Physiological BR promotes cell elongation by inducing GA accumulation through activation of GA3ox-2 expression, whereas exogenously applied high concentrations of BR inhibit cell elongation by repressing GA level. As a feedback mechanism, GA extensively inhibits BR biosynthesis and signaling.
Abstract
Brassinosteroid (BR) and gibberellin (GA) are two predominant hormones regulating plant cell elongation. A defect in either of these leads to reduced plant growth and dwarfism. However, their relationship remains unknown in rice (Oryza sativa). Here, we demonstrated that BR regulates cell elongation by modulating GA metabolism in rice. Under physiological conditions, BR promotes GA accumulation by regulating the expression of GA metabolic genes to stimulate cell elongation. BR greatly induces the expression of D18/GA3ox-2, one of the GA biosynthetic genes, leading to increased GA1 levels, the bioactive GA in rice seedlings. Consequently, both d18 and loss-of-function GA-signaling mutants have decreased BR sensitivity. When excessive active BR is applied, the hormone mostly induces GA inactivation through upregulation of the GA inactivation gene GA2ox-3 and also represses BR biosynthesis, resulting in decreased hormone levels and growth inhibition. As a feedback mechanism, GA extensively inhibits BR biosynthesis and the BR response. GA treatment decreases the enlarged leaf angles in plants with enhanced BR biosynthesis or signaling. Our results revealed a previously unknown mechanism underlying BR and GA crosstalk depending on tissues and hormone levels, which greatly advances our understanding of hormone actions in crop plants and appears much different from that in Arabidopsis thaliana.
INTRODUCTION
Among the plant hormones, brassinosteroid (BR) and gibberellin (GA) are the two most important ones that determine plant height by regulating cell elongation. In rice (Oryza sativa), mutants deficient in either GA or BR display a dwarf stature. Through studies of the corresponding mutants, significant progress has been made in the signaling as well as biosynthetic pathways of both GA and BR in recent years. In rice, GA is perceived by a soluble receptor, GIBBERELLIN-INSENSITIVE DWARF1 (GID1), which binds to GA directly, and also interacts with SLENDER1 (SLR1), the GA-signaling repressor encoding a DELLA family protein (Dill et al., 2001; Ikeda et al., 2001; Ueguchi-Tanaka et al., 2005, 2007). The stable triple complex (GA-GID1-DELLA) is then recognized by the SCFGID2 (for Skp1-Cullin-F-box) E3 ubiquitin ligase complex, leading to the ubiquitination and, consequently, degradation of the DELLA repressor protein, thereby reversing the repression of the GA response (Sasaki et al., 2003; Gomi et al., 2004; Tsuji et al., 2006; Ueguchi-Tanaka et al., 2006; Feng et al., 2008). Most of our knowledge about BR signaling was obtained from studies in Arabidopsis thaliana. A nearly complete primary signaling pathway has been established involving multiple components, including BRASSINOSTEROID-INSENSITIVE1 (BRI1), BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1), BRI1 KINASE INHIBITOR1, CONSTITUTIVE DIFFERENTIAL GROWTH1, BRASSINOSTEROID-SIGNALING KINASE1, bri1-SUPPRESSOR1, PROTEIN PHOSPHATASE 2A, BRASSINOSTEROID-INSENSITIVE2 (BIN2), bri1-EMS-SUPPRESSOR1 (BES1), and BRASSINAZOLE-RESISTANT1 (BZR1) (Li et al., 2001; Wang et al., 2001; He et al., 2002; Li and Nam, 2002; Nam and Li, 2002; Yin et al., 2002; Mora-García et al., 2004; Wang and Chory, 2006; Tang et al., 2008, 2011; Kim et al., 2011; Tong and Chu, 2012; Wang et al., 2012). In rice, several counterparts of the Arabidopsis BR primary signaling components have been identified, including Os BRI1, Os BAK1, Os BIN2 (named GSK2 for GSK3/SHAGGY-LIKE KINASE2 in rice), and Os BZR1, suggesting that rice has a conserved primary BR signaling pathway like Arabidopsis (Yamamuro et al., 2000; Bai et al., 2007; Li et al., 2009; Tong and Chu, 2012; Tong et al., 2012). As a feedback mechanism, activation of both GA and BR signaling inhibits their respective biosynthesis to fine-tune the hormone response in vivo, with BZR1 and BES1 functioning in this process by inhibiting BR biosynthesis (Hedden and Phillips, 2000; He et al., 2005; Sun et al., 2010; Yu et al., 2011).
It is clear that GA interacts with other phytohormones, including BR, to regulate plant growth and development (Weiss and Ori, 2007). While both GA and BR are growth-promoting hormones, an early study found that BR and GA act antagonistically to regulate the expression of a GA-responsive gene, GASA1 (for GA-stimulated transcript in Arabidopsis), as well as a GA biosynthetic gene, GA5 (Bouquin et al., 2001). BR induces the expression of GA5, which encodes a GA 20-oxidase (GA20ox) involved in GA biosynthesis. In another study, BR was also found to induce several GA biosynthetic genes, including GA20ox-1, GA20ox-2, and GA20ox-5, in Arabidopsis (Lilley et al., 2013). These results suggest that BR might act by upregulating GA levels to promote plant growth. However, metabolic studies using pea (Pisum sativum) mutants suggest that BR negatively regulates GA precursors, but the regulation does not lead to correlated changes in bioactive GA (Jager et al., 2005). Several rice genes have been shown to participate in both BR and GA responses, including SPINDLY, GSR1 (for GA-stimulated transcript in rice), and DWARF1 (D1) (Ueguchi-Tanaka et al., 2000; Shimada et al., 2006; Wang et al., 2006, 2009; Oki et al., 2009a). In rice root, BR application appears to repress the levels of active GA by inhibiting the expression of GA20ox-3, a GA biosynthetic gene, and by simultaneously promoting the expression of GA2ox-3, a GA inactivation gene (De Vleesschauwer et al., 2012). These results suggest that BR and GA have a complex crosstalk depending on species or tissue.
Recently, important progress has been made in understanding the crosstalk between GA and BR in Arabidopsis (Bai et al., 2012; Gallego-Bartolomé et al., 2012; Li et al., 2012). A BR-deficient mutant and a BR-insensitive mutant were found to be insensitive to GA, suggesting that both BR biosynthesis and signaling are required for a normal response to GA (Bai et al., 2012; Gallego-Bartolomé et al., 2012). GA-deficient or GA-insensitive mutants are sensitive to BR, whereas a mutant lacking DELLA proteins has greatly enhanced BR sensitivity (Bai et al., 2012). DELLAs can interact directly with BZR1, the downstream transcription factor of BR signaling, leading to inhibition of its DNA binding activity (Bai et al., 2012; Gallego-Bartolomé et al., 2012; Li et al., 2012). As BR-induced BZR1 dephosphorylation is essential for its nuclear accumulation and transcriptional activity, the BZR1-mediated GA response through DELLA derepression ought to require BR biosynthesis and signaling. Thus, both GA and BR can inactivate their respective repressors, DELLAs and BIN2, thereby activating BZR1 to regulate a number of downstream target genes involved in cell elongation. Interestingly, a more recent study showed that the relationship between GA and BR depends on the developmental stages of Arabidopsis photomorphogenesis (Lilley et al., 2013). This study revealed that, at certain stages, BR application induces the accumulation of DELLAs in an opposite way to GA to regulate Arabidopsis growth.
A certain hormone could have opposite effects on plant growth and development, depending on the concentration and tissue. This effect is well documented with BR. In Arabidopsis, low concentrations of BR promote the growth of both the root and hypocotyl, whereas high concentrations of BR inhibit root growth but still promote hypocotyl growth (Müssig et al., 2003). Intriguingly, BR has been found to have opposite effects on the formation of stomata in cotyledons and hypocotyls (Gudesblat et al., 2012; Kim et al., 2012; Serna, 2013). Similarly, in rice, BR significantly promotes coleoptile growth, but a relatively higher concentration of BR would inhibit both root and seedling growth (Tong et al., 2009). Consistent with this phenomenon, mutants or transgenic plants with enhanced BR levels or BR signaling usually have reduced plant height in rice, to a greater or lesser extent, including the D11 activation mutant (Wan et al., 2009), BRASSINOSTEROID UPREGULATED1 (BU1) overexpressor (Tanaka et al., 2009), BAK1 overexpressor (Wang et al., 2007), leaf and tiller angle increased controller (lic) mutant and LIC antisense lines (Wang et al., 2008; Zhang et al., 2012), Os MADS22/47/55 knockdown plants (Lee et al., 2008), DWARF AND LOW-TILLERING (DLT) overexpressor, and GSK2 knockdown lines (Tong et al., 2012).
Correspondingly, plants may use various signaling pathways in response to different hormone concentrations. GA sensitivity tests revealed that the rice dwarf mutant d1 is only less sensitive to low concentrations of GA, but not to high concentrations of GA, compared with the wild type, indicating the existence of a specific pathway in response to high GA levels (Ueguchi-Tanaka et al., 2000). A study of GA metabolic genes in tobacco (Nicotiana tabacum) revealed that the expression levels of these genes have various sensitivities to different GA concentrations (Gallego-Giraldo et al., 2008). In the BR pathway, BZR1 and LIC, both substrates of GSK3-like kinase, have been suggested to act antagonistically in response to low BR and high BR, respectively (Zhang et al., 2012).
In this study, we endeavored to uncover the relationship between BR and GA responses and to decipher how these two growth-promoting hormones affect each other by using a number of BR- and GA-related mutants. We also tried to determine why a relatively high BR level tends to inhibit plant growth in certain tissues. Our results revealed that BR and GA exhibit crosstalk with each other using complex mechanisms, depending on tissue and hormone level. Under low BR levels, BR induces GA biosynthesis and inhibits GA inactivation, leading to increased GA levels and cell elongation, while under high BR levels, GA inactivation is predominantly induced by BR, contributing to the inhibition of cell elongation in certain tissues. On the other hand, under low GA levels, GA represses BR signaling as well as BR biosynthesis through a feedback mechanism, while under high GA levels, GA can somehow activate the primary BR signaling pathway to facilitate cell elongation.
RESULTS
Effects of Exogenous BR on Rice Growth
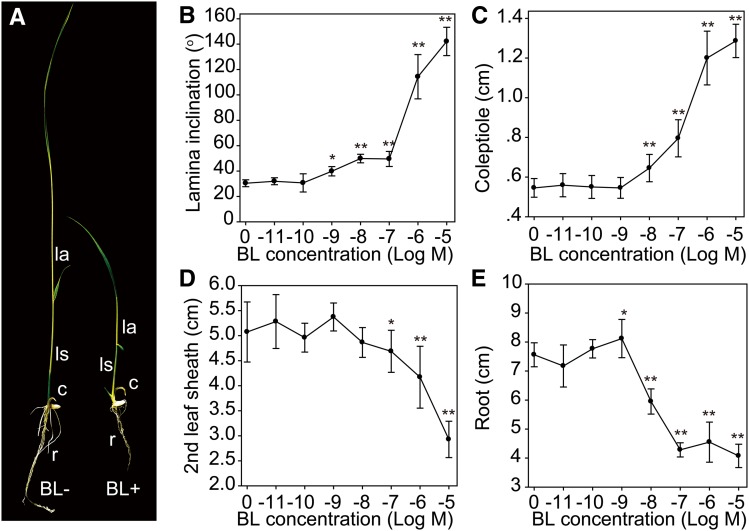
While BR-deficient mutants usually display decreased leaf length, erect leaves, reduced plant height, and shortened roots, exogenous BR application could have inhibitory effects on rice growth and development. We evaluated the effects of exogenous BR on rice seedling growth in terms of plant height, lamina inclination, coleoptile growth, and root elongation (Figure 1), using increasing concentrations of brassinolide (BL), the most active BR. We found that BL greatly promoted coleoptile growth as well as lamina inclination when the concentration reached 10−8 M or greater (Figures 1A to 1C). At 10−5 M BL, the coleoptile elongated to ∼3-fold the length of plants not subjected to exogenous BL application.
