect ATP binding and hydrolysis impact on the property of mouse Atad2 and human ATAD2 to oligomerize, to bind to acetylated histone H4 (Caron et al., 2010) and to co-activate transcription (Zou et al., 2007), indicating that this domain is critical for ATAD2 functions. Since the amino-acid sequence of the AAA+ ATPase domain is highly similar among ATAD2-like proteins, it is likely that the activity and function of this domain are also conserved in other eukaryotes. In agreement with such a possibility, mutations in the first AAA+ ATPase domain of the S. cerevisiae Yta7 protein affect many of its functions (Kurat et al., 2011; Lombardi et al., 2011), as described in the next sections.
ATAD2-like proteins contain also a putative bromodomain, a module known to bind acetylated lysine in histones and other proteins (Filippakopoulos et al., 2012). The binding to acetylated histones was demonstrated for mouse Atad2 and human ATAD2, which show a preferential binding to acetylated histones H3 and H4 (Caron et al., 2010; Revenko et al., 2010). Bromodomains have a three-dimensional structure that consists of four helix bundles (αZ, αA, αB and αC) with two interhelical ZA and BC loops, containing several amino-acids necessary to form an hydrophobic pocket for the interaction with acetylated lysines (Dhalluin et al., 1999). The overall conservation of the bromodomain amino-acid sequence in ATAD2-like proteins suggests that its global architecture and its capacity to bind to histones may be conserved. Nonetheless, alignment of the Yta7 bromodomain with other yeast bromodomains revealed that residues critical for binding acetylated histones are missing, pointing out that the specific binding to acetylated histones is probably not true for all ATAD2-like proteins (Jambunathan et al., 2005). In vitro histone pull-down experiments have indeed shown that Yta7 binds histones but in an acetylation-independent manner (Gradolatto et al., 2009; Jambunathan et al., 2005).
Interestingly, similar pull-down experiments, using truncated forms of Yta7, revealed a second region that binds histones. This region is located in the N-terminal part of Yta7, upstream of the AAA+ ATPase domains (Fig. 1B), and contains a stretch of acidic residues that may be responsible for electrostatic interactions with charged and unmodified lysine and arginine residues in histones (Gradolatto et al., 2009). Remarkably, a patch of acidic residues in the N-terminal part of the protein seems to be a common feature of all ATAD2-like proteins, suggesting that the function of this negatively charged region in binding histones may be conserved. Mutations changing the nature of this region would be informative both in relation to the importance of this domain in contributing to ATAD2-like protein functions, as well as to know whether this region influences the capacity and/or specificity of the bromodomain of Yta7 to bind histones.
Finally, members of the ATAD2 family also have in common a fourth region of around 60 amino-acids located at the extreme C-terminal part of the protein (Fig. 1A, right). This domain, which is highly conserved among ATAD2-like proteins, does not correspond to any annotated domain and is not found in any other type of proteins. The analysis of this newly identified and conserved domain should establish its function within ATAD2 proteins. Interestingly, upstream of this C-terminal domain there is another region conserved in ATAD2B proteins, but not in the ATAD2A/ATAD2 paralogs, indicating that this domain may play an important role in attributing a specific function to the ATAD2B proteins. Here again, the analysis of this newly identified and conserved region, possibly using the powerful yeast genetics, could provide some clues to the function of ATAD2-like proteins.
YTA7, THE UNIQUE BUDDING YEAST ATAD2-LIKE PROTEIN, IS A PROBABLE HISTONE CHAPERONE REGULATING TRANSCRIPTION OF HISTONES AND OTHER GENES
A genome-wide chromatin localization approach using chromatin immunoprecipitation (ChIP)-coupled to chip (ChIP-chip) analysis demonstrated that Yta7 binds to all histone genes (Gradolatto et al., 2008). However, the effects of a deletion of the YTA7 gene on the level of histone transcripts are not clear yet. Gradolatto et al. (2008) reported that Yta7 is a histone gene repressor based on the fact that yta7Δ cells present a precocious increase in mRNAs of all histone genes after α-factor block (G1-phase arrest). In contrast, Fillingham et al. (2009) proposed that Yta7 could be an activator of histone genes transcription, since a deletion of the YTA7 gene correlates with a decrease in HTA1 transcripts that is concomitant with a decreased recruitment of RNA polymerase II to both the promoter region and the ORF of HTA1 (Kurat et al., 2011). More recent studies have shown that the importance of Yta7 in regulating histone transcription may in fact differ depending on the analyzed histone gene. Indeed, in yta7Δ cells HTA1 transcripts are significantly reduced, while in these same cells HHT1, HHF1 or HTA2 transcript levels remain unchanged, despite the fact that Yta7 binds to all these histone loci (Lombardi et al., 2011; Zunder and Rine, 2012).
