Endocytosis-dependent PIN1 localization in leaf primordia determines myrosin cell development specifically along leaf veins, protecting the plant transport system.
Abstract
Myrosin cells, which accumulate myrosinase to produce toxic compounds when they are ruptured by herbivores, form specifically along leaf veins in Arabidopsis thaliana. However, the mechanism underlying this pattern formation is unknown. Here, we show that myrosin cell development requires the endocytosis-mediated polar localization of the auxin-efflux carrier PIN1 in leaf primordia. Defects in the endocytic/vacuolar SNAREs (syp22 and syp22 vti11) enhanced myrosin cell development. The syp22 phenotype was rescued by expressing SYP22 under the control of the PIN1 promoter. Additionally, myrosin cell development was enhanced either by lacking the activator of endocytic/vacuolar RAB5 GTPase (VPS9A) or by PIN1 promoter-driven expression of a dominant-negative form of RAB5 GTPase (ARA7). By contrast, myrosin cell development was not affected by deficiencies of vacuolar trafficking factors, including the vacuolar sorting receptor VSR1 and the retromer components VPS29 and VPS35, suggesting that endocytic pathway rather than vacuolar trafficking pathway is important for myrosin cell development. The phosphomimic PIN1 variant (PIN1-Asp), which is unable to be polarized, caused myrosin cells to form not only along leaf vein but also in the intervein leaf area. We propose that Brassicales plants might arrange myrosin cells near vascular cells in order to protect the flux of nutrients and water via polar PIN1 localization.
INTRODUCTION
Plants have evolved various strategies to defend against herbivores, including the production and release of toxic compounds. The myrosinase-glucosinolate defense system is characteristic of plants of the Brassicaceae and certain other angiosperm families. Myrosin cells in leaves of these plants contain large amounts of myrosinase (thioglucoside glucohydrolase [TGG]) in their vacuoles (Rask et al., 2000; Andréasson et al., 2001; Husebye et al., 2002; Ueda et al., 2006). Glucosinolates substrates are distributed at the periphery of leaves and along veins (Koroleva et al., 2000; Shroff et al., 2008). When herbivores damage tissues and rupture myrosin cells, myrosinase gains access to glucosinolates and hydrolyzes them to produce toxic compounds (Rask et al., 2000; Wittstock and Halkier, 2002; Grubb and Abel, 2006; Halkier and Gershenzon, 2006; Hopkins et al., 2009; Kissen et al., 2009). Myrosin cells develop specifically along veins (Bones et al., 1991; Höglund et al., 1991; Xue et al., 1995; Andréasson et al., 2001; Husebye et al., 2002; Thangstad et al., 2004; Barth and Jander, 2006; Ueda et al., 2006) (Supplemental Figure 1). Recently, we identified the basic helix-loop-helix transcription factor FAMA as a master regulator of myrosin cell development in Arabidopsis thaliana (Shirakawa et al., 2014b). Prior to the expression of TGG2, FAMA is expressed in a subset of ground meristem cells that lack overt signs of myrosin-lineage differentiation (Shirakawa et al., 2014b).
How is the pattern of myrosin cells established? Previously, we reported that SYNTAXIN OF PLANTS22 (SYP22; also known as VAM3/SGR3) is involved in the patterning of myrosin cells (Ueda et al., 2006). syp22 mutants have higher numbers of myrosin cells, resulting in an extensive network structure of these cells (Ueda et al., 2006). Consequently, the myrosinases TGG1 and TGG2 accumulate at higher levels in the rosette leaves, flowers, siliques, and flower stalks of syp22 than in those of wild-type plants (Ueda et al., 2006). The myrosin cell phenotypes (high accumulation levels of TGG1) of the syp22 mutant were enhanced further in multiple mutants between SYP22 and their homologous genes SYP21/PEP12 and SYP23/PLP (Shirakawa et al., 2010). SYP22 is a component of the SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex, which is made up of SYP22 (Qa-SNARE), VTI11 (Qb-SNARE), SYP5 (Qc-SNARE), and VAMP727 (R-SNARE) (Sanderfoot et al., 2001; Yano et al., 2003; Ebine et al., 2008). SYP22 localizes to both the prevacuolar compartment/late endosome/multivesicular body and to the vacuolar membrane (Sanderfoot et al., 1999; Uemura et al., 2004; Ebine et al., 2008). It is involved in vacuolar morphogenesis (Ebine et al., 2008), vacuolar trafficking (Ebine et al., 2008; Shirakawa et al., 2010), and endocytosis (Ebine et al., 2011). However, it is unclear where and when these functions of SYP22 are required for the development of myrosin cells.
Members of the PIN-FORMED (PIN) protein family are auxin-efflux carriers. Their polar localization results in directional auxin flow and mediates various aspects of plant growth and development (Gälweiler et al., 1998; Paponov et al., 2005; Petrásek et al., 2006; Wisniewska et al., 2006; Grunewald and Friml, 2010; Löfke et al., 2013). Although PIN2 proteins are initially transported to the plasma membrane in a nonpolar manner, they show a polarized localization pattern after subsequent endocytosis (Men et al., 2008). Endocytic pathways are required for the establishment and/or maintenance of the polar localization of PIN proteins to the plasma membrane (Goh et al., 2007; Men et al., 2008; Spitzer et al., 2009; Kitakura et al., 2011; Inoue et al., 2013; Ischebeck et al., 2013; Tejos et al., 2014). PIN proteins showed a nonpolarized localization pattern on the plasma membrane in a mutant with a defective VPS9A (RAB5-GEF), a guanine-nucleotide exchange factor that activates small GTPases in the Rab5 subfamily (ARA7/RABF2B and RHA1/RABF2A) (Goh et al., 2007; Inoue et al., 2013). In addition to their involvement in endocytic pathways (Beck et al., 2012; Irani et al., 2012), RAB5 GTPases and VPS9A are required for vacuolar trafficking pathways (Sohn et al., 2003; Kotzer et al., 2004; Ebine et al., 2011). Nonpolar localization of PIN1 has also been observed in the leaves of the syp22-4 mutant (Shirakawa et al., 2009).
In this study, we show that the polarized localization of PIN1 is required for proper development of the myrosin cell. The trafficking pathway mediated by SYP22, conventional-type RAB5s, and RAB5-GEF plays an important role in establishment of this PIN1 polarity. Our findings suggest that the myrosin cell fates are influenced by auxin flow and/or concentration through the PIN1 polarity. We propose that plants arrange defense cells near vascular cells to protect the essential flux of nutrients and water by regulating the polar localization of PIN1.
RESULTS
Leaf Primordia of syp22 Exhibit Increased Number of Ground Meristem Cells that Express FAMA, a Master Regulator for Myrosin Cell Development
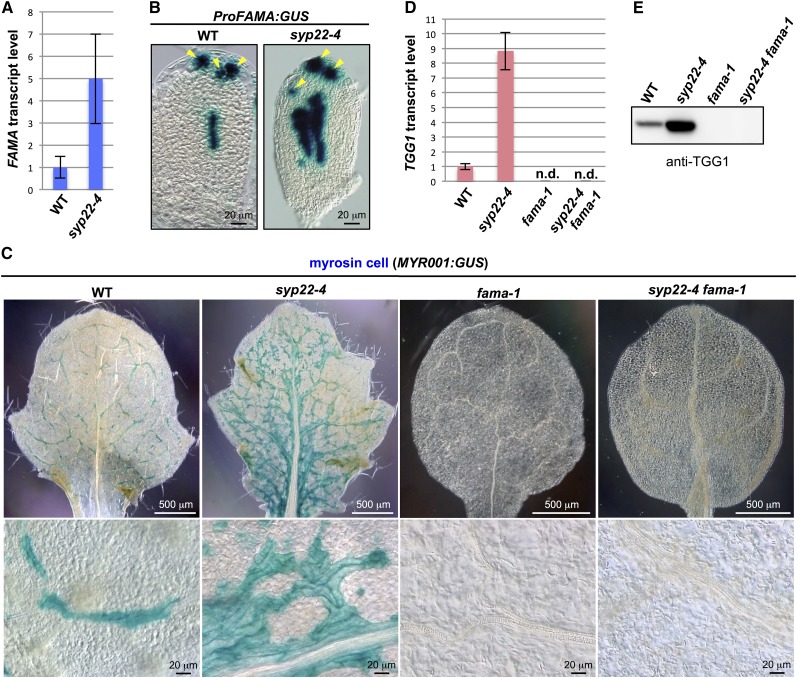
We examined the expression of FAMA in the syp22-4 mutant. Quantitative RT-PCR analysis revealed that the transcript level of FAMA was three to seven times higher in syp22-4 than in wild-type plants (Figure 1A). Compared with ProFAMA:GUS (β-glucuronidase) lines in the wild-type background, syp22-4 ProFAMA:GUS transgenic plants showed a broader GUS-positive area in inner tissues of leaf primordia (Figure 1B), suggesting that more ground meristem cells expressed FAMA in leaf primordia of syp22-4 than in those of the wild type.
Figure 1.
Genetic Interaction between SYP22 and FAMA.
(A) Quantitative RT-PCR of FAMA in 21 DAG plants of the wild type (WT) and syp22-4. Actin2 was used as a control. Error bars indicate sd (n = 3).
(B) GUS staining of rosette leaves of wild-type and syp22-4 plants expressing ProFAMA:GUS. Arrowheads indicate stomatal lineage cells. Bars = 20 μm.