Figure 1.
Effects of BR on Rice Seedling Growth.
(A) One-week-old rice seedlings grown with (BL+) or without (BL−) 10−6 M BL. c, coleoptile; la, leaf angle; ls, leaf sheath; r, root.
(B) to (E) Statistical data of BL effects on leaf angle (B) and coleoptile (C), leaf sheath (D), and root (E) growth. Error bars indicate sd (n = 15). Asterisks indicate P < 0.05 (*) and P < 0.01 (**) in Student’s t test analysis.
[See online article for color version of this figure.]
In leaves and roots, a low concentration of BL (10−9 M) only slightly enhanced leaf sheath and root length, and the increase was insignificant in leaf sheaths (Figures 1D and 1E). Considering that exogenous BL normally rescues the dwarf phenotype of BR-deficient mutants (Hong et al., 2002, 2003; Mori et al., 2002; Tanabe et al., 2005), this result suggests that endogenous BR levels in the wild type might be close to saturated for promoting tissue growth in rice leaves. By contrast, a higher level of the hormone inhibited the growth of both leaf sheaths and roots, resulting in decreased seedling height and root length (Figure 1A). In our analysis, the highest BL application (i.e., 10−5 M) inhibited the lengths of both the second leaf sheath and root to about half of those without the hormone treatment. Microscopy analysis showed that the reduced length of the leaf sheath is attributed to the shortened longitudinal cell length (Supplemental Figure 1).
GA and BR have overlapping roles in promoting cell elongation of various tissues. Compared with BR, GA has a relatively minor role in promoting coleoptile elongation. A concentration of 10−5 M GA3 increased coleoptile length by ∼50% (Supplemental Figure 2A). Exogenous GA (from 10−8 to 10−4 M) greatly promoted leaf sheath elongation, resulting in sheaths that were up to 4-fold longer than those not treated with GA (Ueguchi-Tanaka et al., 2000). GA had little effect on root elongation, with a 20% increase being the greatest effect observed in our analysis (Supplemental Figure 2B).
BR Induces the Expression of GA Biosynthetic Genes
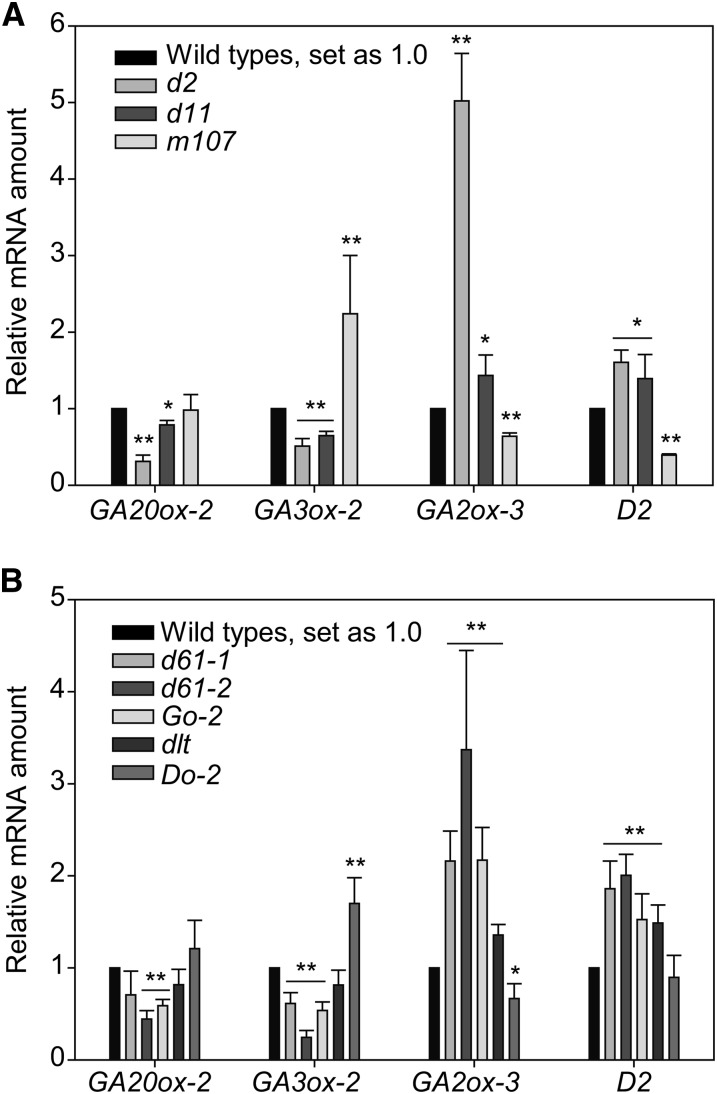
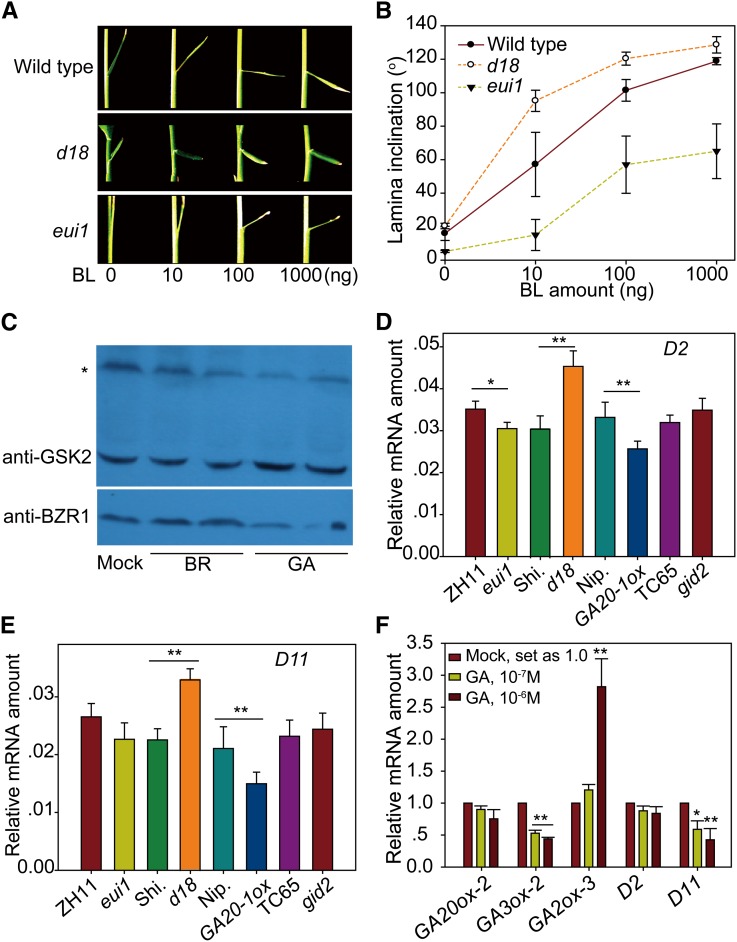
To study the relationship between BR and GA, we collected, identified, and developed various BR- and GA-related plants (Supplemental Table 1). To explore whether BR affects GA biosynthesis, we analyzed the expression levels of several GA metabolic genes in a number of representative BR mutants, including BR-deficient plants (d2 and d11), BR-accumulated plants (m107), decreased BR-signaling plants (d61-1, d61-2, Go-2, and dlt), and enhanced BR-signaling plants (Do-2). Previous studies showed that five GA metabolic genes, GA20ox-1, GA20ox-2, GA3ox-2, GA2ox-1, and GA2ox-3, are the main targets for different biological pathways in rice (Kaneko et al., 2003; Dai et al., 2007). Three of these, GA20ox-2, GA3ox-2, and GA2ox-3, are mainly expressed and functional in seedlings, while GA20ox-1 and GA2ox-1 tend to be expressed and functional in reproductive tissues (Kaneko et al., 2003; Sakamoto et al., 2004). As young rice seedlings were used in this study, we focused on the expression of GA20ox-2, GA3ox-2, and GA2ox-3 in our analysis. Strikingly, quantitative real-time RT-PCR (qRT-PCR) showed that, with rare exceptions, both GA20ox-2 and GA3ox-2 have decreased expression in all of the BR-deficient and decreased BR-signaling plants but increased expression in the BR-accumulated and enhanced BR-signaling plants, compared with their respective wild types (Figure 2). By contrast, GA2ox-3, the GA inactivation gene, has opposite expression patterns compared with GA biosynthetic genes. Thus, we concluded that BR promotes GA biosynthesis and inhibits GA inactivation and that BR signaling components are involved in both processes.
Figure 2.
Expression of GA Metabolic Genes in Various BR-Related Mutants.
(A) Expression of the indicated genes in plants with altered BR biosynthesis.
(B) Expression of the indicated genes in plants with altered BR signaling.
Error bars indicate sd (n = 3). Asterisks indicate P < 0.05 (*) and P < 0.01 (**) in Student’s t test analysis.
