Abstract
This study is to investigate the effect and mechanism of reduced hypoxia-inducible factor (HIF)-1a expression on malignant behavior of MDA-MB-231 cells. HIF-1α expression was interfered by siRNA. Western blot was used to detect protein expression of HIF-1α, active fragments of caspase 3 and vimentin. Cell count, flow cytometry and Hoechst staining were used to evaluate cell growth and apoptosis. Matrigel invasion and wound scratch assay were performed to measure the ability of cell invasion and migration. After MDA-MB-231 cells were transfected with HIF-1α-targeted siRNA, HIF-1α protein expression was successfully interrupted and cell growth was retarded. Compared with random siRNA group, reduced HIF-1α protein expression in HIF-1α-targeted siRNA group facilitated cell apoptosis but had no effect on cell cycle. In addition, cells treated with HIF-1α-targeted siRNA expressed active fragments of caspase 3 (17 and 12 kD) after serum starvation for 0 to 60 h. Caspase 3 activity assay further confirmed the above finding. Reduced HIF-1α expression impaired the migration and invasiveness with a reduction in the expression of vimentin and CK18 protein. Inhibition of HIF-1α protein synthesis or enhancement of its degradation reversed its malignant phenotypes and could probably be a potential means for the treatment of triple-negative breast cancer.
Keywords: Hypoxia-inducible factor-1, triple-negative breast cancer, small-interfering RNA, epithelial mesenchymal transition
Introduction
Hypoxia-inducible factor (HIF) is the main reason for malignant tumor survival in hypoxia. As a transcriptional activator, HIF can induce the expression of hundreds of target genes, which promote angiogenesis, enhance hypoxia tolerance and inhibit apoptosis. Therefore, HIF acts as an important factor in tumor formation, growth and malignant progression [1-3]. HIF is a heterodimer that is composed of a constantly expressed b subunit and an oxygen-regulated a subunit. By now, three a subunits have been identified, among which 1a and 2a subunits both belong to the basic helix-loop-helix (HLH) Per/Arnt/Sim (PAS) (bHLH-PAS) family. The molecular structures of 1a and 2a subunits contain bHLH-PAS (PAS mediates their combination with b subunit to form HIF-1 and HIF-2), oxygen-dependent degradation domain (mediates aerobic degradation) and transcriptional activity domain (responsible for transcriptional activation) [4-6]. The subunit 3a only contains the conserved domains of HLH and PAS but lacks the oxygen-dependent domain. However, it has been discovered that the mRNA of 3a has another spliced variant, inhibitory PAS domain protein (IPAS), which contains N-terminal HLH and PAS but lacks the transcriptional activity domain. The combination of 3a subunit and HIF-1α negatively regulates HIF [7]. Our previous study showed that breast cancer cell line T47D can also express HIF-1α under normal oxygen conditions, and promote the secretion of vascular endothelial growth factor, suggesting that HIF-1 may also play an important role in promoting tumor under aerobic condition [8].
Clinically, triple-negative breast cancer is a type of breast cancer with immune phenotypes of negative estrogen receptor, progesterone receptor and human epidermal growth factor receptor-2. It is characterized by young onset, high histological grade, fast growth, strong invasion, easy metastasis, tolerance to chemotherapy, epithelial mesenchymal transition and lack of targeted therapeutic molecules. Breast cancer cell line MDA-MB-231 that shows triple-negative immune phenotype, is considered to be a cell line demonstrating epithelial mesenchymal transition, which is characterized as negative expression of E-Cadherin and positive expression of N-Cadherin and vimentin [9]. Previous study identified the high expression of HIF-1α protein in MDA-MB-231 cells. This study investigates the effect of decreased expression of HIF-1α protein on the biological behavior of triple-negative breast cancer cell line MDA-MB-231 and its possible mechanism.
Materials and methods
Ethics
The study was approved by the ethics review board of the Affiliated Hospital of Inner Mongolia Medical University and the Inner Mongolia Medical University.