(C) GUS staining of rosette leaves of wild-type, syp22-4, fama-1, and syp22-4 fama-1 plants expressing myrosin cell marker MYR001:GUS. Lower panels are enlarged images of upper panels. See Supplemental Figure 3 for GUS staining of whole plants. Bars = 500 μm (top panels) and 20 μm (bottom panels).
(D) Quantitative RT-PCR of TGG1 in 21 DAG plants of the wild type, syp22-4, fama-1, and syp22-4 fama-1. Actin2 was used as a control. Error bars indicate sd (n = 3).
(E) Immunoblot analysis of rosette leaves of the wild type, syp22-4, fama-1, and syp22-4 fama-1 using anti-TGG1 antibody.
To analyze the genetic interaction between SYP22 and FAMA, we generated syp22-4 fama-1 double mutants harboring the myrosin-cell-specific reporter, MYR001:GUS, which shows the same expression pattern as ProTGG2:VENUS-2sc (Shirakawa et al., 2014a). The plant morphologies of syp22-4 fama-1 were indistinguishable from those of fama-1 (Supplemental Figure 2). Like the leaves of fama-1, syp22-4 fama-1 leaves had no GUS-positive cells (Figure 1C). Also, TGG1 transcript and TGG1 protein were undetectable in syp22-4 fama-1 and in fama-1 (Figures 1D and 1E). These results indicate that FAMA is genetically epistatic to SYP22 in myrosin cell development. Collectively, these results suggest that SYP22 is involved in the cell fate determination process that selects myrosin-lineage cells (i.e., FAMA-positive cells) from the pool of ground meristem cells.
syp-22 Leaves Show a Dramatic Increase in the Number of Mature Myrosin Cells
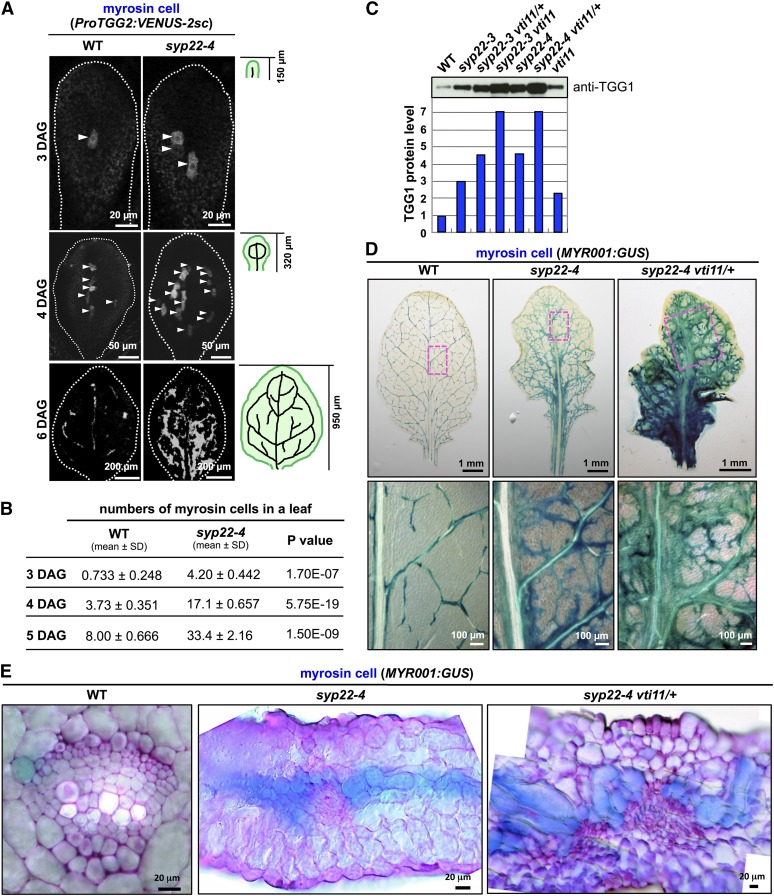
To identify when myrosin cells are generated during leaf development, we examined the time course of myrosin cell development in leaf primordia of the first pair of true leaves using the mature myrosin cell-specific reporter ProTGG2:VENUS-2sc. VENUS-2sc localizes to the endoplasmic reticulum and vacuole (Shirakawa et al., 2014a), and the TGG2 promoter, which is specific for myrosin cells (Barth and Jander, 2006), is activated after the expression of FAMA (Shirakawa et al., 2014b). In wild-type leaves, one reporter-positive cell was first detected at 3 d after germination (DAG) (Figure 2A). In syp22-4 leaves, there were already three myrosin cells at 3 DAG (Figure 2A). At 6 DAG, the pattern of myrosin cell development was nearly the same in mature leaves of the wild type and syp22-4 (Figures 2A and 2D; Supplemental Figure 3). We counted the number of cells expressing ProTGG2:VENUS-2sc from 3 to 5 DAG and found at least 4 times more myrosin cells in syp22-4 leaves than in wild-type leaves (Figure 2B). This result was consistent with the expression level and pattern of FAMA expression in leaf primordia (Figures 1A and 1B). Our observations suggest that a vast increase in the number of mature myrosin cells in syp22-4 is already in progress at 3 DAG and that this results from a greater number of ground meristem cells expressing FAMA.
Figure 2.
Myrosin Cell Development in syp22 vti11.
(A) Fluorescence images of myrosin cells in leaf primordia of wild-type (WT) and syp22-4 plants expressing ProTGG2:VENUS-2sc at 3, 4, and 6 DAG (top to bottom). Arrowheads indicate myrosin cells. The white outline shows the approximate border of the leaf. The small illustrations (right) depict the provascular pattern in first pair of true leaves at indicated DAG. Bars = 20 μm (top panels), 50 μm (middle panels), and 200 μm (bottom panels)
(B) Comparison of myrosin cell numbers in wild-type versus syp22-4 leaves. Cells were counted at 3, 4, and 5 DAG (WT, 3 DAG, n = 15; WT, 4 DAG, n = 23; WT, 5 DAG, n = 10; syp22-4, 3 DAG, n = 10; syp22-4, 4 DAG, n = 10; syp22-4, 5 DAG, n = 10).
(C) Rosette leaves of the wild type and indicated mutants immunoblotted and probed with anti-TGG1 antibody. The immunoblot shows levels of myrosin cell marker protein TGG1. The wild-type level was set to 1, and other genotypes are expressed relatively.
(D) GUS staining of rosette leaves of wild-type, syp22-4, and syp22-4 vti11/+ plants expressing MYR001:GUS, a myrosin cell marker, at 24 DAG. The boxed areas in each image in the top panel are enlarged in corresponding lower panel. Bars = 1 mm (top panels) and 100 μm (bottom panels)
(E) Enlarged images of cross sections. Sections were counterstained with Toluidine Blue O. In the wild type, GUS-positive myrosin cells are adjacent to phloem cells and procambium cells. Myrosin cells overproliferate along vascular bundles in syp22-4 and syp22-4 vti11/+ plants. See Supplemental Figure 5 for details. Bars = 20 μm.
Knockout of VTI11 in syp22 Enhances Myrosin Cell Developmental Phenotype
To investigate the function of SYP22 in the context of the SNARE complex during myrosin cell development, we examined the effect of a defect in VTI11, a Qb-SNARE that interacts with SYP22 (Figure 3A) (Sanderfoot et al., 2001; Yano et al., 2003; Ebine et al., 2008). In the vti11 single mutant, TGG1 accumulated to slightly higher levels than that in the wild type (Figure 2C). However, the introduction of a heterozygous mutation in VTI11 in the syp22-4 background (syp22-4 vti11/+) dramatically increased TGG1 accumulation (Figure 2C), despite the fact that vti11 is a recessive mutation. The syp22-4 vti11 double mutant was not viable (Shirakawa et al., 2009). Heterozygous and homozygous mutations in VTI11 in the syp22-3 background (syp22-3 is a weak allele of syp22; Ueda et al., 2006) also increased TGG1 accumulation (Figure 2C). Introduction of a heterozygous mutation of VTI12, a homolog of VTI11, in the vti11 background (vti11 vti12/+) dramatically increased TGG1 accumulation, even though vti12 is a recessive mutation (Supplemental Figure 4A), further supporting the involvement of VTI11 in myrosin cell development.
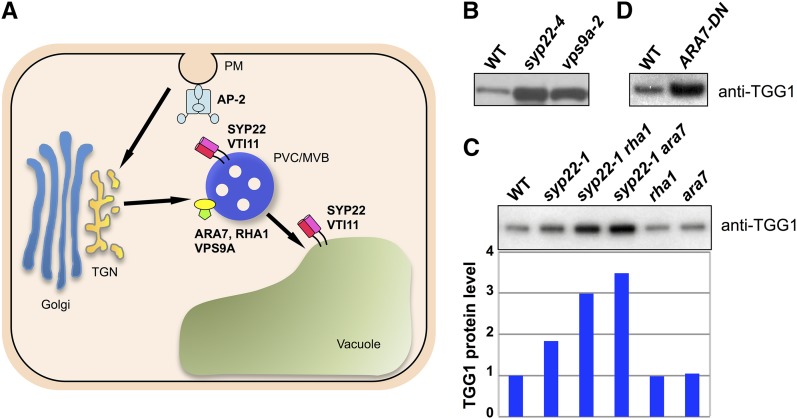
Figure 3.
Levels of TGG1 Accumulation in vps9a-2 and ARA7-DN Leaves.