BR Promotes Cell Elongation through Inducing GA Accumulation
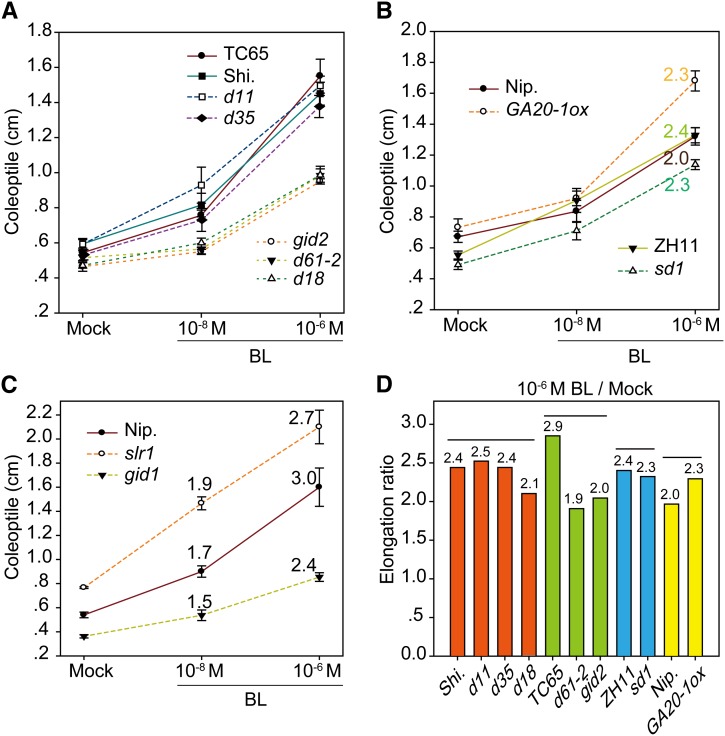
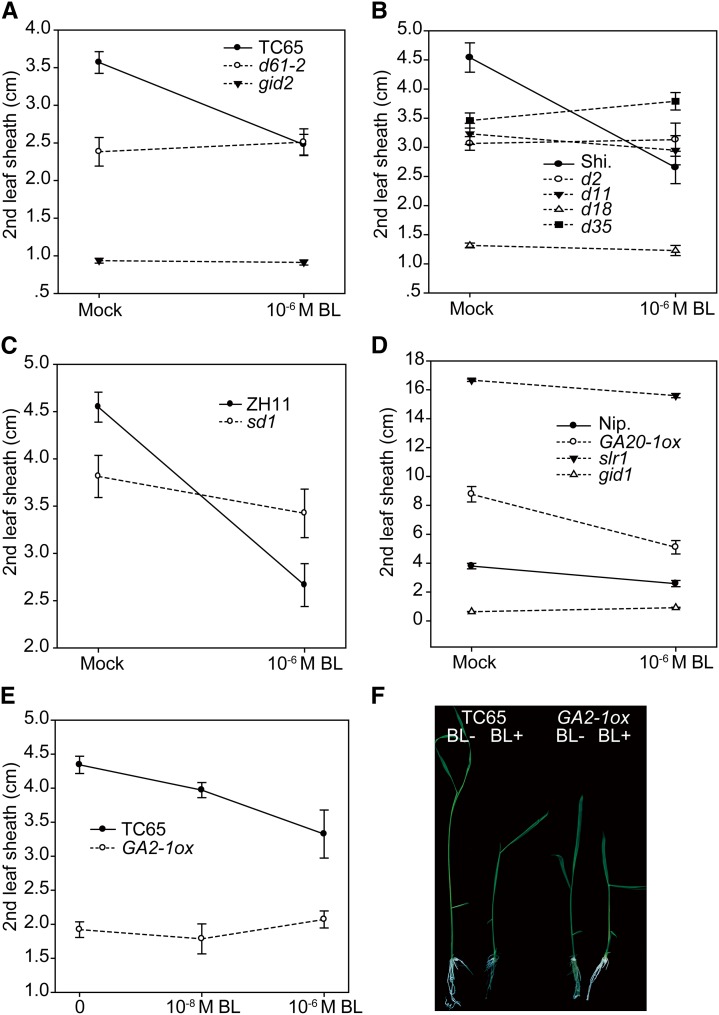
The above results raised the interesting possibility that BR promotes cell elongation by upregulating GA level. To test this hypothesis, we first compared the coleoptile elongation extent of GA-related mutants with their respective wild types in response to exogenous BR. These plants include GA-deficient mutants (sd1, d18, and d35), decreased GA-signaling mutants (gid1 and gid2), an enhanced GA-signaling mutant (slr1), and a GA-accumulated plant (GA20-1ox) (Supplemental Table 1). We found that, while sd1 and d35 had similar elongation curves to the wild types in response to different concentrations of BR, d18, gid1, and gid2 had markedly decreased BR sensitivities (Figures 3A to 3C). For comparison, we calculated the elongation ratio of the coleoptile length under 10−6 M BL compared with those without BL (Figures 3B to 3D). Specifically, d18 had an elongation ratio of 2.1, which is less than that of the wild type (2.4). The ratios for gid1 and gid2 versus their respective wild types were 2.4:2.7 and 2.0:2.9. However, the ratio for sd1 was 2.3, only slightly lower than the 2.4 of its wild type; and for d35, the ratio was 2.4, the same as that of the wild type. Notably, slr1 appeared to have a decreased elongation ratio compared with its wild type (2.7:3.0) under 10−6 M BL. However, under lower concentrations (e.g., 10−8 M), the ratio was 1.9:1.7 (Figure 3C), suggesting the increased BL sensitivity of the mutant. Therefore, slr1 could have reached the maximum possible elongation of coleoptile under 10−6 M BL with 2.1 cm in length, which is the longest in the tests. In addition, GA20-1ox plants had markedly increased BR sensitivities, with the elongation ratio of 2.3 versus 2.0 of the wild type. As both sd1 and d35 have unaltered BR sensitivities, we speculated that D18/GA3ox-2 is one of the main targets for BR regulating cell elongation. Overexpression of GA20ox-1 resulted in the accumulation of its catalyzed product, GA20, the precursor of GA3ox-2, to produce bioactive GA1, leading to the increased sensitivity of the plant to BR-induced GA3ox-2 expression.
Figure 3.
Coleoptile Elongation of GA-Related Mutants in Response to BR.
(A) to (C) Plants were grouped by different backgrounds or independent experiments. Dashed lines indicate mutants. Numbers indicate elongation ratios in (B) and (C). Error bars indicate sd (n = 8 for slr1, gid1, and gid2; n = 15 for others).
(D) Elongation ratios at 10−6 M BL as analyzed in (A) and (B). Top lines and colors indicate groups classified with different backgrounds.
These results suggest that BR promotes coleoptile elongation at least partially by inducing GA accumulation. To confirm that the BR signaling pathway is involved in this process, we tested the coleoptile sensitivity of several BR mutants and transgenic plants in response to BR, including decreased BR-signaling plants (d61-2, Go-2, and dlt), enhanced BR-signaling plants (Gi-2 and Do-1), and a BR-deficient plant (d11) (Supplemental Table 1). Also, we calculated the coleoptile length ratios between BR-treated and nontreated mutants and compared these with their respective wild-type ratios (Figures 3A and 3D). The ratio for d61-2 versus the wild type was 1.7:2.9, for Go-2 was 1.7:2.4, for Gi-2 was 3.1:2.4, for dlt was 2.3:2.5, for Do-1 was 3.1:2.5, and for d11 was 2.3:2.4. These results, combined with previous results obtained from GA mutants, show that D18/GA3ox-2 and GA signaling components act like these BR signaling components, all of which are important for BR-mediated coleoptile elongation.
Altered GA Levels in BR-Related Mutants
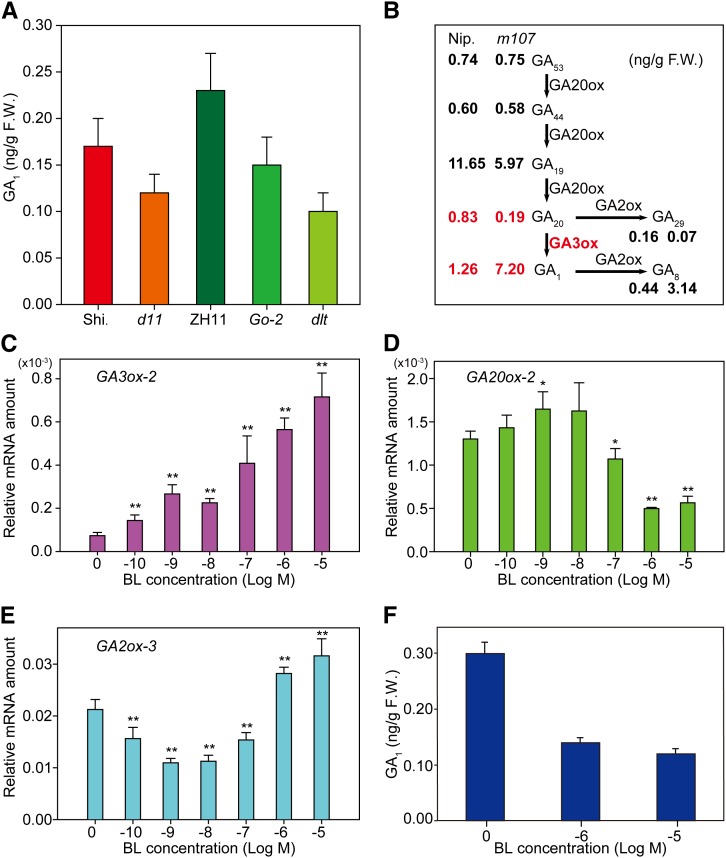
To provide further evidence for BR-induced GA accumulation, we directly analyzed the GA amounts in several representative BR mutants compared with their respective wild types. Consistent with the gene expression data, we found that both a BR-deficient mutant (d11) and decreased BR-signaling plants (Go-2 and dlt) have markedly decreased GA1 levels compared with their respective wild types (Figure 4A). We also quantified the GA1 levels in three Do (for DLT overexpression) lines and found that all of these lines have slightly increased GA1 levels compared with the wild type (Supplemental Table 2). Although the increase is small, the GA1 levels are consistent with the plant heights of the three Do lines (Supplemental Figure 3A).
Figure 4.
Quantification of GA Amount in BR-Related Mutants and BR Effects on GA Metabolic Genes.
(A) Bioactive GA1 levels in the wild type and BR-related mutants. One-month-old seedling shoots were used for the measurements. Error bars indicate sd (n = 3). F.W., fresh weight.
(B) Levels of different GA forms in Nip. and m107. Two-week-old seedling shoots were used for the measurements. Average values of three replicates are shown. See Supplemental Table 3 for sd. GA20ox, GA3ox, and GA2ox and the reaction processes they catalyzed are indicated. Steps with the most prominent changes are marked in red.
(C) to (E) Expression changes of GA and BR metabolic genes in response to BL in rice shoot, including GA3ox-2 (C), GA20ox-2 (D), and GA2ox-3 (E). Error bars indicate sd (n = 3). Asterisks indicate P < 0.05 (*) and P < 0.01 (**) in Student’s t test analysis.
(F) GA1 levels in mock- and high-BL-treated plants. Error bars indicate sd (n = 3).
We further quantified the amounts of different forms of GA in BR accumulation line m107 and its wild type (Figure 4B; Supplemental Table 3). Because of the activated expression of the BR biosynthetic gene D11, m107 has greatly increased BR levels, as shown by direct BR (castasterone [CS], one of the active BRs) measurement (Supplemental Table 4). In contrast with the enhanced BR-signaling plants with inconspicuous height changes at the seedling stage, m107 had greatly increased seedling height (Supplemental Figures 3A and 3B; represented by leaf sheath length) and is thus suitable material for evaluating the effect of endogenous BR application on GA level. Plants were grown in hydroponic culture medium for 2 weeks, and shoots were used for GA measurement. Whereas GA53 and GA44 had comparable levels in both plants, GA1 was greatly elevated in m107 compared with its wild type, with an increase of ∼5.7-fold; by contrast, GA19 and GA20, the precursors of GA1, were largely decreased ∼2- and 4-fold, respectively, with GA20 (the direct substrate of GA3ox-2) being reduced the most (Figure 4B). These results demonstrate that D18/GA3ox-2, which catalyzes GA20 oxidization to produce GA1, is the main target for the BR-mediated upregulation of GA biosynthesis. In addition, the increase of GA8 and the decrease of GA29 are consistent with the altered GA1 and GA19, respectively. We also used 10−6 M BL to treat wild-type plants for 2 d and measured the GA levels at the same time. It should be mentioned that this kind of short-term treatment does not inhibit plant growth (Supplemental Figures 3C and 3D). The GA1 level increased by ∼2.0-fold, while GA20 decreased (Supplemental Table 3). Interestingly, the increase of GA1 was much less than that in m107, suggesting that exogenous application of high levels of BL may have an additional negative role in inducing GA accumulation.