Cell culture
Human breast cancer cell lines MDA-MB-468 and MDA-MB-453 were purchased from the Basic Medical Research Institute of China Medical Academy of Sciences. MDA-MB-231 and MCF-7 were obtained from the Department of Pathology, Peking University Health Science Center, and T47D was a gift from the University of Sheffield. Cells were cultured in RPMI 1640 or DMEM medium supplemented with 10% fetal bovine serum at 37°C in a humidified environment with 21% O2 and 5% CO2.
Western blot
Total cellular proteins were separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane. Then membrane was incubated with mouse anti–human monoclonal antibody HIF-1α (R&D Systems, USA), mouse caspase-3 monoclonal antibody and β-tubulin (Beyotime Institute of Biotechnology, China) or cytokeratin CK18 and Vimentin (Santa Cruz Biotechnology, Inc., USA) antibodies. Next, the membranes were incubated with horseradish peroxidase-conjugated IgG with routine chemiluminescence.
Small-interfering RNA (siRNA) interference
Transfection agents and siRNA duplexes that targeted HIF-1α mRNA or control were purchased from Santa Cruz Biotechnology, Inc., USA. Cells were transfected with either siRNA duplexes that targeted HIF-1α (HIF-1α: sense, 5-UCAAGUUGCUGGUCAUCAGdTdT-3 and antisense, 5-CUGAUGACCAGCAACUUGAdTdT-3) or with control siRNA that did not target any known genes. According to the manufacturer’s protocol, siRNA duplex (80 pmol) and 18 μl siRNA transfection reagent were added into cells with 600 μl siRNA transfection medium in 6-well plate for incubation of 7 h, followed by the replacement with normal culture medium containing 20% normal serum and antibiotics.
Cell proliferation curve
HIF-1α-targeted and randomly interfered Cells (1 × 104/ml) were seeded in 24-well plates respectively and cultured for 9 days with RPMI 1640 containing 10% fetal calf serum. Then, the cells were stained with trypan blue. Cell numbers of four wells were counted for each group every other day. The average cell numbers were used to plot the growth curve.
Flow cytometry
According to the manufacturer’s protocol and after appropriate treatment, cells with either HIF-1α-targeted or randomly interfered siRNA were stained with either propidium iodide (IP) or Annexin V-fluorescent isothiocyanate (FICT) in combination with IP. Cell cycle and apoptosis were analyzed by flow cytometry.
Detection of cell apoptosis
Cells of 70% confluence were cultured for 0, 24, 48, and 60 hours in serum-free medium, and then collected in Eppendorf tubes. Cells were digested with trypsin and prepared into cell suspension in phosphate buffered saline (PBS). Then, binding buffer, Annexin V and propidium iodide were added according to the manufacture’s instruction (Beyotime Institute of Biotechnology, China).
Hoechst staining
The pretreated coverslips were placed in six-well tissue culture plates and the cells were cultured overnight to reach 70-80% confluence. After incubation with serum-free medium for different time periods, the cells were fixed with stationary liquid for 10 minutes. According to the instructions (Beyotime), the cells were washed with PBS, followed by the addition of Hoechst 33258 staining solution before incubation for 5 minutes in dark. After washing with PBS, anti-fluorescence quenching agent was dropped onto the glass slide that was then covered with a cover slip with cells. Then, the blue nuclei were examined using fluorescence microscopy.
Detection of caspase activity
Cells transfected with HIF-1α-targeted siRNA or random interference siRNA in serum-free culture were collected and digested with trypsin. The cells were centrifuged at 600 × g at 4°C for 10-15 minutes and washed with PBS once. Then, cell lysate was added in the ration of 2 million cells in 100 μL lysate according to the kit (Beyotime) and cell precipitation was resuspended. The supernatants were lysed on ice for 15 minutes. Then, cell lysates were centrifuged at 4°C at 16,000-20,000 × g for 10-15 minutes and the supernatant was transferred to a centrifuge tube pre-cooled with ice bath. Next, the detection buffer was added in accordance with the reagent instruction and the sample was added and appropriately mixed. Subsequently, 10 μl 2 mM ice-cold Ac-DEVD-pNA was mixed at 37°C for 60-120 minutes. When the color significantly changed, absorbance at 405 nm was measured.