(A) Model of post-Golgi trafficking pathway in plants. Qa-SNARE SYP22 interacts with Qb-SNARE VTI11. The SYP22-VTI11 SNARE complex is localized at both the prevacuolar compartment/multivesicular body (PVC/MVB) and the vacuole. VPS9A, a guanine-nucleotide exchange factor of the Rab5 subfamily, interacts with the Rab5s ARA7 and RHA1, which localize at the PVC/MVB. The AP-2 complex consists of four subunits: the α-, β-, μ-, and σ-subunits. AP-2 complex localizes at the plasma membrane (PM). In addition to these factors, trans-Golgi network (TGN)-localized COV1 protein is also required for the proper development of myrosin cells. Arrows indicate the endocytic pathway.
(B) Rosette leaves of the wild type (WT), syp22-4, and vps9a-2 immunoblotted with anti-TGG1 antibody.
(C) Rosette leaves of the wild type and indicated mutants immunoblotted with anti-TGG1 antibody. The wild-type level was set to 1; values for other genotypes are expressed relatively.
(D) Rosette leaves of the wild type and ARA7-DN immunoblotted with anti-TGG1 antibody.
Next, we monitored the abnormal development of myrosin cells using the myrosin-cell-specific reporter MYR001:GUS. In syp22-4 vti11/+, GUS-positive cells formed a much denser network than that observed in syp22-4 (Figure 2D). Analyses of leaf cross sections showed that GUS-positive cells formed a band near the center of syp22-4 and syp22-4 vti11/+ leaves (Supplemental Figure 5A). This band was absent from leaves of the wild type (Supplemental Figure 5A). The band of myrosin cells was broader in syp22-4 vti11/+ than in syp22-4 (Supplemental Figure 5A). Magnified images showed that the abaxial side of vascular bundles in syp22-4 and syp22-4 vti11/+ plants was covered with a GUS-positive band continuing to the right and left (Figure 2E; Supplemental Figure 5B). Occasionally, a single GUS-positive cell was surrounded by mesophyll cells in syp22-4 vti11/+ plants (Supplemental Figure 5B). As well as the increased number of myrosin cells, syp22-4 vti11/+ showed abnormal vascular bundles. In particular, we did not observe the ordered vascular cell layer composed of xylem, procambium, and phloem (Figure 2E; Supplemental Figure 5B).
By contrast, mutants of other endocytic/vacuolar SNAREs, such as SYP21 and SYP23, exhibited no genetic interaction with vti11 (Supplemental Figure 4B), indicating a specific and close functional relationship between SYP22 and VTI11. Taken together, these results suggest that mutations in syp22 and vti11 act synergistically and that the SNARE complex containing SYP22 and VTI11 is required for the proper development of myrosin cells.
RAB5 GTPases and the Activator VPS9A Are Required for Proper Myrosin Cell Development
SYP22 functions in both vacuolar trafficking and endocytosis (Figure 3A) (Ebine et al., 2008, 2011; Shirakawa et al., 2010). To determine which pathway is predominantly involved in the development of myrosin cells, we examined myrosin cell development in a variety of vacuolar-trafficking mutants that we isolated previously. These included vsr1 (Shimada et al., 2003), mag1 (Shimada et al., 2006), mag2 (Li et al., 2006), vps35 (Yamazaki et al., 2008), and gfs10 (Fuji et al., 2007). None of these vacuolar-trafficking mutants showed abnormal TGG1 accumulation in their leaves (Supplemental Figure 6A), although some showed growth defects similar to those of the syp22 mutant (Supplemental Figure 6B). Compared with other mutants, vps9a-2 plants accumulated a higher level of TGG1 (Figure 3B) and had more myrosin cells that formed an abnormal network similar to that in the syp22 mutant (Supplemental Figure 7A). VPS9A activates the RAB5 GTPases ARA7 and RHA1, which function in both vacuolar trafficking and endocytosis (Figure 3A) (Goh et al., 2007; Ebine et al., 2011; Beck et al., 2012; Irani et al., 2012; Singh et al., 2014). The levels of TGG1 in ara7 and rha1 single mutants were indistinguishable from those in wild-type plants (Figure 3C), possibly reflecting functional redundancy between ARA7 and RHA1. This is consistent with the fact that pollen carrying either ara7 or rha1 single mutations was fertile, but pollen carrying the ara7 rha1 double mutant was infertile (Ebine et al., 2011). Therefore, we generated transgenic plants expressing a GDP-fixed (S24N) dominant-negative form of ARA7 (ARA7-DN) under the control of the constitutive 35S promoter. Like vps9a-2 plants, ARA7-DN plants accumulated a high level of TGG1 (Figure 3D). ARA7-DN expression was previously reported to affect endocytosis (Beck et al., 2012; Irani et al., 2012). We found that FM4-64, a lipophilic dye that labels membranes, was not internalized as quickly in the syp22 vti11/+ mutant as in wild-type plants (Supplemental Figure 7B), suggesting that endocytosis is impaired or delayed in syp22 vti11/+ plants. Taken together, our results suggest that abnormal myrosin cell development might be triggered by a defect in endocytosis.
Finally, we analyzed the genetic interactions between SYP22 and RAB5 GTPases and their effects on myrosin cell development. Mutations in ARA7 and RHA1 in the syp22-1 background (syp22-1 is a weak allele of syp22; Ohtomo et al., 2005) increased the TGG1 level (Figure 3C). These results indicate that mutations in SYP22 and RAB5 interact synergistically in the same genetic pathway, and disrupt the proper development of myrosin cells.
Expression of ARA7-DN in Leaf Primordia Increases the Number of Myrosin Cells
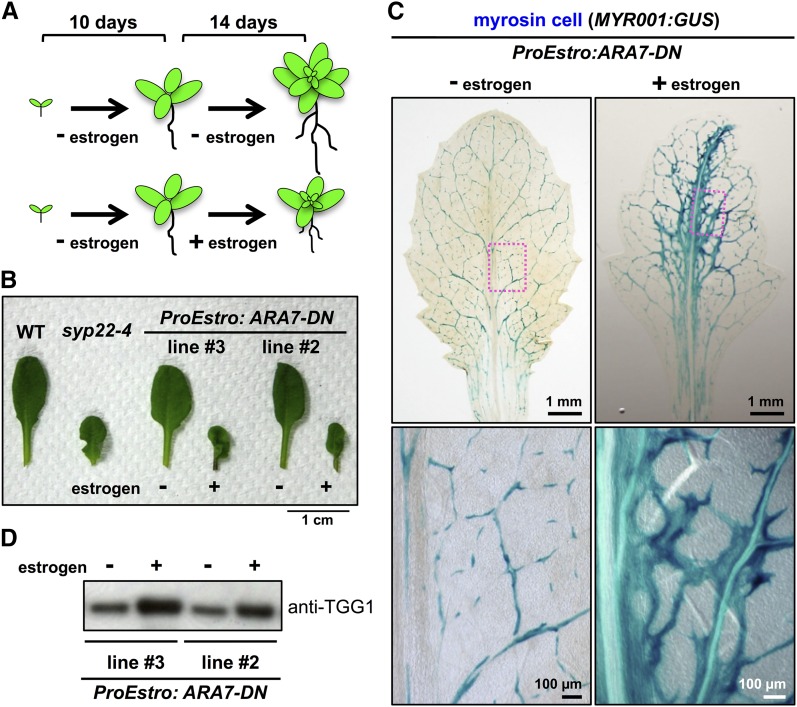
SYP22 and VPS9A are required for embryonic development (Goh et al., 2007; Shirakawa et al., 2010). To exclude the possibility that abnormal myrosin cell development results from embryonic defects during postembryonic development, we generated transgenic plants harboring the ARA7-DN gene under the control of the estrogen-inducible promoter (ProEstro:ARA7-DN). At 10 DAG, transgenic plants grown under normal conditions were transplanted onto induction medium containing 10 μM estrogen and incubated for an additional 2 weeks (Figure 4A). The leaf primordia of the estrogen-treated plants developed abnormally (Supplemental Figure 8A) and had a stunted and wavy morphology similar to that of syp22-4 leaves (Figure 4B). Compared with untreated leaves, the estrogen-treated leaves had more myrosin cells that formed a denser network pattern (Figure 4C; Supplemental Figure 8B) and accumulated higher levels of TGG1 (Figure 4D). Interestingly, the estrogen-treated plants had GUS-positive root cells, similar to those observed in FAMA-overexpressing plants (Supplemental Figure 8C) (Shirakawa et al., 2014b). These results suggest that ARA7 in leaf primordia is crucial for the proper development of myrosin cells.
Figure 4.
Induction of ARA7-DN in Leaf Primordia Leads to Enhanced Development of Myrosin Cells.
(A) Experimental design of estrogen application. Ten-day-old plants were transplanted onto induction medium with or without 10 μM estrogen and incubated for an additional 2 weeks.
(B) The seventh rosette leaves of wild-type (WT), syp22-4, and transgenic plants expressing ARA7-DN under the control of the estrogen-inducible promoter (ProEstro:ARA7-DN). ProEstro:ARA7-DN leaves have a stunted and wavy morphology, similar to that of syp22-4 leaves. Two independent transgenic lines were examined. Bar = 1 cm.
(C) GUS staining of a MYR001:GUS leaf expressing ProEstro:ARA7-DN. Ten-day-old plants were transplanted onto induction medium containing 10 μM estrogen and incubated for 2 weeks. The boxed area is enlarged in the panel on the right. Bars = 1 mm (top panels) and 100 μm (bottom panels).