Expression of GA and BR Metabolic Genes in Response to Exogenous BR
Next, we analyzed the expression levels of GA and BR metabolic genes, including GA20ox-2, GA3ox-2, GA2ox-3, D2, and D11, in response to various exogenous BL concentrations, from 10−10 to 10−5 M. Young rice seedlings were treated for 2 d before sampling for the analysis. GA3ox-2 expression gradually increased with the increased BL concentrations, further strengthening our previous conclusion that BL increases GA levels (Figure 4C). However, very strikingly, all the other genes have a differential but coordinated response to the increasing BL gradient. From 10−10 to 10−9 M BL, expression levels of GA20ox-2, D2, and D11 were gradually increased, but GA2ox-3 decreased, while from 10−8 to 10−5 M, their expression tendencies were reversed (Figures 4D and 4E; Supplemental Figure 4). Notably, the expression of all of these genes had a turning point at the concentration of 10−9 M, in concert with the increase in leaf sheath and root elongation observed at 10−9 M BL (Figures 1D and 1E). Interestingly, this kind of short-term treatment tended to have a more obvious promotional effect on plant growth (Supplemental Figures 3C and 3D). These results suggest that high BR levels inhibit rice growth by repressing both GA and BR biosynthesis.
To test if high BL levels indeed suppress GA levels, we treated wild-type plants with 10−6 and 10−5 M BL for 1 week and then measured the GA levels in the plant shoots. Consistent with the greatly decreased plant height, both the treatments led to marked decreases in GA1 levels, with 10−5 M treatment having a stronger effect (Figure 4F). Interestingly, all of the other detected forms of GAs also exhibited decreased levels (Supplemental Table 5).
Decreased Sensitivity of GA Mutants to BR-Mediated Inhibition of Leaf Growth
If exogenous BR inhibits leaf sheath and root elongation through reducing GA and BR levels, both GA and BR mutants should have altered sensitivity to BL in terms of its inhibitory effect on the growth of both leaf sheath and root. We first analyzed the BR responses of various GA mutants in leaf sheath under 10−6 M BL (Figures 5A to 5D). As expected, we found that while d61-2, the BR receptor mutant, is totally insensitive to BL-mediated inhibition of leaf sheath elongation (Figure 5A), d2 and d11, two BR biosynthetic mutants, also showed insensitivity (Figure 5B). In addition, most of the GA mutants analyzed are nearly completely insensitive to BL-mediated inhibition of leaf sheath elongation, including d18, d35, sd1, gid1, gid2, and slr1.
Figure 5.
Effects of BR on Leaf Sheath Elongation of GA- and BR-Related Mutants.
Plants were grouped by different backgrounds or independent experiments. Dashed lines indicate mutants. Error bars indicate sd (n = 8 for slr1, gid1, and gid2; n = 15 for others). Representative seedling morphology of TC65 and GA2-1ox grown with or without 10−6 M BL is shown in (F).
[See online article for color version of this figure.]
If GA20ox-2 is one of the transcriptional targets of BR-mediated inhibition of cell elongation, overexpressing its homolog GA20ox-1, which functions redundantly with GA20ox-2, should make plants resistant to the inhibitory effect of high levels of BR. However, we found that GA20-1ox plants have increased sensitivity (Figure 5D). Therefore, we suspected that GA2ox-3 is the major target. GA20-1ox has enhanced GA1 levels, resulting in increased sensitivity to BR-induced GA2ox-3 expression. If this is the case, overexpressing its functional redundant homolog, GA2ox-1, should make plants resistant to the growth inhibition caused by high levels of BR. We then generated GA2ox-1 overexpression plants (designated as GA2-1ox), which are dwarfed. Consistent with our speculation, a sensitivity test showed that the leaf sheath of the transgenic plant is totally insensitive to 10−6 M BL application (Figures 5E and 5F). Thus, we concluded that GA2ox-3 is the major transcriptional target for BR-mediated inhibition of cell elongation.
Decreased Sensitivity of GA Mutants to Exogenous BR Inhibition in Root
We observed similar results in the root, but not to the same extent as in the leaf sheath (Supplemental Figure 5). For instance, d61, d18, d35, gid1, gid2, and slr1 also had decreased sensitivity to BL-mediated inhibition of root growth, but the extent was less than that in leaf sheaths. However, unlike the insensitivity of d2 and d11 mutants to BL-mediated inhibition on leaf sheath elongation, both of these mutants had a normal response in root sensitivity tests, suggesting that BL-mediated inhibition of roots does not rely on negative feedback regulation of these BR biosynthetic genes. In addition, sd1 and GA20-1ox had no obvious changes in sensitivity to high levels of BR. Accordingly, we found that sd1 had longer roots while GA20-1ox had shorter roots compared with their respective wild types. Considering the unchanged root length in d2, d11, and even brd1, the most severe BR-deficient mutant (Supplemental Table 1) (Mori et al., 2002), rice root systems may have more complex mechanisms than aerial tissues that regulate growth (González-García et al., 2011; Fàbregas et al., 2013).
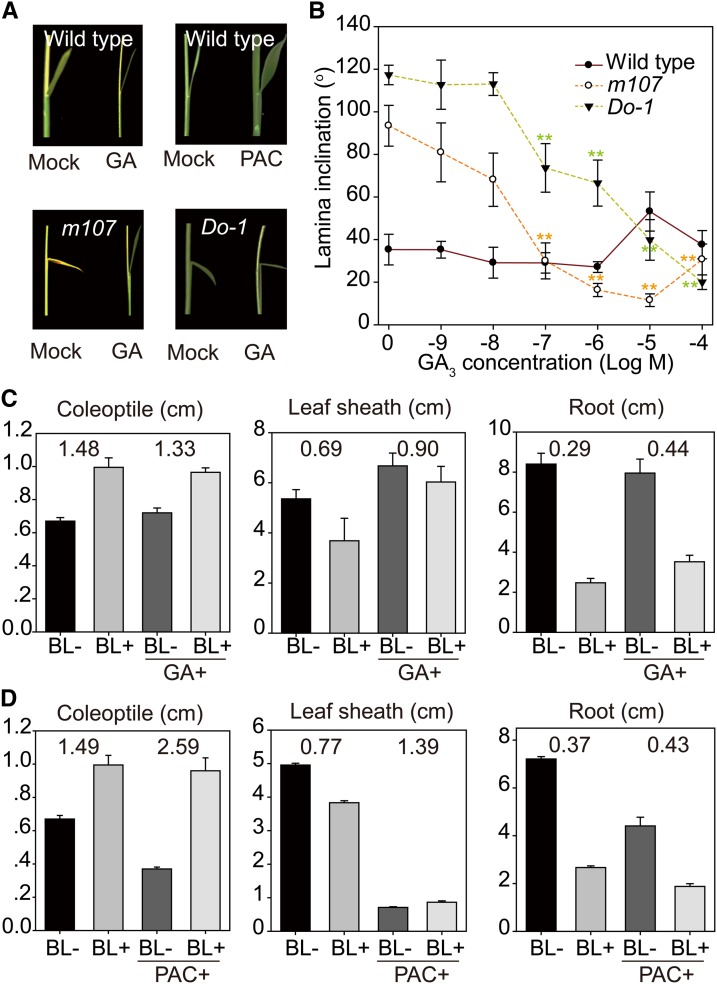
Exogenous GA Inhibits the BR Response
Unexpectedly, we found that GA appears to slightly inhibit wild-type leaf angles, whereas paclobutrazol (PAC), a GA biosynthesis inhibitor, slightly increased the leaf angles (Figures 6A and 6B). The GA-mediated inhibition tended to be reverted when GA concentrations exceeded 10−6 M in the wild type. However, the effects were not significant. Thus, we further used m107 as material to carry out the analysis, as it has greatly enlarged leaf angles due to elevated rates of BR biosynthesis (Wan et al., 2009). Consistently, we found that GA strikingly inhibited lamina bending, even at the very low concentration of ∼10−9 M (Figure 6B). A concentration of 10−6 M GA reduced the second leaf angle from 93° to ∼16° (Figures 6A and 6B), demonstrating that GA is a negative regulator of lamina inclination. We noted that the inhibition was also alleviated when GA concentration reached 10−4 M, higher than the 10−5 M in wild-type treatment. To test whether the inhibition is mediated through the BR-signaling pathway, we performed a similar analysis using Do-1, a weak DLT overexpression line, which also has enlarged leaf angles because of enhanced BR sensitivity (Tong et al., 2012). We found that GA also inhibited Do-1 lamina bending, but only when the concentration reached 10−7 M (Figure 6B). In addition, the inhibition was much less than in m107 at 10−6 and 10−5 M GA (Figures 6A and 6B). Moreover, alleviation of GA-mediated inhibition of lamina inclination was not observed even at 10−4 M GA in Do-1. Thus, compared with m107 with enhanced BR biosynthesis, the enhancement of BR signaling leads to resistance to GA inhibition, demonstrating that GA-mediated inhibition on lamina inclination is mediated through the BR-signaling pathway.
Figure 6.
(A) Lamina inclination of the second leaf in wild-type, m107, and Do-1 plants subjected to GA or PAC treatment.
(B) Responses of lamina inclination in the wild type, m107, and Do-1 to different GA concentrations. Error bars indicate sd (n = 10). Asterisks indicate P < 0.01 (**) in Student’s t test analysis.
(C) and (D) Effects of GA (C) and GA inhibitor (D) on BR sensitivity in various tissues. Length ratios of BL-treated and nontreated tissues were calculated and are shown.
Concentrations of 10−6 M GA, 10−6 M BR, and 10−5 M PAC were used for the treatments in (A), (C), and (D). Error bars indicate sd (n = 15).
[See online article for color version of this figure.]
As lamina bending is generally believed to be a specific BR response in rice, we considered that exogenous GA might also repress other BR responses. To this end, we further analyzed the effects of GA on BR responses in the coleoptile, leaf sheath, and root in the wild type (Figure 6C). Indeed, application of 10−6 M GA decreased the BR effects in all three tissues, either promoting or inhibiting growth (as determined by comparing the length ratio between BL-treated and nontreated plants). We also tested the effects of 10−5 M PAC on BR responses in these tissues. However, PAC greatly increased the BR response in promoting coleoptile growth but repressed the BR sensitivity in inhibiting leaf sheath and root growth (Figure 6D), suggesting that BR functions in different tissues via different mechanisms involving GA biosynthesis regulation. These results are consistent with the decreased sensitivity of GA-deficient mutants to BL-mediated inhibition of leaf sheath elongation (Figure 5). A reduction of any GA precursor by either gene mutation or PAC treatment would lead to decreased sensitivity of plants to BL-repressed GA20ox-2 or induced GA2ox-3 expression. In addition, exogenous GA application also decreased the plant’s sensitivity to BR-mediated inhibition (Figure 6C), because exogenous GA is supplied in great excess while the in vivo GA-modulating process has little effect on the GA level in planta.
