Background: HSV disrupts nuclear lamina for release from nucleus during productive infection.
Results: A cellular protein, p32, contributes to the release of HSV from nucleus.
Conclusion: p32 is hijacked by viral protein ICP34.5 to facilitate HSV nuclear egress and growth.
Significance: The discovery of a novel target for viral protein provides insight for viral propagation.
Keywords: Herpesvirus, Nuclear Lamina, Nuclear Membrane, Protein-Protein Interaction, Viral Protein, HSV, ICP34.5, p32 (gC1qR/HABP1), Viral Egress
Abstract
As a large double-stranded DNA virus, herpes simplex virus type 1 (HSV-1) assembles capsids in the nucleus where the viral particles exit by budding through the inner nuclear membrane. Although a number of viral and host proteins are involved, the machinery of viral egress is not well understood. In a search for host interacting proteins of ICP34.5, which is a virulence factor of HSV-1, we identified a cellular protein, p32 (gC1qR/HABP1), by mass spectrophotometer analysis. When expressed, ICP34.5 associated with p32 in mammalian cells. Upon HSV-1 infection, p32 was recruited to the inner nuclear membrane by ICP34.5, which paralleled the phosphorylation and rearrangement of nuclear lamina. Knockdown of p32 in HSV-1-infected cells significantly reduced the production of cell-free viruses, suggesting that p32 is a mediator of HSV-1 nuclear egress. These observations suggest that the interaction between HSV-1 ICP34.5 and p32 leads to the disintegration of nuclear lamina and facilitates the nuclear egress of HSV-1 particles.
Introduction
Herpes simplex virus type 1 (HSV-1)4 is a human pathogen responsible for human diseases such as keratitis and encephalitis (1). As a member of the Alphaherpesvirinae subfamily, HSV-1 is able to establish latency or productive infection where the virus undergoes entry, viral gene expression, and DNA replication. Upon capsid assembly, viral particles exit the nucleus by budding through the inner nuclear membrane (2–4). In doing so, HSV-1 triggers conformational changes of the nuclear lamina lining the inner nuclear membrane (5), which favors nuclear egress of nucleocapsids (6).
HSV-1 egress is a complex process that requires a variety of viral proteins. Besides viral glycoproteins (such as gB, gK, gD, gE, and gM) (7–10), several tegument proteins are reported to modulate the nuclear egress of HSV-1. For example, UL31 and UL34 are required for conformational changes of nuclear lamina in HSV-1-infected cells (11–14). Of note, UL31 is thought to form a complex with UL34 and interact with lamin A/C at the nuclear rim (15, 16), which disrupts the nuclear lamina by interference with lamin-lamin interactions (5). This process also involves protein kinase C (PKC), lamin B (12), lamin B receptor (LBR), and emerin (6, 17, 18). Furthermore, Us3, UL13, and UL47 are linked to egress of the viral particles via phosphorylation of UL34 and localization of UL31-UL34 complex to the nuclear membrane (19–21). Additionally, UL12, a viral alkaline nuclease, associates with the cellular protein nucleolin, which is needed for efficient nuclear egress (22).
Previous studies suggest that nuclear egress involves HSV-1 ICP34.5 (23, 24), which is an essential protein in viral pathogenesis (25–27). HSV-1 ICP34.5 consists of 263 amino acids with an amino-terminal domain, a linker Ala-Thr-Pro repeat, and a carboxyl-terminal domain. The amino-terminal domain of ICP34.5 contains a nucleolar localization signal and a nuclear export signal, whereas the carboxyl-terminal domain contains a bipartite nuclear import signal (28). These signals direct shuttling of ICP34.5 between the nucleus and the cytoplasm, which is likely required for different functions. The multiple roles of ICP34.5 have emerged from the fact that ICP34.5 associates with various cellular proteins. For instance, ICP34.5 interacts with TBK1 and IκB kinase α/β, suppressing production of interferon (29), antigen presentation, and T cell response (30). ICP34.5 also inhibits autophagy by binding to Beclin-1 (31). In addition, ICP34.5 acts as a scaffold for protein phosphatase 1 (32) and eIF-2α to promote protein synthesis in infected cells (33). Furthermore, ICP34.5 contributes to viral egress, a function independent of its anti-interferon activity (23). Nevertheless, the mechanism of ICP34.5 involved in viral egress has yet to be unveiled.
In this study, we report that ICP34.5 targets a cellular protein, p32, also known as component 1q receptor (C1qR) or hyaluronic acid-binding protein (HABP1). p32 is an abundant protein primarily located in mitochondrial matrix and associates with a number of cellular proteins including LBR (34). We show that ICP34.5 binds to and redirects p32 to the LBR in HSV-1(F)-infected cells. The interaction between ICP34.5 and p32 leads to the redistribution of nuclear lamina and facilitates the nuclear egress of HSV-1. These results suggest that p32 is a novel target for ICP34.5 and plays a significant role in the productive infection of HSV-1.
EXPERIMENTAL PROCEDURES
Cells and Viruses
HeLa, HEK293T, and Vero cells were originally obtained from the American Type Culture Collection (ATCC) and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) penicillin/streptomycin. U251 cells were kindly provided by Prof. S. Yu (Tianjin Medical University Cancer Institute and Hospital, China) and cultured in DMEM supplemented with 10% (v/v) FBS and 1% (v/v) penicillin/streptomycin. HSV-1(F) and its derived γ134.5-null virus, R3616, were described previously (25).
Plasmids
To construct FLAG-ICP34.5, a DNA fragment encoding ICP34.5 was obtained by PCR and cloned into the BamHI/XhoI site of pCMV-Tag2B vector. To construct GFP-p32, p32 coding region was obtained by PCR and cloned into the XhoI/BamHI site of pEGFP-C1 vector. The short hairpin RNA (shRNA) targeting p32 and a random sequence RNA were obtained from Xu et al. (35). Oligonucleotides were synthesized and cloned into the AgeI/EcoRI site of pLKO.1 vector. Control plasmid pLKO.1-shVav2 was from our laboratory.
Reagents
Anti-mouse p32 antibody (sc-271200), anti-rabbit p32 antibody (sc-48795), anti-lamin A/C antibody (sc-7292), agarose conjugated with protein G (sc-2002), goat anti-rabbit IgG-FITC (sc-2012), goat anti-mouse IgG-TRITC (sc-2092), anti-PKCδ antibody (sc-937), the PKC inhibitors RO-31-7549 (sc-24005), rottlerin (sc-3550), and PKCζ pseudosubstrate (sc-3098) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-phospho-lamin A/C (Ser(P)-22) (2026S) antibody was purchased from Cell Signaling Technology (Danvers, MA). Anti-HSV-1 antibody (B0114) was purchased from Dako, Inc. (Carpinteria, CA). Anti-FLAG antibody (F1804), anti-α-tubulin antibody (T6074), and anti-GFP antibody (G6539) were purchased from Sigma. Anti-LBR antibody (E398L) (ab32535) and anti-coilin antibody (ab11822) was purchased from Abcam (Cambridgeshire, UK). MitoTracker Red CMXRos was purchased from Invitrogen.