Transcription of the histone genes is tightly regulated throughout the cell cycle in order to provide the histone supply required for the replication of chromatin during the S-phase while avoiding inappropriate and toxic accumulation of neosynthesized histones during the other phases of the cell cycle (Gunjan and Verreault, 2003). The localization of Yta7 to the histone genes indicates a possible direct role for this protein in the phase-specific regulation of histone transcription. Interestingly, Yta7 binding to the histone gene HTA1 is precisely regulated during the cell cycle (Kurat et al., 2011) (Fig. 2A). After the loading of RNA polymerase II to histone genes in G1/S, Yta7 is phosphorylated at multiple sites located in its N-terminal part by at least two different kinases Cdk1 (Cyclin-Dependent Kinase 1) and CK2 (Casein Kinase 2). Phosphorylation of Yta7 causes its release from HTA1 and correlates with an efficient transcription of HTA1 and other histone genes (Kurat et al., 2011). The molecular mechanism underlying the Yta7-dependent transcriptional gene activation is not completely understood, but it has been proposed that the phosphorylation-mediated removal of Yta7 from a histone gene during the S-phase could be an important step to allow efficient transcriptional elongation along the histone gene. In agreement with this hypothesis, in a mutant of Yta7 that cannot be phosphorylated, the recruitment of RNA polymerase II is markedly decreased within the ORF of HTA1 gene but not at the promoter region (Kurat et al., 2011).
Fig. 2.
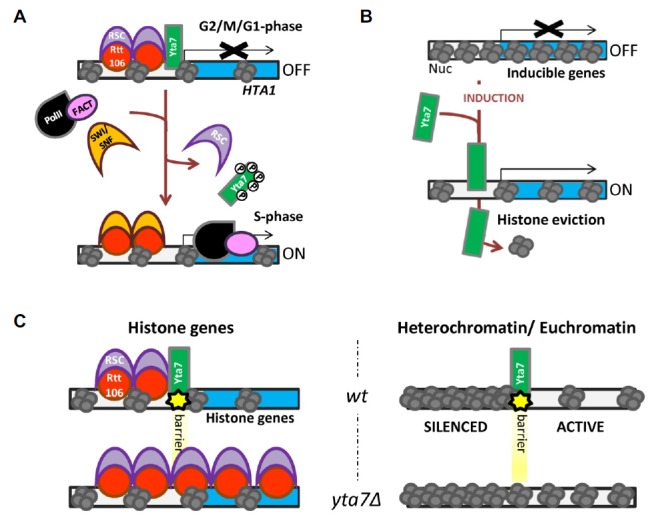
Yta7 is a probable histone chaperone, regulating the transcription of histone and other inducible genes and acting as a barrier element. (A) Yta7 is directly involved in regulating S-phase specific transcription of the HTA1 histone gene. After the binding of RNA polymerase II, the phosphorylation and removal of Yta7 allows an efficient transcriptional elongation along HTA1. The S-phase specific removal of RSC and the recruitment of SWI/SNF by Rtt106 to the histone genes accompany Yta7 departure and HTA1 transcriptional activation. (B) Yta7 is required for the transcription of inducible genes. Yta7 binds to the promoter region of these genes and is believed to promote the eviction or degradation of the histones. (C) Yta7 is a boundary element acting both at the promoter of histone genes (left) and at the border between heterochromatin and euchromatin (right).