(D) Accumulation levels of TGG1 in leaves of ProEstro:ARA7-DN transgenic plants with or without 10 μM estrogen. Two independent transgenic lines expressing ProEstro:ARA7-DN were grown with (+) or without (−) 10 μM estrogen, and rosette leaves were immunoblotted with anti-TGG1 antibody.
Proper Development of Myrosin Cells Requires Endocytic/Vacuolar Trafficking Pathway in PIN1-Expressing Cells
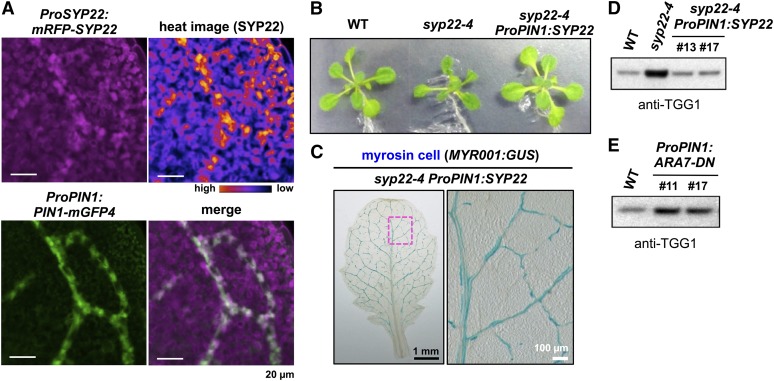
To investigate why the deficiency of SYP22 caused abnormal development of myrosin cells, we monitored the pattern of SYP22 expression in leaf primordia. In transgenic plants expressing both ProPIN1:PIN1-mGFP4 and ProSYP22:mRFP-SYP22, SYP22 was expressed throughout the leaf primordia and at particularly high levels in some cells (Figure 5A). The expression of PIN1 in leaf primordia was reported to be gradually restricted (Scarpella et al., 2006; Wenzel et al., 2007). In the merged mGFP4 and mRFP images, cells with a high expression level of SYP22 largely overlapped with those specifically expressing PIN1 (Figure 5A), suggesting that SYP22 plays a significant role in PIN1-expressing cells. To investigate this hypothesis, we expressed SYP22 in the syp22-4 background under the control of the 4-kb PIN1 promoter. This region of the PIN1 promoter was shown to be sufficient to complement the pin1 phenotype (Xu et al., 2006). The transgenic plants expressing ProPIN1:SYP22 exhibited marked attenuation of syp22-4 phenotypes, such as stunted and wavy leaves and dwarfism (Figure 5B). Myrosin cell development was indistinguishable between leaves of syp22-4 ProPIN1:SYP22 (Figure 5C) and those of the wild type (Figure 2D). Furthermore, the amount of TGG1 in syp22-4 ProPIN1:SYP22 decreased to the same level as that in wild-type plants (Figure 5D). We also expressed ARA7-DN in the wild-type background under the control of the same PIN1 promoter. The ProPIN1:ARA7-DN transgenic plants accumulated high levels of TGG1 (Figure 5E). These results suggest that SYP22 and ARA7 play crucial role(s) in PIN1-expressing cells for the proper development of myrosin cells.
Figure 5.
SYP22 and ARA7 Play Important Roles in Development of Myrosin Cells in PIN1-Expressing Cells.
(A) Expression patterns of ProSYP22:mRFP-SYP22 (magenta) and ProPIN1:PIN1-mGFP4 (green) in leaf primordia. Upper left, RFP fluorescence. Lower left, GFP fluorescence. Upper right, heat image of RFP fluorescence. Signal intensities are shown in black-to-yellow according to increasing intensity levels. Lower right, merged image. Bars = 20 μm.
(B) Images of wild-type (WT), syp22-4, and syp22-4 plants expressing SYP22 under the control of the PIN1 promoter (ProPIN1:SYP22). Images show plants at 19 DAG. Expression of SYP22 under control of PIN1 promoter rescues morphological defects of syp22-4.
(C) GUS staining of a syp22-4 ProPIN1:SYP22 leaf expressing MYR001:GUS, a myrosin cell marker, at 24 DAG. The boxed area is enlarged in panel on the right. Bars = 1 mm (left panel) and 100 μm (right panel).
(D) Rosette leaves of the wild type, syp22-4, and syp22-4 expressing SYP22 under control of the PIN1 promoter and immunoblotted with anti-TGG1 antibody. Numbers 13 and 17 indicate two independent transgenic lines.
(E) Rosette leaves of wild-type and transgenic plants expressing ARA7-DN under the control of the PIN1 promoter (ProPIN1:ARA7-DN) immunoblotted with anti-TGG1 antibody. Numbers 11 and 17 indicate two independent transgenic lines.
Disruption of PIN1 Polarity Triggers Abnormal Development of Myrosin Cells
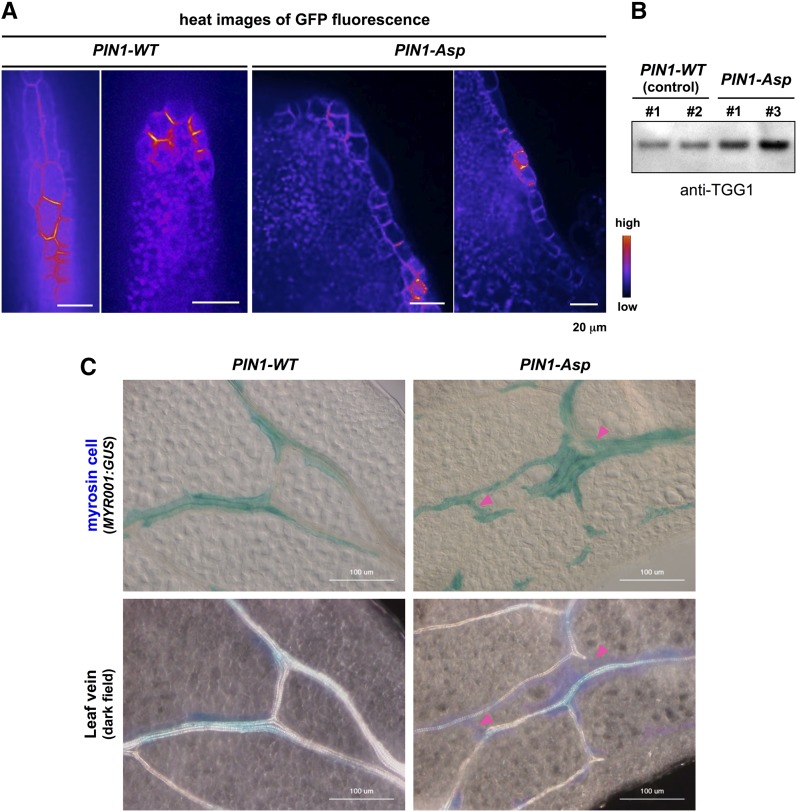
PIN1 is localized to the plasma membrane in a polarized manner; however, PIN1 polarization is impaired in the abaxial epidermis and procambium of syp22-4 leaves (Shirakawa et al., 2009). To determine the effects of disruption of PIN1 polarity on myrosin cell development, we investigated myrosin cell development in transgenic plants expressing a phosphomimic PIN1 fused to GFP under the control of the native PIN1 promoter (PIN1-Asp) in the MYR001:GUS background. PIN1-Asp has a dominant effect on endogenous PIN1 and a portion of cells expressing PIN1-Asp has apolar localization of PIN1-Asp to the plasma membrane in roots (Zhang et al., 2010). In wild-type leaf primordia, the polarity of PIN1 dynamically changes in cell-type-specific and time-dependent manner, but PIN1 polarity is achieved in leaf margin cells (Scarpella et al., 2006; Wenzel et al., 2007). We found that PIN1-Asp had defects in polar localization at the plasma membrane in the subset of leaf margin cells of primordia (Figure 6A). Interestingly, transgenic plants expressing PIN1-Asp accumulated more TGG1 than did control plants expressing PIN1-WT (Figure 6B). Accordingly, compared with PIN1-WT leaves, PIN1-Asp leaves had more myrosin cells that developed not only along the veins but also between veins (Figure 6C). This abnormal distribution pattern of myrosin cells was similar to those observed in the syp22-4 and vps9a-2 mutants, although these phenotypes in PIN1-Asp were weaker than those in the syp22-4 and vps9a-2 (Figure 2; Supplemental Figure 7). Collectively, our results suggest that the polarized localization of PIN1 is required for proper development of myrosin cells.
Figure 6.
Expression of Nonpolarizable PIN1 Driven by PIN1 Promoter Phenocopies Abnormal Development of Myrosin Cells.
(A) Subcellular localization of GFP-tagged wild-type PIN1 (PIN1-WT) and nonpolarizable PIN1 (PIN1-Asp) in the leaf margin cells of primordia. Bars = 20 μm.
(B) Rosette leaves expressing PIN1-WT (control) or PIN1-Asp immunoblotted with anti-TGG1 antibody. Numbers (1 and 2 for PIN1-WT; 1 and 3 for PIN1-Asp) indicate independent transgenic lines.
(C) GUS staining (upper panels) and dark-field images (lower panels) of the same areas of MYR001:GUS leaves expressing either PIN1-WT (control) or PIN1-Asp. Arrowheads indicate regions that are positive for myrosin cells and negative for veins. Bars = 100 μm.