Immunoblotting
Cells were harvested, washed with phosphate-buffered saline (PBS), and lysed with ice-cold radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm EDTA, 1.0% Triton X-100, and protease inhibitor mixture) for 30 min on ice. After centrifugation, supernatants were boiled in 1× loading buffer (50 mm Tris-HCl, pH 6.8, 2% SDS (w/v), 0.1% bromphenol blue, 10% glycerol, and 100 mm β-mercaptoethanol). The protein contents were separated by 12% SDS-PAGE, transferred to PVDF membranes, and identified with the indicated antibodies.
Immunoprecipitation Analysis
To examine protein interactions, transfected or infected HeLa cells were harvested and lysed with ice-cold radioimmune precipitation assay buffer for 30 min on ice. After centrifugation, cell extracts were obtained and incubated with antibody and agarose conjugated with protein G for 2 h at 4 °C. The beads were washed four times with wash buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm EDTA, 0.1% Triton X-100, and protease inhibitor mixture) and boiled in 1× loading buffer. The immobilized proteins were then subjected to immunoblotting analysis.
Virus Growth Assay
HeLa cell monolayers were infected with HSV-1(F) or R3616 at the indicated multiplicity of infection (m.o.i.). The free viral particles in the culture medium and cell-associated viruses were collected at the indicated hours postinfection (hpi), frozen, and thawed three times followed by infection of Vero cells with serial dilution. Virus titers were determined by oncolytic plaques on a monolayer of Vero cells.
Lentivirus-based Transduction
To generate HeLa cells with knockdown of the p32 protein or an unrelated Vav2 protein, HEK293T cells were transfected with plasmids including pLKO.1-shRNA, pCMV-VSV-G, pMDLg/pRRE, and pRSV-REV. At 48 h post-transfection, the viral particles in the supernatants were pelleted by centrifugation at 20,000 × g for 1.5 h, resuspended in DMEM supplemented with 10% (v/v) FBS, and incubated with HeLa cells at 37 °C for 24 h. After selection with 1.0 μg/ml puromycin for 7 days, HeLa cells with knockdown of p32 or Vav2 were obtained.
Confocal Microscopy
Infected or transfected HeLa cells were incubated with 100 nm MitoTracker Red CMXRos in DMEM for 30 min at 37 °C. Cells were then washed with PBS, fixed with ice-cold 4% paraformaldehyde for 30 min, permeabilized with 0.5% Triton X-100 in PBS, and incubated with primary antibodies at 4 °C overnight. Cells were then washed three times and incubated with fluorescence (FITC or TRITC)-conjugated secondary antibodies for 2 h at room temperature. After washing five times with PBS, cells were mounted and visualized with a TCS SP5 confocal microscope.
Cell Fractionation
Infected cells were harvested, washed with PBS, and lysed with 0.2% Nonidet P-40 in PBS and protease inhibitor mixture for 15 min on ice. After centrifugation for 5 min, supernatants were transferred to a new tube. The nuclei were pelleted and resuspended with 0.2% Nonidet P-40 in PBS. The viral particles in cytoplasmic and nuclear fractions were collected, and the yield was determined on Vero cells as described above.
Electron Microscopy Analysis
HeLa cells were infected with HSV-1(F) or R3616 at an m.o.i. of 5. At 16 hpi, cells were harvested and then washed two times with PBS and one time with FBS. Then the samples were fixed in 6% glutaraldehyde for 2 h followed by 1% osmium tetroxide for 1 h. After washing two times with PBS, the samples were dehydrated in a series of 30, 70, 90, and 100% ethanol and embedded in Epon 812. The ultrathin sections were prepared with Leica UC7 and stained with a mixture of uranyl acetate and lead citrate. Thin sections were viewed with an H600 transmission electron microscope. The numbers of viral particles in the nucleus and extranucleus were counted in electron micrographs of 10 randomly sampled HeLa cells. The numbers shown in the tables represent the average numbers of viral particles in different cellular compartments and the total viral particles in 10 randomly sampled HeLa cells. The numbers in parentheses represent the percentage of viral particles per cellular compartment. The quantitative data are presented as mean value ±S.D. of three independent experiments.
Statistical Analysis
The quantitative data are presented as mean value ±S.D. of three independent experiments. Data were analyzed using Student's t test. A p value <0.05 (*) was considered to be statistically significant.
RESULTS
Identification of Cellular p32 as an HSV-1 ICP34.5-interacting Protein
HSV-1 ICP34.5 is a virulence factor that has multiple roles in virus-infected cells, such as reducing interferon β production (29), promoting protein synthesis (36), blocking autophagy (31), and facilitating viral egress (23). To study ICP34.5, we set out to identify cellular proteins that may interact with ICP34.5. In brief, the plasmid encoding FLAG-tagged ICP34.5 was transfected into HEK293T cells, and a pulldown was performed with anti-FLAG antibody. The immunoprecipitates were fractionated by SDS-PAGE (Fig. 1A), and a distinguished band was isolated and identified to be p32/33 by mass spectrophotometer analysis. Human p32/33 (NM_001212), originally designated as gC1qR (receptor for globular head domain of complement component C1q), is ubiquitously expressed in almost all cell types (37). The preprotein, p33, contains 282 amino acids, and the mature form, p32, contains residues 74–282 (38). We verified the interaction by co-immunoprecipitation of p32 with FLAG-ICP34.5 (Fig. 1B). An unrelated FLAG-tagged protein with similar molecular weight as ICP34.5 was used as a negative control (Ctrl) (Fig. 1B, left lane). Furthermore, we confirmed the interaction between p32 and ICP34.5 in virus-infected HeLa cells by co-immunoprecipitation assay. As illustrated in Fig. 1, C and D, the association of ICP34.5 and p32 was detected in cells infected with wild type HSV-1 with either anti-ICP34.5 antibody or anti-p32 antibody. However, such interaction was not seen in cells infected with R3616, an HSV-1 mutant that lacks the γ134.5 gene. Taken together, these results demonstrate a specific interaction between viral protein ICP34.5 and host protein p32 in vitro and in the virus-infected cells.
FIGURE 1.