Transcriptome analyses of yta7Δ cells revealed that the expression of inducible genes is also deregulated and that Yta7 probably acts as a histone chaperone (Lombardi et al., 2011). ChIP experiments have shown that Yta7 localizes to the promoters and 5’ORF of early meiotic genes and galactose genes while they are induced. Moreover, the presence of Yta7 limits the nucleosome density in these genic regions, possibly by promoting histone H3/H4 eviction (Fig. 2B). In agreement with the existence of an Yta7-mediated histone eviction activity, the deficiency in inducing gene expression in yta7Δ cells is rescued by a decrease in the dosage of histones H3 and H4 caused by the deletion of a pair of histone genes (hht1Δhhf1Δ). Interestingly, the AAA+ ATPase domain is required for this potential chaperone activity of Yta7, indicating that Yta7 ATP hydrolysis may be important for the eviction or degradation of histones (Lombardi et al., 2011). In addition, Yta7 was co-purified with several subunits of the RNA polymerase II (Rpb2, Rpb5 and Rpb10), as well as with several histone chaperones, the FACT complex (Spt16 and Pob3) and Rtt106 protein (Kurat et al., 2011; Tackett et al., 2005). These results further reinforce the connection between Yta7 transcriptional regulation and histone chaperone activity. Nonetheless, it should also be pointed out that so far there is no in vitro evidence that Yta7 is a bonafide histone chaperone. Moreover, since in vivo experiments do not exclude the possibility that the change in histone density observed in yta7Δ cells is due to the activity of one of the histone chaperones interacting with Yta7, these findings await further experiments to conclude that Yta7 and, by extension, other ATAD2-like proteins are indeed histone chaperones.
YTA7 PROTEIN AND CHROMATIN BOUNDARY ELEMENTS
Yta7 localization to histone genes promoters was also linked to a boundary activity acting at the promoters of all histone genes. Indeed, at these boundaries, Yta7 prevents the spreading of the histone chaperone Rtt106 from the promoter to their respective coding regions (Fillingham et al., 2009; Kurat et al., 2011; Zunder and Rine, 2012) (Fig. 2C, left). The recruitment of Rtt106 to histone gene promoters depends on Asf1 and the HIR complex and is not regulated through the cell cycle. HIR/Asf1 binding to histone genes and the repression of their transcription rely on a specific DNA sequence, the negative regulatory element, present in every histone pairs except for HTA2-HTB2. Rtt106 was found to bind and act as a repressor of the histone genes targeted by HIR and Asf1 (HTA1-HTB1, HHT1-HHF1, HHT2-HHF2) (Fillingham et al., 2009). Rtt106 is regulating both positively and negatively the expression of histone genes during the cell cycle. Outside S-phase, when histone genes are repressed, Rtt106 recruits the ATP-dependent remodeling complex RSC. On the contrary, during histone gene activation in S-phase Rtt106 recruits the SWI/SNF complex (Ferreira et al., 2011). In yta7Δ cells Rtt106 spreads within the coding sequence of histone genes together with the RSC complex, except for the HTA2-HTB2 histone pair that is not bound by HIR/Asf1 and Rtt106 (Fillingham et al., 2009). The spreading of Rtt106 through the coding regions of histone genes is associated with a clear decrease of HTA1 transcripts, suggesting that Rtt106 and RSC spreading may be responsible for the transcriptional repression of HTA1 gene. However, since this spreading occurs on all his-tone genes even though it is not always associated to a decrease in histone mRNA levels, for instance at HHT1 or HHF1 genes, the potential repressive action of Rtt106 requires further investigations to be validated (Zunder and Rine, 2012). In addition, it is also possible that the regulation of histone genes expression presents a certain degree of locus-specificity that would allow Rtt106 spreading to repress transcription of some, but not all histone genes.