Matrigel invasion assay in vitro
NIH3T3 cells were cultured in serum-free DMEM medium for 24-48 h and the cell culture was centrifuged at 12,000 rpm for 15 min. Then, 200 μl supernatant was separated as chemokine and added to the lower chamber of the Boyden chamber. The 8 µm polycarbonate microporous membrane was placed between the upper and lower chambers. Suspension (400 μl) of cells (2 × 105) at logarithmic growth phase was added into the upper chamber and cultured at 37°C with 5% CO2 for 12 h. Liquid in the upper chamber was removed and the non-invasion cells on the membrane surface were wiped off with wet cotton swab. After being rinsed with physiological saline and dried, the chamber was fixed with methanol for 30 min and then stained routinely with hematoxylin & eosin. Each group had 3 parallel samples and the number of cell in 5 fields was counted under a 200 × microscope. Then, the average cell number was calculated followed by statistical analysis.
Cell scratch migration test
Cells in logarithmic growth phase were cultured in 6-well plates until confluence. Then, 200 μl sterile pipetting tip was used to scratch the well bottom and PBS was used to wash off cell debris for 3 times. Then, medium containing 10% fresh fetal calf serum was added into each well, followed by incubation at 37°C in 5% CO2 for 24 h before being observed under inverted microscope. Scratch width was measured using built-in ruler of the inverted microscope (10 μm). Scratch migration rate = (scratch width at 0 h-scratch width at 12 h)/scratch width at 0 h × 100%.
Statistical analysis
The results were analyzed using SPSS 17.0 software. The difference between two samples was determined using t-test. P < 0.05 was considered statistically significant and P < 0.01 was considered statistically very significant.
Results
Breast cancer cells have universal expression of HIF-1α protein under normal oxygen conditions, the levels of which may be associated with tumor growth and invasion capabilities
To measure the basal expression of HIF-1α in breast cancer cell lines under normal oxygen conditions, MDA-MB-231, MCF-7, SK-3, MDA-MB-468, MDA-MB-453, and T47D cells were cultured in serum-free medium for 24 h before the extraction of total cellular protein and the determination of HIF-1α expression using Western blot. The results showed that all these breast cancer cell lines were found to express HIF-1α protein under normal oxygen condition. Of note, triple-negative cell line MDA-MB-231 that had the strongest growth and invasion capability demonstrated the highest expression level of HIF-1α (Figure 1). These data indicated that breast cancer cells had universal expression of HIF-1α protein under normal oxygen conditions, the levels of HIF-1α expression which might be associated with tumor growth and invasion capabilities.
Figure 1.
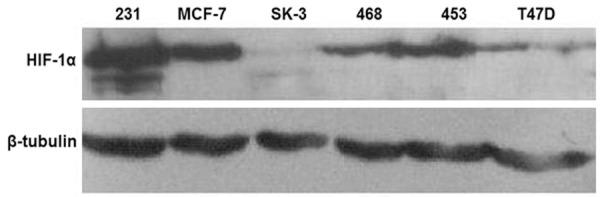
Expression of HIF-1α in breast cancer cell lines under normal oxygen conditions. After serum starvation for 24 h, total cellular protein was extracted from MDA-MB-231, MCF-7, SK-3, MDA-MB-468, MDA-MB-453, and T47D cells, and the protein level of HIF-1α was detected using Western blot. β-tubulin was used as internal reference.
Inhibition of HIF-1α by siRNA suppresses MDA-MB-231 cell growth
To investigate the effect of HIF-1 on the biological behavior of malignant breast cancer cells, HIF-1α-targeted siRNA or random siRNA was used on triple-negative breast cancer cell line MDA-MB-231, followed by the extraction of total cellular protein that was examined using Western blot. In addition, cells treated with random siRNA and HIF-1α-targeted siRNA were cultured for 9 consecutive days, and the number of cells in each group was counted every other day. To elucidate the mechanism of growth inhibition, flow cytometry was used to detect the cell cycle of cells transfected with randomized siRNA or HIF-a-targeted siRNA. Western blot results showed that HIF-1α-targeted siRNA effectively inhibited the expression of HIF-1α protein (Figure 2A). Cell growth curve showed that HIF-a-targeted siRNA significantly decreased the number of cells on each day (Figure 2B). In addition, data of flow cytometry showed no significant difference in cell proliferation ratio between the two groups. These data suggested that inhibition of HIF-1α by siRNA suppressed MDA-MB-231 cell growth.