Interaction between ICP34.5 and p32. A, HEK293T cells were transfected with a plasmid encoding FLAG-ICP34.5 or a control plasmid, and cells were processed for immunoprecipitation with antibody against FLAG. Proteins in precipitations (lanes 1 and 2), supernatants (lanes 3 and 4), and the whole cell lysates (WCL; lanes 5 and 6) were resolved by SDS-PAGE and stained with Coomassie Blue. The specific band co-precipitated with ICP34.5 was subjected to mass spectrophotometer analysis and identified to be p32 protein. B, HeLa cells transfected with plasmid encoding FLAG-Ctrl or FLAG-ICP34.5 were processed for immunoprecipitation (IP) with anti-FLAG antibody. Precipitated proteins (top two panels) and cell lysates (bottom three panels) were analyzed by immunoblotting with antibodies against p32, FLAG, and tubulin, respectively. HeLa cells were infected with HSV-1(F) or R3616 (m.o.i. = 5). At 16 hpi, cells were processed for immunoprecipitation with anti-ICP34.5 antibody (C) or anti-p32 antibody (D). Precipitated proteins (top two panels) and cell lysates (bottom three panels) were analyzed by immunoblotting with antibodies against p32, ICP34.5, and tubulin as a loading control, respectively.
p32 Is Required to Facilitate Release of HSV-1 from Infected Cells
Previous studies have demonstrated that the ICP34.5 protein is essential to promote viral virulence. An HSV-1 mutant lacking the γ134.5 gene exhibits a growth defect in cells and experimental animals (25–27). Therefore, to determine whether the host protein p32, as an interactor of ICP34.5, plays a role in HSV-1 replication, we carried out shRNA knockdown analysis. For this purpose, we established cells stably expressing an shRNA to suppress the endogenous expression of p32 or with a random sequence RNA (shCtrl) as well as shRNA for an unrelated protein, Vav2 (shVav2), as controls. The expression of p32 and Vav2 were significantly reduced as monitored by immunoblotting analysis (Fig. 2A). We next determined viral growth in a time course. As shown in Fig. 2B, p32 knockdown reduced virus yield at 48 hpi. A similar pattern was seen with the yield of cell-associated virus (Fig. 2C). As infection progressed, the yield of cell-free virus was reduced significantly in p32 knockdown cells (Fig. 2D). These results suggest that viral release or egress is impaired in the absence of cellular p32.
FIGURE 2.
Effect of p32 on viral replication and release. A, HeLa cells were transduced with the lentivirus particles incorporated with DNA fragments encoding shRNA for p32 (shP32) or with a random sequence RNA (shCtrl) and shRNA for an unrelated protein, Vav2 (shVav2), as controls, respectively. The protein expression was analyzed by immunoblotting with antibody against p32, Vav2, or tubulin as indicated, respectively. The stable cells, shCtrl (open circles), shVav2 (squares), and shP32 (triangles), were infected respectively with HSV-1(F) at 0.05 m.o.i. At the indicated hpi, the total virus yield (B), intracellular virus yield (C), and extracellular virus yield (D) were determined with Vero cells, respectively. Viral replication is presented as a log value of plaque-forming units (PFU) and plotted on the vertical axis. Data represent the mean value of three independent experiments with the S.D. (error bars) as indicated.
ICP34.5 and p32 Are Necessary for Viral Nuclear Egress
HSV egress from the infected cells includes two steps: 1) budding from the nucleus where the viral capsids are enveloped and 2) release from the host cell (39). Previous studies suggest that ICP34.5 is required for viral nuclear egress in MEF 3T6 cells (23, 24). To further study this phenotype, we examined the nuclear egress of HSV-1 in HeLa cells by electron microscopy (EM) (Fig. 3). Accordingly, HeLa cells were infected with HSV-1(F) (Fig. 3A) or R3616 (Fig. 3B), and the viruses located inside (inset a) or outside (inset b) the nucleus were visualized by EM, respectively. The distribution of viral particles was also quantitated in 10 randomly selected HeLa cells for each infection and is summarized in Table 1. In cells infected with HSV-1(F), approximately equal distribution of viral particles was seen in the nucleus and in the extranucleus. In comparison, in cells infected with R3616, about 79% of viral particles were restrained in the nucleus, and only 21% were seen outside the nucleus. Consistent with the previous observations in MEF 3T6 cells, ICP34.5 is indeed involved in nuclear egress of viral particles in HeLa cells infected with HSV-1(F).
FIGURE 3.
Effect of ICP34.5 and p32 on viral nuclear egress. HeLa cell monolayers were infected with HSV-1(F) (A) or R3616 (B) (m.o.i. = 5). Cells with stably transduced RNA, shCtrl (C) and shP32 (D), were infected with HSV-1(F) at 5 m.o.i. At 16 hpi, cells were collected and processed for electron microscopy analysis as described under “Experimental Procedures.” Insets a and b in A–D are magnified in separate panels at the bottom of the corresponding panel. Scale bars, 500 (A–D) and 250 nm (insets a and b). Nuc, nucleus region; Cyt, cytoplasm region. HeLa cells with shCtrl or shP32 were infected with HSV-1(F) at 5 m.o.i. At 16 hpi, cells were collected, and nuclear (top panel) and cytoplasmic (bottom panel) fractions were analysis by immunoblotting with antibodies against coilin and tubulin, respectively (E), or the nuclear and cytoplasmic viral particles were titrated on Vero cells (F). Quantitation of viral particles in shCtrl cells (solid bars) and shP32 cells (open bars) is presented as the percentage of total pfu in the bar graph. Data represent the mean value of three independent experiments with the S.D. (error bars) as indicated. *, p < 0.05 by Student's t test.
TABLE 1.
Subcellular localization of viral particles in infected cells
| Type of virus | Average no. of viral particlesa (percentage of total particles counted) |
Total no. of particles/total no. of cells | |
|---|---|---|---|
| Nucleus | Extranucleus | ||
| HSV-1(F) | 53.6 ± 10.0 (51%) | 52.4 ± 4.3 (49%) | 1060/10 |
| R3616 | 19.9 ± 5.1 (79%) | 5.4 ± 2.2 (21%) | 253/10 |
a The numbers of viral particles in the nucleus and extranucleus were counted in electron micrographs of 10 randomly sampled HeLa cells infected with the indicated virus.