DISCUSSION
Myrosin Cells Require the Endocytic Pathway for Proper Development
Although membrane trafficking has been suggested to mediate the proper development of myrosin cells (Ueda et al., 2006; Shirakawa et al., 2010, 2014a), the exact trafficking pathways that are required for myrosin cell development remain unknown. The results of this study show that SYP22, VTI11, ARA7, RHA1, and VPS9A affect myrosin cell development. The results of this and other studies show that mutations in these genes lead to trafficking defects in both endocytic (Figure 3A) (Ebine et al., 2011; Beck et al., 2012; Irani et al., 2012) and vacuolar trafficking pathways (Figure 3A) (Sohn et al., 2003; Ebine et al., 2008, 2011; Kotzer et al., 2004; Shirakawa et al., 2010). However, several vacuolar trafficking mutants that we characterized previously did not show abnormal development of myrosin cells (Supplemental Figure 6A), although we cannot completely exclude the possibility that vacuolar trafficking plays a minor role in myrosin cell development. Mutations in a plant-specific RAB5 (ARA6/RABF1) suppressed both the delayed endocytosis of a boron transporter and the high level expression of TGG1 in syp22-1 but did not suppress vacuolar trafficking defects of 12S globulin in syp22-1 rha1 (Ueda et al., 2001; Ebine et al., 2011). Taken together, these results suggest that endocytic pathways specifically play a primary role in the development of myrosin cells.
Next, we examined the role of endocytosis in myrosin cell development. Our results show that myrosin cell development requires membrane trafficking in leaf primordia, where the pattern of SYP22 expression largely overlapped with that of PIN1 (Figure 5A). PIN proteins are initially transported to the plasma membrane uniformly, and their polar localization is established after subsequent endocytosis (Men et al., 2008). Therefore, PIN1 is a possible target of the endocytic pathway mediated by SYP22 and RAB5 GTPases. Consistent with this prediction, nonpolar localization of PIN1 to the plasma membrane has been reported in syp22 and vps9a mutants (Goh et al., 2007; Shirakawa et al., 2009). Our data show that SYP22 and RAB5 GTPases are required for myrosin cell development in PIN1-expressing cells (Figure 5). PIN1-Asp partially lost a polarized localization in leaf margin cells and the expression of PIN1-Asp abnormally enhanced development of myrosin cells (Figure 6), suggesting that PIN1 polarity is important for proper development of myrosin cells. However, vps29, which shows a markedly decreased level of PIN1 but normal PIN1 localization (Jaillais et al., 2007; Kleine-Vehn et al., 2008), accumulated a similar amount of TGG1 as the wild type (Supplemental Figure 6A). These results suggest that the level of PIN1 protein is not a critical factor for myrosin cell development. Taken together, these results indicate that the endocytosis-dependent establishment of PIN1 polarity is required for myrosin cell development in leaf primordia of Arabidopsis. Apolar localization of PIN1 in a part of leaf margin cells of leaf primordia may cause defects in the formation of convergence points of auxin flow in leaf primordial, resulting in a reduction of the amount of auxin in internal tissues of leaf primordia. This hypothesis is supported by the result that exogenous auxin treatment rescued the high accumulation level of TGG1 in syp22 (Supplemental Figure 9A). These results also suggest that auxin accumulation and/or flux levels regulate myrosin cell development. Recently, the activation of an auxin response reporter, ProDR5rev:3XVENUS-N7, was transiently observed in the developing myrosin cells, but not in ground meristem cells (Li and Sack, 2014). This result implies that the polar PIN-dependent transient accumulation of auxin regulates determination of ground meristem cells into myrosin lineage cells through auxin influx from neighboring cells and/or efflux from developing myrosin cells. Local manipulation at the single-cell level (Dubrovsky et al., 2008; Benková et al., 2009) will demonstrate the role of auxin and PIN proteins in myrosin cell specification and differentiation.
Recently, the AP-2 complex has been extensively characterized in Arabidopsis (Bashline et al., 2013; Di Rubbo et al., 2013; Fan et al., 2013; Kim et al., 2013; Yamaoka et al., 2013). The AP-2 complex is evolutionarily conserved among eukaryotes and plays a major role in clathrin-mediated endocytosis (McMahon and Boucrot, 2011). We found that the mutant of the μ-subunit of the AP-2 complex accumulated high levels of TGG1 (Supplemental Figure 9B), suggesting that endocytosis is important for the myrosin cell development. The σ-subunit of the AP-2 complex is required for polar localization of PIN1 (Fan et al., 2013). During endocytic uptake of cargo proteins, the AP-2 complex recognizes a tyrosine motif in the cytoplasmic tails of proteins. This tyrosine motif is also present in PIN1 (Mravec et al., 2009); therefore, it is possible that AP-2 regulates the endocytosis of PIN1 by interacting with the tyrosine motif of PIN1.
Relationship between Myrosin Cell Development and Vascular Cell Development
Recently, we showed that myrosin cell differentiation from ground meristem cells is regulated by the master transcription factor FAMA (Shirakawa et al., 2014b). Vascular precursor cells (preprocambium/procambium) are also derived from ground meristem cells (Kang and Dengler, 2004; Scarpella et al., 2004). These two cell lineages arise independently from ground meristem cells because both ProFAMA:GUS and ProAtHB8:GUS (AtHB8 is a master transcription factor of vascular precursor cells) were expressed simultaneously, but with different spatial patterns, in a subset of ground meristem cells (Baima et al., 1995, 2001; Kang and Dengler, 2004; Scarpella et al., 2004; Li and Sack, 2014; Shirakawa et al., 2014b). Arabidopsis HB8 expression in ground meristem cells is determined by auxin (Mattsson et al., 2003; Donner et al., 2009; Ohashi-Ito and Fukuda, 2010; Krogan et al., 2012).
Compared with mature leaves of the wild type, mature leaves of syp22-4 and ProEstro:ARA7-DN formed more myrosin cells (Figures 2 and 4) (Ueda et al., 2006) and showed a simpler vascular pattern (Supplemental Figures 8D and 10A) (Shirakawa et al., 2009). Consistent with this, syp22-4 leaf primordia contained more cells expressing FAMA than did wild-type leaf primordia (Figure 1B). Also, there was a simpler expression pattern of the preprocambium/procambium reporter ProAtHB8:GUS in syp22-4 leaf primordia than in wild-type leaf primordia (Supplemental Figure 10B). These results suggest that vascular development and myrosin cell development might be closely related to each other. In the syp22 mutants and ProEstro:ARA7-DN transgenic plants, the ground meristem cells that should have differentiated into vascular cells might differentiate into myrosin cells. Our results show that the polar localization of PIN1 is important for the patterning of myrosin cells. The polarity of PIN1 has also been shown to play an important role in vascular development (Scarpella et al., 2006; Wenzel et al., 2007). Consistent with this idea, the expression of SYP22 in PIN1-expressing cells in syp22-4 rescued the phenotypes of myrosin cells (Figure 5) and vascular cells (Supplemental Figure 10C). However, not all of the mutant and transgenic plants with defective myrosin cell development showed vascular defects (Supplemental Table 1 and Supplemental Figures 10D and 10E). Myrosin cell phenotypes appeared to be more sensitive than vascular phenotypes to defects in the endocytic/vacuolar trafficking pathway because only the mutants with severe myrosin cell phenotypes showed vascular defects (Supplemental Table 1).
Recently, live imaging revealed that cells in PIN1 expression domains differentiate into vascular and nonvascular cells in leaf primordia (Marcos and Berleth, 2014). These nonvascular cells subsequently downregulate PIN1 expression and maintain ground meristem cell morphology (isodiametric shape) (Marcos and Berleth, 2014). These ground meristem cells might subsequently differentiate into myrosin cells. To clarify the spatiotemporal dynamics between the polarity of PIN1 and the development of myrosin cells and vascular cells, high-resolution live-cell fluorescence imaging of transgenic plants expressing PIN1 reporter, vascular cell precursor marker, and the myrosin cell precursors marker is required hereafter. Although syp22-4 exhibited wavy leaves, vps9a-2 did not. Therefore, leaf morphologies are not necessarily correlated with myrosin cell development.