GA Inhibits BR Signaling as Well as BR Biosynthesis
Having established that exogenous GA inhibits the BR response, we further investigated whether endogenous GA has a similar effect on the BR response. Thus, we used several GA metabolic mutants to analyze their BR response using a lamina-bending experiment that was thought to detect a BR-specific response (Figures 7A and 7B). In the test, we found that d18, the GA-deficient mutant, had a markedly increased leaf angle as well as enhanced BR sensitivity; by contrast, eui1, a GA-accumulated mutant with defects in a GA inactivation gene (Supplemental Table 1) (Luo et al., 2006; Nomura et al., 2006; Zhu et al., 2006), had the opposite phenotype and decreased BR sensitivity. These results demonstrate that GA indeed inhibits the BR response.
Figure 7.
GA Extensively Inhibits BR Signaling as Well as BR Biosynthesis.
(A) BR response of the wild type, d18, and eui1 in lamina inclination tests.
(B) Statistical analysis of the results shown in (A). Error bars indicate sd (n = 12).
(C) Detection of GSK2 and BZR1 by immunoblot analysis under 10−6 M BR and 10−5 M GA treatment. The asterisk marks an unspecific band that was used as a reference.
(D) and (E) Expression of D2 (D) and D11 (E) in GA-related mutant plants compared with their respective wild types. Error bars indicate sd (n = 3). Asterisks indicate P < 0.05 (*) and P < 0.01 (**) in Student’s t test analysis.
(F) Effects of GA treatment on the expression of GA and BR metabolic genes. Error bars indicate sd (n = 3). Asterisks indicate P < 0.05 (*) and P < 0.01 (**) in Student’s t test analysis.
To verify this result, we quantified the protein level of GSK2, the central negative regulator of BR signaling, and BZR1, the downstream positive regulator of BR signaling, under 10−5 M GA treatment (Figure 7C). In contrast with the effect of BL, GSK2 was induced by GA, whereas BZR1 had decreased levels under GA treatment. A further treatment using a gradient of GA concentrations led to a similar result (Supplemental Figure 6), further proving that GA inhibits the BR response.
To investigate whether GA also inhibits BR biosynthesis, we performed qRT-PCR to determine the expression of two BR biosynthetic genes, D2 and D11, in various GA-related mutants, including a GA-deficient mutant (d18), GA-accumulated plants (eui1 and GA20-1ox), and a decreased GA-signaling mutant (gid2) (Supplemental Table 1), and to compare these values with those in their respective wild types. Both D2 and D11 had consistently increased expression in GA-deficient and decreased GA-signaling mutants (d18 and gid2) but decreased expression in GA-accumulated plants (eui1 and GA20-1ox) (Figures 7D and 7E). We also analyzed the effect of exogenous GA on D2 and D11 expression and found that, similar to the GA biosynthetic genes, including GA20ox-2 and GA3ox-2, both of these genes were repressed by GA application (Figure 7F). Taken together, these results suggest that GA inhibits both BR biosynthesis and the BR response.
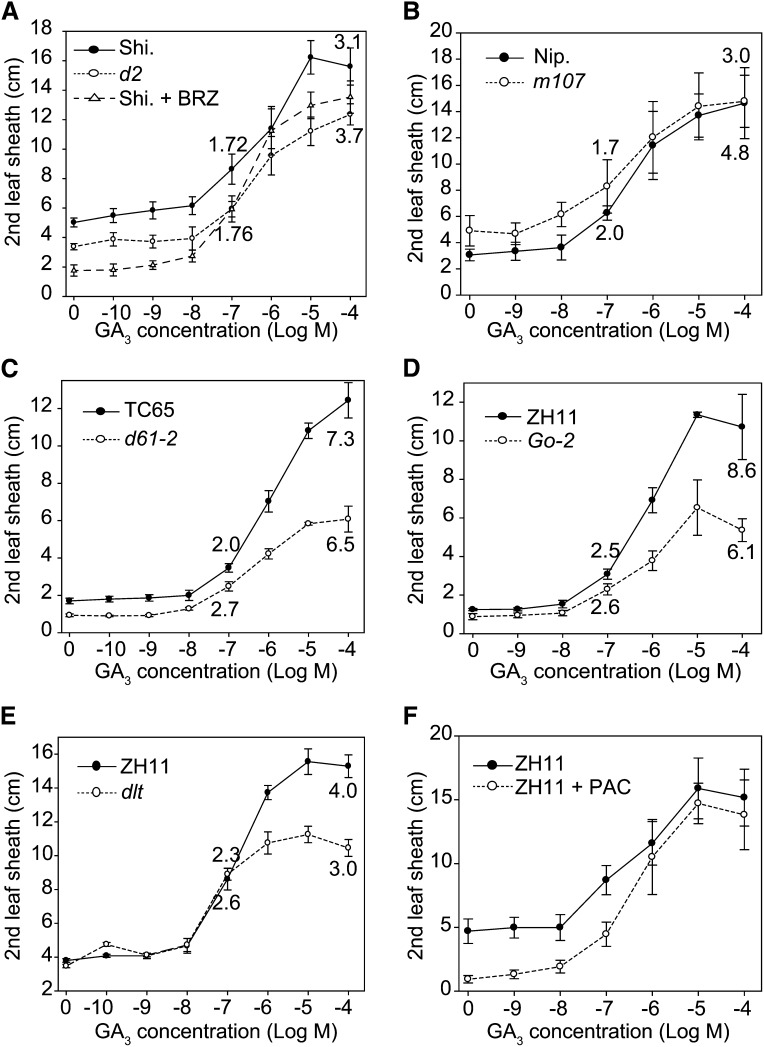
Altered GA Sensitivity of BR-Related Mutants
Next, we sought to determine the effects of BR on the GA response. As exogenous BR usually inhibits leaf sheath growth, we directly used a number of BR-related mutants to carry out the second leaf sheath elongation analysis in response to GA (Figure 8). These plants include a BR-deficient plant (d2), a BR-accumulated plant (m107), and decreased BR-signaling plants (d61-2, Go-2, and dlt) (Supplemental Table 1). Compared with their respective wild types, d2, as well as the brassinazole (BRZ)-treated plants (the BR biosynthesis inhibitor), have increased GA sensitivity (Figure 8A), while m107 has decreased GA sensitivity (Figure 8B). Interestingly, those decreased BR-signaling plants, including d61-2, Go-2, and dlt, have slightly enhanced sensitivity under low concentrations of GA (10−9 to 10−7 M) but have markedly decreased sensitivity under high concentrations of GA (10−6 to 10−4 M) (Figures 8C to 8E). For example, under 10−7 M GA, the second leaf sheath of d61-2 elongated 2.7-fold compared with that not treated with GA, which is more than the 2.0-fold of the wild type. However, under 10−4 M, d61-2 elongated 6.5-fold, less than the 7.3-fold of the wild type. These results suggest that BR signaling components (BRI1-GSK2-DLT) are essential for the response to high GA concentrations but are not needed for the response to low GA concentrations close to physiological GA levels. The increased sensitivities in plants with deficient BR or decreased BR signaling are caused by the decreased GA levels in these plants, as PAC-treated plants also have increased GA sensitivity (Figure 8F).
Figure 8.
Responses of BR-Related Mutants to Different Concentrations of GA.
Plants are as follows: d2 and BRZ-treated plants (A), m107 (B), d61-2 (C), Go-2 (D), dlt (E), and PAC-treated plants (F). The elongation ratios of the leaf sheath lengths between GA-treated and nontreated leaves were calculated and are shown. Error bars indicate sd (n = 10).
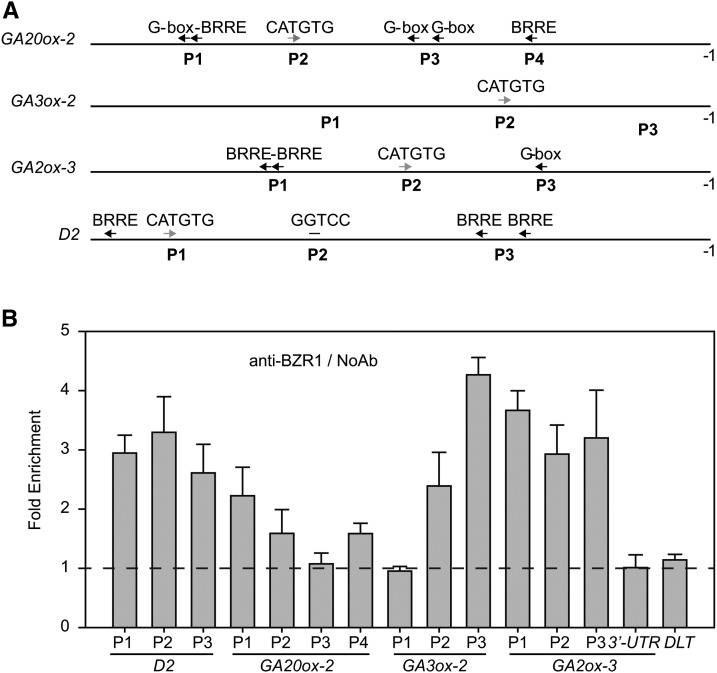
BZR1 Directly Binds to Promoters of GA Biosynthetic Genes to Regulate Their Expression
In Arabidopsis, BZR1/BES1 family proteins were thought to be the primary transcription factors regulating huge numbers of genes involved in BR signal output (Sun et al., 2010; Yu et al., 2011). Rice BZR1 has been suggested to play a conserved role as in Arabidopsis (Bai et al., 2007; Tong et al., 2012). We then considered whether BZR1 could directly bind to GA and BR biosynthetic genes in rice to regulate their expression. According to previous reports in Arabidopsis, BZR1/BES1 binds to BRRE and G-box elements to mostly repress gene expression but binds to CATGTG to primarily stimulate gene expression (Sun et al., 2010; Yu et al., 2011). Interestingly, analysis of the promoters of GA20ox-2, GA3ox-2, GA2ox-3, and D2 revealed that each of these promoters contained one CATGTG element; however, except for the GA3ox-2 promoter, the other promoters also contained several BRREs or G-box elements (Figure 9A). These observations are consistent with the finding that GA3ox-2 was always induced by BL, whereas the other genes are either induced or repressed by BL (Figures 4C to 4E; Supplemental Figure 4), strongly suggesting that rice BZR1 is a direct regulator of these genes. We then conducted a chromatin immunoprecipitation (ChIP) assay to detect BZR1 binding to these elements. Quantitative PCR (qPCR) analysis of the output DNA revealed that most of the regions containing these elements were enriched when rice BZR1 antibody was applied, except for two regions (GA20ox-2 p2 and GA3ox-2 p1) as well as two negative controls (GA2ox-3 3′ untranslated region and DLT coding region), which have no enrichment (Figure 9B). These results support the hypothesis that rice BZR1 binds to the promoters of these genes to regulate their expression.
Figure 9.
Os BZR1 Can Directly Bind to the Promoters of GA20ox-2, GA3ox-2, GA2ox-3, and D2.
(A) Distribution of BZR1 binding cis-elements in the 2-kb region preceding the initial codon (−1) of GA20ox-2, GA3ox-2, GA2ox-3, and D2. Gray and black arrows indicate that BZR1 binding could presumably promote and repress gene expression, respectively, according to a previous report.
(B) Enrichment fold of Os BZR1 binding sites compared with the samples without antibody application (NoAb) by qPCR analysis. The amplified regions are indicated in (A). All values were normalized to an unrelated ACTIN1 intron region, and segments located in the GA2ox-3 3′ untranslated region (UTR) and DLT coding region were used as negative controls.