Because the p32 protein is a host target of ICP34.5, we examined whether p32 was also involved in nuclear egress of viral particles in HSV-1(F)-infected cells. As seen in Fig. 2, knockdown of p32 impeded viral release, so we speculated that one of the steps for viral export was affected. As such, we assessed nuclear egress of HSV-1(F) by EM in shCtrl and shP32 HeLa cells. The representative EM images of the two cells are shown in Fig. 3, C and D, with the zoomed in images for inside (inset a) and outside (inset b) the nucleus, respectively. The distribution of viral particles was counted in 10 randomly selected cells for each infection and is summarized in Table 2. In HeLa-shCtrl cells (Fig. 3C), 54% of HSV-1 particles were in the nucleus, and 46% were in the extranucleus. By contrast, in HeLa-shP32 cells (Fig. 3D), 75% of viral particles were present in the nucleus, whereas only 25% egressed to the extranucleus. Furthermore, we examined the nuclear egress of HSV-1 in different cell fractions of shCtrl or shP32 cells (Fig. 3E). Control proteins coilin and tubulin were used as indicators for the nuclear and cytoplasmic fractions, respectively. Quantitation of viral particles in nuclear and cytoplasmic fractions was analyzed by titration and is presented in the bar graph (Fig. 3F). In HeLa-shP32 cells (open bars), nearly 90% of viral particles were restrained in the nucleus. In comparison, less than 70% of viral particles were in the nuclear fraction of HeLa-shCtrl cells (solid bars). Thus, the host cellular protein p32 is favorable for efficient nuclear egress of viral particles in HSV-1(F)-infected cells.
TABLE 2.
Subcellular localization of viral particles in infected cells
| Type of infected cells | Average no. of viral particlesa (percentage of total particles counted) |
Total no. of particles/total no. of cells | |
|---|---|---|---|
| Nucleus | Extranucleus | ||
| shCtrl | 48.4 ± 7.8 (54%) | 42.0 ± 11.9 (46%) | 904/10 |
| shP32 | 57.6 ± 9.1 (75%) | 18.9 ± 5.5 (25%) | 765/10 |
a The numbers of viral particles in the nucleus and extranucleus were counted in electron micrographs of 10 randomly sampled HeLa-shCtrl or HeLa-shP32 cells infected with HSV-1(F).
ICP34.5 Recruits p32 to LBR
Cellular p32 is a multifunctional protein that disperses in the cytoplasm, mitochondria, and nucleus (40). To assess whether p32 localization is affected by HSV-1 infection, we first performed confocal microscopy analysis (Fig. 4A). In mock-infected cells (left column) and cells infected with R3616 (right column), p32 stain overlapped with the mitochondrial marker, indicating the colocalization of p32 with mitochondria. In contrast, in cells infected with HSV-1(F) (center column), p32 converged profoundly to the nuclear rim as shown in the merge row. Moreover, p32 colocalized to the nuclear rim with ICP34.5 in HSV-1(F)-infected cells (Fig. 4B). The above results implicate that HSV-1 infection results in the relocalization of p32 to the nuclear membrane.
FIGURE 4.
Colocalization of ICP34.5 and p32 with LBR. A, HeLa cells were infected with HSV-1(F) (m.o.i. = 5). At 16 hpi, cells were collected, stained with MitoTracker Red CMXRos (red), p32 antibody (green), and DAPI (blue) prior to confocal microscopy. The merged images are shown in the bottom row. B, mock- (upper row) or HSV-1 (lower row)-infected cells (m.o.i. = 5) were processed for confocal microscopy analysis with the indicated dyes for the viral ICP34.5, endogenous p32, and nuclei, respectively. The merged images are shown in the right column. C, overexpression of GFP-p32 fusion protein alone (upper row) or together with FLAG-ICP34.5 (lower row) in HeLa cells was achieved by transient transfection and analyzed by confocal microscopy. D, HeLa cells were infected with HSV-1(F) or R3616 (m.o.i. = 5). At 16 hpi, cells lysates were prepared for immunoprecipitation (IP) with anti-p32 antibody. Co-precipitated proteins (top three panels) and cell lysates (bottom four panels) were analyzed by immunoblotting with the indicated antibodies, respectively. E, as indicated at the top, HeLa cells transiently transfected with vector alone as a control (left lane), GFP-p32 alone (center lane), or together with FLAG-ICP34.5 (right lane) were processed for immunoprecipitation with anti-GFP antibody. Immobilized proteins (top three panels) and cell lysates (bottom four panels) were analyzed by immunoblotting with antibodies as indicated.
To evaluate whether ICP34.5 mediates p32 redistribution in the absence of other viral proteins, cells transfected with the plasmids encoding ICP34.5 and GFP-p32 were visualized by confocal microscopy (Fig. 4C). Similar to that seen in HSV-1(F)-infected cells, GFP-p32 predominantly localized in the cytoplasm in the absence of ICP34.5 (upper row) and colocalized to the nuclear rim with ICP34.5 or in the nuclei (lower row). These results suggest that when expressed ICP34.5 triggers relocalization of p32.
Human p32 interacts with various cellular and pathogen proteins such as pre-mRNA splicing factor in the nucleus (41), BH3-only protein harakiri in the mitochondria (42), HIV Tat and Rev (43, 44), human cytomegalovirus (CMV) pUL97 at the nuclear membrane (45), and hepatitis C virus core protein on the cell surface (46). Besides, by yeast two-hybrid assay, p32 was identified to interact with ectopically expressed LBR in vitro (47, 48). Because wild type virus, but not the ICP34.5 null mutant, redirected p32 to the nuclear rim, we wondered whether ICP34.5-p32 associated with the nuclear lamina proteins. By co-immunoprecipitation assay, LBR was co-precipitated with p32 in HSV-1(F)-infected cells (Fig. 4D), whereas LBR co-precipitation was not detectable in R3616-infected cells. Moreover, when ectopically expressed, ICP34.5 stimulated the association of p32 and LBR, but such binding was barely detectable in the absence of ICP34.5 (Fig. 4E). We conclude from these experiments that the translocation and association of p32 with LBR require ICP34.5 of HSV-1(F).
p32 Is Indispensable for ICP34.5-induced Phosphorylation and Redistribution of the Nuclear Lamina
An important step in HSV-1 nuclear egress is the disruption of nuclear lamina. To examine the impact of p32 and ICP34.5, we carried out infection analysis with wild type virus and the ICP34.5 null mutant, respectively. The organization of nuclear lamina, represented by lamin A/C, was visualized by confocal microscopy (Fig. 5A). In mock-infected cells (top row) and cells infected with R3616 (bottom row), lamin A/C lined the nuclear rim, whereas in cells infected with HSV-1(F), it appeared diffuse at the nuclear rim and punctate in the nucleus (middle row). These results indicate that ICP34.5 is required to perturb the integrity of nuclear lamina.
FIGURE 5.