Figure 2.
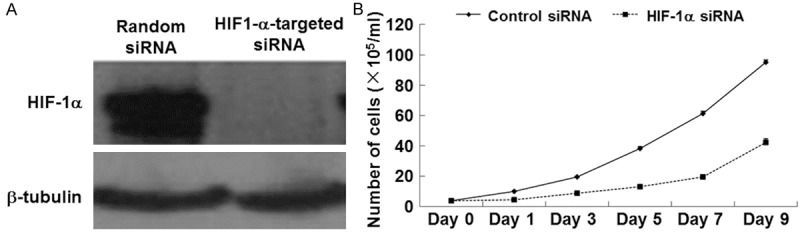
A. Expression of HIF-1α in MDA-MB-231 cells transfected with random siRNA or HIF-1α-targeted siRNA. After serum starvation for 12 h, total cellular protein was extracted and the protein level of HIF-1α was detected using Western blot. β-tubulin was used as internal reference. B. Growth curves of MDA-MB-231 cells transfected with random siRNA or HIF-1α-targeted siRNA. Cells were seeded in 24-well plates and cultured for 9 days. Cell numbers of four wells were counted every other day for each group. The average cell numbers were used to plot the growth curve (n = 3). The data were expressed as means ± SEM. *, P < 0.05 compared with control at respective time point.
Inhibition of HIF-1α by siRNA facilitates the apoptosis of MDA-MB-231 cells
To explore the influence of HIF-1 on MDA-MB-231 cell apoptosis, Hoechst staining, flow cytometry, Western blot and spectrophotometry were used. The results of Hoechst 33258 fluorescence staining showed that the number of apoptotic cells transfected with HIF-1α-targeted siRNA was significantly higher than that of the random siRNA group at 24, 48 and 60 h of serum starvation (Figure 3). Consistent with this observation, flow cytometry using AnnexinV-PI staining on cells treated in the same way demonstrated that the percentage of apoptotic cells in HIF-1α-targeted siRNA group was higher than that in random siRNA group at 48 and 60 h (Table 1). Western blot analysis of the protein level of caspase 3 showed that 35 kD caspase 3 in both groups did not show significant change, but the level of 17 kD active fragments of caspase 3 were identified in HIF-1α-targeted siRNA group after 0, 24, 48 and 60 h of serum starvation and 12 kD active fragment was also identified at 60 h (Figure 4A). Furthermore, detection of caspase 3 activity using spectrophotometer showed that the activity of caspase 3 in HIF-1α-targeted siRNA group was higher than that in random siRNA group (Figure 4B). These data indicated that inhibition of HIF-1α by siRNA facilitated the apoptosis of MDA-MB-231 cells.
Figure 3.
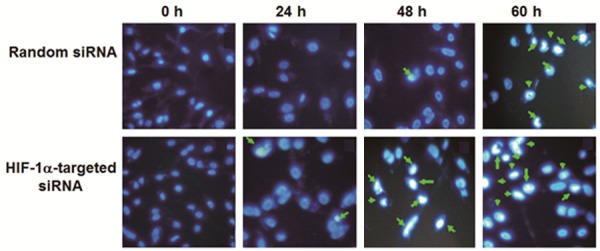
Hoechst 33258 fluorescence staining of cells transfected with random siRNA or HIF-1α-targeted siRNA after serum starvation for 0, 24, 48, and 60 h. Cell apoptosis was detected using Hoechst staining kit and apoptotic cells were indicated with white arrows.
Table 1.