DISCUSSION
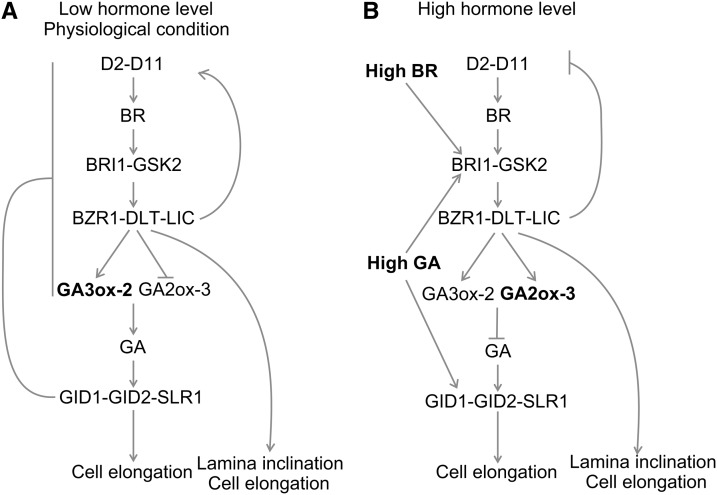
Promotion of cell elongation is the shared function of BR and GA, and dwarfism is the most common phenotype of their deficient mutants in rice. It is possible that BR and GA interact closely to regulate cell elongation. In this study, we explored the BR-GA crosstalk concerning their overlapping roles in promoting cell elongation by genetic, physiological, hormone quantification, and gene expression analyses. Our results demonstrated that BR modulates GA metabolism to regulate cell elongation, and D18/GA3ox-2 could be the major target for endogenous BR to promote GA biosynthesis and to induce cell elongation, while GA2ox-3 could be the target for an exogenous high concentration of BR to inactive GA and to inhibit cell elongation. Physiological GA inhibits both BR signaling and BR biosynthesis, likely in a feedback inhibitory loop. However, high exogenous GA levels activate the primary BR signaling pathway to facilitate cell elongation (Figure 10).
Figure 10.
Proposed Model for BR-GA Crosstalk Based on This Study.
(A) Physiological BR induces GA biosynthesis, inhibits GA inactivation, and stimulates self-biosynthesis to promote cell elongation. As a feedback manner, GA inhibits BR responses and biosynthesis. BR also has other branches for promoting cell elongation or other specific actions.
(B) High BR levels promote GA inactivation and repress self-biosynthesis to inhibit cell elongation. High GA levels facilitate cell elongation via BR signaling.
Arrows indicate activation, and blunt arrows indicate inhibition.
Our results showed that BR-related mutants are tightly associated with GA metabolic gene expression as well as GA levels, strongly suggesting that BR promotes rice cell elongation by upregulating GA biosynthesis. The insensitivity of relative GA mutants to BR further strengthened our conclusion. To our surprise, we showed that GA inhibits lamina inclination, and the inhibition becomes more obvious in plants with accumulated BR or enhanced BR signaling. It is known that defective elongation/division of the abaxial cells at the lamina joint usually leads to erect leaves, a characteristic of BR-deficient mutants in rice (Hong et al., 2004; Duan et al., 2006; Zhao et al., 2010), which is not obvious and rarely reported in GA-deficient mutants. One possibility for the inhibition is likely due to the feedback inhibition of GA on the BR response, as lamina inclination was thought to be a BR-specific response. Despite the decreased cell length, rice BR-deficient mutants also have distorted cell organization, resulting in twisted or frizzled leaves (Hong et al., 2002; Mori et al., 2002). Thus, BR appears to have broader roles than GA in regulating rice growth at the seedling stage. The regulation on GA biosynthesis could be one of the downstream branches of the BR response pathway utilized to regulate cell elongation.
While BR has been widely used as a plant growth modulator in the field for over 30 years (Khripach et al., 2000), its effects on plants are perplexing, and conflicting results have been reported regarding its roles in several processes (Hu et al., 2000; Sasse, 2003; De Vleesschauwer et al., 2012; Wang, 2012; Serna, 2013). In contrast with its growth-promoting effect, exogenous BR application frequently leads to retarded growth; however, the detailed mechanism is unclear. According to our observations, the ability of BR to inhibit or promote plant growth depends on multiple factors, including the treatment time, the hormone concentration used, and endogenous BR levels in a certain tissue. For example, a 2-d short-term treatment using even 10−6 M BL can still upregulate the GA1 level ∼2-fold as well as slightly increase leaf length (Supplemental Table 3 and Supplemental Figures 3C and 3D). In this case, high levels of BR inhibit its own biosynthesis and induce GA inactivation, which antagonizes the increased GA biosynthesis because of elevated GA3ox-2 expression, eventually resulting in a subtle effect on plant growth. A long-term treatment would continuously strengthen the inhibition of both hormones, leading to growth inhibition. As we mainly used young seedlings as material in this study, it will be very interesting to test whether GA2ox-1, the functional homolog of GA2ox-3 at the reproductive stage, is similarly regulated by BR, because reduced culm length has been frequently observed in rice plants that accumulate BR or exhibit enhanced BR signaling, such as m107, Do-2, Gi-2, BU1ox, BAK1ox, and lic (Supplemental Figure 3E). One more intriguing question is whether BR also modulates GA level to affect other processes like flowering and plant fertility.
Our results revealed a previously unknown mechanism underlying the crosstalk between the two hormones, which appears much different from that in Arabidopsis. In Arabidopsis, both the BR-deficient mutant de-etiolated2 and the weak BR-insensitive mutant bri1-119 were found to be nearly completely insensitive to GA in terms of hypocotyl elongation (Bai et al., 2012; Gallego-Bartolomé et al., 2012), whereas in rice, both the BR-deficient plants (d2, d11, and BRZ-treated plants) and the BR-insensitive mutants (d61-1, d61-2, Go-2, and dlt) have normal or even higher sensitivity to physiological GA than the wild type in terms of leaf sheath elongation. Mori et al. (2002) mentioned that brd1, the most severe BR-deficient mutant, has basically no leaf sheath elongation in response to GA but that the leaf blade elongated normally, as in the wild type. Our observation suggested that this is because brd1, as well as another allele termed brd1-1 with similar severity (Hong et al., 2002), originally have no visible leaf sheath development (Supplemental Figures 7A and 7B). These results suggest that a basal BR level is essential for leaf sheath development. When we used brd1-3, a slightly weaker allele than brd1 and brd1-1 (Hong et al., 2002), to carry out a similar analysis, the leaf sheath as well as the leaf blade elongated normally, as in the wild type (Supplemental Figures 7C and 7D), demonstrating that GA function does not require BR biosynthesis. Thus, BR-induced BZR1 activation (including nuclear localization) is essential for the GA response in Arabidopsis but not in rice. Another possibility is that different tissues (hypocotyl and leaf sheath, respectively) were used for evaluation of the GA response, which may lead to different results.
Additionally, in contrast with the observation that GA supplementation enhances BR sensitivity in Arabidopsis (Bai et al., 2012), GA generally inhibits the BR response in rice in terms of lamina inclination, coleoptile elongation, and sheath and root growth. Indeed, we found that GA inhibits BZR1 protein level but enhances GSK2 level, whereas GA induces BZR1 accumulation in Arabidopsis (Li et al., 2012). It should be emphasized that the feedback inhibition of BR response and biosynthesis by GA signaling that act far downstream of BR was thought to be a fine-tuning mechanism for hormone function, thus tending to be relatively weak. Despite these differences, however, our results are consistent with previous reports that BR induces GA biosynthesis in Arabidopsis and GA inhibits BR biosynthesis in Arabidopsis (Li et al., 2012; Lilley et al., 2013). Thus, BR might also promote GA biosynthesis to promote cell elongation in Arabidopsis. Moreover, in rice root, exogenous GA was found to repress BR biosynthesis, whereas BR was found to promote the expression of GA2ox-3, the GA inactivation gene (De Vleesschauwer et al., 2012). The authors speculated that BR regulates GA metabolism to inhibit root growth, which is consistent with our finding. Interestingly, our yeast two-hybrid analysis also showed an interaction between BZR1 and SLR1, the rice DELLA (Supplemental Figure 8), suggesting that BR and GA might exhibit crosstalk at multiple levels. Actually, in our model, the inhibition of BR biosynthetic genes by GA, as well as the involvement of BR-signaling components in response to high GA concentrations, could be partially explained by GA derepression of SLR1 inhibition on BZR1 activity.
Numerous BIN2/GSK2 substrates were identified in recent years, including a number of transcription factors (Vert et al., 2008; Tong et al., 2012; Ye et al., 2012; Zhang et al., 2012, 2014a, 2014b; Cho et al., 2014), suggesting that the negative regulator BIN2/GSK2 acts as the central player in the BR response, similar to most other hormone-signaling pathways (Huq, 2006). Our analysis combined with previous reports implied that these transcription factors, such as BZR1, DLT, and LIC in rice, work together in a complex and dynamic manner to regulate GA homeostasis in response to different BR levels in various tissues (Figure 10). We cannot exclude the possibility that BR has other branched pathways, which are independent of GA biosynthesis and signaling, that promote cell elongation. Several studies have suggested that there is another BR signaling pathway independent of the BRI1 receptor, possibly involving components such as D1, BU1, and TAIHU DWARF1/ERECT LEAF1 (TUD1/ELF1) (Wang et al., 2006; Oki et al., 2009b; Tanaka et al., 2009; Hu et al., 2013; Sakamoto et al., 2013). Interestingly, D1 has also been suggested to be involved only in the high-sensitivity response of GA (i.e., in response to low concentrations of GA) (Ueguchi-Tanaka et al., 2000). By contrast, our study found that the primary BR signaling components (BRI1, GSK2, and DLT) are generally involved in the low-sensitivity response to GA (in response to high concentrations of GA). Thus, it is tempting to speculate that rice utilizes these two complementary pathways, the D1-TUD1-BU1 pathway and the BRI1-GSK2-DLT pathway, to completely fulfill both BR and GA responses.
Exogenous hormone application has been widely used to test plant sensitivity in phytohormone research. However, insufficient attention has been paid to the hormone concentrations used in the studies, partially because it is challenging to determine the incorporation efficiency of the applied hormone to plants. In our study, the assumed BR-specific-deficient mutants were found to have obviously decreased sensitivity to high GA levels, indicating that hormone concentrations are important when defining a hormone-sensitive or -insensitive mutant. In addition, different concentrations of hormone may have opposite effects on plant growth and development, especially when exogenously applied. Moreover, reduced plant height was frequently observed in BR accumulation or enhanced BR-signaling rice mutants. These phenomena strongly suggest that endogenous BR equilibrium has great significance for plant growth and development. However, in the case of GA, plants tend to have much higher tolerance to the hormone, at least in rice. Mutants deficient in DELLA protein (slr1) or transgenic plants that overexpress a GA biosynthetic gene (GA20-1ox) usually exhibit greatly increased stature. In addition, a concentration of 10−4 M exogenous GA still greatly promotes rice growth. In young rice seedlings, this high GA level can even induce internode elongation (Ueguchi-Tanaka et al., 2000). Notably, various biological processes such as auxin and light responses were found to modulate GA metabolism and GA signaling to regulate plant growth (Jiang et al., 2007; Weiss and Ori, 2007; de Lucas et al., 2008; Yamaguchi, 2008; Chapman et al., 2012). Thus, GA-induced cell elongation should have great significance to rice plants, which in turn develop a low-sensitivity pathway in response to high hormone levels. It is very interesting that the available BR signaling components are used to differentially regulate the GA pathway for plant growth under various conditions.