The redistribution and phosphorylation of lamin A/C induced by ICP34.5 and p32. A, HeLa cells were mock-infected or infected with HSV-1(F) or R3616 (m.o.i. = 5) for 16 h and then subjected to confocal microscopy analysis. Nuclei, viral proteins, and lamin were traced with DAPI (left column), anti-HSV-1 antibody (center column), and anti-lamin A/C antibody (right column), respectively. B, HeLa cells (left panels) and neuroglioma U251 cells (right panels) were mock-infected or infected with HSV-1(F) or R3616 at 5 m.o.i. for 16 h. Then cell lysates were analyzed by immunoblotting with antibodies against phosphorylated lamin A/C (Ser(P)-22) and lamin A/C, respectively. Phosphorylated lamin A/C (Ser(P)-22) and lamin A/C were quantitated using a densitometer, and the phosphorylation of lamin A/C (Ser(P)-22) is presented as a ratio to lamin A/C. The ratio in the control group was defined as 1. C, HeLa cells transduced with shCtrl or shP32 were mock-infected (top two panels) or infected with HSV-1(F) (m.o.i. = 5) (bottom two panels). At 16 hpi, cells were processed for confocal microscopy analysis with the indicated markers for nuclei (DAPI) (left column), the viral proteins (center column), and lamin A/C (right column). D, the cells in C were collected; lysates were analyzed by immunoblotting with antibodies against phosphorylated lamin A/C (Ser(P)-22), lamin A/C, p32, and ICP34.5, respectively; and the bands were quantitated using a densitometer. The phosphorylation of lamin A/C is presented as a ratio to total lamin as described in B.
The instability of nuclear lamina is characterized by the phosphorylation of nuclear lamina, and Ser-22 is one of the phosphorylated residues of lamin A/C (49–52). By immunoblotting analysis (Fig. 5B), we noted elevated phosphorylation of lamin A/C (Ser(P)-22) in HSV-1(F)-infected HeLa cells (left panels, lane 2). In contrary, in R3616-infected cells, the phosphorylation of lamin A/C (Ser(P)-22) remained the same as that in mock-infected cells (left panels, lanes 1 and 3). As HSV-1 is a neurotropic virus, we also tested the phosphorylation of lamin A/C (Ser(P)-22) in a human neuroglioblastoma cell line U251 (right panels), and the data showed that HSV-1 ICP34.5 induced the phosphorylation of lamin A/C (Ser(P)-22) as well.
The above results led us to hypothesize that p32 triggers the integration of nuclear lamina. To address this issue, we visualized the organization of lamin A/C by confocal microscopy. As shown in Fig. 5C, lamin A/C lined the nuclear rim in mock-infected cells regardless of the presence of p32 (top two rows). However, when infected by HSV-1 (bottom two rows), lamin A/C appeared diffuse around the nuclear rim and punctate in the nucleus in shCtrl cells, whereas it remained intact in cells lacking p32 (shP32). The data here strongly suggested that p32 influenced the redistribution of nuclear lamina during HSV-1 infection. Furthermore, we tested the phosphorylation of lamin A/C (Ser(P)-22) in shCtrl and shP32 cells. As shown in Fig. 5D, the basal phosphorylation of lamin A/C (Ser(P)-22) was at comparable levels in mock-infected shCtrl and shP32 cells (left two lanes), whereas upon HSV-1(F) infection, the phosphorylated level of lamin A/C (Ser(P)-22) was more elevated in shCtrl cells (lane 3) than that in shP32 cells (lane 4), implying that p32 was involved in the phosphorylation of nuclear lamina induced by HSV-1 infection. Therefore, the disruption of nuclear lamina during HSV-1 infection coincides with the localization of ICP34.5 and p32 at the nuclear rim, which induces the phosphorylation of lamin A/C (Ser(P)-22).
PKCδ Is Responsible for the Phosphorylation of Lamin A/C (Ser(P)-22) Induced by HSV-1
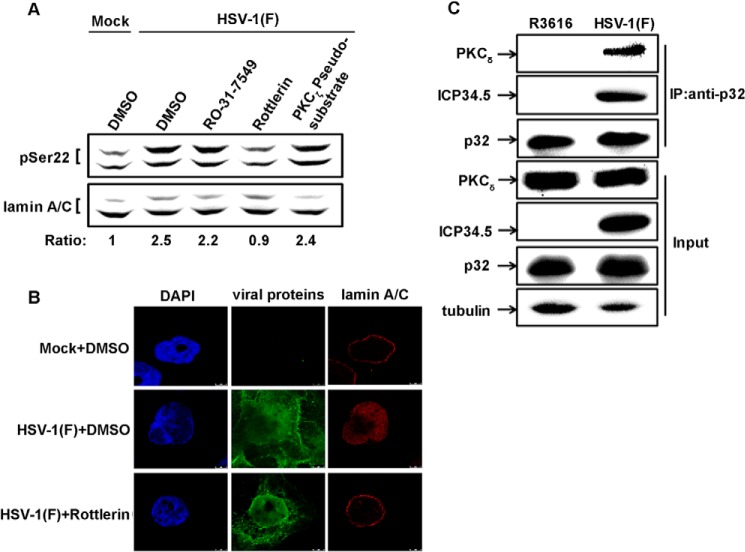
Previous studies reveal that p32 binds to several PKC isoforms in vitro (53). Because neither p32 nor ICP34.5 is a kinase, we hypothesized that p32 may chaperone PKC to the nuclear rim along with ICP34.5. We first attempted to identify the isoform(s) of PKC involved in HSV-1-induced phosphorylation of lamin A/C (Ser(P)-22) by a panel of inhibitors for conventional PKCs, novel PKCs, and atypical PKCs, respectively. As shown in Fig. 6A, compared with HSV-1(F)-infected sample (lane 2 from left), RO-31-7549, which preferentially inhibits PKCα isoform, and PKCζ pseudosubstrate displayed no apparent inhibition toward the phosphorylation of lamin A/C (Ser(P)-22), whereas rottlerin, which is a widely used selective inhibitor for PKCδ, exhibited efficient inhibition toward the phosphorylation of lamin A/C (Ser(P)-22) (lane 4 from left). Furthermore, the distribution of lamin A/C was analyzed by confocal microscopy. As shown in Fig. 6B, in contrast with HSV-1(F)-infected cells in which lamin A/C was in a diffuse pattern in the nucleus (middle row), rottlerin exhibited efficient inhibition toward the redistribution of lamin A/C (bottom row). In addition to this observation, a similar distribution pattern of PKCδ with p32 was observed by confocal microscopy analysis (not shown), and the complex of p32 with PKCδ by co-immunoprecipitation was detected in cells infected with HSV-1(F) but not R3616 (Fig. 6C). These results suggest that ICP34.5 triggers the redistribution of p32 and PKCδ to the nuclear rim where the phosphorylation of lamin A/C (Ser(P)-22) occurs.
FIGURE 6.