Cell apoptosis detected using Annexin V-PI-FITC
| (Annexin V/PI) % ± SD | 0 h | 24 h | 48 h | 60 h |
|---|---|---|---|---|
| Random siRNA | 1.92% ± 0.45 | 7.39% ± 1.51 | 16.1% ± 4.24 | 23.7% ± 4.78 |
| HIF-1α-targeted siRNA | 3.73% ± 1.20 | 9.76% ± 2.12 | 31.39% ± 5.76* | 44.75% ± 6.14* |
Note: After cells were deprived of serum for different time periods, cell apoptosis rate was determined using Annexin V-PI staining.
P < 0.01, compared with randomized control siRNA group.
Figure 4.
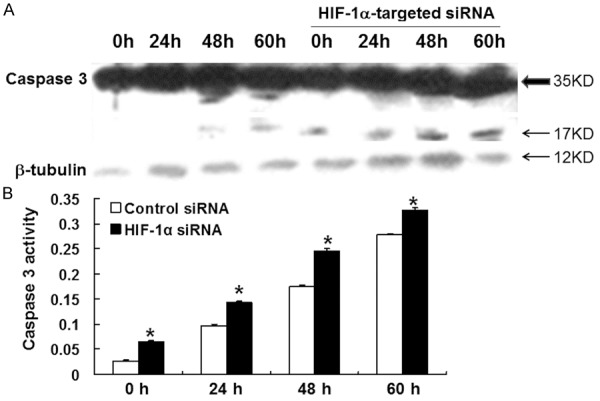
A. Expression of caspase 3 and its active fragments in cells transfected with random siRNA and HIF-1α-targeted siRNA after serum starvation for 24, 48, and 60 h. Total cellular protein was extracted and the expression of caspase 3 was analyzed using Western blot. β-tubulin was used as internal reference. B. Caspase 3 activity detected as absorption intensity (405 nm) from cell lysates from random siRNA and HIF-1α-targeted siRNA groups after serum starvation for 24, 48, and 60 h. Data were means ± SEM. *, P < 0.01 compared with control at respective time point.
Inhibition of HIF-1α suppressed MDA-MB-231 cell invasion and migration
To test the effect of HIF-1α on MDA-MB-231 cell invasion and migration, Matrigel invasion assay, scratch migration test, two-dimensional gel electrophoresis, mass flight spectrum and Western blot were performed. In vitro invasion assay showed that the number of transmembrane cells in HIF-1α-targeted siRNA group (44.13 ± 3.68) was lower than that in random siRNA group (93.13 ± 8.21), with statistically significant difference (P < 0.05) (Figure 5). Results of scratch migration test showed that the migration rate was 25% in cells transfected with HIF-1α-targeted siRNA, which was significantly lower than the migration rate of random siRNA group (50%), suggesting that reduced HIF-1α protein expression significantly weakened the migration ability of the cells (Table 2). Two-dimensional gel electrophoresis and mass flight spectrum showed that CK18 and Vimentin were significantly reduced in cells transfected with HIF-1α-targeted siRNA, which was further verified by Western blot (Figure 6), indicating that HIF-1 interference might induce mesenchymal epithelial transition. These data demonstrated that inhibition of HIF-1α suppressed MDA-MB-231 cell invasion and migration.
Figure 5.
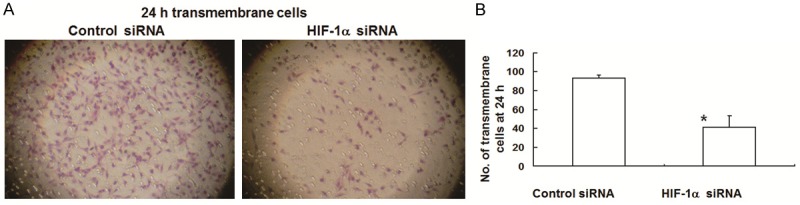
A. MDA-MB-231 cell invasion in random siRNA and HIF-1α-targeted siRNA groups. Cells were added into the upper chamber of Boyden chamber. When cells migrated through Matrigel and polycarbonate membrane after 24 h, the filtration membrane was treated, followed by routine hematoxylin & eosin staining. B. Quantification of the number of transmembrane cells. The data were expressed as means ± SEM. *, P < 0.05 compared with control.