Concluding Remarks
Myrosin cells function in plant defense against herbivores. They accumulate large quantities of myrosinases (TGGs), which produce toxic compounds that protect against insects and pathogens (Rask et al., 2000; Wittstock and Halkier, 2002; Grubb and Abel, 2006; Halkier and Gershenzon, 2006; Hopkins et al., 2009; Kissen et al., 2009). Myrosin cells are distributed close to leaf vascular networks in Arabidopsis (Xue et al., 1995; Andréasson et al., 2001; Husebye et al., 2002; Thangstad et al., 2004; Barth and Jander, 2006; Ueda et al., 2006), Brassica napus (Bones et al., 1991; Höglund et al., 1991; Andréasson et al., 2001), Cardamine schinziana, and Nasturtium officinale (Supplemental Figure 1). Myrosin cells might specifically localize near vascular cells via the polar localization of PIN1 and auxin concentrations, resulting in the expression of FAMA in a subset of ground meristem cells. This arrangement could effectively protect the plant transport system, which is essential for plant survival. Future work will be directed toward identifying the regulators that directly connect the PIN1-auxin module with the transcriptional heterodimer, FAMA-ICE1.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana, ecotype Columbia, was used as the wild-type plant. syp22-1 (Ohtomo et al., 2005), syp23 (Ohtomo et al., 2005), syp22-3 (Ueda et al., 2006), syp22-4 (Ohtomo et al., 2005), syp21 syp23 (Shirakawa et al., 2010), syp22-3 vti11/+ (Shirakawa et al., 2009), syp22-3 vti11 (Shirakawa et al., 2009), syp22-4 vti11/+ (Shirakawa et al., 2009), vsr1-2 (Shimada et al., 2003), mag1-1 (Shimada et al., 2006), mag1-2 (Shimada et al., 2006), mag2-1 (Li et al., 2006), mag2-3 (Li et al., 2006), vps35 b-1c-1 (Yamazaki et al., 2008), vps35 a-1b-2c-1 (Yamazaki et al., 2008), gfs10-1 (Fuji et al., 2007), gfs10-2 (Fuji et al., 2007), GFP-CT24 (Fuji et al., 2007), ap2m (Yamaoka et al., 2013), ap2m AP2-GFPg (Yamaoka et al., 2013), MYR001:GUS (Shirakawa et al., 2014a), ProTGG2:VENUS-2sc (Shirakawa et al., 2014a), ProPIN1:PIN1-mGFP4 ProSYP22:mRFP-SYP22 (Shirakawa et al., 2009), ProFAMA:GUS (Shirakawa et al., 2014b), and fama-1 (Shirakawa et al., 2014b) were previously reported. vti11 syp21 syp23, syp22-4 MYR001:GUS, syp22-4 vti11/+ MYR001:GUS, syp22-4 fama-1 MYR001:GUS, syp22-4 ProFAMA:GUS, syp22-4 ProTGG2:VENUS-2sc, syp22-4 ProAtHB8:GUS, and syp22-4 ProSUC2:GFP were generated in this study. vti11 (Kato et al., 2002) and syp21 (Shirakawa et al., 2010; Uemura et al., 2010) were gifts from M.T. Morita (Nagoya University, Japan). PIN1-WT, PIN1-Asp (Zhang et al., 2010), and ProPIN1:PIN1-mGFP4 (Benková et al., 2003) were provided by J. Friml (Institute of Science and Technology Austria, Austria). vps9a-2 (Goh et al., 2007) was provided by A. Nakano (University of Tokyo, Japan). ara7 (Ebine et al., 2011), rha1 (Ebine et al., 2011), syp22-1 ara7 (Ebine et al., 2011), syp22-1 rha1 (Ebine et al., 2011), and ProSYP22:mRFP-SYP22 (Ebine et al., 2008) were provided by T. Ueda (University of Tokyo, Japan). vti11 vti12/+ (Surpin et al., 2003) was a gift from N. Raikhel (University of California, Riverside). ProAtHB8:GUS (Kang and Dengler, 2002) was provided by N.G. Dengler (University of Toronto). ProSUC2:GFP (Imlau et al., 1999) was provided by N. Sauer (Universitat Erlangen-Nurnberg). Seeds were surface-sterilized with 70% ethanol and then sown onto 0.5% (w/v) Gellan Gum medium (Wako) that contained 1% (w/v) sucrose and Murashige and Skoog medium (Wako). The seeds were incubated at 4°C for 3 to 5 d to break dormancy, followed by growth at 22°C for up to 20 d under continuous light (100 μmol s−1 m−2). The plants were transferred to vermiculite for subsequent growth.
Generation of Multiple Mutants
We crossed a homozygote for vti11 with a homozygote for syp22-3, syp22-4, and syp21 syp23. Multiple mutants were identified by genotyping F2 progeny. We crossed a homozygote for syp22-4 with a homozygote for ProAtHB8:GUS and ProSUC2:GFP. We crossed a homozygote for syp22-4 MYR001:GUS with a heterozygous for fama-1.Transgenic lines were identified by genotyping F2 progeny. Genotyping was performed as described previously (Ueda et al., 2006; Shirakawa et al., 2009, 2010, 2014b).
Plasmid Construction and Transgenic Plants
The Gateway cloning system (Invitrogen) was used to construct ARA7-DN, ProEstro:ARA7-DN, ProPIN1:SYP22, and ProPIN1:ARA7-DN. The TOPO reaction was employed to clone ARA7 coding sequence into pENTR D-TOPO. To generate pENTR D-TOPO ARA7-DN (a dominant-negative form of ARA7), a S24N substitution was introduced into pENTR D-TOPO ARA7 by site-directed mutagenesis. Next, pENTR D-TOPO Ara7-DN was introduced into binary vectors pB2GW7 and pMDC7 (Curtis and Grossniklaus, 2003) using LR reactions to generate ARA7-DN and ProEstro:ARA7-DN, respectively. The 4-kb promoter region of PIN1 was cloned into pENTR D-TOPO using the TOPO reaction. An In-Fusion HD Cloning Kit (Takara Bio) was used to introduce each of the following DNA sequences into the AscI site of the pENTR D-TOPO PIN1pro plasmid: (1) SYP22 coding sequence and the NOS terminator, (2) ARA7-DN and the 35S terminator, (3) the PIN1 genomic region fused to GFP, and (4) the PIN1 genomic region containing two substitutions (S337D and T340D) fused to GFP. The following clones were generated: (1) ProPIN1:SYP22, (2) ProPIN1:ARA7-DN, (3) PIN1-WT, and (4) PIN1-Asp, respectively. Subsequently, each entry clone was introduced into the binary vector FAST-G01 (Shimada et al., 2010) using the LR reaction. Each construct was introduced into Agrobacterium tumefaciens, and plants were subsequently infected with these bacteria using the floral dip method (Clough and Bent, 1998). T1 seeds were selected on medium containing 10 mg/L BASTA or 25 mg/L hygromycin B or 100 mg/L kanamycin. Primer sets used are presented in Supplemental Table 2. ProPIN1:SYP22 was introduced into syp22-4 background. ProEstro:ARA7-DN, ProPIN1:ARA7-DN, PIN1-WT, and PIN1-Asp were introduced into the wild-type background. Crosses were performed to generate syp22-4 MYR001:GUS and syp22-4 vti11/+ MYR001:GUS. Each of ProTGG2:VENUS-2sc and ProFAMA:GUS was introduced in to syp22-4 by Agrobacterium-mediated transformation.
RT-PCR
We prepared total RNA from wild-type and mutant plants at 21 DAG using an RNeasy plant mini kit (Qiagen). After DNase I (Invitrogen) treatment, reverse transcription was performed using a SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) with an oligo(dT)12-18 primer (Invitrogen). Quantitative RT-PCR was performed using a gene-specific primer set (FAMA, At02279294_g1; TGG1, At02185835_g1; Actin2, At02335270_gH; Applied Biosystems) and a TaqMan Gene Expression Assay Kit (Applied Biosystems) in a 7500 Real-Time PCR system (Applied Biosystems). The relative quantity of target mRNA was calculated using Actin2 as a control.
GUS Staining
Samples were first placed into ice-cold acetone for 15 min and then into GUS staining solution containing 0.5 mg/mL X-Gluc, 0.1 M sodium phosphate buffer, pH 7.0, 10 mM EDTA, 0.5 mM potassium ferricyanide, 0.5 to 5 mM potassium ferrocyanide, and 0.1% Triton X-100. Samples in the GUS staining solution were placed under a vacuum and incubated at room temperature for 12 to 24 h.
Histological Analysis
After GUS staining of 19-d-old rosette leaves, samples were fixed in ethanol:water:acetic acid:formalin = 50:35:5:10, dehydrated in a graded ethanol series, and embedded in Technovit 7100 resin according to the manufacturer’s instructions. Sections of 3 to 5 μm were cut using a microtome and then counterstained using Toluidine Blue O.
Coomassie Blue Staining
Rosette leaves were harvested from 12-d-old plants of the indicated genotypes. The leaves were stained using Coomassie Brilliant Blue, as previously described (Ueda et al., 2006).
Analysis of Vein Pattern
Leaves were made transparent by overnight incubation in a chloral hydrate solution (chloral hydrate:water:glycerol, 8:2:1 [w/v/v]) followed by incubation in 60% glycerol. The leaves were mounted onto a glass slide and inspected with a dark-field illumination microscope (model MVX10; Olympus).
SDS-PAGE and Immunoblot Analysis
SDS-PAGE and immunoblot analyses were performed as previously described (Shimada et al., 2003). The antibody used was anti-TGG1 (diluted 5000-fold) (Ueda et al., 2006).
Staining with FM4-64
FM4-64 staining and observations were performed as previously described (Teh et al., 2013). The roots of intact seedlings were incubated in 5 μM FM4-64 solution for 1 min and washed in water. After 6 min, fluorescent images of FM4-64 staining were taken with a confocal laser scanning microscope.
Confocal Laser Scanning Microscopy
Fluorescence micrographs (except Figure 6) were obtained with a confocal laser scanning microscope (LSM510 META; Carl Zeiss) equipped with a water immersion objective (63× 1.20 numerical aperture [NA]) and dry objectives (40× 0.95 NA, 20× 0.80 NA, 10× 0.50 NA). The wavelengths of lasers that were employed include 488 nm (GFP and Venus) and 543 nm (mRFP and FM4-64). The images were analyzed using LSM image-examiner software (Carl Zeiss). Polarized localization of PIN1-GFP (Figure 6) was inspected with a fluorescence microscope (Axio Observer.Z1; Carl Zeiss) equipped with a confocal scanner unit (CSU-X1; Yokogawa Electric) and the 488-nm laser unit (Andor Technology). Fluorescent images of PIN1-GFP were collected by an EM-CCD camera (iXon3; Andor Technology) with a 40× 0.75-NA objective using Andor iQ software, version 2.8 (Andor Technology), and processed using ImageJ and Photoshop software (Adobe Systems).