Phosphorylation of lamin A/C (Ser(P)-22) by PKCδ. A, HeLa cells were mock-infected or infected with HSV-1(F) (m.o.i. = 5) for 12 h followed by incubation in medium with DMSO as a control or the indicated PKC inhibitors for another 4 h. Cells lysates were analyzed by immunoblotting with antibodies against phosphorylated lamin A/C (Ser(P)-22) and lamin A/C, respectively. The level of phosphorylated lamin A/C was quantitated as described in Fig. 5. B, similar to A, HeLa cells were infected and cultured in the presence of DMSO (control) or rotterlin and then observed by confocal microscopy with indicators for nuclei (DAPI), the viral proteins, and lamin A/C, respectively. C, HeLa cells were infected with HSV-1(F) or R3616 at an m.o.i. of 5. At 16 hpi, cell extracts were immunoprecipitated with anti-p32 antibody. Precipitated proteins (top three panels) and cell lysates (bottom four panels) were analyzed by immunoblotting with antibodies against PKCδ, p32, ICP34.5, and tubulin, respectively. IP, immunoprecipitation.
DISCUSSION
Upon productive infection, HSV-1 undergoes viral gene expression, DNA replication, capsid assembly, and egress. In the present study, we found that the viral protein ICP34.5 interacted with cellular p32 to form a complex, and the redistribution of this complex was linked to the viral budding from nuclei. Notably, knockdown of p32 by shRNA caused a restriction of HSV-1 nuclear egress by 75% (Table 2), whereas 79% of R3616 was trapped in the nucleus in normal HeLa cells (Table 1), similar to that in MEF 3T6 cells in which the majority of viruses are retained in the nucleus of cells infected with ICP34.5 deletion mutants (23). These results suggest that p32 is a mediator of HSV-1 nuclear egress by interaction with ICP34.5.
p32 (HABP1) is a ubiquitous protein in most cell types and is reported to interact with pathogen proteins as a surface receptor of immune cells, i.e. human immunodeficiency virus type 1 Rev (44) and Tat (43), hepatitis C virus core protein (46), EBNA-1 of Epstein-Barr virus (54), and open reading frame P of HSV (55). The intracellular localization and function of p32 have been demonstrated primarily as a mitochondrial membrane protein in harakiri-mediated apoptosis (42), a mitochondrial antiviral signaling protein-interacting protein in RNA-induced retinoic acid-inducible gene I signaling (35), an interactor of splicing factor SF2/alternative splicing factor for RNA splicing (41), and an RNA-interacting protein for the translation of mitochondrial RNA (56). To date, the only viral kinase reported to associate with intracellular p32 is human CMV pUL97, a kinase that phosphorylates nuclear lamina at the inner nuclear membrane (45). Kinase pUL97 is recruited to the nuclear rim by p32, directly phosphorylates lamin A/C, and induces nuclear lamina disassembly to promote viral nuclear egress (57). In this context, in both HeLa and human neuroglioblastoma cells infected with HSV-1(F), the similar phenotypes including p32 relocalization to the nuclear rim (Fig. 4) and phosphorylation of nuclear lamina occurred only in the presence of ICP34.5 (Fig. 5). Although viral kinase Us3 has been suggested to phosphorylate lamin A by an in vitro assay (58), the phosphorylation at Ser-22 in our study was well correlated with the presence of ICP34.5 (Fig. 5B). However, ICP34.5 is by no means a kinase. These results suggest that a host kinase must be recruited to the complex at the nuclear rim. Furthermore, the kinase responsible for ICP34.5-mediated phosphorylation of lamin A/C at Ser-22 was identified to be PKCδ.
Our work suggests that p32 forms a complex through interactions with ICP34.5, LBR, and PKCδ at the inner nuclear membrane. In this respect, p32 probably serves as an adaptor to bridge viral and host proteins. This notion is consistent with the observation noted in cytomegalovirus infection (48, 53). Although mostly present in the cytoplasm, p32 was recruited to the nuclear rim by ICP34.5 in HSV-1-infected cells. The precise nature of the p32 complex is unknown. We speculate that the interaction between p32 and ICP34.5 is required for the phosphorylation and reorganization of nuclear lamina.
HSV-1 infection modifies the architecture of nuclear lamina to allow nuclear egress and envelopment of viral particles (6, 11–14). Prior studies demonstrate that UL31 and UL34 translocate to the nuclear rim and induce conformational changes of the nuclear lamina via association with lamin A/C or indirect recruitment of PKCα and PKCδ to the nuclear membrane (12). However, it is unclear whether this process involves other viral proteins. In this study, a cellular protein, p32, was identified to cooperatively recruit PKCδ to disrupt nuclear lamina together with ICP34.5. In this context, ICP34.5 appeared to play a critical role. This was supported by the following evidence. 1) Without ICP34.5, the mutant virus was mostly restrained inside the nucleus, although all other viral proteins including UL31, UL34, Us3, and others were present. 2) The phosphorylation of nuclear lamina and the recruitment of PKCδ relied on the efficient expression of p32 in association with ICP34.5. Based on these data, we speculate that ICP34.5, UL31/UL34, p32, and PKCδ may coordinately work to promote viral egress. Further investigation is needed to address the formation of such complex.
In summary, we identified cellular p32 as a novel HSV-1 virulence factor ICP34.5-interacting protein and revealed that p32 was required for the nuclear egress of HSV-1 capsids. Based on these results, we postulate a model whereby ICP34.5 facilitates viral nuclear egress (Fig. 7). In HSV-1-infected cells, ICP34.5 binds to and induces the redistribution of p32 along with PKCδ to the nuclear rim, forming a complex that phosphorylates the nuclear lamina components. As a result, this dismantles the nuclear lamina, allowing the export of viral capsids. Future work will be directed toward understanding the details of HSV-1 egress.
FIGURE 7.
Proposed model of ICP34.5-p32-mediated HSV-1 nuclear egress. In uninfected cells (left), p32 and PKCδ predominantly localize in the cytoplasm, and LBR lining the nuclear lamina is around the nuclear rim. Upon HSV-1 infection (right), viral protein ICP34.5 binds and recruits p32 to LBR. Meanwhile, p32-associated PKCδ translocates to the nuclear lamina and forms a complex with ICP34.5, p32, and PKCδ, which phosphorylates lamins, leading to the disintegration of the nuclear lamina, promoting HSV nucleocapsid egress from the nucleus (right).
Acknowledgments
We thank Prof. S. Yu (Tianjin Medical University Cancer Institute and Hospital, China) for providing U251 cells and Prof. J. Ji (Peking University, China) for help with the mass spectrophotometer analysis.