Table 2.
MDA-MB-231 cell scratch migration
| MDA-MB-231 | 0 h scratch width (μm) | 12 h scratch width (μm) | Scratch migration rate (%) |
|---|---|---|---|
| Random siRNA | 20 | 10 | 50% |
| HIF-1α-targeted siRNA | 20 | 15 | 25% |
Figure 6.
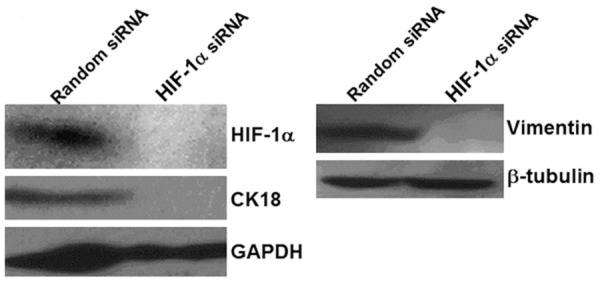
Expression of CK18 and Vimentin in cells transfected with random siRNA and HIF-1α-targeted siRNA after serum starvation or 24 h. Total cellular protein was extracted and CK18 and Vimentin protein expression was analyzed using Western blot. β-tubulin was used as internal reference.
Discussion
Our previous study found that HIF-1α subunits demonstrated basically expression at normal oxygen concentration in breast cancer cell line T47D, and were also regulated by fibroblast growth factor to promote the secretion of vascular endothelial growth factor target genes [8]. This study examined breast cancer cell lines with various immune phenotypes such as estrogen positive, progesterone receptor positive, and human epidermal growth factor receptor-2 positive, as well as triple-negative cell lines with negative estrogen, progesterone receptors and human epidermal growth factor receptor-2. All cell lines showed basal expression of HIF-1α in normal oxygen conditions, with the expression in triple-negative cell line, MDA-MB-231, being the strongest. Clinically, triple-negative breast cancer is a kind of cancer characterized by fast growth, strong invasion ability, rapid progression and lack of targeted treatment, indicating that HIF-1 may play a role in the malignant biological behavior of triple-negative breast cancer.
The expression of HIF-1α protein was significantly interfered using a specific siRNA that targeted HIF-1α, and the growth of MDA-MB-231 cells was obviously inhibited. Since cell growth rate mainly depends on the proportion of proliferating cells and cell apoptosis, flow cytometry showed that reduced expression of HIF-1α did not affect cell cycle. Both flow cytometry and Hoechst staining confirmed that reduced HIF-1α expression enhanced cell apoptosis in serum starvation. In the meantime, active fragments of caspase 3 were identified with improved activity, indicating that HIF-1 might maintain cell survival and growth through the inhibition of cell apoptosis. Recent studies found that HIF-1 or HIF-2 regulated the transcription of miR-210, which induced the loss of mitochondrial ion channel protein complex, thereby increasing cytochrome C release and caspase 3/7 activity [10]. HIF-1 can also induce a variety of apoptosis-related target proteins, including pro-apoptosis proteins, BNIP3 and Noxa [11,12]. Our other experiments showed that HIF-1α-targeted siRNA could increase the molecular weight of Bcl-2. Studies showed that Bcl-2 underwent phosphorylation at multiple sites, leading to the increase of molecular weight and the loss of anti-apoptosis function [13-15]. IPAS is a negative regulator of HIF-1, and leads to mitochondrial depolarization and caspase 3 activation by binding with the Bcl-2 family members, Bcl-xL, Bcl-w and Mcl-1, thereby promoting cell apoptosis [16]. These data confirmed the effect of HIF-1 in inhibiting cell apoptosis, which is mainly completed through the mitochondria/caspase pathway. Recently, in human glioma cell line U87MG, reduced HIF-1 expression by shRNA interference led to substantial loss of live cells blocked at G0-G1 phase, cell apoptosis and the disruption of cell migration and invasion [17].