In conclusion, our discoveries revealed a novel mechanism underlying BR and GA crosstalk depending on tissues and hormone levels, which has greatly advanced our understanding of hormone actions in crop plants and appears much different from that in Arabidopsis. Thus, this work will provide the basis for further agricultural application of the plant growth regulators as well as for future biotechnological breeding.
METHODS
Plant Materials and Culture Conditions
See Supplemental Table 1 for information about the rice (Oryza sativa) and Arabidopsis thaliana plants used in this study. Mutants were kindly provided by others, as follows: brd1 by M. Mori (Mori et al., 2002) and gid2, brd1-1, and brd1-3 by M. Matsuoka (Hong et al., 2002; Sasaki et al., 2003). Mutants were identified or developed in Q.Q.’s laboratory, including d18-Id18h, d35, sd1, d2-2, d11-2, d61-1, and d61-2 (Itoh et al., 2001, 2004; Hong et al., 2003; Sakamoto et al., 2004; Tanabe et al., 2005). Other mutants or plants were identified or produced in our laboratory, including eui1-4, dlt, Do-1, Do-2, Go-2, Gi-2, m107, gid1, slr1, GA20-1ox, and GA2-1ox (Luo et al., 2006; Tong et al., 2009, 2012; Wan et al., 2009). Among them, gid1, slr1, sd1, GA20-1ox, and GA2-1ox were newly identified or produced. gid1 carries a point mutation at 587 (G to T) of the GID1 coding sequence, resulting in an amino acid change of glycine to valine. slr1 carries a 1-bp deletion at 1101 (T) of the SLR1 coding sequence, resulting in a frame shift. sd1 carries a 7-bp deletion at 546 to 552 of the SD1 coding sequence, resulting in a frame shift. Transgenic plants of GA20-1ox and GA2-1ox were produced by introducing the respective genes (GA20ox-1 and GA2ox-1) driven by the maize (Zea mays) Ubiquitin promoter into wild-type plants. Plants were grown in the field under natural conditions or on half-strength Murashige and Skoog (MS) culture medium in a growth chamber at 30°C for 10 h (day/light) and at 28°C for 14 h (night/dark).
Hormone and Hormone Inhibitor Treatments
BL (WAKO) and BRZ (TCI) were resolved in DMSO, and GA3 and PAC (Sigma-Aldrich) were resolved in ethanol, to suitable concentrations as storage solutions. For the treatments, the chemical reagents were first diluted to 1,000-fold of the desired concentrations to a certain volume using their respective solvents and then added to culture medium at 1:1000 dilution to achieve the final concentrations. The identical volume of the blank solvent (DMSO or ethanol) was used as mock treatment. Unless specified, 10−6 M BR, 10−6 M GA, 10−5 M PAC, and 10−5 M BRZ were used for the treatments.
For physiological analysis, seeds were dehusked, and those with identical appearance were selected for further sterilization by immersing in 2% NaClO solution for 0.5 h. For GA sensitivity analysis, the sterilized seeds were first incubated in 10−5 M PAC for 2 d to block the endogenous GA biosynthesis. The seeds were then directly sown on half-strength MS agar medium supplemented with or without hormones or inhibitors and grown for 10 d. Then, images of plants were taken for leaf angle measurement (ImageJ), and lengths of roots, coleoptiles, and leaf sheaths were measured for statistics. For gene expression analysis, germinated seeds were plated in half-strength MS hydroponic medium and grown in a growth chamber for 4 d, and then hormones were added to the medium and the plants were grown for another 2 d. The lamina inclination assay by the microdrop method was performed as described previously (Hong et al., 2003).
Hormone Measurements
For Shi., d11, ZH11, Go-2, and dlt, germinated seeds were sown in field soil and grown for 1 month. Further measurements of ZH11, Do, Nip., and m107 were performed using 2-week-old plants grown in half-strength MS hydroponic medium in a growth chamber. For short-term BL treatment, 10−6 M BL was added to the medium for 2 d before sampling. For long-term BL treatment, 10−6 or 10−5 M BL was supplemented in the culture medium after the plants had been allowed to grow for 2 d after germination and grown for an additional 1 week. About 4 g of shoots was harvested from the rice seedlings for GA measurements. Quantification of endogenous GAs was performed as described (Chen et al., 2012). Quantification of BR (CS) was performed as described previously (Ding et al., 2013).
ChIP and Immunoblotting
One-week-old wild-type plants (ZH11) were used as materials for the ChIP assay according to a previous description with slight modification (Saleh et al., 2008). Briefly, ∼4 g of rice seedling shoots was ground in liquid nitrogen, the powder was suspended in buffer, protein-DNA was cross-linked with formaldehyde, nuclei were isolated, DNA was sheared into 200- to 1000-bp fragments by sonication, protein-DNA complexes were immunoprecipitated with anti-BZR1 polyclonal antibody (1:100) and pulled down using Protein A magnetic beads (Invitrogen), the beads were washed and eluted, and cross-linking was reversed. The precipitated DNA was purified for further qPCR analysis.
Commercial anti-BZR1 and anti-GSK2 polyclonal antibodies (BPI) were used to detect BZR1 and GSK2 protein levels, respectively. Both antibodies were used at a dilution of 1:1000 for the analysis. Plant materials were ground into powder in liquid nitrogen, and then SDS-PAGE sample buffer was added. The samples were boiled and centrifuged, and the supernatants were resolved by SDS-PAGE. Immunoblotting was performed according to a general procedure.
qRT-PCR and ChIP-qPCR
For gene expression analysis, shoots, the second leaf sheaths, and roots from 1-week-old seedlings were collected for RNA isolation using Trizol reagent (Invitrogen). First-strand cDNA was synthesized using a commercial kit (Toyobo). qRT-PCR was performed using SYBR Green-containing PCR mix (Roche) on a real-time PCR detection system (Bio-Rad CFX96). Rice ACTIN1 was used as an internal reference for normalization. For ChIP-qPCR, the prepared DNA in ChIP was used as template and primer pairs designed to amplify fragments of 90 to 150 bp in length were used to detect protein binding. A segment spanning the rice ACTIN1 intron with a 101-bp length was used as an internal reference for normalization, and enrichment fold of the protein binding DNA amount was calculated compared with the sample without antibody application. Three or four repeats were performed for each gene or region analyzed, and average values and sd are shown. Primer sequences were referred to in previous reports (Shimada et al., 2006; Dai et al., 2007; Tong et al., 2009) and are listed in Supplemental Table 6.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AP003244 (D2), AB158759 (D11), AC096690 (GA20ox-1), AP003561 (GA20ox-2), AP002523 (GA3ox-2), AC119288 (GA2ox-1), AP003375 (GA2ox-3), AK106449 (DLT), and X16280 (ACTIN1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Exogenous BR Inhibits Cell Elongation in Leaf Sheath.
Supplemental Figure 2. GA Effect on Wild-Type Growth.
Supplemental Figure 3. Comparison of Plant Height of BR-Related Plants.
Supplemental Figure 4. Expression of D2 and D11 in Response to Various BR Levels in Shoot.
Supplemental Figure 5. Root Response to BR in GA-Related Mutants.
Supplemental Figure 6. The Effect of GA and PAC Application on GSK2 and BZR1 Protein Level.
Supplemental Figure 7. Response of brd1 Alleles to GA Treatment.
Supplemental Figure 8. SLR1 Can Interact with Os BZR1 in Yeast.
Supplemental Table 1. Information of the Mutants and the Transgenic Plants Used in This Study.
Supplemental Table 2. Quantification of GA1 in Wild-Type and Do Plants.
Supplemental Table 3. Quantification of GAs in Wild-Type, m107, and Short-Term BL-Treated Plants.
Supplemental Table 4. Quantification of CS in Wild-Type and m107 Plants.
Supplemental Table 5. Quantification of GAs in Wild-Type and Long-Term BL-Treated Plants.
Supplemental Table 6. Primers Used for qRT-PCR and ChIP-qPCR Analysis.
Supplementary Material
Acknowledgments
We thank M. Mori (National Institute of Agrobiological Sciences, Japan) for providing brd1 seeds and M. Matsuoka (Nagoya University) for providing gid2, brd1-1, and brd1-3 mutants. We thank S. Cao and G. Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for plant transformation and field management. We also thank Yuqi Feng (Wuhan University) and his team for all the GA and BR measurements. This work was supported by the National Natural Science Foundation of China (Grants 31170715 and 91335203) and the State Key Laboratory of Plant Genomics (Grant 2013B0125-01).
AUTHOR CONTRIBUTIONS
H.T. and C.C. designed the study, analyzed the data, and wrote the article. H.T. and Y.X. performed gene and protein expression analysis, physiological analysis, ChIP assay, and transgenic experiment. D.L. and Y.J. assisted in physiological analysis. Y.Y. assisted in ChIP assay, data analysis, and article preparation. L.L., S.G., and Q.Q. provided assistance in mutant identification and transgenic plant development. H.T. performed all the other studies.