This work was supported, in whole or in part, by National Institutes of Health Grant AI092230 from the NIAID (to B. H.). This work was also supported by National Natural Science Foundation of China Grants 81171556 and 31370862 (to Y. C.), Ministry of Science and Technology of China Grant BE091561 (to Y. C.), and Program of Introducing Talents of Discipline to Universities Grant B08011.
- HSV
- herpes simplex virus
- C1qR
- component 1q receptor
- HABP1
- hyaluronic acid-binding protein 1
- LBR
- lamin B receptor
- m.o.i.
- multiplicity of infection
- hpi
- hours postinfection
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1. Whitley R. J., Roizman B. (2001) Herpes simplex virus infections. Lancet 357, 1513–1518 [DOI] [PubMed] [Google Scholar]
- 2. Stackpole C. W. (1969) Herpes-type virus of the frog renal adenocarcinoma. I. Virus development in tumor transplants maintained at low temperature. J. Virol. 4, 75–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnson D. C., Spear P. G. (1982) Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J. Virol. 43, 1102–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wild P., Engels M., Senn C., Tobler K., Ziegler U., Schraner E. M., Loepfe E., Ackermann M., Mueller M., Walther P. (2005) Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J. Virol. 79, 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Senior A., Gerace L. (1988) Integral membrane proteins specific to the inner nuclear membrane and associated with the nuclear lamina. J. Cell Biol. 107, 2029–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scott E. S., O'Hare P. (2001) Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 75, 8818–8830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calistri A., Sette P., Salata C., Cancellotti E., Forghieri C., Comin A., Göttlinger H., Campadelli-Fiume G., Palù G., Parolin C. (2007) Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J. Virol. 81, 11468–11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chouljenko V. N., Iyer A. V., Chowdhury S., Chouljenko D. V., Kousoulas K. G. (2009) The amino terminus of herpes simplex virus type 1 glycoprotein K (gK) modulates gB-mediated virus-induced cell fusion and virion egress. J. Virol. 83, 12301–12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hutchinson L., Johnson D. C. (1995) Herpes simplex virus glycoprotein K promotes egress of virus particles. J. Virol. 69, 5401–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chouljenko D. V., Kim I. J., Chouljenko V. N., Subramanian R., Walker J. D., Kousoulas K. G. (2012) Functional hierarchy of herpes simplex virus 1 viral glycoproteins in cytoplasmic virion envelopment and egress. J. Virol. 86, 4262–4270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bjerke S. L., Roller R. J. (2006) Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347, 261–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park R., Baines J. D. (2006) Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80, 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reynolds A. E., Liang L., Baines J. D. (2004) Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J. Virol. 78, 5564–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simpson-Holley M., Baines J., Roller R., Knipe D. M. (2004) Herpes simplex virus 1 U(L)31 and U(L)34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 78, 5591–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reynolds A. E., Ryckman B. J., Baines J. D., Zhou Y., Liang L., Roller R. J. (2001) U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75, 8803–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reynolds A. E., Wills E. G., Roller R. J., Ryckman B. J., Baines J. D. (2002) Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76, 8939–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leach N., Bjerke S. L., Christensen D. K., Bouchard J. M., Mou F., Park R., Baines J., Haraguchi T., Roller R. J. (2007) Emerin is hyperphosphorylated and redistributed in herpes simplex virus type 1-infected cells in a manner dependent on both UL34 and US3. J. Virol. 81, 10792–10803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morris J. B., Hofemeister H., O'Hare P. (2007) Herpes simplex virus infection induces phosphorylation and delocalization of emerin, a key inner nuclear membrane protein. J. Virol. 81, 4429–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ryckman B. J., Roller R. J. (2004) Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78, 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kato A., Yamamoto M., Ohno T., Tanaka M., Sata T., Nishiyama Y., Kawaguchi Y. (2006) Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 80, 1476–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Z., Kato A., Shindo K., Noda T., Sagara H., Kawaoka Y., Arii J., Kawaguchi Y. (2014) Herpes simplex virus 1 UL47 interacts with viral nuclear egress factors UL31, UL34, and Us3 and regulates viral nuclear egress. J. Virol. 88, 4657–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sagou K., Uema M., Kawaguchi Y. (2010) Nucleolin is required for efficient nuclear egress of herpes simplex virus type 1 nucleocapsids. J. Virol. 84, 2110–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jing X., Cerveny M., Yang K., He B. (2004) Replication of herpes simplex virus 1 depends on the γ134.5 functions that facilitate virus response to interferon and egress in the different stages of productive infection. J. Virol. 78, 7653–7666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brown S. M., MacLean A. R., Aitken J. D., Harland J. (1994) ICP34.5 influences herpes simplex virus type 1 maturation and egress from infected cells in vitro. J. Gen. Virol. 75, 3679–3686 [DOI] [PubMed] [Google Scholar]
- 25. Chou J., Kern E. R., Whitley R. J., Roizman B. (1990) Mapping of herpes simplex virus-1 neurovirulence to γ134.5, a gene nonessential for growth in culture. Science 250, 1262–1266 [DOI] [PubMed] [Google Scholar]
- 26. MacLean A. R., ul-Fareed M., Robertson L., Harland J., Brown S. M. (1991) Herpes simplex virus type 1 deletion variants 1714 and 1716 pinpoint neurovirulence-related sequences in Glasgow strain 17+ between immediate early gene 1 and the 'a' sequence. J. Gen. Virol. 72, 631–639 [DOI] [PubMed] [Google Scholar]
- 27. Whitley R. J., Kern E. R., Chatterjee S., Chou J., Roizman B. (1993) Replication, establishment of latency, and induced reactivation of herpes simplex virus γ1 34.5 deletion mutants in rodent models. J. Clin. Investig. 91, 2837–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng G., Brett M. E., He B. (2002) Signals that dictate nuclear, nucleolar, and cytoplasmic shuttling of the γ134.5 protein of herpes simplex virus type 1. J. Virol. 76, 9434–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Verpooten D., Ma Y., Hou S., Yan Z., He B. (2009) Control of TANK-binding kinase 1-mediated signaling by the γ134.5 protein of herpes simplex virus 1. J. Biol. Chem. 284, 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jin H., Ma Y., Prabhakar B. S., Feng Z., Valyi-Nagy T., Yan Z., Verpooten D., Zhang C., Cao Y., He B. (2009) The γ134.5 protein of herpes simplex virus 1 is required to interfere with dendritic cell maturation during productive infection. J. Virol. 83, 4984–4994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Orvedahl A., Alexander D., Tallóczy Z., Sun Q., Wei Y., Zhang W., Burns D., Leib D. A., Levine B. (2007) HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1, 23–35 [DOI] [PubMed] [Google Scholar]