Yta7 boundary function is not limited to histone promoter regions. Earlier evidences implicated Yta7, together with other proteins, at barriers that demarcate euchromatin from heterochromatin regions (Jambunathan et al., 2005; Tackett et al., 2005) (Fig. 2C, right panel). S. cerevisiae contains heterochromatin-like regions located at the mating type, telomeres and rRNA-encoding DNA loci. The implication of Yta7 in such barrier function was first found in a genetic screen aimed at identifying new genes that, when mutated, allow inappropriate spreading of silent chromatin from the mating type locus (HMR) over the adjacent tRNA gene into the neighboring genomic region which included the ADE2 reporter gene (Jambunathan et al., 2005). The boundary activity of Yta7 was investigated also by using silencing assays testing the sensitivity to 5 FluoroOrotic Acid (5FOA) drug of yeast cells in which the URA3 reporter gene was ectopically incorporated either within the silenced HMR or the transcriptionally competent adjacent region (Tackett et al., 2005). In yta7Δ cells the silent transcriptional state spreads to the neighboring regions. Furthermore, both the bromodomain and the AAA+ ATPase domains are required for Yta7 barrier function (Gradolatto et al., 2009; Lombardi et al., 2011). In agreement with a direct implication of Yta7 in such a barrier function, it was found to co-purify with a complex localizing at the boundaries between euchromatin and heterochromatin-like regions in S. cerevisiae (Tackett et al., 2005). Nonetheless, the exact role of Yta7 at these chromatin barriers remains to be determined. For example, it would be interesting to understand if the probable histone chaperone activity of Yta7 plays any important role at these barriers.
FROM YTA7 TO ATAD2A: FUNCTIONAL ORTHOLOGS?
The high similarity between Yta7 and ATAD2 domains organization (Fig. 1B) supports the hypothesis that at least part of their function is shared. An implication of both proteins in transcriptional regulation is clear. However, although data on Yta7 suggest a role for this factor in the organization of the genome and chromatin dynamics, such indications are scarce for ATAD2. Indeed, only one experiment that measured histone H2A mobility in a lung cancer cell line showed that a decrease in ATAD2 content modifies H2A turnover (Caron et al., 2010). Even in this case there is no indication on the mechanism that links ATAD2 and histone mobility.
Considering the importance of Yta7 in regulating nucleosome density, it is tempting to propose that ATAD2 can also act as a histone chaperone, evicting histones from chromatin to avoid potentially deleterious effects associated to an increase in his-tone density. Therefore ATAD2 and Yta7 could primarily be chromatin remodelers and the above-mentioned effects on gene expression regulation could be a mere consequence of their actions on chromatin organization. Alternatively, these proteins could be dual factors playing a role both in transcriptional regulation as scaffolds or as co-activators, and in chromatin remodeling as histone chaperones. In agreement with such a possibility, the acidic N-terminal region of ATAD2 is a histone-interacting module in the case of Yta7 (as described previously), but has also been shown to interact with the androgen receptor (Zou et al., 2009) and E2F transcription factor in the case of human ATAD2 (Revenko et al., 2010), indicating that this domain may play a central role in the potential dual function of ATAD2-like proteins. It is also tempting to imagine that this domain may be involved in a cellular protective mechanism which would “sense” an increase in nucleosome density and in response would favor the chromatin remodeler functions of Yta7 instead of its transcriptional role. The phosphorylation of ATAD2/Yta7 in its N-terminal region could allow switching towards a chaperone function of the protein by enabling its detachment from chromatin. The histone binding of Yta7 is mediated both by its bromodomain and the N-terminal region (Gradolatto et al., 2009), and we can therefore speculate that a functional inactivation or a lack of the N-terminal part of ATAD2/Yta7 could transform the factor into a pure chromatin regulator. In support of this hypothesis, a spliced form of Atad2 lacking this N-terminal acidic domain is expressed exclusively in mouse spermatogenic cells (Caron et al., 2010), where one of the most extensive chromatin remodeling takes place (Goudarzi et al., 2014). However, neither in human cells nor in yeast is such N-terminally shortened protein found expressed, suggesting that, if the above hypothesis is right, in the cases of human or yeast cells, a regulatory mechanism should inactivate the transcriptional role of the N-terminal domain to transform the protein into a pure chromatin regulator. This regulation could involve changes, such as Yta7 phosphorylation at its N-terminal part, which would lead to the release of the protein from histone genes (Kurat et al., 2011). The data discussed above therefore support the idea that ATAD2 and Yta7 follow similar regulatory rules, since the underlying activity remains conserved (even if their mechanisms of action could be regulated differently due to specific requirements and evolutionary adaptations).