This study further explored the invasion and migration capability of the cells transfected with HIF-1α-targeted siRNA. Matrigel transmembrane invasion and scratch migration tests showed that reduced expression of HIF-1α protein inhibited the invasion and migration ability of triple-negative breast cancer cell lines. Multiple target proteins of HIF-1 are involved in invasion and migration processes, including extracellular matrix degradation target proteins such as urokinase-type plasminogen activator receptor, collagen prolyl hydroxylase, matrix metalloproteinase (e.g. MMP-2), inhibitors of matrix metalloproteinases and intercellular adhesion molecules such as E-cadherin. According to one research, renal cell carcinoma lack of von Hippel-Lindau protein (mediating HIF-1α ubiquitination degradation) had increased expression of HIF-1α protein, which led to decreased expression of E-cadherin [18], suggesting that HIF-1 was involved in epithelial mesenchymal transition. In addition, chemokine-receptor (CXCR4) and its ligand (SDF-1) were an important way to control tumor invasion and metastasis, which was regulated directly by HIF-1 [19]. Cox-2 is one of the target proteins of HIF-1. In breast cancer cell line MCF-7 that lack of Cox-2, transient transfection by Cox-2 down-regulated E-cadherin and β-catenin and up-regulated the expression of N-cadherin, which were accompanied by increased invasion capability [20]. In another study of our group that uses miRNA to interfere with HIF-1α, the protein levels of extracellular matrix metalloproteinase 8, 10, 13 and 17 were significantly up-regulated as detected by real-time quantitative PCR, suggesting that HIF-1 might promote the migration and invasion of cells through the up-regulation of matrix metalloproteinase 8, 10, 13 and 17. This study also found that reduced expression of protein HIF-1α inhibited the expression of cytokeratin CK18 and Vimentin, indicating that the reduced expression of HIF-1α induced mesenchymal epithelial transformation [21,22].
In summary, HIF-1 plays important roles in facilitating angiogenesis, promoting tumor malignant progression and inducing resistance to chemotherapy and radiotherapy. The pro-angiogenetic effect might be useful for the treatment of ischemic diseases and the inhibition of angiogenesis could be equivalently important in the treatment of neoplastic lesions.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 30860331 and 81360395).
Disclosure of conflict of interest
None.
References
- 1.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 2.Doublier S, Belisario DC, Polimeni M, Annaratone L, Riganti C, Allia E, Ghigo D, Bosia A, Sapino A. HIF-1 activation induces doxorubicin resistance in MCF7 3-D spheroids via P-glycoprotein expression: a potential model of the chemo-resistance of invasive micropapillary carcinoma of the breast. BMC Cancer. 2012;12:4. doi: 10.1186/1471-2407-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwab LP, Peacock DL, Majumdar D, Ingels JF, Jensen LC, Smith KD, Cushing RC, Seagroves TN. Hypoxia-inducible factor 1a promotes primary tumor growth and tumor-initiating cell activity in breast cancer. Breast Cancer Research. 2012;14:R6. doi: 10.1186/bcr3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang BH, Rue E, Wang GL Roe R, Semenza GL. DNA binding and transactivation properties of hypoxia-inducible factor 1. J Biol Chem. 1996;271:17771–8. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- 5.Jiang BH, Zheng JZ, Leung SW, Roe R, Semenza GL. Transactivation and inhibitory domains of hypoxia-inducible factor 1 alpha. Modulation of transcriptional activity by oxygen tension. J Biol Chem. 1997;272:19253–60. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- 6.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1 alpha is mediated by an O-2-dependent degradation domain via the ubiquitin- proteasome pathway. Proc Natl Acad Sci U S A. 1998;95:7987–92. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makino Y, Cao R, Svensson K, Bertilsson G, Asman M, Tanaka H, Cao Y, Berkenstam A, Poellinger L. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–4. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- 8.Shi YH, Wang YX, Bingle L, Gong LH, Heng WJ, Li Y, Fang WG. In vitro study of HIF-1 activation and VEGF release by bFGF in the T47D breast cancer cell line under normoxic conditions: involvement of PI-3K/Akt and MEK1/ERK pathways. J of Pathol. 2005;205:530–6. doi: 10.1002/path.1734. [DOI] [PubMed] [Google Scholar]