- 32. He B., Gross M., Roizman B. (1998) The γ134.5 protein of herpes simplex virus 1 has the structural and functional attributes of a protein phosphatase 1 regulatory subunit and is present in a high molecular weight complex with the enzyme in infected cells. J. Biol. Chem. 273, 20737–20743 [DOI] [PubMed] [Google Scholar]
- 33. Li Y., Zhang C., Chen X., Yu J., Wang Y., Yang Y., Du M., Jin H., Ma Y., He B., Cao Y. (2011) ICP34.5 protein of herpes simplex virus facilitates the initiation of protein translation by bridging eukaryotic initiation factor 2α (eIF2α) and protein phosphatase 1. J. Biol. Chem. 286, 24785–24792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simos G., Georgatos S. D. (1994) The lamin B receptor-associated protein p34 shares sequence homology and antigenic determinants with the splicing factor 2-associated protein p32. FEBS Lett. 346, 225–228 [DOI] [PubMed] [Google Scholar]
- 35. Xu L., Xiao N., Liu F., Ren H., Gu J. (2009) Inhibition of RIG-I and MDA5-dependent antiviral response by gC1qR at mitochondria. Proc. Natl. Acad. Sci. U.S.A. 106, 1530–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He B., Gross M., Roizman B. (1997) The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. U.S.A. 94, 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ghebrehiwet B., Peerschke E. I. (2004) cC1q-R (calreticulin) and gC1q-R/p33: ubiquitously expressed multi-ligand binding cellular proteins involved in inflammation and infection. Mol. Immunol. 41, 173–183 [DOI] [PubMed] [Google Scholar]
- 38. Dedio J., Jahnen-Dechent W., Bachmann M., Müller-Esterl W. (1998) The multiligand-binding protein gC1qR, putative C1q receptor, is a mitochondrial protein. J. Immunol. 160, 3534–3542 [PubMed] [Google Scholar]
- 39. Mettenleiter T. C. (2002) Herpesvirus assembly and egress. J. Virol. 76, 1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Leeuwen H. C., O'Hare P. (2001) Retargeting of the mitochondrial protein p32/gC1Qr to a cytoplasmic compartment and the cell surface. J. Cell Sci. 114, 2115–2123 [DOI] [PubMed] [Google Scholar]
- 41. Petersen-Mahrt S. K., Estmer C., Ohrmalm C., Matthews D. A., Russell W. C., Akusjärvi G. (1999) The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 18, 1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sunayama J., Ando Y., Itoh N., Tomiyama A., Sakurada K., Sugiyama A., Kang D., Tashiro F., Gotoh Y., Kuchino Y., Kitanaka C. (2004) Physical and functional interaction between BH3-only protein Hrk and mitochondrial pore-forming protein p32. Cell Death Differ. 11, 771–781 [DOI] [PubMed] [Google Scholar]
- 43. Yu L., Zhang Z., Loewenstein P. M., Desai K., Tang Q., Mao D., Symington J. S., Green M. (1995) Molecular cloning and characterization of a cellular protein that interacts with the human immunodeficiency virus type 1 Tat transactivator and encodes a strong transcriptional activation domain. J. Virol. 69, 3007–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tange T. O., Jensen T. H., Kjems J. (1996) In vitro interaction between human immunodeficiency virus type 1 Rev protein and splicing factor ASF/SF2-associated protein, p32. J. Biol. Chem. 271, 10066–10072 [DOI] [PubMed] [Google Scholar]
- 45. Marschall M., Marzi A., aus dem Siepen P., Jochmann R., Kalmer M., Auerochs S., Lischka P., Leis M., Stamminger T. (2005) Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 280, 33357–33367 [DOI] [PubMed] [Google Scholar]
- 46. Kittlesen D. J., Chianese-Bullock K. A., Yao Z. Q., Braciale T. J., Hahn Y. S. (2000) Interaction between complement receptor gC1qR and hepatitis C virus core protein inhibits T-lymphocyte proliferation. J. Clin. Investig. 106, 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Milbradt J., Auerochs S., Marschall M. (2007) Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen. Virol. 88, 2642–2650 [DOI] [PubMed] [Google Scholar]
- 48. Milbradt J., Auerochs S., Sticht H., Marschall M. (2009) Cytomegaloviral proteins that associate with the nuclear lamina: components of a postulated nuclear egress complex. J. Gen. Virol. 90, 579–590 [DOI] [PubMed] [Google Scholar]
- 49. Heald R., McKeon F. (1990) Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell 61, 579–589 [DOI] [PubMed] [Google Scholar]
- 50. Peter M., Nakagawa J., Dorée M., Labbé J. C., Nigg E. A. (1990) In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell 61, 591–602 [DOI] [PubMed] [Google Scholar]
- 51. Ward G. E., Kirschner M. W. (1990) Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell 61, 561–577 [DOI] [PubMed] [Google Scholar]
- 52. Peter M., Heitlinger E., Häner M., Aebi U., Nigg E. A. (1991) Disassembly of in vitro formed lamin head-to-tail polymers by CDC2 kinase. EMBO J. 10, 1535–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Robles-Flores M., Rendon-Huerta E., Gonzalez-Aguilar H., Mendoza-Hernandez G., Islas S., Mendoza V., Ponce-Castaneda M. V., Gonzalez-Mariscal L., Lopez-Casillas F. (2002) p32 (gC1qBP) is a general protein kinase C (PKC)-binding protein; interaction and cellular localization of p32-PKC complexes in ray hepatocytes. J. Biol. Chem. 277, 5247–5255 [DOI] [PubMed] [Google Scholar]
- 54. Wang Y., Finan J. E., Middeldorp J. M., Hayward S. D. (1997) P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 236, 18–29 [DOI] [PubMed] [Google Scholar]
- 55. Bruni R., Roizman B. (1996) Open reading frame P—a herpes simplex virus gene repressed during productive infection encodes a protein that binds a splicing factor and reduces synthesis of viral proteins made from spliced mRNA. Proc. Natl. Acad. Sci. U.S.A. 93, 10423–10427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yagi M., Uchiumi T., Takazaki S., Okuno B., Nomura M., Yoshida S., Kanki T., Kang D. (2012) p32/gC1qR is indispensable for fetal development and mitochondrial translation: importance of its RNA-binding ability. Nucleic Acids Res. 40, 9717–9737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hamirally S., Kamil J. P., Ndassa-Colday Y. M., Lin A. J., Jahng W. J., Baek M. C., Noton S., Silva L. A., Simpson-Holley M., Knipe D. M., Golan D. E., Marto J. A., Coen D. M. (2009) Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 5, e1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mou F., Forest T., Baines J. D. (2007) US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 81, 6459–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]