Accession Numbers
Sequence data and mutants from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: TGG1 (At5g26000), TGG2 (At5g25980), SYP22 (At5g46860), FAMA (At3g24140), VTI11 (At5g39510), VPS9A (At3g19770), ARA7 (At4g19640), RHA1 (At5g45130), PIN1 (At1g73590), SYP21 (At5g16830), SYP23 (At4g17730), VSR1 (At3g52850), MAG1 (At3g47810), MAG2 (At3g47700), VPS35A (At2g17790), VPS35B (At1g75850), VPS35C (At3g51310), GFS10 (At4g35870), VTI12 (At1g26670), AP2M (At5g46630), ACT2 (At3g18780), syp22-3 (CS68716), syp22-4 (CS68717), syp22-3 vti11 (CS68722), and syp22-4 vti11/+ (CS68723).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Myrosin Cells Are Associated with Vascular Cells.
Supplemental Figure 2. Plant Morphology of syp22-4 fama-1.
Supplemental Figure 3. Patterning of Myrosin Cells in Wild Type and syp22-4.
Supplemental Figure 4. Levels of TGG1 Accumulation in Multiple Mutants.
Supplemental Figure 5. Distribution of MYR001:GUS-Expressing Cells in Wild-Type, syp22-4, and syp22-4 vti11/+ Plants.
Supplemental Figure 6. Levels of TGG1 Accumulation and Morphology of Various Membrane-Trafficking Mutants.
Supplemental Figure 7. Endocytic Pathway and Myrosin Cell Development.
Supplemental Figure 8. Phenotypes of ProEstro:Ara7-DN Transgenic Plants.
Supplemental Figure 9. Levels of TGG1 Accumulation in the ap2m Mutant and Auxin-Treated syp22.
Supplemental Figure 10. Vascular Patterns and Plant Morphologies of Mutants and Transgenic Lines in This Study.
Supplemental Table 1. Myrosin Cell Development, PIN1 Localization, and Vascular Pattern in Mutants and Transgenic Plants.
Supplemental Table 2. Primer Sets Used in This Study.
Supplementary Material
Acknowledgments
We thank Hiroo Fukuda (University of Tokyo) and Tobias Baskin (University of Massachusetts) for critical reading of this article. We thank Akihiko Nakano (University of Tokyo), Hiroshi Kudoh (Kyoto Univeristy), Jiri Friml (Ghent University), Miyo T. Morita (Nagoya University), Nam-Hai Chua (Rockefeller University), Takashi Ueda (University of Tokyo), Natasha Raikhel (University of California, Riverside), Nancy G. Dengler (University of Toronto), and Norbert Sauer (Universitat Erlangen-Nurnberg) for sharing materials. We also thank the ABRC and the Nottingham Arabidopsis Stock Centre for providing seeds of Arabidopsis lines. This work was supported by Specially Promoted Research of Grant-in-Aid for Scientific Research to I.H-.N. (22000014) and Grant-in-Aid for Scientific Research to H.U. (21200065 and 25440132) from the Japan Society for the Promotion of Science (JSPS). M.S. was supported by a research fellowship from JSPS (20002057 and 24005453) and by a Grant-in-Aid for Plant Graduate Students from the Nara Institute of Science and Technology.
AUTHOR CONTRIBUTIONS
M.S., H.U., T.S., and I.H.-N. designed the research. M.S. and H.U. performed all experiments. M.S., H.U., T.S., and I.H.-N. wrote the article. T.K. and I.H.-N. supervised the study.
Glossary
- DAG
days after germination
- NA
numerical aperture
Footnotes
Online version contains Web-only data.
References
- Andréasson E., Bolt Jørgensen L., Höglund A.S., Rask L., Meijer J. (2001). Different myrosinase and idioblast distribution in Arabidopsis and Brassica napus. Plant Physiol. 127: 1750–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima S., Nobili F., Sessa G., Lucchetti S., Ruberti I., Morelli G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121: 4171–4182. [DOI] [PubMed] [Google Scholar]
- Baima S., Possenti M., Matteucci A., Wisman E., Altamura M.M., Ruberti I., Morelli G. (2001). The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126: 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth C., Jander G. (2006). Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46: 549–562. [DOI] [PubMed] [Google Scholar]
- Bashline L., Li S., Anderson C.T., Lei L., Gu Y. (2013). The endocytosis of cellulose synthase in Arabidopsis is dependent on μ2, a clathrin-mediated endocytosis adaptin. Plant Physiol. 163: 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Zhou J., Faulkner C., MacLean D., Robatzek S. (2012). Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24: 4205–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Ivanchenko M.G., Friml J., Shishkova S., Dubrovsky J.G. (2009). A morphogenetic trigger: is there an emerging concept in plant developmental biology? Trends Plant Sci. 14: 189–193. [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Bones A.M., Thangstad O.P., Haugen O.A., Espevik T. (1991). Fate of myrosin cells: characterization of monoclonal antibodies against myrosinase. J. Exp. Bot. 42: 1541–1549. [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rubbo S., et al. (2013). The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell 25: 2986–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner T.J., Sherr I., Scarpella E. (2009). Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 136: 3235–3246. [DOI] [PubMed] [Google Scholar]
- Dubrovsky J.G., Sauer M., Napsucialy-Mendivil S., Ivanchenko M.G., Friml J., Shishkova S., Celenza J., Benková E. (2008). Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 105: 8790–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., et al. (2011). A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13: 853–859. [DOI] [PubMed] [Google Scholar]
- Ebine K., Okatani Y., Uemura T., Goh T., Shoda K., Niihama M., Morita M.T., Spitzer C., Otegui M.S., Nakano A., Ueda T. (2008). A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20: 3006–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L., Hao H., Xue Y., Zhang L., Song K., Ding Z., Botella M.A., Wang H., Lin J. (2013). Dynamic analysis of Arabidopsis AP2 σ subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development 140: 3826–3837. [DOI] [PubMed] [Google Scholar]
- Fuji K., Shimada T., Takahashi H., Tamura K., Koumoto Y., Utsumi S., Nishizawa K., Maruyama N., Hara-Nishimura I. (2007). Arabidopsis vacuolar sorting mutants (green fluorescent seed) can be identified efficiently by secretion of vacuole-targeted green fluorescent protein in their seeds. Plant Cell 19: 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230. [DOI] [PubMed] [Google Scholar]
- Goh T., Uchida W., Arakawa S., Ito E., Dainobu T., Ebine K., Takeuchi M., Sato K., Ueda T., Nakano A. (2007). VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell 19: 3504–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb C.D., Abel S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11: 89–100. [DOI] [PubMed] [Google Scholar]
- Grunewald W., Friml J. (2010). The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 29: 2700–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier B.A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57: 303–333. [DOI] [PubMed] [Google Scholar]
- Höglund A.S., Lenman M., Falk A., Rask L. (1991). Distribution of myrosinase in rapeseed tissues. Plant Physiol. 95: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R.J., van Dam N.M., van Loon J.J. (2009). Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu. Rev. Entomol. 54: 57–83. [DOI] [PubMed] [Google Scholar]
- Husebye H., Chadchawan S., Winge P., Thangstad O.P., Bones A.M. (2002). Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 128: 1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T., et al. (2013). Phosphatidylinositol 4,5-bisphosphate influences PIN polarization by controlling clathrin-mediated membrane trafficking in Arabidopsis. Plant Cell 25: 4894–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlau A., Truernit E., Sauer N. (1999). Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Kondo Y., Naramoto S., Nakano A., Ueda T. (2013). RAB5 activation is required for multiple steps in Arabidopsis thaliana root development. Plant Cell Physiol. 54: 1648–1659. [DOI] [PubMed] [Google Scholar]
- Irani N.G., et al. (2012). Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 8: 583–589. [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Santambrogio M., Rozier F., Fobis-Loisy I., Miège C., Gaude T. (2007). The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell 130: 1057–1070. [DOI] [PubMed] [Google Scholar]
- Kang J., Dengler N. (2002). Cell cycling frequency and expression of the homeobox gene ATHB-8 during leaf vein development in Arabidopsis. Planta 216: 212–219. [DOI] [PubMed] [Google Scholar]
- Kang J., Dengler N. (2004). Vein pattern development in adult leaves of Arabidopsis thaliana. Int. J. Plant Sci. 165: 231–242. [Google Scholar]
- Kato T., Morita M.T., Fukaki H., Yamauchi Y., Uehara M., Niihama M., Tasaka M. (2002). SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Xu Z.Y., Song K., Kim D.H., Kang H., Reichardt I., Sohn E.J., Friml J., Juergens G., Hwang I. (2013). Adaptor protein complex 2-mediated endocytosis is crucial for male reproductive organ development in Arabidopsis. Plant Cell 25: 2970–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissen R., Rossiter J.T., Bones A.M. (2009). The ‘mustard oil bomb’: not so easy to assemble?! Localization, expression and distribution of the components of the myrosinase enzyme system. Phytochem. Rev. 8: 69–86. [Google Scholar]
- Kitakura S., Vanneste S., Robert S., Löfke C., Teichmann T., Tanaka H., Friml J. (2011). Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva O.A., Davies A., Deeken R., Thorpe M.R., Tomos A.D., Hedrich R. (2000). Identification of a new glucosinolate-rich cell type in Arabidopsis flower stalk. Plant Physiol. 124: 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzer A.M., Brandizzi F., Neumann U., Paris N., Moore I., Hawes C. (2004). AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J. Cell Sci. 117: 6377–6389. [DOI] [PubMed] [Google Scholar]