- 9.Shah P, Gau Y, Sabnis G. Expression of Cox-2 in human breast cancer cells as a critical determinant of epithelial-to-mesenchymal transition and invasiveness. Expert Opin Ther Targets. 2014;18:121–35. doi: 10.1517/14728222.2014.860447. [DOI] [PubMed] [Google Scholar]
- 10.Puisségur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K, Cardinaud B, Hofman V, Fourre S, Magnone V, Ricci JE, Pouysségur J, Gounon P, Hofman P, Barbry P, Mari B. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death and Differentiation. 2011;18:465–78. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci U S A. 2000;97:9082–7. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim JY, Ahn HJ, Ryu JH, Suk K, Park JH. BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia-inducible factor 1alpha. J Exp Med. 2004;199:113–24. doi: 10.1084/jem.20030613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amamoto K, Ichijo H, Korsmeyer SJ. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-Terminal protein kinase pathway normally activated at G2/M. Mol Cell Biol. 1999;19:8469–78. doi: 10.1128/mcb.19.12.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo E, Ichijo H, Korsmeyer SJ. Expression of phosphorylated Ser70 of Bcl-2 correlates with malignancy in human colorectal neoplasms. Clin Cancer Res. 2005;11:7255–63. doi: 10.1158/1078-0432.CCR-05-0274. [DOI] [PubMed] [Google Scholar]
- 15.Blagosklonny MV, Schulte T, Nguyen P, Trepel J, Neckers LM. Taxol-induced apoptosis and phosphorylation of Bcl-2 protein involves c-Raf-1 and represents a novel c-Raf-1 signal transduction pathway. Cancer Res. 1996;56:1851–4. [PubMed] [Google Scholar]
- 16.Torii S, Goto Y, Ishizawa T, Hoshi H, Goryo K, Yasumoto K, Fukumura H, Sogawa K. Pro-apoptotic activity of inhibitory PAS domain protein (IPAS), a negative regulator of HIF-1, through binding to pro-survival Bcl-2 family proteins. Cell Death Differ. 2011;18:1711–25. doi: 10.1038/cdd.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong CG, Wu WK, Feng SY, Yu J, Shao JF, He GM. Suppressing the malignant phenotypes of glioma cells by lentiviral delivery of small hairpin RNA targeting hypoxia-inducible factor-1α. Int J Clin Exp Pathol. 2013;6:2323–32. [PMC free article] [PubMed] [Google Scholar]
- 18.Krishnamachary B, Zagzag D, Nagasawa H, Rainey K, Okuyama H, Baek JH, Semenza GL. Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel–Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res. 2006;66:2725–31. doi: 10.1158/0008-5472.CAN-05-3719. [DOI] [PubMed] [Google Scholar]
- 19.Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425:307–11. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- 20.Bocca C, Ievolella M, Autelli R. Expression of Cox-2 in human breast cancer cells as a critical determinant of epithelial-to-mesenchymal transition and invasiveness. Expert Opin Ther Targets. 2014;18:121–35. doi: 10.1517/14728222.2014.860447. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Huang G, Li X, Zhang Y, Jiang Y, Shen J, Liu J, Wang Q, Zhu J, Feng X, Dong J, Qian C. Hypoxia induces epithelial-mesenchymal transition via activation of SNAI1 by hypoxia-inducible factor -1α in hepatocellular carcinoma. BMC Cancer. 2013;13:108. doi: 10.1186/1471-2407-13-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barriga EH, Maxwell PH, Reyes AE, Mayor R. The hypoxia factor Hif-1α controls neural crest chemotaxis and epithelial to mesenchymal transition. J Cell Biol. 2013;201:759–76. doi: 10.1083/jcb.201212100. [DOI] [PMC free article] [PubMed] [Google Scholar]