Glossary
- BR
brassinosteroid
- GA
gibberellin
- BL
brassinolide
- qRT-PCR
quantitative real-time RT-PCR
- CS
castasterone
- PAC
paclobutrazol
- ChIP
chromatin immunoprecipitation
- qPCR
quantitative PCR
- MS
Murashige and Skoog
- BRZ
brassinazole
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 104: 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquin T., Meier C., Foster R., Nielsen M.E., Mundy J. (2001). Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 127: 450–458. [PMC free article] [PubMed] [Google Scholar]
- Chapman E.J., Greenham K., Castillejo C., Sartor R., Bialy A., Sun T.P., Estelle M. (2012). Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS ONE 7: e36210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.L., Fu X.M., Liu J.Q., Ye T.T., Hou S.Y., Huang Y.Q., Yuan B.F., Wu Y., Feng Y.Q. (2012). Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 905: 67–74. [DOI] [PubMed] [Google Scholar]
- Cho H., et al. (2014). A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat. Cell Biol. 16: 66–76. [DOI] [PubMed] [Google Scholar]
- Dai M., Zhao Y., Ma Q., Hu Y., Hedden P., Zhang Q., Zhou D.X. (2007). The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 144: 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D., Van Buyten E., Satoh K., Balidion J., Mauleon R., Choi I.R., Vera-Cruz C., Kikuchi S., Höfte M. (2012). Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 158: 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A., Jung H.S., Sun T.P. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98: 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Mao L.J., Wang S.T., Yuan B.F., Feng Y.Q. (2013). Determination of endogenous brassinosteroids in plant tissues using solid-phase extraction with double layered cartridge followed by high-performance liquid chromatography-tandem mass spectrometry. Phytochem. Anal. 24: 386–394. [DOI] [PubMed] [Google Scholar]
- Duan K., Li L., Hu P., Xu S.P., Xu Z.H., Xue H.W. (2006). A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J. 47: 519–531. [DOI] [PubMed] [Google Scholar]
- Fàbregas N., Li N., Boeren S., Nash T.E., Goshe M.B., Clouse S.D., de Vries S., Caño-Delgado A.I. (2013). The brassinosteroid insensitive1-like3 signalosome complex regulates Arabidopsis root development. Plant Cell 25: 3377–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadí D., Blázquez M.A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 13446–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L., Ubeda-Tomás S., Gisbert C., García-Martínez J.L., Moritz T., López-Díaz I. (2008). Gibberellin homeostasis in tobacco is regulated by gibberellin metabolism genes with different gibberellin sensitivity. Plant Cell Physiol. 49: 679–690. [DOI] [PubMed] [Google Scholar]
- Gomi K., Sasaki A., Itoh H., Ueguchi-Tanaka M., Ashikari M., Kitano H., Matsuoka M. (2004). GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 37: 626–634. [DOI] [PubMed] [Google Scholar]
- González-García M.P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-García S., Russinova E., Caño-Delgado A.I. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859. [DOI] [PubMed] [Google Scholar]
- Gudesblat G.E., Schneider-Pizoń J., Betti C., Mayerhofer J., Vanhoutte I., van Dongen W., Boeren S., Zhiponova M., de Vries S., Jonak C., Russinova E. (2012). SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14: 548–554. [DOI] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Yang Y., Li J., Wang Z.Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P., Phillips A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5: 523–530. [DOI] [PubMed] [Google Scholar]
- Hong Z., Ueguchi-Tanaka M., Matsuoka M. (2004). Brassinosteroids and rice architecture. J. Pestic. Sci. 29: 184–188. [Google Scholar]
- Hong Z., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32: 495–508. [DOI] [PubMed] [Google Scholar]
- Hong Z., Ueguchi-Tanaka M., Umemura K., Uozu S., Fujioka S., Takatsuto S., Yoshida S., Ashikari M., Kitano H., Matsuoka M. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Qian Q., Xu T., Zhang Y., Dong G., Gao T., Xie Q., Xue Y. (2013). The U-box E3 ubiquitin ligase TUD1 functions with a heterotrimeric G α subunit to regulate brassinosteroid-mediated growth in rice. PLoS Genet. 9: e1003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Bao F., Li J. (2000). Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. 24: 693–701. [DOI] [PubMed] [Google Scholar]
- Huq E. (2006). Degradation of negative regulators: A common theme in hormone and light signaling networks? Trends Plant Sci. 11: 4–7. [DOI] [PubMed] [Google Scholar]
- Ikeda A., Ueguchi-Tanaka M., Sonoda Y., Kitano H., Koshioka M., Futsuhara Y., Matsuoka M., Yamaguchi J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Tatsumi T., Sakamoto T., Otomo K., Toyomasu T., Kitano H., Ashikari M., Ichihara S., Matsuoka M. (2004). A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol. Biol. 54: 533–547. [DOI] [PubMed] [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Sentoku N., Kitano H., Matsuoka M., Kobayashi M. (2001). Cloning and functional analysis of two gibberellin 3 beta-hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA 98: 8909–8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager C.E., Symons G.M., Ross J.J., Smith J.J., Reid J.B. (2005). The brassinosteroid growth response in pea is not mediated by changes in gibberellin content. Planta 221: 141–148. [DOI] [PubMed] [Google Scholar]
- Jiang C., Gao X., Liao L., Harberd N.P., Fu X. (2007). Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 145: 1460–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M., Itoh H., Inukai Y., Sakamoto T., Ueguchi-Tanaka M., Ashikari M., Matsuoka M. (2003). Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 35: 104–115. [DOI] [PubMed] [Google Scholar]
- Khripach V., Zhabinskii V., De Groot A. (2000). Twenty years of brassinosteroids: Steroidal plant hormones warrant better crops for the XXI century. Ann. Bot. (Lond.) 86: 441–447. [Google Scholar]
- Kim T.W., Guan S., Burlingame A.L., Wang Z.Y. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43: 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Michniewicz M., Bergmann D.C., Wang Z.Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Choi S.C., An G. (2008). Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J. 54: 93–105. [DOI] [PubMed] [Google Scholar]
- Li D., Wang L., Wang M., Xu Y.Y., Luo W., Liu Y.J., Xu Z.H., Li J., Chong K. (2009). Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol. J. 7: 791–806. [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H., Vafeados D., Chory J. (2001). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127: 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.F., Wang C., Jiang L., Li S., Sun S.S.M., He J.X. (2012). An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 5: ra72. [DOI] [PubMed] [Google Scholar]
- Lilley J.L.S., Gan Y., Graham I.A., Nemhauser J.L. (2013). The effects of DELLAs on growth change with developmental stage and brassinosteroid levels. Plant J. 76: 165–173. [DOI] [PubMed] [Google Scholar]
- Luo A., et al. (2006). EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 47: 181–191. [DOI] [PubMed] [Google Scholar]
- Mora-García S., Vert G., Yin Y., Caño-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18: 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M., Nomura T., Ooka H., Ishizaka M., Yokota T., Sugimoto K., Okabe K., Kajiwara H., Satoh K., Yamamoto K., Hirochika H., Kikuchi S. (2002). Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 130: 1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig C., Shin G.H., Altmann T. (2003). Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 133: 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K.H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212. [DOI] [PubMed] [Google Scholar]
- Nomura T., Hanada A., Zhu Y.Y., He Z.H., Mander L., Kamiya Y., Yamaguchi S. (2006). ELONGATED UPPERMOST INTERNODE epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell Physiol. 47: S144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki K., Inaba N., Kitagawa K., Fujioka S., Kitano H., Fujisawa Y., Kato H., Iwasaki Y. (2009a). Function of the alpha subunit of rice heterotrimeric G protein in brassinosteroid signaling. Plant Cell Physiol. 50: 161–172. [DOI] [PubMed] [Google Scholar]
- Oki K., Kitagawa K., Fujisawa Y., Kato H., Iwasaki Y. (2009b). Function of alpha subunit of heterotrimeric G protein in brassinosteroid response of rice plants. Plant Signal. Behav. 4: 126–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., et al. (2004). An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134: 1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Kitano H., Fujioka S. (2013). An E3 ubiquitin ligase, ERECT LEAF1, functions in brassinosteroid signaling of rice. Plant Signal. Behav. 8: e27117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Jeong D.H., An G., Kitano H., Ashikari M., Matsuoka M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898. [DOI] [PubMed] [Google Scholar]
- Sasse J.M. (2003). Physiological actions of brassinosteroids: An update. J. Plant Growth Regul. 22: 276–288. [DOI] [PubMed] [Google Scholar]
- Serna L. (2013). What causes opposing actions of brassinosteroids on stomatal development? Plant Physiol. 162: 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A., Ueguchi-Tanaka M., Sakamoto T., Fujioka S., Takatsuto S., Yoshida S., Sazuka T., Ashikari M., Matsuoka M. (2006). The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 48: 390–402. [DOI] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S., Ashikari M., Fujioka S., Takatsuto S., Yoshida S., Yano M., Yoshimura A., Kitano H., Matsuoka M., Fujisawa Y., Kato H., Iwasaki Y. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17: 776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., et al. (2009). BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 151: 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Chu C. (2012). Brassinosteroid signaling and application in rice. J. Genet. Genomics 39: 3–9. [DOI] [PubMed] [Google Scholar]
- Tong H., Jin Y., Liu W., Li F., Fang J., Yin Y., Qian Q., Zhu L., Chu C. (2009). DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 58: 803–816. [DOI] [PubMed] [Google Scholar]
- Tong H., Liu L., Jin Y., Du L., Yin Y., Qian Q., Zhu L., Chu C. (2012). DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24: 2562–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H., Ueguchi-Tanaka M., Nakajima M., Ashikari M., Kitano H., Yamaguchi I., Matsuoka M. (2006). Interaction among SLRI1, GID1, and GID2 in the gibberellin signaling pathway in rice cells. Plant Cell Physiol. 47: S125. [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Matsuoka M. (2006). Gibberellin perception and its signaling in rice. Plant Cell Physiol. 47: S6. [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T.Y., Hsing Y.I.C., Kitano H., Yamaguchi I., Matsuoka M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Fujisawa Y., Kobayashi M., Ashikari M., Iwasaki Y., Kitano H., Matsuoka M. (2000). Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc. Natl. Acad. Sci. USA 97: 11638–11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Katoh E., Ohmiya H., Asano K., Saji S., Hongyu X., Ashikari M., Kitano H., Yamaguchi I., Matsuoka M. (2007). Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19: 2140–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Walcher C.L., Chory J., Nemhauser J.L. (2008). Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl. Acad. Sci. USA 105: 9829–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S., et al. (2009). Activation tagging, an efficient tool for functional analysis of the rice genome. Plant Mol. Biol. 69: 69–80. [DOI] [PubMed] [Google Scholar]
- Wang L., Wang Z., Xu Y., Joo S.H., Kim S.K., Xue Z., Xu Z., Wang Z., Chong K. (2009). OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. 57: 498–510. [DOI] [PubMed] [Google Scholar]
- Wang L., Xu Y., Zhang C., Ma Q., Joo S.H., Kim S.K., Xu Z., Chong K. (2008). OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE 3: e3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Xu Y.Y., Li J., Powell R.A., Xu Z.H., Chong K. (2007). Transgenic rice plants ectopically expressing AtBAK1 are semi-dwarfed and hypersensitive to 24-epibrassinolide. J. Plant Physiol. 164: 655–664. [DOI] [PubMed] [Google Scholar]
- Wang L., Xu Y.Y., Ma Q.B., Li D., Xu Z.H., Chong K. (2006). Heterotrimeric G protein alpha subunit is involved in rice brassinosteroid response. Cell Res. 16: 916–922. [DOI] [PubMed] [Google Scholar]
- Wang X., Chory J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313: 1118–1122. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y. (2012). Brassinosteroids modulate plant immunity at multiple levels. Proc. Natl. Acad. Sci. USA 109: 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Bai M.Y., Oh E., Zhu J.Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46: 701–724. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Seto H., Fujioka S., Yoshida S., Chory J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383. [DOI] [PubMed] [Google Scholar]
- Weiss D., Ori N. (2007). Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 144: 1240–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251. [DOI] [PubMed] [Google Scholar]
- Yamamuro C., Ihara Y., Wu X., Noguchi T., Fujioka S., Takatsuto S., Ashikari M., Kitano H., Matsuoka M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Li L., Guo H., Yin Y. (2012). MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 20142–20147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646. [DOI] [PubMed] [Google Scholar]
- Zhang C., Xu Y., Guo S., Zhu J., Huan Q., Liu H., Wang L., Luo G., Wang X., Chong K. (2012). Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet. 8: e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ye H., Guo H., Johnson A., Lin H., Yin Y. (2014a). Transcription factors involved in brassinosteroid repressed gene expression and their regulation by BIN2 kinase. Plant Signal. Behav. 9: e27849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ye H., Guo H., Johnson A., Zhang M., Lin H., Yin Y. (2014b). Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in Arabidopsis. Plant J. 77: 59–70. [DOI] [PubMed] [Google Scholar]
- Zhao S.Q., Hu J., Guo L.B., Qian Q., Xue H.W. (2010). Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res. 20: 935–947. [DOI] [PubMed] [Google Scholar]
- Zhu Y., et al. (2006). ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18: 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.