CONCLUSION
ATAD2 is one of the most conserved proteins in eukaryotes although paradoxically its function remains obscure. The remarkable conservation of its functional domains which, as shown here, goes beyond the AAA+ ATPase and the bromodomain, pleads in favor of conserved activities. An almost systematic up-regulation of ATAD2 in all cancers (Boussouar et al., 2013) also indicates a critical cellular activity for this protein. Therefore, an intriguing question is why, despite these important features, the ATAD2-like proteins have not been identified as critical components in the hundreds of functional screens that have been undertaken in the recent years. One explanation is that the function of ATAD2/Yta7 is so diverse and generalist, involved in many cellular activities, that none of them is dramatically affected by the loss of ATAD2/Yta7, but all work more efficiently in the presence of these proteins. ATAD2 family members could therefore be important auxiliary factors in many chromatin related activities, including transcription.
Finally, as illustrated in this review, the studies done on the yeast S. cerevisiae Yta7 protein are complementary to the studies conducted on ATAD2 in mammals, which are mostly connected to cancer. In this regard, characterizing the function of ATAD2 proteins in the fission yeast Schizosaccharomyces pombe, another yeast model distantly related to budding yeast and often used for studying chromatin biology, could also be informative. This yeast possesses several pathways and his-tones modifications, like RNA interference (RNAi) and the H3K9me (histone H3 methylated on lysine 9) heterochromatin mark, that are present in larger eukaryotes but absent in S. cerevisiae. Similarly, LEX-1 (Lin-48 EXpression abnormal protein 1), the ATAD2 homologue in the worm Caenorhabditis elegans, was found in a screen to control the expression of repeated transgenes, suggesting that repeated sequences may also be genomic targets for ATAD2 proteins (Tseng et al., 2007). Hence, complementary studies on ATAD2 homologues conducted in the yeast S. cerevisiae or other eukaryotic model organisms shall continue to bring more valuable information about the general and conserved functions of ATAD2-like proteins, part of which may be deregulated in cancer.
Acknowledgments
ATAD2 project in SK laboratory is supported by INCa. YM is post-doctoral fellow supported by foundation ARC and MJ is recipient of a PhD fellowship from INCa. Research done in AV laboratory is supported by funds from the French Institut National de la Santé Et de la Recherche Médicale (INSERM), the Université Joseph Fourier (UJF), the Agence National de la Recherche (ANR) and the Association pour la Recherche contre le Cancer (ARC).
REFERENCES
- Boussouar F., Jamshidikia M., Morozumi Y., Rousseaux S., Khochbin S. Malignant genome reprogramming by ATAD2. Biochim. Biophys. Acta. 2013;1829:1010–1014. doi: 10.1016/j.bbagrm.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Caron C., Lestrat C., Marsal S., Escoffier E., Curtet S., Virolle V., Barbry P., Debernardi A., Brambilla C., Brambilla E., et al. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene. 2010;29:5171–5181. doi: 10.1038/onc.2010.259. [DOI] [PubMed] [Google Scholar]
- Ciro M., Prosperini E., Quarto M., Grazini U., Walfridsson J., McBlane F., Nucifero P., Pacchiana G., Capra M., Christensen J., et al. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 2009;69:8491–8498. doi: 10.1158/0008-5472.CAN-09-2131. [DOI] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., et al. Phylogeny.fr: robust phylogenetic analysis for the nonspecialist. Nucleic Acids Res. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Audic S., Claverie J.M., Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C., Carlson J.E., Zeng L., He C., Aggarwal A.K., Zhou M.M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J.M., Taly J.F., Notredame C. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M.E., Flaherty K., Prochasson P. The Saccharomyces cerevisiae histone chaperone Rtt106 mediates the cell cycle recruitment of SWI/SNF and RSC to the HIR-dependent histone genes. PLoS One. 2011;6:e21113. doi: 10.1371/journal.pone.0021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J., Kainth P., Lambert J.P., van Bakel H., Tsui K., Pena-Castillo L., Nislow C., Figeys D., Hughes T.R., Greenblatt J., et al. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell. 2009;35:340–351. doi: 10.1016/j.molcel.2009.06.023. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J.P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Muller S., Pawson T., et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi A., Shiota H., Rousseaux S., Khochbin S. Genome-scale acetylation-dependent histone eviction during spermatogenesis. J. Mol. Biol. 2014 doi: 10.1016/j.jmb.2014.02.023. pii: S0022-2836(14)00122-3. [DOI] [PubMed] [Google Scholar]
- Gradolatto A., Rogers R.S., Lavender H., Taverna S.D., Allis C.D., Aitchison J.D., Tackett A.J. Saccharomyces cerevisiae Yta7 regulates histone gene expression. Genetics. 2008;179:291–304. doi: 10.1534/genetics.107.086520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradolatto A., Smart S.K., Byrum S., Blair L.P., Rogers R.S., Kolar E.A., Lavender H., Larson S.K., Aitchison J.D., Taverna S.D., et al. A noncanonical bromodomain in the AAA ATPase protein Yta7 directs chromosomal positioning and barrier chromatin activity. Mol. Cell. Biol. 2009;29:4604–4611. doi: 10.1128/MCB.00160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan A., Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- Jambunathan N., Martinez A.W., Robert E.C., Agochukwu N.B., Ibos M.E., Dugas S.L., Donze D. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics. 2005;171:913–922. doi: 10.1534/genetics.105.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemena C., Notredame C. Upcoming challenges for multiple sequence alignment methods in the high-throughput era. Bioinformatics. 2009;25:2455–2465. doi: 10.1093/bioinformatics/btp452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat C.F., Lambert J.P., van Dyk D., Tsui K., van Bakel H., Kaluarachchi S., Friesen H., Kainth P., Nislow C., Figeys D., et al. Restriction of histone gene transcription to S phase by phosphorylation of a chromatin boundary protein. Genes Dev. 2011;25:2489–2501. doi: 10.1101/gad.173427.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi L.M., Ellahi A., Rine J. Direct regulation of nucleosome density by the conserved AAA-ATPase Yta7. Proc. Natl. Acad. Sci. USA. 2011;108:E1302–1311. doi: 10.1073/pnas.1116819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., Lopez R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013;41:W597–600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenko A.S., Kalashnikova E.V., Gemo A.T., Zou J.X., Chen H.W. Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol. Cell. Biol. 2010;30:5260–5272. doi: 10.1128/MCB.00484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackett A.J., Dilworth D.J., Davey M.J., O’Donnell M., Aitchison J.D., Rout M.P., Chait B.T. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 2005;169:35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng R.J., Armstrong K.R., Wang X., Chamberlin H.M. The bromodomain protein LEX-1 acts with TAM-1 to modulate gene expression in C. elegans. Mol. Genet. Genomics. 2007;278:507–518. doi: 10.1007/s00438-007-0265-6. [DOI] [PubMed] [Google Scholar]
- UniProt C. Activities at the universal protein resource (UniProt) Nucleic Acids Res. 2014;42:D191–198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J.X., Revenko A.S., Li L.B., Gemo A.T., Chen H.W. ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERalpha, is required for coregulator occupancy and chromatin modification. Proc. Natl. Acad. Sci. USA. 2007;104:18067–18072. doi: 10.1073/pnas.0705814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J.X., Guo L., Revenko A.S., Tepper C.G., Gemo A.T., Kung H.J., Chen H.W. Androgen-induced coactivator ANCCA mediates specific androgen receptor signaling in prostate cancer. Cancer Res. 2009;69:3339–3346. doi: 10.1158/0008-5472.CAN-08-3440. [DOI] [PubMed] [Google Scholar]
- Zunder R.M., Rine J. Direct interplay among histones, histone chaperones, and a chromatin boundary protein in the control of histone gene expression. Mol. Cell. Biol. 2012;32:4337–4349. doi: 10.1128/MCB.00871-12. [DOI] [PMC free article] [PubMed] [Google Scholar]