- Krogan N.T., Ckurshumova W., Marcos D., Caragea A.E., Berleth T. (2012). Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytol. 194: 391–401. [DOI] [PubMed] [Google Scholar]
- Li L., Shimada T., Takahashi H., Ueda H., Fukao Y., Kondo M., Nishimura M., Hara-Nishimura I. (2006). MAIGO2 is involved in exit of seed storage proteins from the endoplasmic reticulum in Arabidopsis thaliana. Plant Cell 18: 3535–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Sack F.D. (2014). Myrosin idioblast cell fate and development are regulated by the Arabidospsis transcription factor FAMA, the auxin pathway, and vesicular trafficking. Plant Cell 26:in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfke C., Luschnig C., Kleine-Vehn J. (2013). Posttranslational modification and trafficking of PIN auxin efflux carriers. Mech. Dev. 130: 82–94. [DOI] [PubMed] [Google Scholar]
- Marcos D., Berleth T. (2014). Dynamic auxin transport patterns preceding vein formation revealed by live-imaging of Arabidopsis leaf primordia. Front. Plant Sci. 5: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J., Ckurshumova W., Berleth T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131: 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H.T., Boucrot E. (2011). Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12: 517–533. [DOI] [PubMed] [Google Scholar]
- Men S., Boutté Y., Ikeda Y., Li X., Palme K., Stierhof Y.D., Hartmann M.A., Moritz T., Grebe M. (2008). Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat. Cell Biol. 10: 237–244. [DOI] [PubMed] [Google Scholar]
- Mravec J., et al. (2009). Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Fukuda H. (2010). Transcriptional regulation of vascular cell fates. Curr. Opin. Plant Biol. 13: 670–676. [DOI] [PubMed] [Google Scholar]
- Ohtomo I., Ueda H., Shimada T., Nishiyama C., Komoto Y., Hara-Nishimura I., Takahashi T. (2005). Identification of an allele of VAM3/SYP22 that confers a semi-dwarf phenotype in Arabidopsis thaliana. Plant Cell Physiol. 46: 1358–1365. [DOI] [PubMed] [Google Scholar]
- Paponov I.A., Teale W.D., Trebar M., Blilou I., Palme K. (2005). The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci. 10: 170–177. [DOI] [PubMed] [Google Scholar]
- Petrásek J., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918. [DOI] [PubMed] [Google Scholar]
- Rask L., Andréasson E., Ekbom B., Eriksson S., Pontoppidan B., Meijer J. (2000). Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42: 93–113. [PubMed] [Google Scholar]
- Sanderfoot A.A., Kovaleva V., Bassham D.C., Raikhel N.V. (2001). Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell 12: 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A.A., Kovaleva V., Zheng H., Raikhel N.V. (1999). The t-SNARE AtVAM3p resides on the prevacuolar compartment in Arabidopsis root cells. Plant Physiol. 121: 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E., Francis P., Berleth T. (2004). Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131: 3445–3455. [DOI] [PubMed] [Google Scholar]
- Scarpella E., Marcos D., Friml J., Berleth T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20: 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Koumoto Y., Li L., Yamazaki M., Kondo M., Nishimura M., Hara-Nishimura I. (2006). AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 47: 1187–1194. [DOI] [PubMed] [Google Scholar]
- Shimada T., Fuji K., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2003). Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 16095–16100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T.L., Shimada T., Hara-Nishimura I. (2010). A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61: 519–528. [DOI] [PubMed] [Google Scholar]
- Shirakawa M., Ueda H., Koumoto Y., Fuji K., Nishiyama C., Kohchi T., Hara-Nishimura I., Shimada T. (2014a). CONTINUOUS VASCULAR RING (COV1) is a trans-Golgi network-localized membrane protein required for Golgi morphology and vacuolar protein sorting. Plant Cell Physiol. 55: 764–772. [DOI] [PubMed] [Google Scholar]
- Shirakawa M., Ueda H., Nagano A.J., Shimada T., Kohchi T., Hara-Nishimura I. (2014b). FAMA is an essential component for the differentiation of two distinct cell types, myrosin cells and guard cells, in Arabidopsis. Plant Cell 26:in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa M., Ueda H., Shimada T., Koumoto Y., Shimada T.L., Kondo M., Takahashi T., Okuyama Y., Nishimura M., Hara-Nishimura I. (2010). Arabidopsis Qa-SNARE SYP2 proteins localized to different subcellular regions function redundantly in vacuolar protein sorting and plant development. Plant J. 64: 924–935. [DOI] [PubMed] [Google Scholar]
- Shirakawa M., Ueda H., Shimada T., Nishiyama C., Hara-Nishimura I. (2009). Vacuolar SNAREs function in the formation of the leaf vascular network by regulating auxin distribution. Plant Cell Physiol. 50: 1319–1328. [DOI] [PubMed] [Google Scholar]
- Shroff R., Vergara F., Muck A., Svatos A., Gershenzon J. (2008). Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc. Natl. Acad. Sci. USA 105: 6196–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M.K., Krüger F., Beckmann H., Brumm S., Vermeer J.E., Munnik T., Mayer U., Stierhof Y.D., Grefen C., Schumacher K., Jürgens G. (2014). Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Curr. Biol. 24: 1383–1389. [DOI] [PubMed] [Google Scholar]
- Sohn E.J., Kim E.S., Zhao M., Kim S.J., Kim H., Kim Y.W., Lee Y.J., Hillmer S., Sohn U., Jiang L., Hwang I. (2003). Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15: 1057–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C., Reyes F.C., Buono R., Sliwinski M.K., Haas T.J., Otegui M.S. (2009). The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 21: 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M., Zheng H., Morita M.T., Saito C., Avila E., Blakeslee J.J., Bandyopadhyay A., Kovaleva V., Carter D., Murphy A., Tasaka M., Raikhel N. (2003). The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15: 2885–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh O.K., Shimono Y., Shirakawa M., Fukao Y., Tamura K., Shimada T., Hara-Nishimura I. (2013). The AP-1 μ adaptin is required for KNOLLE localization at the cell plate to mediate cytokinesis in Arabidopsis. Plant Cell Physiol. 54: 838–847. [DOI] [PubMed] [Google Scholar]
- Tejos R., Sauer M., Vanneste S., Palacios-Gomez M., Li H., Heilmann M., van Wijk R., Vermeer J.E., Heilmann I., Munnik T., Friml J. (2014). Bipolar plasma membrane distribution of phosphoinositides and their requirement for auxin-mediated cell polarity and patterning in Arabidopsis. Plant Cell 26: 2114–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangstad O.P., Gilde B., Chadchawan S., Seem M., Husebye H., Bradley D., Bones A.M. (2004). Cell specific, cross-species expression of myrosinases in Brassica napus, Arabidopsis thaliana and Nicotiana tabacum. Plant Mol. Biol. 54: 597–611. [DOI] [PubMed] [Google Scholar]
- Ueda H., Nishiyama C., Shimada T., Koumoto Y., Hayashi Y., Kondo M., Takahashi T., Ohtomo I., Nishimura M., Hara-Nishimura I. (2006). AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol. 47: 164–175. [DOI] [PubMed] [Google Scholar]
- Ueda T., Yamaguchi M., Uchimiya H., Nakano A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20: 4730–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Morita M.T., Ebine K., Okatani Y., Yano D., Saito C., Ueda T., Nakano A. (2010). Vacuolar/pre-vacuolar compartment Qa-SNAREs VAM3/SYP22 and PEP12/SYP21 have interchangeable functions in Arabidopsis. Plant J. 64: 864–873. [DOI] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R.L., Nakano A., Takeyasu K., Sato M.H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29: 49–65. [DOI] [PubMed] [Google Scholar]
- Wenzel C.L., Schuetz M., Yu Q., Mattsson J. (2007). Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J. 49: 387–398. [DOI] [PubMed] [Google Scholar]
- Wisniewska J., Xu J., Seifertová D., Brewer P.B., Ruzicka K., Blilou I., Rouquié D., Benková E., Scheres B., Friml J. (2006). Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Wittstock U., Halkier B.A. (2002). Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 7: 263–270. [DOI] [PubMed] [Google Scholar]
- Xu J., Hofhuis H., Heidstra R., Sauer M., Friml J., Scheres B. (2006). A molecular framework for plant regeneration. Science 311: 385–388. [DOI] [PubMed] [Google Scholar]
- Xue J., Jørgensen M., Pihlgren U., Rask L. (1995). The myrosinase gene family in Arabidopsis thaliana: gene organization, expression and evolution. Plant Mol. Biol. 27: 911–922. [DOI] [PubMed] [Google Scholar]
- Yamaoka S., Shimono Y., Shirakawa M., Fukao Y., Kawase T., Hatsugai N., Tamura K., Shimada T., Hara-Nishimura I. (2013). Identification and dynamics of Arabidopsis adaptor protein-2 complex and its involvement in floral organ development. Plant Cell 25: 2958–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M., Shimada T., Takahashi H., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2008). Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence. Plant Cell Physiol. 49: 142–156. [DOI] [PubMed] [Google Scholar]
- Yano D., Sato M., Saito C., Sato M.H., Morita M.T., Tasaka M. (2003). A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc. Natl. Acad. Sci. USA 100: 8589–8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nodzynski T., Pencík A., Rolcík J., Friml J. (2010). PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl. Acad. Sci. USA 107: 918–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.